Abstract
Plants utilize extracellular vesicles (EVs) to transport small RNAs (sRNAs) into their fungal pathogens and silence fungal virulence-related genes through a phenomenon called “cross-kingdom RNAi.” It remains unknown, however, how sRNAs are selectively loaded into EVs. Here, we identified several RNA-binding proteins (RBPs) in Arabidopsis, including Argonaute 1 (AGO1), RNA helicases (RHs) and Annexins (ANN), which are secreted by exosome-like EVs. AGO1, RH11 and RH37 selectively bind to EV-enriched sRNAs but not non-EV enriched sRNAs, suggesting that they contribute to the selective loading of sRNAs into EVs. Conversely, ANN1 and ANN2 bind to sRNAs non-specifically. The ago1, rh11rh37 and ann1ann2 mutants showed reduced secretion of sRNAs in EVs, demonstrating that these RBPs play an important role in sRNA loading and/or stabilization in EVs. Furthermore, rh11rh37 and ann1ann2 showed increased susceptibility to Botrytis cinerea, supporting that RH11, RH37, ANN1 and ANN2 positively regulate plant immunity against B. cinerea.
Introduction
Small RNAs (sRNAs) induce RNA interference (RNAi) and play an important role in regulating host immune responses and pathogen virulence 1, 2, 3. In addition to their role in regulating gene expression endogenously, sRNAs can travel between interacting organisms to induce gene silencing in the counterparty in trans, a phenomenon called “cross-kingdom RNAi” 4–7. We discovered that fungal pathogens can deliver sRNAs into host plants and hijacks host RNAi machinery to silence plant immunity genes5. Further, we demonstrated that cross-kingdom RNAi is bidirectional 4, 8, hosts plants can utilize extracellular vesicles (EVs) to transport sRNAs into interacting fungal cells to suppress the expression of fungal virulence-related genes 4.
EVs are secreted membrane-encased vesicular compartments which play important roles in communication between cells and interacting organisms by transporting proteins, lipids, and RNA molecules 9. EVs released by mammalian cells can be divided into three major categories based on their distinct biogenesis pathways and specific protein markers, including exosomes, microvesicles and apoptotic bodies 10. Exosomes are small vesicles derived from multivesicular bodies (MVBs) and are released as a consequence of the fusion of the outer membrane of MVBs with the plasma membrane 9. Microvesicles are formed and released by direct budding from the plasma membrane, and apoptotic bodies are formed during the execution phase of the apoptotic process 10. In mammalian systems, multiple classes of EVs, especially exosomes, have been shown to transport RNAs into specific recipient cells within an organism 11–14. Recently, we discovered that Arabidopsis cells secrete exosome-like EVs to deliver sRNAs into fungal pathogen B. cinerea 4, providing the first example of a host sRNA delivery mechanism in any system. These sRNA-containing EVs are enriched in tetraspanin (TET) 8 and TET9 proteins which are the orthologues of mammalian exosome markers — tetraspanins CD63, CD81, and CD9 15. Meanwhile, TET8 colocalizes with MVB markers inside the cells, indicating that TET8-positive EVs are plant exosome-like EVs. These studies have expanded the known function of EVs to include cross-kingdom trafficking of functional sRNAs. We found that the expression profile of EV-enriched sRNAs is distinct from that of total sRNAs isolated from the same tissue 4. Reports from mammalian systems also determined that EVs had RNA expression profiles distinct from total cellular RNAs. 16–18. These studies suggest that sorting of RNAs into EV is a precisely regulated process. However, how sRNAs are selectively loaded and stabilized in exosomes is largely unknown.
In this study, we identified a group of RNA binding proteins (RBPs) in plant EVs, mainly tetraspanin-positive exosome-like vesicles, including Argonaute protein 1 (AGO1), DEAD-box RNA helicases (RHs) and Annexins (ANNs). AGO proteins complex with sRNAs to induce the silencing of genes with complementary sequences1. The Arabidopsis genome encodes ten AGOs with selective sRNA binding activities 19. AGO1 preferentially binds to 20–22 nt sRNAs with a 5’-terminal uridine, whereas AGO2 and AGO4 favor sRNAs with a 5’-terminal adenosine, AGO5 predominantly binds small RNAs beginning with a cytosine20. Furthermore, the secondary structure of an sRNA duplex can also affect its loading efficiency into AGO proteins21. DEAD-box RNA helicases contain a conserved Asp-Glu-Ala-Asp (DEAD) motif. They bind and rearrange the secondary structure of RNAs in an energy-dependent manner 22,23. Here, we show that EV-associated AGO1, RH11 and RH37 specifically bind to EV-enriched sRNAs and co-localize with MVBs in the cell, which facilitate the packaging of these RBPs and their associated sRNAs into MVB-derived EVs. The ANNs, however, bind to sRNAs non-specifically and may only contribute to the stabilization of EV-encased sRNAs. Moreover, the rh11rh37 and ann1ann2 mutants were more susceptible to B. cinerea than wild-type plants and the fungal target genes of transferred plant sRNAs were de-repressed in B. cinerea purified from infected rh11rh37 and ann1ann2. These findings further support that EV-localized RBPs contribute to sorting and stabilizing plant secreted sRNAs in EVs.
Results
Proteomic analysis reveals a group of RBPs in plant EVs
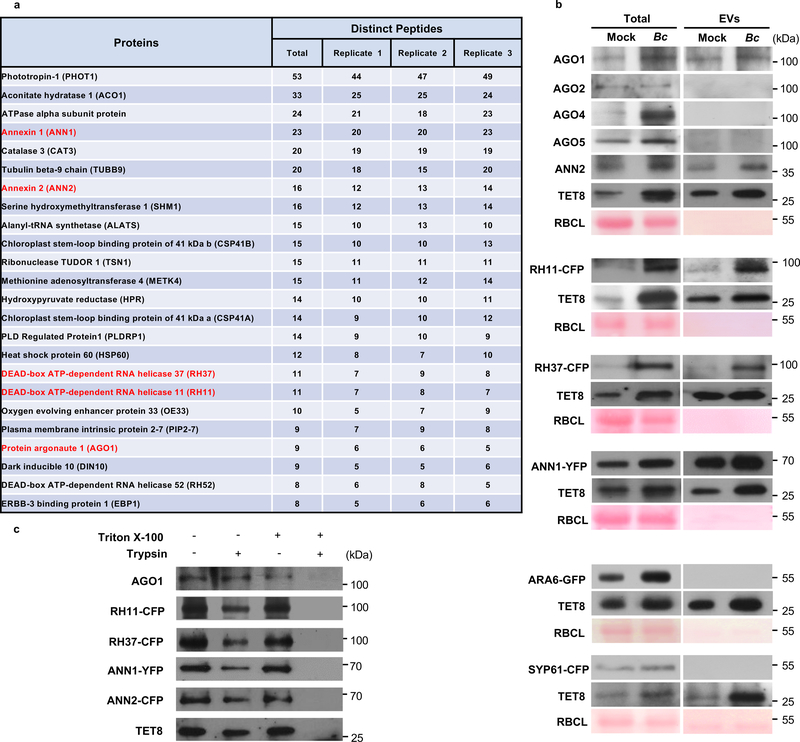
We hypothesize that a set of EV-associated proteins, specifically RBPs, may be responsible for the selective loading of sRNAs into EVs. To identify EV-associated proteins, we isolated EVs from the apoplastic fluid of Arabidopsis rosette leaves and performed proteomic analysis using mass spectrometry (MS) (Extended Data Fig. 1) 4. Specifically, we used B. cinerea infected leaves, and centrifuged at 100,000 g (P100) to increase the yield of isolated TET8-positive EVs, because we previous showed that fungal infection increases EV secretion (Extended Data Fig. 1)4. Proteins detected in all three biological replicates with distinct peptide counts of five or more in each replicate were further analyzed for Gene Ontology (GO) terms related to biological process and molecular function (Supplementary Table 1 and Extended Data Fig. 2). Among the identified proteins, 28.24% were categorized by GO terms as stress response proteins and 14.88% were categorized as response to biotic stimulus proteins, both of which were enriched in EV fractions, suggesting that EVs are involved in defense response and stress adaption processes. Total of 93 RNA binding proteins (RBPs) were detected with at least five distinct peptides in each of the three biological replicates (Supplementary Table 2). We selected the RBPs that have potential sRNA binding activity for further analysis, including AGO1 (9 peptides), DEAD-box RNA helicases RH11 (11 peptides), RH37 (11 peptides), RH52 (8 peptides), Annexin1 (ANN1, 23 peptides), and ANN2 (16 peptides) (Fig. 1a, Supplementary Table 2). RH11, RH37 and RH52 belong to the same clade of DEAD-box RNA helicases, and share more than 80% homology. Because MS analysis identified five peptides specific to RH11, three specific to RH37, and none specific to RH52, we chose to mainly study the function of RH11 and RH37. To confirm the proteomics results experimentally, we conducted Western blot analyses on proteins isolated from EVs. For the RBPs lacking native antibodies, we generated transgenic fluorescence protein-tagged Arabidopsis plants, including RH11-CFP, RH37-CFP and ANN1-YFP lines. AGO1, RH11, RH37, ANN1 and ANN2 were easily detected in isolated Arabidopsis EVs (P100) (Fig. 1b), whereas AGO2, AGO4 and AGO5 were not present in EVs (Fig. 1b). This result is consistent with the proteomics result (Supplemental Table 2). The EV-associated TET8 marker was used as a positive control, and the MVB marker ARA6 and the trans-Golgi marker SYP61 were used as negative controls to ensure that the isolated EVs were not contaminated by plasma membrane or other endomembrane fractions (Fig. 1b). To further confirm that these RBPs were located inside of EVs, not just adhered to the outer surface of EVs, the isolated EVs (P100) from Arabidopsis leaves were subjected to trypsin digestion in the presence or absence of Triton X-100, a detergent that can rupture EVs. These RBPs were still detected in EVs after trypsin treatment, while the combination of Triton X-100 and trypsin completely digested them (Fig. 1c). These results indicate that AGO1, RH11, RH37, ANN1 and ANN2 are secreted by EVs in plants.
Fig. 1. RNA binding proteins present in plant EVs.
a, Candidates of RNA binding proteins detected in plant EVs. Proteins in red were chosen for further experimental analysis. b, RNA binding proteins AGO1, RH11, RH37, ANN1 and ANN2 can be detected from purified EVs by western blot. EVs were isolated from the untreated (Mock) and B. cinerea infected (Bc) wild-type, RH11-CFP, RH37-CFP, ANN1-YFP, ARA6-GFP and CFP-SYP61 plants. AGO1, AGO2, AGO4, AGO5 ANN2 and TET8 were detected by western blot using antibodies against AGO1, AGO2, AGO4, AGO5, ANN2, and TET8, respectively. RH11-CFP, RH37-CFP, ANN1-YFP, ARA6-GFP and CFP-SYP61 were detected by western blot using antibodies against GFP. TET8, ARA6, and SYP61 were used as EV, MVB, and Trans Golgi Network markers, respectively. 20 μg of total and 10 μg of EV proteins were used to perform the western blot. Ponceau-S staining of Rubisco was used as the loading control. c, Trypsin digestion of plant EVs. EVs were isolated from wild-type, RH11-CFP, RH37-CFP, ANN1-YFP and ANN2-CFP plants and incubated with 1% Triton X-100, with 10 μg/ml trypsin, or a combination of both, for 30 min at 37 °C. Samples were analyzed by western blot with antibodies against AGO1, TET8 and GFP, respectively. The experiments in Fig. 1b, c were repeated three times with similar results. Source data are provided as a Source Data file.
Secreted RBPs are enriched in TET8 -positive EVs
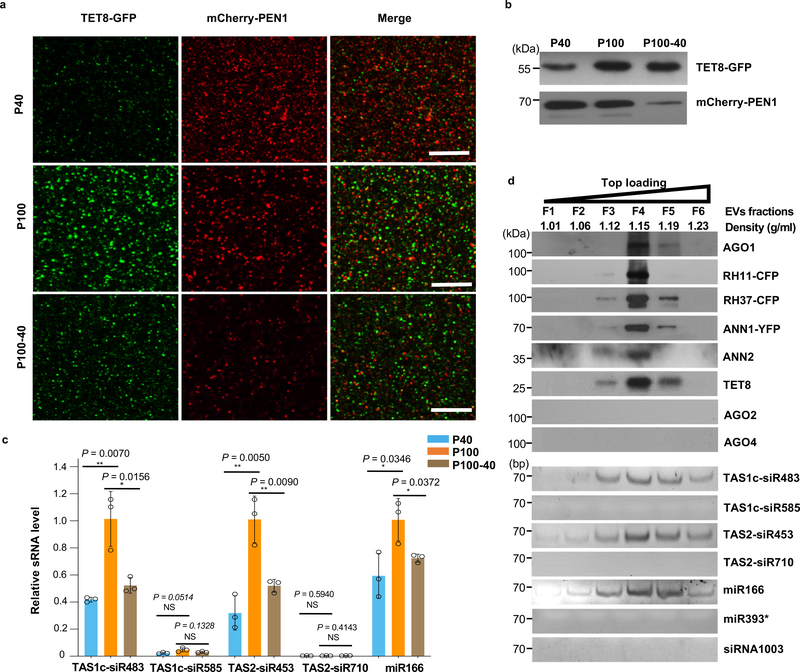
In mammalian systems EVs are divided into multiple subclasses based on the associated protein markers and their biogenesis pathways, and it is likely that plants also possess distinct subclasses of EVs. In addition to TET8-positive EVs 4, Rutter and Innes have identified PENETRATION 1 (PEN1)-positive EVs from Arabidopsis, which are enriched in the 40,000 g (P40) ultracentrifugation fraction 24. To determine whether TET8-positive EVs and PEN1-positive EVs are distinct subclasses of plant EVs, we generated transgenic plants co-expressing two fluorescence-tagged fusion proteins TET8-GFP and mCherry-PEN1. Numerous distinct GFP-labeled EVs and mCherry-labeled EVs were observed in the leaf apoplast fluid from the transgenic plants (Fig. 2a). TET8-GFP labeled EVs were not co-localized with mCherry-PEN1-labeled EVs, indicating that these are two distinct subclasses of plant EVs. Consistent with previous results 4, 24, TET8-positive EVs and PEN1-positive EVs were enriched in the P100 and P40 fractions, respectively (Fig. 2a,b). Furthermore, a significant amount of TET8-positive EVs were isolated after centrifugation of the supernatant of fraction P40 at 100,000 g (P100–40), which was largely depleted of PEN1-positive EVs (Fig. 2a,b). Further, secreted sRNAs were more concentrated in the P100 fraction than the P40 fraction (Fig. 2c). We previously demonstrated that TET8 co-localized with Arabidopsis MVB marker Rab5-like GTPase, ARA6, inside the plant cell, suggesting that TET8-positive EVs are likely derived from MVBs and could be considered plant exosome-like EVs 4. However, our observations indicate that PEN1 does not co-localize with ARA6-marked MVBs (Extended Data Fig. 3), suggesting that the PEN1- and TET8-positive EVs have distinct biogenesis pathways. These results support the hypothesis that plants secrete different subtypes of EVs with distinct biomarkers and biogenesis pathways.
Fig. 2. TET8 and PEN1 label different classes of EVs in Arabidopsis.
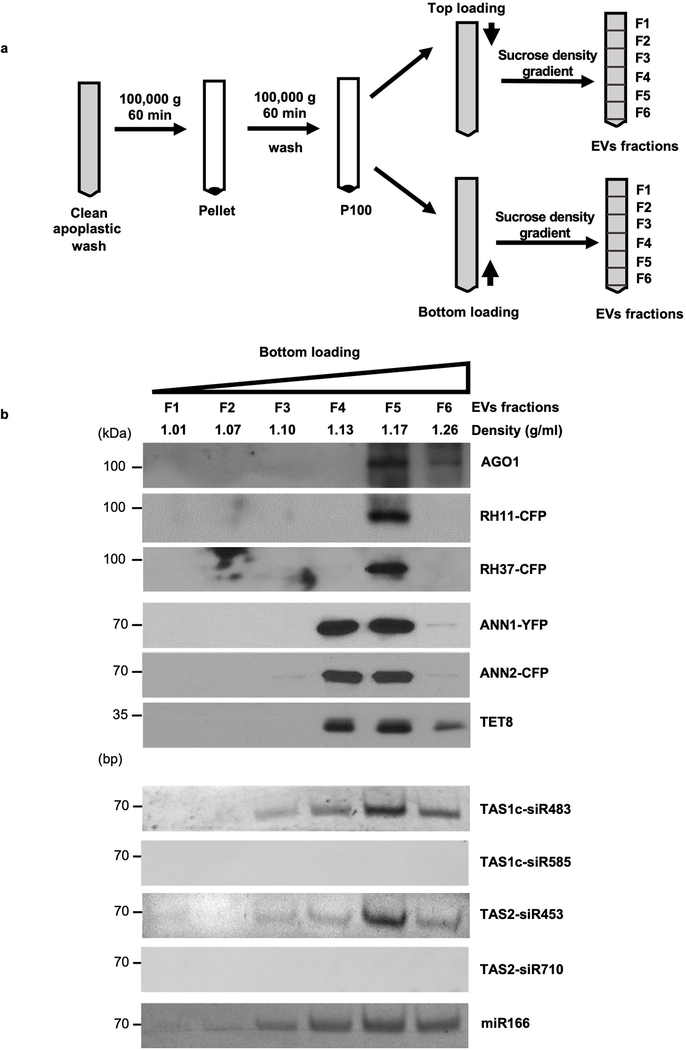
a, Confocal microscopy of EVs isolated from B. cinerea-infected TET8-GFP/mCherry-PEN1 double fluorescence plants by ultracentrifugation at 40,000 g (P40 fraction) and 100,000 g (P100 fraction) for 1 hour. For the P100–40 fraction, the supernatant of P40 fraction was centrifuged at 100,000 g for 1 hour. Scale bars, 10 μm. b, Detection of GFP-labeled TET8 and mCherry-labeled PEN1 in P40, P100, and P100–40 EV fractions by western blot. c, EV-enriched sRNAs were detected in P40, P100, and P100–40 EV fractions by real-time-RT PCR. The data are presented as mean ± s.d., n = 3 biological replicates. The error bars represent standard deviations (s.d.). d, Six fractions were collected from top loading plant EV sucrose gradient centrifugation and analyzed for protein content by western blot and for sRNAs by RT-PCR. AGO1, AGO2, AGO4, RH11, RH37, ANN1, ANN2 and TET8 proteins were detected by western blot. EV-enriched sRNAs (TAS1c-siR483, TAS2-siR453 and miR166), non-EV-enriched sRNAs (TAS1c-siR585 and TAS2-siR710), AGO2-associated miR393* and AGO4- associated siRNA1003 were detected by RT-PCR. All EVs used here were not pretreated with trypsin and RNase. The statistical analysis was performed using analysis of variance (ANOVA) Dunnett’s multiple comparisons test. The small open circles represent the individual values. The asterisks indicate significant differences: *P < 0.05, **P < 0.01. NS, not significant. The experiments in Fig. 2a-b, d were repeated three times independently with similar results. Source data are provided as a Source Data file.
Since plant exosome-like EVs carry sRNAs, we hypothesize that the AGO1 protein and other identified RBPs may be also enriched in TET8-positive EVs. In order to test this hypothesis, we performed density gradient analysis, a classical method used to separate different EVs according to their floatation speed and equilibrium density12, 13, 25, 26. Isolated EVs (P100) were further separated by sucrose gradients centrifugation using both top loading and bottom loading methods (Extended Data Fig. 4a). EV-enriched sRNAs that we previously identified4 and TET8 were used to identify the TET8-positive EV fractions. As shown in Fig. 2d and Extended Data Fig. 4b, plant EV-enriched sRNAs, TAS1c-siR483, TAS2-siR453 and miR166, and TET8 are found in the EV fractions at the density of 1.12–1.19 g/ml, which is consistent with the density of exosomes in animal systems 11. As expected, the RBPs AGO1, RH11, RH37, ANN1 and ANN2 were also detected only in the fraction of TET8-positive EVs, by both top and bottom loading ultracentrifugation procedures (Fig. 2d and Extended Data Fig. 4b). These results suggest that sRNAs and RBPs are likely secreted in TET8-positive EVs.
Exosome-like EVs are the main subclass of plant EVs for sRNA and RBP secretion
Recent advances in methodology development for EV isolation in mammalian systems indicate that the most powerful tool to isolate pure specific subclasses of EVs is immunoaffinity capture 26, 27. This method isolates a specific subclass of EVs using beads coated with an antibody that recognizes the specific protein marker exposed on EV membranes (such as exosome marker CD63) 25, 26, 28, 29. In order to determine whether exosome-like EVs are responsible for sRNA and RBP secretion in plants, we isolated exosome-like EVs using TET8 immunoaffinity capture. Because both the N- and C-termini of the TET8 protein are inside of vesicles (Fig. 3a), it was not possible to use a fused GFP-tag for immunoaffinity capture of EVs. Therefore, we had to generate a native antibody that specifically recognizes the large exposed extra-vesicular loop, EC2 domain, of TET8 (Fig. 3a). This antibody was unable to detect a signal in the tet8 mutant background, demonstrating that it was indeed TET8-specific (Fig. 3b). We successfully isolated TET8-positive EVs by direct immunoaffinity capture using this antibody (Fig. 3c). We could easily detect the EV-enriched sRNAs that we previously identified 4, including TAS1c-siR483, TAS2-siR453 and miR166, in the anti-TET8 antibody-captured EVs (Fig. 3d), further confirming that TET8-positive EVs are an important class of EVs for sRNA secretion. Furthermore, the RBPs AGO1, RH11, RH37, ANN1 and ANN2 were clearly detectable in the TET8-positive EVs (Fig. 3e).
Fig. 3. TET8-positive EVs are an important subclass of EVs for secretion of sRNAs and RBPs.
a, The structures and the topology of plant tetraspanin TET8. The peptide of the large extracellular loop EC2 domain was used for production of TET8 specific antibody. b, Anti-TET8 antibody can detect TET8 in EVs isolated from wild-type plants but not from tet8 mutants. c, TET8-positive EVs can be pulled-down by TET8-specific antibody linked beads. IgG was used as a negative control. d, EV-associated sRNAs are enriched in EVs isolated from TET8 specific antibody pull-down fractions. AGO2- associated miR393* and AGO4-associated siRNA1003 were used as negative control. e, AGO1, AGO2, AGO4, RH11, RH37, ANN1 and ANN2 were detected in anti-TET8 immuno-isolated EV fractions by western blot. AGO2 and AGO4 serve as negative control. Scale bars, 10 μm. The experiments in Fig. 3b, d-e were repeated three times with similar results. Source data are provided as a Source Data file.
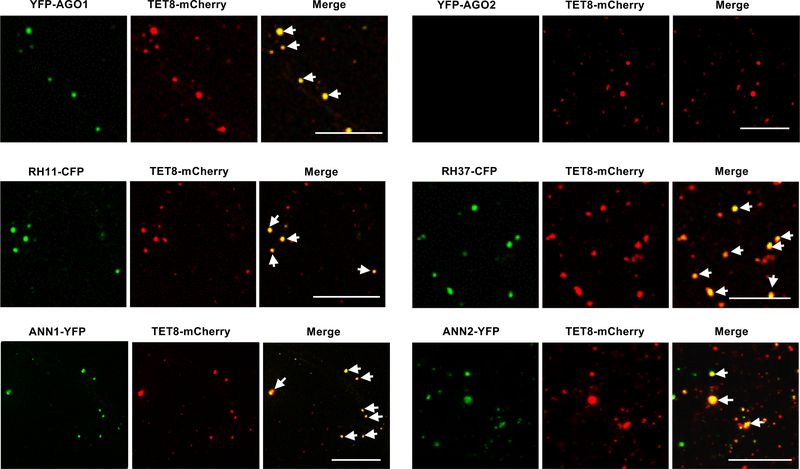
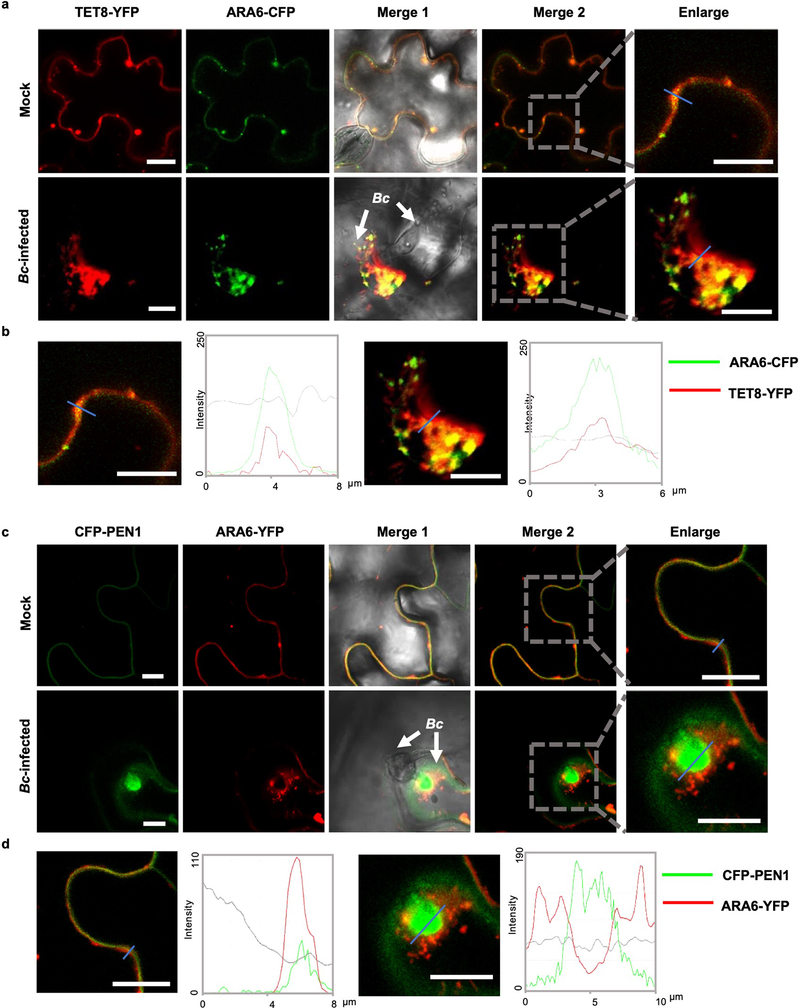
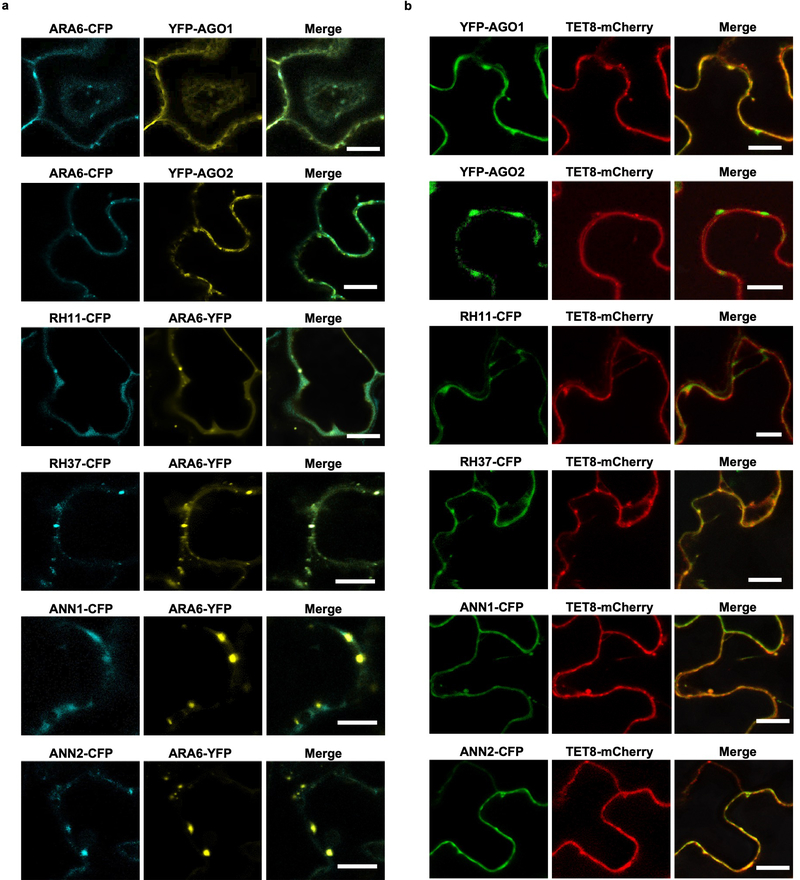
We also confirmed that these RBPs co-localized with TET8 and MVB marker ARA6 (Extended Data Fig. 5) inside the plant cells, and with TET8 in the secreted EVs (Fig. 4). Co-localization of these RBPs with MVBs may facilitate their loading into EVs with their associated sRNAs. This data strongly supports that these RBPs, together with sRNAs, are secreted by MVB-derived EVs, and that the subcellular localization of these RBPs facilitates their loading into exosome-like EVs.
Fig. 4. EV-associated RBPs colocalize with EV marker TET8 in isolated EVs.
Fluorescent protein labeled RBPs were co-expressed transiently with EV marker protein TET8 in Nicotiana benthamiana. Confocal microscopy was used to determine the localization of RBPs with TET8 in the isolated EVs. AGO2 was used as a negative control. Scale bars, 10 μm. Similar results were observed in three biological repeats.
EV-localized RBPs mediate sorting and stabilization of secreted sRNAs
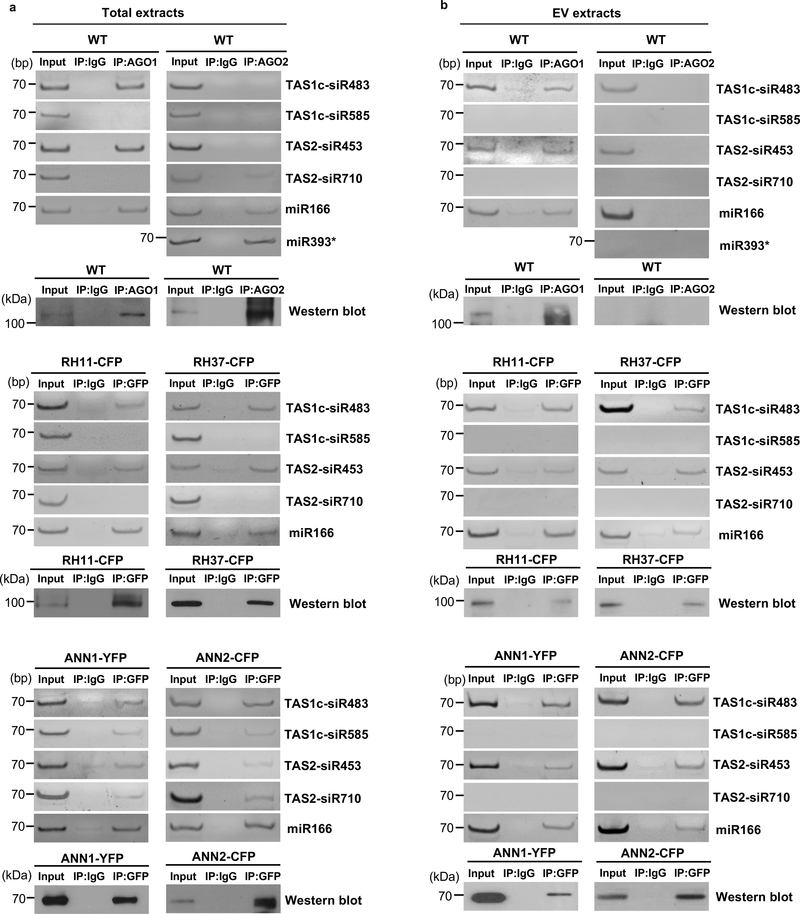
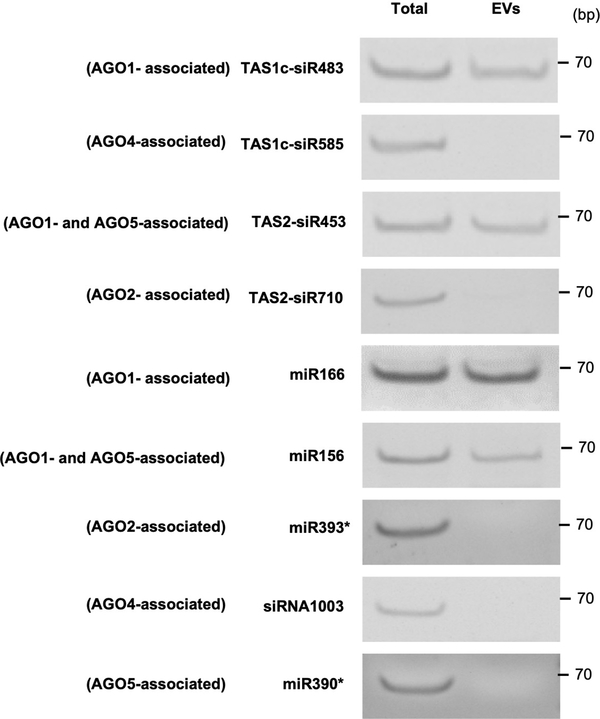
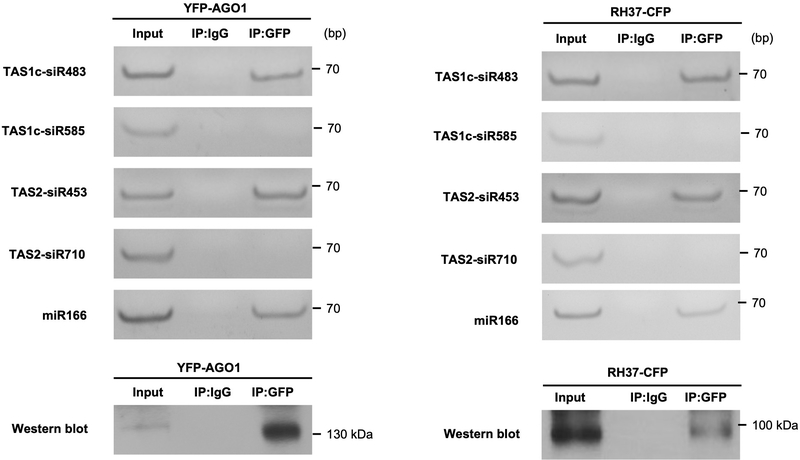
The AGO1-associated tasiRNAs (TAS1c-siR483 and TAS2-siR453) and miRNAs (miR166 and miR156) that we previously identified4 could be easily detected in EVs, whereas AGO2-, AGO4- and AGO5-associated sRNAs were undetectable in the EV fractions (Extended Data Fig. 6). These results led us to hypothesize that EV-localized RBPs, such as AGO1, may play an important role in sRNA selective loading into plant EVs because some of the identified RBPs bind to sRNAs specifically. To determine whether identified EV-encased AGO1 and other RBPs specifically associate with secreted sRNAs, we immunoprecipitated RBPs from total cell extraction and EV extraction and analyzed the co-purified sRNAs. Our results showed that AGO1, RH11 and RH37 could bind specifically to EV-enriched sRNAs 4, such as miR166, TAS1c-siR483 and TAS2-siR453, in both the total cell extraction and the EV extraction (Fig. 5a, b). We previously showed that TAS1c-siR585 and TAS2-siR710, which are derived from the same mRNA precursors as TAS1c-siR483 and TAS2-siR453, respectively, were not enriched in the EVs 4. These non-EV sRNAs were not associated with AGO1, RH11, and RH37 in either the total cell extraction or the EV extraction (Fig. 5a, b). We also co-expressed AGO1 or RH37 with EV-enriched and non-EV sRNAs in Nicotiana benthamiana to confirm the selective binding between AGO1 and RH37 with EV-enriched sRNAs. As shown in Extended Data Fig. 7, both AGO1 and RH37 can only selectively bind EV-enriched sRNAs. Because of the selective binding of EV-enriched sRNAs in the total cell extraction, AGO1, RH11 and RH37 likely bind to a specific set of sRNAs and carry them into EVs for secretion. However, ANN1 and ANN2 had no sRNA binding specificity, they could bind both EV-enriched and non-EV sRNAs in the total cell extraction (Fig. 5a, b). These results suggest that AGO1, RH11 and RH37 function in selectively loading sRNAs into EVs, while ANN1 and ANN2 likely only contribute to the stabilization of RNA molecules once inside EVs.
Fig. 5. EV secreted RBPs mediate sorting and stabilization of sRNAs into EVs.
a, b, sRNAs were immunoprecipitated from plant total extraction (a) and EV extraction (b) using antibodies raised against AGO1, AGO2 and GFP to detect EV-enriched and non-EV-enriched sRNA association with AGO1, AGO2, RH11, RH37, ANN1 and ANN2. AGO1 and AGO2 proteins were detected by western blot using anti-AGO1 and anti-AGO2 antibodies, respectively. RH11, RH37, ANN1, and ANN2 were detected by western blot using anti-GFP antibodies. IgG was used as a negative control. The experiments in Fig. 5a and b were repeated three times with similar results. Source data are provided as a Source Data file.
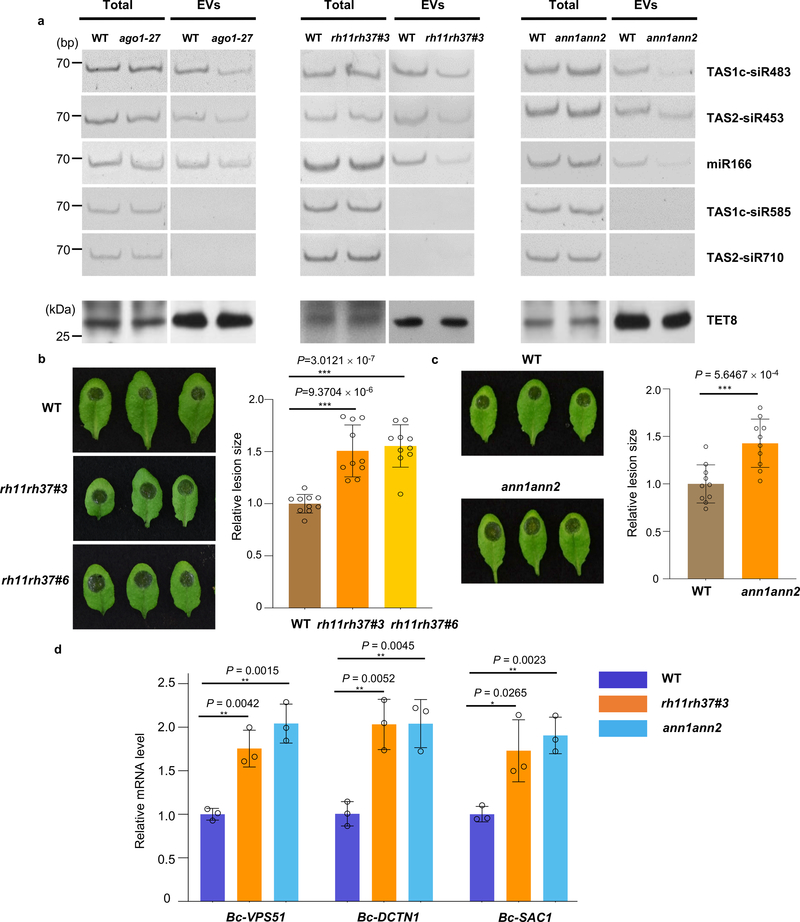
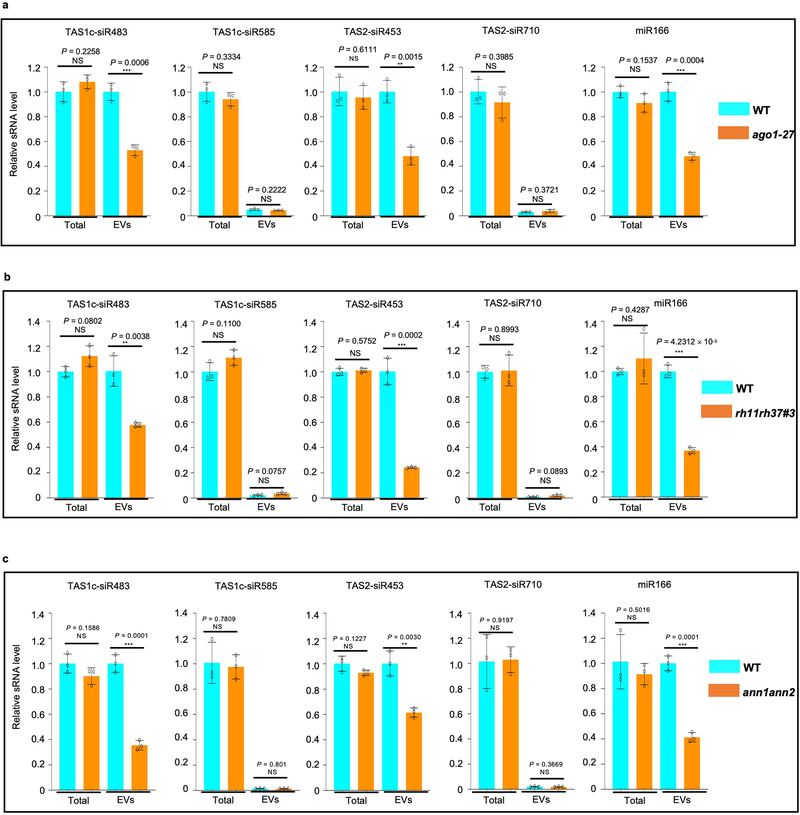
To genetically confirm their function in sRNA secretion, we investigated both total and EV-associated sRNA levels by both sRNA reverse-transcription PCR (RT-PCR) and real-time quantitative PCR in the ago1–27, rh11rh37, and ann1ann2 double mutants. We generated the rh11rh37 double mutants by knocking down RH11 and RH37 expression with an artificial miRNA that can target both genes simultaneously in Col-0 (rh11rh37#3) or by knocking down RH37 expression in the rh11 knockout mutant background (rh11rh37#6) and generated the ann1ann2 double mutants by crossing single knockout mutants ann1 (SALK_095886C) and ann2 (SALK_205024C) (Extended Data Fig. 8). We found that the sRNA level in the EVs isolated from ago1–27, rh11rh37 and ann1ann2 was clearly decreased in comparison to wild-type plants (Fig. 6a and Extended Data Fig. 9), but the total amount of sRNAs from ago1–27, rh11rh37 and ann1ann2 was similar to wild-type plants. Furthermore, the levels of TET8 protein in the EV fractions are similar between RBP mutants and WT (Fig. 6a), indicating that ago1–27, rh11rh37 and ann1ann2 do not affect TET8-positive EV secretion, and the reduction of EV-enriched sRNA accumulation in these RBP mutants is not due to a reduction in the amount of TET8-positive EVs.
Fig. 6. EV-associated RBPs contribute to plant immunity to fungal infection.
a, sRNAs were detected by means of sRNA RT-PCR in EVs isolated from wild-type, ago1–27, rh11rh37 and ann1ann2 mutant plants. The “total” lane indicates total RNAs from leaves. The “EVs” lane indicates total RNAs from isolated EV fractions. TET8 was detected by western blot using anti-TET8 antibodies. Similar results were observed in three biological repeats. b, c, The rh11rh37 (b) and ann1ann2 (c) mutant plants exhibited enhanced susceptibility to B. cinerea in comparison to the wild-type. Relative lesion sizes were measured 2 days after infection. The data are presented as mean ± s.d., n = 10 biological replicates. d, Fungal target genes of transferred sRNAs were de-repressed in B. cinerea collected from the rh11rh37 and ann1ann2 mutants. The data are presented as mean ± s.d., n = 3 biological replicates. The statistical analysis in Fig. 6b and d was performed using ANOVA Dunnett’s multiple comparisons test. The statistical analysis in Fig. 6c was performed using unpaired two-tailed Student’s t-test. The small open circles represent the individual values. The error bars indicate the standard deviation (s.d.). The asterisks indicate significant differences: *P < 0.05, **P < 0.01. Source data are provided as a Source Data file.
As EV-localized RBPs mediate selective loading and stabilization of secreted sRNAs, many of which move into fungal cells to silence fungal virulence-related genes, they are likely contribute to plant immune responses against fungal infection. Indeed, we found that the rh11rh37 and ann1ann2 mutants were more susceptible to B. cinerea in comparison to wild-type plants (Fig. 6b, c). Meanwhile, the expression of fungal virulence-related genes that are targeted by plant secreted sRNAs were derepressed in B. cinerea that collected from rh11rh37 and ann1ann2 mutants (Fig. 6d). These results support that EV-localized RBPs contribute to host immunity, likely by sorting and stabilizing fungal gene-targeting sRNAs in EVs.
Discussion
Previously, we demonstrated that plants can selectively transport sRNAs into fungal pathogens using EVs to induce cross-kingdom RNAi of fungal virulence-related genes 4. However, the mechanisms of selective sRNA loading and stabilization in EVs were unclear. Although mammalian EV studies suggest that some RBPs are involved in sRNA loading into EVs, different RBPs were identified in different cell lines or systems and a conclusive mechanism is still lacking 16, 17, 30–32. In this study, we identified several RBPs in plant EVs, including the AGO1 protein, RNA helicases, and Annexins. They are co-localized with the endosomal MVB marker ARA6 and are likely packaged into MVB originated exosomes with their associated sRNAs. We further demonstrate that AGO1, RH11 and RH37 bind to EV-enriched sRNAs in the total cell extraction. In the ago1–27 and rh11rh37 mutants, the abundance of EV-enriched sRNAs was reduced. These results suggest that these EV-encased RBPs likely contribute to selective sRNA sorting and stabilization within EVs. Annexins are traditionally viewed as Ca2+- and lipid-binding proteins, but there is more and more evidence of their role in RNA binding. Human Annexin A2 (ANXA2) interacts with mRNAs through its C-terminal helices C and D in domain IV 33, it also regulates the loading of miRNAs into EVs in a sequence-independent manner 18. While Arabidopsis ANN1 and ANN2 bind to both EV-enriched and non-EV-enriched sRNAs in total cell extraction but EV-associated sRNAs were reduced in the ann1ann2 double mutant, suggesting that they do not play a role in selective sRNA loading into EVs but stabilizing sRNAs within EVs.
Studies of mammalian EVs exhibit contradictory results on the role of AGOs in exosomal miRNA sorting 34–36. For example, in colon cancer cell lines, AGO2 was found in exosomes and the knockdown of AGO2 leads to the decrease of specific miRNAs in exosomes 34, although mammalian AGOs bind miRNAs and siRNAs indiscriminately of sequence 37–39. Why selective miRNA loading was affected is still not understood. Discordantly, other studies reported that EVs do not contain AGO2, and it was found to carry and stabilize a population of non-vesicle-associated microRNAs in human plasma 40. To date, the function of AGOs in sRNA selection and loading into EVs is still not clear. In plants, however, AGOs have distinct binding preferences to sRNAs with specific 5’ terminal nucleotides and duplex structures 20, 21. AGO1 is secreted in plant EVs and binds specifically to 20–22 nt sRNAs with 5’-terminal U, suggesting that AGO1 is likely responsible for the selective loading of at least some of the sRNAs in EVs. In animals, MVBs associate with components of miRNA effector complexes, including AGO2, and modulate miRNA activity41. Arabidopsis AGO1 was shown to be enriched in the endoplasmic reticulum 42 and MVBs (Extended Data Fig. 5). Its subcellular compartmentation allows AGO1, with its selectively associated sRNAs, to be efficiently loaded into MVB-derived exosomes.
Increasing evidence suggests that animal EVs are made up of heterogeneous populations of vesicles15, including exosomes, microvesicles and apoptotic bodies 9. RNAs and RNA binding proteins have been found in different classes of EVs isolated from different cells and tissues 43,31. Plants have at least three subclasses of EVs: TET8-positive EVs, PEN1-positive EVs, and exocyst positive organelle (EXPO)-derived EVs4, 24, 44, indicating that EV heterogeneity also exists in plants. But whether PEN1-positive EVs and EXPO-derived EVs are also involved in cross-kingdom RNAi remains to be discovered. Here, we used sucrose density gradient centrifugation and immunoaffinity purification methods to isolate TET8-positive EVs and confirmed that RBPs and sRNAs are enriched in TET8-positive EVs. Recently, animal exosomes have been implicated in transporting various sRNAs between cells within an organism 11–14, 45 or between organisms 46, 47. Thus, sRNA secretion through exosomes or exosome-like EVs has evolved in both plant and animal systems and is likely a conserved mechanism for cell-to-cell communication within an organism as well as communications between interacting organisms. This study helps to elucidate the underlying molecular mechanisms of RNA-based communication and sRNA-mediated gene regulation between different organisms. A better understanding of EV-mediated sRNA trafficking will aide in the development of RNA-containing vesicles as new generation of fungicides and antifungal drugs against crop and potential human pathogens.
Methods
Plant materials
Arabidopsis thaliana ecotype Col-0 was used in this study. Arabidopsis marker lines TET8pro::TET8-GFP 4, 48, 35Spro::ARA6-GFP 4, SYP61pro:: CFP-SYP61 49, single mutant ago1–27 50, tet8 (SALK_136039C), were used as described previously. The ann1ann2 double mutant was generated by crossing the respective homozygous single knockout mutants ann1 (SALK_095886C) and ann2 (SALK_205024C), which were ordered from the Arabidopsis Biological Resource Center (ABRC). The rh11rh37 double mutants were generated by suppressing RH11 and RH37 with an artificial miRNA in Arabidopsis Col-0 or by suppressing RH37 in rh11 knockout mutant. The rh11 knockout mutant line CS423310 was ordered from ABRC. For 35Spro::ANN1-YFP, 35Spro::ANN2-CFP, 35Spro::RH11-CFP and 35Spro::RH37-CFP lines, the full length CDS of ANN1, ANN2, RH11 and RH37 were cloned into the pENTR vector (Life Technologies), then into the destination vector pEARLYGATE 101 for YFP tagging and pEARLYGATE 102 for CFP tagging, respectively. All constructs were introduced into Arabidopsis Col-0 separately to generate 35Spro::ANN1-YFP, 35Spro::ANN2-CFP, 35Spro::RH11-CFP and 35Spro::RH37-CFP lines. For TET8/PEN1 double fluorescence lines, fragments expressing mCherry-tagged PEN1 were generated by overlapping PCR. These fragments were cloned separately into the pENTR vector (Life Technologies), then into the destination vector pK2GW7 using Gateway LR clonase (Life Technologies). Constructs for expressing mCherry-tagged PEN1 were introduced into the 35Spro::TET8-GFP background 48 to generate the double fluorescence lines. Primer sequences are provided in Supplementary Table 3.
Plant extracellular vesicle (EV) isolation
Plant EVs were isolated from Arabidopsis or N. benthamiana apoplastic wash. Plant leaves (For Arabidopsis ~100 4-week old plants, for N. benthamiana ~50 4-week old plants ) were harvested and vacuumed with infiltration buffer (20 mM MES hydrate, 2 mM CaCl2, 0.1 M NaCl, pH 6.0) and centrifuged for 10 min at 900 g to collect the apoplastic fluids. The cellular debris in apoplastic fluids were removed by centrifugation at 2,000 g for 30 min, followed by filtration through a 0.45 μm filter. Next, the apoplastic wash was further purified by centrifugation for 30 min at 10,000 g, the supernatants were then transferred to new ultracentrifuge tubes and centrifuged for 1 h at 100,000 g to obtain the P100 pellet or 40,000 g to obtain the P40 pellet. The supernatants of P40 were centrifuged for 1 h at 100,000 g to obtain the P100–40 pellet. The pelleted material was washed with filtered infiltration buffer at 100,000 g for 1 hour to collect the pellet. To detect the protein levels in P40, P100 and P100–40 fractions, two equal parts of apoplastic wash isolated from TET8-GFP/mcherry-PEN1 were centrifuged to obtain (P40, P100–40) and (P100) respectively. After getting each fraction, the same amount of SDS-loading buffer (200 ul) was used for re-suspension of the pellet, and then the same amount of sample (10 ul) was loaded into each lane for Western blots. AGO1, AGO2, AGO4, AGO5 ANN2 and TET8 were detected by Western blot using antibodies against AGO1 (1:2000 dilution), AGO2 (1:2000 dilution), AGO4 (1:2000 dilution), AGO5 (1:1000 dilution), ANN2 (1:2000 dilution), and TET8 (1:2000 dilution), respectively. TET8-GFP, RH11-CFP, RH37-CFP, ANN1-YFP, ARA6-GFP and CFP-SYP61 were detected by western blot using antibodies against GFP (1:2000 dilution). mCherry-PEN1 was detected by western blot using antibodies against mCherry (1:2000 dilution).
Trypsin treatment
EVs (40 μl per treatment) were resuspended in PBS buffer, trypsin solution (Sigma) was added into EVs to the final concentration of 10 μg/ml, or Triton X-100 (Sigma) to the final concentration of 1%, or a combination of both trypsin and Triton X-100. EVs were incubated at 37°C for 30min, samples were analyzed by western blot with antibodies against AGO1, GFP and TET8, respectively.
Sucrose gradient separation of EVs
Plant EVs were purified by discontinuous sucrose density gradient centrifugation. We prepared 10–90% sucrose stocks (w/v), including 10, 16, 22, 28, 34, 40,46, 52, 58, 64, 70, and 90%, using infiltration buffer (20 mM MES hydrate, 2 mM CaCl2, 0.1 M NaCl, pH 6.0). The discontinuous gradient was prepared by layering 1 ml of each solution in the 15 ml ultracentrifuge tube. For top loading, the P100 fraction (in 100 μl infiltration buffer) was premixed with 1 ml of 10% sucrose stock. Afterward, samples were centrifuged in a swinging-bucket rotor for 16 h at 100,000 g, 4 °C and six fractions (2 ml each) were collected. For bottom loading, the P100 fraction was premixed with 1 ml of 90% sucrose stock, then centrifuged in a swinging-bucket rotor for 72 h at 100,000 g, 4 °C and six fractions (2 ml each) were collected. Collected fractions were transferred to new ultracentrifuge tubes and each sample was diluted to 12 ml using infiltration buffer, followed by a final centrifugation for 1 h at 100,000 g, 4 °C to obtain pellet for further analysis.
Immuno-isolation of EVs
For TET8-EV immuno-isolation, protein A beads (Roche) were coupled to antibodies [by either Rabbit polyclonal anti-Arabidopsis TET8 (homemade) or rabbit immunoglobulin G (Thermo Fisher)] in IP buffer (20 mM MES hydrate, 2 mM CaCl2, 0.1 M NaCl, Adjust pH to 7.5). Beads were then washed with IP buffer (containing 0.3% BSA). Plant EVs were resuspended in IP buffer and then added into beads, followed by overnight incubation at 4 °C. Bead-bound EVs were collected and washed by IP buffer. RNA was extracted from the Bead-bound EVs with Trizol reagent (Invitrogen). For detecting proteins, the Bead-bound EVs were boiled with SDS-loading buffer and resolved in an SDS-PAGE gel.
Immunoprecipitation
Anti-GFP (Invitrogen, Rabbit polyclonal antibody) was used to immunoprecipitate RH11, RH37, ANN1 and ANN2 from 35Spro::RH11-CFP, 35Spro::RH37-CFP, 35Spro::ANN1-YFP and 35Spro::ANN2-CFP lines. Anti-AGO1 and AGO2 (home-made, Rabbit polyclonal antibody) were used to immnoprecipitate AGO1 and AGO2 from Arabidopsis Col-0 plants. Rabbit immunoglobulin G (Thermo Fisher) was used as a control. Before being coupled with the antibody, the Protein A beads (Roche) were blocked in IP extraction buffer (20 mM Tris–HCl at pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.5% NP40, proteinase inhibitor cocktail; Sigma) containing 1% BSA and 0.1 mg/ml RNA from yeast (Sigma) for 1 hour at 4 °C. The beads were coupled with antibodies in IP extraction buffer and then washed with IP extraction buffer (containing 1% BSA and 0.1 mg/ml RNA from yeast). Plant total and EV extraction were lysed by IP extraction buffer and then added into antibody coupled beads, followed by overnight incubation at 4 °C. The Bead-bound protein-RNA complex was collected and washed by IP extraction buffer. For immunoprecipitation of ANN1 and ANN2, IP extraction buffer containing 1 mM CaCl2 was used. RNA was extracted from the bead-bound EVs with Trizol reagent (Invitrogen). For detecting proteins, the bead-bound EVs were boiled with SDS-loading buffer and resolved in an SDS-PAGE gel.
Transient co-expression assays
Transient co-expression assays in N. benthamiana were performed by infiltrating 3-week-old N. benthamiana plants with Agrobacterium tumefaciens (OD600 =1.0) strain GV3101 carrying the corresponding cloned binary vectors. Constructs for expressing CFP or YFP-tagged ARA6, AGO1, RH11, RH37, ANN1, and ANN2 were generated using pEarleyGate binary vectors. For the TET8-mCherry construct, fragment expression C-terminal mCherry-tagged TET8 was generated by overlapping PCR. The fragment was cloned into the pENTR vector (Life Technologies) and then into the destination vector pK2GW7 using Gateway LR clonase (Life Technologies). For the CFP tagged PEN1 construct, overlapping PCR was used to tag PEN1 with CFP at the N-terminus and the corresponding fragment was introduced into pK2GW7 using Gateway LR clonase (Life Technologies). TAS1c-siR483, TAS1c-siR585, TAS2-siR453, TAS2-siR710 and miR166 were cloned into the pENTR vector (Life Technologies) and then into the destination vector pEarleyGate 100 to overexpress in N. benthamiana. Primer sequences are provided in Supplementary Table 3.
Confocal microscopy analyses
For visualization of EV-associated fluorescence in ultracentrifuge fractions, pellets were suspended in EV infiltration buffer and examined using a 40x water immersion lens mounted on a Leica TCS SP5 confocal microscope as previously described 4. For visualization of protein localization, N. benthamiana leaves that transiently co-expressed fluorescent protein labeled RBPs with ARA6 or TET8 were examined using a 40x water immersion lens on a Leica TCS SP5 confocal microscope. Lines were drawn to determine a region of interest (ROI) and fluorescence intensity was determined per pixel along the ROI.
sRNA expression analyses
RNA was extracted using the Trizol extraction method. sRNA RT-PCR was performed as previously described5. The PCR products were visualized using a 12% PAGE gel. Quantitative PCR was performed with the CFX96 real-time PCR detection system (Bio-Rad) using the SYBR Green mix (Bio-Rad). Primer sequences are provided in Supplementary Table 3.
Mass spectrometry
EV samples were loaded onto a 12.5% SDS-PAGE gel and, after a very short separation, the gel slices containing the proteins were excised and cut into small pieces. The proteins were then reduced in-gel with dithiothreitol, alkylated with iodoacetamide, and subsequently digested with modified MS-grade trypsin (Thermo Pierce) at an enzyme/substrate ratio was 1:100 in 50 mM NH4HCO3 (pH 8.5) at 37 °C overnight. Subsequently, peptides were extracted from gels with a solution containing 5% acetic acid in H2O and then 50% CH3CN with 2.5% acetic acid in H2O (v/v). The peptide mixture was subsequently dried in a Speed-vac. The resulting peptide samples were desalted by employing OMIX C18 pipet tips (Agilent Technologies, Santa Clara, CA). LC–MS/MS analyses were conducted on an Q Exactive Plus mass spectrometer equipped with an EASY-nLC 1200 system (Thermo Fisher Scientific). Raw files were searched using Maxquant, version 1.5.2.8. Arabidopsis gene accession numbers for all proteins identified in our EVs fraction are provided in Supplementary Table 1. Proteins that were present in all three replicates and had an average peptide count of no less than eight were used for further analysis. The selected proteomes were categorized based on GO annotation using The Arabidopsis Information Resource bulk data retrieval and analysis tool PANTHER 14.1 (https://www.arabidopsis.org/tools/bulk/go/index.jsp).
TET8 polyclonal antibody preparation
A fragment coding 132-amino acid TET8-specific peptide, which consisted of amino acids 99–230 of the EC2 domain protein, was cloned into the pMAL-C5X vector (NEB) to fuse with maltose binding protein (MBP) tag. The recombinant protein was expressed in E. coli (BL21) (Thermo Scientific) and purified using Amylose Resin (NEB). After MBP tag was removed by Factor Xa (NEB), the peptide was injected to produce rabbit polyclonal antibodies (prepared by Covance Antibody). The antibody (1:1,000 dilution) was tested by immunoblot analysis using total proteins and EV proteins extracted from wild-type Arabidopsis, which detected the target band of the expected size at 30.7 kD.
Fungal pathogen assays
The B. cinerea spores were diluted in 1% sabouraud maltose broth buffer to a final concentration of 105 spores/ml for drop inoculation of four-week-old Arabidopsis plants. The lesion sizes of B. cinerea-infected plant materials were measured and calculated using ImageJ 1.52r software.
Statistical analysis
The statistical analyses were performed using ANOVA Dunnett’s multiple comparisons test and unpaired two-tailed Student’s t-test using GraphPad Prism version 9.0.0 software. The statistical tests and n numbers, including sample sizes or biological replications, are described in the figure legends. All the experiments were repeated at least three times independently, with similar results.
Data availability
The data that support the findings of this study are available within the paper, Supplementary Information, and Source Data or from the corresponding authors upon reasonable request. Additional data related to this study are available from the corresponding author upon request. The raw files for LC-MS/MS analyses generated during this study are available at PeptideAtlas with the identifier number of PASS01572 (http://www.peptideatlas.org/PASS/PASS01572).
Extended Data
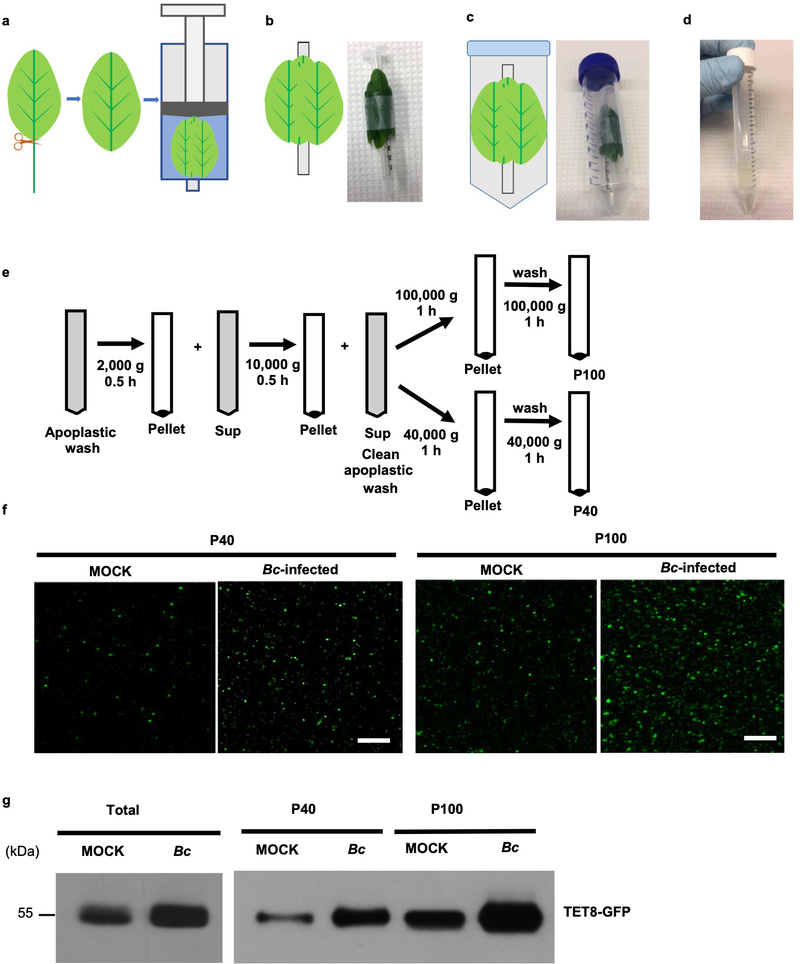
Extended Data Fig. 1. Images show the various steps in plant EVs isolation.
a, Leaves were harvested and rinsed; the petioles were removed. Next, leaves were placed in a syringe with infiltration buffer and vacuumed. b, The leaves were taped onto a 1ml syringe after infiltration buffer was vacuumed into leaves. c, Syringe with taped leaves was placed into a 50 ml conical tube. d, The apoplastic wash was collected by centrifuging the infiltrated leaves at 900 g at 4 °C. e, Scheme of EV isolation by differential ultracentrifugation from apoplastic wash of Arabidopsis. Sup, Supernatant. f, Confocal microscopy images of P40 and P100 fractions isolation by ultracentrifugation from apoplastic wash of TET8pro:TET8-GFP plants. Equivalent amounts of plants were inoculated with B. cinerea for 36 hours before P40 and P100 fraction isolation. Scale bars, 10 μm. g, GFP-labeled TET8 was detectable in both P40 and P100 EV fractions by western blot. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
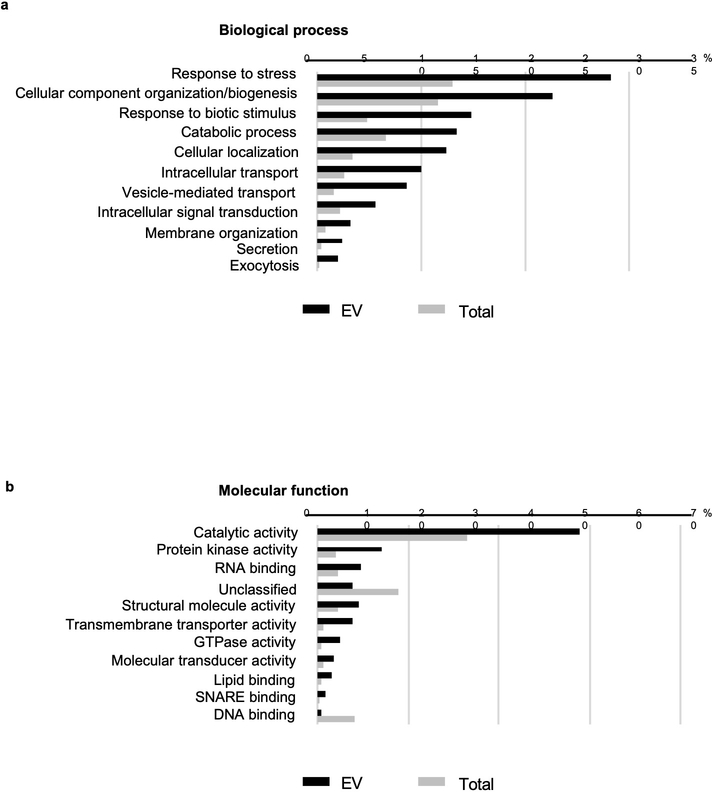
Extended Data Fig. 2. Gene ontology (GO) enrichment analysis of proteins enriched in EVs.
a, b, The plant EV proteome from B. cinerea infected Arabidopsis plants was categorized based on GO terms related to biological process (a) and molecular function (b) through The Arabidopsis Information Resource Web site (www.arabidopsis.org).
Extended Data Fig. 3. TET8 and PEN1 localization.
a, Confocal microscopy images of TET8-YFP and ARA6-CFP at B. cinerea infection site on N. benthamiana. TET8-YFP was partially co-localized with ARA6-CFP signals. b, Fluorescent intensity was quantified for the images used in (a). Transections used for fluorescence intensity measurements are indicated by blue lines. Green and red lines represent histograms of ARA6-CFP and TET8-YFP fluorescent intensities, respectively. c, Confocal microscopy images of CFP-PEN1 and ARA6-YFP at the B. cinerea infection site on N. benthamiana. CFP-PEN1 and ARA6-YFP do not co-localize with ARA6-YFP signals. d, Fluorescence intensity was quantified for the images used in (c). Transections used for fluorescence intensity measurements are indicated by blue lines. Green and red lines represent histograms of CFP-PEN1 and ARA6-YFP fluorescent intensities, respectively. Scale bars, 10 μm.
Extended Data Fig. 4. Bottom loading EV separation by sucrose gradient centrifugation.
a, Pellets obtained from 100,000 g centrifugations (P100) were used to perform sucrose gradient centrifugation by both top and bottom loading. b, Six fractions were collected from bottom loading plant EV sucrose gradient centrifugation. TET8, AGO1, RH11, RH37, ANN1, ANN2 were detected by western blot. EV enriched (TAS1c-siR483, TAS2-siR453 and miR166) and non-EV-enriched (TAS1c-siR585 and TAS2-siR710) sRNAs were detected by RT-PCR. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 5. Colocalization between EV associated RBPs with MVB marker ARA6 and EV markerTET8.
a, b, Fluorescent protein labeled RBPs were co-expressed transiently with MVB marker ARA6 (a) and EV marker TET8 (b) in N. benthamiana. Confocal microscopy was used to determine the localization of RBPs (AGO1, RH11, RH37, ANN1, ANN2) with ARA6 and TET8. AGO2 was used as a control. Scale bars, 10 μm. The experiments were repeated three times independently with similar results.
Extended Data Fig. 6. sRNAs specifically bound by AGO2, AGO4 and AGO5 were absent from plant EVs.
AGO2-associated miR393*, AGO4-associated siR1003, AGO5-associated miR390* and both AGO1 and AGO5-associated miR156 were detected in isolated plant EVs by sRNA RT-PCR. TAS1c-siR483, TAS2-siR453 and miR166 were used as positive control. TAS1c-siR585 and TAS2-siR710 were used as negative controls. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 7. AGO1 and RH37 selectively bind EV-enriched sRNAs in N. benthamiana.
YFP-AGO1 or RH37-CFP were co-expressed with EV-enriched (TAS1c-siR483, TAS2-siR453 and miR166) and non-EV-enriched sRNAs (TAS1c-siR585 and TAS2-siR710) in N. benthamiana, sRNAs were immunoprecipitated from plant total extraction using antibodies against GFP and detected by sRNA RT-PCR. IgG was used as a negative control. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 8. Verification of rh11rh37 and ann1ann2 double mutants.
a, The developmental phenotypes of the rh11rh37 double mutant that both RH11 and RH37 expression was suppressed by artificial miRNA in Col-0. b, The real-time RT-PCR analysis of RH11 and RH37 expression in rh11rh37 mutants. The data are presented as mean ± s.d., n = 3 biologically independent replicates. The error bars indicate the standard deviation (s.d.). c, The developmental phenotypes of the rh11rh37#6 mutant that RH37 expression was suppressed by artificial miRNA in rh11 knockout mutant. d, RT-PCR analysis of RH11 and real-time PCR analysis of RH37 in rh11rh37#6 mutant. The data are presented as mean ± s.d., n = 3 biologically independent replicates. The error bars indicate the standard deviation (s.d.). e, Phenotype of wild-type and ann1ann2 mutant grown for 4 weeks in a growth chamber. f, RT-PCR analysis of the expression levels of ANN1 and ANN2 in wild-type and ann1ann2 double mutant. The statistical analysis was performed using unpaired two-tailed Student’s t-tests. The small open circles represent the individual values. The asterisks indicate significant differences: *P < 0.05, **P < 0.01, ***P < 0.001. The experiments were repeated three times independently with similar results. Source data are provided as a Source Data file.
Extended Data Fig. 9. EV-enriched sRNA amount was reduced in EVs isolated from EV-associated RBP mutants.
The relative level of both EV-enriched and non-EV-enriched sRNAs were examined by real-time RT-PCR in the total fraction and EV fraction from ago1–27 (a), rh11rh37 (b) and ann1ann2 (c) mutants. The data are presented as mean ± s.d., n = 3 biologically independent replicates. The error bars indicate the standard deviation (s.d.). The statistical analysis was performed using unpaired two-tailed Student’s t-tests. The small open circles represent the individual values. The asterisks indicate significant differences: **P < 0.01, ***P < 0.01, NS, not significant.
Supplementary Material
Acknowledgements
We thank N. Raikhel for the 35S pro ::ARA6-GFP and SYP61pro:: CFP-SYP61 seeds, L. Boavida for TET8 pro ::TET8-GFP and 35S pro ::TET8-GFP seeds, and R. Hamby for editing the paper. This work was supported by grants from the National Institute of Health (R01GM093008 & R35GM136379), the National Science Foundation (IOS2017314) and AES-CE (PPA-7517H) to H.J., and the National Institutes of Health (R01 ES025121) to Y.W.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Baulcombe D RNA silencing in plants. Nature 431, 356–363 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Mello CC & Conte D Jr. Revealing the world of RNA interference. Nature 431, 338–342 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Huang CY, Wang H, Hu P, Hamby R & Jin H Small RNAs - big players in plant-microbe interactions. Cell Host Microbe. 26, 173–182 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Cai Q et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiberg A et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai Q, He B, Weiberg A, Buck AH & Jin H Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog. 15, e1008090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiberg A & Jin H Small RNAs--the secret agents in the plant-pathogen interactions. Curr. Opin. Plant Biol. 26, 87–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2, 16151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colombo M, Raposo G & Thery C Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Kowal J, Tkach M & Thery C Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 29, 116–125 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Valadi H et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Lasser C et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med. 9, 9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo SS et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 81, 717–729 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Yan W et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell Biol. 20, 597–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowal J et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 113, E968–E977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santangelo L et al. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 17, 799–808 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S & Schekman R Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5, e19276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagiwara K, Katsuda T, Gailhouste L, Kosaka N & Ochiya T Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 589, 4071–4078 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Vaucheret H Plant Argonautes. Trends Plant Sci. 13, 350–358 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Mi S et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 133, 116–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X et al. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 5, 5468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linder P & Jankowsky E From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol 12, 505–516 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Mingam A et al. DEAD-box RNA helicases in Arabidopsis thaliana: establishing a link between quantitative expression, gene structure and evolution of a family of genes. Plant Biotechnol J. 2, 401–415 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Rutter BD & Innes RW Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 173, 728–741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeppesen DK et al. Reassessment of exosome composition. Cell 177, 428–445 e418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Théry C et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750–1535750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivasan S et al. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell 177, 446–+ (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thery C, Amigorena S, Raposo G & Clayton A Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology / editorial board, Juan S. Bonifacino … [et al.] Chapter 3, Unit 3 22 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Xu R, Greening DW, Rai A, Ji H & Simpson RJ Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87, 11–25 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Chow FW et al. Secretion of an Argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res. 47, 3594–3606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leidal AM et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee K et al. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 17, 1184–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aukrust I et al. The mRNA-binding site of annexin A2 resides in helices C-D of its domain IV. J Mol Biol 368, 1367–1378 (2007). [DOI] [PubMed] [Google Scholar]
- 34.McKenzie AJ et al. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep. 15, 978–987 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldie BJ et al. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 42, 9195–9208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melo SA et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Burroughs AM et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 8, 158–177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Czech B & Hannon GJ Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 12, 19–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arroyo JD et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U S A 108, 5003–5008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gibbings DJ, Ciaudo C, Erhardt M & Voinnet O Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11, 1143–1149 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Li S et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akers JC, Gonda D, Kim R, Carter BS & Chen CC Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. Journal of neuro-oncology 113, 1–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J et al. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22, 4009–4030 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahlgren J et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 40, e130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buck AH et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 5, 5488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z et al. Extracellular vesicle-mediated communication within host-parasite interactions. Frontiers in immunology 9, 3066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boavida LC, Qin P, Broz M, Becker JD & McCormick S Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol. 163, 696–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drakakaki G et al. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res 22, 413–424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaucheret H, Vazquez F, Crété P & Bartel DP The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available within the paper, Supplementary Information, and Source Data or from the corresponding authors upon reasonable request. Additional data related to this study are available from the corresponding author upon request. The raw files for LC-MS/MS analyses generated during this study are available at PeptideAtlas with the identifier number of PASS01572 (http://www.peptideatlas.org/PASS/PASS01572).