Abstract
In the emerging field of 3D bioprinting, cell damage due to large deformations is considered a main cause for cell death and loss of functionality inside the printed construct. Those deformations, in turn, strongly depend on the mechano-elastic response of the cell to the hydrodynamic stresses experienced during printing. In this work, we present a numerical model to simulate the deformation of biological cells in arbitrary three-dimensional flows. We consider cells as an elastic continuum according to the hyperelastic Mooney–Rivlin model. We then employ force calculations on a tetrahedralized volume mesh. To calibrate our model, we perform a series of FluidFM compression experiments with REF52 cells demonstrating that all three parameters of the Mooney–Rivlin model are required for a good description of the experimental data at very large deformations up to 80%. In addition, we validate the model by comparing to previous AFM experiments on bovine endothelial cells and artificial hydrogel particles. To investigate cell deformation in flow, we incorporate our model into Lattice Boltzmann simulations via an Immersed-Boundary algorithm. In linear shear flows, our model shows excellent agreement with analytical calculations and previous simulation data.
Electronic supplementary material
The online version of this article (10.1007/s10237-020-01397-2) contains supplementary material, which is available to authorized users.
Keywords: Hyperelasticity, Cell deformation, Mooney–Rivlin, Atomic force microscopy, Shear flow, Lattice-Boltzmann
Introduction
The dynamic behavior of flowing cells is central to the functioning of organisms and forms the base for a variety of biomedical applications. Technological systems that make use of the elastic behavior of cells are, for example, cell sorting (Shen et al. 2019), real-time deformability cytometry (Otto et al. 2015; Fregin et al. 2019) or probing techniques for cytoskeletal mechanics (Kollmannsberger and Fabry 2011; Gonzalez-Cruz et al. 2012; Huber et al. 2013; Bongiorno et al. 2014; Fischer-Friedrich et al. 2014; Lange et al. 2015; Fischer-Friedrich et al. 2016; Nyberg et al. 2017; Lange et al. 2017; Kubitschke et al. 2017; Jaiswal et al. 2017; Mulla et al. 2019). In most, but not all, of these applications, cell deformations typically remain rather small. A specific example where large deformations become important is 3D bioprinting. Bioprinting is a technology which, analogously to common 3D printing, pushes a suspension of cells in highly viscous hydrogels—so-called bioink—through a fine nozzle to create three-dimensional tissue structures. A major challenge in this process lies in the control of large cell deformations and cell damage during printing. Those deformations arise from hydrodynamic stresses in the printer nozzle and ultimately affect the viability and functionality of the cells in the printed construct (Snyder et al. 2015; Blaeser et al. 2015; Zhao et al. 2015; Paxton et al. 2017; Müller et al. 2020). How exactly these hydrodynamic forces correlate with cell deformation, however, strongly depends on the elastic behavior of the cell and its interaction with the flowing liquid. Theoretical and computational modeling efforts in this area have thus far been restricted to pure fluid simulations without actually incorporating the cells (Khalil and Sun 2007; Aguado et al. 2012; Blaeser et al. 2015) or simple 2D geometries (Tirella et al. 2011; Li et al. 2015). The complexity of cell mechanics and the diversity of possible applications make theoretical modeling of cell mechanics in flow a challenge which, to start with, requires reliable experimental data for large cell deformations.
The most appropriate tool to measure cellular response at large deformations is atomic force microscopy (AFM) (Lulevich et al. 2003; Lulevich et al. 2006; Ladjal et al. 2009; Kiss 2011; Fischer-Friedrich et al. 2014; Hecht et al. 2015; Ghaemi et al. 2016; Sancho et al. 2017; Efremov et al. 2017; Ladjal et al. 2018; Chim et al. 2018). AFM cantilevers with pyramidal tips, colloidal probes, or flat geometries are used to indent or compress cells. Therefore, a common approach to characterize the elasticity of cells utilizes the Hertzian theory, which describes the contact between two linear elastic solids [(Johnson 2003), p. 90–104], but is limited to the range of small deformations (Dintwa et al. 2008). Experimental measurements with medium-to-large deformations typically show significant deviations from the Hertz prediction, e.g., for cells or hydrogel particles (Neubauer et al. 2019). Instead of linear elasticity, a suitable description of cell mechanics for bioprinting applications requires more advanced hyperelastic material properties. While for simple anucleate fluid-filled cells such as, e.g., red blood cells, theoretical models abound (Freund 2014; Závodszky et al. 2017; Mauer et al. 2018; Guckenberger et al. 2018; Kotsalos et al. 2019), the availability of models for cells including a complex cytoskeleton is rather limited. In axisymmetric geometries, Caille et al. (2002) and Mokbel et al. (2017) used an axisymmetric finite element model with neo-Hookean hyperelasticity to model AFM and microchannel experiments on biological cells. In shear flow, approximate analytical treatments are possible (Roscoe 1967; Gao and Hu 2009; Gao et al. 2011; Gao et al. 2012). Computationally, Gao and Hu (2009) carried out 2D simulations while in 3D Lykov et al. (2017) utilized a DPD technique based on a bead-spring model. Furthermore, Villone et al. (2014, 2015) presented an arbitrary Lagrangian-Eulerian approach for elastic particles in viscoelastic fluids. Finally, Rosti et al. (2018) and Saadat et al. (2018) considered viscoelastic and neo-Hookean finite element models, respectively, in shear flow.
In this work, we introduce and calibrate a computational model for fully three-dimensional simulations of cells in arbitrary flows. Our approach uses a Lattice-Boltzmann solver for the fluid and a direct force formulation for the elastic equations. In contrast to earlier works (Caille et al. 2002; Gao et al. 2011; Mokbel et al. 2017; Rosti et al. 2018; Saadat et al. 2018), our model uses a three-parameter Mooney–Rivlin elastic energy functional. To demonstrate the need for this more complex elastic model, we carry out extensive FluidFM indentation experiments for REF52 (rat embryonic fibroblast) cells at large deformation up to 80% (Alexandrova et al. 2008). In addition, our model compares favorably with previous AFM experiments on bovine endothelial cells (Caille et al. 2002) as well as artificial hydrogel particles (Neubauer et al. 2019). Our model provides a much more realistic force-deformation behavior compared to the small-deformation Hertz approximation, but is still simple and fast enough to allow the simulation of dense cell suspensions in reasonable time. Particularly, our approach is less computationally demanding than conventional finite-element methods which usually require large matrix operations. Furthermore, it is easily extensible and allows, e.g., the inclusion of a cell nucleus by the choice of different elastic moduli for different parts of the volume.
We finally present simulations of our cell model in different flow scenarios using an Immersed-Boundary algorithm to couple our model with Lattice Boltzmann fluid calculations. In a plane Couette (linear shear) flow, we investigate the shear stress dependency of single cell deformation, which we compare to the average cell deformation in suspensions with higher volume fractions and show that our results in the neo-Hookean limit are in accordance with earlier elastic cell models (Gao et al. 2011; Rosti et al. 2018; Saadat et al. 2018).
Theory
In general, hyperelastic models are used to describe materials that respond elastically to large deformations [(Bower 2010), p. 93]. Many cell types can be subjected to large reversible shape changes. This section provides a brief overview of the hyperelastic Mooney–Rivlin model implemented in this work.
The displacement of a point is given by
| 1 |
where () refers to the undeformed configuration (material frame) and to the deformed coordinates (spatial frame). We define the deformation gradient tensor and its inverse as [(Bower 2010), p. 14, 18]
| 2 |
Together with the right Cauchy-Green deformation tensor, (material description), we can define the following invariants which are needed for the strain energy density calculation below:
| 3 |
| 4 |
| 5 |
Here,
| 6 |
are the trace of the right Cauchy-Green deformation tensor and its square, respectively. The nonlinear strain energy density of the Mooney–Rivlin model is given by (Mooney 1940; Rivlin 1948)
| 7 |
where , , and are material properties. They correspond—for consistency with linear elasticity in the range of small deformations—to the shear modulus and bulk modulus of the material and are therefore related to the Young’s modulus and the Poisson ratio via [(Bower 2010), p. 74]
| 8 |
Through the choice in (7), we recover the simpler and frequently used (Gao et al. 2011; Saadat et al. 2018) neo-Hookean strain energy density:
| 9 |
As we show later, this can be a sufficient description for some cell types. To control the strength of the second term and quickly switch between neo-Hookean and Mooney–Rivlin strain energy density calculation, we introduce a factor and set
| 10 |
such that , which equals setting in (7), corresponds to the purely neo-Hookean description in (9), while increases the influence of the -term and thus leads to a more pronounced strain hardening as shown in figure S-6 of the Supporting Information.
Tetrahedralized cell model
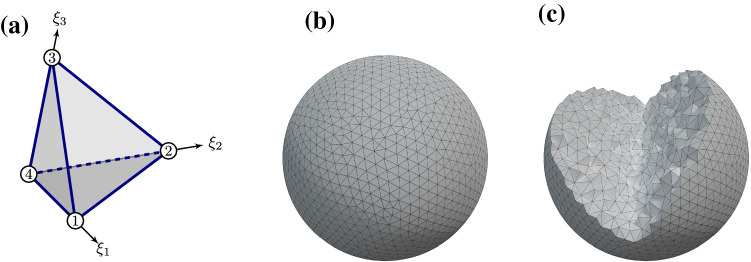
In this section, we apply the hyperelastic theory of Sect. 2 to a tetrahedralized mesh as shown in Fig. 1.
Fig. 1.
a The four noded tetrahedron as mesh element within a local dimensionless coordinate system . b The spherical cell model with its triangulated surface. c Its inner tetrahedralized mesh
Calculation of elastic forces
We consider a mesh consisting of tetrahedral elements as depicted in Fig. 1. The superscript refers to the four vertices of the tetrahedron. The elastic force acting on vertex in direction i is obtained from (7) by differentiating the strain energy density with respect to the vertex displacement as
| 11 |
where is the reference volume of the tetrahedron. In contrast to Saadat et al. (2018), the numerical calculation of the force in our model does not rely on the integration of the stress tensor, but on a differentiation where the calculation of all resulting terms involves only simple arithmetics. Applying the chain rule for differentiation yields:
| 12 |
The evaluation of (12) requires the calculation of the deformation gradient tensor , which is achieved by linear interpolation of the coordinates and displacements inside each tetrahedral mesh element as detailed in the next section. We note that our elastic force calculation is purely local making it straightforward to employ different elastic models in different regions of the cell and/or to combine it with elastic shell models. This flexibility can be used to describe, e.g., the cell nucleus (Caille et al. 2002) or an actin cortex (Bächer and Gekle 2019) surrounding the cell interior.
Interpolation of the displacement field
Following standard methods, e.g., Bower (2010), we start by interpolating a point inside a single tetrahedron using the vertex positions (). The interpolation uses an inscribed, dimensionless coordinate system, denoted by with 1, as depicted in Fig. 1a. One vertex defines the origin while the remaining three indicate the coordinate axes. A set of shape functions, i.e., interpolation functions, is employed to interpolate positions inside the tetrahedron volume. An arbitrary point inside the element is interpolated as
| 13 |
where the shape functions are defined as [(Bower 2010), p. 483]:
| 14 |
| 15 |
| 16 |
| 17 |
According to (1), the displacement of vertex in i-direction is given by
| 18 |
Therefore similar to (13), the displacement at an arbitrary point in the volume can also be expressed in terms of the shape functions and the vertex displacements as
| 19 |
The calculation of the deformation gradient tensor according to (2) requires the spatial derivative of the displacement:
| 20 |
By inserting (19) into (20) and evaluating the shape functions, the components of the matrix are easily determined to be the difference of the displacements between the origin (vertex 4) and the remaining vertices 1, 2 and 3:
| 21 |
Note that due to the linear interpolation is constant inside a given tetrahedron. The matrix is the inverse of the Jacobian matrix, obtained similarly to (21) as
| 22 |
Since refers to the reference coordinates, the calculation of the matrices and has to be performed only once at the beginning of a simulation. With the interpolation of the displacement in each tetrahedron, we can write all derivatives occurring in (12), as listed in the following:
Taylor deformation parameter
As a measure for the cell deformation, we use the Taylor deformation parameter (Ramanujan and Pozrikidis 1998; Clausen and Aidun 2010; Guckenberger et al. 2016; Saadat et al. 2018)
| 23 |
where and are, respectively, the minor and major semi-axis of an ellipsoid corresponding to the inertia tensor of the cell. The Taylor deformation is a good measure for approximately elliptic cell deformations, as they occur in shear flow (cf. Sect. 6).
To calculate , first the components of the inertia tensor
| 24 |
where is a vector inside the volume , are calculated using our discretized cell with tetrahedra as
| 25 |
The vector denotes the center of mass of the tetrahedron and is its current volume. The eigenvalues of can be used to fit the semi axes of the corresponding ellipsoid:
| 26 |
The prefactor contains the mass of the ellipsoid (considering uniform mass density) and drops out in the calculation of .
Comparison of the numerical model to FluidFM measurements on REF52 cells
In this section, we validate compression simulations of our cell model with FluidFM compression experiments of REF52 cells stably expressing paxillin-YFP (Alexandrova et al. 2008). These experiments provide as an output the required force to produce a certain deformation of the cell, which can be directly compared to our model. We start with a detailed description of the experiments and show the suitability of our model to describe the elastic behavior of REF52 cells afterwards.
FluidFM indentation measurements
We perform a series of compression measurements of REF52 cells with a Flex FPM (Nanosurf GmbH, Germany) system that combines the AFM with the FluidFM technology (Cytosurge AG, Switzerland). In contrast to conventional AFM techniques, FluidFM uses flat cantilevers that possess a microchannel connected to a pressure system. By applying a suction pressure, cells can be aspirated and retained at the aperture of the cantilever’s tip. A more detailed description of the setup and its functionality is already reported in Sancho et al. (2017). All experiments are based on a cantilever with an aperture of 8 diameter and a nominal spring constant of 2. In order to measure the cellular deformation, a cell was sucked onto the tip and compressed between the cantilever and the substrate until a setpoint of 100nN was reached. Immediately before the experiment, the cells were detached by using Accutase (Sigma Aldrich) and were therefore in suspension at the time of indentation. In this way, it can be ensured that only a single cell is deformed during each measurement.
An example micrograph of the experiment before compression is shown in Fig. 2. Analogously to AFM, primary data in form of cantilever position (in ) and deflection (in ) has to be converted to force and deformation through the deflection sensitivity (in ) and the cantilevers’ spring constant. The cellular deformation further requires the determination of the contact point, which we choose as the cantilever position where the measured force starts to increase. The undeformed cell size is obtained as mean from a horizontal and vertical diameter measurement using the software imageJ.
Fig. 2.
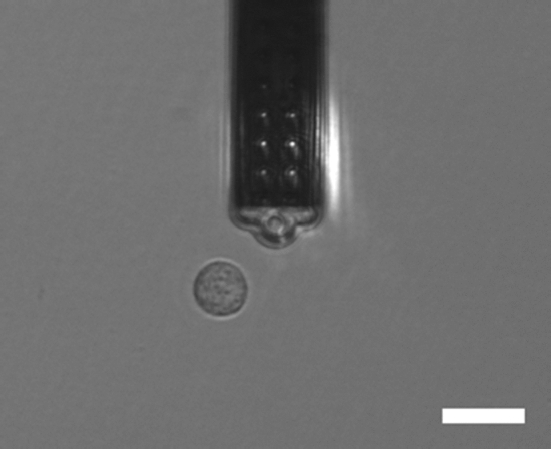
Example micrograph showing the FluidFM cantilever and a cell viewed from the top. Scale bar is 30
Simulation setup
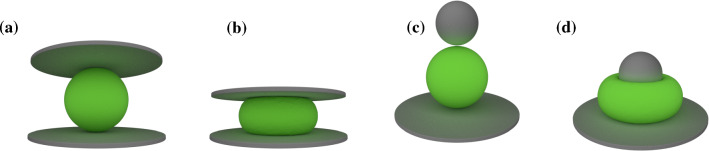
The experimental setup of the previous section is easily transferred and implemented for our cell model: The undeformed spherical cell rests on a fixed plate while a second plate approaches from above to compress the cell as depicted in Fig. 3a and b. In Sect. 5.2, we will also use a slightly modified version where a sphere indents the cell as shown in Fig. 3c and d. A repulsive force prevents the cell vertices from penetrating the plates or the spherical indenter. The elastic restoring forces (cf. Sect. 3) acting against this imposed compression are transmitted throughout the whole mesh, deforming the cell.
Fig. 3.
a and b Cell compression simulations: The cell is compressed between a lower, resting, and an upper, moving, plate. c and d Colloidal probe cell indentation simulations: The cell rests on a plate, while being indented with a sphere
We use meshes consisting of 2000 to 5000 vertices and about 10000 to 30000 tetrahedra to build up a spherical structure. More details of the mesh and its generation (Sect. S-2.4) as well as the algorithm (Sect. S-3) are provided in the SI.
Results
In our FluidFM experiment series with REF52 cells, the cell radii lie between 7.1 and 10.4 with an overall average of 8.6±0.7. In Fig. 4, we depict the force as function of the non-dimensionalized deformation, i.e., the absolute compression divided by the cell diameter. The experimental data curves share general characteristics: The force increases slowly in the range of small deformations up to roughly 40%, while a rapidly increasing force is observed for larger deformations. Although the variation of the cell radius in the different measurements is already taken into account in the deformation, the point of the force upturn differs significantly which indicates a certain variability in the elastic parameters of the individual cells.
Fig. 4.
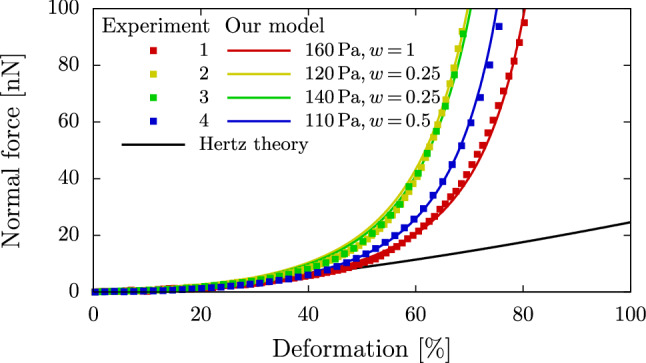
Our numerical model in comparison to our FluidFM measurements on REF52 cells. The labels give the two fit parameters and . We find Young’s moduli in the range of 110Pa to 160Pa. The Hertz theory is shown for a Young’s modulus of 180Pa
We use the compression simulation setup as detailed in Sect. 4.2 to calculate force-deformation curves of our cell model. The Poisson ratio is chosen as . In section S-2.7 of the Supporting Information, we show that variations of do not strongly affect the results. A best fit approach is used to determine the Young’s modulus and the ratio of shear moduli and leads to very good agreement between model prediction and experimental data as shown in Fig. 4 as well as section S-1 of the SI. While the general range of force values is controlled using the Young’s modulus, the Mooney–Rivlin ratio especially defines the point of the force upturn. We find Young’s moduli in the range 110Pa to 160Pa and , 0.5, and 1. For very small deformations, our hyperelastic model produces the same results as would be expected from a linear elastic model according to the Hertz theory. See the SI (section S-2.5) for further details on the calculation of the force-deformation according to the Hertzian theory. For large deformations, the force rapidly increases due to its nonlinear character, showing strain-hardening behavior and huge deviations from the Hertz theory. Overall, we find an excellent match between simulation and our FluidFM measurements with REF52 cells.
Comparison of our numerical model to other micromechanical setups
In this section, we compare our simulations to axisymmetric calculations using the commercial software Abaqus and validate our cell model with further experimental data for bovine endothelial cells from (Caille et al. 2002) and very recent data for hydrogel particles from (Neubauer et al. 2019).
Validation with axisymmetric simulations
To validate our model numerically, we compare our simulated force—deformation curves to calculations using the commercial software Abaqus (Smith 2009) (version 6.14).
In Abaqus, we use a rotationally symmetric setup consisting of a two-dimensional semicircle, which is compressed between two planes, similar to our simulation setup in Sect. 4.2 and the finite element model utilized in (Caille et al. 2002). The semicircle has a radius , a Young’s modulus of and a Poisson ratio of . We choose a triangular mesh and the built-in implementation of the hyperelastic neo-Hookean model. In Fig. 5, we see very good agreement between the results of the two different numerical methods.
Fig. 5.
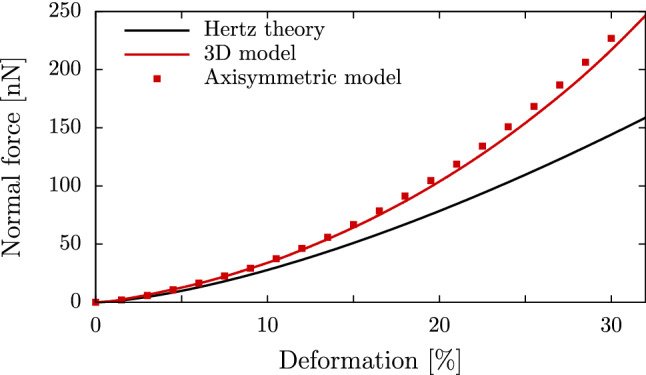
Comparison of force-deformation curves obtained from our model (red line) with the linear elastic Hertz theory (black line) and the two-dimensional simulation with Abaqus (red squares), showing good agreement between our three-dimensional and the axisymmetric model
Validation with AFM experiments
To compare with the AFM experiments of Caille et al. (2002), we simulate a cell with radius 15 using the setup of Sect. 4.2. For the hydrogel particle indentation (Neubauer et al. 2019) we use the setup depicted in Fig. 3c and d with a particle radius of 40 and a radius of the colloidal probe of 26.5. The Poisson ratio is chosen as 0.48 in all simulations and the Young’s modulus is determined using a best fit to the experimental data points. Since the neo-Hookean description appears to be sufficient for these data sets, we further set .
In Fig. 6a, we show the experimental data for suspended, round, bovine endothelial cells of five separate measurements from (Caille et al. 2002) together with the prediction of the Hertz theory for a Young’s modulus of 1000Pa. Fitting our data with Young’s moduli in the range of 550Pa to 2400Pa, we find good agreement between our calculations and the experimental data. We note that Caille et al. (2002) observed similarly good agreement for their axisymmetric incompressible neo-Hookean FEM simulations which, however, cannot be coupled to external flows in contrast to the approach presented here. The same procedure is applied to the colloidal probe indentation data of hydrogel particles from (Neubauer et al. 2019), showing in Fig. 6b the experimental data and the prediction of the Hertz theory from (Neubauer et al. 2019). We find excellent agreement between our model calculations for Young’s moduli in the range of Pa and the experimental data. For both systems, Fig. 6 shows large deviations between the Hertzian theory and the experimental data for medium-to-large deformations. Our model provides a significant improvement in this range.
Fig. 6.
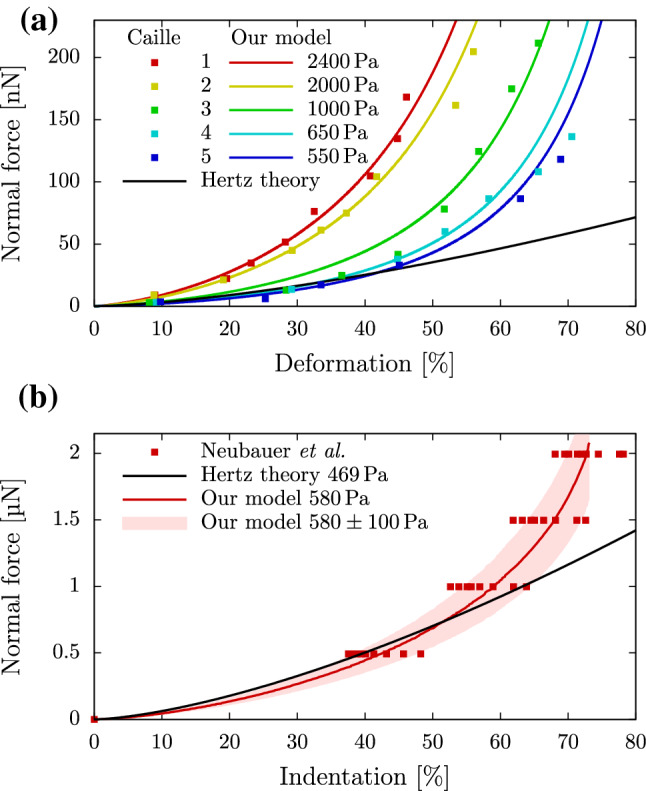
a Our numerical model in comparison to experimental measurements of bovine endothelial cells from (Caille et al. 2002). The black line depicts the prediction of the Hertz theory for a Young’s modulus of 1000Pa. b Our numerical model in comparison to experimental measurements of hydrogel particles from (Neubauer et al. 2019). The indicated range corresponds to the experimentally found range of ± 100Pa for the Young’s modulus according to the depicted Hertz model
Application in shear flow
We now apply our model to study the behavior of cells in a plane Couette (linear shear) flow setup and compare the steady cell deformation to other numerical and analytical cell models of Gao et al. (2011), Rosti et al. (2018) and Saadat et al. (2018). A sketch of the simulation setup is shown in Fig. 7. For simplicity, we choose to reduce the Mooney–Rivlin description (7) to two free parameters and (or and ), obtaining a compressible neo-Hookean form. We use the Lattice Boltzmann implementation of the open source software package ESPResSo (Limbach et al. 2006; Roehm and Arnold 2012). Coupling between fluid and cell is achieved via the immersed-boundary algorithm (Devendran and Peskin 2012; Saadat et al. 2018) which we implemented into ESPResSo (Bächer et al. 2017; Bächer and Gekle 2019). We note here that, in contrast to Saadat et al. (2018), we do not subtract the fluid stress within the particle interior. This leads to a small viscous response of the cell material in addition to its elasticity. To obtain (approximately) the limit of a purely elastic particle, we exploit a recently developed method by Lehmann et al. (2020) to discriminate between the cell interior and exterior during the simulation. Using this technique, we can tune the ratio between inner and outer viscosity with representing a purely elastic particle. For simplicity, we will nevertheless set in the following, except where otherwise noted. Details of the method are provided in the SI (section S-4.1). As measure for the deformation, we investigate the Taylor parameter (23) of our initially spherical cell model in shear flow at different shear rates .
Fig. 7.
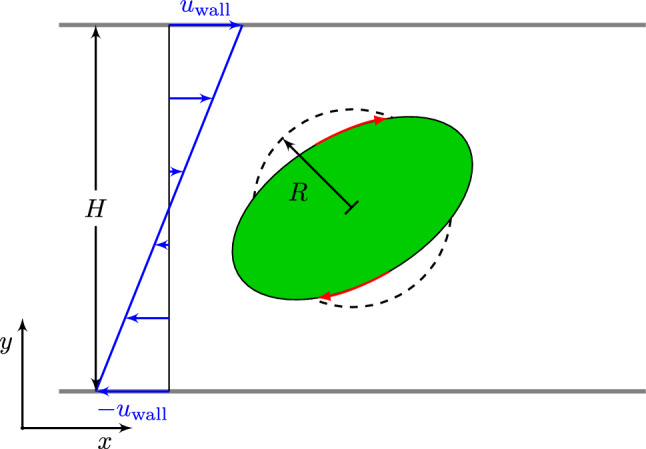
Schematic of the single cell in shear flow. The cell sits in the center of the box and shows an approximately elliptic deformation as well as tank-treading, i.e., a rotation of the membrane around the steady shape in the x-y-plane
Single cell simulation
The first simulation setup, a single cell in infinite shear flow, is realized by choosing a simulation box of the dimensions () in units of the cell radius. The infinite shear flow is approximated by applying a tangential velocity on the x-z-planes at in negative and at in positive x-direction, as depicted in Fig. 7. The tangential wall velocity is calculated using the distance H of the parallel planes and the constant shear rate via
| 27 |
The box is periodic in x and z. A single cell is placed at the center of the simulation box corresponding to a volume fraction of . We choose the following parameters: fluid mass density , dynamic viscosity Pas, and shear rate . The capillary number is defined by (Gao and Hu 2009)
| 28 |
and is used to set the shear modulus of our cell relative to the fluid shear stress . Simulation snapshots of the steady-state deformation of a single cell in shear flow are depicted in dependency of the capillary number in Fig. 8a. We compare the Taylor deformation parameter to previous approximate analytical calculations of Gao et al. (2011) for a three-dimensional elastic solid in infinite shear flow in Fig. 8b and see reasonable agreement for our standard case of . Reducing the inner viscosity by setting , i.e., close to the limit of a purely elastic solid, the agreement is nearly perfect. Finally, we demonstrate that the elastic particle exhibits a tank-treading motion in section S-4.2.
Fig. 8.
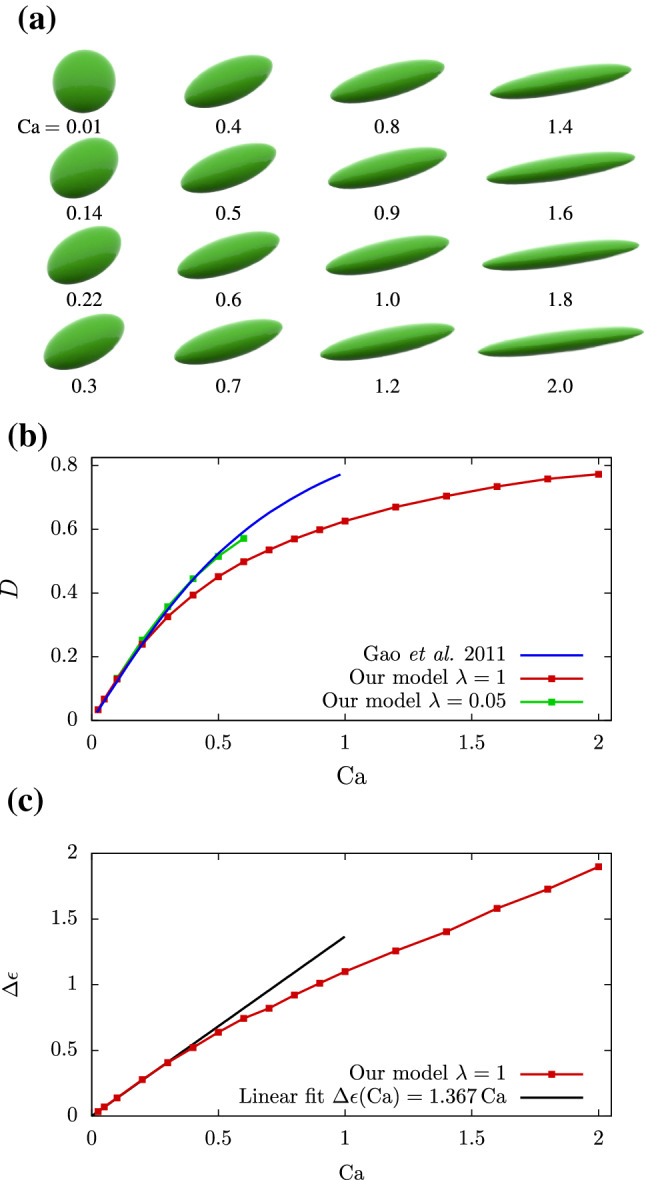
a Converged shapes of a single cell in a () simulation box (in units of the cell radius) with a shear flow in x-direction as function of the capillary number . b Comparison of our model predictions for a single cell in shear flow to the analytical 3D calculations in Fig. 7 of Gao et al. (2011) in the range of . c The relative stretch of our cell model as function of the capillary number . A linear behavior is found for small capillary numbers up to , while increasing stress is required for larger deformations due to the strain-hardening quality of the neo-Hookean model. Lines are a guide to the eye
A possibly even more intuitive way to measure cell deformation is the net strain of the cell which we define as
| 29 |
It describes the relative stretching of the cell using the maximum elongation , i.e., the maximum distance of two cell vertices, and its reference diameter . A strain of thus corresponds to an elongation of the cell by an additional 100% of its original size. In Fig. 8c, we depict the as function of . For small capillary numbers, i.e., small shear stresses, a linear stress-strain dependency is observed. Above , the strain-hardening, nonlinear behavior of the neo-Hookean model can be seen. By stretching the cell up to 280% of its initial size, this plot demonstrates again the capability of our model to smoothly treat large deformations.
Multiple cell simulations
The second simulation setup, implemented to investigate the multiple particle aspect of our model, consists of 4 (8) cells in a simulation box (in units of the cell radius), corresponding to a volume fraction of () occupied by cells. The cells are inserted at random initial positions in the box and the flow parameters are the same as in the first setup (cf. Sect. 6.1).
Figure 9a shows simulation snapshots of the cells in suspensions with volume fraction and for . The Taylor deformation of the suspensions, depicted in Fig. 9b, is calculated as an average over all cells and over time after an initial transient timespan. We find good agreement when comparing the averaged cell deformation in suspension with Rosti et al. (2018), Saadat et al. (2018).
Fig. 9.
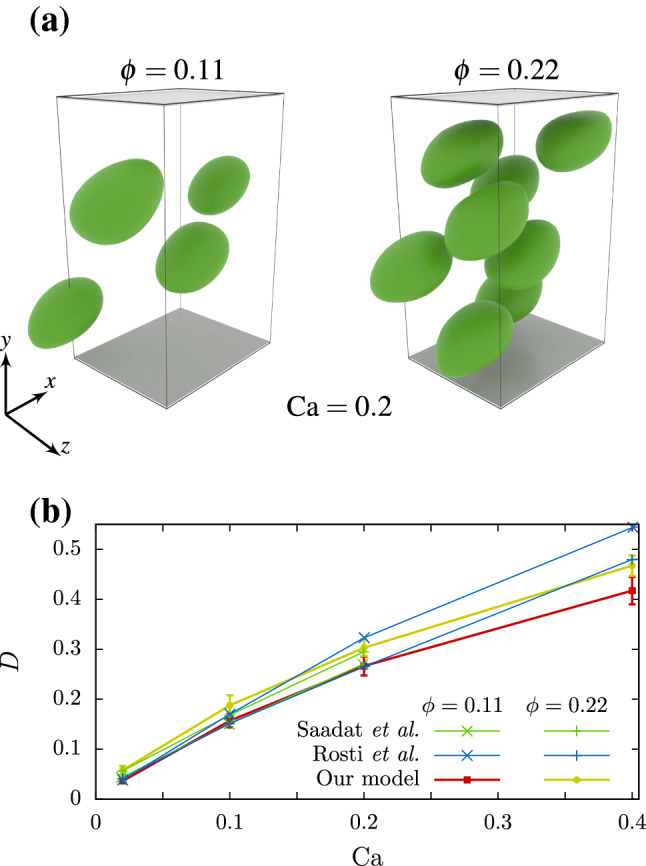
a Multiple cells in a () simulation box (in units of the cell radius) with a confined shear flow in x-direction for a capillary number of and 4 cells corresponding to a volume fraction of , and 8 cells corresponding to . b Averaged deformation of multiple cell simulations with and in comparison to data from Fig. 3 of Rosti et al. (2018) and Fig. 13 of Saadat et al. (2018)
Conclusion
We presented a simple but accurate numerical model for cells and other microscopic particles for the use in computational fluid-particle dynamics simulations.
The elastic behavior of the cells is modeled by applying Mooney–Rivlin strain energy calculations on a uniformly tetrahedralized spherical mesh. We performed a series of FluidFM compression experiments with REF52 cells as an example for cells used in bioprinting processes and found excellent agreement between our numerical model and the measurements if all three parameters of the Mooney–Rivlin model are used. In addition, we showed that the model compares very favorably to force versus deformation data from previous AFM compression experiments on bovine endothelial cells (Caille et al. 2002) as well as colloidal probe AFM indentation of artificial hydrogel particles (Neubauer et al. 2019). At large deformations, a clear improvement compared to Hertzian contact theory has been observed.
By coupling our model to Lattice Boltzmann fluid calculations via the Immersed-Boundary method, the cell deformation in linear shear flow as function of the capillary number was found in good agreement with analytical calculations by Gao et al. (2011) on isolated cells as well as previous simulations of neo-Hookean and viscoelastic solids (Rosti et al. 2018; Saadat et al. 2018) at various volume fractions.
The presented method together with the precise determination of model parameters by FluidFM /AFM experiments may provide an improved set of tools to predict cell deformation— and ultimately cell viability—in strong hydrodynamic flows as occurring, e.g., in bioprinting applications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project number 326998133—TRR 225 ”Biofabrication” (subproject B07). We gratefully acknowledge computing time provided by the SuperMUC system of the Leibniz Rechenzentrum, Garching. We further acknowledge support through the computational resources provided by the Bavarian Polymer Institute. Christian Bächer thanks the Studienstiftung des deutschen Volkes for financial support and acknowledges support by the study program “Biological Physics” of the Elite Network of Bavaria. Furthermore, we thank the laboratory of professor Alexander Bershadsky at Weizmann Insitute of Science in Isreal for providing the REF52 cells stably expressing paxillin-YFP.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict of interest.
Footnotes
(Bower 2010), p. 481, 483] erroneously states a range of for the tetrahedral element.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sebastian J. Müller, Email: sjmueller@uni-bayreuth.de
Stephan Gekle, Email: stephan.gekle@uni-bayreuth.de.
References
- Aguado BA, Mulyasasmita W, Su J, Lampe KJ, Heilshorn SC. Improving viability of stem cells during syringe needle flow through the design of hydrogel cell carriers. Tissue Eng Part A. 2012;18(7–8):806–815. doi: 10.1089/ten.tea.2011.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova AY, Arnold K, Schaub S, Vasiliev JM, Meister JJ, Bershadsky AD, Verkhovsky AB. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS ONE. 2008;3(9):e3234. doi: 10.1371/journal.pone.0003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bächer C, Gekle S. Computational modeling of active deformable membranes embedded in three-dimensional flows. Phys Rev E. 2019;99(6):062418. doi: 10.1103/PhysRevE.99.062418. [DOI] [PubMed] [Google Scholar]
- Bächer C, Schrack L, Gekle S. Clustering of microscopic particles in constricted blood flow. Phys Rev Fluids. 2017;2(1):013102. doi: 10.1103/PhysRevFluids.2.013102. [DOI] [Google Scholar]
- Blaeser A, Duarte Campos DF, Puster U, Richtering W, Stevens MM, Fischer H. Controlling shear stress in 3D bioprinting is a key factor to balance printing resolution and stem cell integrity. Adv Healthcare Mater. 2015;5(3):326–333. doi: 10.1002/adhm.201500677. [DOI] [PubMed] [Google Scholar]
- Bongiorno T, Kazlow J, Mezencev R, Griffiths S, Olivares-Navarrete R, McDonald JF, Schwartz Z, Boyan BD, McDevitt TC, Sulchek T. Mechanical stiffness as an improved single-cell indicator of osteoblastic human mesenchymal stem cell differentiation. J Biomech. 2014;47(9):2197–2204. doi: 10.1016/j.jbiomech.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower AF. Applied Mechanics of Solids. Boca Raton: CRC Press; 2010. [Google Scholar]
- Caille N, Thoumine O, Tardy Y, Meister JJ. Contribution of the nucleus to the mechanical properties of endothelial cells. J Biomech. 2002;35(2):177–187. doi: 10.1016/S0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- Chim YH, Mason LM, Rath N, Olson MF, Tassieri M, Yin H. A one-step procedure to probe the viscoelastic properties of cells by atomic force microscopy. Sci Rep. 2018;8(1):1–12. doi: 10.1038/s41598-018-32704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen JR, Aidun CK. Capsule dynamics and rheology in shear flow: particle pressure and normal stress. Phys Fluids. 2010;22(12):123302. doi: 10.1063/1.3483207. [DOI] [Google Scholar]
- Devendran D, Peskin CS. An immersed boundary energy-based method for incompressible viscoelasticity. J Comput Phys. 2012;231(14):4613–4642. doi: 10.1016/j.jcp.2012.02.020. [DOI] [Google Scholar]
- Dintwa E, Tijskens E, Ramon H. On the accuracy of the Hertz model to describe the normal contact of soft elastic spheres. Granular Matter. 2008;10(3):209–221. doi: 10.1007/s10035-007-0078-7. [DOI] [Google Scholar]
- Efremov YM, Wang WH, Hardy SD, Geahlen RL, Raman A. Measuring nanoscale viscoelastic parameters of cells directly from AFM force-displacement curves. Sci Rep. 2017;7(1):1541–14. doi: 10.1038/s41598-017-01784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Friedrich E, Hyman AA, Jülicher F, Müller DJ, Helenius J. Quantification of surface tension and internal pressure generated by single mitotic cells. Sci Rep. 2014;4:6213. doi: 10.1038/srep06213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer-Friedrich E, Toyoda Y, Cattin CJ, Müller DJ, Hyman AA, Jülicher F. Rheology of the active cell cortex in mitosis. Biophys J. 2016;111(3):589–600. doi: 10.1016/j.bpj.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregin B, Czerwinski F, Biedenweg D, Girardo S, Gross S, Aurich K, Otto O. High-throughput single-cell rheology in complex samples by dynamic real-time deformability cytometry. Nat Commun. 2019;10(1):415. doi: 10.1038/s41467-019-08370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund JB. Numerical Simulation of Flowing Blood Cells. Annu Rev Fluid Mech. 2014;46(1):67–95. doi: 10.1146/annurev-fluid-010313-141349. [DOI] [Google Scholar]
- Gao T, Hu HH. Deformation of elastic particles in viscous shear flow. J Comput Phys. 2009;228(6):2132–2151. doi: 10.1016/j.jcp.2008.11.029. [DOI] [Google Scholar]
- Gao T, Hu HH, Castañeda PP. Rheology of a suspension of elastic particles in a viscous shear flow. J Fluid Mech. 2011;687:209–237. doi: 10.1017/jfm.2011.347. [DOI] [Google Scholar]
- Gao T, Hu HH, Castañeda PP. Shape dynamics and rheology of soft elastic particles in a shear flow. Phys Rev Lett. 2012;108(5):058302–4. doi: 10.1103/PhysRevLett.108.058302. [DOI] [PubMed] [Google Scholar]
- Ghaemi A, Philipp A, Bauer A, Last K, Fery A, Gekle S. Mechanical behaviour of micro-capsules and their rupture under compression. Chem Eng Sci. 2016;142(C):236–243. doi: 10.1016/j.ces.2015.11.002. [DOI] [Google Scholar]
- Gonzalez-Cruz RD, Fonseca VC, Darling EM. Cellular mechanical properties reflect the differentiation potential of adipose-derived mesenchymal stem cells. Proc Nat Acad Sci (USA) 2012;109(24):E1523–E1529. doi: 10.1073/pnas.1120349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckenberger A, Schraml MP, Chen PG, Leonetti M, Gekle S. On the bending algorithms for soft objects in flows. Comput Phys Commun. 2016;207:1–23. doi: 10.1016/j.cpc.2016.04.018. [DOI] [Google Scholar]
- Guckenberger A, Kihm A, John T, Wagner C, Gekle S. Numerical-experimental observation of shape bistability of red blood cells flowing in a microchannel. Soft Matter. 2018;14(11):2032–2043. doi: 10.1039/C7SM02272G. [DOI] [PubMed] [Google Scholar]
- Hecht FM, Rheinlaender J, Schierbaum N, Goldmann WH, Fabry B, Schäffer TE. Imaging viscoelastic properties of live cells by AFM: power-law rheology on the nanoscale. Soft Matter. 2015;11(23):4584–4591. doi: 10.1039/C4SM02718C. [DOI] [PubMed] [Google Scholar]
- Huber F, Schnau J, Rönicke S, Rauch P, Müller K, Fütterer C, Käs J. Emergent complexity of the cytoskeleton: from single filaments to tissue. Adv Phys. 2013;62(1):1–112. doi: 10.1080/00018732.2013.771509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal D, Cowley N, Bian Z, Zheng G, Claffey KP, Hoshino K. Stiffness analysis of 3D spheroids using microtweezers. PLoS One. 2017;12(11):e0188346. doi: 10.1371/journal.pone.0188346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KL. Contact mechanics. 9. Cambridge: Cambridge University Press; 2003. [Google Scholar]
- Khalil S, Sun W. Biopolymer deposition for freeform fabrication of hydrogel tissue constructs. Mater Sci Eng: C. 2007;27(3):469–478. doi: 10.1016/j.msec.2006.05.023. [DOI] [Google Scholar]
- Kiss R. Elasticity of human embryonic stem cells as determined by atomic force microscopy. J Biomech Eng. 2011;133(10):101009. doi: 10.1115/1.4005286. [DOI] [PubMed] [Google Scholar]
- Kollmannsberger P, Fabry B. Linear and nonlinear rheology of living cells. Ann Rev Mater Res. 2011;41(1):75–97. doi: 10.1146/annurev-matsci-062910-100351. [DOI] [Google Scholar]
- Kotsalos C, Latt J, Chopard B. Bridging the computational gap between mesoscopic and continuum modeling of red blood cells for fully resolved blood flow. J Comput Phys. 2019;398:108905. doi: 10.1016/j.jcp.2019.108905. [DOI] [Google Scholar]
- Kubitschke H, Schnauss J, Nnetu KD, Warmt E, Stange R, Kaes J. Actin and microtubule networks contribute differently to cell response for small and large strains. New J Phys. 2017;19(9):093003–13. doi: 10.1088/1367-2630/aa7658. [DOI] [Google Scholar]
- Ladjal H, Hanus JL, Pillarisetti A, Keefer C, Ferreira A, Desai JP (2009) Atomic force microscopy-based single-cell indentation: Experimentation and finite element simulation. In: 2009 IEEE/RSJ International Conference on Intelligent Robots and Systems, IEEE, St. Louis, MO, USA, pp 1326–1332, 10.1109/IROS.2009.5354351
- Ladjal H, Hanus JL, Pillarisetti A, Keefer C, Ferreira A, Desai JP (2018) Atomic force microscopy-based single-cell indentation: Experimentation and finite element simulation. In: 2009 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS 2009), IEEE, pp 1326–1332
- Lange JR, Steinwachs J, Kolb T, Lautscham LA, Harder I, Whyte G, Fabry B. Microconstriction arrays for high-throughput quantitative measurements of cell mechanical properties. Biophys J. 2015;109(1):26–34. doi: 10.1016/j.bpj.2015.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange JR, Metzner C, Richter S, Schneider W, Spermann M, Kolb T, Whyte G, Fabry B. Unbiased high-precision cell mechanical measurements with microconstrictions. Biophys J. 2017;112(7):1472–1480. doi: 10.1016/j.bpj.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Müller SJ, Gekle S. Efficient viscosity contrast calculation for blood flow simulations using the lattice Boltzmann method. Int J Numer Methods Fluids. 2020;103(18):1–15. doi: 10.1002/fld.4835. [DOI] [Google Scholar]
- Li M, Tian X, Kozinski JA, Chen X, Hwang DK. Modeling mechanical cell damage in the bioprinting process employing a conical needle. J Mech Med Biol. 2015;15(05):1550073–15. doi: 10.1142/S0219519415500736. [DOI] [Google Scholar]
- Limbach H, Arnold A, Mann B, Holm C. ESPResSo—an extensible simulation package for research on soft matter systems. Comp Phys Commun. 2006;174(9):704–727. doi: 10.1016/j.cpc.2005.10.005. [DOI] [Google Scholar]
- Lulevich V, Zink T, Chen HY, Liu FT, Gy Liu. Cell mechanics using atomic force microscopy-based single-cell compression. Langmuir. 2006;22(19):8151–8155. doi: 10.1021/la060561p. [DOI] [PubMed] [Google Scholar]
- Lulevich VV, Radtchenko IL, Sukhorukov GB, Vinogradova OI. Deformation properties of nonadhesive polyelectrolyte microcapsules studied with the atomic force microscope. J Phys Chem B. 2003;107(12):2735–2740. doi: 10.1021/jp026927y. [DOI] [Google Scholar]
- Lykov K, Nematbakhsh Y, Shang M, Lim CT, Pivkin IV. Probing eukaryotic cell mechanics via mesoscopic simulations. PLoS Comput Biol. 2017;13(9):e1005726–22. doi: 10.1371/journal.pcbi.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Mendez S, Lanotte L, Nicoud F, Abkarian M, Gompper G, Fedosov DA. Flow-induced transitions of red blood cell shapes under shear. Phys Rev Lett. 2018;121(11):118103. doi: 10.1103/PhysRevLett.121.118103. [DOI] [PubMed] [Google Scholar]
- Mokbel M, Mokbel D, Mietke A, Träber N, Girardo S, Otto O, Guck J, Aland S. Numerical simulation of real-time deformability cytometry to extract cell mechanical properties. ACS Biomater Sci Eng. 2017;3(11):2962–2973. doi: 10.1021/acsbiomaterials.6b00558. [DOI] [PubMed] [Google Scholar]
- Mooney M. A theory of large elastic deformation. J Appl Phys. 1940;11(9):582–592. doi: 10.1063/1.1712836. [DOI] [Google Scholar]
- Mulla Y, MacKintosh FC, Koenderink GH. Origin of Slow Stress Relaxation in the Cytoskeleton. Phys Rev Lett. 2019;122(21):218102. doi: 10.1103/PhysRevLett.122.218102. [DOI] [PubMed] [Google Scholar]
- Müller SJ, Mirzahossein E, Iftekhar EN, Bächer C, Schrüfer S, Schubert DW, Fabry B, Gekle S. Flow and hydrodynamic shear stress inside a printing needle during biofabrication. PLOS ONE. 2020;15(7):e0236371. doi: 10.1371/journal.pone.0236371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer JW, Hauck N, Männel MJ, Seuss M, Fery A, Thiele J. Mechanoresponsive hydrogel particles as a platform for three-dimensional force sensing. ACS Appl Mater Interfaces. 2019;11(29):26307–26313. doi: 10.1021/acsami.9b04312. [DOI] [PubMed] [Google Scholar]
- Nyberg KD, Hu KH, Kleinman SH, Khismatullin DB, Butte MJ, Rowat AC. Quantitative deformability cytometry: rapid, calibrated measurements of cell mechanical properties. Biophys J. 2017;113(7):1574–1584. doi: 10.1016/j.bpj.2017.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Girardo S, Pagliara S, Ekpenyong A, Jacobi A, Wobus M, Töpfner N, Keyser UF, Mansfeld J, Fischer-Friedrich E, Guck J. Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat Methods. 2015;12(3):199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- Paxton N, Smolan W, Böck T, Melchels F, Groll J, Jungst T. Proposal to assess printability of bioinks for extrusion-based bioprinting and evaluation of rheological properties governing bioprintability. Biofabrication. 2017;9(4):044107. doi: 10.1088/1758-5090/aa8dd8. [DOI] [PubMed] [Google Scholar]
- Ramanujan S, Pozrikidis C. Deformation of liquid capsules enclosed by elastic membranes in simple shear flow: large deformations and the effect of fluid viscosities. J Fluid Mech. 1998;361:117–143. doi: 10.1017/S0022112098008714. [DOI] [Google Scholar]
- Rivlin RS. Large elastic deformations of isotropic materials. I. fundamental concepts. Philos Trans R Soc A: Math Phys Eng Sci. 1948;240(822):459–490. doi: 10.1098/rsta.1948.0002. [DOI] [Google Scholar]
- Roehm D, Arnold A. Lattice Boltzmann simulations on GPUs with ESPResSo. Euro Phys J Spec Top. 2012;210(1):89–100. doi: 10.1140/epjst/e2012-01639-6. [DOI] [Google Scholar]
- Roscoe R. On the rheology of a suspension of viscoelastic spheres in a viscous liquid. J Fluid Mech. 1967;28(02):273–21. doi: 10.1017/S002211206700206X. [DOI] [Google Scholar]
- Rosti ME, Brandt L, Mitra D. Rheology of suspensions of viscoelastic spheres: Deformability as an effective volume fraction. Phys Rev Fluids. 2018;3(1):012301. doi: 10.1103/PhysRevFluids.3.012301. [DOI] [Google Scholar]
- Saadat A, Guido CJ, Iaccarino G, Shaqfeh ESG. Immersed-finite-element method for deformable particle suspensions in viscous and viscoelastic media. Phys Rev E. 2018;98(6):063316. doi: 10.1103/PhysRevE.98.063316. [DOI] [Google Scholar]
- Sancho A, Vandersmissen I, Craps S, Luttun A, Groll J. A new strategy to measure intercellular adhesion forces in mature cell-cell contacts. Sci Rep. 2017;7(1):46152–14. doi: 10.1038/srep46152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yalikun Y, Tanaka Y. Recent advances in microfluidic cell sorting systems. Sensors Actuators B: Chem. 2019;282:268–281. doi: 10.1016/j.snb.2018.11.025. [DOI] [Google Scholar]
- Smith M (2009) ABAQUS/Standard User’s Manual, Version 6.9. Dassault Systèmes Simulia Corp, United States
- Snyder J, Rin Son A, Hamid Q, Wang C, Lui Y, Sun W. Mesenchymal stem cell printing and process regulated cell properties. Biofabrication. 2015;7(4):044106. doi: 10.1088/1758-5090/7/4/044106. [DOI] [PubMed] [Google Scholar]
- Tirella A, Vozzi F, Vozzi G, Ahluwalia A. PAM2 (Piston Assisted Microsyringe): a new rapid prototyping technique for biofabrication of cell incorporated scaffolds. Tiss Eng Part C: Methods. 2011;17(2):229–237. doi: 10.1089/ten.tec.2010.0195. [DOI] [PubMed] [Google Scholar]
- Villone MM, Hulsen MA, Anderson PD, Maffettone PL. Simulations of deformable systems in fluids under shear flow using an arbitrary Lagrangian Eulerian technique. Comp Fluids. 2014;90(C):88–100. doi: 10.1016/j.compfluid.2013.11.016. [DOI] [Google Scholar]
- Villone MM, D’Avino G, Hulsen MA, Maffettone PL. Dynamics of prolate spheroidal elastic particles in confined shear flow. Phys Rev E. 2015;92(6):062303. doi: 10.1103/PhysRevE.92.062303. [DOI] [PubMed] [Google Scholar]
- Závodszky G, van Rooij B, Azizi V, Hoekstra A. Cellular level in-silico modeling of blood rheology with an improved material model for red blood cells. Front Physiol. 2017;8:061006–14. doi: 10.3389/fphys.2017.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li Y, Mao S, Sun W, Yao R. The influence of printing parameters on cell survival rate and printability in microextrusion-based 3D cell printing technology. Biofabrication. 2015;7(4):045002. doi: 10.1088/1758-5090/7/4/045002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.