Abstract
Recent advances in single-cell sequencing technologies enable the generation of large-scale data sets of paired TCR sequences from patients with autoimmune disease. Methods to validate and characterize patient-derived TCR data are needed, as well as relevant model systems that can support the development of antigen-specific tolerance inducing drugs. We have generated a pipeline to allow streamlined generation of ‘artificial’ T cells in a robust and reasonably high throughput manner for in vitro and in vivo studies of antigen-specific and patient-derived immune responses. Hereby chimeric (mouse-human) TCR alpha and beta constructs are re-expressed in three different formats for further studies: (i) transiently in HEK cells for peptide-HLA tetramer validation experiments, (ii) stably in the TCR-negative 58 T cell line for functional readouts such as IL-2 production and NFAT-signaling, and lastly (iii) in human HLA-transgenic mice for studies of autoimmune disease and therapeutic interventions. As a proof of concept, we have used human HLA-DRB1∗04:01 restricted TCR sequences specific for a type I diabetes-associated GAD peptide, and an influenza-derived HA peptide. We show that the same chimeric TCR constructs can be used in each of the described assays facilitating sequential validation and prioritization steps leading to humanized animal models.
Keywords: HLA, TCR, Cell lines, Humanized animal models, Tolerance
Abbreviations: APC, antigen presenting cells; BM, bone marrow; Ca2+, calcium; GAD, glutamic acid decarboxylase; GFP, green fluorescent protein; GWAS, Genome-wide association studies; HA, Influenza hemagglutinin; hCD4, human CD4; HLA, Human leukocyte antigen; HSCs, hematopoietic stem cells; hTCR, human TCR; MHC, major histocompatibility complex; NFAT, Nuclear factor of activated T-cells; RA, Rheumatoid arthritis; RAG, Recombination-activating genes; T1D, Type-1 diabetes; TCR, T cell receptor; TCRa, TCR alpha; TCRb, TCR beta; TMR, HLA tetramer
Highlights
-
•
Peptide-HLA-TCR interactions are central to autoimmune disease and can be targeted.
-
•
Experimental model systems are needed to study patient peptide-HLA-TCR interactions.
-
•
Large scale TCR sequencing efforts need assays to validate and prioritize patient TCRs.
-
•
Chimeric TCR expression constructs can be used as a versatile discovery platform.
1. Introduction
Genome-wide association studies (GWAS) have linked specific HLA alleles to several autoimmune diseases. For example, versions of the class II HLA-DRB1∗04 allele are strongly linked to an increased risk of developing rheumatoid arthritis (RA) and type 1 diabetes (T1D) [[1], [2], [3], [4]]. A common model explaining the increased risk relates to the ability of different class II HLA proteins to present peptides linked to disease development, and consequently activation of autoreactive CD4+ T cells [[5], [6], [7]]. In line with this concept, increasing numbers of peptides have been identified as activating T cells in patients with different HLA-restricted autoimmune diseases [[8], [9], [10], [11], [12], [13], [14]]. The fact that RA and T1D patients demonstrate reduced symptoms of disease when treated with Abatacept (a CTLA4-Ig complex that reduces T cell activation) further highlights the role of interactions between antigen presenting cells and T cells in the disease pathogenesis [[15], [16], [17]]. Existing animal models also support this concept where specific α/β TCR – MHC/HLA combinations are sufficient to cause full-blown autoimmune disease [[18], [19], [20]].
Current treatments for autoimmune diseases are predominately comprised of non-specific immunosuppressive drugs [21]. These drugs are often effective but do not provide a cure, and thus need to be chronically administered to the patients. A great hope within the field of autoimmunity is to develop curative immunomodulatory drugs that specifically target disease-causing peptide-HLA-TCR interactions. To achieve this, experimental systems allowing for detailed interrogation of relevant peptide-HLA-TCR interactions and their consequences are needed. Importantly, recent advances allowing for large scale TCR repertoire sequencing in patients will greatly facilitate the identification of relevant TCR sequences for therapeutic interventions [[22], [23], [24], [25]]. Still, TCR validation and prioritization tools are a pre-requisite for exploiting the translational potential of patient repertoire sequencing. Given this, we here present a platform to validate and prioritize TCRs cloned from patients. We believe that such a platform can be central for preclinical studies of antigen-specific tolerance therapies for autoimmune diseases.
2. Material and methods
(See supplementary table 1 for more detailed ordering information for reagents.)
2.1. Cell lines
The HEK293T line was purchased from ATCC and used at a passage number below 15. The cells were cultured in DMEM (Sigma-Aldrich) with 10% heat-inactivated bovine serum, and 1% penicillin-streptomycin-glutamine (100x, Gibco), from here on referred to as complete DMEM (cDMEM). The 58 alpha-beta- TCR negative cell line [26], from here on referred to as 58 cells, was a gift from Dr. Bernard Malissen (Centre d’Immunologie de Marseille Luminy, France). The 58 cell line was cultured in RPMI-1640 (Sigma-Aldrich) with 10% heat-inactivated bovine serum, and 1% penicillin-streptomycin-glutamine (100x, Gibco), from here on referred to as complete RPMI (cRPMI). For activation assays with 58 cells, 2mM Ca2+ was typically added to the medium. Cells were incubated in a humidified incubator with 5% CO2 at 37 °C and handled in laminar flow hoods using standard sterile techniques.
2.2. Mice
DRAG mice (human DRB1∗04:01/DR4 transgene, Rag1−/−, IL2Rγc−/− on NOD background; Stock No: 017914 [27,28]), and B6.Tcrb (Tcrb−/− on C57BL/6 background; Stock No: 002118) were obtained from the Jackson Laboratories. Mice were genotyped with primers specified in supplementary table 1 using genomic DNA extracted from ear punch biopsies with the REDExtract-N-Amp Kit (modified using 20 μl lysis and neutralization buffers, and 5 μl tissue preparation buffer per sample) (Sigma-Aldrich). Strains were crossed to generate RAG.DR4 mice (Rag1−/−, I–Ag7+/+, DR4Tg/0; lacking B and T cells, unable to recombine the TCR locus, and hemizygous for the human DR4 transgene), and TCR.DR4 mice (Tcrb−/−, I–Ag7+/+, DR4Tg/0; lacking α/β T cells, and hemizygous for the human DR4 transgene) on a mixed NOD/C57BL/6 background. Strains were maintained by breeding pairs where one parent was hemizygous for the DR4 transgene, and the other did not express the transgene. All experiments were approved by the local ethical committee at Karolinska Institute, Sweden. Sex-, and age-matched mice aged between 8 and 12 weeks old were used for experiments. Mice were housed in individually ventilated cages (IVC) in a specific pathogen-free environment in a 12/12 h light-dark cycle with standard diet ad libitum.
2.3. Peptides and proteins
The Hemagglutinin306-318 (HA306-318: PKYVKQNTLKLAT), Glutamate decarboxylase115-127 (GAD115-127: MNILLQYVVKSFD), and control (ATIKAEFVXAETPYM) peptides were synthesized by GenScript, reconstituted in DMSO at a concentration of 10 mg/ml, and stored at −20 °C in aliquots until being used. Influenza A H3N2 (A/Aichi/2/1968) and Influenza A H1N1 (A/California/07/2009) Hemagglutinin proteins (SinoBiological) were reconstituted at 50 μg/ml in PBS and kept at −20 °C in aliquots for short term storage.
2.4. TCR expression construct
The GAD65 reactive TCR expression construct was described in Ref. [29]. The sequence for the H3N2 HA reactive TCR HA1.7 was obtained from the NCBI nucleotide database: GenBank accession number X63455.1 (TCRα), and X63456.1 (TCRβ), as deposited by Hewitt et al. [30]. The variable TCRα and TCRβ chains were ordered as separate gBlocks (IDT), or with both the TCRα and TCRβ sequence in the same gBlock, containing 5′ and 3′ adaptors with respective restriction sites, and sequentially cloned into the TCR-pMSCVII-Ametrine (TCR-pMIA) plasmid [29]. Briefly, the TCRα gBlock and vector were digested with SnaBI and SacII (both New England Biolabs), and the digested gBlock product was purified with a DNA clean and concentrate kit (Zymo Research), while the digested TCR-pMIA plasmid was run on an agarose gel and the correct band purified with a gel purification kit (Zymo Research). The purified insert and plasmid backbone were ligated, and the product expanded in DH5α competent cells (Invitrogen). The insert was confirmed by sequencing, and subsequently the TCRβ was cloned into the vector by restriction digestion using MfeI and BstbI (both New England Biolabs), using the same setup as for the cloning of the TCRα chain.
2.5. Transient transfection of HEK293T cells with TCR, CD3, and CD4
HEK293T cells were plated in 24-well plates at a density of ~5 x104 cells/well in 1 ml cDMEM. The next day, cells were transfected with 0.5 μg TCR-pMIA plasmid [29], 0.5 μg mouse CD3 WTdelta-F2A-gamma-T2A-epsilon-P2A-zeta pMIG II plasmid (Addgene [31]) and 0.5 μg pMX human CD4 plasmid (Addgene [32]) in different combinations using Lipofectamine LTX transfection reagent (Invitrogen) and cDMEM lacking antibiotics. Cells were incubated for 24h after transfection before analysis by flow cytometry for surface expression, staining with anti-mouse CD3-APC (Invitrogen), anti-mouse TCRβ-biotin, Streptavidin-PE or Streptavidin-PeCy7 (all three from BD Bioscience), and anti-human CD4-PE (Miltenyi Biotech) antibodies, as well as 7-AAD viability dye (BD Pharmingen).
2.6. Tetramer staining of TCR transfected HEK293T cells
HEK293T cells were harvested, filtered through a 70 μm cell strainer, and stained with anti-mouse CD3-BV421 and anti-human CD4-BUV737 (both BD Biosciences) antibodies. Cells were washed and then stained with near IR fixable viability dye (ThermoFischer Scientific) and washed with sterile room temperature PBS. The cell pellet was then stained with HLA-DRB1∗04:01 HA (PE) and GAD65 (APC) tetramers (1:50 dilution) (generated as described in Ref. [33]). The cells were stained for 1 h in a humidified incubator with 5% CO2 at 37 °C, with intermittent mixing after 30 min. The cells were washed and analyzed on a BD LSRFortessa flow cytometer, using appropriate single-color controls for compensation. The FCS files were analyzed using FlowJo 10.7.1.
2.7. T cell stimulation
For 58 cell co-culture assays, antigen-presenting cells (single-cell suspensions of red blood cell lysed splenocytes from TCR.DR4+ or TCR.DR4-mice) and 58 cells were co-cultured in 200 μl cRPMI/well in U-bottom 96-well plates at a concentration of 2x105 58 cells/well and 2x105 splenocytes/well in cRPMI. Peptides or proteins were added to cultures as indicated in the figures, and cells and supernatant were collected after 24h (peptide, anti-CD3/28, PMA/ionomycin), or 72h (protein).
For HLA/peptide stimulation of 58 cells, HLA-DRB1∗04:01 monomeric protein (500 μg/ml, produced as described in Ref. [34]) was incubated for 72 h at 37 °C with peptide (400 μg/ml, a ~20–30 M excess depending on the peptide) in sodium phosphate buffer (pH 6.0) containing n-octyl β-d-Glucopyranoside (Sigma-Aldrich, 2.5 mg/ml) and Pefabloc SC (Sigma-Aldrich, 1 mM), and then stored at 4 °C until used. Loaded HLA monomers were subsequently coated (0.5–2 μg/well) onto U-bottom 96-well plates in 50 μl PBS for 4 h at 37 °C. The HLA/peptide solution was subsequently flicked off the plate and specific T cells (2x105) and anti-CD28 (1 μg/well) were added to the monomer-coated wells and incubated for 48 h at 37 °C before collecting cells and supernatant.
For assays with splenocytes from retrogenic mice, single-cell suspensions were generated using 40 μm cell strainers (Fisher Scientific), and red blood cells lysed using RBC lysis buffer (ThermoFisher Scientific) following the suggested protocol. Cells were plated in a U-bottom 96-well plate at a concentration of 2x105 cells/well in cRPMI and stimulated as indicated in the figures.
As positive controls, anti-mouse CD3/28 beads (Gibco), anti-CD3 (BioLegend), anti-CD28 (BioLegend), or PMA/Ionomycin (Sigma-Aldrich, stock solution 1.6 mM and 1mM, respectively, in DMSO) were used. Tofacitinib, a JAK inhibitor, was purchased from Sigma-Aldrich, dissolved in DMSO at 5 mg/ml (9.9 mM) concentration, and used at concentrations indicated in the figure.
To evaluate T cell activation, for 58 cells, GFP signal was assessed in cells gated on viable, singlets, and for retrogenic mice, anti-CD44 (Biolegend) and anti-CD25 (Invitrogen) signal was assessed on viable, CD3+, CD4+ singlets using a BD B6 Accuri or a BD FACSVerse. In co-cultures with splenocytes and 58 cells, the splenocytes were removed by gating on the FSC high 58 cells. IL-2 secretion was assessed by ELISA according to manufacturer instructions (BioLegend).
2.8. Retroviral production and transduction
For retroviral production, HEK293T cells were seeded at 3x105 cells/well in 6-well plates with 2 ml cDMEM/well. The next day, the medium was replaced with cDMEM lacking antibiotics, including a brief wash of the cells to remove residual antibiotics. The cells were subsequently transfected using Lipofectamine LTX (Invitrogen) as suggested by the manufacturer, with 2 μg transfer plasmid and 2 μg EcoPak gag-pol-env plasmid (the EcoPak plasmid was a kind gift from Dr. Mark P. Kamps, University of California San Diego, CA, USA). The medium was changed 24h and 48h post-transfection with 2 ml cDMEM/well. The 0–24h medium was discarded, while the 24–48h, and 48–72h medium were collected, centrifuged at 360 g for 10 min, and the cell-free medium was then immediately used to transduce the target cells. For the transduction, 3x105 cells were plated in 500 μl cDMEM per well in 6-well plates, and 1.5–2 ml fresh retroviral supernatant and lipofectamine 2000 (Invitrogen, 1:1000 dilution) was added to the cells. Plates were then centrifuged at 815 g for 2h at room temperature and incubated for 24h in a humidified incubator with 5% CO2 at 37 °C. A second round of transduction was subsequently carried out with supernatant collected 72h post-transfection. The medium of the transduced cells was changed 24h after the last transduction, and cells cultured for 5–6 days, with 2–3 changes of the medium, before using the cells for sorting or assays.
2.9. Generation of 58 cells with stable expression of NFAT-GFP reporter, hCD4, and TCR
The NFAT-GFP reporter cell line (58.NFAT-GFP) was generated by transducing 58 cells with retroviral particles carrying the pSIRV-NFAT-eGFP transfer plasmid (Addgene plasmid) [35]. Five-six days after transduction, cells were plated one cell/well in 96-well plates and expanded for 2 weeks until visible colonies appeared. Colonies were then stimulated with PMA and Ionomycin (100 nM each) for 24h and analyzed by flow cytometry for GFP signal. 58.NFAT-GFP clones were selected based on having a low GFP signal in unstimulated conditions and a strong GFP signal following PMA/Ionomycin stimulation.
58.NFAT-GFP cells expressing human CD4 (58.NFAT-GFP.CD4) were generated by transducing the 58.NFAT-GFP line with retroviral particles carrying the pMX hCD4 transfer plasmid (Addgene [32]) and subsequent sorting of cells based on high CD4 expression using the SH800 cell sorter (Sony Biotechnology). Importantly, human CD4 interacts well with both human and mouse MHC class II molecules [36].
TCR expressing versions of the 58 cell line were generated by transducing 58 cells with the TCR-pMSCVII-Ametrine (hTCR-pMIA) plasmid [29], and sorting cells based on strong Ametrine signal or strong staining with anti-mouse CD3, rabbit anti-TCR alpha/TRAC Picoband polyclonal antibody (Bosterbio), and PE donkey anti-rabbit (BioLegend).
2.10. Generation of retrogenic TCR mice
Bone marrow (BM) cells were isolated by flushing the femurs and tibias from RAG.DR4 mice using a 10 ml syringe and cRPMI. Mononuclear cells were isolated using Histopaque-1077 (Sigma-Aldrich), collecting a generous part of the medium and gradient adjacent to the interphase. Cells were subsequently washed with cRPMI and resuspended in cRPMI with mouse cytokines (50 ng/ml SCF, 20 ng/ml IL-3, and 50 ng/ml TPO; all from PeproTech). Cells were cultured for 2 days before transduction using undiluted TCR retroviral supernatant with the addition of Lipofectamine 2000 (Invitrogen, 1:1000) and mouse cytokines. Plates were spin infected for 2h at 815g, and cultured for 24h, before being spin infected again with new, fresh, TCR retroviral supernatant. The cell culture medium was changed 24h after the last transduction and the cells cultured for another 4–6 days with medium changes (cRPMI and cytokines) every 2–3 days before transplantation. On the day of the transplantation, TCR.DR4+ mice were irradiated with 900 rads, and 4–5h later injected i.v. into the tail vein with the TCR transduced RAG.DR4 BM cells together with fresh BM cells from TCR.DR4 cells at a 1:1 ratio (6–8x105 cells of each). Engraftment was assessed 6–7 weeks later by analyzing peripheral blood for the presence of TCRβ+ T cells which are not formed unless the retrogenic TCR is expressed, allowing for T cell development.
2.11. Statistics
Statistical tests were performed as indicated in the respective figure legend using GraphPad Prism 8.
3. Results
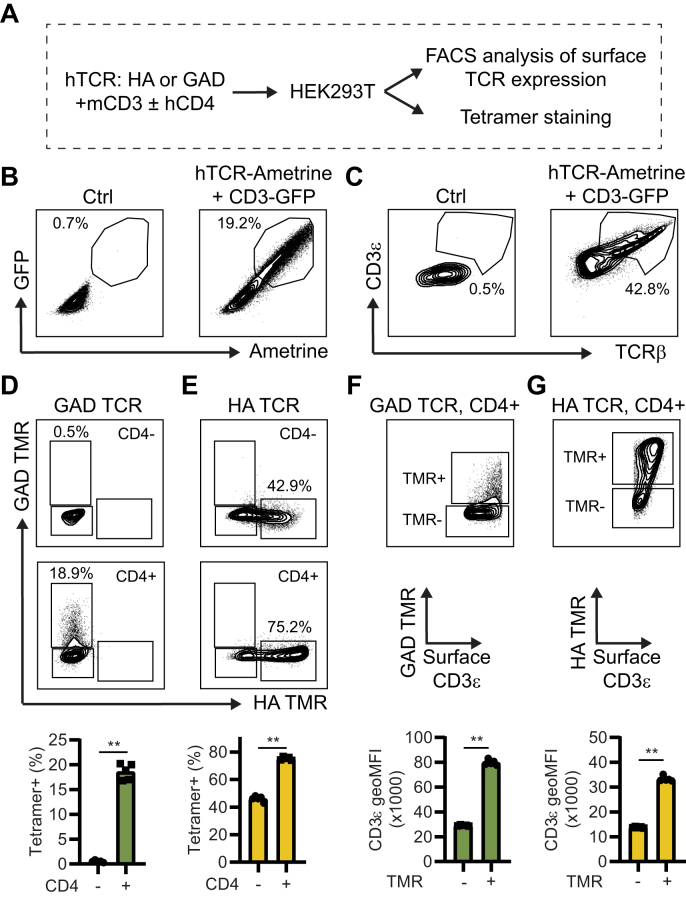
3.1. CD4 and TCR expression levels are important for HLA tetramer binding to HEK293T cells transiently transfected with the TCR complex
To establish a broad experimental pipeline studying HLA-DRB1∗04:01 restricted TCRs, we first aimed to develop a system to study tetramer binding to cloned TCRs. To this end, we used a GAD65 reactive hTCR (GAD), isolated from a type 1 diabetes patient [29], as well as the “HA1.7” influenza (subtype A/H3N2) hemagglutinin (HA) reactive hTCR [30] that we cloned into the retroviral hTCR-pMSCVII-Ametrine (pMIA) plasmid [29]. The generated expression plasmids have the cloned human variable domains (TRAV/TRBV) fused to the mouse constant domains (TRAC/TRBC). This plasmid, thus, encodes a chimeric TCR with specificity defined by the human sequence, yet functional interaction with the mouse TCR signaling interactome via the constant domain, as described in Sprouse et al. [29]. As overviewed in Fig. 1A, the TCR plasmids were transiently transfected into HEK293T cells, together with a plasmid encoding the mouse CD3 complex (CD3ε, CD3γ, CD3δ, CD3ζ, as well as GFP), and a plasmid encoding for human CD4, in different combinations. As expected, a robust but variable uptake of the CD3-GFP and TCR-Ametrine plasmids can be seen 24h post-transfection based on GFP and Ametrine signal (Fig. 1B), and the cells showed proportional surface expression of both TCR and CD3 proteins (Fig. 1C). Next, the TCR (HA and GAD) expressing cells, transfected with the hCD4 plasmid or control, were simultaneously stained with human HLA-DRB1∗04:01 tetramers (TMRs) loaded with either the HA peptide or the GAD peptide. We found that the HA and GAD hTCR cells were positively stained specifically by the anticipated TMRs, and importantly that simultaneous expression of hCD4, expected to stabilize the HLA-TCR interaction, resulted in increased TMR binding (Fig. 1D and E, and Supplementary Fig. 1). This was especially obvious for the GAD TCR, where the hCD4 negative version of the cells was essentially not recognized by the TMR, while the hCD4+ version of the cells, showed clear staining. We also observed that the TMR staining was predominately seen in cells with the highest surface expression of the TCR complex, as indicated by surface CD3ε expression. Efficient transfection can, thus, be a limiting factor for robust tetramer staining (Fig. 1F and G). In conclusion, we show the feasibility of transient transfection of cloned hTCRs in HEK293T cells for TMR binding studies and identify that CD4 and TCR expression levels are important parameters for the sensitivity of the assay.
Fig. 1.
CD4 and TCR expression levels are important for HLA tetramer binding to HEK293T cells transiently transfected with a TCR complex. (A) Schematic representation of transient transfection of HEK293T cells with combinations of different plasmids. (B) HEK293T cells were transiently transfected with mouse CD3-GFP and TCR-Ametrine. 24h later cells were analyzed for GFP and Ametrine signal by flow cytometry. (C) Cells in B were also stained for surface expression of CD3 and TCRb and analyzed by flow cytometry. Data shows cells gated on viable, GFP+, Ametrine + singlets. (D-E) HEK293T cells were transfected with GAD (D), or HA (E) TCR plasmid together with CD3 ± hCD4 plasmids and stained simultaneously with both GAD-specific and HA-specific tetramers (TMR). Flow cytometry data shows cells gated on viable, GFP+, Ametrine+, CD3+ singlets. (F-G) Flow cytometry plots of CD4+ GAD (F), and CD4+ HA (G) TCR expressing HEK 293T cells, gated on viable, GFP+, Ametrine+, CD3+ singlets, comparing tetramer binding to surface expression of the TCR complex (CD3e). ∗∗P < 0.01 by Mann-Whitney test (n = 5/group). Data is representative of three independent experiments.
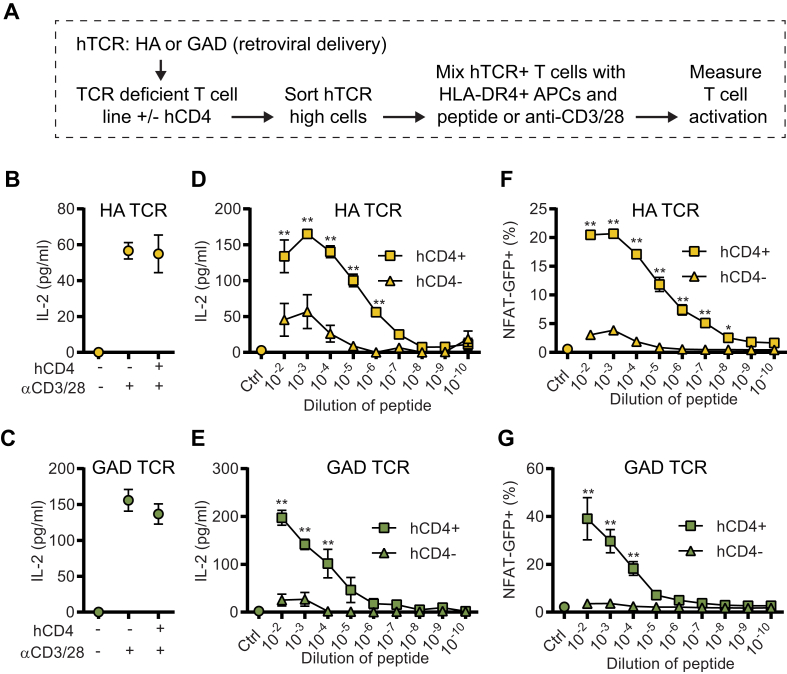
3.2. Overexpression of hCD4 in hTCR 58 cell lines enhances sensitivity to peptide stimulation
Next, we set out to generate an in vitro experimental model system where we could assay T cell activation following crosslinking of the cloned hTCRs. This allows for testing the ability of different peptides, protein preparations, and HLA alleles to trigger the specific TCR. To this end, we tested different TCR-deficient T cell lines and had the greatest success transducing the 58 cell line [26] with the chimeric TCR expression construct (Fig. 2A). Initially, we used IL-2 secretion by ELISA as a readout and found that TCR crosslinking with anti-CD3/28 (Fig. 2B and C), as well as by cognate peptides (Fig. 2D and E) resulted in robust IL-2 secretion. We also wanted to develop complementary readouts for TCR crosslinking. Therefore, we generated an NFAT-GFP reporter version of the 58 cell line, which enables a rapid flow cytometry-based readout for TCR cross-linking (Fig. 2F and G). Based on the increased binding of tetramer to cells in the presence of CD4, as described in Fig. 1, we also generated a stable CD4 expressing version of the 58 cell line. This allowed us to compare how CD4 expression affected the activation of TCR expressing 58 cells. Incubating CD4+ and CD4 negative versions of HA and GAD TCR 58 cells with the cognate peptide presented by splenocytes from TCR.DR4 (TCR-deficient HLA-DRB1∗04:01+) mice identified that CD4 expressing versions of the cells show a strong activation while CD4 negative versions showed a significantly lower response (Fig. 2D–G). In line with the role of CD4 stabilizing TCR-HLA interactions, we did not observe any difference in the response comparing CD4+ and CD4-versions of TCR expressing 58 cells with anti-CD3/28 stimulation, since it does not require CD4 interactions (Fig. 2B and C). Based on the important role of calcium ions (Ca2+) in the cellular activation mediated by TCR crosslinking [37,38], we tested if increased levels of Ca2+, beyond the levels found in the medium, could further increase the sensitivity of the 58 cell assays. While we noted a slightly increased peak response in both the NFAT-GFP and IL-2 readout when increasing the Ca2+ concentration, we did not observe an increased sensitivity in these assays, as defined by the lowest concentration of peptide where a significant activation is seen (Supplementary Fig. 2). Furthermore, we noted that the NFAT-GFP and the IL-2 readouts often correlate, as expected by the direct role of the NFAT transcription factor in the transcription of IL-2 [39]. Still, the correlation was not always perfect, likely influenced by several factors, including the kinetics of the NFAT-GFP signal and the IL-2 secretion. In line with this, we observed a significant suppression of the IL-2 secretion but minor effects on the NFAT-GFP signal by the addition of the JAK inhibitor Tofacitinib [40] to the culture, suggesting that the IL-2 secretion and the NFAT-GFP signal have partial differences in their regulation (Supplementary Fig. 3). We concluded that IL-2 secretion and NFAT-GFP signal are robust complementary readouts that can be used with the 58.NFAT-GFP cells to assess the crosslinking of cloned hTCRs. We also found that CD4 expression is critical to increase the sensitivity of the assay.
Fig. 2.
Overexpression of hCD4 in 58 hTCR cell lines enhances sensitivity to peptide stimulation. (A) Schematic representation of in vitro co-culture of stable hTCR expressing 58.NFAT-GFP cells (+/− hCD4) with DR4+ antigen-presenting cells (APCs; splenocytes from T cell deficient DR4+ mice) stimulated with cognate peptide (stock solution 10 mg/ml) or anti-CD3/28. (B–C) HA-58.NFAT-GFP (+/− hCD4) cell line (B), and GAD-58.NFAT-GFP (+/− hCD4) cell line (C) stimulated with anti-CD3/28 for 24h and IL-2 measured in the supernatant by ELISA. (D-E) Co-culture of HA-58.NFAT-GFP (+/− hCD4) cell line (D), or GAD-58.NFAT-GFP (+/− hCD4) cell line (E) with DR4+ APCs stimulated with different dilutions of the respective cognate peptide and IL-2 measured in the supernatant by ELISA after 24h stimulation. (F-G) Same experiments as D-E, but instead of IL-2, NFAT-GFP signal was recorded by flow cytometry. ∗P < 0.05, and ∗∗P < 0.01 by Two-way ANOVA with Sidak’s multiple comparison test (n = 4/group). Data is representative of three independent experiments.
3.3. Peptide and HLA specific activation of hTCR expressing 58 cells
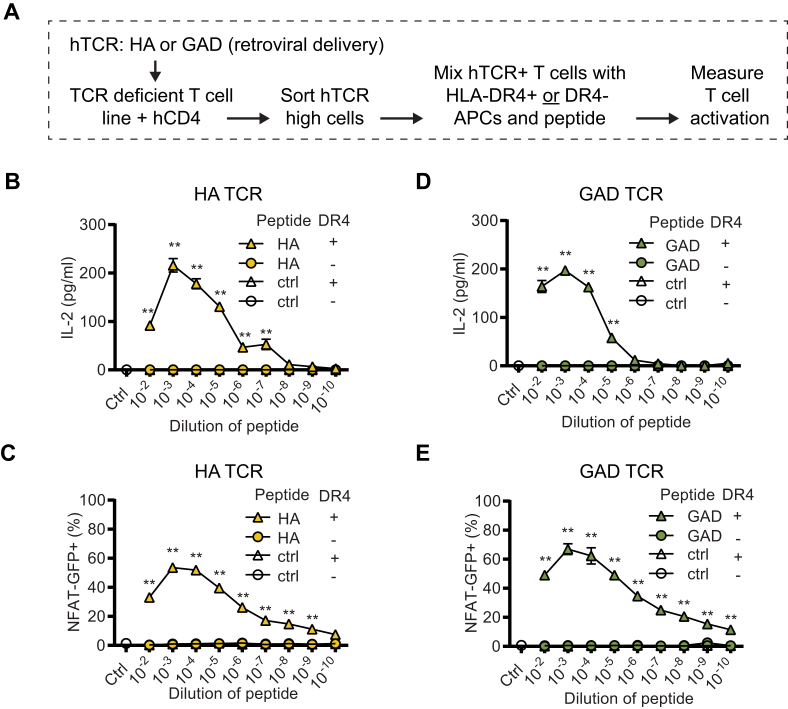
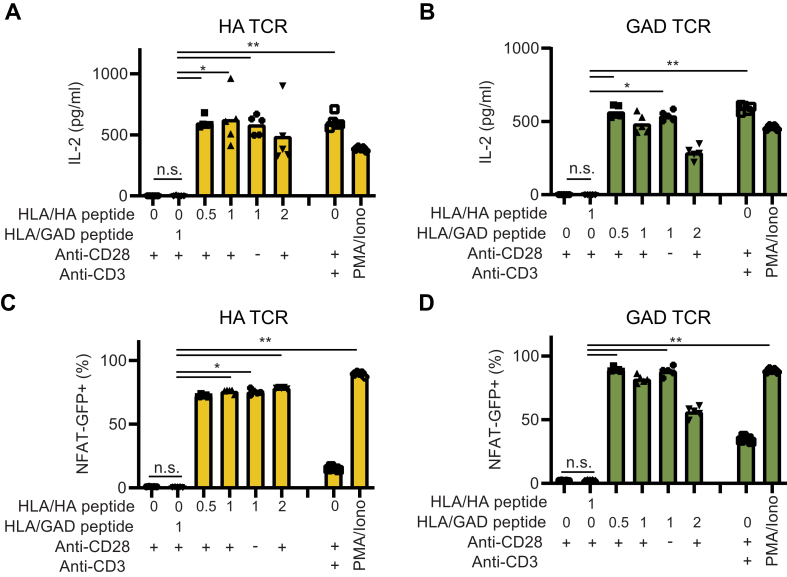
The induction of antigen-specific tolerance in patients with autoimmune disease could be a curative treatment for the patient, similar to what can be achieved by specific immunotherapy (SIT) performed in allergic patients [41,42]. In this regard, assays, where antigen preparations can be tested in diverse HLA context, could be instrumental for the design of novel tolerance-inducing therapeutic preparations. To this end, we wanted to confirm the peptide- and HLA-specificity of our experimental setup. First, we compared the activation of HA or GAD TCR expressing 58.NFAT-GFP.CD4 cells by cognate peptide or control peptide, presented by DR4+ or DR4-antigen-presenting cells (APCs). Similar to the data presented in Fig. 2, we noted a robust activation (both NFAT-GFP signal and IL-2 secretion) of hTCR 58.NFAT-GFP.CD4 cells when co-cultured with DR4+ APCs in the presence of their cognate peptide antigen. Importantly, neither DR4- APCs with cognate peptide, nor DR4+ APCs with control peptide were able to activate the hTCR 58 cells, thus supporting the specificity of the experimental system (Fig. 3A–E). Next, we tested whether an experimental setup that did not depend on using the DR4 transgenic animals as a source for DR4+ APCs could be developed. To this end, we tested if the monomer version of the HLA-DRB1∗04:01 protein preparation, used to assemble the HLA tetramers in Fig. 1, could also be used to present peptides to the hTCR expressing 58 cells in culture. We found that both the HA and GAD TCR expressing 58 cells were readily activated by the specific peptide/HLA combinations, but not by an irrelevant control peptide (GAD peptide for HA TCR, and HA peptide for the GAD TCR) (Fig. 4A–D). We included an anti-CD28 antibody in the monomer stimulations but noticed that the anti-CD28 stimulation was not needed to activate these specific cells, instead, TCR crosslinking is sufficient for activation with the used readouts (Fig. 4A–D, and Supplementary Fig. 4A). In Fig. 2, Fig. 3, Fig. 4, we typically observed that the highest concentration of peptide gave less activation compared to stimulations with slightly lower peptide concentrations, suggesting that too strong stimulation is not optimal for the used readouts. Supporting this observation, adding an anti-CD3 antibody to the hTCR 58 HLA/peptide cultures, thus specifically increasing the TCR signaling, lowered the activation of the hTCR 58 cells (Supplementary Fig. 4B). This could potentially be explained by activation-induced cell death (AICD) [43], where excessive TCR signaling results in cell death [44]. However, including Fas-Ig protein, to block Fas-FasL interactions involved in AICD, did not increase the activation (Supplementary Fig. 4C and D). We concluded that the experimental setup showed good characteristics allowing for assessing specific peptide-HLA-TCR interactions and that HLA monomers could be used to generate an animal-free version of the assay.
Fig. 3.
58.NFAT-GFP CD4+ HA and GAD TCR cell lines are activated by cognate peptide in HLA-DRB1∗04:01 (DR4). (A) Schematic of in vitro stimulation assay. T cell lines were co-cultured with peptide and splenocytes from T cell deficient DR4+ or DR4-mice. (B) HA-58. NFAT-GFP CD4+ cells co-cultured with DR4+ or DR4- APCs stimulated with different dilutions of HA and control peptide. IL-2 was measured in supernatant 24h after the addition of peptides. (C) GAD-58.NFAT-GFP CD4+ cells co-cultured with DR4+ or DR4- APCs stimulated with different dilutions of GAD and control peptide. IL-2 was measured in supernatant 24h after the addition of peptides. (D-E) Same experiments as B–C, but instead of IL-2, NFAT-GFP signal was recorded by flow cytometry. ∗∗P < 0.01 by Two-way ANOVA with Sidak’s multiple comparison test (n = 4), comparing DR4+ and DR4-conditions with cognate peptide. Data is representative of two independent experiments.
Fig. 4.
DRB1∗04:01 monomers loaded with cognate peptide activate TCR expressing 58.NFAT-GFP hCD4+ cells. (A-B) Different amounts (μg/well) of peptide-loaded HLA-DRB1∗04:01 monomers (HLA) were coated on plates for 4h before cells were added and incubated for 48h. IL-2 section was analyzed by ELISA. In (A) cells express the HA TCR, and in (B) GAD TCR. Combinations of anti-CD3 and anti-CD28 (1 μg/well), as well as PMA and ionomycin (100nM each), were included as positive controls. (C-D) The same setup as in (A–B) but instead of IL-2 secretion, NFAT-GFP signal was recorded by flow cytometry. ∗P < 0.05, ∗∗P < 0.01, and n.s. = no significance by Kruskal-Wallis test with Dunn’s multiple comparison correction (n = 5/group) comparing all samples to the HLA/control peptide sample using GraphPad Prism 8. Data is representative of two independent experiments.
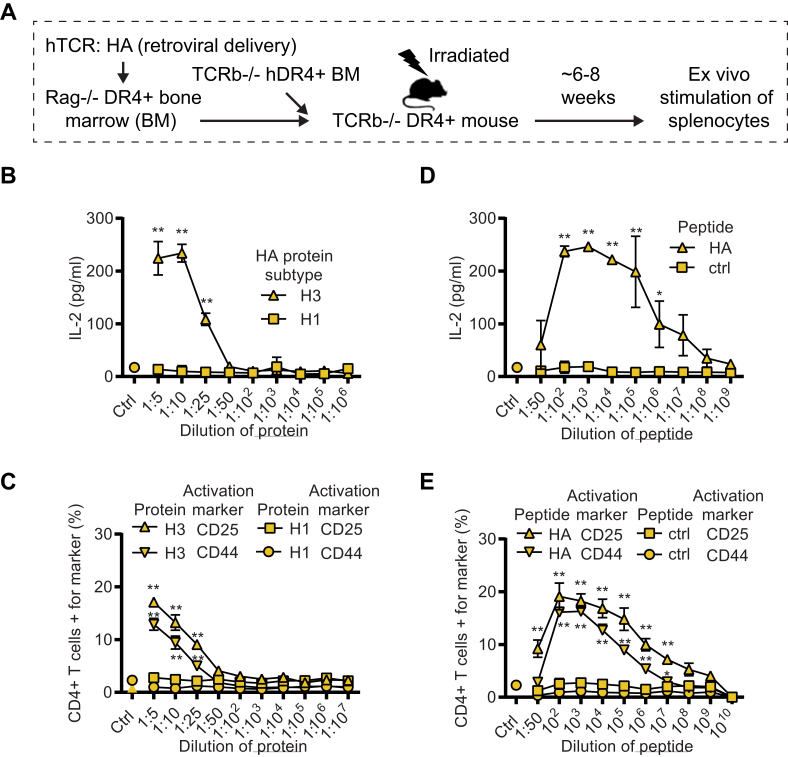
3.4. Splenocytes from HA TCR retrogenic mice activated by cognate protein and peptide
Finally, we wanted to confirm whether we could use the same TCR construct to generate retrogenic mice, thus supporting broad applications of the same TCR expression construct. To this end, we used a similar but partially modified protocol as used by Sprouse et al. [29]. In this, we transduced bone marrow cells from RAG.DR4 (Rag1−/−HLA-DRB1∗04:01+) mice with the retroviral construct, cultured the cells for 5–6 days to remove any residual viral particles and grafted the cells together with TCR.DR4 BM cells into lethally irradiated TCR.DR4 recipient mice (Fig. 5A). 8 weeks after the transplantation we collected splenocytes from the mice and incubated them ex vivo with different concentrations of full-length HA protein (of the relevant H3 or irrelevant H1 subtype, Fig. 5B and C) or peptide (cognate HA peptide, or control peptide, Fig. 5D and E). We found robust IL-2 secretion, as well as upregulation of activation markers (CD44 and CD25) on CD4+ T cells as assessed by flow cytometry (Fig. 5B–E). Importantly, we noted that the cells exclusively responded to the H3 subtype of the HA protein, but not to the H1 subtype (Fig. 5B and C), in line with the fact that the TCR recognizes a peptide in the H3 subtype, not found in the H1 version. We concluded that we could generate retrogenic mice with the modified protocol and that traditional T cell activation markers (CD44 and CD25 upregulation, as well as IL-2 secretion) could be used as readouts for TCR specificity. We also concluded that the retroviral chimeric TCR construct used can be applied seamlessly in all three different assays: Tetramer staining, in vitro activation, and to generate retrogenic mice (Fig. 6).
Fig. 5.
Splenocytes from HA (H3 subtype) TCR retrogenic mice activated by Flu-specific proteins and peptide. (A) Schematic of retrogenic mice generation expressing HA TCR. Rag.DR4+ bone marrow (BM) cells were transduced with the retroviral TCR construct, mixed with TCR.DR4+ BM, and grafted into TCR.DR4+ mice. Spleens were harvested for ex vivo stimulation with proteins and peptides 6–8 weeks after BM transplantation. (B) Splenocytes from retrogenic mice were isolated and stimulated with different dilutions of influenza HA proteins (H3 and H1 subtype, stock solution 50 μg/ml) and IL-2 was measured in the supernatant 3 days after the addition of the proteins. (C) The cells in (B) were analyzed for expression of the activation markers CD25 and CD44 on CD4+ T cells by flow cytometry 3 days after the addition of proteins. Cells were gated on viable, CD3+, CD4+ singlets. (D) Splenocytes from the retrogenic mice were isolated and stimulated with different dilutions of HA or control peptide (stock solution 10 mg/ml) and IL-2 was measured in the supernatant 24h after the addition of the peptide. (E) The cells in (D) were analyzed for expression of the activation markers CD25 and CD44 on CD4+ T cells by flow cytometry. Cells were gated on viable, CD3+, CD4+ singlets. ∗P < 0.05, and ∗∗P < 0.01 by Two-way ANOVA with Sidak’s multiple comparison test using GraphPad Prism 8, comparing H3 and H1 response in (B–C), and HA and ctrl in (D–E). Data is representative of two independent experiments.
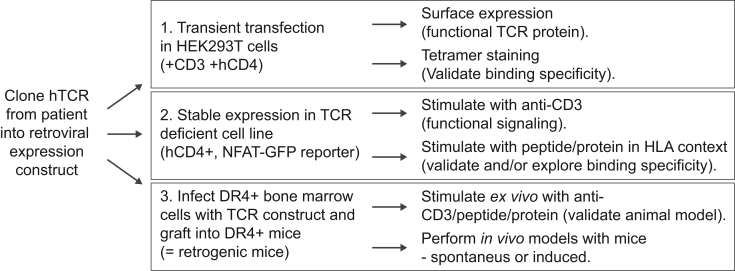
Fig. 6.
Model describing the setup of the presented hTCR discovery platform.
4. Discussion
The development of therapeutic modalities that induce antigen-specific T cell tolerance in patients with autoimmune disease would represent a major medical breakthrough. The concept of specifically suppressing the response to a limited set of autoantigens, while leaving the rest of the immune system intact, should be compared to the current long-term, non-curative, broadly immunosuppressive treatments used for these patients. Multiple tolerance strategies have been suggested and proven in various pre-clinical model systems [41,42,[45], [46], [47], [48]]. Often such strategies are based on providing the autoantigen in a context that induces tolerance (anergy, cell death, or T-reg induction of autoreactive clones), or modify the response in order to cause less tissue damage (e.g. polarization of T helper cells, or modification of antibody subclass or glycoform) [41,42,[49], [50], [51]]. However, no approved drug for antigen-specific tolerance for autoimmune patients exists on the market, despite the positive results from pre-clinical models. In contrast, tolerance induction for patients with allergic diseases has proven to be much more feasible [52,53]. In fact, this therapeutic concept was demonstrated more than 100 years ago [54]. Several explanations for the discrepancy between the allergic and autoimmune tolerance induction can be envisaged. Nevertheless, the absence of relevant preclinical model systems serves as a major bottleneck for developing antigen-specific tolerance inducing strategies.
It is well described that specific HLA alleles are associated with the development of many autoimmune diseases, providing a foundation for peptide-HLA-TCR based therapeutic interventions. Recent technical developments enabling high throughput identification of paired α/β TCR sequences in patients will likely resolve part of the challenges hampering discovery in the field of peptide-HLA-TCR based therapies. Still, this emerging field suffers from the same difficulties as other fields generating large scale data sets, namely how to transition from arguably descriptive OMICS data sets to fundamental functional understanding. Here we set out to solve this disconnect by developing quick assays to functionally test TCR sequences cloned from HLA-DRB1∗04:01 individuals. For these studies, we used a retroviral TCR expression construct where human variable TCRα and TCRβ domains were cloned into a plasmid backbone containing mouse TCR constant domains [29]. This results in a chimeric expression construct that retains the binding specificity of the human TCR but can interact with the mouse CD3 signaling complex, allowing for the subsequent generation of retrogenic mouse models following initial characterization and prioritization assays [55,56]. Compared to traditional transgenic TCR animal models, the retrogenic approach is faster, cheaper, and more versatile [57]. When performed with mice expressing human HLA, it has the potential of generating a novel class of highly relevant humanized animal models [58].
In summary, we here describe an optimized human TCR discovery platform, using the same TCR expression construct in multiple assays allowing for initial screening experiments, all the way to humanized animal models where patient TCRs can be combined with HLA alleles and antigens linked to disease. We see major potential in integrating these assays as part of a high throughput TCR discovery setup, to validate and prioritize patient TCRs from repertoire sequencing experiments. In this context, pooled TCR expression libraries could, for example, be generated from repertoire sequence data, and cells expressing TCRs that are activated by specific stimuli could be rapidly isolated from the pool and prioritized. The development of tolerizing treatments for patients is hampered by the absence of relevant model systems that include patient-derived TCRs, disease-relevant HLA alleles, and antigens. The combination of explorative studies in patient material to identify and prioritize TCRs with making humanized TCR retrogenic mice have good potential to generate such models. Importantly, the retrogenic mice can also easily be generated to carry multiple different TCRs, both autoreactive and control TCRs. This allows for more complex immune interactions, as well as the generation of a heterogenous T cell pool better mimicking a true biological setting [59].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We are grateful to Dr. Robert Harris for sharing reagents, Dr. Karine Chemin for suggestions, and the staff working at the Karolinska Institute Department of Comparative Medicine and Preclinical Laboratory animal facilities for the help provided. The 58 alpha-beta- cell line [26], was a gift from Dr. Bernard Malissen. The EcoPak plasmid was a gift from Dr. Mark P. Kamps. The mouse CD3 WTdelta-F2A-gamma-T2A-epsilon-P2A-zeta pMIG II was a gift from Dr. Dario Vignali (Addgene plasmid # 52092 [31]). pMX hCD4 was a gift from Dan Littman (Addgene plasmid # 14614 [32]). pSIRV-NFAT-eGFP was a gift from Peter Steinberger (Addgene plasmid # 118031 [35]). This research was partly funded by grants from the Swedish Research Council, Karolinska Institutet, The Swedish Rheumatism Association, King Gustaf V’s 80-year Foundation, Stiftelsen Professor Nanna Svartz Fond (to FW), The Börje Dahlin Foundation (to SVB, and YS), Karolinska Institutets Foundation Grants (to SVB), the China Scholarship Council (to YS), and the Nanyang Technological University–Karolinska Institutet Joint PhD Programme (VSI). This work has received support from Pfizer Inc. and the European Union/European Federation of Pharmaceutical Industries and Associations (EU/EFPIA) Innovative Medicines Initiative Joint Undertaking (RTCure Grant 777357) (SVB, RKS, AD, BR, CG, YS, VSI, ZK, ARW, LK, VM, and FW are participants of the program).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtauto.2021.100087.
Authors’ contributions
SVB, and FW designed experiments. SVB, RKS, BR, CG, YS, VSI, and ZK Performed experiments. AD, WWK, and MB generated and/or provided unique reagents. SVB, RKS, AW, LK, VM, and FW analyzed data. SVB, and FW wrote the manuscript. RKS, AD, BR, CG, YS, VSI, ZK, AW, LK, VM, and MB helped to revise the manuscript. The final manuscript was read and approved by all the authors.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Todd J.A., Acha-Orbea H., Bell J.I., Chao N., Fronek Z., Jacob C.O. A molecular basis for MHC class II--associated autoimmunity. Science. 1988;240:1003–1009. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- 2.Nepom G.T., Erlich H. MHC class-II molecules and autoimmunity. Annu. Rev. Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 3.Terao C., Brynedal B., Chen Z., Jiang X., Westerlind H., Hansson M. Distinct HLA associations with rheumatoid arthritis subsets defined by serological subphenotype. Am. J. Hum. Genet. 2019;105:616–624. doi: 10.1016/j.ajhg.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klareskog L., Forsum U., Scheynius A., Kabelitz D., Wigzell H. Evidence in support of a self-perpetuating HLA-DR-dependent delayed-type cell reaction in rheumatoid arthritis. Proc. Natl. Acad. Sci. U. S. A. 1982;79:3632–3636. doi: 10.1073/pnas.79.11.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raychaudhuri S., Sandor C., Stahl E.A., Freudenberg J., Lee H.S., Jia X. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson K.M., Roark C.L., Portas M., Aubrey M.T., Rosloniec E.F., Freed B.M. A molecular analysis of the shared epitope hypothesis: binding of arthritogenic peptides to DRB1∗04 alleles. Arthritis Rheum. 2016;68:1627–1636. doi: 10.1002/art.39636. [DOI] [PubMed] [Google Scholar]
- 7.Klareskog L., Forsum U., Wigren A., Wigzell H. Relationship between HLA-DR-expressing cells and T lymphocytes of different subsets in rheumatoid synovial tissue. Scand. J. Immunol. 1981;15:501–507. doi: 10.1111/j.1365-3083.1982.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 8.Galindo-Feria A.S., Albrecht I., Fernandes-Cerqueira C., Notarnicola A., James E.A., Herrath J. Proinflammatory histidyl-transfer RNA synthetase-specific CD4+ T cells in the blood and lungs of patients with idiopathic inflammatory myopathies. Arthritis Rheum. 2020;72:179–191. doi: 10.1002/art.41075. [DOI] [PubMed] [Google Scholar]
- 9.Pieper J., Dubnovitsky A., Gerstner C., James E.A., Rieck M., Kozhukh G. Memory T cells specific to citrullinated alpha-enolase are enriched in the rheumatic joint. J. Autoimmun. 2018;92:47–56. doi: 10.1016/j.jaut.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerstner C., Turcinov S., Hensvold A.H., Chemin K., Uchtenhagen H., Ramwadhdoebe T.H. Multi-HLA class II tetramer analyses of citrulline-reactive T cells and early treatment response in rheumatoid arthritis. BMC Immunol. 2020;21:27. doi: 10.1186/s12865-020-00357-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Jelcic I., Muhlenbruch L., Haunerdinger V., Toussaint N.C., Zhao Y. Cell; 2020. HLA-DR15 Molecules Jointly Shape an Autoreactive T Cell Repertoire in Multiple Sclerosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delong T., Wiles T.A., Baker R.L., Bradley B., Barbour G., Reisdorph R. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pieper J., Dubnovitsky A., Gerstner C., James E.A., Rieck M., Kozhukh G. Memory T cells specific to citrullinated α-enolase are enriched in the rheumatic joint. J. Autoimmun. 2018;92:47–56. doi: 10.1016/j.jaut.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimoda S., Miyakawa H., Nakamura M., Ishibashi H., Kikuchi K., Kita H. CD4 T-cell autoreactivity to the mitochondrial autoantigen PDC-E2 in AMA-negative primary biliary cirrhosis. J. Autoimmun. 2008;31:110–115. doi: 10.1016/j.jaut.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Kremer J.M., Westhovens R., Leon M., Di Giorgio E., Alten R., Steinfeld S. Treatment of rheumatoid arthritis by selective inhibition of T-cell activation with fusion protein CTLA4Ig. N. Engl. J. Med. 2003;349:1907–1915. doi: 10.1056/NEJMoa035075. [DOI] [PubMed] [Google Scholar]
- 16.Orban T., Bundy B., Becker D.J., DiMeglio L.A., Gitelman S.E., Goland R. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet. 2011;378:412–419. doi: 10.1016/S0140-6736(11)60886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumitomo S., Nagafuchi Y., Tsuchida Y., Tsuchiya H., Ota M., Ishigaki K. A gene module associated with dysregulated TCR signaling pathways in CD4(+) T cell subsets in rheumatoid arthritis. J. Autoimmun. 2018;89:21–29. doi: 10.1016/j.jaut.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Ji H., Korganow A.S., Mangialaio S., Hoglund P., Andre I., Luhder F. Different modes of pathogenesis in T-cell-dependent autoimmunity: clues from two TCR transgenic systems. Immunol. Rev. 1999;169:139–146. doi: 10.1111/j.1600-065x.1999.tb01312.x. [DOI] [PubMed] [Google Scholar]
- 19.Anderson A.C., Chandwaskar R., Lee D.H., Sullivan J.M., Solomon A., Rodriguez-Manzanet R. A transgenic model of central nervous system autoimmunity mediated by CD4+ and CD8+ T and B cells. J. Immunol. 2012;188:2084–2092. doi: 10.4049/jimmunol.1102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berlo S.E., van Kooten P.J., Ten Brink C.B., Hauet-Broere F., Oosterwegel M.A., Glant T.T. Naive transgenic T cells expressing cartilage proteoglycan-specific TCR induce arthritis upon in vivo activation. J. Autoimmun. 2005;25:172–180. doi: 10.1016/j.jaut.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Smolen J.S., Landewe R.B.M., Bijlsma J.W.J., Burmester G.R., Dougados M., Kerschbaumer A. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2019 update. Ann. Rheum. Dis. 2020;79:685–699. doi: 10.1136/annrheumdis-2019-216655. [DOI] [PubMed] [Google Scholar]
- 22.Benichou J., Ben-Hamo R., Louzoun Y., Efroni S. Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012;135:183–191. doi: 10.1111/j.1365-2567.2011.03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Simone M., Rossetti G., Pagani M. Single cell T cell receptor sequencing: techniques and future challenges. Front. Immunol. 2018;9:1638. doi: 10.3389/fimmu.2018.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bagaev D.V., Vroomans R.M.A., Samir J., Stervbo U., Rius C., Dolton G. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 2020;48:D1057–D1062. doi: 10.1093/nar/gkz874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corrie B.D., Marthandan N., Zimonja B., Jaglale J., Zhou Y., Barr E. iReceptor: a platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol. Rev. 2018;284:24–41. doi: 10.1111/imr.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letourneur F., Malissen B. Derivation of a T cell hybridoma variant deprived of functional T cell receptor alpha and beta chain transcripts reveals a nonfunctional alpha-mRNA of BW5147 origin. Eur. J. Immunol. 1989;19:2269–2274. doi: 10.1002/eji.1830191214. [DOI] [PubMed] [Google Scholar]
- 27.Ito K., Bian H.J., Molina M., Han J., Magram J., Saar E. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J. Exp. Med. 1996;183:2635–2644. doi: 10.1084/jem.183.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danner R., Chaudhari S.N., Rosenberger J., Surls J., Richie T.L., Brumeanu T.D. Expression of HLA class II molecules in humanized NOD.Rag1KO.IL2RgcKO mice is critical for development and function of human T and B cells. PloS One. 2011;6 doi: 10.1371/journal.pone.0019826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sprouse M.L., Blahnik G., Lee T., Tully N., Banerjee P., James E.A. Rapid identification and expression of human TCRs in retrogenic mice. J. Immunol. Methods. 2016;439:29–36. doi: 10.1016/j.jim.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewitt C.R., Lamb J.R., Hayball J., Hill M., Owen M.J., O’Hehir R.E. Major histocompatibility complex independent clonal T cell anergy by direct interaction of Staphylococcus aureus enterotoxin B with the T cell antigen receptor. J. Exp. Med. 1992;175:1493–1499. doi: 10.1084/jem.175.6.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holst J., Wang H., Eder K.D., Workman C.J., Boyd K.L., Baquet Z. Scalable signaling mediated by T cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nat. Immunol. 2008;9:658–666. doi: 10.1038/ni.1611. [DOI] [PubMed] [Google Scholar]
- 32.Landau N.R., Warton M., Littman D.R. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature. 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 33.Gerstner C., Turcinov S., Hensvold A.H., Chemin K., Uchtenhagen H., Ramwadhdoebe T.H. Multi-HLA class II tetramer analyses of citrulline-reactive T cells and early treatment response in rheumatoid arthritis. BMC Immunol. 2020;21:1–14. doi: 10.1186/s12865-020-00357-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novak E.J., Liu A.W., Nepom G.T., Kwok W.W. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J. Clin. Invest. 1999;104:R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jutz S., Leitner J., Schmetterer K., Doel-Perez I., Majdic O., Grabmeier-Pfistershammer K. Assessment of costimulation and coinhibition in a triple parameter T cell reporter line: simultaneous measurement of NF-kappaB, NFAT and AP-1. J. Immunol. Methods. 2016;430:10–20. doi: 10.1016/j.jim.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Law Y.M., Yeung R.S., Mamalaki C., Kioussis D., Mak T.W., Flavell R.A. Human CD4 restores normal T cell development and function in mice deficient in murine CD4. J. Exp. Med. 1994;179:1233–1242. doi: 10.1084/jem.179.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timmerman L.A., Clipstone N.A., Ho S.N., Northrop J.P., Crabtree G.R. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- 38.Imboden J.B., Stobo J.D. Transmembrane signalling by the T cell antigen receptor. Perturbation of the T3-antigen receptor complex generates inositol phosphates and releases calcium ions from intracellular stores. J. Exp. Med. 1985;161:446–456. doi: 10.1084/jem.161.3.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chow C.W., Rincon M., Davis R.J. Requirement for transcription factor NFAT in interleukin-2 expression. Mol. Cell Biol. 1999;19:2300–2307. doi: 10.1128/mcb.19.3.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz D.M., Kanno Y., Villarino A., Ward M., Gadina M., O’Shea J.J. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov. 2017;16:843–862. doi: 10.1038/nrd.2017.201. [DOI] [PubMed] [Google Scholar]
- 41.Miller S.D., Turley D.M., Podojil J.R. Antigen-specific tolerance strategies for the prevention and treatment of autoimmune disease. Nat. Rev. Immunol. 2007;7:665–677. doi: 10.1038/nri2153. [DOI] [PubMed] [Google Scholar]
- 42.Sabatos-Peyton C.A., Verhagen J., Wraith D.C. Antigen-specific immunotherapy of autoimmune and allergic diseases. Curr. Opin. Immunol. 2010;22:609–615. doi: 10.1016/j.coi.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green D.R., Scott D.W. Activation-induced apoptosis in lymphocytes. Curr. Opin. Immunol. 1994;6:476–487. doi: 10.1016/0952-7915(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 44.Varadhachary A.S., Perdow S.N., Hu C., Ramanarayanan M., Salgame P. Differential ability of T cell subsets to undergo activation-induced cell death. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5778–5783. doi: 10.1073/pnas.94.11.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srivastava A., Arlian B.M., Pang L., Kishimoto T.K., Paulson J.C. Tolerogenic nanoparticles impacting B and T lymphocyte responses delay autoimmune arthritis in K/BxN mice. bioRxiv. 2020 doi: 10.1101/2020.07.02.185140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perera J., Liu X., Zhou Y., Joseph N.E., Meng L., Turner J.R. Insufficient autoantigen presentation and failure of tolerance in a mouse model of rheumatoid arthritis. Arthritis Rheum. 2013;65:2847–2856. doi: 10.1002/art.38085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian J., Clare-Salzler M., Herschenfeld A., Middleton B., Newman D., Mueller R. Modulating autoimmune responses to GAD inhibits disease progression and prolongs islet graft survival in diabetes-prone mice. Nat. Med. 1996;2:1348–1353. doi: 10.1038/nm1296-1348. [DOI] [PubMed] [Google Scholar]
- 48.Krienke C., Kolb L., Diken E., Streuber M., Kirchhoff S., Bukur T. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science. 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- 49.Matzinger P., Kamala T. Tissue-based class control: the other side of tolerance. Nat. Rev. Immunol. 2011;11:221–230. doi: 10.1038/nri2940. [DOI] [PubMed] [Google Scholar]
- 50.Akamatsu M., Mikami N., Ohkura N., Kawakami R., Kitagawa Y., Sugimoto A. Conversion of antigen-specific effector/memory T cells into Foxp3-expressing Treg cells by inhibition of CDK8/19. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw2707. [DOI] [PubMed] [Google Scholar]
- 51.Shade K.C., Conroy M.E., Washburn N., Kitaoka M., Huynh D.J., Laprise E. Sialylation of immunoglobulin E is a determinant of allergic pathogenicity. Nature. 2020;582:265–270. doi: 10.1038/s41586-020-2311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durham S.R., Walker S.M., Varga E.M., Jacobson M.R., O’Brien F., Noble W. Long-term clinical efficacy of grass-pollen immunotherapy. N. Engl. J. Med. 1999;341:468–475. doi: 10.1056/NEJM199908123410702. [DOI] [PubMed] [Google Scholar]
- 53.Anagnostou K., Islam S., King Y., Foley L., Pasea L., Bond S. Assessing the efficacy of oral immunotherapy for the desensitisation of peanut allergy in children (STOP II): a phase 2 randomised controlled trial. Lancet. 2014;383:1297–1304. doi: 10.1016/S0140-6736(13)62301-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noon L., Prophylactic Inoculation Against Hay F.E.V.E.R. Lancet. 1911;177:1572–1573. [Google Scholar]
- 55.Holst J., Szymczak-Workman A.L., Vignali K.M., Burton A.R., Workman C.J., Vignali D.A. Generation of T-cell receptor retrogenic mice. Nat. Protoc. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- 56.Burton A.R., Vincent E., Arnold P.Y., Lennon G.P., Smeltzer M., Li C.S. On the pathogenicity of autoantigen-specific T-cell receptors. Diabetes. 2008;57:1321–1330. doi: 10.2337/db07-1129. [DOI] [PubMed] [Google Scholar]
- 57.Bettini M.L., Bettini M., Vignali D.A. T-cell receptor retrogenic mice: a rapid, flexible alternative to T-cell receptor transgenic mice. Immunology. 2012;136:265–272. doi: 10.1111/j.1365-2567.2012.03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schinnerling K., Rosas C., Soto L., Thomas R., Aguillon J.C. Humanized mouse models of rheumatoid arthritis for studies on immunopathogenesis and preclinical testing of cell-based therapies. Front. Immunol. 2019;10:203. doi: 10.3389/fimmu.2019.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pacheco Y., Acosta-Ampudia Y., Monsalve D.M., Chang C., Gershwin M.E., Anaya J.M. Bystander activation and autoimmunity. J. Autoimmun. 2019;103:102301. doi: 10.1016/j.jaut.2019.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.