Graphic abstract
The recent emergence of novel coronavirus (SARS-CoV-2) has been a major threat to human society, as the challenge of finding suitable drug or vaccine is not met till date. With increasing morbidity and mortality, the need for novel drug candidates is under great demand. The investigations are progressing towards COVID-19 therapeutics. Among the various strategies employed, the use of repurposed drugs is competing along with novel drug inventions. Based on the therapeutic significance, the chemical constituents from the extract of Tinospora cordifolia belonging to various classes like alkaloids, lignans, steroids and terpenoids are investigated as potential drug candidates for COVID-19. The inhibition potential of the proposed compounds against viral spike protein and human receptor ACE2 were evaluated by computational molecular modeling (Auto dock), along with their ADME/T properties. Prior to docking, the initial geometry of the compounds were optimized by Density functional theory (DFT) method employing B3LYP hybrid functional and 6–311 + + G (d,p) basis set. The results of molecular docking and ADME/T studies have revealed 6 constituents as potential drug candidates that can inhibit the binding of SARS-CoV-2 spike protein with the human receptor ACE2 protein. The narrowed down list of constituents from Tinospora cordifolia paved way for further tuning their ability to inhibit COVID-19 by modifying the chemical structures and by employing computational geometry optimization and docking methods.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13337-021-00666-7.
Keywords: SARS-CoV-2; COVID-19, Therapeutics, Inhibitors, Tinospora cordifolia
Introduction
The emergence of COVID-19 pandemic [1, 2] in December 2019, caused by the severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2] [3, 4] has imposed severe social and economic burden in countries across the globe. Initially this viral infection was diagnosed to cause severe acute respiratory syndrome (SARS) in humans [5], and later reports revealed the susceptability of cats and ferrets also to have acquired COVID-19 infection. [6, 7]. It is believed that the viral spread is not air/water borne or through insects/animals. It has spread to many countries round the globe mainly through societal interactions including transmission from human-to-human through droplets, contaminated hand or surface contacts. Now there has been a daunting task for the scientists to not only control the morbidity but also the steeply raising mortality [8].
Though the mode of infection by SARS-CoV-2 is known to be very similar to SARS-CoV [9, 10], there exists the challenge of controlling the rate of infection and rapid treatment methods for the infected patients [11, 12]. Till date no antiviral drugs with proven efficacy nor are there vaccines for its prevention. Several clinically available antiviral drugs are reported in the literature and have been in use to suppress different viral particles [13, 14]. For the past 10–15 years, researchers are working on the development of antiviral drugs for SARS-CoV and MERS-CoV [15, 16] and till date no successful results appear. Now SARS-CoV-2 is added to the list.
Detailed investigations on various phytoconstituents of Tinospora cordifolia as potent drugs targeting the main protease (Mpro) of the virus was carried out recently [17]. The present focus of investigations resides on two other potential targets: 1. Virus (Receptor binding motifs—spike (S), envelope (E) and nucleocapsid (N) proteins, RNA dependent RNA polymerases and 2. Receptor motif on human ACE2 (angiotensin converting enzyme) and its associated functional proteins like TMPRSS2 and B0AT1. It is difficult to have a complete evaluation of small molecular drug candidates for therapies directed towards the host with the inadequately available knowledge on the molecular details of the infection caused by SARS-CoV-2 [18, 19]. Recently, several research works have been published with novel and refurbished drug candidates to tackle the situation [14].
Until recently, there was a speculation that hydroxychloroquine could inhibit the viral infection [20]. But there was no solid proof on the method of inhibition. With the current status on the spread of infection, it is mandatory on emergency basis to develop strategies to control the morbidity and mortality. A systematic understanding on the host dependencies of the SARS-CoV-2 virus to identify other host proteins is the need of the hour. Many therapeutic strategies target the host-virus interface, but such drugs are prone to induced severe side effects [20]. It is very unfortunate that we have very minimal knowledge on the molecular details of SARS-CoV-2 infection to further proceed with a comprehensive evaluation of small molecular therapeutic candidates directed towards the host. Several mathematical models [21, 22] and computational strategies [23] are being currently under investigation to identify the interactions at the interface. Moreover, to devise therapeutic strategies, it is important to know how the virus invades the humans during infection and this knowledge can be applied to develop new drugs and to repurpose the existing ones [24]. There are also reports on various constituents from plants [25] of medicinal values as potential inhibitors and anti-viral drugs [26–28].
Recently, Government of India has released an advisory from the ministry of Ayurveda, to meet the challenges caused by the rapid spread of COVID-19 in India [29]. The major focus of this system was to bring lifestyle modifications and prophylactics to improve the immunity in humans. In this context, it was reported that an ayurvedic medicine Samshamani Vati (aqueous extract of Tinospora cordifolia), when administered at 500 mg, twice a day for 15 days, could serve as prophylaxis [30]. The same is also reported to induce immunomodulatory effect [31–33] in human immuno-deficiency virus positive patients. [34] The various constituents of Tinospora cordifolia are known to exhibit a broad spectrum of therapeutic activities including anticancer, antimicrobial, antitoxic, antidiabetic, hypolipidermic, wound healing, immunomodulation, etc. and 31 different constituents (or chemical compounds) of Tinospora cordifolia were reported in literature [35]. It belongs to the family of Menispermaceae and is known for the pharmacological activities exhibited by the chemical constituents like glycosides, terpenoids, alkaloids, essential oils, fatty acids, etc., present in different parts of the plant like root and stem. The plant possesses various medicinal properties [36] like anti-diabetic, anti-allergic, anti-stress, anti-leprotic, anti-malarial, anti-neoplastic, hepatoprotective, immunomodulatory, etc.
With the available scientific approaches and computational facilities to model proteins and investigate protein–ligand interactions it becomes more supportive to predict the binding of small molecular drugs to protein targets [28, 37, 38]. Employing density functional theory [39] the geometry of all the molecules proposed as COVID-19 drug candidates, were optimized to understand structural features and hence their contribution to the inhibition or druggable potential. For the first time in literature, herein we are reporting the inhibitory effects of selected constituents of Tinospora cordifolia on human ACE2 protein and the main protease of SARS-CoV-2 using molecular docking and pharmacokinetic studies. In this manuscript, we have investigated the various constituents of Tinospora cordifolia for their potency to inhibit the host receptor for SARS-CoV-2 by molecular docking interactions.
Materials and methods
Docking calculations
The X-ray structure were obtained from Brookhaven Protein Data Bank for SARS coronavirus spike receptor-binding domain complexed with its receptor (PDB id: 2AJF) and the main protease of COVID-19 in complex with an inhibitor (PDB ID: 6LU7) and were considered for docking studies. The protein structure coordinates for docking were obtained from the above by removing the bound receptors, inhibitors, water molecules and other hetero atoms. Further, using AutoDockTools-1.5.6, the Kollman charges [40] were assigned to the proteins after adding the hydrogen atoms. Employing molecular mechanics, the 3D coordinates of the proposed inhibitors were constructed and optimized. Auto Tors was used to define the possible torsions associated with the inhibitors. All the inhibitors were treated as flexible throughout the docking procedures. The already reported binding pocket in the proteins was used to generate a grid box to encapsulate the active site. The auto grid program was used to pre-calculate the grid maps of interaction energies between protein and various atom types present in the inhibitors. The conformational states of the flexible inhibitors were explored using Lamarckian genetic algorithm coupled with energy assessments based on AMBER force field. Docking calculations were performed with default parameters. The binding energy was evaluated using the following scoring function.
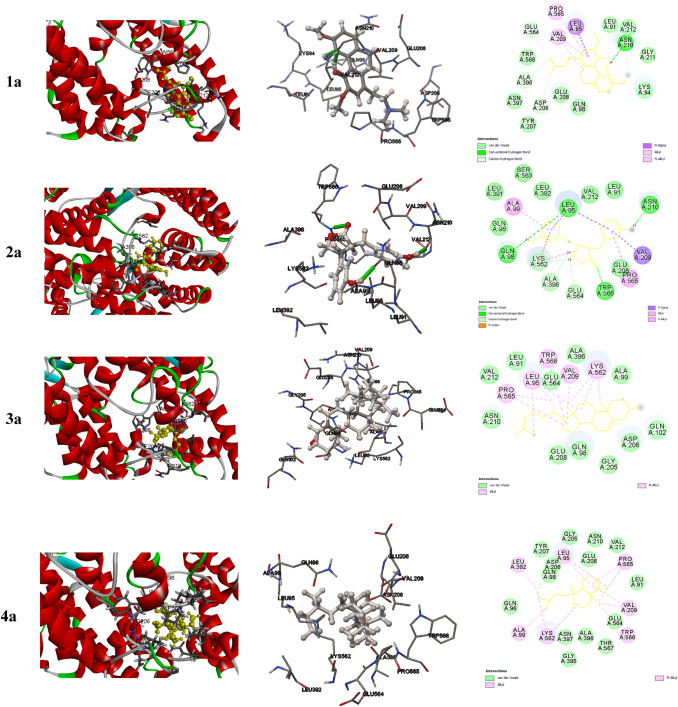
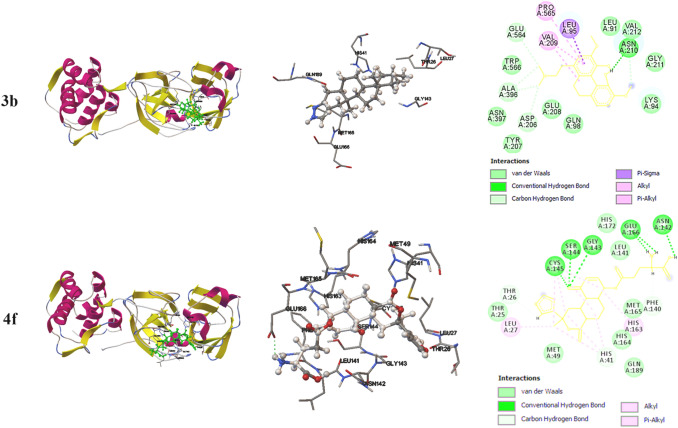
The free energy upon binding of the flexible ligand to the rigid target could be calculated using the equation that includes parameters like ΔGvdw (dispersion/repulsion), ΔGelec (electrostatic interaction), ΔGhbond (hydrogen bonding), ΔGtor (torsional constraints) and ΔGsol (desolvation effects). The 3-dimensional interactions were generated using the visualizer associated with AutoDockTools-1.5.6 while the 2-dimensional interactions were generated using BIOVIA Discovery Studio visualizer (Figs. 1, 2 and ESI Fig. 1).
Fig.1.
Representative docked conformation of various constituents of Tinospora cordifolia with 2AJF. The interaction of 3 dimensionally oriented molecules with the active site of 2ACE is shown (left); the interacting residues in the active site and the localization of the inhibitors is shown (middle); The 2 dimensional interactions with various residues in the active site and the nature of interactions are depicted (Right)
Fig.2.
Docked conformation of various constituents of Tinospora cordifolia with 6LU7. The interaction of 3 dimensionally oriented molecules with the active site of 6LU7 is shown (left); the interacting residues in the active site and the localization of the inhibitors are shown (middle); The 2 dimensional interactions with various residues in the active site and the nature of interactions are depicted (Right)
ADMET predictions
The pharmaco-kinetic properties such as absorption, distribution, metabolism, excretion and toxicity (ADMET) were predicted using the pkCSM/ADMET [41, 42]. This method employs graph-based signatures to develop predictive models for generating central ADMET properties for drug development. In absorption process, the drug reaches the blood stream from the site of the drug administration. The absorption of drugs depends on factors including polar surface area (PSA), membrane permeability (LogP), cell-based methods such as Caco-2, intestinal absorption, skin permeability levels, P-glycoprotein substrate or inhibitor. This approach uses various parameters such as the blood–brain barrier (logBB), CNS permeability (logPB), and the volume of distribution (VDss) are evaluating the distribution of drugs. Using CYP models for substrate (CYP2D6, CYP1A2, CYP2C19, CYP2C9, and CYP3A4) the metabolisms of the drug molecules are evaluated. Excretion, a process where the body eliminates an unchanged drug or its metabolite, is predicted based on the total clearance model and renal OCT2 substrate. The toxicity of drugs is predicted based on AMES toxicity, hERG inhibition, hepatotoxicity, and skin sensitization [43, 44]. The PSA value relates the absorption properties of the inhibitor drugs. The PSA value of compounds greater than 140 indicates that the compounds have strong polarity and were poorly absorbed. The lipophilicity values are predicted by the LogP value. The LogP values less than 5 shows that the compound can easily permeable into cell membrane. Apart from these two parameters PSA and Log P, the following parameters such as Caco-2 permeability, intestinal absorption (human), skin permeability, and P-glycoprotein substrate or inhibitor were used to predict the absorption properties of the compounds. When the compounds exhibit the predicted value of > 0.90 indicated that the compound has high Caco-2 permeability and easily absorbed. The intestinal absorption (human), less than 30% is considered as poor absorption. The logKp value of greater than − 2.5 is considered as poor skin permeability. The permeability glycoprotein (P-glycoprotein) also known as multidrug resistant protein or ATP binding Cassette (ABC) is an important protein in the cell membrane that eliminates the toxins and foreign substances from the cells. After the drug is absorbed from the membrane, the drug should be distributed to various tissues in the body to produce the pharmacological effects. The distribution volume (VDss) is a parameter which predicts the distribution of drugs in various tissues in the body. When the log VDss < − 0.15 is considered as low distribution and higher than > 0.45 is considered as high distribution. After the drug absorbed in the body, the circulating drug exist in either bound or unbound state with the serum proteins in the blood. The efficacy of the drug may be affected by the fraction of drug binds with proteins within blood; the more bound state leads to poor efficacy. The blood brain barriers (BBB) protect the brain from exogenous compounds and selectively transport various nutrients, ions and other molecules that are crucial for neural functions. The BBB parameter is the important factor which is measured in vivo in animal models as logBB. The logBB value greater than 0.3 shows that the compounds can easily cross the BBB and logBB < − 1 suggest that the compounds can’t easily cross the BBB. Compounds with logPB > − 2 are considered to penetrate the CNS, and logPB < − 3 are considered unable to penetrate the CNS. The central nervous system (CNS) permeability is obtained by direct measurement of blood–brain permeability-surface area product. The CNS permeability between − 3 to − 2 is considered as penetrable to CNS. Cytochrome P450s plays important role in drug metabolism. Cytochrome P450s are classified into several groups depends on their biological functions. The major cytochromes subtypes involved in the drug metabolism are CYP2D6 and CYP3A4. The amount of the drug that is eliminated from the body is predicted by the total clearance.
Results
Composition of Tinospora cordifolia
Tinospora cordifolia also known as Guduchi or Amrita is a medicinal plant possessing various therapeutic properties like jaundice, rheumatism, urinary tract infections, dermal diseases, anemia, inflammation, diabetes, etc. The constituents of this plant is known to support the immune system by increasing the body’s resistance to infections and also to support the structure, function and levels of white blood cells. The observed pharmacological properties [32, 35, 45–47] of this plant is due to the presence of various constituents like alkaloids, steroids, terpenoids, fatty acid mixtures, polysaccharides, etc. It is also reported that the extract of Tinospora cordifolia possesses broad-spectrum antiviral [17] and protease inhibitors [48]. Based on all the above pharmacological significance of these plant constituents, here in we have tried to investigate the interactions of various constituents with human host receptors for SARS-CoV-2, ACE2 [49–51] and main protease [52–54] by molecular docking [55].
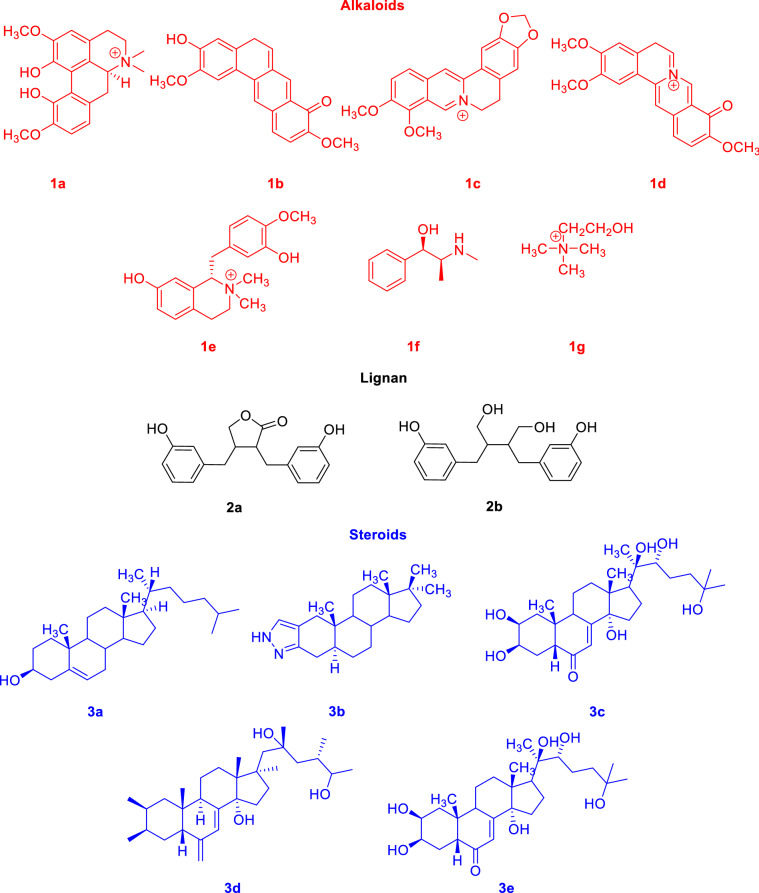
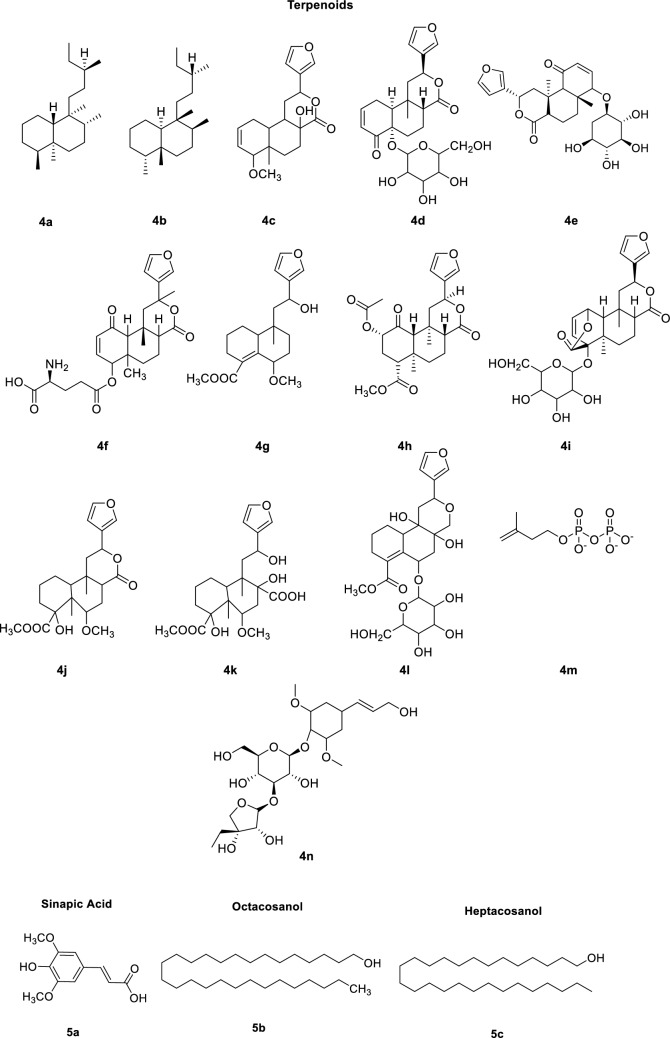
The extract of Tinospora cordifolia consists of constituents that belong to different classes including alkaloids, steroids, terpenoids, lignans, glycosides, polysaccharides, aliphatic compounds, etc. [35]. The major and active chemical constituents along with their chemical structure are shown in Scheme 1. Structures 1a–g belong to alkaloids, 2a–b are lignans, 3a–e are steroids, 4a–n are terpenoids and 5a–c belong to other categories. Most of these molecules are optically active and their stereochemistry plays a major role in their pharmacological properties. It is known that enantiomeric drugs possess different pharmacological properties.
Scheme 1.
Chemical constituents of Tinospora cordifolia
Docking simulation
Recently, many natural products [28, 56] and refurbished drugs are reported in the literature as possible drug candidates for COVID-19. The broad spectrum therapeutic significance of Tinospora cordifolia has paved way for their investigation as potent inhibitors for SARS-CoV-2. To the best of our knowledge, till date there are no results related to SARS-CoV-2 inhibition through theoretical and experimental studies of compounds in Tinospora cordifolia. Herein we have chosen two targets to study the interaction of the constituents of Tinospora cordifolia: angiotensin converting enzyme (ACE2), an integral membrane glycoprotein, which serves as the human receptor for SARS-CoV-2 and the main protease (Mpro), which is involves in processing the polyprotein that is translated from viral RNA. With the above two targets we have carried out the molecular docking studies with 31 different constituents of Tinospora cordifolia. The results (ESI Table 1) indicate that 1a, 2a, 3a–b and 4a–e exhibit strong interaction while 1b–f, 2b, 3c–d, and 4f–n exhibit moderate interaction with 2AJF [57] (human ACE2 protein). In order to narrow down, the molecules that contribute significantly towards destabilizing the binding interactions are only considered and are listed in Table 1. The inhibitory effect on 6LU7 [58] (main protease) as evaluated from various interaction energy parameters (ESI Table 3) indicate that 2a and 4a interact strongly while 3a–b and 4b–e show moderate inhibition. The strong binding of proposed candidates to the active site of the enzyme leads to activity inhibition. The corresponding interacting residues with 2AJF and 6LU7 are given in ESI Table 2 and ESI table 4 respectively.
Table 1.
Energy parameters (kCal/mol) associated with docking interactions of various compounds in Tinospora cordifolia with human ACE2 (PDB id. 2AJF): A, Binding energy; B, van der Waals & hydrogen bond energy, C, Electrostatic energy, D, Torsional energy
| 2AJF | A | B | C | D | Interacting residues |
|---|---|---|---|---|---|
| Alkaloids | |||||
| 1a | − 5.5 | − 6.63 | − 0.95 | 2.09 | L91, K94, L95, Q98, D206, E208, V209, N210, V212, P565, W566 |
| 1e | − 4.2 | − 6.51 | − 0.08 | 2.39 | L91, L95, A99, Q98, E208, V209, N210, L391, L392, A396, K562, E564, P565, W566 |
| Lignans | |||||
| 2b | − 4.81 | − 7.71 | − 0.37 | 3.28 | L91, L95, Q98, A99, E208, V209, N210, V212, K562, P565, W566 |
| Steroids | |||||
| 3a | − 5.83 | − 7.63 | 0.01 | 1.79 | L95, Q98, Q102, G205, D206, E208, V209, N210, A396, K562, E564, P565, W566 |
| 3c | − 4.84 | − 7.53 | − 0.29 | 2.98 | L91, K94, L95, Q98, D206, E208, V209, N210, A396, E564, P565, W566 |
| 3d | − 4.62 | − 6.8 | − 0.21 | 2.39 | L85, K94, L95, Q98, N194, H195, Y196, V209, N210, V212, R219, P565 |
| 3e | − 3.83 | − 6.55 | − 0.56 | 3.28 | L91, L95, Q98, Q102, D206, E208, N210, A396, K562 |
| Terpenoids | |||||
| 4d | − 5.05 | − 7.18 | − 0.25 | 2.39 | L91, L95, Q98, G205, D206, E208, V209, N210, V212, A396, K562, P565, W566 |
| 4e | − 4.88 | − 7.09 | 0.12 | 2.09 | L91, L95, Q98, Q102, Y202, G205, D206, V209, A396, K562, E564, P565, W566 |
| 4f | − 4.08 | − 6.31 | − 0.45 | 2.68 | L95, Q98, G205, D206, E208, V209, A396, K562, P565, W566 |
| 4 g | − 4.03 | − 6.09 | − 0.03 | 2.09 | L91, K94, L95, Q98, E208, V209, N210, V212, E564, P565, W566 |
| 4i | − 3.43 | − 5.82 | 0.01 | 2.39 | Q86, E87, I88, L91, K94, L95, N210 |
| 4 k | − 2.2 | − 5.52 | 0.04 | 3.28 | L91, L95, Q98, G205, D206, E208, V209, A396, K562, E564, P565, W566 |
| 4 l | − 2.06 | − 5.37 | − 0.27 | 3.58 | L95, Q98, A99, Q102, Y202, G205, G206, E208, A396, K562, E564, P565, W566 |
| 4 m | − 1.84 | − 4.17 | − 0.36 | 2.68 | L95, D206, V209, N210, A396, K562, P565, W566 |
| 4n | 0.98 | − 4.12 | − 0.57 | 5.67 | Q102, Y196, Y202, G205, D206, N394, N397, G395, E398, K562 |
| Others | |||||
| 5b | − 1.04 | − 9.03 | − 0.07 | 8.05 | L91, K94, L95, Q98, A99, D206, E208, V209, N210, A396, K562, E564, P565, W566 |
| 5c | − 0.59 | − 8.3 | − 0.04 | 7.76 | L91, K94, L95, Q98, D206, E208, V209, N210, K562, E564, P565 |
*Non-polar residues shown in bold
The interacting resides from the binding pocket of 2AJF with various constituents are given in (ESI Table 2). The dominantly interacting compounds with 2AJF are depicted in Fig. 1.
The interaction parameters of selected compounds from the extract of Tinospora cordifolia with the main protease (6LU7) of SARS-CoV-2 are given Table 2 along with the interacting residues (ESI tables 3 & 4). Unlike the interactions with human ACE2, compounds 3b and 4f were found to exhibit strong interaction while moderate interactions were observed in the cases of 3a, 3d, 4a, 4b, 4e and 4 h. No significant interactions were observed with alkaloid class of compounds as such. The interacting residues are tabulated in (ESI table 4) and the significant inhibitory interactions from the selected compounds are depicted in Fig. 2.
Table 2.
Energy parameters (kCal/mol) associated with docking interactions of various compounds in Tinospora cordifolia with Mpro (PDB id. 6LU7): A, Bindind energy; B, van der Waals & hydrogen bond energy, C, Electrostatic energy, D, Torsional energy
| 6LU7 | A | B | C | D | Interacting residues |
|---|---|---|---|---|---|
| 3b | − 5.05 | − 5 | − 0.05 | 0 | T26, L27, H41, G143, M165, E166, Q189 |
| 4f | − 5.09 | − 6.22 | − 1.55 | 2.68 | T26, L27, H41, M49, F140, L141, N142, G143, S144, C145, H163, H164, M165, E166 |
*Non-polar residues shown in bold
Absorption, distribution, metabolism, excretion and toxicity studies [59–61]
Prediction of ADMET properties of alkaloids
The pharmacokinetic [30] parameters play important role in drug discovery process [62]. The predicted ADME/T properties are given in ESI table 5. It could be observed that compounds 1a, 1c and 1d are poorly absorbed due to their high polar nature, though they have high potential to penetrate the cell membrane. The absorption properties of the alkaloids are listed in ESI table 5. The listed alkaloids show high distribution, expect for 1 g. All the listed alkaloids possess the ability to penetrate through the CNS except 1c. All alkaloids can be easily cleared from the system except 1b. The toxicology prediction shows that all alkaloids exhibit hepatotoxicity except 1a, 1e and 1f while 1e exhibits cardiotoxicity.
Prediction of ADMET properties of lignans and steroids
The predicted ADME/T properties of lignans and steroids are given in ESI table 6. The results indicate that lignans possess better absorption ability than steroids. Among the lignans and steroids only 3b shows high distribution potential. The compounds 3a and 3d can easily penetrate through the CNS. 2a, 2b and 3d show moderate levels for excretion. The compounds 2a, 2b, 3a, 3b exhibit cardiotoxicity whereas hepatotoxicity is observed only for 3b and 3d.
Prediction of ADMET properties of terpenoids
The predicted ADME/T properties of terpenoids are given in ESI table 7. The results indicate that all terpenoids except 4a, 4b and 4n are highly polar and show poor absorption. The compounds 4a and 4b possess the ability to cross blood brain barrier while all terpenoids exhibit similar tendency to permeate CNS. Except 3f all the terpenoids can be easily excreted from the body. 4 g, 4j and 4 k show hepatotoxicity.
Prediction of ADMET properties of others
The predicted ADME/T properties of other constituents are given in ESI table 8. Compound 5a exhibits very good absorption and membrane permeation. 5b and 5c possess the ability to cross the BBB easily while compound 5a can penetrate the CNS. 5a induces hepatotoxicity while compounds 5b and 5c are cardiotoxic. The predicted ADME/T properties of selected compounds are listed in Table 3.
Table 3.
Predicted ADME/T properties
| Compounds | Polar surface area | LogP | BBB permeability | CNS permeability | Total clearance | Hepatotoxicity |
|---|---|---|---|---|---|---|
| 1e | 139.63 | 3.0607 | − 0.221 | − 2.403 | 0.743 | No |
| 3a | 174.309 | 7.3887 | 0.777 | − 1.746 | 0.589 | No |
| 4b | 127.66 | 6.6914 | 0.853 | − 1.399 | 0.986 | No |
| 4n | 206.31 | − 0.559 | − 1.742 | − 4.092 | 1.576 | No |
| 5b | 185.387 | 10.1412 | 1.029 | − 1.066 | 2.152 | No |
| 5c | 179.022 | 9.7511 | 1.004 | − 1.054 | 2.13 | No |
Discussions
The results of molecular docking and pharmacokinetic analyses for the interactions with human receptor protein ACE2 have revealed that among the selected candidates listed in Table 1, the major destabilizing contribution is from the torsional energy arising from the presence of freely rotating single bonds. This could be the reason for 1 g to have very poor interaction among the alkaloids. The same can be extended for the case of lignans and steroids (2a and 3a-b respectively) to have comparatively stronger affinity for the considered active site than 2b and 3c-d respectively. In case of terpenoids, 4a-d binds strongly to the active site than the moderately binding 4e-g. It could be observed from Table 1 and ESI Table 2 that the major interacting residues are non-polar in nature. In the case of 4d, the stabilization caused by non-polar interactions is nullified by the high torsional energy of the molecule. Though there is a negligible difference in the intermolecular energy for all the above preferred molecules, the moderately binding ones have high torsional energy, which reduces their target interaction, while the imparted stability has its major contribution from hydrogen bond interactions and van der Waals interactions. 5b-c, bind weakly to the target 2AJF, as the intermolecular stabilizing energy is neutralized by the high torsional energies.
In the case of interactions with viral main protease (Mpro), the results of molecular docking and pharmacokinetic analyses have revealed that the stabilizing interactions have its contribution from polar and acidic amino acids along with non-polar interactions. In compound 4f, though the torsional energy is very high, the stabilizing effect is observed from the low internal energy of the molecule as shown in Table 2 and ESI table 4.
The docking results have revealed that compounds 1a, 1e, 2b, 3a, 3c–e, 4d–g, 4i, 4k–n, 5b and 5c possess the ability to bind to the proposed targets 2AJF and 6LU7. Some of the above candidates have poor binding affinities, by the fact that these molecules exhibit very high torsional energies that could potentially destabilize the interactions with target residues. But still their candidacy is considered, as these molecules could serve as lead compounds to engineering molecules with reduced torsions.
The pharmacokinetic analyses revealed that among the alkaloids (ESI table 5), the polar surface area of 1b, 1e, 1f and 1g were less than 140 and have high potential to get absorbed, while in the case of lignans (ESI table 6), both 2a and 2b were found to be more potent. Among terpenoids (ESI table 7), 4a and 4b and with other miscellaneous compounds listed in ESI table 8, 5a was highly polar. Based on the estimated lipophilicity, all alkaloids (1a–g) and lignans (2a–b) can easily penetrate the cell membrane. While among the steroids, (3a and 3d) and terpenoids (4a and 4b) and 5b and 5c does not possess the ability to penetrate cell membrane. All alkaloids except 1g, 3b, 4a and 4b possess very high drug distribution, while others exhibit moderate or poor distribution. Most of the alkaloids (1a, 1c–d, 1f) and steroids (3b and 3d) were hepatotoxic, while terpenoids and other compounds were non-toxic. The potentiality of various chemical constituents from Tinospora cordifolia to inhibit SARS-CoV-2 was evaluated through computational methods. These compounds have been reported to possess numerous pharmacological activities. The docking results indicate that among all the constituent compounds 1a 1e, 2a–c and 3a–d possess the ability to interact strongly with human ACE2 protein and 3b and 4f with the main protease of SARS-CoV-2. The above conclusion is based on the extent of interaction or their potential as lead compounds for further investigations to reduce their torsional energies. It is believed that strong interactions with human receptor for SARS-CoV-2 could prevent the entry of the virus and thus could act as a prophylactic for COVID-19. Further the evaluated pharmaco-kinetic properties narrowed down the above candidates based on their druggability. From the overall ADMET properties, it could be concluded that compounds 1a, 1e, 2a, 2b, 4a, 4g and 5a as potential drug candidates for COVID-19. Overall observation from combined results of docking analysis and ADMET properties revealed compounds 1e, 3a, 4b, 4n, 5b and 5c could be potential drug candidates out of 31 constituents of the extract from Tinospora cordifolia. The druggable potential of the above six constituents can be tuned by engineering the molecules with less torsional energies and by lowering their cardiotoxicity.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the Chancellor and Vice Chancellor of Vellore Institute of Technology for providing opportunity to carry out this study.
Funding
None.
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Balamurali MM, Email: mmbala@gmaill.com.
Kaushik Chanda, Email: chandakaushik1@gmail.com.
References
- 1.Ciotti M, Angeletti S, Minieri M, Giovannetti M, Benvenuto D, Pascarella S, et al. COVID-19 outbreak: an overview. Chemotherapy. 2019;64(5–6):215–223. doi: 10.1159/000507423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patil AM, Gothert JR, Khairnar V. Emergence, transmission, and potential therapeutic targets for the COVID-19 pandemic associated with the SARS-CoV-2. Cellul Physiol Biochem Int J Exp Cellula Physiol Biochem Pharmacol. 2020;54(4):767–790. doi: 10.33594/000000254. [DOI] [PubMed] [Google Scholar]
- 3.Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens. 2020 doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y, Shin WI, Pang YX, Meng Y, Lai J, You C, et al. The first 75 days of novel coronavirus (SARS-CoV-2) outbreak: recent advances, prevention, and treatment. Int J Environ Res Publ Health. 2020 doi: 10.3390/ijerph17072323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muldoon KM, Fowler KB, Pesch MH, Schleiss MR. SARS-CoV-2: Is it the newest spark in the TORCH? J Clin Virol Official Publ Pan Am Soc Clin Virol. 2020;127:104372. doi: 10.1016/j.jcv.2020.104372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2020 doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patterson EI, Elia G, Grassi A, Giordano A, Desario C, Medardo M et al. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. bioRxiv : the preprint server for biology. 2020. 10.1101/2020.07.21.214346. [DOI] [PMC free article] [PubMed]
- 8.Abd El-Aziz TM, Stockand JD. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2)—an update on the status. Inf Gen Evol J Mol Epidemiol Evol Gen Inf Dis. 2020;83:104327. doi: 10.1016/j.meegid.2020.104327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halaji M, Farahani A, Ranjbar R, Heiat M, Dehkordi FS. Emerging coronaviruses: first SARS, second MERS and third SARS-CoV-2: epidemiological updates of COVID-19. Le infezioni in medicina. 2020;28(suppl 1):6–17. [PubMed] [Google Scholar]
- 10.Singh A, Singh RS, Sarma P, Batra G, Joshi R, Kaur H, et al. A comprehensive review of animal models for coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV. Virol Sin. 2020;35(3):290–304. doi: 10.1007/s12250-020-00252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasoksuz M, Kilic S, Sarac F. Coronaviruses and SARS-COV-2. Turkish journal of medical sciences. 2020;50(SI-1):549–56. 10.3906/sag-2004-127. [DOI] [PMC free article] [PubMed]
- 12.Ludwig S, Zarbock A. Coronaviruses and SARS-CoV-2: a brief overview. Anesth Analg. 2020;131(1):93–96. doi: 10.1213/ANE.0000000000004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12(541):1. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tulgar S, Ahiskalioglu A, Kok A, Thomas DT. Possible old drugs for repositioning in COVID-19 treatment: combating cytokine storms from haloperidol to anti-interleukin agents. Turk J Anaesthesiol Reanim. 2020;48(3):256–257. doi: 10.5152/TJAR.2020.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley BT, Bryan A. Emerging respiratory infections: the infectious disease pathology of SARS, MERS, pandemic influenza, and Legionella. Semin Diagn Pathol. 2019;36(3):152–159. doi: 10.1053/j.semdp.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019 doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhury P. In silico investigation of phytoconstituents from Indian medicinal herb 'Tinospora cordifolia (giloy)' against SARS-CoV-2 (COVID-19) by molecular dynamics approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1803968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bein B, Bachmann M, Huggett S, Wegermann P. SARS CoV-2/COVID-19: evidence-based recommendation on diagnosis and therapy. Anasth Intens Notfallmedizin Schmerztherapie AINS. 2020;55(4):257–265. doi: 10.1055/a-1146-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobo-Galo N, Terrazas-Lopez M, Martinez-Martinez A, Diaz-Sanchez AG. FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1764393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klimke A, Hefner G, Will B, Voss U. Hydroxychloroquine as an aerosol might markedly reduce and even prevent severe clinical symptoms after SARS-CoV-2 infection. Med Hypotheses. 2020;142:109783. doi: 10.1016/j.mehy.2020.109783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S, Choi S, Ko Y, Ki M, Jung E. Risk estimation of the SARS-CoV-2 acute respiratory disease outbreak outside China. Theor Biol Med Model. 2020;17(1):9. doi: 10.1186/s12976-020-00127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, An G, Becker A, Cockrell C, Collier N, Craig M et al. Rapid community-driven development of a SARS-CoV-2 tissue simulator. bioRxiv: the preprint server for biology. 2020. 10.1101/2020.04.02.019075.
- 23.Rensi S, Altman RB, Liu T, Lo YC, McInnes G, Derry A, et al. Homology modeling of TMPRSS2 yields candidate drugs that may inhibit entry of SARS-CoV-2 into human cells. ChemRxiv: the preprint server for chemistry. 2020 [Google Scholar]
- 24.Lisi L, Lacal PM, Barbaccia ML, Graziani G. Approaching coronavirus disease 2019: mechanisms of action of repurposed drugs with potential activity against SARS-CoV-2. Biochem Pharmacol. 2020;180:114169. doi: 10.1016/j.bcp.2020.114169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idrees M, Khan S, Memon NH, Zhang Z. Effect of the phytochemical agents against the SARS-CoV and selected some of them for application to COVID-19: a mini-review. Curr Pharm Biotechnol. 2020 doi: 10.2174/1389201021666200703201458. [DOI] [PubMed] [Google Scholar]
- 26.Jahan I, Onay A. Potentials of plant-based substance to inhabit and probable cure for the COVID-19. Turkish journal of biology = Turk biyoloji dergisi. 2020;44(3):228–41. 10.3906/biy-2005-114. [DOI] [PMC free article] [PubMed]
- 27.Rosales-Mendoza S. Will plant-made biopharmaceuticals play a role in the fight against COVID-19? Expert Opin Biol Ther. 2020;20(6):545–548. doi: 10.1080/14712598.2020.1752177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma A, Tiwari V, Sowdhamini R. Computational search for potential COVID-19 drugs from FDAapproved drugs and small molecules of natural origin identifies several anti-virals and plant products. J Biosci. 2020;45. [DOI] [PMC free article] [PubMed]
- 29.Priya R, Sujatha V. AYUSH for COVID-19: science or superstition? Indian J Publ Health. 2020;64(Supplement):S105–S107. doi: 10.4103/ijph.IJPH_500_20. [DOI] [PubMed] [Google Scholar]
- 30.Bhapkar V, Sawant T, Bhalerao S. A critical analysis of CTRI registered AYUSH studies for COVID- 19. J Ayurveda Integr Med. 2020 doi: 10.1016/j.jaim.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alsuhaibani S, Khan MA. Immune-stimulatory and therapeutic activity of tinospora cordifolia: double-edged sword against salmonellosis. J Immunol Res. 2017;2017:1787803. doi: 10.1155/2017/1787803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhama K, Sachan S, Khandia R, Munjal A, Iqbal HMN, Latheef SK, et al. Medicinal and beneficial health applications of tinospora cordifolia (guduchi): a miraculous herb countering various diseases/disorders and its immunomodulatory effects. Recent Pat Endocr, Metab Immune Drug Discov. 2017;10(2):96–111. doi: 10.2174/1872214811666170301105101. [DOI] [PubMed] [Google Scholar]
- 33.Kaushik A, Husain A, Awasthi H, Singh DP, Khan R, Mani D. Antioxidant and hepatoprotective potential of swaras and hima extracts of tinospora cordifolia and boerhavia diffusa in swiss albino mice. Pharmacognosy Mag. 2017;13(Suppl 3):S658–S662. doi: 10.4103/pm.pm_448_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalikar MV, Thawani VR, Varadpande UK, Sontakke SD, Singh RP, Khiyani RK. Immunomodulatory effect of Tinospora cordifolia extract in human immuno-deficiency virus positive patients. Indian J Pharmacol. 2008;40(3):107–110. doi: 10.4103/0253-7613.42302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma P, Dwivedee BP, Bisht D, Dash AK, Kumar D. The chemical constituents and diverse pharmacological importance of Tinospora cordifolia. Heliyon. 2019;5(9):e02437. doi: 10.1016/j.heliyon.2019.e02437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saha S, Ghosh S. Tinospora cordifolia: one plant, many roles. Ancient Sci Life. 2012;31(4):151–159. doi: 10.4103/0257-7941.107344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J Chem Inf Model. 2020;60(6):3277–3286. doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zeng T, Chen L, Ding S, Huang T, Cai YD. Identification of COVID-19 infection-related human genes based on a random walk model in a virus-human protein interaction network. Biomed Res Int. 2020;2020:4256301. doi: 10.1155/2020/4256301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng YD, Zhou LS, Chen LL, Ma L, Zhao Y, Zhang WW, et al. Ferrocene-isocoumarin conjugated molecules: synthesis, structural characterization, electronic properties, and DFT-TDDFT computational study. Dalton Trans. 2015;44(32):14465–14474. doi: 10.1039/c5dt02169c. [DOI] [PubMed] [Google Scholar]
- 40.Kollman UCSPA. An approach to computing electrostatic charges for molecules. J Comput Chem. 1984;5(2):129–145. [Google Scholar]
- 41.Han Y, Zhang J, Hu CQ, Zhang X, Ma B, Zhang P. In silico ADME and toxicity prediction of ceftazidime and its impurities. Front Pharmacol. 2019;10:434. doi: 10.3389/fphar.2019.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pires DE, Blundell TL, Ascher DB. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norinder U, Bergstrom CA. Prediction of ADMET properties. ChemMedChem. 2006;1(9):920–937. doi: 10.1002/cmdc.200600155. [DOI] [PubMed] [Google Scholar]
- 44.Sanders JM, Beshore DC, Culberson JC, Fells JI, Imbriglio JE, Gunaydin H, et al. Informing the selection of screening hit series with in silico absorption, distribution, metabolism, excretion, and toxicity profiles. J Med Chem. 2017;60(16):6771–6780. doi: 10.1021/acs.jmedchem.6b01577. [DOI] [PubMed] [Google Scholar]
- 45.Singh D, Chaudhuri PK. Chemistry and pharmacology of tinospora cordifolia. Nat Prod Commun. 2017;12(2):299–308. [PubMed] [Google Scholar]
- 46.Stanca MH, Nagy A, Tosa M, Vlad L. Hepatoprotective effects of orally administered melatonin and tinospora cordifolia in experimental jaundice. Chirurgia. 2011;106(2):205–210. [PubMed] [Google Scholar]
- 47.Upadhyay AK, Kumar K, Kumar A, Mishra HS. Tinospora cordifolia (Willd.) Hook. f. and Thoms. (Guduchi)—validation of the Ayurvedic pharmacology through experimental and clinical studies. Int J Ayurveda Res. 2010;1(2):112–21. 10.4103/0974-7788.64405. [DOI] [PMC free article] [PubMed]
- 48.Gala VC, John NR, Bhagwat AM, Datar AG, Kharkar PS, Desai KB. Attenuation of quorum sensing-regulated behaviour by Tinospora cordifolia extract & identification of its active constituents. Indian J Med Res. 2016;144(1):92–103. doi: 10.4103/0971-5916.193295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armijos-Jaramillo V, Yeager J, Muslin C, Perez-Castillo Y. SARS-CoV-2, an evolutionary perspective of interaction with human ACE2 reveals undiscovered amino acids necessary for complex stability. Evol Appl. 2020 doi: 10.1111/eva.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H, Gai S, Wang X, Zeng J, Sun C, Zhao Y, et al. Single-cell analysis of SARS-CoV-2 receptor ACE2 and spike protein priming expression of proteases in the human heart. Cardiovasc Res. 2020;116(10):1733–1741. doi: 10.1093/cvr/cvaa191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu J, Sun PD. High affinity binding of SARS-CoV-2 spike protein enhances ACE2 carboxypeptidase activity. bioRxiv: the preprint server for biology. 2020. 10.1101/2020.07.01.182659. [DOI] [PMC free article] [PubMed]
- 52.Ibrahim MAA, Abdeljawaad KAA, Abdelrahman AHM, Hegazy MF. Natural-like products as potential SARS-CoV-2 M(pro) inhibitors: in-silico drug discovery. J Biomol Struct Dyn. 2020:1–13. 10.1080/07391102.2020.1790037. [DOI] [PMC free article] [PubMed]
- 53.Lokhande KB, Doiphode S, Vyas R, Swamy KV. Molecular docking and simulation studies on SARS-CoV-2 M(pro) reveals Mitoxantrone, Leucovorin, Birinapant, and Dynasore as potent drugs against COVID-19. J Biomol Struct Dyn. 2020:1–12. 10.1080/07391102.2020.1805019. [DOI] [PMC free article] [PubMed]
- 54.Sacco MD, Ma C, Lagarias P, Gao A, Townsend JA, Meng X et al. Structure and inhibition of the SARS-CoV-2 main protease reveals strategy for developing dual inhibitors against M(pro) and cathepsin L. bioRxiv: the preprint server for biology. 2020. 10.1101/2020.07.27.223727. [DOI] [PMC free article] [PubMed]
- 55.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williamson G, Kerimi A. Testing of natural products in clinical trials targeting the SARS-CoV-2 (Covid-19) viral spike protein-angiotensin converting enzyme-2 (ACE2) interaction. Biochem Pharmacol. 2020;178:114123. doi: 10.1016/j.bcp.2020.114123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 58.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 59.Eddershaw PJ, Beresford AP, Bayliss MK. ADME/PK as part of a rational approach to drug discovery. Drug Discov Today. 2000;5(9):409–414. doi: 10.1016/s1359-6446(00)01540-3. [DOI] [PubMed] [Google Scholar]
- 60.Ekins S, Waller CL, Swaan PW, Cruciani G, Wrighton SA, Wikel JH. Progress in predicting human ADME parameters in silico. J Pharmacol Toxicol Methods. 2000;44(1):251–272. doi: 10.1016/s1056-8719(00)00109-x. [DOI] [PubMed] [Google Scholar]
- 61.Thompson TN. Early ADME in support of drug discovery: the role of metabolic stability studies. Curr Drug Metab. 2000;1(3):215–241. doi: 10.2174/1389200003339018. [DOI] [PubMed] [Google Scholar]
- 62.Ortega SS, Cara LC, Salvador MK. In silico pharmacology for a multidisciplinary drug discovery process. Drug Metab Drug Interact. 2012;27(4):199–207. doi: 10.1515/dmdi-2012-0021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.