Significance
While zinc (Zn2+) is a vital ion for cell function and human health, little is known about the role it plays in regulating cell signaling. Here, we use fluorescent tools to study the interaction between Zn2+ and cell signaling pathways that play a role in cell growth and proliferation. Importantly, we use small, non-toxic Zn2+ concentrations to ensure that our Zn2+ changes are closer to what cells would experience in the body and not stress inducing. We also demonstrate that these signaling changes are driven by Ras activation, which contradicts one of the major hypotheses in the field. Our sensors shed light on how cells respond to an important micronutrient in real time.
Keywords: zinc, kinase signaling, ERK, Akt, Ras
Abstract
Zinc (Zn2+) is an essential metal in biology, and its bioavailability is highly regulated. Many cell types exhibit fluctuations in Zn2+ that appear to play an important role in cellular function. However, the detailed molecular mechanisms by which Zn2+ dynamics influence cell physiology remain enigmatic. Here, we use a combination of fluorescent biosensors and cell perturbations to define how changes in intracellular Zn2+ impact kinase signaling pathways. By simultaneously monitoring Zn2+ dynamics and kinase activity in individual cells, we quantify changes in labile Zn2+ and directly correlate changes in Zn2+ with ERK and Akt activity. Under our experimental conditions, Zn2+ fluctuations are not toxic and do not activate stress-dependent kinase signaling. We demonstrate that while Zn2+ can nonspecifically inhibit phosphatases leading to sustained kinase activation, ERK and Akt are predominantly activated via upstream signaling and through a common node via Ras. We provide a framework for quantification of Zn2+ fluctuations and correlate these fluctuations with signaling events in single cells to shed light on the role that Zn2+ dynamics play in healthy cell signaling.
Zinc (Zn2+) is an essential metal in biology, with approximately 10% of the proteins encoded by the human genome predicted to bind Zn2+ (1). All cells maintain and regulate a small pool of labile Zn2+ that can be exchanged among Zn2+-binding proteins and Zn2+ biosensors. The concentration of labile Zn2+ in the cytosol, measured in the hundreds of picomolar range (2–5), falls within the affinity range of many Zn2+ binding proteins, suggesting that under normal conditions many of these proteins will bind Zn2+ and function properly. However, some Zn2+ binders may need higher Zn2+ concentrations in order to function (6). Furthermore, there is growing evidence that mammalian cells experience fluctuations in available Zn2+, and these dynamics have been shown to be important for cell physiology (7–11).
In addition to serving as an important biological cofactor (12), there are increasing examples that Zn2+ also plays a role in biological signaling. Crosstalk has been observed between Zn2+ dynamics and calcium signaling where increases in cytosolic Zn2+ lead to decrease in endoplasmic reticulum (ER) calcium, and conversely, increases in cytosolic calcium change Zn2+ homeostasis in the ER3. Zn2+ sequestration has been shown to block cell cycle progression in both meiotic oocytes (13) and mitotic cells (14–16). At a molecular level, picomolar concentrations of Zn2+ potentiate the response of the ryanodine receptor in cardiomyocytes (17). Zn2+ has also been implicated in metabotropic signaling via the G protein–coupled receptor 39 (GPR39 (18)), direct modulation of protein kinase C activity (19), and activation of MAPK kinase signaling pathways in neurons (20), cardiomyocytes (21), and mast cells (7). While the above studies demonstrate that Zn2+ fluctuations influence cellular processes, in many cases the molecular details of how Zn2+ interacts with canonical signaling pathways, second messengers, or serves as a signal itself are unclear. This is especially true for the MAPK pathway.
MAPK signaling plays a role in cell proliferation, differentiation, and development and is one of the most well-studied signaling pathways (22). A connection between MAPK signaling and Zn2+ was first reported in 1996 when it was observed that addition of 300 μM ZnCl2 to 3T3 fibroblasts led to increased phosphorylation of ERK1/2 kinases in the MAPK pathway (23). Early studies used epithelial cell lines to study the connection between Zn2+ and ERK signaling (23, 24). More recently, Zn2+ elevation has been demonstrated to increase ERK phosphorylation in dissociated neurons and transformed HT22 cells, where ERK signaling has been linked to synaptic plasticity and memory consolidation (20, 25, 26). The mechanism of ERK activation by Zn2+ remains enigmatic. The leading hypothesis has been that Zn2+ inhibits protein phosphatases, leading to sustained ERK activation. This idea is supported by the observation that ERK-directed phosphatase PP2A activity is reduced when Zn2+ is added to cell lysates (20, 25). Furthermore, it has been demonstrated that certain phosphatases are inhibited by nano- and picomolar concentrations of Zn2+ in vitro, although these phosphatases are not known to directly interact with ERK1/2 (27, 28). However, it is unclear how these bulk in vitro analyses relate to the role of Zn2+ fluctuations in living cells.
The connection between Zn2+ and modulation of the MAPK pathway is even more perplexing when examining how Zn2+ influences Ras activity, which acts upstream of Raf-MEK-ERK. Two studies that involved acute perturbation of Zn2+ by adding high concentrations of Zn2+ concluded that Zn2+ promotes Ras activation (29, 30). On the other hand, two genetic screens in Caenorhabditis elegans suggested that Zn2+ inhibits Ras activity (31, 32). While these studies involved different model systems (cell lines versus C. elegans) and different means of altering Zn2+ (acute elevation versus chronic manipulation), it is important to note that there is a lack of consensus on how Zn2+ influences the Ras-Raf-MEK-ERK pathway.
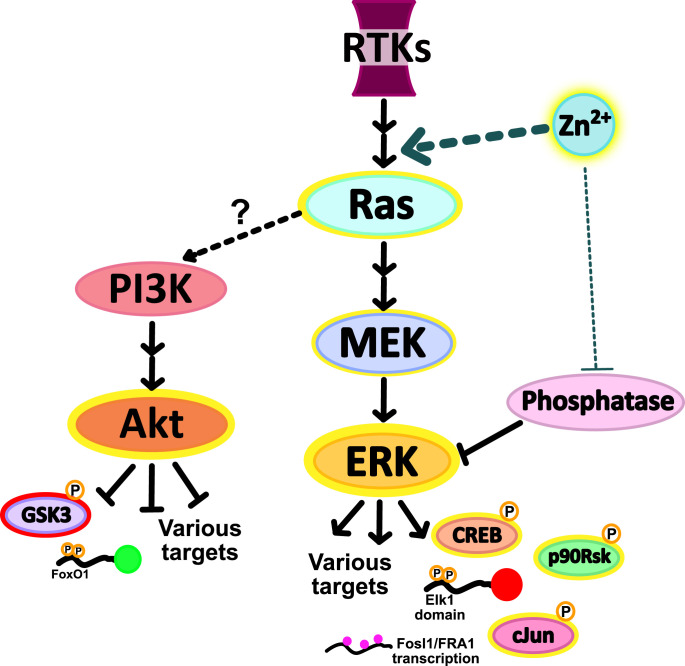
In this work we set out to dissect the connection between Zn2+ and ERK in an effort to elucidate the mechanism of activation. Using a combination of kinase translocation reporters and a Förster resonance energy transfer (FRET)-sensor for Zn2+, we quantified the changes in intracellular Zn2+ in response to subtle extracellular perturbations and correlated them directly with changes in kinase activity at the single cell level. We found that while elevated Zn2+ broadly inhibits phosphatase activity to some extent in vitro, in live cells, Zn2+ primarily activates ERK via upstream signaling, suggesting that ERK phosphatase inhibition can’t fully account for the Zn2+-induced increase in ERK activity. Finally, we demonstrate that our Zn2+ conditions activate Ras and Akt signaling along with ERK but that few other kinases are activated, including stress-response kinases JNK, p38, and p53. We therefore propose a mechanism of action where Zn2+ activates ERK and Akt pathways upstream of Ras, while the specific Zn2+-protein interaction remains elusive.
Results
Quantification of Zn2+ Manipulations.
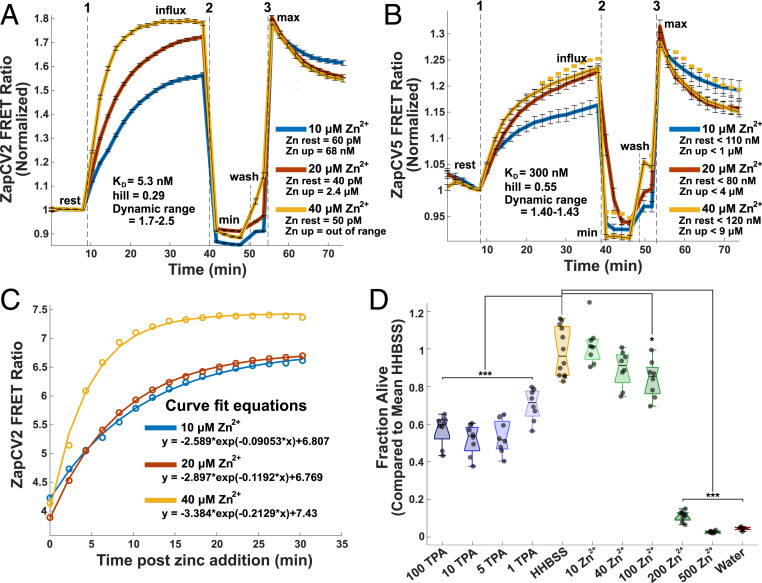
One limitation of previous studies was the treatment of cells with high concentrations of Zn2+ and the lack of quantification of how these extracellular perturbations altered the intracellular Zn2+ concentration. Therefore, we used genetically encoded FRET-based Zn2+ sensors to quantify the changes in intracellular Zn2+ in response to a series of applied extracellular Zn2+ solutions. The FRET ratios of these sensors are proportional to the concentration of labile Zn2+ so that a higher ratio corresponds to higher Zn2+. Addition of ZnCl2 extracellularly to HeLa cells, without the use of ionophores, causes a rapid and dose-dependent increase in intracellular Zn2+ that saturates in ∼40 min (Fig. 1). After measuring the FRET ratio upon addition of extracellular Zn2+, cells were subjected to perturbations that deplete or saturate the sensor (minimum and maximum FRET ratio, respectively). As described in Materials and Methods, parameters obtained from this in situ calibration can be used to approximate the concentration of labile Zn2+ under resting conditions and after Zn2+ addition. We used two cytosolic sensors with different apparent dissociation constants for Zn2+ (NES-ZapCV2, Kd = 5.3 nM and NES-ZapCV5, Kd = 300 nM) (33, 34). Resting Zn2+ in the cytosol was in the low pM range, consistent with previous measurements (2, 4, 5). Addition of 10 to 40 μM Zn2+ caused intracellular Zn2+ to increase to the low micromolar range (Fig. 1). The higher affinity ZapCV2 sensor became fully saturated upon addition of 40 μM Zn2+ (Fig. 1A), so to quantify the range of Zn2+ concentrations in cells, the lower affinity ZapCV5 sensor was also used to measure Zn2+ influx (Fig. 1B). ZapCV5 has a low dynamic range (∼1.4 versus ∼2 for ZapCV2), which can lead to overestimation of Zn2+ concentrations due to the inverse relationship between dynamic range and fractional saturation of the sensor regardless of apparent KD (35). The ZapCV5 data have therefore been reported as “less than” the calculated value. Calculations were made using the asymptote of the curve fit equations for each condition (Fig. 1C) to provide a reliable Zn2+ estimate but minimize the overall time cells were exposed to treatment and light. These results demonstrate that increasing the concentration of extracellular Zn2+ leads to a titratable increase in intracellular Zn2+ levels. To put these perturbations in perspective, the concentration of Zn2+ in human serum is ∼15 μM (36), and cell culture medias vary from 1 to 40 μM (37), with most of the Zn2+ being supplied by the serum.
Fig. 1.
Characterization of Zn2+ perturbations. Quantification of cytosolic Zn2+ influx from extracellular addition in HeLa cells using (A) ZapCV2 and (B) ZapCV5. The FRET ratio of each trace was normalized by dividing by the FRET ratio at the frame immediately prior to Zn2+ addition. Dotted lines indicate media manipulation as follows: 1) addition of 10, 20, or 40 μM ZnCl2; 2) addition of 50 μM TPA (minimum); or 3) addition of 81.6 μM buffered Zn2+ + 0.002% saponin + 750 nM pyrithione (maximum). Traces are representative of three experiments on different days with n ≥ 20 cells per trace. Curve fits (C) were used to estimate maximum for calculations of the ZapCV2 (C) and ZapCV5 data. Fit parameters are in SI Appendix, Table S1. Cell death (D) was measured using CellTiter-Glo. Cells were incubated for 4 h in HHBSS with added micrometer concentrations of Zn2+ or TPA. Each point represents an individual well from a single experiment. Box plot shows the median, and whiskers represent 25th and 75th percentiles. One-way ANOVA results for deviation from HHBSS control represented by *P < 0.05, **P < 0.005, ***P < 0.0005.
We want to clearly distinguish between our goal of understanding the impacts of small Zn2+ changes on cell signaling processes in healthy cells compared to the study of Zn2+ toxicity following traumatic brain injuries, epilepsy, and stroke (26, 38, 39). Therefore, we measured whether Zn2+ perturbations induce cell toxicity using a CellTiter-Glo assay. As demonstrated in Fig. 1D, Zn2+ manipulations up to 40 μM do not cause significant levels of cell death in HeLa cells. Interestingly, we did find that the Zn2+ chelator Tris(2-pyridylmethyl)amine (TPA) can be toxic to cells at concentrations as low as 1 μM, suggesting that HeLa cells are better able to withstand Zn2+ increases than decreases.
Zn2+ Activates Select Kinases Including ERK and Akt.
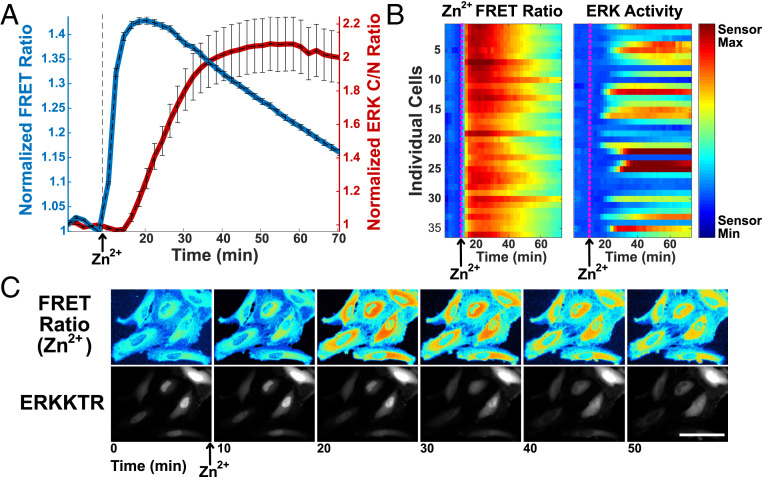
We next sought to measure kinase activation by Zn2+ in single cells and populations of cells to examine dynamics, heterogeneity, and breadth of kinase activation. Using a combination of genetically encoded reporters, cytosolic Zn2+ and ERK activity can be simultaneously monitored in live cells in response to cellular perturbations. The ZapCV2 sensor was used to monitor changes in the labile Zn2+ pool and an ERK kinase translocation reporter (KTR) (40) was used to monitor ERK activity in live cells. Epidermal growth factor (EGF) was used as a positive control for ERK activation via receptor tyrosine kinases. Simultaneous imaging of Zn2+ and ERK activity reveals that upon treatment with 40 μM Zn2+, cytosolic Zn2+ rises immediately and precedes the increase in ERK activity (Fig. 2A and Movies S1 and S2). We verified that the converse is not true; treatment with EGF does not alter labile Zn2+ (SI Appendix, Fig. S1 A and D). There is variability from cell to cell in both the magnitude of Zn2+ increase and ERK activation, but all cells with increased ERK activity exhibited an increase in Zn2+ (Fig. 2 B and C). There was not a strong correlation between the variability in the magnitude of the Zn2+ signal and variability in magnitude of the ERK KTR signal (SI Appendix, Fig. S2A). These results suggest that low micromolar Zn2+ influx into cells leads to a rapid increase in ERK activity in live cells.
Fig. 2.
Simultaneous imaging of Zn2+ and ERK activity demonstrates that in individual HeLa cells, Zn2+ increase precedes the increase in ERK activity. (A) Normalized mean fluorescence signal (FRET ratio for ZapCV2 [blue] and C/N intensity ratio for ERKKTR [red]) and SD from 28 cells plotted against time. Fluorescence signal is normalized to the frame before Zn2+ addition. (B) Heatmap of individual cells compares the magnitude of Zn2+ increase and ERK activation side by side as a function of time. (Scale bar, 0.9 to 1.5 for Zn2+ FRET ratio; scale bar, 0.4 to 3.5 for ERK activity). (C) Snapshot images over the course of the experiment showing the pseudocolor FRET ratio image (ZapCV2 Zn2+ sensor) and mCherry fluorescence image (ERK translocation reporter movement from the nucleus to cytosol). (Scale bar, 100 μm.) Time-course videos are available as Movie S1 (ERKKTR) and Movie S2 (NES-ZapCV2 FRET).
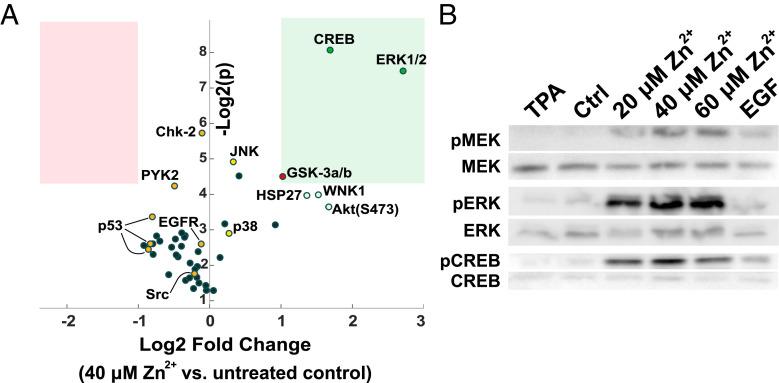
To better understand the breadth of the cell signaling response to Zn2+, we then performed a kinase phospho array on cells where 40 μM Zn2+ was added for 30 min. We found that low micromolar increases in Zn2+ led to robust phosphorylation of a few cell signaling proteins, including ERK1/2, the transcription factor CREB that acts downstream of ERK, and GSK-3α/β, which is phosphorylated and inactivated by Akt. Notably, proteins that participate in cell stress pathways, including JNK, p38, Chk-2, and p53, are not activated by Zn2+ under these conditions. JNK was confirmed to be insensitive to Zn2+ via live-cell imaging with a JNK translocation sensor (SI Appendix, Fig. S3). Furthermore, MAPK pathway stimulating proteins PYK2, Src, and EGFR also do not appear to be activated by Zn2+ (Fig. 3A and SI Appendix, Table S2). When analyzed by Western blot, cell populations treated with different concentrations of Zn2+ demonstrate robust phosphorylation of proteins both upstream (MEK) and downstream (CREB) of ERK (Fig. 3B). Combined, the phospho array and Western blot results suggest that Zn2+ elevation activates a number of kinases in the MAPK pathway, both upstream and downstream of ERK. However, Zn2+ does not induce widespread nonspecific kinase activation, arguing against broad spectrum inhibition of phosphatases by Zn2+ under these conditions. Finally, the Zn2+ treatments used here do not activate stress-response–related kinases.
Fig. 3.
Kinase activation upon Zn2+ addition. (A) Log2 fold change in phosphorylation versus Log2 (P) for a series of human kinases upon Zn2+ treatment of HeLa cells. A Proteome Profiler Human Phospho-Kinase Array was used to evaluate changes in phosphorylation for a panel of kinases from cells either untreated or treated with 40 μM Zn2+ for 30 min. Two biological replicates were run per condition. The pink box represents Log2 fold change < −1 and P < 0.05. The green box represents Log2 fold change > 1 and P < 0.05. Details are in SI Appendix, Table S2. (B) Western blots for total and phosphorylated protein of MEK, ERK, and CREB show activation after treatment with different concentrations of Zn2+ for 30 min. Individual antibody blots were run on separate gels, cut at ∼48 kDa to compare with β-actin loading control (Fig. S4), and contrast was adjusted individually using ImageJ. Blots are representative of at least four separate experiments.
To further explore whether Zn2+ activation of ERK is transduced through the signaling pathway, we carried out RNA fluorescence in situ hybridization using probes against the messenger RNA (mRNA) of the ERK-target Fosl1 (also called Fos-related antigen 1, FRA1). As shown in SI Appendix, Fig. S5, treatment with EGF or Zn2+ significantly increased the expression of Fosl1 mRNA compared to control cells, demonstrating that Zn2+-activation of ERK leads to propagation of the signal through the pathway. We also performed immunoblotting with antibodies against phosphorylated targets of ERK, which revealed that Zn2+ leads to phosphorylation of ERK targets CREB, p90RSK, and cJun (SI Appendix, Figs. S6 and S7).
To test whether Zn2+ activates calcium signaling pathways that in turn lead to ERK activation, we expressed the ERK translocation sensor in conjunction with the D3cpV FRET-based Ca2+ biosensor (41). Zn2+, but not treatments that elevate calcium (Ca2+/ionomycin and histamine), activated ERK. Conversely, Zn2+ did not lead to changes in cytosolic calcium levels (SI Appendix, Fig. S8). These experiments demonstrate that elevation of cytosolic Zn2+ does not lead to detectable changes in cytosolic Ca2+, and hence, Zn2+-induced ERK activation does not occur by indirect activation of Ca2+ signaling.
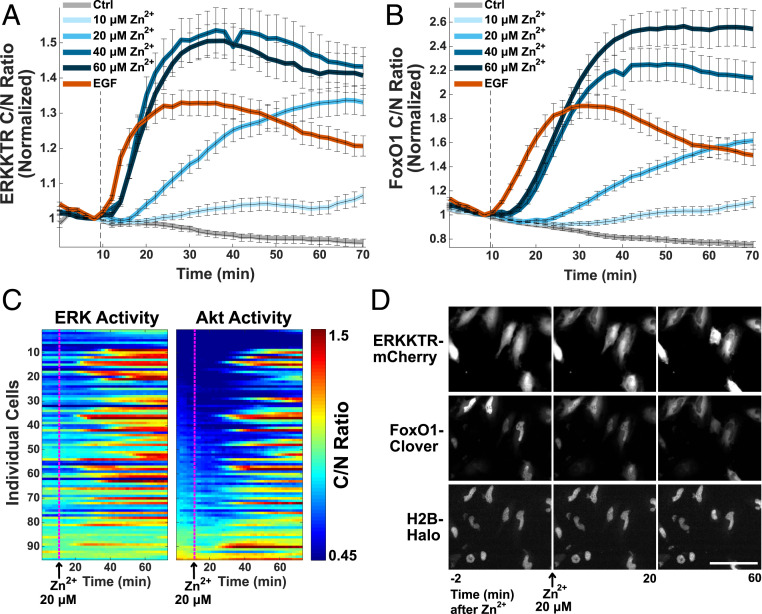
Zn2+ leads to activation of both ERK and Akt, as demonstrated by cells expressing the Akt translocation sensor FoxO1-Clover (42), ERK-KTR-mCherry, and the nuclear marker H2B-Halo imaged using JF646-Halo dye. Both Akt and ERK are stimulated by Zn2+ in a titratable manner, and sensor signals appear to saturate between 20 and 40 μM Zn2+ (Fig. 4 A and B and Movies S3 and S4). While the activation of kinases by Zn2+ varies from cell to cell, the pattern of activation of ERK and Akt is similar, suggesting the possibility of a common activation mechanism or pathway crosstalk (Fig. 4C and SI Appendix, Fig. S2B). Data are sorted by initial Akt activity to highlight that activation by Zn2+ is independent of initial kinase activity.
Fig. 4.
Zn2+ activates both ERK and Akt in a concentration-dependent manner. Averaged ERK activity (A) and corresponding averaged Akt activity (B) when indicated Zn2+ concentration or 20 nM EGF is added to HeLa cells at the dotted line. Both sensors were expressed in the same cell population and are represented by the mean cytosol/nuclear intensity ratio and SD error bars from at least 50 cells plotted against time, normalized to the frame before Zn2+ addition. (C) Representative heat map of non-normalized ERK and Akt activity in cells treated with 20 μM Zn2+ at 10 min. (Heat map scale bar, ratio: 0.45 to 1.5.) (D) Snapshot images over the course of the experiment showing the ERK (ERK-KTR-mCherry) and Akt (FoxO1-Clover) translocation sensors and nuclear marker H2B-Halo upon treatment with 20 μM Zn2+. (Image scale bar, 100 μM.) Time-course videos are available as Movie S3 (ERKKTR-mCherry) and Movie S4 (FoxO1-Clover).
While experiments in this paper were conducted in HeLa cells for ease of use, similar patterns of ERK activation by 40 μM Zn2+ were seen in noncancer mammary epithelial MCF10A cells and the mouse hippocampal neuronal cell line HT-22 (SI Appendix, Fig. S1). These results demonstrate that activation of ERK by Zn2+ is a general phenomenon in multiple mammalian cell types.
Mechanistic Insight into ERK Activation by Zn2+.
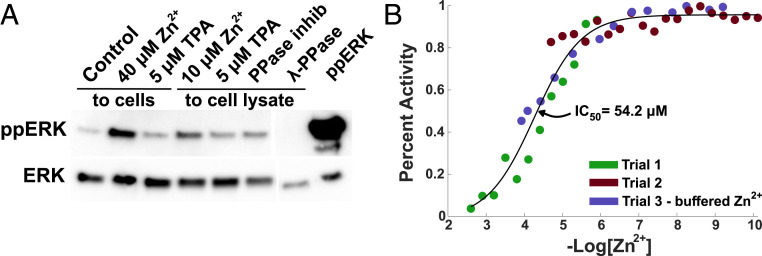
Previous research has suggested that the increase in ERK activity upon Zn2+ treatment may result from Zn2+ inhibition of phosphatases (20, 25). However, many of these studies were performed with cell lysates or in vitro. To explore whether this mechanism could explain ERK activation in our system, we carried out an ERK phosphatase assay on cells treated with Zn2+ under conditions analogous to our imaging experiments, followed by lysis. As a positive control, we also measured phosphatase activity in lysed cells. Briefly, His-tagged dual-phosphorylated ERK was incubated with whole cell lysate from cells treated with TPA or Zn2+ for 30 min. Alternately, His-ppERK was incubated with lysate from untreated cells to which we added Zn2+, TPA, phosphatase inhibitor (BCI-hydrochloride), or λPPase after lysis (Fig. 5A). His-ERK was then removed by nickel beads, and the extent of de-phosphorylation was determined via Western blot for total and phosphorylated ERK. Zn2+ added to cells pre- and postlysis resulted in a decrease of ERK de-phosphorylation, suggesting that Zn2+ can inhibit ERK-directed phosphatases in vitro. Our results are in line with previous research (25), namely that treatment of cell lysates with Zn2+ reduces phosphatase activity. Furthermore, we now show that the low micromolar increases in cytosolic Zn2+ upon treatment of intact cells also reduce phosphatase activity. While this assay demonstrates that phosphatase inhibition can contribute to increased ERK phosphorylation, it does not implicate ERK phosphatases directly. This is especially the case given that we observe robust activation of upstream kinases (e.g., MEK) and similar activation patterns for ERK and Akt, suggesting the possibility of a common upstream activator.
Fig. 5.
ERK phosphatases are inhibited by Zn2+ in vitro. (A) ERK phosphatase assay using ppERK-His and nickel column purification before Western blotting. Zn2+ or chelator (TPA) were either added to cells for 30 min before lysis or added directly to HeLa cell lysate. BCI-hydrochloride and λ-PPase controls were added to cell lysate, and ppERK lane indicates the amount of ppERK added to each sample. Blot is representative of four separate experiments. (B) In vitro inhibition studies of MKP-3 by Zn2+ through detection of phosphate group released from 4-methylumbelliferyl phosphate. Colors indicate data points from three separate experiments with a variety of Zn2+ concentrations. Purple dots use a variety of chelators and counter ions to buffer the Zn2+ concentration for more accurate measurement (SI Appendix, Table S3).
To determine whether Zn2+ can inhibit ERK-directed phosphatases, we performed an in vitro inhibition assay with the ERK-selective dual-specificity phosphatase DUSP6 (MKP-3) (43). Three separate experiments with Zn2+ concentrations from 76 pM to 2.5 mM (SI Appendix, Table S3) were overlaid and converged on an IC50 of 54.2 μM, which demonstrates that DUSP6 activity is inhibited by Zn2+ but at concentrations well above cellular Zn2+ levels (Fig. 5B). Together the data suggest that while phosphatase inhibition by Zn2+ may play a role in ERK activation, the concentration of Zn2+ required for inhibition of an ERK-directed phosphatase is higher than the intracellular concentration in our studies. Therefore, it seems unlikely that direct inhibition of ERK phosphatases is the dominant mechanism in cells.
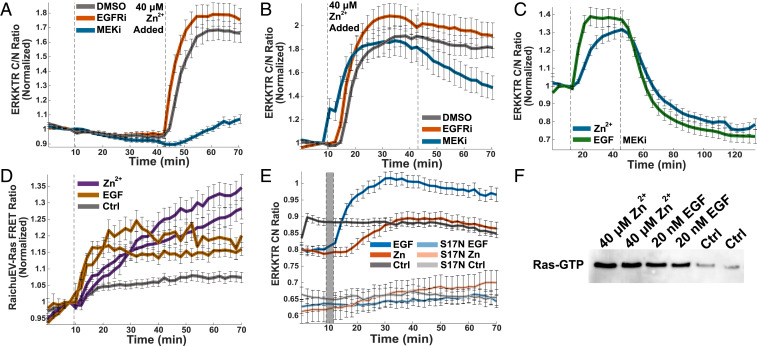
While phosphatases can be inhibited by Zn2+ in vitro, the work with DUSP6 suggests that ERK-directed phosphatases may be inhibited at supraphysiological concentrations, and therefore, phosphatase inhibition may only play a small role in ERK activation by Zn2+ in cells. Further, bulks assays (Western blot and phospho array) suggested activation of the MAPK pathway upstream of ERK. To further characterize the extent of activation at the single cell level, we used a variety of kinase inhibitors and the ERK translocation sensor in imaging experiments. MEK was shown to be activated by Zn2+ in Fig. 3B, so first we sought to explore the effect of MEK inhibition on ERK activation by Zn2+. We show that MEK inhibition by CI-1040 prior to Zn2+ treatment greatly decreases the extent of ERK activation by Zn2+ (Fig. 6A). ERK activation is not totally abolished, suggesting that either inhibition was not complete or that downstream phosphatase inhibition also plays a small role in increasing ERK activity. Inhibition of the upstream EGFR with Gefitinib, however, had no effect of Zn2+-induced ERK activation (Fig. 6A). Furthermore, when cells are stimulated with Zn2+, followed by inhibitors, MEK inhibition leads to a decrease in Zn2+-activated ERK signaling, whereas EGFR inhibition does not (Fig. 6B), indicating that a significant portion of Zn2+ activation of the pathway occurs upstream of MEK but downstream of activation of the receptor tyrosine kinase EGFR. In an extended time course, cells activated by both Zn2+ and EGF experience similar rates of ERK signal decay upon addition of a MEK inhibitor, suggesting a similar upstream signaling process for both Zn2+ and growth factors (Fig. 6C).
Fig. 6.
Kinases upstream of ERK are activated by Zn2+. MEK inhibition (1 μM CI-1040) either before (A) or after (B) Zn2+ addition reduces ERK activation, whereas EGFR inhibition (1 μM Gefitinib) does not. Dotted lines indicate addition of drug or Zn2+. Normalized mean and SD of at least 35 HeLa cells are plotted against time. Traces are representative of at least three separate experiments. (C) HeLa cells stimulated by Zn2+ and EGF (first dotted line) exhibit similar rates of ERK decay upon MEK inhibition (second dotted line). Normalized mean and SD of at least 38 cells are plotted against time. Traces are representative of three separate experiments. (D) Ras is activated by both Zn2+ and EGF when indicated Zn2+ concentration or 20 nM EGF is added to HeLa cells at the dotted line, measured via RaichuEV-Ras FRET sensor (Kazuhiro Aoki). The normalized mean and SD from at least 30 cells are plotted against time. Traces are representative of three out of four separate biological replicates. (E) Dominant negative Ras (S17N) blocks both Zn2+ and EGF activation of ERK. Non-normalized mean and SD of at least 29 HeLa cell traces are plotted against time. S17N traces are representative of ERK activity in cells with at least 500 fluorescence counts in the GFP channel, indicative of HRas-S17N expression. Data are not normalized to show the shift in ERK activity when the dominant negative HRas-S17N is present. Due to large shift in background upon media change, the difference between frames five and six were subtracted from all traces after frame six, and that frame was itself omitted from representation (indicated with a gray bar). (F) Ras GTP pull-down shows that treatment with 20 mM Zn2+ or EGF increases the amount of active GTP-bound Ras compared to treatment with buffer. Experiments in this figure were carried out in HeLa cells.
To further explore activation mechanisms upstream of both ERK and Akt, we used the RaichuEV-Ras FRET sensor (44) to determine whether Zn2+ activates the Ras GTPase. We demonstrated that Zn2+ activates Ras to an extent similar to EGF activation (Fig. 6D). The Ras FRET sensor exhibits a low dynamic range, making it difficult to infer further about magnitude or dynamics of Ras activation. However, transfection with a dominant negative Ras (GFP-HRas-S17N) (45) blocked activation of ERK by Zn2+, revealing that Ras activation is a critical step in Zn2+ activation of the MAPK pathway (Fig. 6E and SI Appendix, Fig. S9). Furthermore, on a population level, GFP-HRas-S17N diminished activation of ERK along with MEK, CREB, p90RSK, cJun, and GSK3β (SI Appendix, Fig. S6), suggesting that targets downstream of ERK and Akt are affected by the dominant negative Ras. Finally, treatment with Zn2+ (or EGF) leads to an increase in the amount of GTP-bound Ras, comparable to that observed upon EGF activation, as demonstrated by Ras GTP pull-down (Fig. 6F). Our results demonstrate that acute increase in Zn2+ in the low micromolar range leads to activation of Ras, which is upstream of both ERK and Akt. Combined with results showing that the dominant negative Ras inhibits Zn2+-induced ERK activation at the single cell level and decreases activation of ERK and Akt downstream targets, these results implicate Ras in the Zn2+-dependent activation of the MAPK and likely Akt signaling pathways.
Discussion
The traditional model of zinc in biology is that Zn2+ functions as a cofactor for a number of proteins, either to maintain their structure or facilitate catalysis, and these proteins are thought to constitutively bind Zn2+ (1, 12, 46). Recently, we have seen an increase in examples of Zn2+ dynamics in cells (3, 7, 8, 11, 47) and evidence that Zn2+ can play a regulatory role for a variety of cell processes (7, 48–50). Although Zn2+ has long been suggested to serve as a cellular signal, much like calcium, the precise molecular details of how Zn2+ accomplishes this task remains elusive for most systems.
One of the challenges in defining how changes in Zn2+ influence cellular processes is that, like many trace essential elements, too little or too much Zn2+ can activate stress pathways and induce toxicity (51–53). It has been difficult to define what constitutes physiological zinc fluctuations. A widely used approach to elevate cellular zinc is to use the ionophore pyrithione to shuttle zinc into cells. However, pyrithione is not an innocent ligand. Pyrithione inhibits the growth of yeast and can act as a copper ionophore, perturbing both copper and iron homeostasis (54). Pyrithione with Zn2+ can increase susceptibility of cells to oxidative stress (55) and inhibit the growth of human skin cells, including DNA synthesis (56), and has been implicated in several cell death pathways including canonical apoptosis (57), p53-independent apoptosis via ERK activation (29), and nonapoptotic cell death via adenosine triphosphate (ATP) depletion and bio-energetic collapse (58). Furthermore, the field has suffered from lack of quantification of how cellular perturbations of Zn2+ alter intracellular Zn2+ levels. Recent work has shown that subtle, more physiological changes in zinc can influence oocyte maturation (10), epigenetic chromatin covalent modifications (59), gene expression in neurons (34), and the mammalian proliferation–quiescence decision (16).
The connection between Zn2+ and kinase signaling has been studied in a variety of systems. Both Zn2+ excess (25, 26) and depravation (60, 61) have been linked to ERK-dependent cell death in neurons and neuronal cell lines. In myogenic cells (skeletal muscle precursor), Zn2+ addition was shown to promote proliferation and prevent differentiation through both ERK and Akt signaling (62). High concentrations of Zn2+ or zinc pyrithione have also been shown to activate other kinase signaling pathways in cells including JNK and p38 stress-related kinases (63, 64), Src family kinases (65–67), protein kinase C (19), and the zinc-sensing receptor GPR39 (18, 68). Finally, we demonstrate that three different cell lines exhibit Zn2+-dependent ERK activation, suggesting this may be a widespread phenomenon in a variety of cell types.
In this study, we induce acute Zn2+ elevation in the cytosol of cells in the absence of ionophores or chelators. Our Zn2+ conditions elevate Zn2+ into the low micromolar range. While influx of Zn2+ from extracellular media is the logical explanation for cytosolic Zn2+ elevation, it is also possible that elevation of extracellular Zn2+ induces release of Zn2+ from an intracellular store. This zinc elevation is several orders of magnitude larger than the low nanomolar Zn2+ transient seen upon stimulation of dissociated neurons (11), suggesting that our results likely amplify the cell signaling response to physiological Zn2+ fluxes. However, using 40 μM Zn2+, we were able to detect changes in multiple kinase signaling pathways while preventing activation of stress pathways and cell death. We demonstrated that ERK and Akt signaling are activated by addition of as little as 10 μM extracellular Zn2+ (∼70 nM cytosolic labile Zn2+) and that this signal is propagated to downstream targets (phosphorylation of CREB, p90RSK, and cJun and an increase in mRNA expression of Fosl1 for ERK and phosphorylation of GSK3β for Akt). As noted in Figs. 2 and 4, there is heterogeneity in the extent of kinase activation in response to Zn2+, and the origin of this heterogeneity is unknown. It would be intriguing in the future to explore whether the heterogeneity depends on cell cycle state or prior activation of kinase signaling pathways. We demonstrate that the upstream signaling proteins MEK and Ras are activated by Zn2+ addition, but that EGFR is not, thus honing in on Ras as a signaling node through which Zn2+ activates cell signaling pathways (Fig. 7). The mechanism by which Ras is activated by Zn2+ remains elusive. It is intriguing that Zn2+ activation of the MAPK pathway appears to be very different from copper activation, as copper has been shown to bind directly to MEK, stimulating activity and leading to increased ERK phosphorylation (69, 70).
Fig. 7.
The model we propose for Zn2+ activation of kinase pathways involves primarily activation of kinases upstream of ERK, in particular both MEK and Ras, as well as a potentially smaller component of inhibition of phosphatases that sustain ERK signaling. Activation of the MAPK pathway at the Ras node would explain the similar activation patterns of both Akt and ERK.
Four previous studies have drawn a connection between Ras and Zn2+, but with conflicting conclusions. Two of these studies found that acute treatment with high Zn2+ (100 to 500 μM Zn2+) led to Ras activation as well as activation of stress response and apoptosis (29, 30). While our study also demonstrates Ras activation, in contrast to the prior studies, we demonstrate that subtle elevation of cytosolic Zn2+ ranging from 70 nM to ∼1 μM leads to biochemical activation of Ras-MEK-ERK without activating stress pathways or inducing cell death. Two other studies involved genetic manipulation of cation diffusion facilitator (cdf) transporters in C. elegans and inferred that elevation of Zn2+ antagonized Ras signaling (31, 32). There are some key differences between those studies and the work we present here. Both C. elegans papers identified loss-of-function mutations in a cdf transporter that suppressed a vulval developmental phenotype. Because cdf transports Zn2+ out of the cytosol, it was inferred that loss-of-function mutants in cdf increase cytosolic Zn2+. Furthermore, because vulval development is a Ras-Raf-MEK-ERK dependent process, it was reasoned that Zn2+ must inhibit Ras. However, direct biochemical evidence of Zn2+ elevation and Ras inhibition were not presented. One possible interpretation is that chronic disruption of cdf-mediated Zn2+ transport could lead to compensatory changes in the Zn2+ regulatory homeostasis network such that cdf loss of function doesn’t alter cytosolic Zn2+ in the expected way. In contrast to chronic changes in Zn2+ homeostasis, our study reports how acute elevation of Zn2+ influences cell signaling in a short period of time (30 min). It is also possible that there are cell-type or model system specific responses that are not fully understood. While we demonstrate that three different cell lines exhibit Zn2+-dependent ERK activation, there is still much to learn about whether certain cell systems respond to Zn2+ in unique ways.
This work provides context for understanding the origin and breadth of kinase activation in cells that experience physiological Zn2+ fluctuations. Much like Ca2+, defining how Zn2+ acts as a signaling ion is a critical step in determining how Zn2+ influences cell biology and understanding how disruptions in Zn2+ (deficiency or overdose) may impact cellular systems. This study provides a framework for Zn2+ manipulation in which cytosolic Zn2+ changes are quantified and correlated with signaling events in single cells. Our work suggests that targeting Ras signaling may be effective in systems that experience Zn2+ dysregulation and that broad nonspecific phosphatase inhibition by Zn2+ is not a strong driver of Zn2+-dependent signaling changes when the Zn2+ perturbations don’t induce stress-response pathways. As the landscape of fluorescent biosensors and chemical probes expands, hopefully more pieces of this signaling pathway will fall into place, and we will gain an even fuller understanding of the role Zn2+ plays in kinase signaling.
Materials and Methods
Key Resources Table.
See Dataset S1.
Molecular Cloning.
pLentiCMV-Puro-DEST-ERKKTRClover and pLentiPGK-Blast-DEST-JNKKTRmRuby2 were purchased from Addgene (plasmid 59150 and 59154, respectively), and translocation sensor domains were subcloned into the pcDNA3.1-mCherry backbone to create mCherry fusions. KTR sequences were PCR amplified using primers listed in the resources table, with Nhe1 overhang on the forward primer and Age1 overhang on the reverse primer. Sensors were then cloned into pcDNA3.1-mCherry using restriction digest upstream of the mCherry fluorescent protein using restriction enzymes Nhe1 and Age1.
To generate ERKKTR-mCherry adenovirus for transduction of difficult-to-transfect cells, primers in the resources table were used to PCR amplify ERKKTR-mCherry out of the pcDNA3.1 backbone with overhangs matching the pShuttle recipient plasmid. This insert was then cloned into pShuttle using an In-Fusion HD Cloning Kit (Clontech/Takara) according to manufacturer’s instruction.
Mammalian Cell Culture.
HeLa cells (ATCC CCL-2) were maintained in full growth Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% bovine serum albumin and 1% Pen/strep antibiotics. MCF10A cell line (ATCC) was maintained in full growth DMEM/F12 medium (FGM) supplemented with 5% horse serum, 1% Pen/strep antibiotics, 20 ng/mL EGF, 0.5 μg/mL hydrocortisone, 100 ng/mL cholera toxin, and 10 μg/mL insulin. The HT-22 cell line was obtained from Xuedong Liu Lab (University of Colorado Boulder), who obtained it as a gift from Toni Pak (Loyola University), and was maintained in full growth DMEM supplemented with 10% bovine serum albumin and 1% Pen/strep antibiotics. All cells were grown in a humidified incubator at 37 °C and 5% CO2. Cells were passaged with trypsin-ethylenediaminetetraacetic acid (EDTA) and routinely tested for mycoplasma.
HeLa cell lines expressing PB-NES ZapCV2 (33) and/or PB-H2B-Halo (71) were generated using the PiggyBac transposon system via TransIT-LT1 (Mirus Bio) according to the manufacturer’s instructions. HeLa cell lines expressing pLenti-FoxO1-Clover were generated by transient transfection of HEK293T cells with pLenti-FoxO1-Clover and Lenti-X fourth-generation lentiviral packaging plasmids, pRev, pMDL, and pVSV-G (Takara Bio), followed by viral amplification in cells for 72 h and the addition of viral particles to HeLa cells. Stable cell lines used for imaging were generated by antibiotic selection (G418: PB-H2B-Halo, blasticidin: PB-NES-ZapCV2, and puromycin: pLenti-FoxO1-Clover) followed by fluorescence-activated cell sorting enrichment for positive fluorescent cells. All transiently transfected sensors (pcDNA3.1-ERKKTR-mCherry, pcDNA3.1-JNKKTR-mCherry, pBSR-RaichuEV-Ras, and pcDNA3.1-ZapCV5) and the Ras mutant EGFP-HRas-S17N (Addgene #18665) were transfected using TransIT-LT1 per manufacturer’s instructions, and cells were imaged 48 to 72 h posttransfection. MCF10A cells were transiently transduced via addition of adenovirus particles to cells at a multiplicity of infection = 10 (adenovirus generation in SI Appendix, Supplemental Methods).
For imaging and growth experiments, cells were transferred to phosphate-free Hepes-buffered Hanks’ Balanced Salt Solution (HHBSS) buffer and incubated at 37 °C and 0% CO2 for at least 30 min prior to imaging.
Live-Cell Imaging.
For live imaging of cells stably expressing H2B-Halo, cells were incubated with 10 nM Halo-tagged Janelia Fluor 646 (JF646) dye (Janelia Research Campus) in phosphate-free HHBSS imaging media for 10 min at 37 °C and 0% CO2. Cells were then washed twice and incubated in phosphate-free HHBSS at 37 °C and 0% CO2 for 20 to 30 min prior to imaging. For imaging of cells without H2B-Halo tag (Fig. 2), cells were instead incubated with 20 μg/mL Hoechst in phosphate-free HHBSS imaging media for 30 min at 37 °C and 0% CO2, and cells were transferred into fresh phosphate-free HHBSS prior to imaging.
Fluorescence microscopy was performed on a Nikon Ti-E inverted microscope with a Lumencor SPECTRA X light engine and Hamamatsu ORCA-Flash4.0 V2 CMOS camera. Images were collected every 1, 2, or 5 min with a 20× 0.8 numerical aperture Plan-Apo objective lens (Nikon Instruments). Cells were kept in an environmental chamber surrounding the microscope (Okolab Cage Incubator) at 37 °C, 0% CO2, and 90% humidity. Several regions of interest in the same dish were imaged in a single cycle with light exposure of cells being 100 ms per timepoint at 50% light-emitting diode light power. Filter sets (excitation = Ex, emission = Em) used for live-cell imaging were as follows: CFP Ex: 440, Em: 460 to 500; GFP Ex: 470, Em: 500 to 545; YFP Ex: 508, Em: 520 to 550; CFPYFP FRET Ex: 440, Em: 520 to 550; mCherry Ex: 555, Em: 590 to 650; and Cy5 Ex: 640, Em: 663 to 738.
Image Analysis.
A diagram of our MATLAB image processing pipeline is in SI Appendix, Fig. S10. Briefly, the Nikon ND2 image file is imported into MATLAB and cells are segmented by nuclear marker in either the Cy5 (H2B-Halo + JF646) or BFP (Hoechst nuclear dye) channel by using the watershed method to generate nuclear masks. Automatic image registration happens at any pause in the experiment to account for small shifts of imaging dishes during media manipulation, and nuclei are tracked throughout the experiment. A nuclear mask and cytosolic ring (4 pixels dilated from the nucleus) are generated, and all channels are subjected to local background subtraction. The average intensity in the nucleus and cytosol of each cell is measured. Cells with very low or high sensor fluorescence were omitted as well as cells that die during imaging and cells that lose tracking during the course of the experiment. For EGFP-HRas-S17N experiments, cells with very low green fluorescence were omitted. For a more detailed look at image processing, please see supplemental information in Lo et al. (16).
Translocation sensors use the average cytosolic fluorescence divided by average nuclear fluorescence to yield a cytosol/nuclear (C/N) ratio. For FRET sensors, average intensity in the cytosol for the acceptor channel (CFP ex, YFP em) is divided by the average intensity for the donor channel (CFP) to yield a FRET ratio. YFP fluorescence is also tracked over time to monitor photobleaching. Normalization of each trace was performed by dividing the FRET or C/N ratio at each timepoint by the FRET or C/N ratio at the frame prior to Zn2+ or EGF addition to facilitate visualization of the changes resulting from perturbation. From this, the mean and SEM were taken for each sensor and plotted over time.
FRET Sensor Calibration.
Zn2+ sensor calibrations were performed in phosphate-free HHBSS, pH 7.4, to prevent Zn2+ precipitation. To collect Rrest (rest, in Fig. 1), cells were incubated in HHBSS for at least 30 min and then imaged for 10 min. Cells were then treated with either 10, 20, or 40 μM ZnCl2 for 30 min by adding 1 mL of 2× concentrated Zn2+ solutions in HHBSS to imaging dishes containing 1 mL HHBSS to get Rinflux (influx, in Fig. 1). To collect Rapo (min, in Fig. 1), 1 mL was removed from imaging dishes and 100 μM TPA in 1 mL HHBSS was added to cells (50 μM TPA final). After 8 min, cells were washed with HHBSS three times, and two frames were imaged before adding Rmax solution of 81.6 μM buffered Zn2+ (Zn2+ buffered with the chelator ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) and counter ion CaCl2, SI Appendix, Table S2), 750 nM pyrithione, and 0.002% saponin (added as 2× concentrated solution in 1 mL HHBSS). The average FRET ratio for rest was calculated by averaging across the timepoints collected, min FRET ratio was the minimum over the timeframe after TPA addition, and max FRET ratio was taken as the maximum FRET ratio over the timeframe after Zn2+/pyrithione/saponin addition. To find FRET ratio max after Zn2+ influx, data from the timeframe after Zn2+ addition was fit to the equation using the MATLAB curve fitting tool. Fit parameters in SI Appendix, Table S2.
To convert FRET ratios into approximate Zn2+ concentrations, the equations and were used, with each R value being the mean of each cell and Rinflux coming from the curve fit asymptote. ZapCV2: KD = 5.3 nM, Hill = 0.29 (34); ZapCV5: KD = 300 nM, Hill = 0.55 (33).
Cell Death Assay.
To assess the toxicity of Zn2+ treatment conditions, cells in a 96-well clear-bottom plate were incubated in HHBSS with a variety of Zn2+ or TPA conditions for 3.5 h at 37 °C and 0% CO2 and 30 min at room temperature. The 4-h timepoint was chosen to represent the longest possible time cells would be in HHBSS for imaging experiments, and each condition was represented by at least six individual wells. CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used to measure the average ATP content of each well, which is representative of the number of metabolically active cells. Cells were plated at equal density, and the signal of each well was divided by average signal in background wells with no cells. The mean HHBSS signal was set as 1, and fraction alive was calculated for each individual well by dividing the well signal by mean HHBSS signal. A one-way ANOVA test comparing HHBSS control to each other experimental condition was conducted in MATLAB.
Kinase Array.
Proteome Profiler Human Phospho-Kinase Array Kit (R&D Systems) was used to measure changes in phosphorylation of 43 kinases upon 40 μM Zn2+ treatment. Cells were plated at equal density in 10-cm dishes, and cells were incubated in HHBSS for 30 min at 37 °C and 0% CO2. Media was then replaced with HHBSS with and without 40 μM Zn2+ and further incubated at 37 °C and 0% CO2 for 30 min. For cell lysis, cells were washed twice with cold phosphate-buffered saline; then, 900 μL radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulphate [SDS], 50 mM Tris pH 8.0, and 5 mM EDTA) was added to each dish, and cells were scraped into a microfuge tube and rocked at 4 °C for 30 min to lyse followed by centrifugation and collection of supernatant. Protein concentration was measured by the Pierce BCA Protein Assay Kit (Thermo Fisher), and 192 μg protein was used for each condition (2× HHBSS control, 2× 40 μM Zn2+).
Kinase array kits were used according to manufacturer’s recommendation. Briefly, membranes were blocked for an hour at room temperature, and then protein lysate was added and incubated at 4 °C overnight. Membranes were washed three times before primary antibody mixtures were added and incubated for 2 h at room temperature. Membranes were washed three times before the addition of Streptavidin horseradish peroxidase (HRP) for 30 min at room temperature. Membranes were again washed three times, detection reagent was added, and membranes were imaged on an ImageQuant LAS 4000 imaging system (GE Healthcare Life Sciences). Dot intensity data are in SI Appendix, Table S2.
Immunoblots.
Cells in 6-well plates were incubated in HHBSS for 30 min at 37 °C and 0% CO2, and then media was replaced with HHBSS with indicated EGF and Zn2+ concentrations incubated 30 more minutes at 37 °C and 0% CO2. Total cell lysates were collected with RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0, and 5 mM EDTA). Two wells of each 6-well plate were combined for each condition to have high enough protein concentration of immunoblotting. Proteins were separated using 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels and transferred to polyvinylidene difluoride (PVDF). Blots were blocked with 5% milk and probed with primary antibodies in the resources table. Secondary antibody Goat anti-Rabbit IgG (HRP, Novus Biologicals) was reacted with Amersham ECL Prime Western Blotting Detection Reagent (Thomas Scientific) and imaged on an ImageQuant LAS 4000 imaging system (GE Healthcare Life Sciences). Antibody dilutions are reported in the resource table.
ERK Phosphatase Assay.
To determine the impact of Zn2+ on ERK phosphatase activity in whole cell extracts, an ERK phosphatase assay was adapted from Levinthal and DeFranco (72). Six dishes of cells were transferred to phosphate-free HHBSS; one dish was treated with 40 μM ZnCl2 and one was treated with 5 μM TPA. After 30 min at 37 °C and 0% CO2, 100 μg protein from whole cell lysate was diluted into phosphatase assay buffer (10 mM MgCl2, 10 mM Hepes pH 7.5, and 2 μM MEK inhibitor CI-1040); to samples of untreated cell lysate, 10 μM ZnCl2, 5 μM TPA, or 200 μM phosphatase inhibitor BCI-hydrochloride was added. For a positive control, 1,200 units λ-protein phosphatase (New England Biolabs) were diluted into phosphatase assay buffer without cell lysate. A total of 20 ng recombinant dual-phosphorylated His-ERK2 (generous gift from the laboratory of Natalie Ahn) was added to each sample and incubated at 37 °C for 40 min. The reaction was stopped by adding 8 M urea, pH 8.6, with 10 mM imidazole, and 20 μL Ni-nitrilotriacetic acid beads were added to each sample to precipitate the His-tagged ERK. Samples were incubated for 90 min at 4 °C. Samples were then washed twice in the urea/imidazole mixture and twice in 300 mM NaCl, 25 mM Tris, pH 7.5 buffer. Beads were then resuspended in 20 μL NaCl/Tris buffer + 25 μL 2× Laemmli sample buffer and boiled for 5 min. Samples were loaded onto a 4 to 20% Mini-PROTEAN gradient polyacrylamide gel (Bio-Rad), transferred to PVDF, and blotted for total and phosphorylated ERK as above (in Immunoblots).
MKP3 Inhibition.
To find IC50 of Zn2+ inhibition of MKP3/DUSP6, we performed an in vitro plate-based fluorescence assay using recombinant human MKP-3 (Enzo Life Sciences) supplied in 20 mM Tris/HCl, pH 8.0, 200 mM NaCl, 5 mM dithiothreitol (DTT), 0.1% Tween-20, and 10% glycerol. Due to the Zn2+-chelating capacity of DTT (73), the assay buffer we used contains Tris(2-carboxyethyl)phosphine (TCEP) as the reducing agent—50 mM Tris/HCl, pH 7.4, 100 mM NaCl, 100 μM TCEP, and 0.01% Tween-20. Methylumbelliferyl phosphate (MUP; Fisher Scientific), the fluorogenic phosphatase substrate (ex: 386, em: 448 nm), was diluted 1:10 into TCEP/Tween20 assay buffer, and 110 μL was plated into each well of a 96-well glass-bottom plate. A variety of Zn2+ concentrations was added to the plate. Briefly, for trials one and two, a dilution series of Zn2+ was used to establish Zn2+ concentrations ranging from 1 to 2,500 μM and 1 nM to 120 μM, respectively. For trial three, a variety of Zn2+ chelators and counter ions (SI Appendix, Table S3) were used to create a spectrum of buffered Zn2+ from 80 pM to 20 μM. Phosphatase inhibitor sodium orthovanadate, 1 mM, was used as a negative control. A total of 250 nM MKP3 (10 μL volume added) was added to each well, and the plate was immediately scanned for MUP fluorescence, with data points taken every 30 s for 40 min with 360 to 20 nm excitation and 450 to 30 nm emission. The fluorescence signal for each sample was fit via linear regression, and slope was used to approximate phosphatase activity with “no Zn2+” wells set at percent activity = 1.
Active Ras Pull-Down.
The Active Ras Pull-Down and Detection Kit (Fisher Scientific) was used to detect Ras activity in cells treated with EGF or Zn2+. The procedure (described in ref. 74) uses the Raf1 Ras-binding domain coupled to glutathione beads to pull down only GTP-bound Ras—exploiting the specificity of this protein–protein interaction (74). Briefly, cells were cultured to equal density in 10-cm dishes and then incubated in HHBSS for 30 min at 37 °C and 0% CO2. Media was then replaced with control (HHBSS alone) media or HHBSS with 40 μM Zn2+ or 20 nM EGF and further incubated at 37 °C and 0% CO2 for 30 min. Cells were lysed according to kit protocol, and protein was purified by incubation with GST-Raf1-RBD agarose resin and subsequent filtration and eluted in SDS sample buffer with β-mercaptoethanol. The sample was run on a 10% SDS-PAGE gel and transferred to PVDF membrane, and protein was detected using the provided anti-Ras antibody. Secondary antibody Goat anti-Rabbit IgG (Novus Biologicals) was reacted with Amersham ECL Prime Western Blotting Detection Reagent (Thomas Scientific) and imaged on an ImageQuant LAS 4000 imaging system (GE Healthcare Life Sciences).
Supplementary Material
Acknowledgments
We thank the following for financial support: NIH DP1 to A.E.P. (GM114863), NIH Molecular Biophysics Training Grant T32 to K.J.A. (GM065103), and NSF Graduate Research Fellowships Program to K.J.A. (DGE1650115). We would like to acknowledge the BioFrontiers Institute Advanced Light Microscopy Core, where data analysis and microscope support was provided by Drs. Joseph Dragavon and Jian Tay, supported by the BioFrontiers Institute and the Howard Hughes Medical Institute. We would also like to acknowledge the University of Colorado Biochemistry Cell Culture Core Facility, especially Theresa Nahreini, for providing resources and support for all our cell work. We would like to thank Dr. Johannes Rudolph for assistance with in vitro phosphatase inhibition assays and helpful discussions and Dr. Stephen Langers for helping us create adenoviral versions of our sensors. We would also like to acknowledge Dr. Natalie Ahn, Dr. Luke Lavis, and Dr. Xuedong Liu for their generous assistance with experimental materials.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015786118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Andreini C., Banci L., Bertini I., Rosato A., Counting the zinc-proteins encoded in the human genome. J. Proteome Res. 5, 196–201 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Vinkenborg J. L., et al., Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat. Methods 6, 737–740 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin Y., Dittmer P. J., Park J. G., Jansen K. B., Palmer A. E., Measuring steady-state and dynamic endoplasmic reticulum and Golgi Zn2+ with genetically encoded sensors. Proc. Natl. Acad. Sci. U.S.A. 108, 7351–7356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin Y., et al., Direct comparison of a genetically encoded sensor and small molecule indicator: Implications for quantification of cytosolic Zn(2+). ACS Chem. Biol. 8, 2366–2371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hessels A. M., et al., eZinCh-2: A versatile, genetically encoded FRET sensor for cytosolic and intraorganelle Zn(2+) imaging. ACS Chem. Biol. 10, 2126–2134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maret W., Zinc biochemistry: From a single zinc enzyme to a key element of life. Adv. Nutr. 4, 82–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamasaki S., et al., Zinc is a novel intracellular second messenger. J. Cell Biol. 177, 637–645 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim A. M., et al., Zinc sparks are triggered by fertilization and facilitate cell cycle resumption in mammalian eggs. ACS Chem. Biol. 6, 716–723 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vergnano A. M., et al., Zinc dynamics and action at excitatory synapses. Neuron 82, 1101–1114 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Que E. L., et al., Quantitative mapping of zinc fluxes in the mammalian egg reveals the origin of fertilization-induced zinc sparks. Nat. Chem. 7, 130–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanford L., Palmer A. E., Dissociated hippocampal neurons exhibit distinct Zn2+ dynamics in a stimulation-method-dependent manner. ACS Chem. Neurosci. 11, 508–514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCall K. A., Huang C., Fierke C. A., Function and mechanism of zinc metalloenzymes. J. Nutr. 130(suppl. 5), 1437S–1446S (2000). [DOI] [PubMed] [Google Scholar]
- 13.Kim A. M., Vogt S., O’Halloran T. V., Woodruff T. K., Zinc availability regulates exit from meiosis in maturing mammalian oocytes. Nat. Chem. Biol. 6, 674–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesters J. K., Petrie L., A possible role for cyclins in the zinc requirements during G1 and G2 phases of the cell cycle. J. Nutr. Biochem. 10, 279–290 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Yan M., Song Y., Wong C. P., Hardin K., Ho E., Zinc deficiency alters DNA damage response genes in normal human prostate epithelial cells. J. Nutr. 138, 667–673 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo M. N., Damon L. J., Wei Tay J., Jia S., Palmer A. E., Single cell analysis reveals multiple requirements for zinc in the mammalian cell cycle. eLife 9, e51107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodier J., Rainbow R. D., Stewart A. J., Pitt S. J., Intracellular zinc modulates cardiac ryanodine receptor-mediated calcium release. J. Biol. Chem. 290, 17599–17610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besser L., et al., Synaptically released zinc triggers metabotropic signaling via a zinc-sensing receptor in the hippocampus. J. Neurosci. 29, 2890–2901 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard S. R., et al., Identification and characterization of zinc binding sites in protein kinase C. Science 254, 1776–1779 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Sindreu C., Palmiter R. D., Storm D. R., Zinc transporter ZnT-3 regulates presynaptic Erk1/2 signaling and hippocampus-dependent memory. Proc. Natl. Acad. Sci. U.S.A. 108, 3366–3370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang Y., et al., NO mobilizes intracellular Zn2+ via cGMP/PKG signaling pathway and prevents mitochondrial oxidant damage in cardiomyocytes. Cardiovasc. Res. 75, 426–433 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis T. S., Shapiro P. S., Ahn N. G., “Signal transduction through MAP kinase cascades” in Advances in Cancer Research, Vande Woude G. F., Klein G., Eds. (Elsevier, 1998), 74, pp. 49–139. [DOI] [PubMed] [Google Scholar]
- 23.Hansson A., Extracellular zinc ions induces mitogen-activated protein kinase activity and protein tyrosine phosphorylation in bombesin-sensitive Swiss 3T3 fibroblasts. Arch. Biochem. Biophys. 328, 233–238 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Samet J. M., et al., Activation of MAPKs in human bronchial epithelial cells exposed to metals. Am. J. Physiol. 275, L551–L558 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Ho Y., et al., Selective inhibition of mitogen-activated protein kinase phosphatases by zinc accounts for extracellular signal-regulated kinase 1/2-dependent oxidative neuronal cell death. Mol. Pharmacol. 74, 1141–1151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He K., Aizenman E., ERK signaling leads to mitochondrial dysfunction in extracellular zinc-induced neurotoxicity. J. Neurochem. 114, 452–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haase H., Maret W., Intracellular zinc fluctuations modulate protein tyrosine phosphatase activity in insulin/insulin-like growth factor-1 signaling. Exp. Cell Res. 291, 289–298 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Wilson M., Hogstrand C., Maret W., Picomolar concentrations of free zinc(II) ions regulate receptor protein-tyrosine phosphatase β activity. J. Biol. Chem. 287, 9322–9326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein C., et al., Zinc induces ERK-dependent cell death through a specific Ras isoform. Apoptosis 11, 1933–1944 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Wu W., Graves L. M., Gill G. N., Parsons S. J., Samet J. M., Src-dependent phosphorylation of the epidermal growth factor receptor on tyrosine 845 is required for zinc-induced Ras activation. J. Biol. Chem. 277, 24252–24257 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Bruinsma J. J., Jirakulaporn T., Muslin A. J., Kornfeld K., Zinc ions and cation diffusion facilitator proteins regulate Ras-mediated signaling. Dev. Cell 2, 567–578 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Morita K., Han M., Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 25, 5794–5804 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiedler B. L., et al., Droplet microfluidic flow cytometer for sorting on transient cellular responses of genetically-encoded sensors. Anal. Chem. 89, 711–719 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanford L., Carpenter M. C., Palmer A. E., Intracellular Zn2+ transients modulate global gene expression in dissociated rat hippocampal neurons. Sci. Rep. 9, 9411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J. G., Qin Y., Galati D. F., Palmer A. E., New sensors for quantitative measurement of mitochondrial Zn(2+). ACS Chem. Biol. 7, 1636–1640 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess S. Y., National risk of zinc deficiency as estimated by national surveys. Food Nutr. Bull. 38, 3–17 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Glassman A. B., Rydzewski R. S., Bennett C. E., Trace metal levels in commercially prepared tissue culture media. Tissue Cell 12, 613–617 (1980). [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Hawkins B. E., DeWitt D. S., Prough D. S., Maret W., The relationship between transient zinc ion fluctuations and redox signaling in the pathways of secondary cellular injury: Relevance to traumatic brain injury. Brain Res. 1330, 131–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCranor B. J., et al., Quantitative imaging of mitochondrial and cytosolic free zinc levels in an in vitro model of ischemia/reperfusion. J. Bioenerg. Biomembr. 44, 253–263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regot S., Hughey J. J., Bajar B. T., Carrasco S., Covert M. W., High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157, 1724–1734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer A. E., Tsien R. Y., Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 1, 1057–1065 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Gross S. M., Rotwein P., Akt signaling dynamics in individual cells. J. Cell Sci. 128, 2509–2519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groom L. A., Sneddon A. A., Alessi D. R., Dowd S., Keyse S. M., Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 15, 3621–3632 (1996). [PMC free article] [PubMed] [Google Scholar]
- 44.Komatsu N., et al., Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell 22, 4647–4656 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yasuda R., et al., Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat. Neurosci. 9, 283–291 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Kochańczyk T., Drozd A., Krężel A., Relationship between the architecture of zinc coordination and zinc binding affinity in proteins–Insights into zinc regulation. Metallomics 7, 244–257 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Anand A., et al., The extracellular Zn2+ concentration surrounding excited neurons is high enough to bind Amyloid-β revealed by a nanowire transistor. Small 14, e1704439 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Pan E., et al., Vesicular zinc promotes presynaptic and inhibits postsynaptic long-term potentiation of mossy fiber-CA3 synapse. Neuron 71, 1116–1126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelleher S. L., McCormick N. H., Velasquez V., Lopez V., Zinc in specialized secretory tissues: Roles in the pancreas, prostate, and mammary gland. Adv. Nutr. 2, 101–111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Que E. L., et al., Zinc sparks induce physiochemical changes in the egg zona pellucida that prevent polyspermy. Integr. Biol. 9, 135–144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oteiza P. I., Clegg M. S., Zago M. P., Keen C. L., Zinc deficiency induces oxidative stress and AP-1 activation in 3T3 cells. Free Radic. Biol. Med. 28, 1091–1099 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Lemire J., Mailloux R., Appanna V. D., Zinc toxicity alters mitochondrial metabolism and leads to decreased ATP production in hepatocytes. J. Appl. Toxicol. 28, 175–182 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Song Y., Leonard S. W., Traber M. G., Ho E., Zinc deficiency affects DNA damage, oxidative stress, antioxidant defenses, and DNA repair in rats. J. Nutr. 139, 1626–1631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reeder N. L., et al., Zinc pyrithione inhibits yeast growth through copper influx and inactivation of iron-sulfur proteins. Antimicrob. Agents Chemother. 55, 5753–5760 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oyama T. M., Saito M., Yonezawa T., Okano Y., Oyama Y., Nanomolar concentrations of zinc pyrithione increase cell susceptibility to oxidative stress induced by hydrogen peroxide in rat thymocytes. Chemosphere 87, 1316–1322 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Imokawa G., Okamoto K., The effect of zinc pyrithione on human skin cells in vitro. J. Soc. Cosmet. Chem. 34, 1–11 (1982). [Google Scholar]
- 57.Truong-Tran A. Q., Ho L. H., Chai F., Zalewski P. D., Cellular zinc fluxes and the regulation of apoptosis/gene-directed cell death. J. Nutr. 130 (suppl. 5), 1459S–1466S (2000). [DOI] [PubMed] [Google Scholar]
- 58.Forcina G. C., Conlon M., Wells A., Cao J. Y., Dixon S. J., Systematic quantification of population cell death kinetics in mammalian cells. Cell Syst. 4, 600–610.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krapivinsky G., Krapivinsky L., Manasian Y., Clapham D. E., The TRPM7 chanzyme is cleaved to release a chromatin-modifying kinase. Cell 157, 1061–1072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pang W., et al., Depletion of intracellular zinc induced apoptosis in cultured hippocampal neurons through Raf/MEK/ERK pathways. Nutr. Neurosci. 15, 18–24 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Pang W., et al., Depletion of intracellular zinc induces apoptosis of cultured hippocampal neurons through suppression of ERK signaling pathway and activation of caspase-3. Neurosci. Lett. 552, 140–145 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Ohashi K., et al., Zinc promotes proliferation and activation of myogenic cells via the PI3K/Akt and ERK signaling cascade. Exp. Cell Res. 333, 228–237 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Hönscheid A., Dubben S., Rink L., Haase H., Zinc differentially regulates mitogen-activated protein kinases in human T cells. J. Nutr. Biochem. 23, 18–26 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Nimmanon T., Ziliotto S., Morris S., Flanagan L., Taylor K. M., Phosphorylation of zinc channel ZIP7 drives MAPK, PI3K and mTOR growth and proliferation signalling. Metallomics 9, 471–481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pernelle J.-J., Creuzet C., Loeb J., Gacon G., Phosphorylation of the lymphoid cell kinase p56lck is stimulated by micromolar concentrations of Zn2+. FEBS Lett. 281, 278–282 (1991). [DOI] [PubMed] [Google Scholar]
- 66.Manzerra P., et al., Zinc induces a Src family kinase-mediated up-regulation of NMDA receptor activity and excitotoxicity. Proc. Natl. Acad. Sci. U.S.A. 98, 11055–11061 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y. Z., Pan E., Xiong Z.-Q., McNamara J. O., Zinc-mediated transactivation of TrkB potentiates the hippocampal mossy fiber-CA3 pyramid synapse. Neuron 57, 546–558 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Hershfinkel M., Moran A., Grossman N., Sekler I., A zinc-sensing receptor triggers the release of intracellular Ca2+ and regulates ion transport. Proc. Natl. Acad. Sci. U.S.A. 98, 11749–11754 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turski M. L., et al., A novel role for copper in Ras/mitogen-activated protein kinase signaling. Mol. Cell. Biol. 32, 1284–1295 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brady D. C., et al., Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 509, 492–496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grimm J. B., et al., A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods 14, 987–994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Levinthal D. J., Defranco D. B., Reversible oxidation of ERK-directed protein phosphatases drives oxidative toxicity in neurons. J. Biol. Chem. 280, 5875–5883 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Kr zel A., et al., Coordination of heavy metals by dithiothreitol, a commonly used thiol group protectant. J. Inorg. Biochem. 84, 77–88 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Taylor S. J., Resnick R. J., Shalloway D., “Nonradioactive determination of Ras-GTP levels using activated ras interaction assay” in Methods in Enzymology, Balch W. E., Der C. J., Hall A., Eds. (Elsevier, 2001), 333, pp. 333–342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.