Abstract
After the outbreak of SARS and MERS, the world is now in the grip of another viral disease named COVID-19 caused by a beta Coronavirus – SARS COV-2 which appears to be the only one with a pandemic potential. The case of COVID-19 was reported in the Hubei province of Wuhan city in Central China at the end of December 2019 and it is suspected that the sea food market played a role in this outbreak which was closed abruptly. Subsequently, a Public Health Emergency of International Concern was declared on 30 January 2020 by the World Health Organization. Both SARS and MERS corona viruses had its reservoir in bats and were transferred to humans from palm civets and camels respectively. This virus can be transmitted through airborne droplets. Natural reservoir and intermediate host of COVID-19 is yet to be identified. This paper reviews the occurrences of viral diseases in the recent times including SARS and MERS. As an addition to this, the paper will contain a detailed examination of the COVID-19 Pandemic.
Keywords: SARS-COV, MERS-COV, COVID-19, Coronavirus
Introduction
A virus is a small parasite that cannot undergo self-reproduction and once it infects a susceptible cell, the virus can direct the cell machinery to produce more viruses.1 The Coronaviridae family comprising the genera Corona Virus and Torovirus has been classified as members of the Order Nidovirales.2,3 They mutate and also recombine frequently. Laboratory diagnosis is best accomplished by finding viral RNA through polymerase chain reaction. These are enveloped viruses with club-like spikes projecting from their surface.4 Coronaviruses (COVs) commonly bring about mild but occasionally more severe community-acquired acute respiratory infections in humans. They also affect multifarious animals and several have even crossed the barrier, producing outbreaks of severe human respiratory disease. As of 10th March, 2019, 2374 laboratory cases were confirmed. The earliest isolation of this virus was from the Wuhan sea food market in China in the first week of January 2020.5
Community acquired Coronavirus infections cause about 15% of common cold. They are typically epidemic in the winter. There are no accepted effective antiviral drugs. Prevention is done through epidemiologic methods and the use of appropriate respiratory precautions in hospital settings. The SARS epidemic and MERS outbreaks were controlled through careful case identification, quarantine and by the use of barrier precautions. WHO advised staying away from public places and avoiding close contact with infected persons and animals in order to prevent the rapid spread of the disease.6
Dromedary camels in Saudi Arabia encamp for 3 different HCOV species including the MERS HCOv.7 The spread of MERS-COV from dromedary camels, the high case-casualty rates, emphasize the need for preventive and therapeutic measures.8
The structure of this virus holds a helical nucleocapsid with 30 kb, plus-sense RNA genome coiled within an envelope that contains the S, M and E glycoproteins. Some coronaviruses have an additional membrane glycoprotein HE in them.
The name Corona viruses (COVs) was derived from the Greek word meaning Crown referring to the crown or corona like appearance of COV viruses when observed under an electron microscope.10 The Coronaviridae and Arteriviridae are viruses with monopartite plus-stranded RNA genomes that replicate using a nested (nido) set of overlapping sub genomic mRNAs.11
The spikes of coronaviruses, typically described as club-like or petal-shaped, emerge from the virion surface as stalks with bulb-like distal termini. Some of the variation in particle size and shape was likely attributable to stresses exerted by virion purification or distortions introduced by negative staining of samples for electron microscopy. Through sequencing and antigenicity, the pioneer divisions of the animal and human coronaviruses discovered were as follows: group 1, containing HCOV-229E; group 2, enclosing HCOV-OC43; group 3, including the infectious bronchitis virus. The family Coronaviridae, is divided into 4 genera alpha-, beta-, gamma- and deltacoronavirus (α-, β-, γ- and δ-COV).12
During the winter of 2002 to 2003, there was an outbreak of a new disease, severe acute respiratory syndrome (SARS), where the intermediate hosts, were probably the Palm civet (Paguma larvata) and the Raccoon dog (Nyctereutes procyonoides).13,14
The last known case of the SARS epidemic occurred in mid-2004. More recently there was an outbreak of a related but different COV, the Middle East respiratory syndrome coronavirus (MERS-COV).15 A total of 2374 individuals were infected – a majority with acute respiratory symptoms – that were arduous in most and fatal in 823 (as of 10th March, 2019).16,17 The dromedary camels are the reservoir of MERS COV, although evidence suggests that bats may be infected with related viruses.18
Betacoronavirus genus consists of 4 lineages: A, B, C and D.19,20 Alpha and Beta coronavirus descend from the bat gene pool.21-23 The group 2 coronavirus has a smaller protein called hemagglutinin esterase (HE) which is functionally similar to S protein.24 In humans, COV infections include mild diseases, such as the common cold, to more severe manifestations, like bronchitis and pneumonia with renal involvements.25 The first human coronavirus (HCOV) was isolated from the nasal discharge of patients with common cold.26 Currently, 6 different COV strains are known to infect humans.27
229E and OC43 are the 2 main HCOVs (from lineage Alpha and Beta) that cause 15% to 29% of all common colds.28 SARS-COV caused the outbreak of severe respiratory disease in China during 2002 to 200329 and MERS-COV is responsible for an ongoing outbreak of severe respiratory disease in the Middle East since 2012.30 The epidemiology and pathogenesis of HCOVs are discussed in the following sections and are summarized in Table 1.
Table 1.
CoV-2019 cases reported (as per the information collected on 18-05-2020).
| Country | Total cases | Total deaths |
|---|---|---|
| USA | 7 637 066 | 214 615 |
| India | 6 626 291 | 102 746 |
| Brazil | 4 915 289 | 146 375 |
| Russia | 1 225 889 | 21 475 |
| Colombia | 855 052 | 26 712 |
| Peru | 828 169 | 32 742 |
| Spain | 810 807 | 32 086 |
| Argentina | 798 486 | 21 018 |
| Mexico | 761 665 | 79 088 |
| South Africa | 681 289 | 16 976 |
| France | 619 190 | 32 230 |
| UK | 502 978 | 42 350 |
| Iran | 471 772 | 26 957 |
| Chile | 470 179 | 12 979 |
| Iraq | 379 141 | 9399 |
| Bangladesh | 370 132 | 5375 |
| Saudi Arabia | 336 389 | 4875 |
| Italy | 325 329 | 35 986 |
| Philippines | 324 762 | 5840 |
| Turkey | 324 443 | 8441 |
| Pakistan | 315 260 | 6517 |
| Indonesia | 307 120 | 11 253 |
| Germany | 301 571 | 9602 |
| Israel | 268 175 | 1719 |
| Ukraine | 230 236 | 4430 |
| Canada | 166 156 | 9481 |
| Ecuador | 141 034 | 11 647 |
| Romania | 137 491 | 5048 |
| Bolivia | 136 868 | 8101 |
| Netherlands | 135 892 | 6454 |
| Belgium | 130 235 | 10 064 |
| Poland | 102 080 | 2659 |
| Oman | 101 814 | 985 |
| UAE | 99 733 | 429 |
| China | 85 470 | 4634 |
| Australia | 27 149 | 894 |
| S. Korea | 24 164 | 422 |
| Croatia | 17 797 | 300 |
| Srilanka | 3471 | 13 |
In general, 229E, OC43 and NL63 are distributed globally, transmitted predominantly during the winter season in temperate countries,31 while NL63 showed a spring–summer peak of activity.32 Today n-COVs are recognized as one of the most rapidly evolving viruses moving to its higher genome nucleotide substitution rates and recombination.33 The distribution of n-COVs varies according to its geographical area and season.34 Although the corona virus was identified 60 years ago, they received attention at the time of the SARS outbreak.
Severe Acute Respiratory Syndrome (SARS)
SARS is a viral disease associated with an outbreak of a typical pneumonia in Guangdong Province, China. A newly identified group of coronavirus, the SARS Coronavirus, was the causative agent of this life threatening pneumonia in humans.35-39 Typical clinical presentations of SARS are viral pneumonia with rapid respiratory deterioration, fever, chills, myalgia, malaria and non-productive cough being the major presentations showing symptoms, whereas sore throats are less the frequently seen one, with an incubation period of 2 to 7 days.40,41 SARS like viruses were isolated from a few Himalayan palm civets (Paguma larvata) and the Raccoon dog (Nyctereutes procyonoides) at Shenzhen sea food market.42,43
It has been discovered that humans were introduced to this epidemic with the spread of a closely related bat virus among palm civets or other animals that were for sale in the live wild game markets and then to humans. The virus then adopted itself through wild action and recombined until it transmitted readily among humans. RT-PCR method was adopted for the diagnosis of this viral disease.44 Ribavirin Drug: Interferon and Lopinavir/ritonavir are the antiviral agents used in the therapy of SARS. Ribavirin is a ribonucleoside analogue that prevents the replication of a large number of RNA and DNA Viruses.45,46
Even though improvements were seen in some patients, some other reports failed to identify this; there were numerous adverse effects associated with ribavirin or other therapies, particularly the transaminase elevation.47 IFN are a multigene family of inducible cytokines, possessing antiviral activity with Interferon β being far more effective against SARS.48 Lopinavir and Ribavirin are combined and used for the treatment of human immunodeficiency virus (HIV) and have limited side effects.49
Middle East Respiratory Syndrome (MERS)
The first case was identified in a patient with acute pneumonia and renal failure in Jeddah, Kingdom of Saudi Arabia (KSA) in June 2012.50-53 Sequencing was done at the Erasmus Medical Centre (EMC) in Rotterland, the Netherlands, where the virus was named ‘Human Coronavirus EMC (HCOVEMC)’ and further, renamed by the International Committee on Taxonomy of Virus as the Middle East Respiratory Syndrome Coronavirus (MERS COV).54-56 The virus was propagated through African green monkeys and rhesus macaque kidney cells.57,58 The incubation period is 2 to 14 days [medium of approximately 5.5 to 6.5 days].59,60 Clinical symptoms include fever, cough, sore throat, shortness of breath, myalgia, chest pain, malaise and gastro-intestinal symptoms, such as diarrhea, vomiting and abdominal pain. A large proportion of severely ill patients required mechanical ventilation. Interestingly, many of the reported secondary cases showed mild respiratory symptoms or were even asymptomatic.61-63
Chest radiographs of a large percentage of patients admitted to the hospital showed airspace and interstitial opacities, with subtle to extensive, unilateral to bilateral and focal to diffuse distribution; Moreover, the air space opacities vary in their distribution, as reticular or reticulonodular, and demonstrate thickening of broncho-vascular areas.64-67 Gastrointestinal symptoms such as diarrhea and vomiting are sometimes seen in patients.68 The virus acquired by the dromedary camels subsequently spread to other animals in the Middle East region.69,70 No effective antiviral drug against MERS has been discovered, which is another factor contributing to the huge death rate.71
Various IFN regimens in combination with nitro viruses have been intermittently administered to severely ill patients, although typically in an ad hoc manner and in the absence of systematic evolution.72
COVID-19
On 31 December 2019, the WHO China Country Office was informed about a pneumonia of unknown cause in Wuhan City, Hubei Province-China. As of 3 January 2020, a total of 44 patients with pneumonia had been reported to the WHO by the national authorities in China. The cluster was initially reported when the Chinese authorities identified a new coronavirus (COV) as the causative agent of SARS-CoV2 which was named as COVID-19 by the World Health Organization, which is an acronym of ‘coronavirus disease 2019”.73
SARS-COV2, designated by The International Committee on Taxonomy of Viruses (ICTV) on 11 February 2020 is the most recent human pneumonia virus with high outbreak potential. Cases were reported in other cities, leading to a globular outbreak.74-76 The first SARS related Corona virus was discovered in the horseshoe bat species (Rhinolophus sinicus).77 Moreover, they belong to a large family of single-stranded RNA viruses (+ssRNA) that are isolated from different animal species.78 There are speculations, about their animal origin.79 Available evidence suggests that the outbreak was associated with exposures to the seafood market and hence, concerned markets in Wuhan were closed on first January 2020 for environmental sanitation and disinfection.80,81 In March 2019, this outbreak was characterized as a pandemic by the WHO.82-84 This is the first pandemic known to be caused by a new coronavirus; the third zoonotic human coronavirus (COV) of the century has features typical of the coronavirus family and was placed in the beta coronavirus 2b lineage. COVID-19 was declared as the sixth Public Health Emergency of International Concern by the WHO, following HIN1 (2009), polio (2014), Ebola in West Africa (2014) Zika (2016) and Ebola in the Democratic Republic of Congo (2019). The incubation period is the same to other known human coronaviruses, including SARS and MERS.85 2019 n-COV is the seventh member of the family of corona viruses that are infectious to humans.
The Common symptoms are fever, cough, myalgia sputum production, headache, haemoptysis and diarrhea.86 In the early stages, respiratory infections in patients developed to Acute Respiratory Distress Syndrome (ARDS), acute respiratory failure and other serious complications.87 This disease might badly affect older patients with comorbidities and ARDS.88,89 Based on some studies conducted in Hong Kong, in and around Wuhan as well as Shenzhen has revealed that group O, people had more resistivity towards SARSCOV-2, because of the ABO antibodies. In fact, those with blood group A had much more risk of being affected by this virus.90 The vital signs were stable in most cases while leukopenia and lymphopenia were common.91 Mortality has been found to be remarkably higher in patients with increased TnT (Troponium T) level. The further elevation of IP-10, MCP-3 and IL-1ra, shows the severity and the fatal outcome of this viral disease.92,93 Researches to identify the source of 2019 n-COV including possible intermediate animal vectors are going on.94,95 On January 10, the genome sequence of 2019 COV was released on Gen bank of Yong Zhen Zhang’s group at Fudan University.96
Phylogenetic analysis of viral genome uncovered the relatedness of the virus to SARS like coronavirus, genus beta coronavirus, sub genus Sarbeco virus that had previously been found in bats.97,98 Moreover, phylogenetic analysis of corona virus of different Species indicated that 2019 n-COV might have originated from bats.99,100 The viral sequence shows 79.6% identity to SARS COV and 96% identical at the whole genome level to a bat coronavirus.101,102 Moreover, they are closely related to a bat derived corona virus – bat SL COV ZC44, bat SL COV ZX C2.103
The virus which causes COVID-19 most probably has its ecological reservoir in bats. Since there is a very limited close contact between humans and bats, transmission of virus to humans might have occurred through an intermediate animal host like a domestic animal, a wild animal or a domesticated wild animal which has not yet been identified104. Asymptomatic patients can still infect the other which means that COVID-19 is transmittable even during the incubation period.105 The nosocomial transmission of COVID-19, facilitated by the mobile phones of health care workers and hospital equipment, cause severe problems and further accelerate the rate of transmission of this already hiking viral disease.106 The vertical transmission of COVID-19; put pregnant women and fetuses at a higher risk of being infected by the virus. This is primarily due to the high expression of ACE2 receptors in the human maternal interphase. Transmission through organ transplantation and surgical operation; as the Corona viruses home in the respiratory tract and its secretions. These pose a risk to health care providers if the patients require surgery.107
SARS COV-2, can remain for hours on different surfaces and was more stable and last longer on plastic as well as stainless steel which were comparatively low in copper and cardboard surfaces.108-111 The basic reproductive number (R0), has been used as a parameter to calculate the infectivity index. In MERS and SARS epidemic, the value was found be approximately 2, which in turn indicate that each infected person could infect 2 others in an effective contact. But for COVID-19 this index value was slightly higher. The data calculated from Wuhan was 2.2, further studies on this have had received different values (1.95-6.47).112
Ensuring early diagnosis and appropriate quarantine measures are the key factors to check the rate of transmission of the virus considerably. The current method for the diagnosis of COVID-19 includes detection of the virus by genomic techniques like polymerase chain reaction (PCR) – or deep sequencing which relies on the presence of the viral genomes in sufficient amounts.113,114 This method often fails to detect the viral infection, if collection procedure is not optimal or if the patient has low viral load, typically, during the early stage of the disease or if it is suppressed by host immunity. If the sample is obtained at a later stage, a supplementary 18 m test can provide a better sensitivity mean of PCR based method alone.
However, new sensitive PCR essays can even further improve the detection method when combined with IgM ELISA arrays. The 2 diagnostic tests for emergency use are the Genesig real time PCR coronavirus, an open system more suitable for laboratories with moderate sample testing capacity115 and the cobas(R) SARS cov-2, core for use in the Cobas (R) 6800/8500 system is a closed system array for larger laboratories.116
Common laboratory findings include lymphopenia and bilateral ground glass opacity. At present there are no vaccines or antiviral treatments for animal coronavirus.117 It is observed that in more than 20% of patients, the viral RNA remained positive in feces even though it turned negative in the respiratory tract. This shows that the viral gastro intestinal infection and potential fecal, oral transmission can last even after the viral clearance in the respiratory tract of the infected person. Hence the rRT-PCR testing from feces is also an important process to be carried out in the COVID-19 treatment plan.118,119
However various research efforts are under way and number of vaccine candidates are under work which includes inactivated vaccine, adenoviral vector vaccine, recombinant subunits vaccine etc. The S protein, remains as the main factor for the vaccine development. The Cryo-EM structure of SARS COV-2 S Trimer has also accelerated the vaccine development process.120
Patients admitted to ICUs had a high concentration of cytokines in plasma which shows that the cytokine storm is associated with the severity of this disease. In a group of patients by the end of the first week, there is a chance for the progression of the disease to pneumonia, respiratory failure and death. This step up is due to the increased inflammatory cytokines (IL2, IL7, IL10, GCSF, IP10, MCP1, MIP1A and TNFa). The GM-CSF, IL-6 CD4T cells isolated from such patients were typically high compared to the non-ICU patients. This holds that the inhibition of excessive inflammatory response might act as an adjacent therapy for COVID-19.121-123 In most cases, it takes repeated attempts to accurately spot the presence of viral RNA from the deep oral swab samples, hence analysis of deep airway samples [like sputum, bronchoalveolar lavage, tracheal secretions] or stool, would be more effective in determining the viral presence than the prior one.124,125 No therapeutics and pharmaceutical products have yet been shown to be safe and effective.126 Lopinavir and Ritonavir are licensed treatment for n-COV, evidence for COVID-19, MERS and SARS has yet to show whether it can improve clinical outcomes or prevent infection. Interferon beta Ia is used to treat multiple sclerosis. Chloroquine and hydroxychloroquine are very closely related and used to treat malaria and rheumatology conditions respectively.127 Nucleoside analysis such as ribavirin may be potentially beneficial for the treatment of COVID-19. Ribavirin is approved for treating respiratory syncytial virus (RSV) infection and used extensively during SARS and MERS outbreak.128 Ribavirin has been recognized as a promising antiviral drug against RNA virus infections in cultured cells in mice and nonhuman primates (NHP) models.129
The new antiviral drug ribavirin as well as chloroquine at cc50 of 1 mm is found to be effective in preventing replication of this Virus.130 Chloroquine and hydroxychloroquine are 2 of the most fascinating drugs developed in the last 60 years.131 Studies have suggested that chloroquine, an immunomodulant drug traditionally used to treat malaria, is effective in reducing viral replications and other infections. COVID-19 pandemic has spread very quickly taking only 30 days to expand from Hubei province to the rest of the mainland; however rapid rate of increase in illness among people and evidence of human to human transmission suggest that 2019 n-COV is more infectious, spreading with a staggering speed.132-134 Immunomodulators like Baricitinib, eculizumab, interferons; multistem cell therapy, autologous adipose derived mesenchymal stem cell therapy, corticosteroids, teicoplanin, ivermectin, convalescent plasma therapy, were administered on patients with COVID-19.135,136
Teicoplanin is a glycopeptide antibiotic used to treat bacterial infections caused by staphylococcus aureus and streptococcus, which has been adopted to treat COVID-19. Moreover, it is identified that teicoplanin could inhibit the entry of HIV-1-2019-NCoV-S pseudoviruses with the IC50 value of 1.66 µm. They even prevent the S protein activation by directly inhibiting the enzymatic activity of cathepsin L. They are often recommended in the early stage of this viral disease.137-139 Ivermectin; belonging to the family avermectin, with a molecular weight of 870 kDa, possess strong antiviral property and has the potential to convert RT-PCR to negative quickly. It can be combined with other molecules and are safe as well as affordable.140-144 Convalescent plasma treatment; has been proposed as a principle treatment method where the CP, is obtained from a donor who has already recovered by developing a humoral immunity against SARS COV-2.145 The aptamer, selected by the systemic evolution of ligands by exponential enrichment technology, are the DNA or RNA molecules that are capable of binding to a wide range of molecules with high affinity and specificity.146 It seems that the aptamer molecules might be effective in treating the corona virus to some extent. They could be an effective anti-corona virus agent in both diagnosis and treatment. Aptamer-based biosensors and drugs can be used to diagnose and treat coronavirus.147 The SARS COV-2 isolated from the urine and feces of infected person would then enter into the waste water treatment system. Hence analyzing virus especially in waste water would thereby help in tracking out the source of this viral disease through sewage pipe networks and determine the potential carriers in certain areas. Paper based device has emerged as a powerful tool that helps in the rapid diagnosis of pathogen as well as in the determination of infection transmission. This device is used to detect the SARS COV-2 in waste water, which would provide accurate information that can lead to timely treatment and prevent the spread of COVID-19.148,149
Natural products and their derivatives have potential to treat the viral infections. Treatment carried out using Chinese, Indian as well as Iranian herbal medicine solely and in combination with western medicines had a positive impact, though their clinical trials remained to be known.150 Herbal traditional medicines have been used in china since the very beginning of SARSCOV-2, (Lianhuanqingwen). Some of this have even prevented the infections of a healthy person and thereby has contributed towards the improvement in health status of patients with mild or severe symptoms.151 Some of the herbal agents that might be helpful in treatment of COVID-19 include, Echinceae purpurea, Curcumin, Cinchona SP, Xanthorrhizol etc.152
Preventive measures have been discussed to reduce the transmission and to tackle this viral disease.153-156 Social distancing is imperative to run down the number of cases. Furthermore, individuals’ abidance to prescriptions and reduction of social activity should be applied on all social groups for an effective result. Moreover, wearing of mask and eye protection could tackle the disease to some extent.157-159
Figure 1.9.
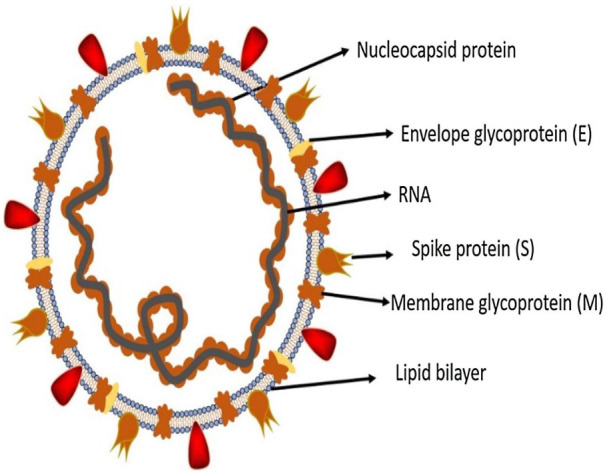
Structure of respiratory syndrome causing human coronavirus.
Conclusion
The spread of the novel coronavirus is declared as a pandemic by the World Health Organization (WHO). The spread of this disease is possible even during the incubation period from an asymptomatic person and this entirely poses a serious threat which can result in the death of thousands. The investigation to find the natural reservoir, intermediate host and vaccines are in progress. Further, there are many unanswered questions with regard to COVID-19 – like the root cause of outbreak, medium of transmission, effective treatment, prevention method, duration of transmission etc. MERS and SARS coronaviruses have shown that these viruses are able to cross the species barrier. In view of the new virological studies, coronaviruses are pathogens infecting a wide range of mammals that are often in contact with humans, thus providing the basis for future zoonotic outbreaks.
Footnotes
Funding:The author(s) received no financial support for the research, authorship and/or publication of this article.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
References
- 1. Lodish H, Berk A, Lawrence Zipursky S, et al. Molecular Cell Biology. 4th ed. W. H. Freeman and Co; 2000. [Google Scholar]
- 2. Peiris JSM. Corona Viruses. In: Douglas D, Richman Whitley RJ, Hayden FG. eds. Clinical Virology. 4th ed. ASM Press; 2012. [Google Scholar]
- 3. Gonzalez JM, Gomez-Puertas P, Cavanagh D, Gorbalenya AE, Enjuanes L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch Virol. 2003;148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hunter RH, Sykes K, Lowman SG, Duncan R, Satariano WA, Belza B. Environmental and policy change to support healthy aging. J Aging So Policy. 2011;23(4):354–371. [DOI] [PubMed] [Google Scholar]
- 6. Kumar D, Malviya R, Kumar Sharma P. Corona virus: a review of COVID-19. EJMO 2020;4:8–25. [Google Scholar]
- 7. Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol 2016;24(6):490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baharoon S, Ziad A., Memish. MERS-CoV as an emerging respiratory illness: a review of prevention methods. Travel Med Infect Dis. 2019;32:101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muhammad Adnan Shereen Suliman Khan Abeer KazmiNadia BashirRabeea Siddique. A Covid-19 infection: origin, transmission and characteristics of human coronaviruses. J Adv Res. 2020;24:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woo PCY, Huang Y, Lau SKP, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kathryn v holmes. Coronaviruses (Coronaviridae). In: Granoff A, Webster RG. eds. Encyclopedia of Virology. Elsevier; 1999:291–298. [Google Scholar]
- 12. Hulswit RJG, De Haan CAM, Boseh BJ. Coronaviruses. In: Ziebuhr J. ed. Advances in Virus Research. vol. 96. Academic Press; 2016:2–295. [Google Scholar]
- 13. Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003;302:276-278. [DOI] [PubMed] [Google Scholar]
- 14. Ge XY, Li J-L, Yang X-L, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814-1820. [DOI] [PubMed] [Google Scholar]
- 16. Assiri A, McGeer A, Perl TM, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oboho IK, Tomczyk SM, Al-Asmari AM, et al. 2014 MERS-CoV outbreak in Jeddah–a link to health care facilities. N Engl J Med. 2015;372: 846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sabir JS, Lam TT-Y, Ahmed MMM, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. [DOI] [PubMed] [Google Scholar]
- 19. “Phylogeny of SARS-like betacoronaviruses”. nextstrain. Retrieved 18 January 2020. [Google Scholar]
- 20. Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. Family cluster of middle east respiratory syndrome coronavirus infections. N E J Med 2013;368:2487–2494. [DOI] [PubMed] [Google Scholar]
- 21. Woo PC, Wang M, Lau SK, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lau SK, Woo PC, Yip CC, et al. Isolation and characterization of a novel Betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol 2012;86:5481-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lau SK, Poon RW, Wong BH, et al. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J Virol. 2010;84:11385-11394. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Langereis MA, Van Vliet AL, Boot W, De Groot RJ. Attachment of mouse heatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. J Virol. 2010:84:8970-8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wevers BA, van der Hoek L. Recently discovered human coronaviruses. Clin Lab Med. 2009;29:715-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tyrrell DA, Bynoe ML. Cultivation of a novel type of common-cold virus in organ cultures. Br Med J. 1965;1:1467-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kin N. Genomic analysis of 15 human coronaviruses OC43 (HCoV-OC43s) circulating in France from 2001 to 2013 reveals a high intra-specific diversity with new recombinant genotypes. Viruses. 2015;7:2358-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Monto AS. Medical reviews. Coronaviruses. Yale J Biol Med. 1974;47:234-251. [PMC free article] [PubMed] [Google Scholar]
- 29. Peiris JSM. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raj VS. MERS: emergence of a novel human coronavirus. Curr Opin Virol. 2014;5:58-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hendley JO. Coronavirus infections in working adults. Eight-year study with 229 E and OC 43. Am Rev Respir Dis. 1972;105:805-811. [DOI] [PubMed] [Google Scholar]
- 32. Chiu SS. Human coronavirus NL63 infection | other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong. China. Clin Infect Dis. 2005;40:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. vabret A, Freymuth F, Dina J, Brison E. Coronavirus humains(ncov) human corona viruses. Pathologie-biologie 2008:57(2):149-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perlman S, Netland J. Coronaviruses post sars: update on replication and pathogenesis. Nat Rev Microbiol. 2009:7(6):439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He J, Wen Peng G, Yus DW, et al.The molecular evolution of the SARS coronavirus, during the course of the SAARS the epidemic in China. Science 2004;303:1666-1669. [DOI] [PubMed] [Google Scholar]
- 37. Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome corona virus Helicase. J Virol. 2004;78(11):5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu D, Tu C, Xin C, et al. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome corona virus isolates. J Virol 2005;79(4):2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guan Y, Zheng BJ, He YQ, et al. Isolation and characterization of viruses Related to the SARS coronavirus from animals in southern China. Science 2003;302:276-278. [DOI] [PubMed] [Google Scholar]
- 40. Vincent CCC, Lau SKP, Woo PCY, Yuen KY. Severe acute respiratory syndrome corona virus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hui DSC, Chan M, Wu AK, Ng PL. Severe acute respiratory syndrome (SARS): epidemiology and clinical features. Postgrad Med J. 2004;80:373-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song H-D, Tu C-C, Zhang G-W, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palmcivet and Human. Proc Natl Acad Sci USA 2005;102:2430-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hawkey PM, Bhagani S, Gillespie SH. Severe acute respiratory syndrome (SARS): Breath taking progress. J Med Microbiol 2003;52:609-613. [DOI] [PubMed] [Google Scholar]
- 44. He Y, Li J, Li W, Lustigman S, Farzan M, Jiang S. Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J Immunol 2006;176:6085-6092. [DOI] [PubMed] [Google Scholar]
- 45. Emery SL, Erdman DD, Bowen MD, et al. Real time reverse transcription – polymerase chain reaction assay for SARS associated coronavirus. Emerg Infect Dis. 2004;10(2):311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peiris JSM, Chu CM, Cheng VCC, et al. Clinical progression and viral load in a community outbreak of coronavirus – associated SARS Pneumonia: a prospective study. Lancet 2003;361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. N Engl J Med 2003;348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 48. Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 2003;289:2801-2809. [DOI] [PubMed] [Google Scholar]
- 49. Samuel CE. Anti-viral actions of interferons.was also been used for the treatment of SARS. Clin Microbiol Rev 2001;14(4):778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet 2003;362:293-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arabi YM, Asiri AY. Saudi critical care trials group et al. Treatment of Middle East respiratory syndrome with a combination of lopinavir/ritonavir and interferon-β1b (MIRACLE trial): statistical analysis plan for a recursive two-stage group sequential randomized controlled trial. Trials 2020;21:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. WHO: MERS Cov research group state of knowledge and data Gaps of Middle East respiratory syndrome coronavirus (MERS COV) in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de groot RJ, Baker SC, Ziebutir J, et al. Middle east respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. 2003. J Virol. 2013; 87(14):7790–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Khan G. A novel corona virus capable of lethal human infections an emerging picture. Virol J 2013;10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alagaili AN, Briese T, Mishra N, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio 2014;5:e00884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aleanizy FS, Mohmed N, Alqahtani FY, El Hadi Mohamed RA. Outbreak of Middle East respiratory syndrome coronavirus in Saudi Arabia: a retrospective study. BMC Infect Dis 2017;17:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Durai PV, Batool M, Shah M, Choi S. Experimental & molecular medicine, Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med 2015;47:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Banik GR, Khandaker H, Rashid. Middle East respiratory syndrome coronavirus “MERS-CoV”: current knowledge gaps. Paediatr Respir Rev. 2015;16:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shirato K, Kawase M, Matsuyama S. Middle east respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 2013;87:12552-12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. de Wit E, Rasmussen AL, Falzarano D, et al. Middle East respiratory syndrome coronavirus (MERS-COV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci USA. 2013;110:16598-16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Assiri A, Al-Tawfiq JA, Al-Rabeeah AA, et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect Dis 2013;13:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Al-Abdallat MM, Payne DC, Alqasrawi S, et al. Hospital-associated outbreak of Middle East respiratory syndrome coronavirus: a serologic, epidemiologic, and clinical description. Clin Infect Dis. 2014;59:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Al-Tawfiq JA, Hinedi K, Ghandour J, et al. Middle East respiratory syndrome coronavirus: a case-control study of hospitalized patients. Clin Infect Dis 2014;59:160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Memish ZA, Al-Tawfiq JA, Assiri A, et al. Middle East respiratory syndrome coronavirus disease in children. Pediatr Infect Dis J 2014;33:904-906. [DOI] [PubMed] [Google Scholar]
- 65. Arabi YM, Arifi AA, Balkhy HH, et al. Clinical course and outcomes of critically ill patients with Middle East respiratory syndrome coronavirus infection. Ann Intern Med. 2014;160:389-397. [DOI] [PubMed] [Google Scholar]
- 66. WHO. Middle East respiratory syndrome coronavirus (MERS-CoV) summary and literature update – as of 11 June 2014. [cited 25 september 014]: http://www.who.int/csr/disease/coronavirus_infections/MERS-CoV_summary_update_20140611.pdf?ua=1
- 67. Ajlan AM, Ahyad RA, Jamjoom LG, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) infection: chest CT findings. Am J Roentgenol. 2014;203:782-787. [DOI] [PubMed] [Google Scholar]
- 68. Interim Guidance – WHO. Investigation of cause of human infection with middle east respiratory syndrome coronavirus (MERSCOV). [Google Scholar]
- 69. Virlogeux V, Park M, Wu JT, Cowling BJ. Association between severity of MERS – COV infection and incubation period. Emerg Infect Dis 2016;22:526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Milne-Price S, Miazgowicz KL, Munster VJ, et al. The emergence of the middle east respiratory syndrome coronavirus. Pathog Dis 2014;71:121-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. V Stalin Raj, Osterhaus ADME, Ron A M Fouchier 1, Bart L, Haagmans 2, et al. MERS: Emergence of a novel Human coronavirus. Curr Poin Virol 2014;5:58-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Abdul Moneim AS. Middle east respiratory syndrome coronavirus (MERS COV): Evidence and speculations. Arch Virol. 2014;159:1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kim J-M, Chung Y-S, Jo J, et al. Identification of Coronavirus Isolated from a Patient in Korea with COVID-19. Osong Public Health Res Perspect. 2020;11:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fuk-Woo Chan J, Yuan S, Kok K-H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395:514-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang L-F, Shi Z, Zhang S, Field H, Daszak P, Bryan T. Eaton review of bats and SARS. Emerg Infect Dis. 2006;12(12):1834-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ren S-Y, Gao R-D, Chen Y-L. Fear can be more harmful than the severe acute respiratory syndrome coronavirus 2 in controlling the corona virus disease 2019 epidemic. World J Clin Cases 2020;8:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Whiteworth J, Train RSOC. COVID-19: a fast evolving pandemic. Trans R Soc Trop Med Hyg 2020;114:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, Evaluation and Treatment Coronavirus (COVID-19) Treasure Island (FL). StatPearls Publishing, 2020. [PubMed] [Google Scholar]
- 80. Malik YS, Sircar S, Bhat S, et al. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40(1):68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.WHO: novel corona virus china. http://www.who.int/crr/don/12-january-2020-novelcoronavirus-china/un/
- 82. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Decaroetal N. A Novel Human Coronavirus (SaRS-COV-2); A lesson from Animal Corona Virus. 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Inten Med 2020;172:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tan W, Xhao Z, Ma X, et al. A novel corona virus genome identified in a cluster of pneumonia cases Wuhan China 2019-2020. China CDC Weekly. 2020;2:61-62. [PMC free article] [PubMed] [Google Scholar]
- 88. Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents 2020;55:105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mohammadpour S, Torshizi Esfahani A, Halaji M, Lak M, Ranjbar R. An updated review of the association of host genetic factors with susceptibility and resistance to COVID-19. J Cell Physiol 2021;236:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cowling BJ, Aiello AE. Public health measures to slow community spread of coronavirus disease 2019. 2020;221:1749-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chauhan S. Comprehensive review of coronavirus disease 2019 (COVID-19). Biomed J 2020;43:334-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Abdul Azeez A, Anjorin. The coronavirus disease 2019 (COVID-19) pandemic: A review and an update on cases in Africa. Asian Pacific J Tropical Med. 2020;13:199. [Google Scholar]
- 94. Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respiratory Med 2020;8:475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kao K-C, Wang C-H, Hsieh M-J, et al. Survival predictors in elderly patients with acute respiratory distress syndrome: a prospective observational cohort study. Sci Rep 2018;8:13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hu ds azhar. China national health commission update on the novel corona virus pneumonia outbreak(January 24 2020) Beijing, clinical national health commission. 2020. [Google Scholar]
- 97. McCreary EK, Pogue JM. Coronavirus disease 2019 treatment: a review of early and emerging options. 2020;7:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Brown EG, Tetro JA. Comparative analysis of the SARS coronavirus genome: a good start to a long journey. Lancet 2003;361:1756-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gao K, Nguyen DD, Wang R, Wei G-W. Machine intelligence design of 2019-nCoV drugs. bioRxiv 2020;2020.01.30.927889. doi: 10.1101/2020.01.30.927889. [DOI] [Google Scholar]
- 100. Gandhi RT. 2019-novel corona virus-1st report published. 2020. [Google Scholar]
- 101. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579:265-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lau SKP, Luk HKH, Wong ACP, et al. Possible bat origin of severe acute respiratory syndrome corona virus 2. Emerg Infect Dis. 2020;26(7):1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tianyi Qiu, Mao T, Wang Y, et al. Identification of potential cross protective epitope between a new type of corona virus {2019 n cov) and severe acute respiratory syndrome virus. J Genet Genom 2020;47:115-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sun Z, Thilakavathy K, Suresh Kumar S, He G, Liu SV. Potential Factors Influencing Repeated SARS Outbreaks in China. Int J Environ Res Public Health. 2020;17:1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Halaji M, Farahani A, Ranjbar R, Heiat M, Dehkordi FS. Emerging Coronaviruses: first SARS, second MERS and third SARS-CoV-2: epidemiological updates of COVID-19. Infez Med. 2020;28:6-17. [PubMed] [Google Scholar]
- 107. Rahman HS, Aziz S, Hussein RH, et al. The transmission modes and sources of COVID-19. Int J Surg Open. 2020; 26:125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382:1564-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chin AWH, Chu JTS, Perera MRA, et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Aboubakr HA, Sharafeldin Goyal SM. Stability of SARS-CoV-2 and other coronaviruses in the environment and on common touch surfaces and the infuence of climatic conditions: A review. Transbound Emerg Dis. Published online June 30, 2020. doi:10.1111/tbed.13707 [DOI] [PMC free article] [PubMed]
- 111. Meselson M. Droplets and aerosols in the transmission of SARS-CoV-2. N Engl J Med 2020;382:2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kolifarhood G, Aghaali M, Mozafarm Saadati H, et al. Epidemiological and Clinical Aspects of COVID-19: a narrative review. Arch Acad Emerg Med. 2020;8:e41. [PMC free article] [PubMed] [Google Scholar]
- 113. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Li Q, Guan X, Wu P, et al. Early transmission dynamics in wuhan, china, of novel coronavirus-infected pneumonia. N Engl J Med 2020;382:1199-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Primerdesign TM Ltd Coronavirus (COVID-19) genesigReal-Time PCR assay Instructions for Use (IFU) Issue 2.0. [Google Scholar]
- 116. WHO: origin of sars cov-2(26 MARCH 2020). [Google Scholar]
- 117. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel corona virus disease. Clin Infect Dis. 2020;71:778-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterol. 2020;158:1831-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lamers MM, Beumer J, van der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020;369:50-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chu DK, Akl EA, Duda S, et al. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Singhal T. A review of coronavirus disease - 2019 (COVID-19). Indian J Pediatr 2020;87(4):281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Harapan H, Yufika A, Winardif W, et al. Review coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020;13(5):667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Dhama K, Khan S, Tiwari R, et al. Coronavirus disease 2019-COVID-19. Clin Microbiol Rev 2020; 33(4): e00028–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hornuss D, Laubner K, Monasterio C, Thimme R, Wagner D. COVID -19 associated pneumonia despite repeatedly negative PCR-analysis from oropharyngeal swabs. Dtsch Med Wochenschr. 2020;145:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Fakheran O, Dehghannejad M, Khademi A. Saliva as a diagnostic specimen for detection of SARS-CoV-2 in suspected patients: a scoping review. Infect Dis Poverty 2020;9(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lu H. Drug treatment options for the 2019 new corona virus (2019 n cov). Bious Trends. 2020;14:69-71. [DOI] [PubMed] [Google Scholar]
- 127. Cao B, Yeming Wang MD, Wen D, et al. A trial of Lopinavir–Ritonavir in adults hospitalized with severe COVID-19. 2020;382:1787-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. WHO: Off label use of medicine for covid-19. [Google Scholar]
- 129. WHO: Solidarity clinical trial for covid-19 treatments. [Google Scholar]
- 130. Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen K-Y. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov 2016;15:327-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Yang Y, Islam M, Wang J, Li Y, Chen X. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci 2020;16:1708-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Colson P, Rolain J-M, Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents 2020;55(3):105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Md. Abdul Alim Al-Bari. Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J Antimicrob Chemother. 2015;70(6):1608-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cortegiani A, Ingoglia G, Ippolito M, Giarratano A, Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J Crit Care. 2020;57:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Mahalmani VM, Mahendru D, Semval A, et al. COVID-19 pandemic: A review based on current evidence. Indian J Pharmacol. 2020;52(2):117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Rizk JG, Kalantar-Zadeh K, Mehra MR. Pharmaco-immunomodulatory therapy in COVID-19. Springer link. Drugs. 2020;80(13):1267-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Baron SA, Devaux C, Colson P, Raoult D, Rolain JM. Teicoplanin: an alternative drug for the treatment of COVID-19? Int J Antimicrob Agents. 2020;55(4):105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ceccarelli G, Alessandri F, d’Ettorre G, et al. Is teicoplanin a complementary treatment option for COVID-19? The question remains. Int J Antimicrob Agents. 2020;56(2):106029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zhang J, Ma X, Yu F, et al. Teicoplanin potently blocks the cell entry. biorxiv. 2020. DOI: 10.1101/2020.02.05.935387. [DOI] [Google Scholar]
- 140. Vora A, Arora VK, Behera D, Tripathy SK. White paper on Ivermectin as a potential therapy for COVID-19. Indian J Tuberc 2020;67(3):448-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Sharun K, Dhama K, Kumar Patel S, et al. Ivermectin, a new candidate therapeutic against SARS-CoV-2/COVID-19. Ann Clin Microbiol Antimicrob 2020;19:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Banerjee K, Nandy M, Dalai CK, Newaz Ahmed S. The battle against COVID 19 pandemic: what we need to know before we “test fire” ivermectin. Drug Res. 2020;70(8):337-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Choudhary R, Sharma AK. Potential use of hydroxychloroquine, ivermectin and azithromycin drugs in fighting COVID-19: trends, scope and relevance. New Microbes New Infect 2020;22;100684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Li H, Zhou Y, Zhang M, Wang H, Zhao Q, Liu J. Updated approaches against SARS-CoV-2. Antimicrob Agents Chemother 2020;64:e00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Chen Z, Wu Q, Chen J, Ni X, Dai J. A DNA aptamer based method for detection of SARS-CoV-2 nucleocapsid protein. Virol Sin. 2020;35(3):351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Torabi R, Ranjbar R, Halaji Md, Heiat M. Aptamers, the bivalent agents as probes and therapies for coronavirus infections: a systematic review. Mol Cell Probes 2020;53:101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Mao K, Zhang H, Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ Sci Technol 2020;54:3733-3735. [DOI] [PubMed] [Google Scholar]
- 149. Kitajima M, Ahmed W, Bibby K, et al. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci Total Environ. 2020;739:139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Mirzaie A, Halaji M, Dehkordi SF, Ranjbar R, Noorbazargan H. A narrative literature review on traditional medicine options for treatment of corona virus disease 2019 (COVID-19). Complement Ther Clin Pract. 2020;40:101214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Benarba B, Pandiella A. Medicinal plants as sources of active molecules against COVID-19. Front Pharmacol. 2020;11:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Nugraha RV, Ridwansyah H, Ghozali M, Khairani AF, Atik N. Traditional herbal medicine candidates as complementary treatments for COVID-19: a review of their mechanisms, pros and cons. Evi Based Complement Alternat Med 2020;2020:2560645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Basile C, Combe C, Pizzarelli F, et al. Recommendations for the prevention, mitigation and containment of the emerging SARS-CoV-2 (COVID-19) pandemic in haemodialysiscentres. Nephrol Dial Transplant 2020;35:737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Adhikari SP, Meng S, Wu Y-J, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) Outbreak in China summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA 2020;323:1239-1242. [DOI] [PubMed] [Google Scholar]
- 156. WHO: corona virus disease (COVID 19) advices for the public. [Google Scholar]
- 157. Courtemanche C, Garuccio J, Le A, Pinkston J, Yelowitz A. Strong social distancing measures in the United States reduced the COVID-19 growth rate. Health Aff (Millwood) 2020;39(7):1237-1246. [DOI] [PubMed] [Google Scholar]
- 158. Muscillo A, Pin P, Razzolini T. Covid19: Unless one gets everyone to act, policies may be ineffective or even backfire. PLoS One 2020;15:e0237057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. De Vos J. The effect of COVID-19 and subsequent social distancing on travel behavior. Interdisciplinary Perspect. 2020;5:100121. [DOI] [PMC free article] [PubMed] [Google Scholar]


