Abstract
Introduction
Obesity is a main global health issue and an outstanding cause of morbidity and mortality. Exploring miRNA profiling may help further studies on obesity.
Methods
Three morbidly obese and 5 normal-weight Chinese women were enrolled in the microarray testing group. Abdominal subcutaneous adipose tissue (SAT) samples were excised. Total RNAs including miRNAs were extracted. Affymetrix GeneChip miRNA 4.0 Array was used to compare the expression profiles of miRNAs between the 2 groups. Two algorithms, miRanda and TargetScan, were used to predict target messenger RNAs (mRNAs). Bioinformatics analysis was then done based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The sample sizes were further expanded to 8 morbidly obese and 9 normal-weight subjects, and quantitative real-time PCR (qRT-PCR) was utilized to verify the expression of differential miRNAs and target genes.
Results
As per the microarray assay, 58 miRNAs were differentially expressed in the SAT from the morbidly obese and normal-weight groups (Fold >4, p < 0.01, FDR <0.05); 54 of these were downregulated and 4 were upregulated in morbidly obese subjects. A total of 1,333 target genes were jointly predicted by miRanda and TargetScan. Further bioinformatics analysis showed that the differential miRNAs were involved in 269 significant biological functions and 89 significant signaling pathways. The validation experiment by qRT-PCR showed that the expression levels of miRNA-143-5p, miRNA-143-3p, miRNA-145-5p, and let-7a-5p were downregulated in morbidly obese subjects, consistent with the microarray detection. High-mobility group A2 (HMGA2), a target gene of the downregulated miRNA let-7a-5p, was first found to be upregulated 3.19-fold in the SAT of morbidly obese Chinese women when compared to normal-weight controls.
Conclusions
MiRNA downregulation is a hallmark of intact SAT in a morbidly obese state. Transcription (DNA-dependent), small-molecule metabolic processes, the MAPK signaling pathway, and cancer-related pathways may play important roles in the occurrence and development of obesity. For the first time, we proved that HMGA2, a target gene of let-7a-5p, is upregulated in the SAT of morbidly obese Chinese women.
Keywords: Morbid obesity, Subcutaneous adipose tissue, MicroRNA, Let-7a-5p, HMGA2 (high-mobility group A2)
Introduction
Obesity is defined by the World Health Organization (WHO) as a disease [1] and has become a significant public health problem. In 2014, it was found that obesity affected 10.8% of the world's adult males and 14.9% of adult females, with 0.64% (0.46–0.86) of the men and 1.6% (1.3–1.9) of the women being morbidly obese [2]. In parallel with the current global scenario, obesity has become one of the most severe problems in China. The number of obese people in China is the largest in the world, affecting >89.6 million Chinese people [2]. The outcomes of obesity are widespread, and the disease contributes substantially to cardiovascular diseases, type 2 diabetes mellitus, and certain types of cancer [3]. Compared with the normal-weight population, the mortality rate of obesity has increased by at least 50% [4]. As a pathophysiological disorder, obesity is characterized by excess accumulation of adipose tissue in the body. Adipose tissue contains several different cell populations including adipocytes, preadipocytes, mesenchymal stem cells (MSCs), macrophages, and lymphocytes [5]. Adipose tissue is a highly responsive endocrine organ that influences and interacts with metabolic homeostasis and inflammation [6]. It is the main lipid storage depot in our body and plays a crucial role in buffering the daily influx of dietary fat entering the circulation. Under the condition of obesity, subcutaneous adipose tissue (SAT) may fail to expand appropriately to store energy surpluses. The expandability of SAT thus seems to be a critical factor in the development of obesity-associated comorbidities [7].
MicroRNAs (miRNAs), encoded by the genome DNA, are a kind of small endogenous non-coding RNA that is widely expressed in the body [8]. MiRNAs are crucial elements in cell transcriptional regulation and are estimated to regulate >60% of human messenger RNAs (mRNAs) [9], either via degradation of target mRNAs or by translational repression [10]. MiRNAs are useful for early diagnosis, risk assessment, and monitoring disease progression because they are more sensitive, more stable, and more slowly degraded than protein and mRNAs [11, 12]. Integrative miRNA-gene-pathway networks provide tools for the identification of disease-related miRNA signatures [13]. A variety of pathophysiological processes, such as the metabolism, have previously been proven to be affected by miRNAs [14]. Thus, exploring miRNA profiling may help with further studies on obesity. However, there is a shortage of in-depth analyses of obesity-related miRNAs alterations. Most previous studies focused on circulating miRNAs, or used animal models, or fat cell lines rather than human adipose tissue [15, 16, 17]. Also, most past studies are characterized by significant inconsistency, and focused on the differentiation of adipocytes and adipogenesis [14, 18] which can only reveal a small part of the pathophysiological process of obesity.
Few studies have analyzed the expression pattern of miRNAs in human fatty tissues in an obese state. In addition, the limited studies have all been conducted in Western populations. Arner et al. [19] identified 20 altered miRNAs (2 upregulated and 18 downregulated) in the white adipose tissue of obese Swedish patients. Ortega et al. [20] revealed that 11 miRNAs were significantly deregulated in the SAT of obese Spanish women. Martinelli et al. [21] showed that the expression of miRNA-519d was altered in the SAT of obese Italian patients. All these studies used microarray chips to study the miRNA expression patterns; an important caveat is that the functional roles of the identified miRNAs were not well established [22]. The lack of miRNA profile studies on adipose tissues in Eastern populations and the absence of bioinformatics analysis of the identified miRNAs limit our understanding of the role played by miRNAs in obesity in Easterners.
In this study, we used the Affymetrix GeneChip miRNA 4.0 Array to compare the miRNA expression signatures in the SAT of 3 morbidly obese and 5 normal-weight Chinese women. We also dissected the role of miRNAs via GO and KEGG bioinformatic analysis. The levels of differential miRNAs and the interested transcription genes were further validated using quantitative real-time PCR (qRT-PCR), after the enlargement of the sample size to 8 morbidly obese and 9 normal-weight subjects. This study may help provide unique data on the SAT miRNA expressing profiles and regulatory networks concerning oriental morbid obesity.
Material and Methods
General Characteristics of Enrolled Subjects
Three morbidly obese female patients (42.00 ± 7.57 years of age; BMI 47.53 ± 1.53) who received laparoscopic adjustable gastric banding (LAGB), and 5 normal-weight females (aged 47.20 ± 2.58 years; BMI 22.03 ± 0.78) admitted for elective abdominal surgical procedures (4 for uterus myoma and 1 for hepatic hemangioma), were enrolled in the microarray testing group. We further expanded the sample size for the purpose of validating the differentially expressed miRNAs and the target genes. Eight morbidly obese female patients (including the 3 obese patients in the microarray group) aged 34.88 ± 3.42 years (BMI 45.45 ± 2.40) and 9 normal-weight females (including 2 of the normal-weight patients in the microarray group) aged 42.11 ± 2.23 years (BMI: 22.42 ± 0.54) were enrolled. A comprehensive nutritional assessment was performed before surgery. All participants were in approximately the middle of their menstrual cycle and reported that their body weight had been stable for at least 3 months. Baseline anthropometric and blood pressure were measured. After an overnight fast, blood samples were obtained from all participants and then measured using routine automated methods in our clinical laboratory. All enrolled subjects were required to have relatively normal hepatic-renal functions (aspartate transaminase <80 U/L, creatinine <132 µmol/L, and blood urea nitrogen <7.14 mmol/L). No history of cancer, no acute inflammatory diseases, and no chronic diseases other than obesity and its comorbidities were allowed. Subjects reporting intake of continuous medication were excluded. All enrolled patients also fasted overnight before surgery.
Preparation of SAT Samples
Abdominal SAT samples (approx. 1–2 cm3 of fat) were extracted from all subjects by experienced surgeons. These fresh SAT specimens were washed with cold saline solution, and then immediately frozen in liquid nitrogen and stored at −80°C until analysis.
RNA Extraction
Total RNAs, including miRNAs, were extracted from SAT using the miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. The purity and quantity of RNA were assessed with the NanoDrop2000 instrument (Thermo Fisher Scientific, Boston, MA, USA).
RNA Profiling
Experiments of microarray chips were conducted by Shanghai Qiming Information Technology Co., Ltd. The steps were as follows: RNA was labeled with the flash tag biotin HSR kit (Genisphere, Hatfield, PA, USA) according to the manufacturer's instructions for poly(A)-tailing. Labeled samples were then hybridized with Affymetrix GeneChip miRNA 4.0 Array (Affymetrix, Santa Clara, CA, USA). This microarray contains 30,424 total mature miRNA probe sets, including 2,578 mature human miRNAs and miRNAs from 202 other organisms [23]. Samples were washed and stained with the Affymetrix GeneChip2 hybridization wash and stain kit, and then scanned with the Affymetrix GeneChip Scanner 3000 7G to generate fluorescent images, as described in the manufacturer's protocol [24].
For miRNA array analysis, CEL-files of the raw data were produced with Affymetrix GeneChip command console software v4.0. Partek Genomics Suite software (St. Louis, MO, USA) was used for further analysis. The RVM t test was used to filter the differentially expressed miRNAs because this test can raise degrees of freedom effectively in the cases of small samples [25]. p values were adjusted for multiple testing using Benjamini-Hochberg method with a false discovery rate (FDR) of 0.05 [26]. Fold changes were calculated by comparing gene expression levels between morbidly obese and normal-weight samples, and then expressed as the ratio between the averages of normalized intensities of the 2 groups. MiRNAs with a fold-change threshold >4 were considered differentially expressed [27].
Target Gene Prediction
Target genes of differential miRNAs were predicted by 2 algorithms: miRanda (http://www.microrna.org/microrna/home.do) and TargetScan (http://www.targetscan.org/). The parameters of miRanda were set as alignment score ≥140 and free energy ≤–20 [28]. The parameters of TargetScan were set as context score percentile >90 [29].
Gene Ontology Category
Gene ontology (GO) analysis was employed to analyze the functional enrichment of the differentially expressed miRNAs (http://geneontology.org/). Fisher's exact test and the χ2 test were used to classify the GO categories. GO enrichment calculation helped to provide a measure of the functional significance. p < 0.001 with an FDR <0.05 was used as the threshold of significance.
Pathway Analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was employed to analyze the significant pathways associated with the differential miRNAs (http://www.genome.jp/kegg/). Fisher's exact test and the χ2 test were used to classify the significant pathways. KEGG enrichment calculation helped to provide a measure of the pathway significance. p < 0.001 with an FDR <0.05 was used as the threshold of significance.
The Validation of Differential miRNA Levels by qRT-PCR Methods
Differential miRNAs were validated using qRT-PCR after RNA-tailing. Reverse transcription was performed using the Mir-XTM miRNA first-strand synthesis kit (Clontech, Dalian, China) according to the user manual. U6 was used as an endogenous control. It was previously used as internal reference in the studies of abdominal adipose-derived MSCs [30] and human visceral tissue and SAT [31, 32]. cDNA was amplified in a single reaction with the forward primers, reverse primers, and QuantiTect SYBR Green PCR Master Mix (Qiagen, Valencia, CA, USA). The primer sequences used in our experiment are shown in online supplementary Table S1 (for all online suppl. material, see www.karger.com/doi/10.1159/000511772). PCR amplification was performed on the ABI ViiA 7 real-time PCR system (Applied Biosystems, Carlsbad, CA, USA). The 2-ΔΔCT method was applied to determine the relative abundance of each miRNA. The specificity of SYBR Green RT-PCR was determined by monitoring the amplification profile and the dissociation curve of the target amplicons [33].
The Validation of Differential miRNA Target Gene Levels by qRT-PCR Methods
Total RNAs in SAT were reverse-transcripted by using the PrimeScriptTM RT reagent kit (Takara, Bio, Inc., Shiga, Japan). GAPDH was used as the endogenous control gene. cDNA was amplified in a single reaction with the forward primers, reverse primers, and QuantiTect SYBR Green PCR Master Mix (Qiagen). Primer sequences used in our experiment are shown in online supplementary Table S2. PCR amplification was performed on the Applied Biosystems ABI ViiA 7 real-time PCR system. The 2-ΔΔCT method was applied to determine the relative abundance of each mRNA. The specificity of amplification was also analyzed by using the amplification profile and the dissociation curve of the target genes [33].
Statistical Analysis
Patients' anthropometric data, laboratory measurements, qRT-PCR results, and Pearson's correlation were analyzed using SPSS v22.0 (SPSS Inc. Chicago, IL, USA). Sample sizes in microarray and qRT-PCR analysis were determined using R statistical software v4.0.2 and the statistical power was 0.75 [34, 35]. The Student t test or Mann-Whitney U test was used for group comparison. Normally distributed data are presented as mean ± SEM. Nonnormally distributed parameters are reported as medians (range). p < 0.05 was considered statistically significant.
Results
Anthropometric and Laboratory Measurements
Anthropometric measurements and biochemical characteristics of the 3 morbidly obese and 5 normal-weight individuals are listed in Table 1. As expected, obese women had significantly greater body weight, BMI, and waist circumference (p all <0.001). Fasting blood glucose (FBG) and triglyceride (TG) levels of morbidly obese patients were markedly higher than those of normal-weight controls (both p < 0.05). There was no considerable difference in age, height, and blood pressure between the 2 groups. Serum levels of total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) were also not significantly different. As an inflammatory marker, the level of circulating high-sensitivity C-reactive protein (hs-CRP) in morbidly obese subjects was much higher than the upper normal limit (3 mg/L). The hs-CRP level was not available for the normal-weight controls.
Table 1.
Anthropometric characteristics of the study groups
| Normal-weight (n = 5) | Morbidly obese (n = 3) | p value | |
|---|---|---|---|
| Age, years | 47.20±2.58 | 42.00±7.57 | 0.455 |
| Height, m | 1.63±0.02 | 1.66±0.04 | 0.454 |
| Weight, kg | 58.30±1.87 | 131.33±4.38 | <0.001 |
| BMI | 22.03±0.78 | 47.53±1.53 | <0.001 |
| WC, cm | 74.40±1.21 | 138.83±4.85 | <0.001 |
| SBP, mm Hg | 110 (103–160) | 160 (160–220) | 0.071 |
| DBP, mm Hg | 72.40±7.62 | 115.0±22.55 | 0.070 |
| FBG, mmol/L | 5.34±0.13 | 7.47±0.24 | <0.001 |
| TC, mmol/L | 5.21±0.20 | 5.93±0.83 | 0.555 |
| TG, mmol/L | 0.61±0.03 | 2.47±0.10 | 0.001 |
| HDL-C, mmol/L | 2.14±0.33 | 0.99±0.25 | 0.065 |
| LDL-C, mmol/L | 2.63±0.21 | 4.06±0.58 | 0.158 |
| hs-CRP, mg/L | − | 10.00 (4.81–10.00) | − |
BMI, body mass index; WC, waist circumference; SBP, systolic blood pressure; DBP, diastolic blood pressure; FBG, fasting blood glucose; TC, total cholesterol; TG, triglyceride; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; hs-CRP, high-sensitivity C-reactive protein.
Microarray Analysis
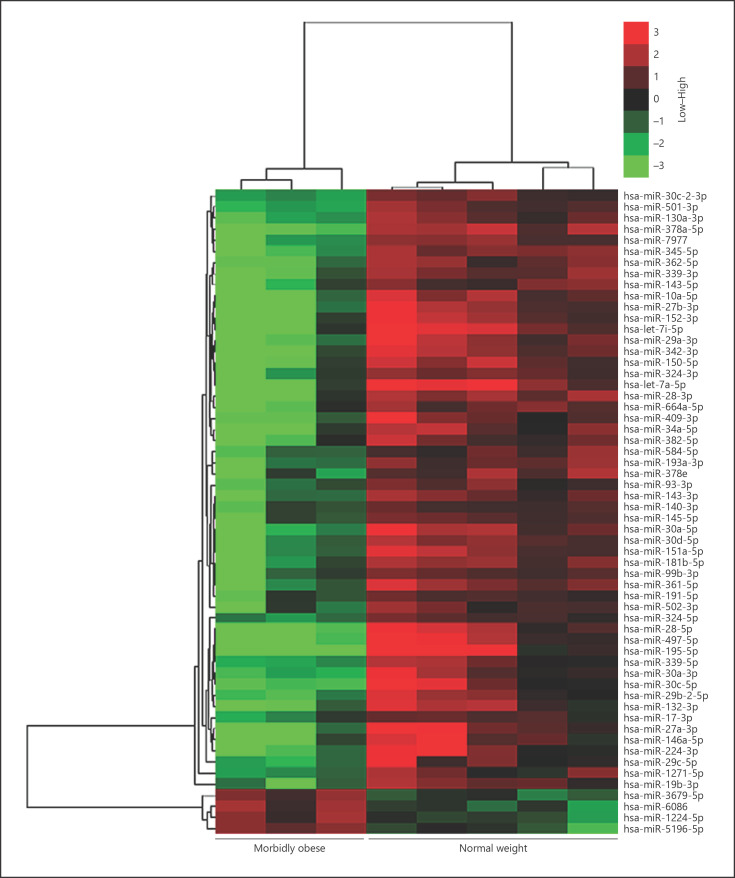
As presented in Table 2, 58 miRNAs were differentially expressed (>4-fold, p < 0.01, FDR <0.05) in the SAT of obese patients and normal-weight controls as per the Affymetrix GeneChip miRNA 4.0 Array. Of these, 54 miRNAs were downregulated and 4 were upregulated in the morbidly obese group. The range of differential downregulation was 0.014–0.23, while that of upregulation was 4.05–4.81. A hierarchical clustering algorithm was used to cluster the differentially expressed miRNAs, and the results showed that the hierarchical clustering of all covered human miRNAs could clearly separate the SAT of morbidly obese patients from that of controls (Fig. 1).
Table 2.
The differentially expressed miRNAs in the SAT of the study groups
| miRNA | O/N | p value | FDR | Style |
|---|---|---|---|---|
| miR-195-5p | 0.014 | 0.0027633 | 0.0291 | Down |
| let-7a-5p | 0.014 | 0.0068512 | 0.0412 | Down |
| let-7i-5p | 0.017 | 0.0052647 | 0.0353 | Down |
| miR-378a-5p | 0.033 | 0.0000367 | 0.0147 | Down |
| miR-497-5p | 0.034 | 0.0010554 | 0.0236 | Down |
| miR-27a-3p | 0.034 | 0.0032048 | 0.0311 | Down |
| miR-29a-3p | 0.037 | 0.0022679 | 0.0266 | Down |
| miR-30a-5p | 0.037 | 0.0033689 | 0.0315 | Down |
| miR-152-3p | 0.040 | 0.0034639 | 0.0315 | Down |
| miR-28-5p | 0.041 | 0.0004003 | 0.0211 | Down |
| miR-342-3p | 0.041 | 0.0029938 | 0.0306 | Down |
| miR-28-3p | 0.042 | 0.0030938 | 0.0311 | Down |
| miR-27b-3p | 0.045 | 0.0010409 | 0.0236 | Down |
| miR-361-5p | 0.048 | 0.0091530 | 0.0470 | Down |
| miR-151a-5p | 0.049 | 0.0091041 | 0.0470 | Down |
| miR-150-5p | 0.051 | 0.0041417 | 0.0326 | Down |
| miR-10a-5p | 0.052 | 0.0008831 | 0.0236 | Down |
| miR-345-5p | 0.056 | 0.0001748 | 0.0147 | Down |
| miR-181b-5p | 0.056 | 0.0085548 | 0.0470 | Down |
| miR-30c-5p | 0.060 | 0.0055179 | 0.0358 | Down |
| miR-146a-5p | 0.063 | 0.0089737 | 0.0470 | Down |
| miR-339-3p | 0.070 | 0.0009224 | 0.0236 | Down |
| miR-34a-5p | 0.071 | 0.0046445 | 0.0330 | Down |
| miR-224-3p | 0.071 | 0.0092876 | 0.0472 | Down |
| miR-30d-5p | 0.074 | 0.0043428 | 0.0330 | Down |
| miR-132-3p | 0.075 | 0.0050523 | 0.0343 | Down |
| miR-362-5p | 0.078 | 0.0005181 | 0.0211 | Down |
| miR-409-3p | 0.084 | 0.0039087 | 0.0324 | Down |
| miR-7977 | 0.087 | 0.0002723 | 0.0170 | Down |
| miR-378e | 0.089 | 0.0069746 | 0.0412 | Down |
| miR-664a-5p | 0.091 | 0.0047292 | 0.0333 | Down |
| miR-193a-3p | 0.095 | 0.0027373 | 0.0291 | Down |
| miR-143-3p | 0.096 | 0.0042270 | 0.0326 | Down |
| miR-29b-2-5p | 0.097 | 0.0025800 | 0.0288 | Down |
| miR-324-3p | 0.098 | 0.0055162 | 0.0358 | Down |
| miR-130a-3p | 0.099 | 0.0002611 | 0.0170 | Down |
| miR-30a-3p | 0.099 | 0.0021607 | 0.0266 | Down |
| miR-19b-3p | 0.100 | 0.0049551 | 0.0340 | Down |
| miR-143-5p | 0.110 | 0.0023759 | 0.0273 | Down |
| miR-382-5p | 0.110 | 0.0081096 | 0.0455 | Down |
| miR-501-3p | 0.120 | 0.0000513 | 0.0147 | Down |
| miR-339-5p | 0.130 | 0.0031182 | 0.0311 | Down |
| miR-30c-2-3p | 0.150 | 0.0000695 | 0.0147 | Down |
| miR-99b-3p | 0.150 | 0.0071791 | 0.0419 | Down |
| miR-145-5p | 0.150 | 0.0080244 | 0.0454 | Down |
| miR-29c-5p | 0.150 | 0.0087899 | 0.0470 | Down |
| miR-502-3p | 0.160 | 0.0089931 | 0.0470 | Down |
| miR-584-5p | 0.170 | 0.0025082 | 0.0284 | Down |
| miR-93-3p | 0.170 | 0.0037939 | 0.0324 | Down |
| miR-1271-5p | 0.190 | 0.0091395 | 0.0470 | Down |
| miR-324-5p | 0.210 | 0.0001372 | 0.0147 | Down |
| miR-140-3p | 0.210 | 0.0044805 | 0.0330 | Down |
| miR-191-5p | 0.220 | 0.0086791 | 0.0470 | Down |
| miR-17-3p | 0.230 | 0.0041569 | 0.0326 | Down |
| miR-3679-5p | 4.050 | 0.0016929 | 0.0236 | Up |
| miR-1224-5p | 4.260 | 0.0044494 | 0.0330 | Up |
| miR-5196-5p | 4.710 | 0.0086927 | 0.0470 | Up |
| miR-6086 | 4.810 | 0.0041345 | 0.0326 | Up |
O/N, obese/normal-weight; FDR, false discovery rate.
Fig. 1.
Hierarchical clustering of the differential miRNAs in the SAT of the study groups. Fifty-eight differentially regulated miRNAs of morbidly obese (n = 3) and normal-weight (n = 5) subjects were hierarchically clustered and represented using TreeView. In the SAT of morbidly obese subjects, 54 miRNAs were downregulated and 4 were upregulated. Red, high expression; green, low expression. The intensity represents the magnitude of the expression difference. The 2 closest remaining items (those with the smallest distance) are joined by a node/branch of a tree, with the length of the branch set to the distance between the joined items.
Biological Analysis of the Differential miRNAs
Target Gene Prediction
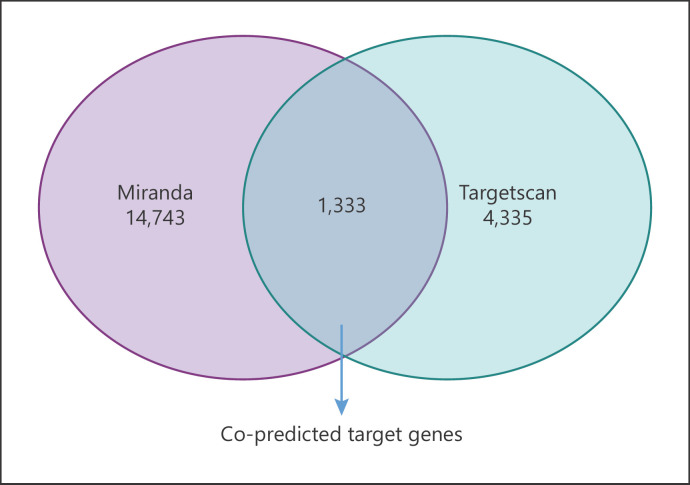
The miRanda and TargetScan computational methods were combined to predict the target genes of the 58 differentially expressed miRNAs. As presented in Figure 2 and online supplementary Table S3, miRanda predicted 14,743 and TargetScan predicted 4,335 target genes. A total of 1,333 target genes were jointly predicted by miRanda and TargetScan.
Fig. 2.
Target gene prediction of the differential miRNAs. Venn diagrams show the number of target genes predicted by the miRanda and TargetScan algorithms.
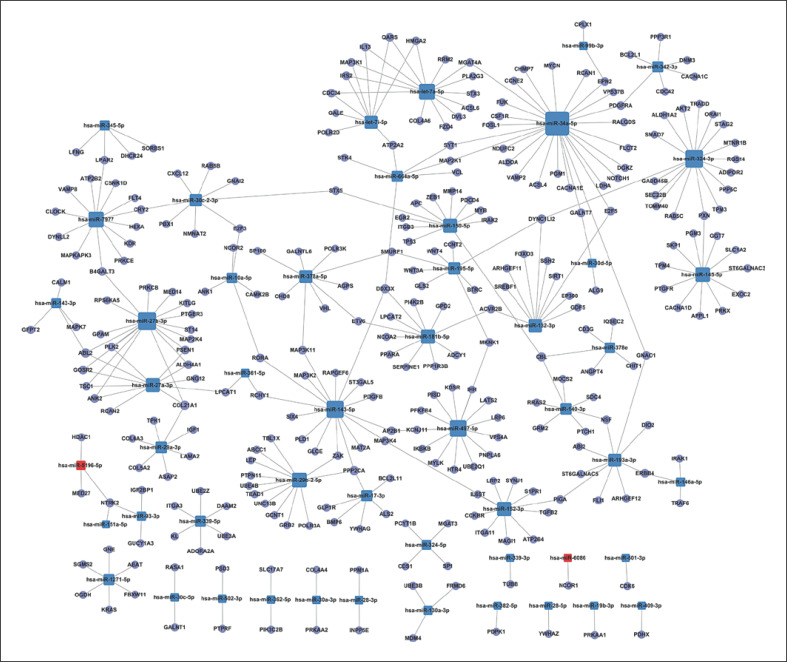
The miRNA-Gene-Network
The differential miRNAs and predicted target genes form a regulatory network. In Figure 3, the circles represent target genes and the squares represent miRNAs. The relationships between them were represented by one edge. The network was denoted by degree. Degree refers to the contribution of 1 miRNA to the genes around or the contribution of 1 gene to the miRNAs around. The key miRNAs and genes were in the network and had the most significant degrees. In this miRNA-gene-network, miRNA-34a-5p, miRNA-27b-3p, miRNA-324-3p, miRNA-143-5p, miRNA-497-5p, let-7a-5p, miRNA-7977, and miRNA-145-5p were the key miRNAs with strong regulatory capacities, and were located at the center of the regulatory network. Some predicted target genes, namely ABL2, CBL, COL21A1, DDX3X, DYNC1LI2, NTRK2, and PPP2CA, had the highest regulatory degrees.
Fig. 3.
The miRNA-gene-network. The network generated by Cytoscape displays the interactions between the key differential miRNAs and predicted target genes. Blue, downregulated miRNAs; red, upregulated miRNAs. The miRNAs are depicted by squares and the predicted genes as circles. Their interaction is represented by one edge.
GO Analysis of the Differential miRNAs
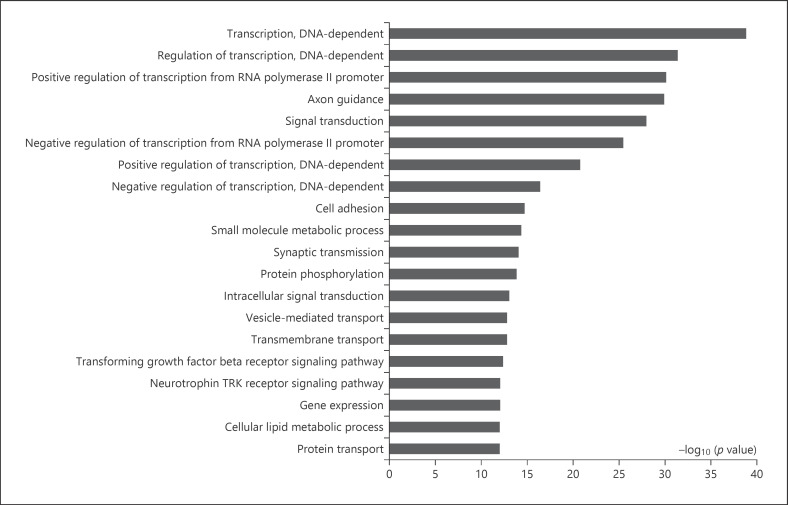
GO analysis indicated that 269 GO terms were enriched (p < 0.001 and FDR <0.05). The top 20 significant GO categories by the differential miRNAs were transcription (DNA-dependent), regulation of transcription (DNA-dependent), positive regulation of transcription from RNA polymerase II promoter, axon guidance, signal transduction, negative regulation of transcription from RNA polymerase II promoter, positive regulation of transcription (DNA-dependent), negative regulation of transcription (DNA-dependent), cell adhesion, small-molecule metabolic processes, synaptic transmission, protein phosphorylation, intracellular signal transduction, vesicle-mediated transport, transmembrane transport, the transforming growth factor (TFG)-β receptor signaling pathway, the neurotrophin TRK receptor signaling pathway, gene expression, cellular lipid metabolic process, and protein transport (Fig. 4; online suppl. Table S4).
Fig. 4.
Top-ranked significant biological functions identified by GO categories. The bar plot represents the −log10 (p value) of the GO terms.
The miRNA-GO-Network
The differential miRNAs and GO functions formed a regulatory network. In online supplementary Figure S1, the circles represent GO functions and the squares represent miRNAs. The relationships between them were represented by one edge. The key miRNAs and GO functions were in the network and had the most significant degrees. In the miRNA-GO-network, 15 miRNA including miRNA-34a-5p, miRNA-27b-3p, miRNA-150-5p, let-7a-5p, miRNA-7977, miRNA-143-5p, miRNA-664a-5p, let-7i-5p, miRNA-132-3p, miRNA-497-5p, miRNA-324-3p, miRNA-29b-2-5p, miRNA-143-3p, miRNA-146a-5p, and miRNA-145-5p were involved in most biological functions. They were located at the network center with larger representative areas and more network lines. Five GO categories including small-molecule metabolic processes, negative regulation of the apoptotic process, the epidermal growth factor (EGF) receptor signaling pathway, protein phosphorylation, and the fibroblast growth factor (FGF) receptor signaling pathway were the ones with the highest regulatory degrees.
Pathway Analysis of the Differential miRNAs
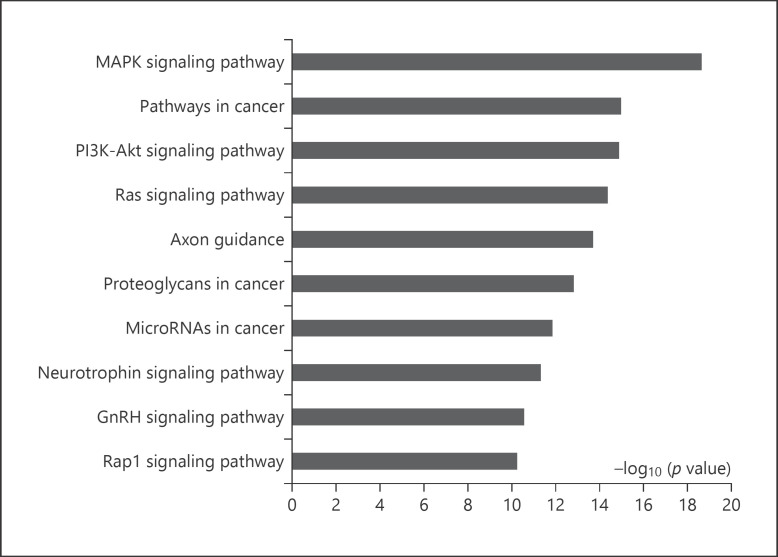
As per the KEGG pathway analysis of predicted target genes, 89 signaling pathways were enriched (p < 0.001 and FDR <0.05). As presented in Figure 5 and online supplementary Table S5, the top 10 ranked significant pathways were the MAPK signaling pathway, pathways in cancer, the PI3K-Akt signaling pathway, the Ras signaling pathway, axon guidance, proteoglycans in cancer, microRNAs in cancer, the neurotrophin signaling pathway, the GnRH signaling pathway, and the Rap1 signaling pathway.
Fig. 5.
Top-ranked significant pathways identified by KEGG. The bar plot represents the −log10 (p value) of the canonical pathways.
The Validation of the Differential miRNA and Target Gene Levels in the SAT of the Validation Groups
Anthropometric and Laboratory Measurements
The sample sizes were expanded to 8 morbidly obese and 9 normal-weight women for validation. Anthropometric measurements and biochemical characteristics are listed in online supplementary Table S6. Body weight, BMI, and waist circumference of morbidly obese women were significantly greater (p all <0.001). Serum TG level was significantly higher, but HDL-C level was significantly lower in morbidly obese women (both p < 0.05). There was no difference in age, height, blood pressure, FBG, TC, or LDL-C. The circulating hs-CRP level in morbidly obese subjects was also much higher than the upper normal limit (3 mg/L).
qRT-PCR Validation of Differential miRNA Levels in the SAT of the Validation Groups
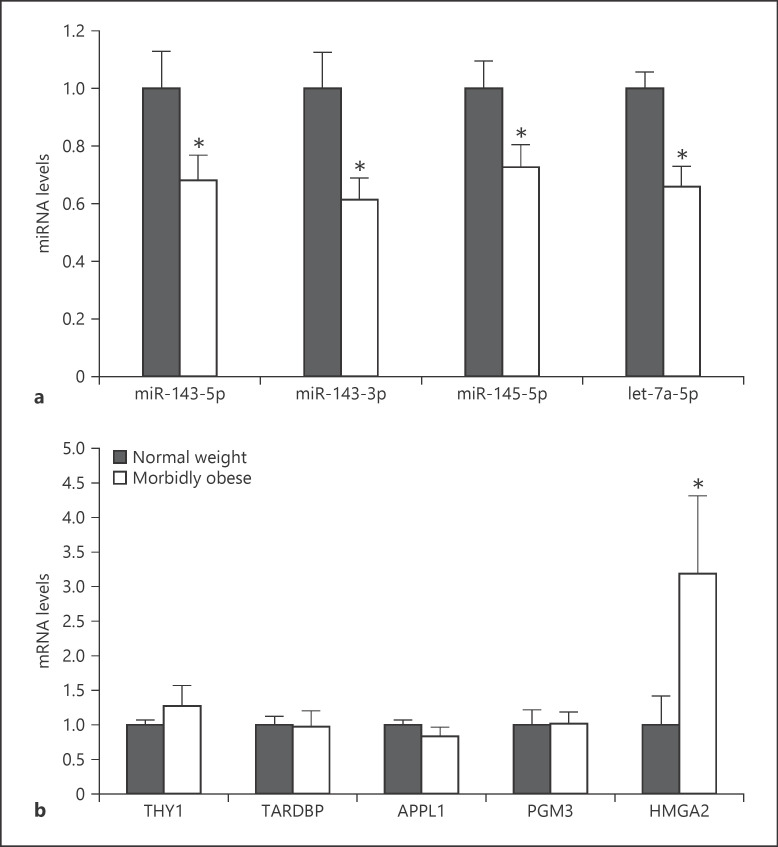
To validate the differential miRNAs, 4 differentially expressed miRNAs, namely miRNA-143-5p, miRNA-143-3p, miRNA-145-5p, and let-7a-5p were validated by qRT-PCR. MiRNA-143-5p, miRNA-143-3p, miRNA-145-5p, and let-7a-5pwere significantly downregulated in the SAT of morbidly obese women, and their expression levels were, respectively, 68% (p = 0.046), 61% (p = 0.016), 73% (p = 0.042), and 66% (p = 0.006) of those of normal-weight controls (Fig. 6a). These results were consistent with the findings from the Affymetrix GeneChip miRNA 4.0 Array.
Fig. 6.
Validation of differential miRNAs and predicted target genes. a Expression of 4 differential miRNAs, miR-143-5p, miR-143-3p, miR-145-5p, and let-7a-5p, was validated using qRT-PCR. b Expression of 5 predicted target genes, THY1, TARDBP, APPL1, PGM3, and HMGA2, was confirmed using qRT-PCR. Error bars indicate SEM (Student's t test, * p < 0.05 vs. normal-weight group).
The Validation of Predicted Target Gene Levels in the SAT of the Validation Groups
Five predicted target genes, namely THY1 (Thy-1 cell surface antigen, a predicted target gene of miRNA-143-5p), TARDBP (TAR DNA-binding protein, a predicted target gene of miRNA-143-3p), APPL1 (adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1, a predicted target gene of miRNA-145-5p), PGM3 (phosphoglucomutase 3, a predicted target gene of miRNA-145-5p), and HMGA2 (high-mobility group A2, a predicted target gene of let-7a-5p), were validated using qRT-PCR. Our results showed that HMGA2, a target gene of let-7a-5p, was 3.19-fold higher in the SAT of morbidly obese women than that of normal-weight controls (p = 0.025). Other predicted target genes, including THY1, TARDBP, APPL1 and PGM3, were not significantly different between the 2 groups (Fig. 6b). The relative expression levels and Pearson's correlation of the validated miRNAs and their predicted target genes were analyzed (online suppl. Fig. S2). The relative expression levels of let-7a-5p and HMGA2 presented a tendency of inverse correlation (r = −0.2) with no significant difference (p = 0.441) (online suppl. Fig. S2E), and there was no correlation between other microRNAs and predicted target genes.
Discussion
Our study first showed that 58 miRNAs were differentially expressed in the SAT of morbidly obese Chinese women, with 54 (93%) of them downregulated and 4 (7%) upregulated. Further analysis demonstrated that these differential miRNAs were related to 269 biological functions and 89 signaling pathways. The validation experiment showed that the expression levels of miRNA-143-5p, miRNA-143-3p, miRNA-145-5p, and let-7a-5p were downregulated in morbidly obese subjects, consistent with the microarray detection. HMGA2, a target gene of let-7a-5p, was first found to be 3.19-fold upregulated in the SAT of morbidly obese Chinese women.
According to our findings, the vast majority (93%) of the differential miRNAs were downregulated in the SAT of morbidly obese Chinese women. Likewise, other studies focusing on human abdominal SAT in insulin-resistant patients versus an insulin-sensitive population showed that 94% of the differentially expressed miRNAs were downregulated in the nanoString nCounter human v1 microRNA expression assay [36]. The studies performed by Zhao et al. [37] found that many more miRNAs were decreased than increased in mouser adipose tissue in response to obesity. Xie et al. [38] revealed that functionally important adipocyte miRNAs were downregulated in obesity because of the inflammatory environment, especially the tumor-necrosis factor (TNF)-α secreted by macrophages. MiRNAs induced during adipogenesis that accelerate fat cell development are downregulated in adipocytes from obese mice and in blood samples from obese patients [38, 39]. Some other studies also found the decreased expression of genes normally involved in adipogenesis in the fat pads of mice with obesity, although these genes normally increase during adipogenesis [40]. Our study was able to compare miRNA expression in the SAT of morbidly obese and normal-weight subjects. We found that most of the miRNAs, including the 4 validated miRNAs, were downregulated. These results suggest that obesity leads to a loss of miRNAs in the SAT, which may characterize fully differentiated and metabolically active adipose tissue [38].
Four of the differential miRNAs, namely miRNA-143-5p, miRNA-143-3p, miRNA-145-5p, and let-7a-5p, were selected to be verified in the expanded samples by the qRT-PCR method. These miRNAs are all highly abundant in adipose tissue [41, 42]. Our results demonstrated that these 4 miRNAs were all downregulated in morbidly obese women, consistent with the microarray detection. The selection of verified miRNAs was based on both our study and previous research [39, 43, 44]. On the one hand, our bioinformative analysis showed that miRNA-143-5p, miRNA-145-5p, and let-7a-5p were key miRNAs with strong regulatory capacities in the miRNA-gene-network, and that miRNA-143-5p, miRNA-143-3p, miRNA-145-5p, and let-7a-5p were located at the network center and involved in most biological functions in the miRNA-GO-network. On the other hand, previous research has proved that these 4 miRNAs participate in adipocyte differentiation, metabolic disorders, and cancer pathways [45, 46, 47, 48, 49].
It has been reported that miRNA-143 is one of the regulators of white adipocyte differentiation [46]. MiRNA-143 levels were increased in human and mouse 3T3-L1 differentiating preadipocytes, and the inhibition of miRNA-143 effectively inhibited the preadipocyte differentiation [46, 50, 51]. Takanabe et al. [52] demonstrated that the expression of miRNA-143 was upregulated in the mesenteric fat of high-fat diet (HFD)-induced obese mice, and that increased miRNA-143 expression was associated with elevated body weight and mesenteric fat mass. According to our analysis using human SAT, miRNA-143was downregulated in morbidly obese Chinses women. Further studies showed that free fatty acids, leptin, and resistin could inhibit miRNA-143 expression in human adipocytes [53]. Likewise, Viesti et al. [43] showed that miRNA-143displayed a reduced tendency in intact SAT in obese patients. As for plasma miRNA levels, Kilic et al. [39] found lower levels of plasma miRNA-143 in obese and morbidly obese patients compared to normal or overweight subjects. Besides, low levels of circulating miRNA-143 may be responsible for dyslipidemia in obesity [54].
Another miRNA related to cell differentiation and proliferation is miRNA-145. La Rocca et al. [47] demonstrated that miRNA-145 expression was increased during cell differentiation. A study to determine miRNA expression by microarray demonstrated that miRNA-145 expression was increased in differentiating human fat cells [20]. However, miRNA-145 expression is downregulated in human adipose tissue in insulin-resistant patients versus insulin-sensitive patients by the microarray method [36]. Viesti et al. [43] also found that miRNA-145 had a reduced tendency in the SAT in obese patients. There was a negative correlation between the expression of miRNA-145 and leptin receptors in the omentum of obese patients, indicating the metabolic dysregulation potential of miRNA-145 [43].
Let-7 is a well-conserved miRNA across species [55] and is upregulated during mouse adipocyte differentiation [45]. It proved to be a negative regulator of adipose tissue development by suppressing the adipogenesis of MSCs [56]. Let-7 transgenic mice were found to be thin and small with little white adipose tissue [57]. Let-7 also has substantial potential as a biomarker of metabolic disease. It is a potent regulator of glucose metabolism and peripheral insulin resistance [55]. When comparing plasma exosomes from obese diabetic patients naive to treatment and normal patients, let-7a was found to be lower in the obese cohort. Interestingly, after receiving antidiabetic treatment, let-7 levels increased in the diabetic patients to levels not significantly different from those of normal controls [58]. For the first time, we proved that the let-7a-5pexpression level in the SAT of obese Chinese patients was decreased, which indicated that the dysregulation of let-7a-5p in the SAT may participate in the development of obesity and its comorbidities.
We also validated the expression of 5 predicted target genes using qRT-PCR. Our study proved, for the first time, that HMGA2, a target gene of let-7a-5p, was 3.19-fold upregulated in the SAT of morbidly obese Chinese women. In previous studies, HMGA2 was confirmed to be a target gene of let-7a-5p, and the expression of HMGA2 was upregulated by arresting endogenous let-7a-5p [59]. In addition, the transfection of let-7 into 3T3-L1 cells could lead to a >3-fold decreased expression of HMGA2 [45]. Let-7 regulates adipogenesis through the repression of HMGA2 expression [56]. HMGA2 is a transcription factor which can colocalize with STAT3 and cooperatively promote adipogenesis [60]. Once HMGA2 is knocked down, the differentiation of 3T3-L1 preadipocytes would be inhibited [45]. Investigations have demonstrated that HMGA2 knockout mice have fewer fat cells, lower fat content, and are not susceptible to HFD-related secondary obesity [61]. In contrast, HMGA2-overexpressing mice are associated with increased body weight and a high level of body fat [62]. In overweight dogs, an elevated level of HMGA2 in SAT was noted [63]. Markowski et al. [64] found significantly higher HMGA2 expression in the SAT of obese Germany individuals than in nonobese patients. Furthermore, HMGA2 expression in patients with type 2 diabetes is significantly higher than in nondiabetic patients [64]. Obesity is not only a metabolic problem, but also a chronic inflammatory disease [65]. HMGA2 overexpression is accompanied by inflammatory changes in adipose tissues [62]. Previous literature has shown that adipose tissue macrophages are the primary sources of obesity-associated inflammation [66]. HMGA2 plays an essential role in macrophage activation, and it could enhance the expression of many proinflammatory cytokines, including TNF-α, interleukin (IL)-6, and IL-1β [67]. According to our laboratory findings, the level of circulating hs-CRP, a hepatic-derived acute-phase reactant stimulated by inflammation, was elevated to above the upper normal limit in obese subjects (Table 1; online suppl. Table S6), indicating that an inflammatory state existed in our obese patient cohort. Gentile et al. [68] also proved that high (>1.5 mg/L, median) hs-CRP is a major biochemical counterpart of increased adipose tissue mass. Taken together, our study proved that let-7a-5p decreased and its target gene HMGA2 increased in the SAT of obese patients; this might be what promotes adipogenesis in the early phase and contribute to a chronic low-grade inflammatory state in mature adipose tissue.
The MAPK pathway was, as a result, proven to be the most significantly changed one. Shi et al. [44] revealed that the MAPK pathway particularly contained meta-signature and adipogenic miRNAs. As an intracellular signaling pathway, the MAPK pathway, including the ERK, p38, and JNK pathways, plays a pivotal role in many essential cellular processes [44, 69]. It can regulate adipogenesis at each step of the process, from stem cells to adipocytes [69]. The inhibition of MAPK phosphorylation led to reduced weight gain in the mice with HFD-induced obesity, and this phenomenon was most evident by the suppression of fat accumulation in the abdomen [70]. Research into human obesity has drawn a similar conclusion. Childhood obesity has been proved to be associated with the MAPK signaling pathway [71]. The MAPK pathway can also regulate glucose metabolism, and its activation could lead to a high level of blood glucose [8]. By inhibiting the ERK/MAPK pathway, HFD-induced obesity, adipose tissue inflammation, metabolic abnormalities, and insulin resistance would be improved [72]. Due to its pleiotropic functions, the MAPK pathway (in this case, the most significantly changed pathway) represents a potential therapeutic target for morbid obesity and the resulting metabolic problems encountered by obese patients.
Obesity is an established risk and progression factor for many types of cancers (e.g., breast, colorectal, endometrial, prostate, esophageal), but the underlying mechanisms are incompletely understood [73]. The possible mechanism linking obesity to cancer involves alterations in the levels of adipokines and sex hormones [74]. Recently, in the literature, miRNA alterations have been linked to the initiation and the progression of human cancer, and the expression of miRNAs is being used as an important tool for cancer diagnosis, staging, progression, prognosis, and response to treatment [75, 76].
In our study, the 4 miRNAs chosen for validation all participate in tumor regulation. The miRNA-143/145 cluster has been described as having tumor-suppressive functions in several tumor types [48]. In miRNA-143/miRNA-145-deficient mice, active neoangiogenesis was reduced [77]. Let-7 miRNA was described as a negative regulator of the oncogenic family of Ras guanosine triphosphatases in both Caenorhabditis elegans and human tumor cell lines [49]. Let-7 expression deregulation was reported in several types of cancers [49]. By means of the KEGG analytical method, our study identified that 3 of the top 10 significant pathways were related to cancer. They were pathways in cancer, proteoglycans in cancer, and microRNAs in cancer. The “overlap” of cancer-related pathways and the differential pathways highlighted the importance of miRNA analyses in the tumor microenvironment in an obese state. Interestingly, many anticancer drugs are also beneficial for treating obesity, and vice versa [74]. Looking to the future, miRNA profiling might develop into a useful method for the assessment of and therapeutic approach towards obesity-linked cancers.
In addition to microarray and RT-qPCR based arrays, next-generation-sequencing (NGS) is the other available high-throughput method for miRNA profiling (miRNome). Compared to microarrays and PCR-based methods, deep-sequencing does not require predesigned probes, thereby allowing for the simultaneous discovery of new miRNAs and the confirmation of known miRNAs [78]. Mutations in miRNAs could also be identified by ultra-deep sequencing, even if these mutations occur in only a small fraction of the sample [79]. The NGS-based approach is becoming more and more popular and it has been used for the analysis of miRNomes in many tissues [80, 81, 82]. Kuryłowicz et al. [80] conducted NGS-based miRNome analysis in adipose tissue which characterized changes in miRNA profile resulting from weight loss. However, NGS technology applied so far to miRNA expression profiling has its limitations. Sample preparation involves many steps that can introduce biases and sequencing errors [83]. The computational tools for analysis are in their infancy [84]. Some NGS library preparation methods and the sequencing technology are not developed for short (<35 bp) sequences [78]. With the rapid increase in miRNAs being discovered and deposited in public databases, NGS can offer another comprehensive view of the miRNA transcriptome and provide a useful complement to microarray assays [84].
Although our study can be considered exploratory, there are some limitations. The number of subjects was small, especially for correlation analysis, which needs a larger sample (n ≥ 84) to achieve a reasonable correlation coefficient [85]. The research was cross-sectional, and we only validated target genes at the mRNA level. In the future, longitudinal cohort studies involving other ethnic populations are warranted. Nevertheless, our research has some strengths. First, by studying intact SAT, we were able to compare miRNA expression in a specific pathophysiological microenvironment. Moreover, the functional role and regulatory network of identified miRNAs were established by using the GO and KEGG bioinformatic tools. To our knowledge, this is the first comprehensive study of miRNAs in the SAT of obese Chinese women. We found that miRNA downregulation is a hallmark of intact SAT in a morbidly obese state. Transcription (DNA-dependent), small-molecule metabolic processes, the MAPK signaling pathway, and cancer-related pathways may play important roles in the occurrence and development of obesity. Our study also extended the understanding of miRNA expression and regulatory networks to obesity in Easterners. We proved, for the first time, that HMGA2 is upregulated in the SAT of morbidly obese Chinese women.
Statement of Ethics
Written informed consent was obtained from the participants to take part in this study. The study protocol was approved by the Ethics Committee of Peking Union Medical College Hospital, Beijing, China (No. JS-1093).
Conflict of Interest Statement
The authors have no conflicts of interest.
Funding Sources
The study was supported by grants from the Beijing Natural Science Foundation (No. 7182130 to F.G.), the China Diabetes Young Scientific Talent Research Project (2020-N-01-10 to L.W.), the National Natural Science Foundation of China (No. 81370898 to F.G. and No. 81471024 to H.Z.), the Non-Profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2017PT32020, No. 2018PT32001, and No. 2019PT320007), the National Key Program of Clinical Science (WBYZ2011-873 to F.G. and H.Z.), and the PUMCH Foundation (2013-020 to F.G.).
Author Contributions
F.G. and H.Z. conceived and designed the experiments. L.W., C.S., and H.Y. collected the clinical samples and performed the experiments. H.P. and F.G. analyzed the data and interpreted the results. C.S., H.Z., and F.G. wrote and revised the paper.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
We are grateful to all study participants and investigators.
References
- 1.Wollner M, Paulo Roberto BB, Alysson Roncally SC, Jurandir N, Edil LS. Accuracy of the WHO's body mass index cut-off points to measure gender- and age-specific obesity in middle-aged adults living in the city of Rio de Janeiro, Brazil. J Public Health Res. 2017 Sep;6((2)):904. doi: 10.4081/jphr.2017.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collaboration NC, NCD Risk Factor Collaboration (NCD-RisC) Trends in adult body-mass index in 200 countries from 1975 to 2014: a pooled analysis of 1698 population-based measurement studies with 19·2 million participants. Lancet. 2016 Apr;387((10026)):1377–96. doi: 10.1016/S0140-6736(16)30054-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietz WH, Solomon LS, Pronk N, Ziegenhorn SK, Standish M, Longjohn MM, et al. An Integrated Framework For The Prevention And Treatment Of Obesity And Its Related Chronic Diseases. Health Aff (Millwood) 2015 Sep;34((9)):1456–63. doi: 10.1377/hlthaff.2015.0371. [DOI] [PubMed] [Google Scholar]
- 4.Poirier P, Després JP. Exercise in weight management of obesity. Cardiol Clin. 2001 Aug;19((3)):459–70. doi: 10.1016/s0733-8651(05)70229-0. [DOI] [PubMed] [Google Scholar]
- 5.Martyniak K, Masternak MM. Changes in adipose tissue cellular composition during obesity and aging as a cause of metabolic dysregulation. Exp Gerontol. 2017 Aug;94:59–63. doi: 10.1016/j.exger.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coelho M, Oliveira T, Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Arch Med Sci. 2013 Apr;9((2)):191–200. doi: 10.5114/aoms.2013.33181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goossens GH. The Metabolic Phenotype in Obesity: Fat Mass, Body Fat Distribution, and Adipose Tissue Function. Obes Facts. 2017;10((3)):207–15. doi: 10.1159/000471488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClelland AD, Kantharidis P. microRNA in the development of diabetic complications. Clin Sci (Lond) 2014 Jan;126((2)):95–110. doi: 10.1042/CS20130079. [DOI] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009 Jan;19((1)):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008 Sep;455((7209)):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 11.Goguet-Rubio P, Klug RL, Sharma DL, Srikanthan K, Puri N, Lakhani VH, et al. Existence of a Strong Correlation of Biomarkers and miRNA in Females with Metabolic Syndrome and Obesity in a Population of West Virginia. Int J Med Sci. 2017 Apr;14((6)):543–53. doi: 10.7150/ijms.18988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008 Oct;18((10)):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Zeng X, Zhang X, Zou Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Brief Bioinform. 2016 Mar;17((2)):193–203. doi: 10.1093/bib/bbv033. [DOI] [PubMed] [Google Scholar]
- 14.Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015 May;11((5)):276–88. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 15.Ortega FJ, Mercader JM, Catalán V, Moreno-Navarrete JM, Pueyo N, Sabater M, et al. Targeting the circulating microRNA signature of obesity. Clin Chem. 2013 May;59((5)):781–92. doi: 10.1373/clinchem.2012.195776. [DOI] [PubMed] [Google Scholar]
- 16.Scheideler M. MicroRNAs in adipocyte formation and obesity. Best Pract Res Clin Endocrinol Metab. 2016 Oct;30((5)):653–64. doi: 10.1016/j.beem.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Oger F, Gheeraert C, Mogilenko D, Benomar Y, Molendi-Coste O, Bouchaert E, et al. Cell-specific dysregulation of microRNA expression in obese white adipose tissue. J Clin Endocrinol Metab. 2014 Aug;99((8)):2821–33. doi: 10.1210/jc.2013-4259. [DOI] [PubMed] [Google Scholar]
- 18.Engin AB. MicroRNA and Adipogenesis. Adv Exp Med Biol. 2017;960:489–509. doi: 10.1007/978-3-319-48382-5_21. [DOI] [PubMed] [Google Scholar]
- 19.Arner E, Mejhert N, Kulyté A, Balwierz PJ, Pachkov M, Cormont M, et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012 Aug;61((8)):1986–93. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, Ferrer A, et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One. 2010 Feb;5((2)):e9022. doi: 10.1371/journal.pone.0009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinelli R, Nardelli C, Pilone V, Buonomo T, Liguori R, Castanò I, et al. miR-519d overexpression is associated with human obesity. Obesity (Silver Spring) 2010 Nov;18((11)):2170–6. doi: 10.1038/oby.2009.474. [DOI] [PubMed] [Google Scholar]
- 22.Lorente-Cebrián S, González-Muniesa P, Milagro FI, Martínez JA. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: emerging roles as biomarkers and therapeutic targets. Clin Sci (Lond) 2019 Jan;133((1)):23–40. doi: 10.1042/CS20180890. [DOI] [PubMed] [Google Scholar]
- 23.Fricke A, Ullrich PV, Heinz J, Pfeifer D, Scholber J, Herget GW, et al. Identification of a blood-borne miRNA signature of synovial sarcoma. Mol Cancer. 2015 Aug;14((1)):151. doi: 10.1186/s12943-015-0424-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohrt-Nissen S, Døssing KB, Rossing M, Lajer C, Vikeså J, Nielsen FC, et al. Characterization of miRNA expression in human degenerative lumbar disks. Connect Tissue Res. 2013;54((3)):197–203. doi: 10.3109/03008207.2013.781594. [DOI] [PubMed] [Google Scholar]
- 25.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003 Dec;19((18)):2448–55. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 26.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990 Jul;9((7)):811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 27.Rajeevan MS, Vernon SD, Taysavang N, Unger ER. Validation of array-based gene expression profiles by real-time (kinetic) RT-PCR. J Mol Diagn. 2001 Feb;3((1)):26–31. doi: 10.1016/S1525-1578(10)60646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Kim J, Jiang X, Brunner SF, Ohno-Machado L., GAMUT GPU accelerated microRNA analysis to uncover target genes through CUDA-miRanda. BMC Med Genomics. 2014;7((Suppl 1)):S9. doi: 10.1186/1755-8794-7-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharya A, Ziebarth JD, Cui Y, Brunner SF, Ohno-Machado L. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014 Jan;42((Database issue)):D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang KH, Kao AP, Singh S, Yu SL, Kao LP, Tsai ZY, et al. Comparative expression profiles of mRNAs and microRNAs among human mesenchymal stem cells derived from breast, face, and abdominal adipose tissues. Kaohsiung J Med Sci. 2010 Mar;26((3)):113–22. doi: 10.1016/S1607-551X(10)70017-6. [DOI] [PubMed] [Google Scholar]
- 31.Ishida M, Shimabukuro M, Yagi S, Fukuda D, Soeki T, Sato H, et al. MicroRNA miR-378 regulates adipocytokine fate by targeting transcriptional factors in human visceral and subctaneous adipose tissue. European Heart Journal. 2013;34((suppl_1)) [Google Scholar]
- 32.Ishida M, Shimabukuro M, Yagi S, Fukuda D, Soeki T, Sato H, et al. MicroRNA-100 regulates a cluster of adipocytokine expression: A human biopsy study in subcutaneous and visceral adipose tissue. European Heart Journal. 2013;34((suppl_1)) [Google Scholar]
- 33.Dhar AK, Roux MM, Klimpel KR. Quantitative assay for measuring the Taura syndrome virus and yellow head virus load in shrimp by real-time RT-PCR using SYBR Green chemistry. J Virol Methods. 2002 Jun;104((1)):69–82. doi: 10.1016/S0166-0934(02)00042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei C, Li J, Bumgarner RE. Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics. 2004 Nov;5((1)):87. doi: 10.1186/1471-2164-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemeshow S, Hosmer DW, Klar J, Lwanga SK, Organization WH. Chichester: Wiley; 1990. Adequacy of sample size in health studies. [Google Scholar]
- 36.Kirby TJ, Walton RG, Finlin B, Zhu B, Unal R, Rasouli N, et al. Integrative mRNA-microRNA analyses reveal novel interactions related to insulin sensitivity in human adipose tissue. Physiol Genomics. 2016 Feb;48((2)):145–53. doi: 10.1152/physiolgenomics.00071.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao E, Keller MP, Rabaglia ME, Oler AT, Stapleton DS, Schueler KL, et al. Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mamm Genome. 2009 Aug;20((8)):476–85. doi: 10.1007/s00335-009-9217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009 May;58((5)):1050–7. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilic ID, Dodurga Y, Uludag B, Alihanoglu YI, Yildiz BS, Enli Y, et al. MicroRNA −143 and −223 in obesity. Gene. 2015 Apr;560((2)):140–2. doi: 10.1016/j.gene.2015.01.048. [DOI] [PubMed] [Google Scholar]
- 40.Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, Attie AD. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci USA. 2000 Oct;97((21)):11371–6. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ugras S, Brill E, Jacobsen A, Hafner M, Socci ND, Decarolis PL, et al. Small RNA sequencing and functional characterization reveals MicroRNA-143 tumor suppressor activity in liposarcoma. Cancer Res. 2011 Sep;71((17)):5659–69. doi: 10.1158/0008-5472.CAN-11-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G, Li Y, Li X, Ning X, Li M, Yang G. MicroRNA identity and abundance in developing swine adipose tissue as determined by Solexa sequencing. J Cell Biochem. 2011 May;112((5)):1318–28. doi: 10.1002/jcb.23045. [DOI] [PubMed] [Google Scholar]
- 43.Viesti A Collares R, Salgado W, Jr, Pretti da Cunha Tirapelli D, dos Santos JS. The expression of LEP, LEPR, IGF1 and IL10 in obesity and the relationship with microRNAs. PLoS One. 2014 Apr;9((4)):e93512. doi: 10.1371/journal.pone.0093512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi C, Huang F, Gu X, Zhang M, Wen J, Wang X, et al. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget. 2016 Jun;7((26)):40830–45. doi: 10.18632/oncotarget.8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009 Jun;23((6)):925–31. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004 Dec;279((50)):52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 47.La Rocca G, Shi B, Audia A, Ferrari-Amorotti G, Mellert HS, Calabretta B, et al. Regulation of microRNA-145 by growth arrest and differentiation. Exp Cell Res. 2011 Feb;317((4)):488–95. doi: 10.1016/j.yexcr.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Almeida MI, Calin GA. The miR-143/miR-145 cluster and the tumor microenvironment: unexpected roles. Genome Med. 2016 Mar;8((1)):29. doi: 10.1186/s13073-016-0284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jérôme T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Curr Genomics. 2007 Jun;8((4)):229–33. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Hou J, Ye L, Chen Y, Cui J, Tian W, et al. MicroRNA-143 regulates adipogenesis by modulating the MAP2K5-ERK5 signaling. Sci Rep. 2014 Jan;4((1)):3819. doi: 10.1038/srep03819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kajimoto K, Naraba H, Iwai N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA. 2006 Sep;12((9)):1626–32. doi: 10.1261/rna.7228806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H, et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun. 2008 Nov;376((4)):728–32. doi: 10.1016/j.bbrc.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 53.Zhu L, Shi C, Ji C, Xu G, Chen L, Yang L, et al. FFAs and adipokine-mediated regulation of hsa-miR-143 expression in human adipocytes. Mol Biol Rep. 2013 Oct;40((10)):5669–75. doi: 10.1007/s11033-013-2668-2. [DOI] [PubMed] [Google Scholar]
- 54.Can U, Buyukinan M, Yerlikaya FH. The investigation of circulating microRNAs associated with lipid metabolism in childhood obesity. Pediatr Obes. 2016 Jun;11((3)):228–34. doi: 10.1111/ijpo.12050. [DOI] [PubMed] [Google Scholar]
- 55.Deiuliis JA. MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int J Obes. 2016 Jan;40((1)):88–101. doi: 10.1038/ijo.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei J, Li H, Wang S, Li T, Fan J, Liang X, et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014 Jul;23((13)):1452–63. doi: 10.1089/scd.2013.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011 Dec;108((52)):21075–80. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santovito D, De Nardis V, Marcantonio P, Mandolini C, Paganelli C, Vitale E, et al. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: effect of glycemic control. J Clin Endocrinol Metab. 2014 Sep;99((9)):E1681–5. doi: 10.1210/jc.2013-3843. [DOI] [PubMed] [Google Scholar]
- 59.Wu WY, Tao SQ, Wang XN, Lobie PE, Wu ZS. XIAP 3′-untranslated region serves as a competitor for HMGA2 by arresting endogenous let-7a-5p in human hepatocellular carcinoma. Tumour Biol. 2017 Jul;39((7)):1010428317719578. doi: 10.1177/1010428317719578. [DOI] [PubMed] [Google Scholar]
- 60.Yuan Y, Xi Y, Chen J, Zhu P, Kang J, Zou Z, et al. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem Biophys Res Commun. 2017 Jan;482((4)):1360–6. doi: 10.1016/j.bbrc.2016.12.042. [DOI] [PubMed] [Google Scholar]
- 61.Berg JP. Pygmy mouse gene mutation protects against obesity. Eur J Endocrinol. 2000 Sep;143((3)):317–8. doi: 10.1530/eje.0.1430317. [DOI] [PubMed] [Google Scholar]
- 62.Arlotta P, Tai AK, Manfioletti G, Clifford C, Jay G, Ono SJ. Transgenic mice expressing a truncated form of the high mobility group I-C protein develop adiposity and an abnormally high prevalence of lipomas. J Biol Chem. 2000 May;275((19)):14394–400. doi: 10.1074/jbc.m000564200. [DOI] [PubMed] [Google Scholar]
- 63.Thies HW, Nolte I, Wenk H, Mertens F, Bullerdiek J, Markowski DN. Permanent activation of HMGA2 in lipomas mimics its temporal physiological activation linked to the gain of adipose tissue. Obesity (Silver Spring) 2014 Jan;22((1)):141–50. doi: 10.1002/oby.20137. [DOI] [PubMed] [Google Scholar]
- 64.Markowski DN, Thies HW, Gottlieb A, Wenk H, Wischnewsky M, Bullerdiek J. HMGA2 expression in white adipose tissue linking cellular senescence with diabetes. Genes Nutr. 2013 Sep;8((5)):449–56. doi: 10.1007/s12263-013-0354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest. 2017 Jan;127((1)):1–4. doi: 10.1172/JCI92035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeyda M, Stulnig TM. Obesity, inflammation, and insulin resistance—a mini-review. Gerontology. 2009;55((4)):379–86. doi: 10.1159/000212758. [DOI] [PubMed] [Google Scholar]
- 67.Huang H, Li H, Chen X, Yang Y, Li X, Li W, et al. HMGA2, a driver of inflammation, is associated with hypermethylation in acute liver injury. Toxicol Appl Pharmacol. 2017 Aug;328:34–45. doi: 10.1016/j.taap.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 68.Gentile M, Panico S, Rubba F, Mattiello A, Chiodini P, Jossa F, et al. Obesity, overweight, and weight gain over adult life are main determinants of elevated hs-CRP in a cohort of Mediterranean women. Eur J Clin Nutr. 2010 Aug;64((8)):873–8. doi: 10.1038/ejcn.2010.69. [DOI] [PubMed] [Google Scholar]
- 69.Bost F, Aouadi M, Caron L, Binétruy B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie. 2005 Jan;87((1)):51–6. doi: 10.1016/j.biochi.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 70.Poudel B, Lim SW, Ki HH, Nepali S, Lee YM, Kim DK. Dioscin inhibits adipogenesis through the AMPK/MAPK pathway in 3T3-L1 cells and modulates fat accumulation in obese mice. Int J Mol Med. 2014 Nov;34((5)):1401–8. doi: 10.3892/ijmm.2014.1921. [DOI] [PubMed] [Google Scholar]
- 71.Chen R, Xin G, Zhang X. Long non-coding RNA HCP5 serves as a ceRNA sponging miR-17-5p and miR-27a/b to regulate the pathogenesis of childhood obesity via the MAPK signaling pathway. J Pediatr Endocrinol Metab. 2019 Dec;32((12)):1327–39. doi: 10.1515/jpem-2018-0432. [DOI] [PubMed] [Google Scholar]
- 72.Ohkura T, Yoshimura T, Fujisawa M, Ohara T, Marutani R, Usami K, et al. Spred2 Regulates High Fat Diet-Induced Adipose Tissue Inflammation, and Metabolic Abnormalities in Mice. Front Immunol. 2019 Jan;10:17. doi: 10.3389/fimmu.2019.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Himbert C, Delphan M, Scherer D, Bowers LW, Hursting S, Ulrich CM. Signals from the Adipose Microenvironment and the Obesity-Cancer Link-A Systematic Review. Cancer Prev Res (Phila) 2017 Sep;10((9)):494–506. doi: 10.1158/1940-6207.CAPR-16-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teoh SL, Das S. Tumour biology of obesity-related cancers: understanding the molecular concept for better diagnosis and treatment. Tumour Biol. 2016 Nov;37((11)):14363–80. doi: 10.1007/s13277-016-5357-7. [DOI] [PubMed] [Google Scholar]
- 75.Wang H. Predicting MicroRNA Biomarkers for Cancer Using Phylogenetic Tree and Microarray Analysis. Int J Mol Sci. 2016 May;17((5)):E773. doi: 10.3390/ijms17050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006 Nov;6((11)):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 77.Dimitrova N, Gocheva V, Bhutkar A, Resnick R, Jong RM, Miller KM, et al. Stromal Expression of miR-143/145 Promotes Neoangiogenesis in Lung Cancer Development. Cancer Discov. 2016 Feb;6((2)):188–201. doi: 10.1158/2159-8290.CD-15-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Darvasi O, Szabo PM, Nemeth K, Szabo K, Spisak S, Liko I, et al. Limitations of high throughput methods for miRNA expression profiles in non-functioning pituitary adenomas. Pathol Oncol Res. 2019 Jan;25((1)):169–82. doi: 10.1007/s12253-017-0330-3. [DOI] [PubMed] [Google Scholar]
- 79.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012 Apr;13((5)):358–69. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuryłowicz A, Wicik Z, Owczarz M, Jonas MI, Kotlarek M, Świerniak M, et al. NGS Reveals Molecular Pathways Affected by Obesity and Weight Loss-Related Changes in miRNA Levels in Adipose Tissue. Int J Mol Sci. 2017 Dec;19((1)):E66. doi: 10.3390/ijms19010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swierniak M, Wojcicka A, Czetwertynska M, Stachlewska E, Maciag M, Wiechno W, et al. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013 Aug;98((8)):E1401–9. doi: 10.1210/jc.2013-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wojcicka A, Swierniak M, Kornasiewicz O, Gierlikowski W, Maciag M, Kolanowska M, et al. Next generation sequencing reveals microRNA isoforms in liver cirrhosis and hepatocellular carcinoma. Int J Biochem Cell Biol. 2014 Aug;53:208–17. doi: 10.1016/j.biocel.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 83.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009 Sep;10((5)):490–7. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tam S, de Borja R, Tsao MS, McPherson JD. Robust global microRNA expression profiling using next-generation sequencing technologies. Lab Invest. 2014 Mar;94((3)):350–8. doi: 10.1038/labinvest.2013.157. [DOI] [PubMed] [Google Scholar]
- 85.Bujang MA, Baharum N. Sample Size Guideline for Correlation Analysis. World Journal of Social Science Research. 2016;3((1)):37. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data