Abstract
Introduction
DNA methylation constitutes one important epigenetic mechanism that regulates gene expression in human cells. With regard to obesity, bariatric surgery-induced weight loss has been associated with promoter methylation changes in several genes. Hyperleptinemia is a characteristic feature of obesity. The underlying regulating mechanisms have not yet been completely elucidated.
Methods
We investigated the methylation of the promoters of the leptin gene (LEP) and the leptin receptor gene (LEPR) as well as leptin expression in pre- and postbariatric surgery patients using a comparative cross-sectional design.
Results
Our results revealed significantly higher LEP promoter methylation patterns in prebariatric surgery patients compared to postoperatively. DNA methylation of the LEPR promoter was significantly higher in the postoperative group. Moreover, we found significantly higher leptin serum levels in patients before the bariatric surgery than afterwards.
Discussion
These findings strengthen the suggestion that there is an association between LEP expression and LEP methylation in obesity. We suggest that the epigenetic profile of LEP might be influenced by leptin serum levels in the form of a regulating feedback mechanism.
Keywords: LEP promoter methylation, LEPR promoter methylation, Leptin, Bariatric surgery, Obesity
Introduction
Obesity is a chronic disease that constitutes one of the most serious global health burdens, and its occurrence is steadily increasing around the world [1, 2]. Obesity is a major public health problem since it is associated with serious consequences including type 2 diabetes mellitus, hypertension, and cardiovascular disease [3]. Moreover, it lessens life expectancy markedly, especially in younger age groups [4]. The imbalance between energy intake and energy expenditure leads to an excess of body fat which is regulated by the interaction of environmental and genetic factors [5].
Genes are known to adapt to environmental influences [6]. Epigenetic modification is one molecular mechanism regulating gene expression without affecting the DNA sequence [7]. In this context, DNA methylation constitutes an important epigenetic modification that regulates gene expression in humans [8]. DNA methylation occurs mainly within the CpG nucleotides which appear grouped in clusters called “CpG islands”; it includes the addition of a methyl group to the 5′ position of the cytosine pyrimidine ring [9]. CpG methylation is known to be affected by different environmental influencing factors [10, 11, 12].
The adipose-derived hormone, leptin (encoded by LEP) is substantially involved in the regulation of energy balance and body weight [13]. Its expression is markedly influenced by the proportion of body adipose mass, indicating that hyperleptinemia is a typical feature of obesity, and it is assumed that there is a resistance to the action of leptin in severely overweight subjects [14].
The findings of different studies suggest LEP methylation to be involved in the pathophysiology of obesity. Leptin DNA methylation levels have been found to be negatively correlated with body mass index (BMI) in obese individuals [15]. Lower methylation frequencies of the LEP promoter have been reported in obese adolescents with insulin resistance [16].
An association between LEP promoter methylation and leptin expression has been studied in vitro during adipose conversion [17]. In this investigation, a reduction in methylation levels was detected coming from human preadipocytes (which do not express leptin) in their conversion to adipocytes expressing leptin, suggesting that the methylation of the leptin promoter region plays an important role in leptin expression.
Another study investigating methylation-dependent mechanisms in leptin expression found a cell type-specific inverse relation between the methylation status of the LEP promoter and leptin expression in obese individuals who had maintained a stable body weight in the previous 12 months [18]. The longitudinal investigation studied obese individuals before and after bariatric surgery-induced weight loss and found a decrease in white-adipose-tissue leptin expression, although, interestingly, this did not correspond with changes in the promoter methylation status. The authors concluded that methylation density in the LEP promoter represents a control mechanism for cell type-specific leptin expression, while changes in leptin expression due to weight loss may not be methylation-dependent.
Assuming that methylation patterns might regulate gene expression, we investigated LEP and LEPR promoter methylation as well as leptin serum levels in obese patients. We expected to find significant differences in LEP and LEPR promoter methylation patterns and leptin expression between extremely obese patients prior to bariatric surgery and patients who lost weight after bariatric surgery.
Material and Methods
Study Design
This was part of a larger cross-sectional study investigating physical activity and cognitive performance in bariatric surgery patients [19, 20]. Data were obtained between March 2013 and August 2014. All investigated subjects were recruited at the routine preoperative evaluation at Hannover Medical School or the postoperative evaluation at Herzogin Elisabeth Hospital Braunschweig, Brunswick, Germany. The investigation was conducted as a cross-sectional design with 2 study groups: preoperative bariatric surgery candidates versus postoperative bariatric surgery patients who showed a relevant excess weight loss after the intervention which was conducted via Roux-en-Y gastric bypass (RYGB) or laparoscopic sleeve gastrectomy. To ensure that the postsurgery patients had undergone substantial metabolic changes associated with weight loss, the study protocol included only patients with at least 40% excess weight loss after intervention.
The analysis included 98 patients (24 males and 74 females). The 2 study groups were well matched concerning age, sex, and presurgery BMI (Table 1). Main inclusion criteria were preoperative obesity grade 2 or 3 (BMI ≥35.0) and an age of 18–65 years. A history of previous bariatric surgery, current substance abuse, intellectual disability, psychosis, neurological disorders (e.g., multiple sclerosis and stroke), dementia, a history of severe head trauma, and insufficient German language skills were defined as the exclusion criteria.
Table 1.
Demographic characteristics
| Before surgery |
After surgery |
||||||
|---|---|---|---|---|---|---|---|
| n | n | χ2 | p value | ||||
| Gender | |||||||
| Female | 35 | 39 | 0.053 | 0.818 | |||
| Male | 12 | 12 | |||||
| Type of intervention | |||||||
| Gastric bypass | 45 | ||||||
| Sleeve | 6 | ||||||
| Mean | SD | Mean | SD | t | df | p value | |
|---|---|---|---|---|---|---|---|
| Age, years | 45.04 | 10.67 | 41.67 | 11.27 | 1.51 | 96.00 | 0.13 |
| BMI at assessment | 46.92 | 7.42 | 32.21 | 4.43 | 11.79 | 73.91 | <0.001 |
| BMI before surgery | 46.92 | 7.42 | 47.80 | 7.02 | 0.61 | 96.00 | 0.55 |
| Weeks after surgery | 30.80 | 12.34 | |||||
| Leptin, ng/mL | 32.69 | 12.08 | 10.25 | 9.18 | 10.40 | 96.00 | <0.001 |
| LEP methylation, % | 37.97 | 13.27 | 14.73 | 13.27 | 8.58 | 94.00 | <0.001 |
| LEPR methylation, % | 7.83 | 3.29 | 8.79 | 3.50 | 1.39 | 96.00 | 0.17 |
In the pre- and postoperative participants, taking blood was scheduled after the routine pre- and postoperative evaluation between 8:00 and 10:00 a.m. All blood samples were centrifuged and stored at −20°C immediately after collection and later transferred to a −80°C storage. Leptin serum levels were measured using a commercial enzyme-linked immunosorbent assay (ELISA) kit (HÖLZEL leptin ELISA kit, Hölzel Biotech, Cologne, Germany) according to the manufacturer's description (assay range 1–100 ng/mL, intra-assay coefficients of variation [CV] approx. 6%, interassay CV approx. 10%).
Evaluation of Gene Promoter Methylation
LEP and LEPR promoter methylation were measured in both study groups. We used the MN Nucleo Mag Blood 200-μL Kit (ref. 744501.4, Macherey Nagel, Düren, Germany) for genomic DNA extraction. Genomic DNA was modified with sodium-bisulfite using the EpiTect® bisulfite kit (Qiagen AG, Hilden, Germany) to deaminate cytosines in CpG dinucleotides to uracils without modifying methylated CpG islands.
Primers were designed to amplify a region covering a fragment of 703 base pairs (bp) for LEP (from −572 bp prior to the starting base pair of exon 1 of leptin to +131 bp following the first bp of the leptin exon) containing 62 CpG sites within the 5′ upstream promoter region of LEP and 460 bp for LEPR (from −234 to +226 bp). As described before [21], we performed a nested PCR for amplification. Conditions of the PCR are described in the online supplementary material (see www.karger.com/doi/10.1159/000511918) in detail.
The automatic clean-up of amplification products was done with Biomek® NXP using Agencourt® AmPure® XP magnetic beads (Beckman Coulter, A63881).
We used a standard 2.0% agarose gel to visualize the PCR product to measure the product size. A BigDye® Terminator v3.1 cycle sequencing kit and 3500xl genetic analyzer (both from Applied Biosystems, Foster City, CA, USA) were employed for sequencing.
We applied the Epigenetic Sequencing Methylation analysis software (ESME) package to analyze the obtained sequences. ESME identifies the DNA methylation status based on the sequence trace files. Moreover, it normalizes the signals, ensures quality control, corrects for incomplete bisulfite conversion, and aligns the generated bisulfite sequence and reference sequence to compare C to T values (forward sequence) and G to A values (reverse sequence) peaks at CpG sites.
Statistical Analyses
The variables did not deviate from the normal distribution according to the Kolmogorov-Smirnov test, so parametric methods were applied. All data are given as mean (SD). All statistical tests were 2-tailed, a significance level of α = ≤0.05 was considered significant. The confidence interval (CI) and p value of post hoc tests were corrected for multiple comparisons according to the method of Bonferroni. Differences between the pre- and postbariatric group were analyzed using Student's t test, Fisher's exact test, or the χ2 test. For analyzing DNA methylation differences between groups, we performed linear mixed models using a restricted estimated maximum likelihood approach with the 2 hierarchical levels of subjects and CpG sites within subjects. DNA methylation of LEP or LEPR were set as the dependent variable, and group and CpG position were computed as fixed factors. We also included CpG × group interaction in the model, to test for evenly distributed effects across the promoter region. Age and gender were used as covariates in the model. Correlations between mean promoter methylation and leptin serum levels were calculated using Pearson's test. Multiple linear regression analyses were used to further validate correlative findings. Statistical analysis was performed employing SPSSTM for Windows v21.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 6 for Windows (GraphPad Software, Inc., San Diego, CA, USA).
Results
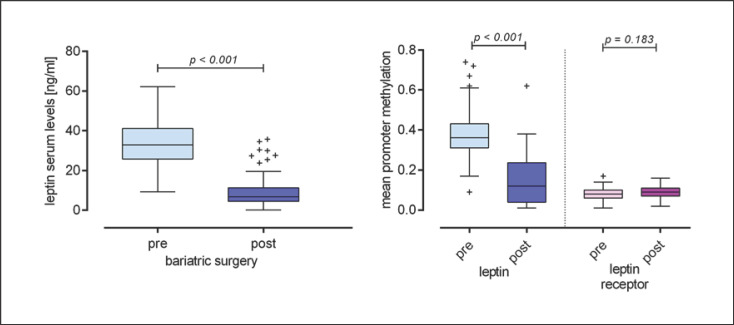
Demographic characteristics of the study population are given in Table 1. Patients after bariatric surgery had significantly lower serum levels of leptin (t[97] = 10.47; p < 0.001; Fig. 1). DNA methylation of LEP promoter was significantly lower in postbariatric patients (fixed effect: F[1;5, 973] = 1,326.3; p < 0.001) while the CpG position had no influence (F[64; 5, 973] = 0.51; p = 0.99) and no CpG × group interaction occurred (F[64; 5, 973] = 0.57; p = 0.998).
Fig. 1.
Difference in leptin serum levels in the pre- and post-bariatric surgery groups; difference in the mean promoter methylation of leptin and leptin receptor in the pre- and post-bariatric surgery groups.
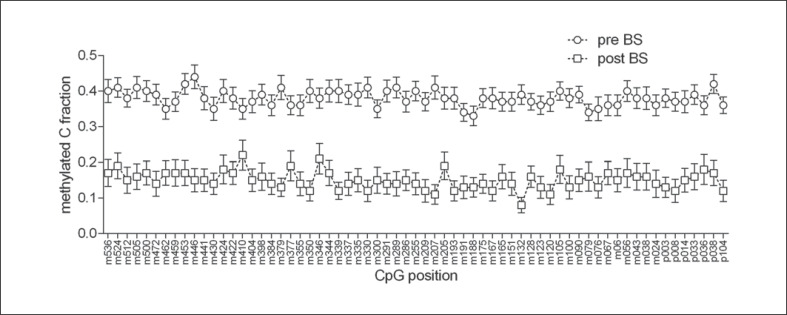
Figure 2 shows the LEP promotor methylation profile with regard to the specific CpG positions in the pre- and postbariatric groups. Gender had an independent main effect on DNA methylation (F[1; 5,973] = 31.14; p < 0.001), with male participants showing slightly lower methylation levels (estimated marginal mean [EMM] 0.237 and standard error [SE] 0.006 vs. female participants EMM 0.273 and SE 0.003) while no group × gender interaction was observed (F[1; 5, 973] = 0.634; p = 0.43). Age did not affect the methylation (F[1; 5, 937] = 1.53; p = 0.22). Comparing mean methylation between groups by Student's t test, a significant difference between pre- and postbariatric surgery patients was also observed (t[95] = 8.59; p < 0.001; Fig. 1).
Fig. 2.
LEP promotor methylation profile with regard to the specific CpG positions in the pre- and post-bariatric groups. pre-BS, study group before undergoing bariatric surgery; post-BS, study group after undergoing bariatric surgery.
DNA methylation of LEPR was marginally, but significantly, higher in the postsurgery group (F[1; 3, 536] = 7.27; p = 0.007). No CpG × group interaction was observed (F[37; 3, 536] = 0.82; p = 0.78) even though we found a significant main effect of the CpG position (F[37; 3, 536] = 55.38; p < 0.001). Age had a significant effect on DNA methylation (F[1; 3, 536] = 18.12; p < 0.001) while gender did not show any effect (F[1; 3, 536] = 0.78; p = 0.38). Mean methylation of LEPR was significantly correlated with age (Pearson's r = 0.288; p = 0.004). Comparing mean methylation between groups by Student's t test, no significant difference was observed for LEPR (t[97] = 1.34; p = 0.184; Fig. 1).
We found significant correlations between leptin serum levels and the methylation of both genes. Mean methylation of LEP was positively associated with leptin levels (Pearson's r = 0.571; p < 0.001), while a negative association was found between serum levels and LEPR methylation (Pearson's r = −0.238; p = 0.018). Mean methylation of both genes was not correlated (Pearson's r = 0.078; p = 0.45). We performed multiple linear regression analysis to test whether the observed association was merely driven by the group (pre- vs. postoperative), and found that mean methylation of both genes and also group were independently associated with leptin serum levels (whole model: R2adj. = 0.57; F[3] = 42.38; p < 0.001; group: β = −0.562; t = −6.01; p < 0.001; LEP methylation: β = 0.221; t = 2.39; p = 0.019; LEPR methylation: β = −0.181; t = −2.59; p = 0.011). No significant partial correlation between leptin or leptin receptor methylation and BMI at assessment was observed when controlling for group (data not shown).
Discussion
DNA methylation patterns, modified by environmental factors, have been suggested to be involved in the pathophysiological mechanisms of obesity and related disorders [15, 16, 22, 23]. The impact of LEP promoter methylation on leptin expression in humans has not completely been elucidated up to now. Some studies conclude that this epigenetic mechanism might act as a tool for regulating leptin expression. It has been suggested that, in most cases, DNA methylation exerts a repressive effect on gene expression [24]. Our study group was able to show that in alcohol-dependent patients undergoing withdrawal, low LEP promoter methylation is associated with increasing serum leptin levels and an increased craving for alcohol [10]. Another investigation into diet-induced obese rats revealed significantly higher leptin promoter methylation levels at one of the studied CpG sites than in the normal-weight control rats [22]. This high methylation status was associated with lower circulating leptin levels. Melzner et al. [17], an in vitro investigation of maturing human (pre-) adipocytes, found increased leptin expression to be associated with reduced methylation of the promoter region. In this context, they concluded that demethylation at distinct CpG sites in the LEP promoter of human adipocytes is required for its expression.
At a glance, our results are not in line with the above finding that a demethylation pattern of the LEP promoter is associated with higher leptin expression. Our cross-sectional study revealed significantly higher LEP methylation patterns at all investigated CpG sites in the patients preoperatively when compared to postoperatively (with an excess weight loss of at least 40% after surgery). We found significantly higher leptin serum levels in pre- versus postoperative patients. Additionally, DNA methylation of LEPR was marginally but significantly higher in the postsurgery group. These results are in contrast to those of a recent study which analyzed leptin expression longitudinally in 8 patients before and after bariatric surgery [18]. The authors found significantly decreased leptin levels postoperatively in bariatric surgery patients but no changes in the corresponding methylation pattern and concluded that weight loss-induced changes in leptin expression are not methylation-dependent. Negating a dynamic regulation of leptin levels in white adipose tissue through methylation patterns in patients undergoing gastric surgery-induced weight loss, they described a tissue-specific inverse correlation between methylation status and leptin expression in patients under stable weight conditions. According to the authors, these findings imply that steady-state and dynamic-state leptin expression are differently regulated.
One limitation of our study was the use of a bisulfite sequencing method which relies on the chemical modification of unmethylated cytosines and PCR, which may have introduced a relevant bias into the measurements. Our research group has used this method (bisulfite treatment followed by direct cycle capillary sequencing) in several studies that yielded valid and reproducible results, but all bisulfite-based methods (e.g., pyrosequencing and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry [MALDI-TOF]) are flawed by the relevant DNA degradation due to this chemical reaction. Future studies should therefore employ methods with direct measurement of methylation, e.g., target enrichment and nanopore sequencing. Another limitation was the cross-sectional study design (rather than longitudinal). Being aware that DNA methylation is affected by cell-type heterogeneity, genetic variants, and aging, it will be necessary to review the results by means of a longitudinal analysis, and, moreover, by analyzing the methylation of a third gene and also a control group in future studies.
We displayed a causal relation between LEP promoter methylation patterns and leptin serum levels in our patients' sample. Even though there are other mechanisms that influence leptin expression in humans, like the function of the androgen receptor as a transcription factor [25], the finding of significantly higher leptin levels in extremely obese patients prior to bariatric surgery is in line with recent data showing that leptin expression is substantially determined by obesity and related conditions. An increase in adipose mass leads to significant leptin induction while leptin expression is reduced during weight loss [14]. There are 2 different ways in which our results could further be interpreted. As stated above, leptin expression, among other mechanisms, might be regulated by the methylation profile of its promoter. Therefore, our results might hint towards a positive association between methylation patterns of the LEP promoter and leptin expression, with the high methylation status in obese patients probably resulting in leptin induction. Taking into account that most of the studies we have cited found an opposite effect, in fact, an inverse relation between LEP methylation status and leptin expression, this interpretation of our results does not seem very likely even though we studied promoter methylation in blood cells and not adipose tissue.
Assuming that hypermethylation of the LEP promoter leads to decreased leptin expression, the other more probable interpretation of our findings is that the well-known condition of elevated leptin levels in obese subjects leads to attempts to counterregulate its epigenetic profile. We thus suggest that the epigenetic pattern of LEP might be influenced by leptin serum levels, in the form of a regulating feedback mechanism.
A weakness of our study is that we investigated blood cells instead of metabolically active adipose tissue since leptin is mainly produced in adipocytes. Therefore, it might be assumed that a correlation between leptin serum levels and LEP promoter methylation in the blood cells may not be associated with obesity or weight loss via bariatric surgery, but rather (for example) to alterations in blood cell types. In this context, 2 recent studies by Houde et al. [15, 26] appear to be of great interest. The objective of the first study was to compare LEP epigenetic profiles in the blood as well as subcutaneous and visceral adipose tissue samples [26]. They found that LEP methylation levels at some of the investigated CpG sites (different from those in our study) were significantly correlated in the blood and subcutaneous adipose tissue and partially significantly correlated in the blood and visceral adipose tissue. In their study mean LEP methylation was also correlated in the visceral adipose tissue and the blood. This might lead to the conclusion that blood cells may reflect the DNA methylation profile of metabolically active tissues and can eventually serve as a biomarker in this context. Their other investigation showed an association between LDL-C levels and LEP methylation, suggesting that LDL-C might regulate its epigenetic profile in adipose tissues. Interestingly, similar correlations were found between LDL-C and LEP methylation in blood cells, suggesting a common regulatory pathway for DNA methylation in the blood and adipose tissues [15].
Conclusions
Our results suggest that bariatric surgery-induced weight loss is associated with changes in leptin serum levels as well as with LEP and LEPR promoter methylation, and, moreover, that there is an association between LEP expression and LEP methylation. We hypothesize that the epigenetic profile of LEP might be influenced by leptin serum levels via a regulating feedback mechanism. In all respects, further and particularly longitudinal investigations, that also consider probable tissue-specific features, are needed to precisely interpret our results and elucidate the complex relationship between epigenetic mechanisms and gene expression in metabolic pathways.
Statement of Ethics
The study protocol was approved by the institute's committee on human research (number of the Ethics Committee vote: 6318). All study participants gave written informed consent for the participation according to procedures approved by the Institutional Ethics Committee.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
The main study investigating physical activity and cognitive performance in bariatric surgery patients was supported by a grant of the Federal Ministry of Education and Research within the German Obesity Competence Network (01GI1323). The funder had no role in study design, collection, analysis and interpretation of data, writing of the report, or the decision to submit the article for publication.
Author Contributions
J.W., A.B., A.M., H.K., and H.F. were responsible for study concept and design. A.B., V.B., S.A.A., K.G.-L., and R.L. performed the analysis and interpretation of data. J.W., T.H., and H.F. performed the statistical analysis. J.W., A.B., and H.F. wrote the first draft and A.M., K.G.K., M.Z., A.G., K.J., T.H., and S.B. critically revised the manuscript. All authors critically reviewed content and approved the final version for publication.
Supplementary Material
Supplementary data
References
- 1.Imes CC, Burke LE. The Obesity Epidemic: The United States as a Cautionary Tale for the Rest of the World. Curr Epidemiol Rep. 2014 Jun;1((2)):82–8. doi: 10.1007/s40471-014-0012-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2014 Aug;384((9945)):766–81. doi: 10.1016/S0140-6736(14)60460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hruby A, Manson JE, Qi L, Malik VS, Rimm EB, Sun Q, et al. Determinants and Consequences of Obesity. Am J Public Health. 2016 Sep;106((9)):1656–62. doi: 10.2105/AJPH.2016.303326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fontaine KR, Redden DT, Wang C, Westfall AO, Allison DB. Years of life lost due to obesity. JAMA. 2003 Jan;289((2)):187–93. doi: 10.1001/jama.289.2.187. [DOI] [PubMed] [Google Scholar]
- 5.O'Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009 Nov;462((7271)):307–14. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 6.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003 Mar;33((S3 Suppl)):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 7.Henikoff S, Matzke MA. Exploring and explaining epigenetic effects. Trends Genet. 1997 Aug;13((8)):293–5. doi: 10.1016/s0168-9525(97)01219-5. [DOI] [PubMed] [Google Scholar]
- 8.Kouzarides T. Chromatin modifications and their function. Cell. 2007 Feb;128((4)):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, et al. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005 Nov;33((20)):e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillemacher T, Weinland C, Lenz B, Kraus T, Heberlein A, Glahn A, et al. DNA methylation of the LEP gene is associated with craving during alcohol withdrawal. Psychoneuroendocrinology. 2015 Jan;51:371–7. doi: 10.1016/j.psyneuen.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Houde AA, Guay SP, Desgagné V, Hivert MF, Baillargeon JP, St-Pierre J, et al. Adaptations of placental and cord blood ABCA1 DNA methylation profile to maternal metabolic status. Epigenetics. 2013 Dec;8((12)):1289–302. doi: 10.4161/epi.26554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling C, Rönn T. Epigenetic adaptation to regular exercise in humans. Drug Discov Today. 2014 Jul;19((7)):1015–8. doi: 10.1016/j.drudis.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Sáinz N, Barrenetxe J, Moreno-Aliaga MJ, Martínez JA. Leptin resistance and diet-induced obesity: central and peripheral actions of leptin. Metabolism. 2015 Jan;64((1)):35–46. doi: 10.1016/j.metabol.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 14.Maffei M, Halaas J, Ravussin E, Pratley RE, Lee GH, Zhang Y, et al. Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat Med. 1995 Nov;1((11)):1155–61. doi: 10.1038/nm1195-1155. [DOI] [PubMed] [Google Scholar]
- 15.Houde AA, Légaré C, Biron S, Lescelleur O, Biertho L, Marceau S, et al. Leptin and adiponectin DNA methylation levels in adipose tissues and blood cells are associated with BMI, waist girth and LDL-cholesterol levels in severely obese men and women. BMC Med Genet. 2015 May;16((1)):29. doi: 10.1186/s12881-015-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Cardona MC, Huang F, García-Vivas JM, López-Camarillo C, Del Río Navarro BE, Navarro Olivos E, et al. DNA methylation of leptin and adiponectin promoters in children is reduced by the combined presence of obesity and insulin resistance. Int J Obes. 2014 Nov;38((11)):1457–65. doi: 10.1038/ijo.2014.30. [DOI] [PubMed] [Google Scholar]
- 17.Melzner I, Scott V, Dorsch K, Fischer P, Wabitsch M, Brüderlein S, et al. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J Biol Chem. 2002 Nov;277((47)):45420–7. doi: 10.1074/jbc.M208511200. [DOI] [PubMed] [Google Scholar]
- 18.Marchi M, Lisi S, Curcio M, Barbuti S, Piaggi P, Ceccarini G, et al. Human leptin tissue distribution, but not weight loss-dependent change in expression, is associated with methylation of its promoter. Epigenetics. 2011 Oct;6((10)):1198–206. doi: 10.4161/epi.6.10.16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartsch M, Langenberg S, Gruner-Labitzke K, Schulze M, Köhler H, Crosby RD, et al. Physical Activity, Decision-Making Abilities, and Eating Disturbances in Pre- and Postbariatric Surgery Patients. Obes Surg. 2016 Dec;26((12)):2913–22. doi: 10.1007/s11695-016-2208-y. [DOI] [PubMed] [Google Scholar]
- 20.Langenberg S, Schulze M, Bartsch M, Gruner-Labitzke K, Pek C, Köhler H, et al. Physical activity is unrelated to cognitive performance in pre-bariatric surgery patients. J Psychosom Res. 2015 Aug;79((2)):165–70. doi: 10.1016/j.jpsychores.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Muschler MA, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. J Neural Transm (Vienna) 2010 Apr;117((4)):513–9. doi: 10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- 22.Milagro FI, Campión J, García-Díaz DF, Goyenechea E, Paternain L, Martínez JA. High fat diet-induced obesity modifies the methylation pattern of leptin promoter in rats. J Physiol Biochem. 2009 Mar;65((1)):1–9. doi: 10.1007/BF03165964. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson EK, Ernst B, Voisin S, Almén MS, Benedict C, Mwinyi J, et al. Roux-en Y gastric bypass surgery induces genome-wide promoter-specific changes in DNA methylation in whole blood of obese patients. PLoS One. 2015 Feb;10((2)):e0115186. doi: 10.1371/journal.pone.0115186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986 May;321((6067)):209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 25.Lenz B, Frieling H, Jacob C, Heberlein A, Kornhuber J, Bleich S, et al. The modulating effect of the androgen receptor on craving in alcohol withdrawal of men is partially mediated by leptin. Pharmacogenomics J. 2010 Jun;10((3)):226–31. doi: 10.1038/tpj.2009.56. [DOI] [PubMed] [Google Scholar]
- 26.Houde AA, Légaré C, Hould FS, Lebel S, Marceau P, Tchernof A, et al. Cross-tissue comparisons of leptin and adiponectin: DNA methylation profiles. Adipocyte. 2014 Apr;3((2)):132–40. doi: 10.4161/adip.28308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data