Abstract
Total hip arthroplasty (THA) is a cost-effective treatment for osteoarthritis (OA), and osteolysis is a common complication of THA. This study was aimed at exploring the relevant molecular biomarkers for osteolysis after THA. We performed RNA sequence to identify and characterize expressed mRNAs and lncRNAs in OA and osteolysis. Differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs) in OA and osteolysis were acquired, as well as shared DEmRNAs/DElncRNAs in OA and osteolysis and osteolysis-specific DEmRNAs/DElncRNAs. Then, shared and osteolysis-specific DElncRNA-DEmRNA coexpression networks were constructed to further investigate the function of DElncRNAs and DEmRNAs in OA and osteolysis. In total, 343 DEmRNAs and 25 DElncRNAs in OA, 908 DEmRNAs and 107 DElncRNAs in osteolysis, and 406 DEmRNAs and 46 DElncRNAs between OA and osteolysis were acquired. A total of 136 shared DEmRNAs and 9 shared DElncRNAs in OA and osteolysis and 736 osteolysis-specific DEmRNAs and 103 osteolysis-specific DElncRNAs were acquired. Then, 128 shared DElncRNA-DEmRNA coexpression pairs and 522 osteolysis-specific DElncRNA-DEmRNA coexpression pairs were identified. The present study highlighted the roles of four interaction pairs, including two shared lncRNA-mRNA interaction pairs in OA and osteolysis (AC111000.4 and AC016831.6), which may function in the immune process of OA and osteolysis by regulating CD8A and CD8B, respectively, and two osteolysis-specific interaction pairs (AC090607.4-FOXO3 and TAL1-ABALON), which may play an important role in osteoclastogenesis.
1. Introduction
Osteoarthritis (OA) is a leading cause of chronic disability in old people. For the treatment, the total hip arthroplasty (THA) is a cost-effective way, which can reduce joint pain, restore joint function, and increase the quality of life of patients [1]. Despite the improvement of the quality of polyethylene, osteolysis remains a risk for older designs and younger, active patients. Osteolysis is a progressive, active, biologic cascade, a phenomenon due to a foreign body response to particulate wear debris from the prosthetic joint [2]. Increased wear particles activate osteoclast formation, and overweight osteoclasts caused much bone resorption, which eventually led to osteolysis [3]. Osteolysis, as a complication of THA, leads to prosthesis failure and bringing about additional suffering and burden for patients [4].
Long noncoding RNAs (lncRNAs), as a type of noncoding RNA, have been recognized as key regulatory molecules with diverse roles in gene expression, epigenetic modification, and protein activity [5]. Recently, lncRNA has been revealed to be involved in osteolysis. lncRNA TSIX was involved in particle-induced osteolysis by regulating miR-30a-5p to promote osteoblast apoptosis [6]. lncRNA DANCR inhibits osteoblast differentiation in osteolysis after THA through regulating FOXO1 [7]. lncRNA KCNQ1OT1 could ameliorate particle-induced osteolysis, by inhibiting miR-21a-5p to induce macrophage polarization [8]. However, more studies focused on the role of lncRNAs in osteolysis after THA need to be performed.
In this study, we, respectively, investigated the gene expression profiles of lncRNAs and mRNAs in patients with osteolysis after THA and OA, attempting to screen out differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs) associated with osteolysis after THA and OA. The objective of this study was to explore the underlying mechanism and the relevant molecular biomarkers for osteolysis after THA.
2. Materials and Methods
2.1. Subjects and Samples
The cohort subjected to RNA-Seq comprised 3 patients with OA, 3 patients with osteolysis after THA, and 3 healthy individuals. Inclusion criteria for patients with osteolysis after THA: (1) with current radiographic evidence of osteolysis and (2) received a THA after failure to improve function and pain after at least 6 months of conservative treatment. Subjects were excluded if they had any history of inflammatory arthropathy or known secondary causes of hip arthritis such as trauma, avascular necrosis, or developmental or childhood hip disease. Subjects were also excluded if they had taken courses of immunosuppressant agents or bisphosphonates for a continuous period of greater than 6 months since THA. OA was diagnosed according to the criteria of the American College of Rheumatology. Healthy individuals with no personal or family history of OA, no symptoms or signs of OA, or any other type of arthritis, or any painful condition of the joints, were included as controls. The participants with history of joint diseases, including inflammatory arthritis (rheumatoid arthritis or any other autoimmune disease), posttraumatic or postseptic arthritis, poliomyelitis, and skeletal dysplasia, were excluded. Table 1 described the characteristics of all these participants. All samples were collected after obtaining written informed consent from every participant. This study was approved by the Ethics Committee of Shandong Provincial Hospital (No. 2020-123) and performed in accordance with the Declaration of Helsinki. Peripheral whole blood (2.5 mL) drawn from each subject was collected in PAXgene® RNA blood tubes and stored at -80°C prior to processing. RNA isolation was performed with PAXgene blood RNA kit. RNA integrity and concentration were evaluated with an Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit). Total RNA samples used in subsequent experiments fulfilled the following requirements: RNA integrity number (RIN) > 7.0 and 28S/18S ≥ 1. In brief, total RNA was subjected to ribosomal RNA (rRNA) removal using Ribo-Zero. A total amount of 3 μg RNA was used for library preparation. Libraries for sequencing were constructed according to the manufacturer's protocol. The quality of the libraries was determined using an Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System. Based on the BGIseq platform, sequencing was performed. The raw sequencing data were submitted to sequencing quality control by FastQC to assess whether they will be used for subsequent data analysis. Reads with low quality (adaptor sequences, sequences with a quality score < 20, and sequences with an N base rate of raw reads > 10%) were removed. Clean reads were aligned with the human reference genome, Ensemble GRCh38.p7.
Table 1.
Patient characteristics.
| Group | Age (years) | p value | Gender | p value | BMI (kg/m2) | p value | Years after THA | Kellgren-Lawrence grade (n) | WBC (cells/mL) | p value | PLT (cells/mL) | p value | ESR (mm/h) | p value | CRP (mg/L) | p value | Part |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Osteolysis | 59 | 0.06 | Male | 0.64 | 24.2 | 0.05 | 15 | — | 7.3 | 0.11 | 276 | 0.56 | 19 | 0.03 | 4.2 | 0.41 | Left |
| 42 | Male | 25.4 | 15 | — | 5.89 | 210 | 19 | 4.2 | Right | ||||||||
| 70 | Female | 25.4 | 17 | — | 7.42 | 278 | 22 | 9.5 | Right | ||||||||
| OA | 55 | Female | 30.5 | — | 3 | 4.1 | 225 | 10 | 2.1 | Right | |||||||
| 40 | Female | 28.1 | — | 4 | 5.77 | 270 | 10 | 2.2 | Right | ||||||||
| 67 | Male | 29.8 | — | 3 | 5.65 | 187 | 10 | 6.6 | Right | ||||||||
| Controls | 30 | Male | 26.1 | — | — | 6.62 | 233 | 15 | 7.7 | Left | |||||||
| 29 | Male | 27.1 | — | — | 4.45 | 221 | 10 | 2.1 | Right | ||||||||
| 18 | Female | 30 | — | — | 6.85 | 253 | 11 | 1.3 | Right |
BMI: body mass index; THA: total hip arthroplasty; WBC: white blood cell; PLT: blood platelet; ESR: erythrocyte sedimentation ratio; CRP: C-reactive protein; OA: osteoarthritis.
2.2. Identification of DEmRNAs and Functional Annotation
StringTie software was used to compare the results to the known transcriptome and calculate the transcriptional abundance. Then, Ballgown was used for quantification of gene expression levels, as well as analysis of intergroup differential expression of genes. DESeq2 was applied to identify DEmRNAs in OA vs. control and osteolysis after THA vs. control with a p value < 0.05. DEmRNAs between OA and osteolysis after THA were obtained with a p value < 0.05 as well. Hierarchical clustering analysis of DEmRNAs was performed with R package “pheatmap.” Then, shared DEmRNAs in OA and osteolysis after THA and osteolysis-specific DEmRNAs (DEmRNAs in osteolysis after THA but no differences in OA) were further identified with Venny 2.1.0. The Database for Annotation, Visualization and Integrated Discovery (DAVID), which is a web-based tool, provides integrated solutions for the annotation and analysis of genome-scale datasets from high-throughput sequencing. DAVID 6.8 was used to perform GO and KEGG enrichment analysis for shared DEmRNAs in OA and osteolysis after THA and osteolysis-specific DEmRNAs. The lower the p value, the more significant are the GO term and the pathway. A value of p < 0.05 was considered to be represented statistically significant.
2.3. Identification of DElncRNAs
DESeq2 was applied to identify DElncRNAs in OA vs. control and osteolysis after THA vs. control with a p value < 0.05 and ∣log2FC | >1.5. DElncRNAs between OA and osteolysis after THA were obtained with a p value < 0.05 and ∣log2FC | >1.5 as well. Hierarchical clustering analysis of DElncRNAs was performed with R package “pheatmap.” Then, shared DElncRNAs in OA and osteolysis after THA and osteolysis-specific DElncRNAs (DElncRNAs in osteolysis after THA but no differences in OA) were further identified with Venny 2.1.0.
2.4. DElncRNA-DEmRNA Coexpression Network and Functional Annotation
The shared and osteolysis-specific DElncRNA-DEmRNA coexpression networks were constructed to further investigate the potential functions of lncRNAs and mRNAs in OA and osteolysis. Pearson correlation coefficients were calculated between the expression values of DElncRNAs and DEmRNAs. The pairs with ∣PCC | >0.8 and p < 0.01 in shared DElncRNA-DEmRNA pairs and pairs with ∣PCC | >0.95 and p < 0.01 in osteolysis-specific DElncRNA-DEmRNA pairs were defined as coexpressed DElncRNA-DEmRNA pairs, respectively. Then coexpressed networks were visualized by using Cytoscape. DAVID 6.8 was used to perform GO and KEGG enrichment analysis for DEmRNAs in shared and osteolysis-specific DElncRNA-DEmRNA coexpression network. A value of p < 0.05 was considered to be represented statistically significant.
2.5. Statistical Analysis
Statistical analyses were performed using R software v3.5.3 (R Foundation for Statistical Computing, Vienna, Austria). All tests were two-tailed, and p values < 0.05 were considered statistically significant. In addition, the GraphPad Prism Software, version 7.0, was used for the statistical analysis of experimental data. The results are expressed as means ± standard deviations (SDs). In addition, we used Shapiro-Wilk to test the normal distribution of data. However, the statistical analysis of clinical data showed that the data are not normal distribution. Therefore, we choose the Kruskal-Wallis test for statistical analysis. Pearson correlation coefficients were calculated between the expression values of DElncRNAs and DEmRNAs. p < 0.05 was considered to indicate a significant difference between the groups.
3. Results
3.1. Identification of DEmRNAs and Functional Annotation
DESeq2 was applied to identify DEmRNAs, and our results showed that a total of 343 DEmRNAs (184 up- and 159 downregulated) in OA vs. control, 908 DEmRNAs (429 up- and 479 downregulated) in osteolysis vs. control, and 406 DEmRNAs (112 up- and 294 downregulated) in OA vs. osteolysis were identified. Of these, HIST2H4A and CD8A, ZNF778 and TMEM56-RWDD3, and EPHB4 and MTURN were the most up- and downregulated DEmRNAs in OA vs. control, osteolysis vs. control, and OA vs. osteolysis, respectively (Table 2). The heatmap of the top 100 up- and downregulated DEmRNAs was shown in Figures 1(a)–1(c). The volcano plots of DEmRNAs are shown in Figures S1A–C. A total of 136 shared DEmRNAs (71 up- and 65 downregulated) in OA vs. control and osteolysis vs. control and 736 osteolysis-specific DEmRNAs (381 up- and 355 downregulated) were acquired (Figure 1(d)).
Table 2.
Top 10 up- and downregulated DEmRNAs.
| Symbol | log2FC | p value | FDR | Regulation |
|---|---|---|---|---|
| OA vs. control | ||||
| HIST2H4A | 2.017903 | 1.45E-12 | 1.86E-08 | Up |
| ZNF778 | 2.30578 | 2.34E-09 | 1.50E-05 | Up |
| RAB8A | 1.084187 | 4.61E-07 | 0.001473 | Up |
| MYOM2 | 3.614517 | 1.16E-05 | 0.021164 | Up |
| PI3 | 1.77021 | 4.16E-05 | 0.05915 | Up |
| VASH1 | 1.031837 | 0.000207 | 0.203576 | Up |
| SNHG28 | 1.440766 | 0.000309 | 0.263506 | Up |
| LILRB3 | 1.133152 | 0.000349 | 0.27883 | Up |
| UPRT | 1.107994 | 0.000428 | 0.322295 | Up |
| UNC13B | 1.407436 | 0.000467 | 0.331741 | Up |
| CD8A | -1.49304 | 2.03E-07 | 0.000865 | Down |
| AL157935.2 | -3.11394 | 2.71E-06 | 0.006946 | Down |
| AL121594.1 | -3.12822 | 4.34E-06 | 0.00926 | Down |
| EPHB4 | -2.33741 | 1.40E-05 | 0.022395 | Down |
| F11R | -1.30037 | 6.96E-05 | 0.080899 | Down |
| YPEL2 | -1.41993 | 0.000184 | 0.196205 | Down |
| CHKB-CPT1B | -1.48977 | 0.00028 | 0.255821 | Down |
| ZNF80 | -1.53824 | 0.000671 | 0.393542 | Down |
| EEF1AKMT3 | -1.76553 | 0.000677 | 0.393542 | Down |
| CD8B | -1.42735 | 0.000767 | 0.426828 | Down |
| Osteolysis vs. control | ||||
| ZNF778 | 2.261961 | 6.77E-08 | 0.000222 | Up |
| LILRB3 | 1.396428 | 1.23E-06 | 0.003224 | Up |
| MYOM2 | 3.476227 | 3.33E-06 | 0.00575 | Up |
| BCORL1 | 2.051727 | 1.40E-05 | 0.018296 | Up |
| RTL5 | 1.812327 | 1.85E-05 | 0.022092 | Up |
| FOSL2 | 1.834524 | 3.50E-05 | 0.026994 | Up |
| CNTNAP3 | 2.346929 | 7.26E-05 | 0.047612 | Up |
| MGAM2 | 2.714451 | 9.47E-05 | 0.049807 | Up |
| ERV3-1 | 1.398861 | 9.50E-05 | 0.049807 | Up |
| TMEM164 | 1.378476 | 0.000107 | 0.053473 | Up |
| TMEM56-RWDD3 | -3.54683 | 9.81E-12 | 1.29E-07 | Down |
| AC013489.1 | -4.17415 | 3.51E-06 | 0.00575 | Down |
| PLVAP | -2.61008 | 2.39E-05 | 0.026097 | Down |
| PPDPF | -2.88183 | 2.75E-05 | 0.026397 | Down |
| RPS26 | -2.22182 | 2.82E-05 | 0.026397 | Down |
| HBB | -2.40451 | 3.29E-05 | 0.026994 | Down |
| RGS6 | -2.33305 | 4.89E-05 | 0.035625 | Down |
| EVPL | -3.08781 | 5.28E-05 | 0.036452 | Down |
| RIOK3 | -2.15799 | 7.94E-05 | 0.049573 | Down |
| LRRN3 | -2.13595 | 8.98E-05 | 0.049807 | Down |
| Osteolysis vs. OA | ||||
| EPHB4 | 3.187856 | 9.59E-07 | 0.003135 | Up |
| RASSF5 | 1.444879 | 1.47E-05 | 0.023969 | Up |
| FAM118A | 1.406201 | 4.97E-05 | 0.064931 | Up |
| AL354822.1 | 2.9104 | 0.000364 | 0.250326 | Up |
| AL121594.1 | 2.129941 | 0.0004 | 0.259591 | Up |
| ZNF587 | 1.181929 | 0.000736 | 0.320854 | Up |
| SERPING1 | 1.589099 | 0.002698 | 0.608048 | Up |
| LRP1 | 1.363794 | 0.004148 | 0.796847 | Up |
| CASP10 | 1.098776 | 0.004527 | 0.796847 | Up |
| REC8 | 1.074025 | 0.004665 | 0.796847 | Up |
| MTURN | -1.58117 | 1.09E-05 | 0.021921 | Down |
| HBB | -2.3627 | 1.17E-05 | 0.021921 | Down |
| CISD2 | -1.29668 | 6.10E-05 | 0.07251 | Down |
| AC093668.3 | -2.78122 | 6.93E-05 | 0.073252 | Down |
| DDA1 | -1.3542 | 8.91E-05 | 0.08323 | Down |
| AC013489.1 | -3.94451 | 0.000133 | 0.11567 | Down |
| ARL4A | -1.83919 | 0.00018 | 0.147286 | Down |
| EPB41 | -1.62564 | 0.000288 | 0.221659 | Down |
| CR1L | -1.61874 | 0.000353 | 0.250326 | Down |
| DLGAP5 | -2.34576 | 0.000426 | 0.259591 | Down |
FC: fold change; FDR: false discovery rate; OA: osteoarthritis.
Figure 1.
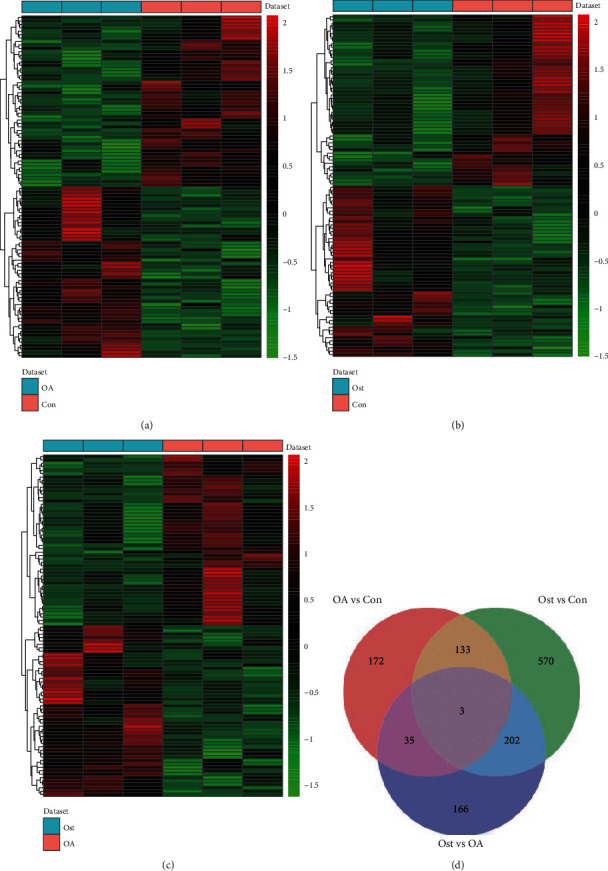
Identification of DEmRNAs: (a) heatmap of top 100 DEmRNAs between OA and normal controls; (b) heatmap of top 100 DEmRNAs between osteolysis after THA and normal controls; (c) heatmap of top 100 DEmRNAs between OA and osteolysis after THA; (d) Venn diagram of DEmRNAs in OA and osteolysis after THA. Con: controls; Ost: osteolysis.
To investigate the functions of shared and osteolysis-specific DEmRNAs, DAVID 6.8 was used to perform GO and KEGG enrichment analysis. For shared DEmRNAs, cell activation (p = 2.81E − 03), leukocyte activation (p = 5.20E − 03), integral to plasma membrane (p = 3.17E − 04), and protein homodimerization activity (p = 1.93E − 02) were several significantly enriched GO terms, and mTOR signaling pathway (p = 1.89E − 02) and cell cycle (p = 3.81E − 02) were significantly enriched KEGG pathways (Figures S2, S4 A and B). For osteolysis-specific DEmRNAs, intracellular signaling cascade (p = 1.80E − 02), cell adhesion (p = 8.45E − 04), plasma membrane (p = 9.54E − 05), and cytoskeletal protein binding (p = 1.08E − 02) were several significantly enriched GO terms, and hematopoietic cell lineage (p = 1.35E − 02), cell adhesion molecules (CAMs) (p = 2.29E − 02), porphyrin and chlorophyll metabolism (p = 3.40E − 02), and systemic lupus erythematosus (p = 4.86E − 02) were significantly enriched KEGG pathways (Figures S3, S4 C–F).
3.2. Identification of DElncRNAs
DESeq2 was applied to identify DElncRNAs, and our results showed that a total of 25 DElncRNAs (15 up- and 10 downregulated) in OA vs. control, 107 DElncRNAs (58 up- and 49 downregulated) in osteolysis vs. control, and 46 DElncRNAs (17 up- and 29 downregulated) in OA vs. osteolysis were identified. Of these, NEAT1 and AC005726.2, FLJ42393 and AC123912.4, and AC016737.1 and AC123912.4 were the most up- and downregulated DElncRNAs in OA vs. control, osteolysis vs. control, and OA vs. osteolysis, respectively (Table 3). The heatmap of DElncRNAs is shown in Figures 2(a)–2(c). The volcano plots of DEmRNAs are shown in Figure S1D-F. A total of 9 shared DElncRNAs (6 up- and 3 downregulated) in OA vs. control and osteolysis vs. control and 103 osteolysis-specific DElncRNAs (52 up- and 51 downregulated) were acquired (Figure 2(d)).
Table 3.
Top 10 up- and downregulated DElncRNAs.
| Symbol | log2FC | p value | FDR | Regulation |
|---|---|---|---|---|
| OA vs. control | ||||
| NEAT1 | 5.1159 | 1.85E-18 | 1.12E-14 | Up |
| AL512625.3 | 2.570376 | 3.24E-05 | 0.065224 | Up |
| AC016831.6 | 1.728574 | 5.90E-05 | 0.088885 | Up |
| AC111000.4 | 2.064848 | 0.001215 | 0.563632 | Up |
| LINC01341 | 2.02527 | 0.005032 | 0.92186 | Up |
| AC018445.3 | 1.654176 | 0.010542 | 0.999047 | Up |
| AC004231.1 | 1.894872 | 0.01745 | 0.999047 | Up |
| AC114752.2 | 1.692437 | 0.020267 | 0.999047 | Up |
| AP003117.2 | 1.771314 | 0.023032 | 0.999047 | Up |
| AC096642.1 | 1.589585 | 0.025643 | 0.999047 | Up |
| AC005726.2 | -1.66485 | 4.64E-08 | 0.00014 | Down |
| PITRM1-AS1 | -2.72439 | 0.000346 | 0.348099 | Down |
| OVCH1-AS1 | -2.18336 | 0.000714 | 0.478621 | Down |
| AC005082.1 | -2.31905 | 0.002001 | 0.670439 | Down |
| AL162457.1 | -1.71468 | 0.002791 | 0.701277 | Down |
| AC126755.7 | -1.50226 | 0.004787 | 0.92186 | Down |
| AP006545.1 | -1.89154 | 0.011714 | 0.999047 | Down |
| AC020659.1 | -1.60564 | 0.013616 | 0.999047 | Down |
| AC007541.1 | -1.69387 | 0.031639 | 0.999047 | Down |
| AC068870.2 | -1.90393 | 0.039154 | 0.999047 | Down |
| Osteolysis vs. control | ||||
| FLJ42393 | 1.62292 | 7.39E-05 | 0.067195 | Up |
| AC007272.1 | 2.182036 | 0.000975 | 0.279887 | Up |
| AC016831.6 | 1.690991 | 0.001091 | 0.285274 | Up |
| AL359532.1 | 1.86552 | 0.001529 | 0.335756 | Up |
| AL359183.1 | 1.921891 | 0.001606 | 0.340963 | Up |
| AC091185.1 | 1.772238 | 0.001807 | 0.359718 | Up |
| AC018946.1 | 1.549455 | 0.002757 | 0.427746 | Up |
| AC015871.4 | 1.542564 | 0.00288 | 0.427746 | Up |
| AC100788.2 | 1.613121 | 0.003089 | 0.427746 | Up |
| LINC02288 | 1.673173 | 0.003444 | 0.429946 | Up |
| AC123912.4 | -3.00991 | 2.78E-10 | 1.77E-06 | Down |
| AC010615.2 | -3.72122 | 1.34E-06 | 0.004253 | Down |
| DDIT4-AS1 | -1.59273 | 1.65E-05 | 0.033738 | Down |
| AC092490.1 | -2.67119 | 2.42E-05 | 0.033738 | Down |
| AC100810.1 | -2.2893 | 5.65E-05 | 0.059947 | Down |
| SLC2A1-AS1 | -2.12909 | 0.000292 | 0.191562 | Down |
| AC100835.2 | -2.25064 | 0.000369 | 0.195639 | Down |
| AC097478.1 | -2.43982 | 0.000553 | 0.242589 | Down |
| LINC00570 | -2.3716 | 0.000559 | 0.242589 | Down |
| MIR9-3HG | -2.61167 | 0.000581 | 0.242589 | Down |
| Osteolysis vs. OA | ||||
| AC016737.1 | 2.007544 | 0.000286 | 0.459454 | Up |
| AC124319.2 | 1.591514 | 0.003026 | 0.999986 | Up |
| AL357078.3 | 1.935801 | 0.005568 | 0.999986 | Up |
| AL139020.1 | 2.119999 | 0.007539 | 0.999986 | Up |
| AC004263.1 | 1.824237 | 0.015321 | 0.999986 | Up |
| AC018653.3 | 1.519922 | 0.020607 | 0.999986 | Up |
| AC099521.2 | 1.851515 | 0.024567 | 0.999986 | Up |
| AC110801.1 | 2.016041 | 0.0264 | 0.999986 | Up |
| AL022328.1 | 1.56358 | 0.02739 | 0.999986 | Up |
| AL844908.2 | 1.529839 | 0.03055 | 0.999986 | Up |
| AC123912.4 | -3.68915 | 3.62E-08 | 0.000233 | Down |
| AC010615.2 | -3.84396 | 5.84E-06 | 0.018749 | Down |
| AL356489.2 | -3.8055 | 0.000598 | 0.654803 | Down |
| AC092821.3 | -1.68773 | 0.001213 | 0.866219 | Down |
| KRT73-AS1 | -1.77092 | 0.002183 | 0.999986 | Down |
| FP236383.2 | -1.99999 | 0.002243 | 0.999986 | Down |
| AL159978.1 | -2.32724 | 0.003423 | 0.999986 | Down |
| TPM1-AS | -1.77588 | 0.003634 | 0.999986 | Down |
| MIR9-3HG | -1.88441 | 0.004272 | 0.999986 | Down |
| LINC00570 | -1.98324 | 0.005975 | 0.999986 | Down |
FC: fold change; FDR: false discovery rate; OA: osteoarthritis.
Figure 2.
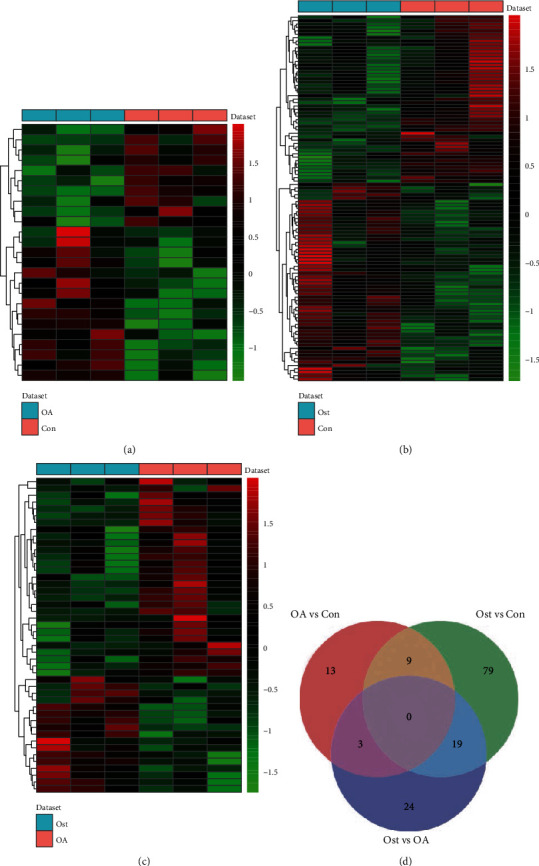
Identification of DElncRNAs: (a) heatmap of DElncRNAs between OA and normal controls; (b) heatmap of DElncRNAs between osteolysis after THA and normal controls; (c) heatmap of DElncRNAs between OA and osteolysis after THA; (d) Venn diagram of DElncRNAs in OA and osteolysis after THA. Con: controls; Ost: osteolysis.
3.3. DElncRNA-DEmRNA Coexpression Network and Functional Annotation
To further investigate the potential functions of lncRNAs and mRNAs in OA and osteolysis, the shared and osteolysis-specific DElncRNA-DEmRNA coexpression networks were constructed. A total of 128 shared DElncRNA-DEmRNA coexpression pairs including 9 DElncRNAs and 81 DEmRNAs were obtained (Figure 3(a)). Among them, AC018761.2 (degree = 49), AC090607.4 (degree = 46), and ABALON (degree = 42) were the top 3 DElncRNAs that covered the most DEmRNAs. Then, to investigate the functions of DEmRNAs in shared and osteolysis-specific DElncRNA-DEmRNA coexpression network, DAVID 6.8 was used to perform GO and KEGG enrichment analysis. For DEmRNAs in shared DElncRNA-DEmRNA coexpression network, T cell activation (p = 1.53E − 02), leukocyte differentiation (p = 1.69E − 02), leukocyte activation (p = 1.77E − 02), immune system development (p = 2.72E − 02), T cell differentiation (p = 2.96E − 02), and mTOR signaling pathway (p = 2.85E − 02) were several significantly enriched pathways (Figure 3(b)).
Figure 3.
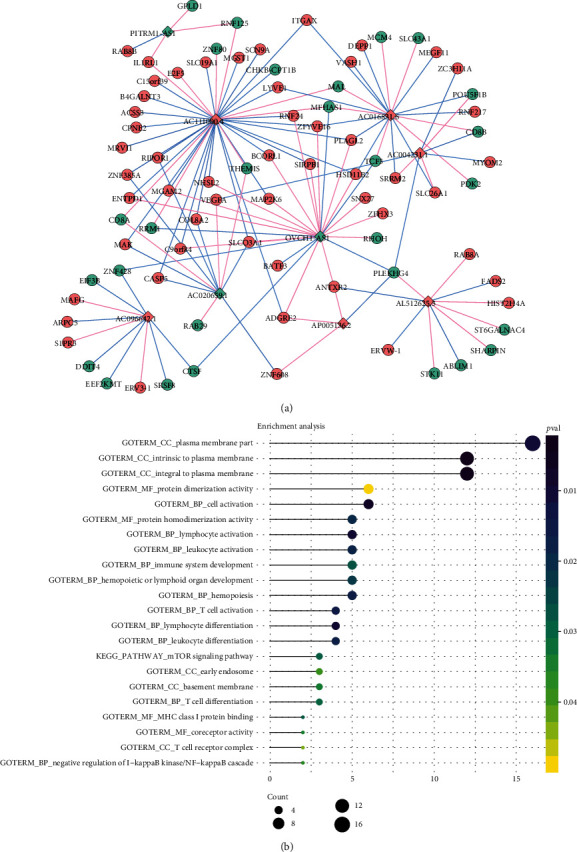
Shared DElncRNA-DEmRNA coexpression network and functional annotation. (a) Shared DElncRNA-DEmRNA co-expression network in OA and osteolysis after THA. The rhombus and ellipses represent DElncRNAs and DEmRNAs, respectively. Red and blue colors represent up- and downregulation, respectively. Red and blue edges represent positively and negatively correlated lncRNA-mRNA pairs, respectively. (b) Function enrichment.
A total of 522 osteolysis-specific DElncRNA-DEmRNA coexpression pairs including 36 DElncRNAs and 194 DEmRNAs were obtained (Figure 4(a)). Among them, AC111000.4 (degree = 38), OVCH1-AS1 (degree = 24), and AC016831.6 (degree = 19) were the top 3 DElncRNAs that covered the most DEmRNAs. For DEmRNAs in osteolysis-specific DElncRNA-DEmRNA coexpression network, myeloid cell differentiation (p = 4.50E − 04), immune system development (p = 7.00E − 04), cell proliferation (p = 2.27E − 03), and positive regulation of myeloid cell differentiation (p = 4.73E − 02) were several significantly enriched pathways (Figure 4(b)).
Figure 4.
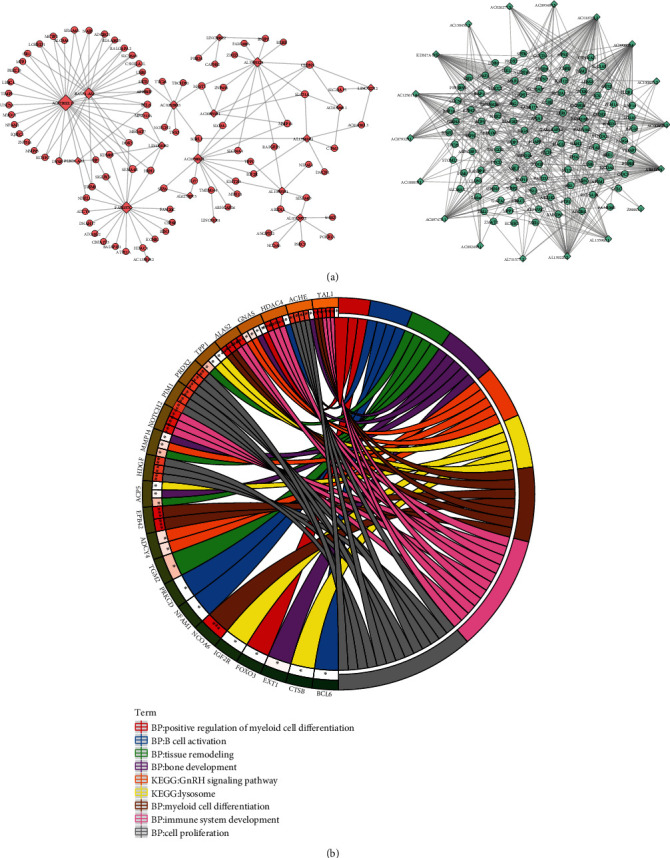
Osteolysis-specific DElncRNA-DEmRNA coexpression network and functional annotation. (a) Osteolysis-specific DElncRNA-DEmRNA coexpression network. The rhombus and ellipses represent DElncRNAs and DEmRNAs, respectively. Red and blue color represent up- and downregulation, respectively. (b) Function enrichment.
4. Discussion
Osteolytic lesions may develop after THA from a biologic reaction to particulate debris [9]. It is a major complication of THA, causing additional suffering and burden for patients. Aggravating evidence indicates that lncRNAs regulate gene expression via cis- and/or transregulation mechanisms and participate in various biological processes, including chromatin modification, DNA synthesis, cell proliferation, differentiation, and apoptosis [10]. In this present study, we screened out critical genes and lncRNAs correlated with OA and osteolysis by RNA sequencing and bioinformatics analysis.
Due to the close interaction between immune cells and bone cells, immune disorders can lead to abnormal bone metabolism [11]. T lymphocyte differentiation could be made by the “clusters of differentiation” (CD), and the common receptors are CD4 and CD8. CD8, a cell surface glycoprotein, is predominantly expressed on the surface of cytotoxic T killer cells and implicated in the immune system. The CD8 antigen is a specifically recognized antigen displayed by the class major histocompatibility complex I molecules. Long et al. identified a 16-gene biomarker panel to differentiate RA from OA and indicated the lower expression level of CD8A in OA than in RA [12]. Landgraeber et al. reported that the CD4+/CD8+ ratio was associated with the stage of osteolysis in aseptic loosening [13]. In this study, both CD8A and CD8B were significantly decreased in OA vs. control and osteolysis vs. control. Functional annotation revealed that CD8A and CD8B were enriched in immune-related pathways, such as T cell activation, leukocyte differentiation, leukocyte activation, immune system development, and T cell differentiation. In addition, CD8A was coexpressed with AC111000.4, and CD8B was coexpressed with AC016831.6 in shared the DElncRNA-DEmRNA coexpression network. Taken together, we speculated that AC111000.4 and AC016831.6 may function in the immune process of OA and osteolysis by regulating CD8A and CD8B, respectively.
The FOXO proteins are an evolutionarily conserved family of transcription factors which comprised FOXO1, FOXO3, FOXO4, and FOXO6 in mammals [14]. The four FOXO members share obvious sequence homology and are ubiquitously expressed in various organs, including bone [15]. FOXOs regulate diverse cellular processes, including oxidative stress, metabolism, apoptosis, and inflammation [16]. In addition, FOXOs have been revealed to regulate bone cell functions, osteoblast differentiation, and the maintenance of skeletal homeostasis [17]. Bartell et al. suggested that FOXO3 regulate receptor activator of NF-κB ligand- (RANKL-) induced osteoclast differentiation [18]. Miller et al. reported that FOXO3 play important inhibitory roles in TNF-α-mediated osteoclastogenesis and bone resorption [19]. Ambrogini et al. found that overexpression of FOXO3 in osteoblasts may reduce osteoclast numbers [20]. Increased osteoclastogenesis leads to osteolysis. Here, FOXO3 was determined to be a significantly decreased osteolysis-specific DEmRNA and coexpressed with AC090607.4, which indicated the importance of AC090607.4 in osteolysis by targeting FOXO3.
TAL1, also known as SCL, plays a crucial role in hematopoiesis and is causally connected to T cell acute lymphatic leukemia [21, 22]. The fact that embryonic stem cells of Tal1-knockout mice did not develop into osteoclasts in vitro supported that Tal1 plays a role in osteoclast differentiation [23]. The maintenance of bone homeostasis mainly depends on the balance between bone-forming osteoblasts and bone-resorbing osteoclasts. The transcription factor TAL1 was of great importance in cell cycle progression and proliferation in differentiating murine bone marrow monocyte precursors [24]. Knockdown of Tal1 in osteoclast progenitors leads to larger osteoclasts, containing more nuclei, and altered expression of a large number genes [25]. Wang et al. found that TAL1 was a differentially expressed transcription factor in osteoporosis [26]. In this study, TAL1 was coexpressed with ABALON in the osteolysis-specific DElncRNA-DEmRNA coexpression network. Therefore, the role of TAL1-BALON in osteolysis should be paid attention.
5. Conclusions
In conclusion, we highlighted the roles of four interaction pairs, including two shared lncRNA-mRNA interaction pairs in OA and osteolysis (AC111000.4-CD8A and AC016831.6-CD8B) and two osteolysis-specific interaction pairs (AC090607.4-FOXO3 and TAL1-BALON) in this present work. Taken as a whole, our work made contribution to understanding the pathophysiology of osteolysis. Our study also had a limitation. The sample size for RNA sequencing in this study was small, and further studies with larger sample size are warranted to confirm these results.
Acknowledgments
This work was supported by the Natural Science Foundation of Shandong (No. ZR2020QH073) and National Natural Science Foundation of China (No. 81972056).
Contributor Information
Guang Yang, Email: mayayangguang@163.com.
Shui Sun, Email: sunshui@sdfmu.edu.cn.
Data Availability
The clinical data used to support the findings of this study are restricted by the Ethics Committee of Shandong Provincial Hospital (No. 2020-123) in order to protect patient privacy. Data are available from Guang Yang (mayayangguang@163.com) for researchers who meet the criteria for access to confidential data.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Supplementary Materials
Figure S1: the volcano plots of DEmRNAs in OA (A), osteolysis (B), OA vs osteolysis (C), DElncRNAs in OA (D), osteolysis (E), and OA vs osteolysis (F). Figure S2: significantly enriched GO terms (A) and KEGG (B) pathways of shared DEmRNAs in OA and osteolysis after THA. Figure S3: significantly enriched GO terms (A) and KEGG (B) pathways of osteolysis-specific DEmRNAs. Figure S4: mTOR signaling pathway and cell cycle were significantly enriched KEGG pathways of shared DEmRNAs (A, B). Hematopoietic cell lineage, cell adhesion molecules (CAMs), porphyrin and chlorophyll metabolism, and systemic lupus erythematosus were significantly enriched KEGG pathways of osteolysis-specific DEmRNAs (C–F).
References
- 1.Wilkinson J. M., Wilson A. G., Stockley I., et al. Variation in the TNF gene promoter and risk of osteolysis after total hip arthroplasty. Journal of Bone and Mineral Research. 2003;18(11):1995–2001. doi: 10.1359/jbmr.2003.18.11.1995. [DOI] [PubMed] [Google Scholar]
- 2.Sheth N. P., Rozell J. C., Paprosky W. G. Evaluation and treatment of patients with acetabular osteolysis after total hip arthroplasty. The Journal of the American Academy of Orthopaedic Surgeons. 2019;27(6):e258–e267. doi: 10.5435/JAAOS-D-16-00685. [DOI] [PubMed] [Google Scholar]
- 3.Roato I., Caldo D., D’Amico L., et al. Osteoclastogenesis in peripheral blood mononuclear cell cultures of periprosthetic osteolysis patients and the phenotype of T cells localized in periprosthetic tissues. Biomaterials. 2010;31(29):7519–7525. doi: 10.1016/j.biomaterials.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Attinger M., Siebenrock K. Total hip replacement: between normal rehabilitation and complication. Praxis (Bern 1994) 2014;103(24):1439–1446. doi: 10.1024/1661-8157/a001853. [DOI] [PubMed] [Google Scholar]
- 5.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Reviews. Genetics. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 6.Bu Y., Zheng D., Wang L., Liu J. LncRNA TSIX promotes osteoblast apoptosis in particle-induced osteolysis by down-regulating miR-30a-5p. Connective Tissue Research. 2018;59(6):534–541. doi: 10.1080/03008207.2017.1413362. [DOI] [PubMed] [Google Scholar]
- 7.Tang Z., Gong Z., Sun X. LncRNA DANCR involved osteolysis after total hip arthroplasty by regulating FOXO1 expression to inhibit osteoblast differentiation. Journal of Biomedical Science. 2018;25(1):p. 4. doi: 10.1186/s12929-018-0406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao X., Ge J., Li W., Zhou W., Xu L. LncRNA KCNQ1OT1 ameliorates particle-induced osteolysis through inducing macrophage polarization by inhibiting miR-21a-5p. Biological Chemistry. 2018;399(4):375–386. doi: 10.1515/hsz-2017-0215. [DOI] [PubMed] [Google Scholar]
- 9.Ries M. D., Link T. M. Monitoring and risk of progression of osteolysis after total hip arthroplasty. The Journal of Bone and Joint Surgery. American Volume. 2012;94(22):2097–2105. [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J. T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Wang Y., Zhang N., et al. Heterozygous deletion of LRP5 gene in mice alters profile of immune cells and modulates differentiation of osteoblasts. Bioscience Trends. 2018;12(3):266–274. doi: 10.5582/bst.2018.01013. [DOI] [PubMed] [Google Scholar]
- 12.Long N. P., Park S., Anh N. H., et al. Efficacy of integrating a novel 16-gene biomarker panel and intelligence classifiers for differential diagnosis of rheumatoid arthritis and osteoarthritis. Journal of Clinical Medicine. 2019;8(1):p. 50. doi: 10.3390/jcm8010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landgraeber S., von Knoch M., Löer F., et al. Association between apoptotis and CD4(+)/CD8(+) T-lymphocyte ratio in aseptic loosening after total hip replacement. International Journal of Biological Sciences. 2009;5(2):182–191. doi: 10.7150/ijbs.5.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salih D. A., Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Current Opinion in Cell Biology. 2008;20(2):126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsitsipatis D., Klotz L. O., Steinbrenner H. Multifaceted functions of the forkhead box transcription factors FoxO1 and FoxO3 in skin. Biochimica et Biophysica Acta (BBA) - General Subjects. 2017;1861(5):1057–1064. doi: 10.1016/j.bbagen.2017.02.027. [DOI] [PubMed] [Google Scholar]
- 16.Xu C., Vitone G. J., Inoue K., Ng C., Zhao B. Identification of a novel role for Foxo3 isoform2 in osteoclastic inhibition. Journal of Immunology. 2019;203(8):2141–2149. doi: 10.4049/jimmunol.1900707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siqueira M. F., Flowers S., Bhattacharya R., et al. FOXO1 modulates osteoblast differentiation. Bone. 2011;48(5):1043–1051. doi: 10.1016/j.bone.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartell S. M., Kim H.-N., Ambrogini E., et al. FoxO proteins restrain osteoclastogenesis and bone resorption by attenuating H2O2 accumulation. Nature Communications. 2014;5(1) doi: 10.1038/ncomms4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller C. H., Smith S. M., Elguindy M., et al. RBP-J-regulated miR-182 promotes TNF-α–induced osteoclastogenesis. Journal of Immunology. 2016;196(12):4977–4986. doi: 10.4049/jimmunol.1502044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambrogini E., Almeida M., Martin-Millan M., et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metabolism. 2010;11(2):136–146. doi: 10.1016/j.cmet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloor A. J., Sanchez M. J., Green A. R., Gottgens B. The role of the stem cell leukemia (SCL) gene in hematopoietic and endothelial lineage specification. Journal of Hematotherapy & Stem Cell Research. 2002;11(2):195–206. doi: 10.1089/152581602753658402. [DOI] [PubMed] [Google Scholar]
- 22.Lecuyer E., Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Experimental Hematology. 2004;32(1):11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Yamane T., Kunisada T., Yamazaki H., Nakano T., Orkin S. H., Hayashi S. I. Sequential requirements for SCL/tal-1, GATA-2, macrophage colony-stimulating factor, and osteoclast differentiation factor/osteoprotegerin ligand in osteoclast development. Experimental Hematology. 2000;28(7):833–840. doi: 10.1016/S0301-472X(00)00175-2. [DOI] [PubMed] [Google Scholar]
- 24.Dey S., Curtis D. J., Jane S. M., Brandt S. J. The TAL1/SCL transcription factor regulates cell cycle progression and proliferation in differentiating murine bone marrow monocyte precursors. Molecular and Cellular Biology. 2010;30(9):2181–2192. doi: 10.1128/MCB.01441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Courtial N., Smink J. J., Kuvardina O. N., Leutz A., Göthert J. R., Lausen J. Tal1 regulates osteoclast differentiation through suppression of the master regulator of cell fusion DC-STAMP. The FASEB Journal. 2012;26(2):523–532. doi: 10.1096/fj.11-190850. [DOI] [PubMed] [Google Scholar]
- 26.Wang C., Yu T., Tan L., Cheng J. Bioinformatics analysis of gene expression profile in callus tissues of osteoporotic phenotype mice induced by osteoblast-specific Krm2 overexpression. International Journal of Rheumatic Diseases. 2016;19(12):1263–1271. doi: 10.1111/1756-185X.12840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: the volcano plots of DEmRNAs in OA (A), osteolysis (B), OA vs osteolysis (C), DElncRNAs in OA (D), osteolysis (E), and OA vs osteolysis (F). Figure S2: significantly enriched GO terms (A) and KEGG (B) pathways of shared DEmRNAs in OA and osteolysis after THA. Figure S3: significantly enriched GO terms (A) and KEGG (B) pathways of osteolysis-specific DEmRNAs. Figure S4: mTOR signaling pathway and cell cycle were significantly enriched KEGG pathways of shared DEmRNAs (A, B). Hematopoietic cell lineage, cell adhesion molecules (CAMs), porphyrin and chlorophyll metabolism, and systemic lupus erythematosus were significantly enriched KEGG pathways of osteolysis-specific DEmRNAs (C–F).
Data Availability Statement
The clinical data used to support the findings of this study are restricted by the Ethics Committee of Shandong Provincial Hospital (No. 2020-123) in order to protect patient privacy. Data are available from Guang Yang (mayayangguang@163.com) for researchers who meet the criteria for access to confidential data.


