Abstract
Low-frequency 1q21.1 distal deletion and duplication copy number variant (CNV) carriers are predisposed to multiple neurodevelopmental disorders, including schizophrenia, autism and intellectual disability. Human carriers display a high prevalence of micro- and macrocephaly in deletion and duplication carriers, respectively. The underlying brain structural diversity remains largely unknown. We systematically called CNVs in 38 cohorts from the large-scale ENIGMA-CNV collaboration and the UK Biobank and identified 28 1q21.1 distal deletion and 22 duplication carriers and 37,088 non-carriers (48% male) derived from 15 distinct magnetic resonance imaging scanner sites. With standardized methods, we compared subcortical and cortical brain measures (all) and cognitive performance (UK Biobank only) between carrier groups also testing for mediation of brain structure on cognition. We identified positive dosage effects of copy number on intracranial volume (ICV) and total cortical surface area, with the largest effects in frontal and cingulate cortices, and negative dosage effects on caudate and hippocampal volumes. The carriers displayed distinct cognitive deficit profiles in cognitive tasks from the UK Biobank with intermediate decreases in duplication carriers and somewhat larger in deletion carriers—the latter potentially mediated by ICV or cortical surface area. These results shed light on pathobiological mechanisms of neurodevelopmental disorders, by demonstrating gene dose effect on specific brain structures and effect on cognitive function.
Subject terms: Psychiatric disorders, Clinical genetics, Molecular neuroscience
Introduction
Inter-individual differences in brain structure are highly heritable1, but identifying the genes that contribute to brain development is challenging. Genome-wide association studies (GWAS) of brain anatomical structures indicate the influence of many single-nucleotide polymorphisms (SNPs) with small effect sizes2,3, but the links to brain function remain weak. Evidence is emerging that some rare copy number variants (CNVs)—that is, regions of the genome that are either deleted or duplicated—are associated with both substantial brain size and shape differences; for example, the 7q11.234,5, 22q11.26,7, 15q11.28–11 and 16p11.2 proximal12–14 and distal CNVs15. Many of these CNVs also have a wide-ranging phenotypic impact, including poorer cognitive abilities8,16–18 and increased risk of neurological or neurodevelopmental disorders. The strong impact of these CNVs on brain structure and behaviour make them valuable for studies of the molecular mechanisms contributing to aberrant human neurodevelopment.
The 1q21.1 distal CNV has a known large effect on head circumference, as evident from a high prevalence of micro- and macrocephaly in deletion and duplication carriers, respectively19–21. This, along with its position in a region that is rich in genes unique to the human lineage (i.e. absent in primates)22,23, makes the 1q21.1 distal CNV particularly interesting for the study of aberrations in human brain structure. However, its relatively low frequency, 1 in ~3400, (deletions) and 1 in 2100 (duplications)8,16, has hampered the study of its effects on brain structure.
1q21.1 distal deletion and duplication carriers are both at higher risk for several neurodevelopmental disorders including schizophrenia24,25, intellectual disability (ID), developmental delay, speech problems, autism spectrum disorders, motor impairment19,26–28 and epilepsy26,29, in addition to the separate risk for the duplication carriers for ADHD30, bipolar disorder and major depression31,32. Further, general cognitive ability (IQ) was lower in carriers in a small clinical study19 and in the UK Biobank33. In addition, 1q21.1 distal CNVs display a positive dose response on head circumference19–21, height and weight34,35 and are associated with various somatic diseases and traits, including bone and muscle deviations34 and cataract36 (deletion only), diabetes36 (duplication only) and heart disease36–39 (both). Conversely, several studies report carriers without any clinically evident phenotypes19,38 and considerable heterogeneity40,41, suggesting incomplete penetrance and variable expressivity. The Df(h1q21)+/− mouse, deleted in the syntenic 1q21.1 distal region, displays some phenotypes similar to human CNV carriers, including reduced head-to-tail length and altered dopamine transmission in response to psychostimulants, as seen in people with schizophrenia42.
The 1q21 region in humans is rich in low copy number repeats20,43 and contains several recurrent CNVs with differing breakpoints21,37. Thus, gene estimates vary, but the core interval encompasses at least 12 protein-coding genes including several human-specific genes such as HYDIN221,37, NOTCH2NLs22,23 and the DUF1220/Olduvain domain-containing NBPF-encoding genes44–46—the two latter were recently shown to have evolved as a two-gene unit47. Particularly interesting in the context of brain development are the recently characterized NOTCH2NL genes, absent in human’s closest living relatives and shown to prolong cortical neurogenesis22,23.
Despite the strong effects on neurodevelopmental traits and disorders, the impact of the 1q21.1 CNVs on human brain structure is largely unknown. Here, we present the first large-scale systematic neuroimaging study of 1q21.1 distal CNV carriers, investigating brain structure in >37,000 individuals including 28 deletion and 22 duplication carriers. We mapped the effect of the 1q21.1 distal CNV on subcortical volumes, intracranial volume (ICV) and global and regional measures of mean cortical thickness and surface area. We investigated variation in cognitive task performance and supplemented with exploratory mediation analysis of the brain on cognition in the UK Biobank. Given prior findings19–21,48, we explored a dose-dependent effect of copy number on brain structures and decreased cognitive performance for both 1q21.1 distal deletion and duplication carriers in comparison to non-carriers.
Materials and methods
Sample description
The brain structural sample comprises a total of 39 cohorts with genotyping and magnetic resonance imaging (MRI) data—38 from the ENIGMA-CNV consortium in addition to a subsample of the UK Biobank49 (project ID #27412). Demographic characteristics for each cohort are described in Supplementary Table 1 with a reference to participants’ collection and datasets including individual inclusion and exclusion parameters. Extended information on diagnosis and family information can be found in Supplementary Note 1 and age distribution of the cohorts in Supplementary Fig. 1. All participants gave written informed consent and sites involved obtained ethical approvals. The main 1q21.1 distal sample consisted of 28 deletion carriers, 22 duplication carriers and 37,088 non-carriers (Table 1) from 13 different datasets and 15 scanner sites with various ascertainments (family, clinical and population studies, case–control study for psychiatric disease) collected up until 30 September 2019. Non-carriers were defined as having no CNVs known to cause neurodevelopmental diseases (as defined in Supplementary Table 2). In the meta-analysis, an independent Icelandic sample from deCODE Genetics consisting of two deletion carriers and five duplication carriers in addition to 1150 non-carriers was added.
Table 1.
Demographic data.
| ENIGMA-CNV | deCODE | |||||||
|---|---|---|---|---|---|---|---|---|
| del | nc | dup | P | del | nc | dup | P | |
| n | 28 | 37,088 | 22 | 2 | 1150 | 5 | ||
| Sex, male (%) | 15 (54%) | 17,912 (48%) | 9 (41%) | 1 (50%) | 511 (44%) | 2 (33%) | ||
| Age (mean (SD)) | 41.7 (19.0) | 61.1 (12.8) | 55.4 (12.7) | <0.001 | 53.5 (2.1) | 44.8 (12.4) | 46.4 (16.5) | |
| Children (age <18 years) | 4 (14%) | 665 (1.8%) | <0.001 | 0 | 0 | 0 | ||
| Known diagnosis (%) | 11 (39.3%) | 2424 (6.5%) | 7 (32%) | <0.001 | 238 (21%) | 2 (40%) | ||
| Disease type (%) | ||||||||
| ADHD | 1 (~0%) | 181 (16%) | 2 (40%) | |||||
| Autism | 2 (0.2%) | |||||||
| Bipolar disorder | 7 (0.6%) | |||||||
| Clinically recruited (no diagnosis) | 6 (21.4%) | 4 (18%) | ||||||
| Dyslexia | 1 (3.6 %) | |||||||
| F-ICD-10 diagnosis (UK Biobank) | 858 (2.3%) | 1 (4%) | ||||||
| G-ICD10 diagnosis (UK Biobank) | 1 (3.0%) | 1439 (3.8%) | 1 (4%) | |||||
| MDD | 1 (~0%) | |||||||
| Multiple diagnosesa | 2 (7.2%) | 1 (4.5%) | ||||||
| Persistent depressive disorder | 1 (~0%) | |||||||
| SCZ | 1 (3.6) | 124 (0.3) | 48 (4.2%) | |||||
| Scanner sites | 11 | 15 | 8 | 2 | 2 | 1 | ||
| Datasets | 9 | 13 | 7 | 1 | 1 | 1 | ||
ADHD attention deficit disorder, clinically recruited in clinical NDD study but without a diagnosis, MDD major depressive disorder, SCZ schizophrenia, del deletion carrier, nc non-carriers, dup duplication carrier, P P value, AvPD avoidant personality disorder, OCD obsessive-compulsive disorder, DPD dependent personality disorder, STPD schizotypal personality disorder, NS non-significant.
P value is based on a χ2 test for categorical values and ANOVA for continuous values.
aFirst deletion carrier: agoraphobia, AvPD, OCD, DPD, other substance-related disorder, conduct disorder. Second deletion carrier: specific phobia, social phobia, MDD, AvPD, STPD. Duplication carrier: social phobia, OCD, MDD, AvPD.
Genotyping and QC
The genotypes were obtained by genotyping with commercially available platforms, performed at participating sites for each cohort (Supplementary Table 1). Individuals were excluded exclusively based on quality control (QC) parameters from the CNV calling. No exclusion was done due to ancestry in the primary analysis, but the effect of ancestry was evaluated in a separate analysis (see below).
CNV calls and validation in the core ENIGMA-CNV sample
Almost all cohorts had CNVs called and identified in a unified manner as described previously15. In brief, CNVs were called using PennCNV50 and appropriate population frequency (PFB) files and GC (content) model files (Supplementary Table 3 and Supplementary Notes2 and 3). Samples were filtered and CNVs identified based on standardized QC metrics15 (Supplementary Notes 2 and 3). The 1q21.1 distal region was well covered by all arrays (Supplementary Fig. 2). CNVs overlapping the region of interest (1q21.1 distal and 1q21.1 distal and proximal) were identified with the R package iPsychCNV, visualized and manually inspected.
Image acquisition and processing
All brain measures were obtained from structural T1-weighted MRI data collected at participating sites around the world and analysed with the standardized image analysis, FreeSurfer, quality assurance and statistical methods as per the harmonized neuroimaging protocols developed within ENIGMA23 and ENIGMA3 (http://enigma.ini.usc.edu/protocols/imaging-protocols/). Further detail on data processing is provided in Supplementary Note 4. Details on study, scanner, vendor, field strength, sequence, acquisition parameters and FreeSurfer versions used are outlined in Supplementary Table 4.
Statistical analysis
Imaging data processing and CNV calling were performed locally and de-identified CNV and imaging data were provided for a central mega-analysis. One of a pair of duplicates was kept. Relatives were removed from the sample used for the main analysis. In addition, we conducted a number of sensitivity analyses to test the robustness of the results (Supplementary Note 5 and Supplementary Tables 5–8). Individuals with a minimum overlap of 0.4 to regions with known pathogenic CNVs (Supplementary Table 2) were excluded from the analysis regardless of copy number status as were individuals from scanner sites without 1q21.1 distal CNV carriers.
Brain measures were normalized in R v3.3.2 by an inverse normal transformation of the residual of a linear regression on the phenotype correcting for covariates as done previously15. For the primary analysis, covariates were age, age2, sex, scanner site and ICV. In the analysis of ICV, ICV was not included as a covariate. These final covariance-corrected values were used in downstream analysis and are reported for each measure. For comparison between groups, normalization was carried out including only the groups addressed (deletion and non-carriers, duplications and non-carriers) except for the deletion versus duplication comparison, where values from normalization of the entire dataset were used due to the low numbers.
For the copy number dosage effect analysis (i.e. the effect on brain structure of 1q21.1 distal copy number variation), a linear regression on the copy number status of the individuals (deletion = 1, normal = 2, duplication = 3) was performed using the following model: covariance-corrected, normalized brain measure ~ copy number (deletion = 1, non-carrier = 2, duplication = 3). For comparison between groups, a two-sample, two-sided t test assuming equal variance in all carrier/non-carrier groups was employed (R v3.3.2) where deletion or duplication carriers were compared either to each other or to non-carriers. To correct for the multiple comparisons, we calculated the number of independent outcome measures through the spectral decomposition of a correlation matrix using MatSpDlite (https://neurogenetics.qimrberghofer.edu.au/matSpDlite/) of the three global, seven subcortical and 68 regional cortical measures. Based on the ratio of observed eigenvalue variance to its theoretical maximum, the estimated equivalent of independent measures was 36. Thus, we set the significance threshold at α = 0.05/36 = 0.0014. We report the uncorrected P values throughout the manuscript.
Effect size is calculated as the absolute effect size (the difference in mean between the two copy number groups in the t test—which, in this case, equals Cohen’s D as the standard deviation of the normalized brain measures is one) and the estimate of beta in the linear regression. Plots were generated using R library ggplot2 v2.2.151. Regional cortical visualization was done with the R package ggseg v1.5.1.
In a novel analysis, the independent Icelandic data were processed and analysed as the main dataset. We meta-analysed the results using the R package metafor v2.0.0, as previously15.
Cognitive task performance data
We downloaded behavioural performance measures on seven cognitive tests (the pairs matching task, the reaction time task, reasoning and problem-solving tests, the digit span test, the symbol digit substitution test and the trail making A and B tests) from the UK Biobank repository, performed by at least 10% of the participants. The results were processed following the general approach by Kendall et al.16. For more details, see Supplementary Note 6. For the analysis of the seven cognitive measures, we set the significance threshold to α = 0.05/7 = 0.007.
Mediation analysis
Mediation analyses were done with the R package mediation v4.4.7. Brain measures were normalized as described above and cognitive tasks were corrected for age, age2 and sex prior to input into the analysis. We report the proportion of the total effect of the CNV on cognitive task performance mediated by the brain measures (‘path ab’/‘path c’), with P values calculated through quasi-Bayesian approximation using 5000 simulations. We set the significance threshold at α = 0.05/((2 + 4) × 6) = 1.4 × 10−3 given the test of two structures for deletion and four for duplication carriers on six cognitive tests. The digit span test was excluded since no 1q21.1 CNV carriers had results from both this cognitive test and brain structural data.
Results
Sample characteristics
The main 1q21.1 distal (146.5–147.4 Mb, hg19) brain structural dataset consisted of 28 deletion and 22 duplication carriers and 37,088 non-carriers (derived from the same scanner sites as the CNV carriers) from ENIGMA-CNV and UK Biobank (Table 1, separate demographics in Supplementary Table 9). The age of CNV carriers was lower (41.7 ± 19.0 (deletions), 55.4 ± 12.7 (duplications), respectively) than that of non-carriers (61.1 ± 12.1) (Table 1). Eleven deletion carriers and seven duplication carriers had a known neurological, neurodevelopmental or psychiatric diagnosis or had been recruited in a clinical CNV study. The remaining carriers either did not have an established diagnosis or were recruited in studies from which diagnostic information was unavailable (Table 1 and Supplementary Table 10). Of the 37,088 non-carriers, 6.5% (2425) had an established neurological, neurodevelopmental or psychiatric disorder.
1q21.1 distal CNV associated with global cortical surface structures
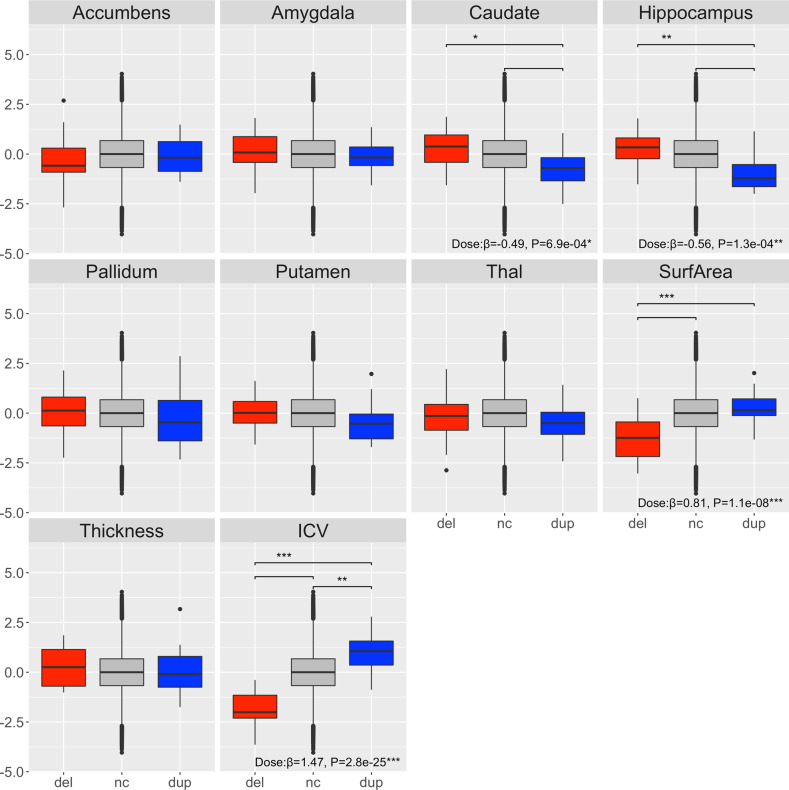
For our main dataset, there was a significant positive association between the number of 1q21.1 distal copies and ICV (β = 1.47, P = 2.8 × 10−25) as well as cortical surface area (β = 0.81, P = 1.1 × 10−8) (Fig. 1 and Supplementary Table 5) at a significance threshold of P < 0.0014 after correction for age, age2, sex, scanner site and ICV. In contrast, a significant negative copy number dosage effect was identified for the caudate (β = −0.49, P = 6.9 × 10−4) and hippocampal volumes (β = −0.56, P = 1.3 × 10−4). T tests indicated a decrease in ICV (Cohen’s D = −1.84 (−17%), P = 1.6 × 10−22) for deletion carriers and an increase for duplication carriers (Cohen’s D = 0.90 (+10%), P = 2.3 × 10−5), respectively, compared to non-carriers (Supplementary Table 6). For a raw value plot of ICV, see Supplementary Fig. 3. The cortical surface area dosage effect was primarily driven by the deletion carriers with a significantly lower total cortical surface area (Cohen’s D = −1.13 (−23%), P = 2.1 × 10−9) and the dosage effect on caudate and hippocampus was primarily driven by duplication carriers with significantly smaller caudate (Cohen’s D = −0.71 (−16%), P = 0.0012) and hippocampal (Cohen’s D = −0.92 (−15%), P = 4.1 × 10−5) volumes than non-carriers (Fig. 1 and Supplementary Table 7). Adding an independent Icelandic dataset with two deletions, five duplications and 1150 non-carriers (Table 1) in a meta-analysis strengthened the majority of the dosage results (Supplementary Fig. 4 and Supplementary Tables 11 and 12) and revealed additional significant between-group differences in nucleus accumbens, caudate and putamen (Supplementary Table 12).
Fig. 1. Cortical surface area and ICV show a positive dosage effect and caudate and hippocampus a negative dosage effect to copy number in the 1q21.1 distal region in our main sample (ENIGMA-CNV and UK Biobank).
Boxplots of subcortical volumes, cortical surface area and mean cortical thickness and ICV are shown. Deletion carriers (del) in red, non-carriers (nc) in grey and duplication carriers (dup) in blue, respectively. The normalized brain values are presented. Boxplots represent the mean. Copy number dosage effect is noted at the bottom of each panel. Significant differences after correction between groups are noted as *P < 0.0014, **P < 0.00014, ***P = 0.000014. Centre line represents the median, box limits are the upper and lower 25% quartiles, whiskers the 1.5 interquartile range and the points are the outliers. All analyses were corrected for age, age squared, sex, scanner site and ICV (except for ICV).
A number of sensitivity analyses were run on the main dataset, namely:
Matching each carrier with one non-carrier for age, sex, scanner site and ICV or age, sex, scanner site;
including only: (i) non-affected individuals (i.e. excluding individuals with a known neurodevelopmental or neurological disorder diagnosis; (ii) adults (age ≥ 18); (iii) non-affected adults; (iv) children (age < 18); (v) ENIGMA-CNV or (vi) UK Biobank;
controlling for ancestry;
excluding ICV as a covariate or;
including first- and second-degree relatives (see Supplementary Note 5 for methods).
These analyses validated the overall effects (Supplementary Tables 5 and 6).
The 1q21.1 distal CNV is associated with regional brain structures
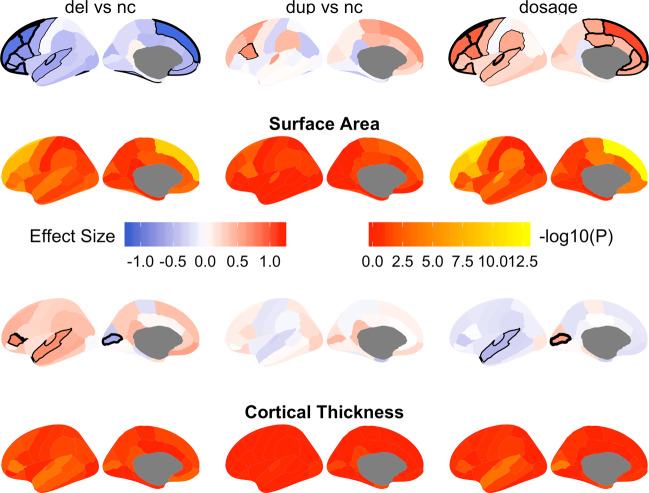
The largest dosage effects for the regional cortical surface area were found in the frontal lobes followed by the cingulate cortex—with additional significant effects in three regions of the parietal and temporal lobes (Fig. 2 and Supplementary Table 7). Likewise, through t tests, the largest effects in both deletion and duplication carriers in comparison to non-carriers were observed in the frontal and cingulate cortices (Fig. 2 and Supplementary Table 8).
Fig. 2. Results from the t tests and linear regression of 1q21.1 copy number variation on regional cortical surface area and cortical thickness.
First and third rows: Effect sizes (Cohen’s d for the t tests, beta coefficient for the dosage/linear regression). Second and fourth rows: Statistical significance in –log 10 of the P value. Significant areas in rows 1 and 3 are marked with black lines with increasing thickness for increasing significance (P < 0.0014). The column names indicate the comparisons with del = deletion carriers, nc = non-carriers, dup = duplication carriers. All measures were corrected for age, age2, sex, scanner site and ICV.
For regional cortical mean thickness, we identified significant negative dosage effects in the superior temporal region and significant positive dosage effects for the pericalcarine region (Fig. 2 and Supplementary Tables 7 and 8). Similarly, significant increases in mean cortical thickness were observed in deletion carriers versus non-carriers in the pars triangularis and superior temporal regions and a significant decrease in the pericalcarine region (Fig. 2 and Supplementary Table 8). All regional results were corrected for age, age2, sex, scanner site and ICV. Sensitivity analyses similar to those performed for subcortical regions confirmed the robustness of the results (Supplementary Tables 7 and 8).
1q21.1 distal CNV associated with cognitive performance and mediation by brain structures
Deletion and duplication carriers had different cognitive profiles in comparison to non-carriers when testing for association in seven different neuropsychological tests available from the full UK Biobank sample: deletion carriers had significantly poorer performance in three tests: symbol digit substitution, trail making B and pairs matching, while duplication carriers had significantly poorer performance in two tests: reaction time and the reasoning and problem-solving task (Table 2).
Table 2.
1q21.1 CNV deletion and duplication carriers show deficits in specific cognitive functions.
| Test | Suggested domain | n | del vs. nc | dup vs. nc | ||||
|---|---|---|---|---|---|---|---|---|
| del | nc | dup | Cohen’s D (SE) | P | Cohen’s D (SE) | P | ||
| Pairs matching | Working memory | 119 | 468,709 | 186 | −0.36 (0.09) | 7.3E − 05** | 0.03 (0.01) | 0.7 |
| Reaction time | Simple processing speed | 115 | 464,648 | 181 | −0.12 (0.06) | 0.18 | −0.23 (0.07) | 2.1E − 03 |
| Reasoning and problem solving | Fluid intelligence | 29 | 154,490 | 71 | −0.48 (0.19) | 9.2E − 03 | −0.33 (0.12) | 5.3E − 03 |
| Digit span | Numeric memory | 12 | 47,569 | 27 | −0.27 (0.14) | 0.36 | 0.14 (0.07) | 0.47 |
| Symbol digit substitution | Complex processing speed | 24 | 111,900 | 28 | −0.78 (0.2) | 1.4E − 04** | 0.04 (0.02) | 0.83 |
| Trail making A | Visual attention | 23 | 98,495 | 27 | −0.29 (0.15) | 0.16 | −0.14 (0.07) | 0.45 |
| Trail making B | Visual attention | 23 | 98,494 | 27 | −0.87 (0.21) | 3.1E − 05*** | −0.19 (0.1) | 0.33 |
n sample size, del deletion carriers, dup duplication carriers, nc non-carriers, SE standard error, P P value.
Multiple comparison-corrected significant findings (P < 0.007) are indicated in bold and with *<0.007, **<0.0007 and ***<0.00007.
Testing the effect of brain structures on cognitive tests in UK Biobank participants, larger ICV and total surface area were associated with better performance on almost all tests (Table 3 and see Supplementary Table 13 for sample size details). A larger hippocampus was associated with better performance for symbol digit substitution, trail making A and B (Table 3) and a larger caudate was associated with higher performance on the trail making A (Table 3).
Table 3.
Mediation analysis of brain structures over the association between 1q21.1 distal CNV carrier status and performance in the cognitive tasks in the UK Biobank.
| Path B—effect of brain structure on cognition | Deletion | Duplication | ||||
|---|---|---|---|---|---|---|
| Estimate (SE) | P | Prop. mediated | P | Prop. mediated | P | |
| Pairs matching | ||||||
| Caudate | 0.0023 (0.0053) | 0.66 | 3.5E − 03 | 0.85 | ||
| Hippocampus | 0.005 (0.0052) | 0.34 | 9.8E − 03 | 0.68 | ||
| SurfArea | 0.031 (0.0055) | 1.9E − 08 | −0.07 | 0.65 | −4.4E − 03 | 0.9 |
| ICV | 0.027 (0.0054) | 4.3E − 07 | −0.12 | 0.64 | −0.07 | 0.51 |
| Reaction time | ||||||
| Caudate | −0.0016 (0.0054) | 0.77 | −2.3E − 03 | 0.67 | ||
| Hippocampus | 0.01 (0.0053) | 0.053 | 0.01 | 0.04 | ||
| SurfArea | −0.0095 (0.0056) | 0.091 | 0.02 | 0.13 | 7.3E − 04 | 0.78 |
| ICV | 0.029 (0.0055) | 2.4E − 07 | −0.1 | 0.07 | −0.03 | 2.4E − 03 |
| Reasoning and problem solving | ||||||
| Caudate | −0.0059 (0.0091) | 0.51 | 5.7E − 03 | 0.55 | ||
| Hippocampus | 0.0031 (0.0089) | 0.73 | −9.6E − 05 | 0.95 | ||
| SurfArea | 0.052 (0.0094) | 2.6E − 08 | 0.06 | 0.250 | −7.4E − 04 | 0.97 |
| ICV | 0.15 (0.0092) | 3.7E − 59 | 0.25 | 0.24 | 0.18 | 0.04 |
| Symbol digit substitution | ||||||
| Caudate | 0.0011 (0.0077) | 0.88 | −4.2E − 03 | 0.83 | ||
| Hippocampus | 0.04 (0.0075) | 6.5E − 08 | −0.01 | 0.82 | ||
| SurfArea | 0.055 (0.0079) | 3.8E − 12 | 0.05 | 2.4E − 03 | 6.9E − 04 | 0.99 |
| ICV | 0.066 (0.0079) | 3.6E − 17 | 0.1 | 4.0E − 04 | 0.13 | 0.68 |
| Trail making A | ||||||
| Caudate | 0.034 (0.0084) | 5.7E − 05 | 4.4E − 04 | 1 | ||
| Hippocampus | 0.04 (0.0081) | 1.0E − 06 | 3.0E − 03 | 0.97 | ||
| SurfArea | 0.046 (0.0086) | 1.1E − 07 | 0.09 | 0.19 | 1.1E − 03 | 0.98 |
| ICV | 0.059 (0.0085) | 6.1E − 12 | 0.21 | 0.20 | −0.01 | 0.99 |
| Trail making B | ||||||
| Caudate | 0.021 (0.0083) | 0.012 | −0.01 | 0.79 | ||
| Hippocampus | 0.04 (0.008) | 6.9E − 07 | −0.01 | 0.86 | ||
| SurfArea | 0.082 (0.0085) | 6.4E − 22 | 0.07 | 8.0E − 04 | 8.9E − 03 | 0.92 |
| ICV | 0.11 (0.0084) | 1.2E − 36 | 0.17 | 1.2E − 03 | 0.16 | 0.73 |
Path B is the effect of the brain structure on cognition overall including all 1q21.1 deletion and duplication carriers (4–13 CNV carriers in each group) and non-carriers (n = 10,501–30,924; for exact numbers, see Supplementary Table 13). Each calculation included 5000 simulations.
The significance value for multiple comparisons (1.4 × 10−3) are in bold
Next, we tested whether the brain structures significantly associated with 1q21.1 distal CNV carriers might mediate the effect of the CNV on cognition. For two of the three tests associated with deletion carrier status, there were significant mediation effects (significance threshold 1.4 × 10−3): cortical surface area and ICV accounted for 5 and 10%, respectively, of the poorer performance of deletion carriers on symbol digit substitution, and 7 and 17%, respectively, of their poorer performance on the trail making B test (Table 3).
Discussion
Our main finding was a significant positive dosage effect in humans of 1q21.1 distal copy number on ICV and cortical surface area, with the largest differences in frontal and cingulate cortical surface area. We also identified a significant negative dosage effect on caudate and hippocampal volumes. A number of sensitivity analyses confirmed the robustness of the results. Both 1q21.1 distal deletion and duplication carriers showed poorer cognitive performance, although on different tests, with an indication that decreased ICV/cortical surface area might mediate the effect in deletion carriers.
The 1q21.1 distal CNV causes copy dosage effect on brain structures
We found a strong effect of the 1q21.1 distal CNV on the total cortical surface area, while no overall effect on mean cortical thickness was observed. A specific increase in the size of the cortical surface area with little effect on cortical thickness is observed throughout mammalian evolution including the primate lineage leading to humans52. This possibly reflects that cortical thickness and surface area appear to be driven by distinct genetic processes53. This pattern may be the result of an increased number of symmetric or self-renewing cell-division cycles, leading to an expansion of the neural progenitor pool and subsequently to an increase in the number of cortical neurons—in line with the radial unit hypothesis52. Interestingly, although not significant, mean cortical thickness tended to decrease in deletion carriers in the frontal cortical surface areas with the highest effect sizes, resembling a pattern found in lissencephaly54. This could suggest that large regional decreases in cortical surface area correlate inversely with mean cortical thickness.
The biomechanical forces of brain growth are thought to form the expansion of the cranium so that the skull grows in harmony with the expanding brain55. Thus, the positive copy number dosage effect on cortical surface area may directly trigger the effect on head circumference19–21 and ICV of 1q21.1 distal carriers due to modifications in pressure. Altered mechanical pressure might also cause the negative copy number dosage effect on the hippocampus and caudate volumes, effects on subcortical volumes also observed in a UK Biobank exploratory study on six individuals with a 1q21.1 distal duplication56.
Human-specific genes may affect the cortical surface area and cross-species effects
The positive copy number dosage effect on brain structure with the same direction as for weight and height34,35 likely results from altered gene expression as observed in 1q21.1 distal CNV cell lines48. In an independent experiment on fetal tissue, we also observed dynamic expression patterns of the genes in the 1q21.1 interval consistent with potential roles in cortical neurogenesis and development (Supplementary Note 7 and Supplementary Figs. 5 and 6).
GWAS based on the hg19 genome assembly have not identified hits in the 1q21.1 genomic region for ICV57, total cortical or regional surface area53,58. Assembly of the 1q21.1 region59 and thus gene discovery is complicated due to the presence of numerous low copy number repeats20,43 and has been faulty until the GRCh38 genome assembly. This may explain the lack of GWAS hits in the region.
Candidates for a dosage-dependent amplifier of the CNV-associated brain phenotypes are the recently identified human-specific NOTCH2NL genes that confer delayed neuronal differentiation and increased progenitor self-renewal22,23—in line with the radial unit hypothesis52. The areas with the highest regional effect sizes overlap with the areas of the highest expression of NOTCH2NLA and C in utero22 in concordance with an early developmental effect such as the macrocephaly observed in utero in a 1q21.1 distal duplication carrier38. Our observations of a 2% reduced skull diameter in the 1q21.1 deletion mouse (Supplementary Fig. 7 and Supplementary Notes 8 and 9) and recent findings of decreased total brain volume focused on the temporo-parietal and subcortical areas in the deletion mouse60 suggest that genes overlapping between human and mice (nine of ten mice genes are syntenic to the human region42) and not specific to humans are also involved in the altered skull and brain morphology. However, although diameter and volume are not directly comparable, the 17% decrease in ICV in human 1q21.1 deletion carriers would still point towards a substantial role of human-specific genes or genes with altered functions in comparison to mice. This underlines the need for additional data to disentangle which specific genes are involved in the skull and brain structural phenotypes. Of note, we also observed shorter bones overall in the 1q21.1 deletion mice (Supplementary Fig. 8 and Supplementary Note 9), expanding on previous head-to-tail length data42, and lower bone mineral density in female mice (Supplementary Fig. 9 and Supplementary Note 9), which mirror bone characteristics from human deletion carriers34 increasing the number of observed cross-species effects between the 1q21.1 mice and human 1q21.1 deletion carriers.
1q21.1 distal CNV deletion and duplication carriers show deficits in different cognitive functions
Our findings of widespread lower performance across several tests in different domains for both carrier groups in the volunteer-based UK Biobank sample are in line with cognitive results from a recent study33 and support that cognitive function in CNV carriers largely without a neurodevelopmental diagnosis may still be compromised8,16. Interestingly, the frontal and cingulate regions61, with the greatest cortical effect sizes for distal 1q21.1, correlate particularly with cognitive function and have gone through the greatest expansion during human development and evolution62. Our analyses indicated that the decreases in cognitive task performance are partially mediated by the observed differences in ICV and cortical surface area, reflecting the positive correlation between brain volume and intellectual function in line with previous findings63. The decrease in performance for several cognitive tasks in duplication carriers despite a larger ICV and cortical surface area suggests that the positive correlations may only be applicable within a certain narrower range. Interestingly, recent genetic analysis of NOTCH2NL in archaic and modern humans revealed ongoing adaptive evolution towards a lower dosage of the protein64, suggesting negative effects of excessive NOTCH2NL protein.
Our brain structural findings in 1q21.1 distal CNV carriers overlap with brain alterations in associated disorders: for example, ADHD65, autism spectrum disorders66, schizophrenia67, bipolar disorder68, major depressive disorder69 and subtypes of epilepsy70, but the exact overlaps differ between carrier groups. Of note, 1q21.1 distal deletion and duplication carriers display direct, opposite effects on several brain structures, while at risk for the same neurodevelopmental diseases. Other pathogenic CNVs also display overlapping disease risk and similar opposite copy number effects6,8–15 including effects on the cortical surface area in 22q11 and 16p11.2 proximal CNV carriers6,12–14. These CNVs impact different genes, but may converge on the same downstream pathways altering cortical surface area formation, similar to what has been reported for behavioural and neurocognitive phenotypes28.
This also suggests that other risk factors interplay to cause disease. It also supports that subgroups within neurodevelopmental disorders can be defined based on genetic profile and brain structural differences.
We demonstrate large effects of 1q21.1 distal CNVs on brain structure and cognition in humans including a mediation effect. These findings provide insight into molecular mechanisms involved in critical stages of human brain development and mapping of gene dosages to brain structural fingerprints.
Supplementary information
Acknowledgements
1000BRAINS: The 1000BRAINS study was funded by the Institute of Neuroscience and Medicine, Research Center Juelich, Germany. We thank the Heinz Nixdorf Foundation (Germany) for the generous support of the Heinz Nixdorf Recall Study on which 1000BRAINS is based. We also thank the scientists and the study staff of the Heinz Nixdorf Recall Study and 1000BRAINS. Funding was also granted by the Initiative and Networking Fund of the Helmholtz Association (Caspers) and the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement 945539 (Human Brain Project SGA3; Amunts, Caspers, Cichon).
Brainscale: The Brainscale study was supported by the Netherlands Organization for Scientific Research MagW 480-04-004 (Dorret I. Boomsma), 51.02.060 (Hilleke E. Hulshoff Pol), 668.772 (Dorret I. Boomsma and Hilleke E. Hulshoff Pol); NWO/SPI 56-464-14192 (Dorret I. Boomsma), the European Research Council (ERC-230374) (Dorret I. Boomsma), High Potential Grant Utrecht University (Hilleke E.Hulshoff Pol) and NWO Brain and Cognition 433-09-220 (Hilleke E.Hulshoff Pol).
Betula: The Betula study was funded by the Knut and Alice Wallenberg (KAW) foundation (Nyberg). The Freesurfer segmentations on the Betula sample were performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at HPC2N (in Umeå, Sweden), partially funded by the Swedish Research Council through grant agreement no. 2018-05973.
Brain Imaging Genetics (BIG): This work makes use of the BIG database, first established in Nijmegen, The Netherlands, in 2007. This resource is now part of Cognomics (www.cognomics.nl), a joint initiative by researchers from the Donders Centre for Cognitive Neuroimaging, the Human Genetics and Cognitive Neuroscience departments of the Radboud University Medical Centre and the Max Planck Institute for Psycholinguistics in Nijmegen. The Cognomics Initiative has received support from the participating departments and centres and from external grants, that is, the Biobanking and Biomolecular Resources Research Infrastructure (Netherlands) (BBMRI-NL), the Hersenstichting Nederland and the Netherlands Organization for Scientific Research (NWO). The research leading to these results also receives funding from the NWO Gravitation grant ‘Language in Interaction’, the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement nos. 602450 (IMAGEMEND), 278948 (TACTICS) and 602805 (Aggressotype), as well as from the European Community’s Horizon 2020 programme under grant agreement no. 643051 (MiND) and from ERC-2010-AdG 268800-NEUROSCHEMA. In addition, the work was supported by a grant for the ENIGMA Consortium (grant number U54 EB020403) from the BD2K Initiative of a cross-NIH partnership.
deCODE genetics: deCODE genetics acknowledges support from the Innovative Medicines Initiative Joint Undertaking under grant agreement nos. 115008 (NEWMEDS) and 115300 (EUAIMS), of which resources are composed of EFPIA in-kind contribution and financial contribution from the European Union’s Seventh Framework Programme (EU-FP7/2007-2013), EU-FP7-funded grant agreement no. 602450 (IMAGEMEND) and EU-funded FP7-People-2011-IAPP grant agreement no. 286213 (PsychDPC).
Dublin: This work was supported by Science Foundation Ireland (SFI grant 12/IP/1359 to Gary Donohoe and grant SFI08/IN.1/B1916-Corvin to Aidan C. Corvin).
ECHO-DEFINE: The ECHO study acknowledges funding from a Medical Research Council (MRC) Centre Grant to Michael J. Owen (G0801418), the Wellcome Trust (Institutional Strategic Support Fund (ISSF) to van den Bree and Clinical Research Training Fellowship to Joanne L. Doherty), the Waterloo Foundation (WF 918-1234 to van den Bree), the Baily Thomas Charitable Fund (2315/1 to van den Bree), National Institute of Mental Health (NIMH 5UO1MH101724 to van den Bree and Michael J. Owen), the IMAGINE-2 study (funded by the MRC (MR/T033045/1) to van den Bree, Jeremy Hall and Michael J. Owen), the IMAGINE-ID study (funded by MRC (MR/N022572/1) to Jeremy Hall, van den Bree and Owen). The DEFINE study was supported by a Wellcome Trust Strategic Award (100202/Z/12/Z) to Michael J. Owen.
ENIGMA: ENIGMA is supported in part by NIH grants U54 EB20403, R01MH116147 and R56AG058854. NIA T32AG058507; NIH/NIMH 5T32MH073526.
EPIGEN-Dublin: The EPIGEN-Dublin cohort was supported by a Science Foundation Ireland Research Frontiers Programme award (08/RFP/GEN1538).
EPIGEN-UK (Sisodiya): The work was partly undertaken at UCLH/UCL, which received a proportion of funding from the UK Department of Health’s NIHR Biomedical Research Centres funding scheme. We are grateful to the Wolfson Trust and the Epilepsy Society for supporting the Epilepsy Society MRI scanner.
GAP: This work was supported by the National Institute for Health Research (NIHR) Mental Health Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and the Institute of Psychiatry, Psychology and Neuroscience, King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
GOBS: The GOBS study data collection was supported in part by the National Institutes of Health (NIH) grants: R01 MH078143, R01 MH078111 and R01 MH083824, with work conducted in part in facilities constructed under the support of NIH grant C06 RR020547.
GSP: Data were in part provided by the Brain Genomics Superstruct Project (GSP) of Harvard University and Massachusetts General Hospital (MGH) (Principal Investigators: Randy Buckner, Jordan Smoller and Joshua Roffman), with support from the Center for Brain Science Neuroinformatics Research Group, Athinoula A. Martinos Center for Biomedical Imaging, Center for Genomic Medicine and Stanley Center for Psychiatric Research. Twenty individual investigators at Harvard and MGH generously contributed data to the overall project. We would like to thank Randy Buckner for insightful comments and feedback on this work.
HUBIN: The HUBIN study was financed by the Swedish Research Council (K2010-62X-15078-07-2, K2012-61X-15078-09-3, 521-2014-3487 K2015-62X-15077-12-3, 2017-00949), the regional agreement on medical training and clinical research between Stockholm County Council and the Karolinska Institutet.
HUNT: The HUNT study is a collaboration between HUNT Research Centre (Faculty of Medicine and Movement Sciences, NTNU—Norwegian University of Science and Technology), Nord-Trøndelag County Council, Central Norway Health Authority and the Norwegian Institute of Public Health. HUNT-MRI was funded by the Liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology, and the Norwegian National Advisory Unit for functional MRI.
IMAGEN: This work received support from the following sources: the European Union-funded FP6 Integrated Project IMAGEN (reinforcement-related behaviour in normal brain function and psychopathology) (LSHM-CT- 2007-037286), the Horizon 2020 funded ERC Advanced Grant ‘STRATIFY’ (Brain network based stratification of reinforcement-related disorders) (695313), ERANID (Understanding the Interplay between Cultural, Biological and Subjective Factors in Drug Use Pathways) (PR-ST-0416-10004), BRIDGET (JPND: BRain Imaging, cognition Dementia and next generation GEnomics) (MR/N027558/1), Human Brain Project (HBP SGA 2, 785907),the FP7 projects IMAGEMEND(602450; IMAging GEnetics for MENtal Disorders) and MATRICS (603016), the Innovative Medicine Initiative Project EUAIMS (115300-2), the Medical Research Council Grant ‘c-VEDA’ (Consortium on Vulnerability to Externalizing Disorders and Addictions) (MR/N000390/1), the Swedish Research Council FORMAS, the Medical Research Council, the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London, the Bundesministeriumfür Bildung und Forschung (BMBF grants 01GS08152, 01EV0711; eMED SysAlc01ZX1311A; Forschungsnetz AERIAL 01EE1406A, 01EE1406B), the Deutsche Forschungsgemeinschaft (DFG grants, SM 80/7-2, SFB 940/2), the Medical Research Foundation and Medical Research Council (grants MR/R00465X/1 and MR/S020306/1). Further support was provided by grants from: ANR (project AF12-NEUR0008-01—WM2NA, ANR-12-SAMA-0004), the Eranet Neuron (ANR-18-NEUR00002-01), the Fondation de France (00081242), the Fondation pour la Recherche Médicale (DPA20140629802), the Mission Interministérielle de Lutte-contre-les-Drogues-et-les-Conduites-Addictives (MILDECA), the Assistance-Publique-Hôpitaux-de-Paris and INSERM (interface grant), Paris Sud University IDEX 2012, the Fondation de l’Avenir (grant AP-RM-17-013), the Fédération pour la Recherche sur le Cerveau; the National Institutes of Health, Science Foundation Ireland (16/ERCD/3797), USA (Axon, Testosterone and Mental Health during Adolescence; RO1 MH085772-01A1) and by NIH Consortium grant U54 EB020403, supported by a cross-NIH alliance that funds Big Data to Knowledge Centres of Excellence.
Lifespan: The study is funded by the Research Council of Norway (230345, 288083 and 223273).
NCNG: NCNG sample collection was supported by grants from the Bergen Research Foundation and the University of Bergen, the Dr Einar Martens Fund, the Research Council of Norway, to le Hellard, Steen and Espeseth. The Bergen group was supported by grants from the Western Norway Regional Health Authority (Grant 911593 to Arvid Lundervold, Grant 911397 and 911687 to Astri Johansen Lundervold).
NTR: The NTR cohort was supported by the Netherlands Organization for Scientific Research (NWO) and The Netherlands Organisation for Health Research and Development (ZonMW) grants 904-61-090, 985-10-002, 912-10-020, 904-61-193, 480-04-004,463-06-001, 451-04-034, 400-05-717, Addiction-31160008, 016-115-035, 481-08-011, 056-32-010, Middelgroot-911-09-032, OCW_NWO Gravity programme—024.001.003, NWO-Groot 480-15-001/674, Center for Medical Systems Biology (CSMB, NWO Genomics), NBIC/BioAssist/RK(2008.024), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL, 184.021.007 and 184.033.111); Spinozapremie (NWO-56-464-14192), KNAW Academy Professor Award (PAH/6635) and University Research Fellow grant (URF) to Dorret I. Boomsma; Amsterdam Public Health research institute (former EMGO+), Neuroscience Amsterdam research institute (former NCA); the European Science Foundation (ESF, EU/QLRT-2001-01254), the European Community’s Seventh Framework Programme (FP7- HEALTH-F4-2007-2013, grant 01413: ENGAGE and grant 602768: ACTION); the European Research Council (ERC Starting 284167, ERC Consolidator 771057, ERC Advanced 230374), Rutgers University Cell and DNA Repository (NIMH U24 MH068457-06), the National Institutes of Health (NIH, R01D0042157-01A1, R01MH58799-03, MH081802, DA018673, R01 DK092127-04, Grand Opportunity grants 1RC2 MH089951 and 1RC2 MH089995); the Avera Institute for Human Genetics, Sioux Falls, South Dakota (USA). Part of the genotyping and analyses were funded by the Genetic Association Information Network (GAIN) of the Foundation for the National Institutes of Health. Computing was supported by NWO through grant 2018/EW/00408559, BiG Grid, the Dutch e-Science Grid and SURFSARA.
OATS: The OATS study has been funded by a National Health & Medical Research Council (NHMRC) and Australian Research Council (ARC) Strategic Award Grant of the Ageing Well, Ageing Productively Programme (ID No. 401162) and NHMRC Project Grants (ID Nos. 1045325 and 1085606). This research was facilitated through Twins Research Australia, a national resource in part supported by an NHMRC Centre for Research Excellence Grant (ID No.: 1079102). We thank the participants for their time and generosity in contributing to this research. We acknowledge the contribution of the OATS research team (https://cheba.unsw.edu.au/project/older-australian-twins-study) to this study. OATS genotyping was partly funded by a Commonwealth Scientific and Industrial Research Organization Flagship Collaboration Fund Grant.
Osaka: Osaka study was supported by the Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS: Grant Number JP18dm0207006), Brain/MINDS& beyond studies (Grant Number JP20dm0307002) and Health and Labour Sciences Research Grants for Comprehensive Research on Persons with Disabilities (Grant Number JP20dk0307081) from the Japan Agency for Medical Research and Development (AMED), Grants-in-Aid for Scientific Research (KAKENHI; Grant Numbers JP25293250 and JP16H05375). Some computations were performed at the Research Center for Computational Science, Okazaki, Japan.
PAFIP: The PAFIP study was supported by Instituto de Salud Carlos III, FIS 00/3095, 01/3129, PI020499, PI060507, PI10/00183, the SENY Fundació Research Grant CI2005-0308007 and the FundaciónMarqués de Valdecilla API07/011. Biological samples from our cohort were stored at the Valdecilla Biobank and genotyping services were conducted at the Spanish ‘Centro Nacional de Genotipado’ (CEGEN-ISCIII).
MCIC/COBRE: The study is funded by the National Institutes of Health studies R01EB006841, P20GM103472 and P30GM122734 and Department of Energy DE-FG02-99ER62764.
PING: Data collection and sharing for the Paediatric Imaging, Neurocognition and Genetics (PING) Study (National Institutes of Health Grant RC2DA029475) were funded by the National Institute on Drug Abuse and the Eunice Kennedy Shriver National Institute of Child Health & Human Development. A full list of PING investigators is at http://pingstudy.ucsd.edu/investigators.html.
QTIM: The QTIM study was supported by the National Institute of Child Health and Human Development (R01 HD050735) and the National Health and Medical Research Council (NHMRC 486682, 1009064), Australia. Genotyping was supported by NHMRC (389875). Medland is supported in part by an NHMRC fellowship (APP1103623).
SHIP: SHIP is part of the Community Medicine Research net of the University of Greifswald, Germany, which is funded by the Federal Ministry of Education and Research (grant nos. 01ZZ9603, 01ZZ0103 and 01ZZ0403), the Ministry of Cultural Affairs and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide single-nucleotide polymorphism typing in SHIP and MRI scans in SHIP and SHIP-TREND have been supported by a joint grant from Siemens Healthineers, Erlangen, Germany and the Federal State of Mecklenburg-West Pomerania.
StrokeMRI: StrokeMRI was supported by the Norwegian ExtraFoundation for Health and Rehabilitation(2015/FO5146), the Research Council of Norway (249795, 262372), the South-Eastern Norway Regional Health Authority (2014097, 2015044, 2015073) and the Department of Psychology, University of Oslo.
Sydney MAS: The Sydney Memory and Aging Study (Sydney MAS) is funded by National and HealthMedical Research Council (NHMRC) Programme and Project Grants (ID350833, ID568969 and ID109308). We also thank the Sydney MAS participants and the Research Team.
SYS: The SYS Study is supported by Canadian Institutes of Health Research.
TOP: Centre of Excellence: RCN #23273 and RCN #226971. Part of this work was performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT) (tsd-drift@usit.uio.no). The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7-PEOPLE-2013-COFUND) under grant agreement no. 609020—Scientia Fellows; the Research Council of Norway (RCN) #276082—A lifespan perspective on mental illness: toward precision medicine using multimodal brain imaging and genetics. Ida E. Sønderby and Rune Bøen are supported by South-Eastern Norway Regional Health Authority (#2020060). Ida E. Sønderby and Ole A. Andreassen have received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Grant agreement no. 847776 (CoMorMent project) and the KG Jebsen Foundation (SKGJ-MED-021).
UCLA_UMCU: The UCLA_UMCU cohort comprises of six studies which were supported by National Alliance for Research in Schizophrenia and Affective Disorders (NARSAD) (20244 to Prof. Hillegers), The Netherlands Organisation for Health Research and Development (ZonMw) (908-02-123 to Prof. Hulshoff Pol), and Netherlands Organisation for Scientific Research (NWO 9120818 and NWO-VIDI 917-46-370 to Prof. Hulshoff Pol). The GROUP study was funded through the Geestkracht programme of the Dutch Health Research Council (ZonMw, grant number 10-000-1001), and matching funds from participating pharmaceutical companies (Lundbeck, AstraZeneca, Eli Lilly and Janssen Cilag) and universities and mental health care organizations (Amsterdam: Academic Psychiatric Centre of the Academic Medical Center and the mental health institutions: GGZ inGeest, Arkin, Dijk en Duin, GGZ Rivierduinen, Erasmus Medical Centre, GGZ Noord-Holland-Noord. Groningen: University Medical Center Groningen and the mental health institutions: Lentis, GGZ Friesland, GGZ Drenthe, Dimence, Mediant, GGNet Warnsveld, Yulius Dordrecht and Parnassia Psycho-medical Center, The Hague. Maastricht: Maastricht University Medical Centre and the mental health institutions: GGzE, GGZ Breburg, GGZ Oost-Brabant, Vincent van Gogh, voor Geestelijke Gezondheid, Mondriaan, Virenzeriagg, Zuyderland GGZ, MET ggz, Universitair Centrum Sint-JozefKortenberg, CAPRI University of Antwerp, PC Ziekeren Sint-Truiden, PZ Sancta Maria Sint-Truiden, GGZ Overpelt, OPZ Rekem. Utrecht: University Medical Center Utrecht and the mental health institutions: Altrecht, GGZ Centraal and Delta.).
UK Biobank: This work made use of data sharing from UK Biobank (under project code 27412).
Others: Work by Pierre Vanderhaeghen was funded by Grants of the European Research Council (ERC Adv Grant GENDEVOCORTEX), the EOS Programme, the Belgian FWO, the AXA Research Fund and the Belgian Queen Elizabeth Foundation. Ikuo K. Suzuki was supported by a postdoctoral fellowship of the FRS/FNRS.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary information files. The data were gathered from various resources, and material requests will need to be placed with individual PIs. I.E.S. can provide additional detail upon correspondence. Data from PING are available at NIMH Data Archive: https://ndar.nih.gov/edit_collection.html?id=2607
Conflict of interest
H.B. is an advisory board member for Nutricia Australia. He has received research funding from the EU ‘Joint Programme Neurodegenerative Disorders’ (JPND) and the National Health and Medical Research Council, Australia. H.J.G. has received travel grants and speakers honoraria from Fresenius Medical Care, Servier, Neuraxpharm and Janssen Cilag. He has received research funding from the German Research Foundation (DFG), the German Ministry of Education and Research (BMBF), the DAMP Foundation, Fresenius Medical Care, the EU ‘Joint Programme Neurodegenerative Disorders’ (JPND) and the European Social Fund (ESF). C.R.K.C., P.M.T. and N.J. received partial grant support from Biogen Inc. (Boston), for research unrelated to the topic of this manuscript. B.C.-F. has received honoraria from Janssen Cilag, Otsuka and Lundbeck for educational and advisory/consultant activities unrelated with the present research. A.M.D. is a Founder of and holds equity in CorTechs Labs Inc., and serves on its Scientific Advisory Board. He is also a member of the Scientific Advisory Board of Human Longevity Inc., and receives funding through a research agreement with General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. G.B.W., O.G., M.O.U., H.S. and K.S. are employees of deCODE genetics (Amgen). D.P.H. is an employee of Genentech Inc. O.A.A. has received speakers honorarium from Lundbeck, and is a consultant to HealthLytix. A.J.L. has received speaker’s honorarium from Shire. M.J.O., J.H. and v.d.B. report grants from Takeda Pharmaceuticals outside of the submitted work. D.J.S. has received research grants and/or consultancy honoraria from Lundbeck and Sun. D.E.J.L. receives book royalties from Springer Nature and Oxford University Press. T.F. is on the Novo Nordisk advisory board. P.S.S. is on the Expert Advisory Committee of Biogen Australia. All other authors declare no competing financial interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ida E. Sønderby, Email: i.e.sonderby@medisin.uio.no
for the ENIGMA-CNV working group:
Dennis van der Meer, Eco J. C. de Geus, Greig I. de Zubicaray, Sonja M. C. de Zwarte, Stephanie Le Hellard, Dennis van ‘t Ent, and Marianne B. M. van den Bree
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01213-0.
References
- 1.Toga AW, Thompson PM. Genetics of brain structure and intelligence. Annu. Rev. Neurosci. 2005;28:1–23. doi: 10.1146/annurev.neuro.28.061604.135655. [DOI] [PubMed] [Google Scholar]
- 2.Hibar DP, et al. Novel genetic loci associated with hippocampal volume. Nat. Commun. 2017;8:13624. doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hibar DP, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meda SA, Pryweller JR, Thornton-Wells TA. Regional brain differences in cortical thickness, surface area and subcortical volume in individuals with Williams syndrome. PLoS ONE. 2012;7:e31913. doi: 10.1371/journal.pone.0031913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan CC, et al. Williams syndrome-specific neuroanatomical profile and its associations with behavioral features. Neuroimage Clin. 2017;15:343–347. doi: 10.1016/j.nicl.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin A, et al. Mapping 22q11.2 gene dosage effects on brain morphometry. J. Neurosci. 2017;37:6183–6199. doi: 10.1523/JNEUROSCI.3759-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun D, et al. Large-scale mapping of cortical alterations in 22q11.2 deletion syndrome: convergence with idiopathic psychosis and effects of deletion size. Mol. Psychiatry. 2020;25:1822–1834. doi: 10.1038/s41380-018-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefansson H, et al. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2014;505:361–366. doi: 10.1038/nature12818. [DOI] [PubMed] [Google Scholar]
- 9.Silva AI, et al. Reciprocal white matter changes associated with copy number variation at 15q11.2 BP1-BP2: a diffusion tensor imaging study. Biol. Psychiatry. 2019;85:563–572. doi: 10.1016/j.biopsych.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulfarsson MO, et al. 15q11.2 CNV affects cognitive, structural and functional correlates of dyslexia and dyscalculia. Transl. Psychiatry. 2017;7:e1109. doi: 10.1038/tp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Meer D, et al. Association of copy number variation of the 15q11.2 BP1-BP2 region with cortical and subcortical morphology and cognition. JAMA Psychiatry. 2019;77:420–430. doi: 10.1001/jamapsychiatry.2019.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maillard AM, et al. The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol. Psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qureshi AY, et al. Opposing brain differences in 16p11.2 deletion and duplication carriers. J. Neurosci. 2014;34:11199–11211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin-Brevet S, et al. Quantifying the effects of 16p11.2 copy number variants on brain structure: a multi-site genetic-first study. Biol. Psychiatry. 2018;84:253–264. doi: 10.1016/j.biopsych.2018.02.1176. [DOI] [PubMed] [Google Scholar]
- 15.Sonderby IE, et al. Dose response of the 16p11.2 distal copy number variant on intracranial volume and basal ganglia. Mol. Psychiatry. 2018;25:584–602. doi: 10.1038/s41380-018-0118-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kendall KM, et al. Cognitive performance among carriers of pathogenic copy number variants: analysis of 152,000 UK Biobank subjects. Biol. Psychiatry. 2016;82:103–110. doi: 10.1016/j.biopsych.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Huguet G, et al. Measuring and estimating the effect sizes of copy number variants on general intelligence in community-based samples. JAMA Psychiatry. 2018;75:447–457. doi: 10.1001/jamapsychiatry.2018.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hippolyte L, et al. The number of genomic copies at the 16p11.2 locus modulates language, verbal memory, and inhibition. Biol. Psychiatry. 2016;80:129–139. doi: 10.1016/j.biopsych.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Bernier R, et al. Clinical phenotype of the recurrent 1q21.1 copy-number variant. Genet. Med. 2016;18:341–349. doi: 10.1038/gim.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunetti-Pierri N, et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenfeld JA, et al. Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur. J. Hum. Genet. 2012;20:754–761. doi: 10.1038/ejhg.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki IK, et al. Human-specific NOTCH2NL genes expand cortical neurogenesis through Delta/Notch regulation. Cell. 2018;173:1370–1384.e1316. doi: 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiddes IT, et al. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173:1356–1369.e1322. doi: 10.1016/j.cell.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.PGC PCaSWGot. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haldeman-Englert, C. R. & Jewett, T. in GeneReviews((R)) (eds Adam, M. P. et al.) (University of Washington, Seattle University of Washington, 1993).
- 27.Mefford HC, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chawner S, et al. Genotype-phenotype associations in children with copy number variants associated with high neuropsychiatric risk in the UK (IMAGINE-ID): a case-control cohort study. Lancet Psychiatry. 2019;6:493–505. doi: 10.1016/S2215-0366(19)30123-3. [DOI] [PubMed] [Google Scholar]
- 29.Gourari I, Schubert R, Prasad A. 1q21.1 Duplication syndrome and epilepsy: case report and review. Neurol. Genet. 2018;4:e219. doi: 10.1212/NXG.0000000000000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudmundsson OO, et al. Attention-deficit hyperactivity disorder shares copy number variant risk with schizophrenia and autism spectrum disorder. Transl. Psychiatry. 2019;9:258. doi: 10.1038/s41398-019-0599-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green EK, et al. Copy number variation in bipolar disorder. Mol. Psychiatry. 2016;21:89–93. doi: 10.1038/mp.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kendall KM, et al. Association of rare copy number variants with risk of depression. JAMA Psychiatry. 2019;76:818–825. doi: 10.1001/jamapsychiatry.2019.0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kendall KM, et al. Cognitive performance and functional outcomes of carriers of pathogenic copy number variants: analysis of the UK Biobank. Br. J. Psychiatry. 2019;214:297–304. doi: 10.1192/bjp.2018.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owen D, et al. Effects of pathogenic CNVs on physical traits in participants of the UK Biobank. BMC Genomics. 2018;19:867. doi: 10.1186/s12864-018-5292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mace A, et al. CNV-association meta-analysis in 191,161 European adults reveals new loci associated with anthropometric traits. Nat. Commun. 2017;8:744. doi: 10.1038/s41467-017-00556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crawford K, et al. Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J. Med. Genet. 2018;56:131–138. doi: 10.1136/jmedgenet-2018-105477. [DOI] [PubMed] [Google Scholar]
- 37.Dolcetti A, et al. 1q21.1 Microduplication expression in adults. Genet. Med. 2013;15:282–289. doi: 10.1038/gim.2012.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhagen JM, et al. Phenotypic variability associated with a large recurrent 1q21.1 microduplication in a three-generation family. Mol. Syndromol. 2015;6:71–76. doi: 10.1159/000431274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soemedi R, et al. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Hum. Mol. Genet. 2012;21:1513–1520. doi: 10.1093/hmg/ddr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buse M, et al. Expanding the phenotype of reciprocal 1q21.1 deletions and duplications: a case series. Ital. J. Pediatr. 2017;43:61. doi: 10.1186/s13052-017-0380-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet. Med. 2013;15:478–481. doi: 10.1038/gim.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen J, et al. A mouse model of the schizophrenia-associated 1q21.1 microdeletion syndrome exhibits altered mesolimbic dopamine transmission. Transl. Psychiatry. 2017;7:1261. doi: 10.1038/s41398-017-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 44.O’Bleness M, et al. Finished sequence and assembly of the DUF1220-rich 1q21 region using a haploid human genome. BMC Genomics. 2014;15:387. doi: 10.1186/1471-2164-15-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumas LJ, et al. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am. J. Hum. Genet. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Florio M, et al. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife. 2018;7:e32332. doi: 10.7554/eLife.32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiddes IT, Pollen AA, Davis JM, Sikela JM. Paired involvement of human-specific Olduvai domains and NOTCH2NL genes in human brain evolution. Hum. Genet. 2019;138:715–721. doi: 10.1007/s00439-019-02018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harvard C, et al. Understanding the impact of 1q21.1 copy number variant. Orphanet J. Rare Dis. 2011;6:54. doi: 10.1186/1750-1172-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller KL, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci. 2016;19:1523–1536. doi: 10.1038/nn.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
- 52.Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-P. [DOI] [PubMed] [Google Scholar]
- 53.Grasby KL, et al. The genetic architecture of the human cerebral cortex. Science. 2020;367:eaay6690. doi: 10.1126/science.aay6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Donato N, et al. Lissencephaly: expanded imaging and clinical classification. Am. J. Med. Genet. A. 2017;173:1473–1488. doi: 10.1002/ajmg.a.38245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Libby J, et al. Modelling human skull growth: a validated computational model. J. R. Soc. Interface. 2017;14:20170202. doi: 10.1098/rsif.2017.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warland A, Kendall KM, Rees E, Kirov G, Caseras X. Schizophrenia-associated genomic copy number variants and subcortical brain volumes in the UK Biobank. Mol. Psychiatry. 2019;25:854–862. doi: 10.1038/s41380-019-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams HH, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hofer E, et al. Genetic correlations and genome-wide associations of cortical structure in general population samples of 22,824 adults. Nat. Commun. 2020;11:4796. doi: 10.1038/s41467-020-18367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinberg KM, et al. Single haplotype assembly of the human genome from a hydatidiform mole. Genome Res. 2014;24:2066–2076. doi: 10.1101/gr.180893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reinwald JR, et al. Separable neural mechanisms for the pleiotropic association of copy number variants with neuropsychiatric traits. Transl. Psychiatry. 2020;10:93. doi: 10.1038/s41398-020-0771-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill J, et al. Similar patterns of cortical expansion during human development and evolution. Proc. Natl Acad. Sci. USA. 2010;107:13135. doi: 10.1073/pnas.1001229107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fjell AM, et al. High-expanding cortical regions in human development and evolution are related to higher intellectual abilities. Cereb. Cortex. 2015;25:26–34. doi: 10.1093/cercor/bht201. [DOI] [PubMed] [Google Scholar]
- 63.Deary IJ, Penke L, Johnson W. The neuroscience of human intelligence differences. Nat. Rev. Neurosci. 2010;11:201–211. doi: 10.1038/nrn2793. [DOI] [PubMed] [Google Scholar]
- 64.Lodewijk GA, Fernandes DP, Vretzakis I, Savage JE, Jacobs FMJ. Evolution of human brain-size associated NOTCH2NL genes proceeds towards reduced protein levels. Mol. Biol. Evol. 2020;37:2531–2548. doi: 10.1093/molbev/msaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoogman M, et al. Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry. 2017;4:310–319. doi: 10.1016/S2215-0366(17)30049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Rooij D, et al. Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: results from the ENIGMA ASD Working Group. Am. J. Psychiatry. 2018;175:359–369. doi: 10.1176/appi.ajp.2017.17010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Erp TG, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol. Psychiatry. 2016;21:547–553. doi: 10.1038/mp.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hibar DP, et al. Subcortical volumetric abnormalities in bipolar disorder. Mol. Psychiatry. 2016;21:1710–1716. doi: 10.1038/mp.2015.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmaal L, et al. Subcortical brain alterations in major depressive disorder: findings from the ENIGMA Major Depressive Disorder working group. Mol. Psychiatry. 2016;21:806–812. doi: 10.1038/mp.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whelan CD, et al. Structural brain abnormalities in the common epilepsies assessed in a worldwide ENIGMA study. Brain. 2018;141:391–408. doi: 10.1093/brain/awx341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary information files. The data were gathered from various resources, and material requests will need to be placed with individual PIs. I.E.S. can provide additional detail upon correspondence. Data from PING are available at NIMH Data Archive: https://ndar.nih.gov/edit_collection.html?id=2607