Abstract
The SARS-CoV-2 Variant of Concern 202012/01 (VOC-202012/01) is rapidly spreading worldwide owing to its substantial transmission advantage. The variant has changes in critical sites of the spike protein with potential biological significance. Moreover, VOC-202012/01 has a mutation that inactivates the ORF8 protein, whose absence can change the clinical features of the infection. Why VOC-202012/01 is more transmissible remains unclear, but spike mutations and ORF8 inactivation stand out by their known phenotypic effects. Here I show that variants combining relevant spike mutations and the absence of ORF8 occurred in SARS-CoV-2 and related viruses circulating in other host species. A truncated ORF8 (Q23stop) occurred in a SARS-CoV-2-related virus from a pangolin seized in China in 2017, also with several mutations in critical spike sites. Strikingly, I found that variants without ORF8 (E19stop) and with the N501T spike mutation circulated in farmed mink and humans from Denmark. Although with differences to VOC-202012/01, the identification of these variants highlights the danger of having reservoirs of SARS-CoV-2 and related viruses where more transmissible variants may occur and spill over to humans.
Keywords: COVID-19, Coronaviruses, Transmissibility, Nonsense mutations
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease 2019 (COVID-19), has spread rapidly throughout the world following its emergence in Wuhan, China (Zhou et al., 2020). New sequence variants constantly arise during virus replication, some of which increase in frequency locally or worldwide. The spread of new variants over the course of an outbreak most often results from chance events, like when a limited number of individual viruses establish a new population during transmission (Grubaugh et al., 2020). However, the increase in frequency can result from a competitive advantage due to differences in viral infectivity, transmissibility or reactivity with neutralizing antibodies (Duffy et al., 2008).
The SARS-CoV-2 Variant of Concern 202012/01 (VOC-202012/01), lineage B.1.1.7, is the first for which there is compelling evidence that it has a substantial transmission advantage (Chand et al., 2020, Davies et al., 2020, Rambaut et al., 2020b). Epidemiological evidence points to an estimated difference in reproduction numbers between VOC and non-VOC ranging between 0.4 and 0.7 (Volz et al., 2021). VOC-202012/01 originated in the UK in late Summer to early Autumn 2020 and rapidly spread worldwide. The variant has several mutations changing important sites in the interaction between the virus spike protein and the host angiotensin I converting enzyme 2 (ACE2). Moreover, VOC-202012/01 has a mutation (C27972T; Q27stop) that truncates the ORF8 protein or renders it to be inactive (Chand et al., 2020). It has been previously shown that SARS-CoV-2 can persist without a functional ORF8 protein (Gong et al., 2020, Pereira, 2020, Pereira, 2021, Su et al., 2020, Young et al., 2020). Patients infected with a SARS-CoV-2 variant lacking ORF8 had better clinical outcomes than those infected with wild-type variants, showing a longer duration of symptoms and a possible later onset (Young et al., 2020).
SARS-CoV-2 is thought to have been transmitted to humans from bats, although it is possible that other mammalian species may have acted as “intermediate” hosts in the transmission (Andersen et al., 2020, Zhang and Holmes, 2020). The Malayan pangolins (Manis javanica) are likely candidates for intermediates by the discovery of SARS-CoV-2-related viruses in several animals illegally imported into the Chinese provinces of Guangdong and Guangxi (Han, 2020, Lam et al., 2020). Several species can potentially be infected by SARS-CoV-2 via their ACE2 proteins (Damas et al., 2020). During the COVID-19 pandemic, SARS-CoV-2 has been transmitted from humans to pets and other wild and domestic animals (Oreshkova et al., 2020, Sailleau et al., 2020, Sit et al., 2020). At least in the case of farmed mink, viruses had spilled back to humans (Munnink et al., 2021). The circulation of SARS-CoV-2 in other species increases the emergence of new variants, some of which may have significant differences to the ancestral state resulting from the adaptation to the new host. Therefore, the transmission back to humans may introduce variants with higher transmissibility or with the potential to escape the protection resulting from existing vaccines. In this regard, several mutations have been detected in the spike protein during the passage of SARS-CoV-2 through mink populations (Koopmans, 2021, Munnink et al., 2021). It is therefore important to identify variants with mutations of biological relevance, such as in the spike or accessory genes, which may have circulated in domestic or wild animals. Those variants may pose a risk for the ongoing COVID-19 pandemic. Here I show that SARS-CoV-2 variants without the ORF8 occurred in both pangolin and farmed mink, always in the background of mutations in spike sites of biological relevance. These results suggest that mutations as those observed in VOC-202012/01 may occur in other host species, introducing an additional risk to the ongoing pandemic.
2. Materials and methods
2.1. Genomic sequences from SARS-CoV-2 and related viruses
Genomic sequences from SARS-CoV-2 and related viruses were obtained from the GISAID Initiative (https://www.gisaid.org/) on January 11, 2021. I searched for ORF8 nonsense mutation is all available sequences from Canis lupus familiaris, Chlorocebus sabaeus, Felis catus, Manis javanica, Manis pentadactyla, Mus musculus, Mustela lutreola, Neovison vison, Panthera leo, Panthera tigris jacksoni, Rhinolophus affinis, Rhinolophus malayanus and Rhinolophus shameli. The sequences from pangolin (M. javanica) and mink (N. vison) with ORF8 nonsense mutations were classified into clades using the Nextclade tool (https://clades.nextstrain.org/) and the Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN) tool running in GISAID (Rambaut et al., 2020a).
2.2. SARS-CoV-2 related virus from pangolin (M. javanica) sample P1E
The raw sequence reads from the pangolin sample P1E were extracted from the SRA database, BioProject accession number PRJNA606875, experiment SRX7732093 (Lam et al., 2020). A blast analysis (Altschul et al., 1990) was performed to identify reads including the ORF8 position 27,960, where a mutation causes the premature stop codon Q23stop. The reads were assembled to the SARS-CoV-2 reference using the Geneious mapper under the ‘Highest sensitivity’ setting, running on Geneious Prime 2019.0.4 (https://www.geneious.com). Reads matching position 27,960 close to their ends were excluded to avoid sequencing artifacts. The final dataset included 152 reads (mean length=300.1; SD=0.5) holding the position 27,960.
2.3. SARS-CoV-2 in farmed mink (N. vison) from Denmark
All SARS-CoV-2 complete genomic sequences from Denmark with mutation ORF8 E19stop were downloaded from GISAID. The sequences were aligned using the MAFFT version 7 online service (Katoh et al., 2019), with the light-weight option for MSA of full-length SARS-CoV-2 genomes. The SARS-CoV-2 genome with accession number NC_045512.2 was used as reference. The maximum-likelihood phylogenetic tree was constructed using the PhyML 3.0 (Guindon et al., 2010) running in the ATGC bioinformatics platform (http://www.atgc-montpellier.fr). The GTR + I substitution model was selected using the SMS: Smart Model Selection in PhyML (Lefort et al., 2017), with option AIC (Akaike Information Criterion). The branch support was estimated using a bootstrap of 100 trees. The 3D structure of the spike glycoprotein (PDB: 6acj, EM 4.2 Angstrom) in complex with host cell receptor ACE2 were obtained using the CoVsurver enabled by GISAID.
3. Results
3.1. A SARS-CoV-2 variant with a truncated ORF8 (Q23stop) occurred in a pangolin (M. javanica) from Guangx, China
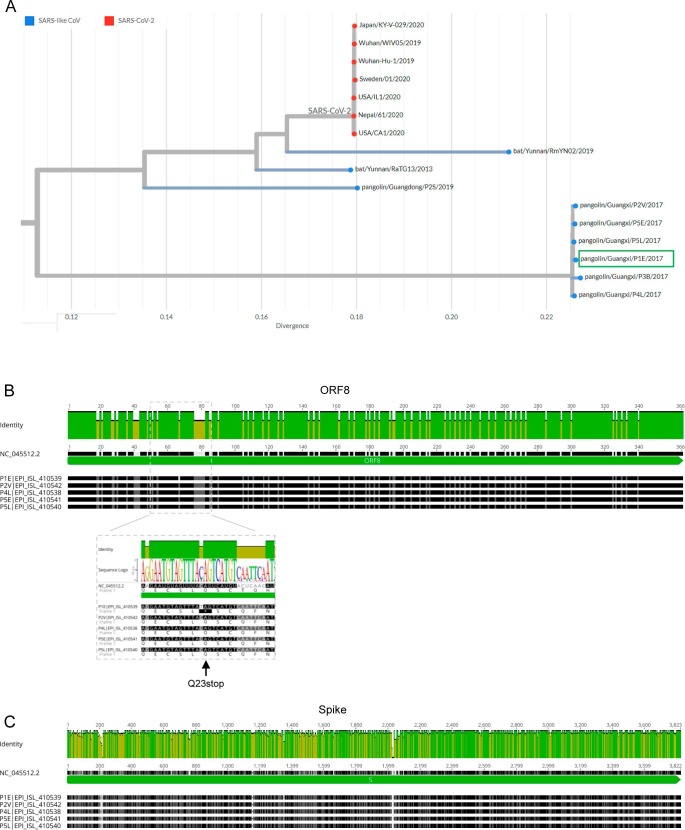
SARS-CoV-2-related viruses have been identified in Malayan pangolins (M. javanica), but is still unknown if they were at the origin of the COVID-19 pandemic (Lam et al., 2020, Dimonaco et al., 2021). I found that a SARS-CoV-2-related virus from a pangolin sample (sample P1E; EPI_ISL_410539) had a truncated ORF8 resulting from a Q23stop mutation (Table 1 ). This pangolin was seized by the Guangxi Customs, China, during their routine anti-smuggling operations in 2017 (Lam et al., 2020).
Table 1.
SARS-CoV-2 and related viruses with variants lacking ORF8 identified in wild or domestic species.
| Sequence | GISAID Accession ID | Collection date | Location | Clade (Nextclade) | Lineage (Pangolin) | ORF8 nonsense mutation | Spike amino acid substitutions | |
|---|---|---|---|---|---|---|---|---|
| Pangolin (Manis javanica) | Guangxi/P1E | EPI_ISL_410539 | 2017 | China/Guangxi | 19B | B.15 | Q23stop | 92 including V483Q, F490Y, N501T, P681M |
| Mink (Neovison vison) | mDK-28 | EPI_ISL_641421 | 16/10/2020 | Denmark | 20A | B.1 | E19stop | T95I, D614G |
| mDK-29 | EPI_ISL_641422 | 16/10/2020 | D614G | |||||
| mDK-30 | EPI_ISL_641423 | 16/10/2020 | D614G | |||||
| mDK-154 | EPI_ISL_683005 | 05/11/2020 | D614G | |||||
| mDK-158 | EPI_ISL_683009 | 05/11/2020 | D614G | |||||
| mDK-159 | EPI_ISL_683010 | 05/11/2020 | D614G | |||||
| mDK-172 | EPI_ISL_683023 | 04/11/2020 | N501T, D614G | |||||
| mDK-173 | EPI_ISL_683024 | 04/11/2020 | N501T, D614G | |||||
| mDK-174 | EPI_ISL_683025 | 04/11/2020 | N501T, D614G |
The pangolin sample P1E and human SARS-CoV-2 reference genomes had 80.3% of identical sites in the ORF8 gene, in line with previous studies (Mohammad et al., 2020, Pereira, 2020). Both lineages diverge by 16 out of 121 amino acids in the ORF8 protein (86.9% of identity). I aligned the ORF8 gene sequences from all pangolin samples reported in Lam et al. with the SARS-CoV-2 reference sequence (Fig. 1 A). After excluding a sequence with a large sequencing gap in ORF8, I found that all pangolin sample had an equal ORF8 gene, with exception of the Q23stop mutation at sample P1E. The similarity among samples from different pangolins suggests that the Q23stop mutation was recent, perhaps even within the sampled individual. If the lineage of sample P1E had acquired the ORF8 Q23stop mutation before, one should expect the accumulation of other mutations in a gene encoding a non-functional protein that was not under selection. The possibility of Q23stop resulting from a sequencing error should be considered in old samples stored in unknown conditions for a few years. However, I found that 151 out of 152 sequencing reads for ORF8 had a T that causes the premature stop codon, with a single read showing the reference C nucleotide. It is therefore unlikely that errors would generate such a clear pattern in obtained sequences.
Fig. 1.
SARS-CoV-2-related viruses in Malayan pangolins (Manis javanica). A) Phylogeny of SARS-CoV-2 and related viruses sampled in bats and pangolins. SARS-CoV-2 from the COVID-19 epidemic are coloured in red, while related SARS-like coronaviruses are coloured in blue. The sample P1E (EPI_ISL_410539) is highlighted by a green box. Tree obtained from the Nextstrain website. B) and C) Identity plot for the alignment of ORF8 (B) and spike (C) genes from SARS-CoV-2-related viruses sequenced from five pangolin samples collected in Guangxi, China. Highlighted in B) is the ORF8 gene region with the mutation C27960T causing the premature stop codon Q23stop observed in sample P1E. The alignments include the reference SARS-CoV-2 genome (NC_045512.2). The most conserved positions are indicated by green bars.
In terms of the spike gene, sample P1E and human SARS-CoV-2 had 83.1% of identical sites, resulting in 99 out of 1274 amino acids differences (Fig. 1B). Among those differences, the pangolin sample P1E had amino acid changes in spike sites V483Q, F490Y, N501T and P681M. On the contrary to ORF8, all pangolin samples diverge in the spike gene, but all share the spike mutations of sample P1E analysed here.
3.2. SARS-CoV-2 variants combining ORF8 E19stop and spike N501T mutations circulated in farmed mink (N. vison)
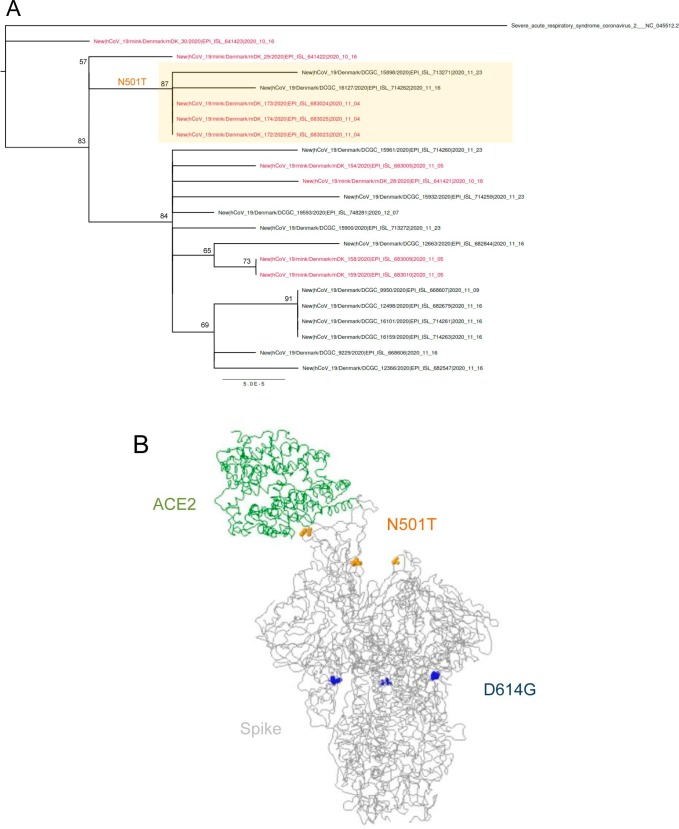
The infection of farmed mink (N. vison) with SARS-CoV-2 that subsequently spilled back into humans prompted the Ministry of Environment and Food of Denmark the culling of all mink in the country on November 5, 2020 (Koopmans, 2021). Full-length virus genome sequencing revealed novel mutations in the spike protein during the passage through the mink population (Hammer et al., 2021, van Dorp et al., 2020). I found nine SARS-CoV-2 sequences with ORF8 E19stop mutation in mink from Denmark (Table 1). The sequences belong to clade 20A (Nextclade) or lineage B.1 that dominated the European outbreak in March 2020 (Rambaut et al., 2020a). The three sequences with the oldest collection date (2020–10-16) were all from mink (EPI_ISL_641421, EPI_ISL_641422 and EPI_ISL_641423). The phylogeny built with all SARS-CoV-2 sequences with the ORF8 E19stop from Denmark showed that one of the 2020–10-16 sequences splits before all the others (Fig. 2 A). It is therefore possible that the ORF8 E19stop mutation occurred in the mink population, and then spread to humans.
Fig. 2.
SARS-CoV-2 variants with truncated ORF8 observed in farmed mink (Neovison vison). A) Maximum-likelihood phylogenetic tree with all SARS-CoV-2 complete genome sequences with the ORF8 E19stop mutation detected in Denmark. Sequences obtained from mink are shown in red. The cluster of sequences with the spike N501T mutation is highlighted by an orange box. Bootstrap support values are shown in main branches. B) Predicted 3D structure of the spike glycoprotein (grey ribbon) in complex with host cell receptor ACE2 (green ribbon), with amino acids 501 (yellow circles) and 614 (blue circles) highlighted. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Five sequences had a mutation (N501T) in a spike site also mutated in the VOC-202012/01 variant (N501Y), three from mink and two from humans (Fig. 2A). The five sequences formed a cluster in the phylogenetic analysis, suggesting a single origin of the mutation. The three mink sequences were equal and diverge from the sequences found in humans by two and three mutations, all outside the spike and ORF8 genes (Table 2 ). In addition to spike N501T, these five sequences had the D614G mutation. Among sequences with the spike N501T mutation, the mink sequences had a collection date (2020–11-04) which is 12 and 18 days before the collection date of samples from humans (Fig. 2A), suggesting that N501T had an origin in the mink population. The N501Y mutation is located at the receptor binding domain (RBD) of the spike protein (Fig. 2B) and helped the adaptation of SARS-CoV-2 in BALB/c mice, being potentially associated with the increased virulence (Gu et al., 2020). Mutation N501T gives rise to a hydrogen bond to lysine 353 (van Dorp et al., 2020) and was shown to enhance ACE2 affinity (Starr et al., 2020). According to GISAID, spike mutation N501T occurred 47 times (0.02% of all samples with Spike sequence) in 9 countries.
Table 2.
Number of differences among the five SARS-CoV-2 complete genome sequences reported in mink and human with both ORF8 E19stop and spike N501T.
| Mink EPI_ISL_683023 | Mink EPI_ISL_683024 | Mink EPI_ISL_683025 | Human EPI_ISL_713271 | |
|---|---|---|---|---|
| Mink EPI_ISL_683024 | 0 | |||
| Mink EPI_ISL_683025 | 0 | 0 | ||
| Human EPI_ISL_713271 | 3 | 3 | 3 | |
| Human EPI_ISL_714262 | 2 | 2 | 2 | 5 |
4. Discussion
The ORF8 accessory gene is believed to interfere with immune responses and many putative cellular interactors have been identified, but its precise function remains unknown (Gordon et al., 2020, Flower et al., 2021). Expression of ORF8 is not essential for the virus replication, as suggested by the several variants lacking this gene identified worldwide (Pereira, 2020, Pereira, 2021, Zinzula, 2021, Alkhansa et al., 2021). This fact was first noticed in a lineage with an ORF8 382-nucleotide deletion detected in Singapore and Taiwan (Gong et al., 2020, Su et al., 2020). Curiously, middle and late phases of the 2002/2003 SARS epidemic were characterized by the spread of SARS-CoV with either partial or complete deletions of the ORF8 gene (Guan et al., 2003, C.S.M.E. Consortium, 2004). It remains unclear if the truncated ORF8 may have changed the SARS-CoV replication capacity or virulence in a way that favoured its adaptation to humans and/or its spread during the SARS epidemic (Guan et al., 2003; C.S.M.E. Consortium, 2004; Lau et al., 2005, Chen et al., 2007). Whatever the case, deletions at ORF8 were the most remarkable changes observed during the SARS epidemic. The same may be happening in SARS-CoV-2, as more transmissible lineages become dominant as the pandemic progresses.
Patients infected with variants lacking ORF8 had less severe symptoms and a possible prolonged infection period that may increase the opportunities for the virus spread (Young et al., 2020). Interestingly, the VOC-202012/01 lacks ORF8 in addition to several spike mutations. It remains to be determined if the absence of ORF8 contributes to the increases transmissibility. The longer duration of symptoms resulting from the absence of ORF8 may increase the opportunity for transmission, and the milder symptoms may increase undiagnosed cases and the number of transmissions.
The identification of a pangolin harbouring a SARS-CoV-2-related virus without a functional ORF8 suggests that loosing this accessory gene may be a common event. In addition to SARS-CoV, deletion of accessory genes have been observed in the Middle East respiratory syndrome related coronavirus (MERS-CoV) (Lamers et al., 2016), feline coronavirus (FCoV) (Kennedy et al., 2001), porcine respiratory coronavirus (PRCV) (Chen et al., 2019) and mouse hepatitis virus (MHV) (de Haan et al., 2002), often leading to an attenuation of virulence. It is clear that accessory genes play a dynamic role in such viruses, often varying along the progression of the pandemics.
The pangolin sample P1E has 92 spike amino acid substitutions in relation to the reference SARS-CoV-2, including V483Q, F490Y, N501T and P681M. The spike protein consists of 2 subunits (S1 and S2) that mediate infection of host cells, with S1 contains a RBD responsible for recognizing and binding ACE2 (Zhou et al., 2020). The RBD region includes the positions 483, 490, 501, where pangolin and human viruses diverge. Changes in these RBD critical sites may influence the virus capacity to infect cells, and therefore be subject to intense selection. Although difficult to perform, a study of SARS-CoV-2-related viruses in pangolins lacking ORF8 could help to understand how ORF8 and spike mutation affect the transmissibility of these viruses.
The detection of ORF8-deficient lineages in farmed mink is also remarkable considering the low frequency of such lineages in the human population, at least before the spread of VOC-202012/01. It is therefore likely that the loss of ORF8 occurred within the mink population, suggestive of adaptation of the virus to the new host or simple by random events. The identification of sequences from humans and mink clustering together suggests that lineages without ORF8 can spill over from one species to another, although it is not clear which species infected the other in this case. The mink population could also have been a good model to study how variants with and without ORF8 may have different transmissibility rates.
Overall, the co-occurrence of ORF8 and spike mutations with phenotypic consequences is particularly problematic, as it may give rise to variants with higher transmissibility, as observed with VOC-202012/01. The data provided here suggests that such variants already occurred in other host species, revealing the risk of having reservoirs of SARS-CoV-2 in species close to humans.
Declaration of Competing Interest
The author declare that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
I would like to thank all the researches who have kindly shared genomes on public databases.
Funding
This work was supported by the Fundação para a Ciência e a Tecnologia [RESEARCH 4 COVID-19 project No. 029] and the EOSCsecretariat.eu COVID-19 Fast Track Funding. EOSCsecretariat.eu has received funding from the European Union's Horizon Programme call H2020-INFRAEOSC-05-2018-2019, grant Agreement number 831644.
References
- Alkhansa A., Lakkis G., El Zein L. Mutational analysis of SARS-CoV-2 ORF8 during six months of COVID-19 Pandemic. Gene Reports. 2021;23:101024. doi: 10.1016/j.genrep.2021.101024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand, M., Hopkins, S., Dabrera, G., Achison, C., Barclay, W., Ferguson, N., Volz, E., Loman, N., Rambaut, A., Barrett, J. 2020. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01. Public Health England.
- Chen C.-Y., Ping Y.-H., Lee H.-C., Chen K.-H., Lee Y.-M., Chan Y.-J., Lien T.-C., Jap T.-S., Lin C.-H., Kao L.-S. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196(3):405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Knutson T.P., Rossow S., Saif L.J., Marthaler D.G. Decline of transmissible gastroenteritis virus and its complex evolutionary relationship with porcine respiratory coronavirus in the United States. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-40564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C.S.M.E. Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 303. 1666–1669. [DOI] [PubMed]
- Damas J., Hughes G.M., Keough K.C., Painter C.A., Persky N.S., Corbo M., Hiller M., Koepfli K.-P., Pfenning A.R., Zhao H., Genereux D.P., Swofford R., Pollard K.S., Ryder O.A., Nweeia M.T., Lindblad-Toh K., Teeling E.C., Karlsson E.K., Lewin H.A. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. 2020;117:22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, N.G., Barnard, R.C., Jarvis, C.I., Kucharski, A.J., Munday, J., Pearson, C.A., Russell, T.W., Tully, D.C., Abbott, S., Gimma, A. 2020. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. medRxiv.
- de Haan C.A.M., Masters P.S., Shen X., Weiss S., Rottier P.J.M. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology. 2002;296(1):177–189. doi: 10.1006/viro.2002.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimonaco N.J., Salavati M., Shih B.B. Computational Analysis of SARS-CoV-2 and SARS-Like Coronavirus Diversity in Human Bat and Pangolin Populations. Viruses. 2021;13(1):49. doi: 10.3390/v13010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 2008;9(4):267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Flower, T.G., Buffalo, C.Z., Hooy, R.M., Allaire, M., Ren, X., Hurley, J.H. 2021. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proceedings of the National Academy of Sciences 118. [DOI] [PMC free article] [PubMed]
- Gong Y.-N., Tsao K.-C., Hsiao M.-J., Huang C.-G., Huang P.-N., Huang P.-W., Lee K.-M., Liu Y.-C., Yang S.-L., Kuo R.-L. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes. Infect. 2020;9(1):1457–1466. doi: 10.1080/22221751.2020.1782271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5(4):529–530. doi: 10.1038/s41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B., He Y., Liu X., Zhuang Z., Cheung C., Luo S., Li P., Zhang L., Guan Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hammer A.S., Quaade M.L., Rasmussen T.B., Fonager J., Rasmussen M., Mundbjerg K., Lohse L., Strandbygaard B., Jørgensen C.S., Alfaro-Núñez A. SARS-CoV-2 Transmission between Mink (Neovison vison) and Humans Denmark. Emerg. Infect. Dis. 2021;27(2):547–551. doi: 10.3201/eid2702.203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.-Z. Pangolins harbor SARS-CoV-2-related coronaviruses. Trends Microbiol. 2020;28(7):515–517. doi: 10.1016/j.tim.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 2019;20(4):1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M., Boedeker N., Gibbs P., Kania S. Deletions in the 7a ORF of feline coronavirus associated with an epidemic of feline infectious peritonitis. Vet. Microbiol. 2001;81(3):227–234. doi: 10.1016/S0378-1135(01)00354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet. Infect. Dis. 2021;21(1):18–19. doi: 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.-Y., Jia N.a., Zhang Y.-W., Shum M.-H., Jiang J.-F., Zhu H.-C., Tong Y.-G., Shi Y.-X., Ni X.-B., Liao Y.-S. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583(7815):282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lamers M.M., Raj V.S., Shafei M., Ali S.S., Abdallh S.M., Gazo M., Nofal S., Lu X., Erdman D.D., Koopmans M.P., Abdallat M. Deletion variants of Middle East respiratory syndrome coronavirus from humans, Jordan, 2015. Emerg. Infect. Dis. 2016;22(4):716–719. doi: 10.3201/eid2204.152065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.-W., Wong B.H.L., Wong S.S.Y., Leung S.-Y., Chan K.-H., Yuen K.-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V., Longueville J.-E., Gascuel O. SMS: smart model selection in PhyML. Mol. Biol. Evol. 2017;34(9):2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad S., Bouchama A., Mohammad Alharbi B., Rashid M., Saleem Khatlani T., Gaber N.S., Malik S.S. SARS-CoV-2 ORF8 and SARS-CoV ORF8ab: Genomic divergence and functional convergence. Pathogens. 2020;9(9):677. doi: 10.3390/pathogens9090677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnink B.B.O., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oreshkova N., Molenaar R.J., Vreman S., Harders F., Munnink B.B.O., Hakze-van der Honing R.W., Gerhards N., Tolsma P., Bouwstra R., Sikkema R.S. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance. 2020;25(23) doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infect. Gen. Evol. 2020;85:104525. doi: 10.1016/j.meegid.2020.104525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira F. SARS-CoV-2 variants combining spike mutations and the absence of ORF8 may be more transmissible and require close monitoring. Biochem. Biophys. Res. Commun. 2021;550:8–14. doi: 10.1016/j.bbrc.2021.02.080. https://www.sciencedirect.com/science/article/pii/S0006291X21002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A., Loman, N., Pybus, O., Barclay, W., Barrett, J., Carabelli, A., Connor, T., Peacock, T., Robertson, D.L., Volz, E. 2020b. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. COVID-19 Genomics Consortium UK (CoG-UK).
- Sailleau C., Dumarest M., Vanhomwegen J., Delaplace M., Caro V., Kwasiborski A., Hourdel V., Chevaillier P., Barbarino A., Comtet L. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound. Emerg. Dis. 2020;67(6):2324–2328. doi: 10.1111/tbed.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit T.H., Brackman C.J., Ip S.M., Tam K.W., Law P.Y., To E.M., Veronica Y., Sims L.D. Infection of dogs with SARS-CoV-2. Nature. 2020;586(7831):776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y.C., Anderson, D.E., Young, B.E., Linster, M., Zhu, F., Jayakumar, J., Zhuang, Y., Kalimuddin, S., Low, J.G. Tan, C.W. 2020. Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2. mBio 11. [DOI] [PMC free article] [PubMed]
- van Dorp, L., Tan, C.C., Lam, S.D., Richard, D., Owen, C., Berchtold, D., Orengo, C. and Balloux, F. 2020. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv.
- Volz, E., Mishra, S., Chand, M., Barrett, J.C., Johnson, R., Geidelberg, L., Hinsley, W.R., Laydon, D.J., Dabrera, G. and O’Toole, Á. 2021. Transmission of SARS-CoV-2 Lineage B. 1.1. 7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2020.12. 30.20249034.
- Young B.E., Fong S.-W., Chan Y.-H., Mak T.-M., Ang L.W., Anderson D.E., Lee C.-P., Amrun S.N., Lee B., Goh Y.S. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396(10251):603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020:1–4. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L. Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophy. Res. 2021;538:116–124. doi: 10.1016/j.bbrc.2020.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]