Abstract
Purpose:
To describe a large, publicly available dataset comprising computed tomography (CT) projection data from patient exams, both at routine clinical doses and simulated lower doses.
Acquisition and Validation Methods:
The library was developed under local ethics committee approval. Projection and image data from 299 clinically performed patient CT exams were archived for three types of clinical exams: noncontrast head CT scans acquired for acute cognitive or motor deficit, low-dose noncontrast chest scans acquired to screen high-risk patients for pulmonary nodules, and contrast-enhanced CT scans of the abdomen acquired to look for metastatic liver lesions. Scans were performed on CT systems from two different CT manufacturers using routine clinical protocols. Projection data were validated by reconstructing the data using several different reconstruction algorithms and through use of the data in the 2016 Low Dose CT Grand Challenge. Reduced dose projection data were simulated for each scan using a validated noise-insertion method. Radiologists marked location and diagnosis for detected pathologies. Reference truth was obtained from the patient medical record, either from histology or subsequent imaging.
Data Format and Usage Notes:
Projection datasets were converted into the previously developed DICOM-CT-PD format, which is an extended DICOM format created to store CT projections and acquisition geometry in a nonproprietary format. Image data are stored in the standard DICOM image format and clinical data in a spreadsheet. Materials are provided to help investigators use the DICOM-CT-PD files, including a dictionary file, data reader, and user manual. The library is publicly available from The Cancer Imaging Archive (https://doi.org/10.7937/9npb-2637).
Potential Applications:
This CT data library will facilitate the development and validation of new CT reconstruction and/or denoising algorithms, including those associated with machine learning or artificial intelligence. The provided clinical information allows evaluation of task-based diagnostic performance.
Keywords: CT projection data, iterative reconstruction, low-dose CT, machine learning, patient data
1. INTRODUCTION
Introduced to the world in 1971, x-ray computed tomography (CT) remains an invaluable medical technology that continues to undergo significant hardware and algorithmic advances. Iterative reconstruction (IR) methods, which were used in the EMI Mark I CT system invented by Sir Godfrey Hounsfield,1,2 were quickly replaced by much faster to perform filtered back projection (FBP) methods, which have been the primary method for reconstructing clinical CT images for decades. With the advent of helical (spiral) and multidetector row CT technologies, analytical CT reconstruction approaches evolved to take into consideration new data acquisition geometries, including cone beam geometries. Between approximately 1990 and 2010, iterative approaches to CT image reconstruction began to emerge that demonstrated improved spatial resolution, decreased image noise, or both.3–7
In 2003, Thibault et al. used multislice helical CT reconstruction projection data from a clinical CT exam to compare images reconstructed with a model-based statistical iterative reconstruction approach to those reconstructed using a commercial FBP-based approach, demonstrating both improved in-plane spatial resolution and decreased image noise.8 Since then, new reconstruction algorithms have been routinely evaluated by comparing the results from the new algorithm to those from an established high-quality reconstruction approach, where each method uses the same input projection data. Radiologist preferences were initially used to evaluate the clinical acceptability of new iterative reconstruction algorithms, which gave way to more rigorous evaluation methods, which have demonstrated, for example, that these new nonlinear reconstruction and denoising approaches have contrast-dependent spatial resolution and change the shape of the noise power spectrum.9–11 To evaluate the impact of these effects on the ability of human observers to perform clinically relevant tasks, multireader, multicase observer performance studies and model observer performance studies have become essential to adequately demonstrate the ability of a new algorithm to maintain or exceed a desired level of diagnostic performance under the condition of reduced patient radiation dose. A subset of cases from this data library has been successfully used for such studies.12–19 Since conducting the 2016 Low Dose CT Grand Challenge,16 in conjunction with the American Association of Physicists in Medicine and support from NIH awards EB017095 and EB017185, over 500 investigators from over 40 countries have requested access to the 30 abdominal CT studies used in the Grand Challenge. In the first 2 weeks after the data library was made public, over 22 TB of data consisting of nearly 7000 image series have been downloaded (one scan results in one image series and one projection data series).
Investigators from a wide range of disciplines have expertise in image reconstruction or noise reduction methods, but to date have been unable to apply their knowledge to medical CT imaging due to the lack of availability of the necessary patient data. This is because access to clinical CT projection data has been extremely limited due to the proprietary information and formatting of manufacturer-specific projection data files. The purpose of this data library is to make patient CT projection data, and reference information regarding type and location of pathology, publicly available to accelerate development of high impact approaches to increasing diagnostic performance as patient dose is decreased. The availability of this CT data library will facilitate the development and validation of new CT reconstruction and/or denoising algorithms, including those associated with machine learning or artificial intelligence while the provided clinical information will allow assessment of task-based diagnostic performance.
2. ACQUISITION AND VALIDATION METHODS
2.A. Overview of dataset
The library consists of CT patient scans from three common exam types: noncontrast head CT scans acquired for acute cognitive or motor deficit, low-dose noncontrast chest scans acquired to screen high-risk patients for pulmonary nodules, and contrast-enhanced CT scans of the abdomen acquired to look for metastatic liver lesions. A large majority of the head CT scans were performed using our default clinical head CT protocol to rule evaluate acute neurologic deficit, bleeding/hemorrhage. All patients had suspected acute neurologic deficit. A few patients were scanned using a higher dose setting as part of a trauma protocol in our Emergency Department. Similarly, contrast-enhanced abdominal CT scans were all portal phase at the dose level corresponding to our default contrast-enhanced abdominal CT dose level. The majority of scans were obtained at the routine dose level; however, some multiphase scans were performed at higher dose levels. In these cases, the noise insertion tool was used to standardize the radiation dose level of the “full” dose cases to that of the default abdominal CT protocol. The data for each patient include CT projection data at the acquired (full) dose and a simulated reduced dose, reconstructed image data, and the location and diagnostic for positive findings. With assistance of the participating scanner manufacturers, Siemens Healthineers (Forchheim, Germany) and GE Healthcare (Waukesha, WI), projection data were converted from the manufacturer’s proprietary format into the previously described DICOM-CT-PD data format.20 Reconstructed images are provided using the DICOM-CT image storage standard.21 Clinical reports provide the location and diagnosis for positive findings, including snapshots of the identified findings delineated by radiologist-drawn regions of interest. These data are provided in a non-DICOM spreadsheet file.
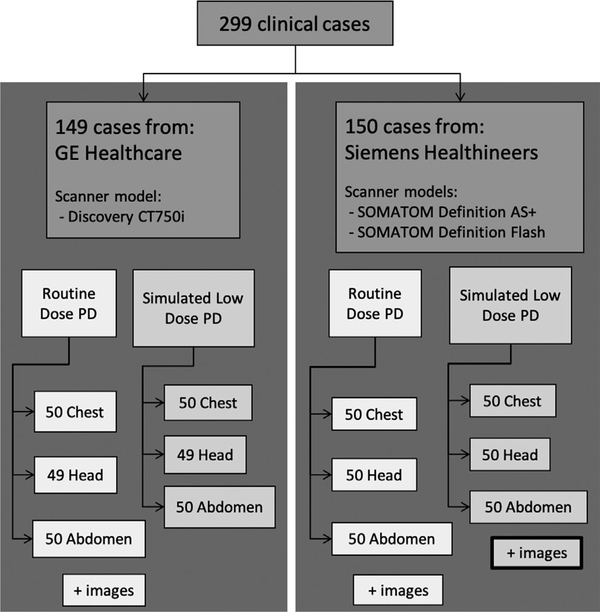
After approval from Mayo Clinic’s Institutional Review Board, patient data were collected at two Mayo Clinic locations (Rochester MN and Scottsdale AZ) using each practice’s routine clinical protocols. A total of 299 adult patient cases were collected, which included CT scans of the head, chest, and abdomen (Fig. 1). Approximately 50% of the data are negative for disease. Each case includes projection data, image data, and clinical findings.
FIG. 1.
Breakdown of the patient cases included in the Low Dose computed tomography (CT) and Projection Data library, outlining how many cases are provided for each manufacturer, anatomical region, and dose level. Reconstructed images are only provided for the simulated low-dose projection data (PD).
2.B. Data acquisition, modification, and reconstruction
Both of the manufacturers whose CT systems were used in the acquisition of patient data granted permission to share projection data using the vendor-neutral DICOM-CT-PD format. The DICOM-CT-PD files were generated using a MATLAB (MathWorks, MATLAB version R2016a) script. The headers of the DICOM-CT-PD files provide the geometric information required for image reconstruction, which were provided by each manufacturer. The attenuation information for each projection was written into the DICOM-CT-PD pixel data matrix using the developed MATLAB script. All DICOM-CT-PD header tags and conventions used to describe the acquisition geometry are detailed in the provided user manual. A DICOM-CT-PD data dictionary and reader script are also provided.
Projection data for each patient were obtained from either a GE Discovery CT750i, SOMATOM Definition AS+or SOMATOM Definition Flash CT system. The projection data were taken from right before image reconstruction, after all preprocessing and the logarithm operation; data without preprocessing such as beam hardening corrections were not available for use in this work.
Acquisition and reconstruction parameters (Table I), which varied by scanner model and anatomic region, were dictated by the routine clinical protocols for each of the three clinical indications studied, but occasionally were adapted according to the clinical situation by the supervising radiologist. The specific acquisition parameters used for each patient case are recorded in the header tags of both the DICOM-CTPD projection data and DICOM image files; reconstruction parameters are recorded only in the header tags of the DICOM image files.21 The acquired data are referred to as the full-dose data.
Table I.
Key data acquisition parameters for each exam type.
| Manufacturer (Scanner) | Scan parameter | Head CT for acute cognitive or motor deficit | Chest CT for lung cancer screening* | Abdomen CT for metastatic liver lesion detection |
|---|---|---|---|---|
| GE Healthcare (Discovery CT750i) | Scan geometry | Axial and Helical | Helical | Helical |
| Contrast enhanced | No | No | Yes | |
| Manual tube current (mA) | 300 | – | – | |
| Noise index for Smart mA | – | Variable based on patient size, tube potential, and thickness of first image series reconstructed | ||
| Tube potential (kV) | 120 | 80 – 120 | 80 – 120 | |
| Rotation time (s) | 1 | 0.5 | 0.5 | |
| Mean CTDIvol (mGy) | 56.8 | 6.4 | 12.2 | |
| Siemens Healthineers (SOMATOM Definition AS+, SOMATOM Definition Flash) | Scan geometry | Helical | Helical | Helical |
| Contrast enhanced | No | No | Yes | |
| Manual effective tube current time product (effective mAs) | 250 | – | – | |
| Quality reference mAs for CareDose4D | – | 70 | 200 at reference of 120 kV | |
| Tube potential (kV) | 120 | 120 | 100 – 120 | |
| Rotation time (s) | 1 | 0.28 or 0.3 | 0.5 | |
| Mean CTDIvol (mGy) | 43.7 | 6.6 | 15.6 | |
To provide a better ground truth, cases were collected using a chest CT protocol that used twice the radiation dose than was used in the National Lung Screening Trial.
CTDIvol = Volume CT Dose Index
A second set of projection data was generated for each scan by inserting noise into the full-dose data to simulate a low-dose scan. Noise was inserted using a previously validated photon counting model that incorporates the effect of the bowtie filter, automatic exposure control, and electronic noise.22 To account for the difference in detector and bowtie filter, the noise insertion model was validated for each scanner model and exam type in a similar way as in the Ref. [22]. All the Siemens exams used in this dataset were acquired with the single source mode on Flash scanners or the single-source AS+scanners, the noise insertion model described in Ref. [22] was modified slightly (a calibration factor and electronic noise parameter) to accommodate the slight difference in detector among these scanner models. For GE scanners, the bowtie profiles were remeasured and determined in the same way as in Ref. [22]. Head and abdomen projection data were modified to simulate an exam acquired at 25% of the full dose; the low-dose chest projection data simulated an exam acquired at 10% of the full dose. These are referred to as the simulated low-dose datasets.
For patient data from both manufacturers, automatic tube current modulation was used in scans of the chest and abdomen but not used in scans of the head. For Siemens data, the tube current information for each projection was directly taken from the respective field in their proprietary data format using decoding tools provided to us by Siemens. For GE data, only the mean tube current across the entire scan was provided to us by GE. As we did not have access to their data format, we could not read the per-projection tube current information, and so we needed to empirically infer the tube current modulation information. We accomplished this using the prelog signal at the peripheral detectors, where patient attenuation was absent. In the few datasets where some patient attenuation was observed at the first and last detector channels, we used interpolation from neighboring projections to estimate the unobstructed detector signal, which is directly proportional to the tube current. The resulting per-projection tube current data were then normalized using the provided mean tube current.
Because some reconstruction algorithms require statistical information, we calculated and provided noise maps for both manufacturers, expressed as an array describing the spatial distribution of noise equivalent quanta along the direction of the detector columns. The noise map takes into account the shape of the bowtie filter and automatic tube current modulation, but neglects the variation across detector rows; the calculation methodology has been previously described by Yu et al.22 The noise maps for the GE data used the empirically derived per-projection tube current values.
Images were reconstructed from the full-dose projection data on the scanner used for each patient exam (Table II). A second image series was generated with the simulated low-dose projection data for patients scanned on Siemens scanners, where it was possible to return the modified projection data to the scanner for reconstruction using the commercial weighted FBP algorithm.23 Images created using IR are not provided for any datasets.
Table II.
Key data reconstruction parameters for each exam type.
| Manufacturer (Scanner) | Reconstruction parameter | Head CT for acute cognitive or motor deficit | Chest CT for lung cancer screening | Abdomen CT for metastatic liver lesion detection |
|---|---|---|---|---|
| GE Healthcare (Discovery CT750i) | Field of view (mm) | 200–260 | 282–423 | 315–500 |
| Reconstruction algorithm | Standard | Standard | Standard | |
| Slice / Increment (mm) | 5 / 5 | 1.25 / 1 | 5 / 3 m | |
| Siemens Healthineers (SOMATOM Definition AS+, SOMATOM Definition Flash) | Field of view (mm) | 250 | 300–500 | 300–500 |
| Reconstruction kernel | H40 | B50 | B30 | |
| Slice / Increment (mm) | 5 / 5 | 1.5 / 1 | 5 / 3 | |
2.C. Clinical information
In addition to the clinical image interpretation performed for each patient, board-certified subspecialist radiologists reviewed all patient cases, including the patient medical record. A region of interest was drawn around each finding (e.g., pulmonary nodule, liver metastasis) and recorded in a custom database, along with the pixel coordinates of the finding, the diagnosis, the diagnostic reference (source of truth), patient age and gender, and a hyperlinked snapshot of each finding.
2.D. Validation studies
Accuracy of the conversion from the manufacturer’s proprietary data format to DICOM-CT-PD was confirmed on the ACR phantom scans by using in-house and open-source software to reconstruct images and comparing them to the commercial reconstructions.20 A subset of the data (13 cases) were successfully used in the 2016 Low Dose CT Grand Challenge.16 The rest of the cases were also tested by reconstructing images from the converted DICOM-CT-PD data format. [Correction added on January 30, 2021, after first online publication: The 30 cases have been changed to 13 cases.]
The accuracy of the noise insertion method used in this work has also been previously demonstrated.22 Additionally, after noise was inserted into each projection dataset, the amount of noise in the reconstructed images was confirmed by measuring the ratio of noise (standard deviation of pixel values) in the simulated low-dose images to that in full-dose images and comparing to the predicted values, which were calculated assuming a Poisson noise distribution (i.e., the inverse square relationship between dose and noise). The average percent difference between the noise inserted images and the theoretically predicted values are −4.2% ± 6.2%; −2.6% ± 4.8%; and 16.1% ± 8.9% for Siemens head, abdomen, and chest exams, respectively. The differences in noise levels in head and abdomen exams were within a reasonable range. The higher differences observed in the chest exams were expected due to the much lower radiation dose used in those cases (70 QRM for the full dose and 7 QRM for the low dose), which results in electronic noise becoming a non-negligible factor that can significantly increase the noise in lower-dose images compared with the value predicted based only on the inverse square relation (i.e., Poisson noise). Because of this, the noise insertion algorithm took into account the increased contribution of electronic noise. To determine approximately how much of the 16.1% difference might be due to the effects of electronic noise, scans were acquired of an anthropomorphic chest phantom at these low dose levels. The data showed that the differences in noise between measured and theoretically predicted values were about 11%. This baseline difference in noise relative to that predicted based on Poisson noise alone existed at very low doses due to the presence of electronic noise. Therefore, because our noise insertion method addressed electronic noise but the differences noted above (e.g., 16.1%) did not, the deviation of the noise in simulated low-dose chest exams compared with a real data acquisition is estimated to be much smaller, within approximately 5–6%.
Each case was de-identified using a custom MATLAB script and removal of all protected health information (PHI) confirmed prior to transferring data to the data repository (The Cancer Imaging Archive, TCIA), where de-identification was confirmed prior to moving the data to the data archive.24 Final verification of the data and evaluation for PHI was performed by re-downloading the cases from TCIA using the National Biomedical Imaging Archive (NBIA) data retriever and comparing the retrieved header and pixel data to the original data.
During case selection, inclusion of positive cases required confirmation of the radiological diagnosis with an independent source of truth (Table III). Findings in the head and liver were confirmed with clinical or imaging evidence of disease stability, progression, or regression after treatment, or histological evidence from resection or biopsy. However, the diagnostic task selected for the chest exams was identification of indeterminate pulmonary nodules, which by definition are neither actionable nor clearly negative. For these cases, the initial clinical interpretation was reviewed and confirmed by un-blinded subspecialized thoracic radiologist.
Table III.
Information used to confirm radiological diagnosis.
| Head | Chest | Abdomen |
|---|---|---|
| Source of truth | ||
| •Additional imaging •Clinical correlation with symptoms or physical findings •Pathologic diagnosis •Surgical correlation •Stability or progression |
•Unblinded interpretation by a second subspecialized thoracic radiologist | •Histology •Similar proven lesion •Stabile > 6 months •Progression •Response to therapy |
3. DATA FORMAT AND USAGE NOTES
All data collected for this data library are in compliance with the Health Insurance Portability and Accountability Act (HIPAA) de-identification standards and are stored at TCIA.24
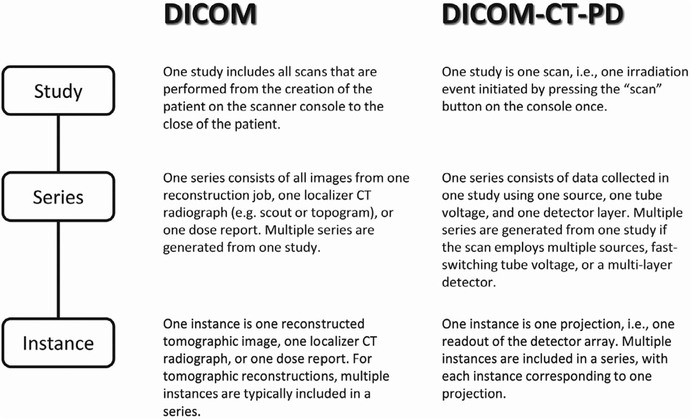
The DICOM-CT-PD format stores attenuation information in the pixel data section of the file and stores the parameters and geometry necessary for image reconstruction in the DICOM-CT-PD header section. It is an extended DICOM class and the study, series, and instance definitions were altered from the standard DICOM definition to accommodate having images and two projection datasets associated with a single “scan/irradiation event” (Fig. 2). Additionally, a sequence of private tags is incorporated into each DICOM-CT-PD file. A data dictionary file is provided for interpreting these tags, as well as a user manual to describe the function of each tag (Table IV). It is important to note that the definitions of some tags differ from those given in our previous publication.20 Each DICOM-CT-PD file is an individual projection (i.e., one view) or one readout of the complete detector array. Therefore, there are large numbers of DICOM-CT-PD files per scan, all of which fall under one study unique identifier (UID) and series UID. This approach substantially decreases algorithm development time as reconstructions can be initiated using only several rotations worth of projections; this would not be possible if all projections were contained in a single data file.
FIG. 2.
The definitions of study, series, and instance for the standard DICOM image format and the modified definitions necessary for the DICOM-CT-PD format.
Table IV.
Private tags in the header of DICOM-CT-PD format.
| Tag | Attribute Name | Description |
|---|---|---|
| (7029,1010) | NumberofDetectorRows | The number of detector rows. |
| (7029,1011) | NumberofDetectorColumns | The number of detector columns. |
| (7029,1002) | DetectorElementTransverseSpacing | The width of each detector column, measured at the detector (mm). |
| (7029,1006) | DetectorElementAxialSpacing | The width of each detector row, measured at the detector (mm). |
| (7029,100B) | DetectorShape | The shape of the detector, such as “CYLINDRICAL,” “SPHERICAL,” or “FLAT.” |
| (7031,1001) | DetectorFocalCenterAngularPosition | ϕ0, the azimuthal angles of the detector’s focal center (rad). |
| (7031,1002) | DetectorFocalCenterAxialPosition | z0, the z location of the detector’s focal center (mm). |
| (7031,1003) | DetectorFocalCenterRadialDistance | ρ0, the in-plane distances between the detector’s focal center and the isocenter (mm). |
| (7031,1031) | ConstantRadialDistance | d0, the distance between the detector’s focal center and the detector element specified in Tag(7031,1033) (mm) |
| (7031,1033) | DetectorCentralElement | (Column X,Row Y), the index of the detector element aligning with the isocenter and the detector’s focal center. |
| (7033,100B) | SourceAngularPositionShift | Δϕ, the ϕ offset from the focal spot to the detector’s focal center (rad). |
| (7033,100C) | SourceAxialPositionShift | Δz, the z offset from the focal spot to the detector’s focal center (mm). |
| (7033,100D) | SourceRadialDistanceShift | Δρ, the ρ offset from the focal spot to the detector’s focal center (mm). |
| (7033,100E) | FlyingFocalSpotMode | The mode of flying focal spot (FFS). “FFSNONE” means no flying focal spot; “FFSZ” means flying focal spot along axial direction; “FFSXY” means in-plane flying focal spot; and “FFSXYZ” means flying focal spot in-plane and along axial direction. |
| (7033,1013) | NumberofSourceAngularSteps | The number of projections per complete rotation. |
| (7033,1061) | NumberofSpectra | The number of sources/tube voltages/detector layers/energy thresholds/energy bins used in the data acquisition. |
| (7033,1063) | SpectrumIndex | The index of the source/tube voltage/detector layer/energy threshold/energy bins. |
| (7033,1065) | PhotonStatistics | An array describing the spatial distribution of photons along the direction of the detector columns, from Column 1 to Column M (neglecting the variation across detector rows). Each element of the array corresponds to a detector column. |
| (7033,1067) | Timestamp | The timestamp in absolute time (ms). |
| (7037,1009) | TypeofProjectionData | “AXIAL” or “HELICAL” |
| (7037,100A) | TypeofProjectionGeometry | “FANBEAM” for third generation CT geometry. |
| (7039,1003) | BeamHardeningCorrectionFlag | A flag used to define whether the projection data have been corrected for beam hardening effects. “YES” or “NO.” |
| (7039,1004) | GainCorrectionFlag | A flag used to define whether the projection data have been calibrated for detector response with respect to the dynamic range available. “YES” or “NO.” |
| (7039,1005) | DarkFieldCorrectionFlag | A flag used to define whether the background signals prior to the x-ray exposure has been subtracted from the projection data. “YES” or “NO.” |
| (7039,1006) | FlatFieldCorrectionFlag | A flag used to define whether the gradient of flood field introduced by the heel effect and the bowtie filter has been compensated in the projection data. “YES” or “NO.” |
| (7039,1007) | BadPixelCorrectionFlag | A flag used to define whether abnormal pixels have been removed from the projection data by interpolation. “YES” or “NO.” |
| (7039,1008) | ScatterCorrectionFlag | A flag used to define whether the projection data has been corrected for scattered radiation. “YES” or “NO.” |
| (7039,1009) | LogFlag | A flag used to define whether the projection data has been logarithmically transformed. “YES” or “NO.” |
| (7041,1001) | WaterAttenuationCoefficient | A calibration factor μ’ for the conversion of measured linear attenuation coefficients μ to CT numbers (mm−1): CT numbers = 1000 * (μ− μ’) / μ’ |
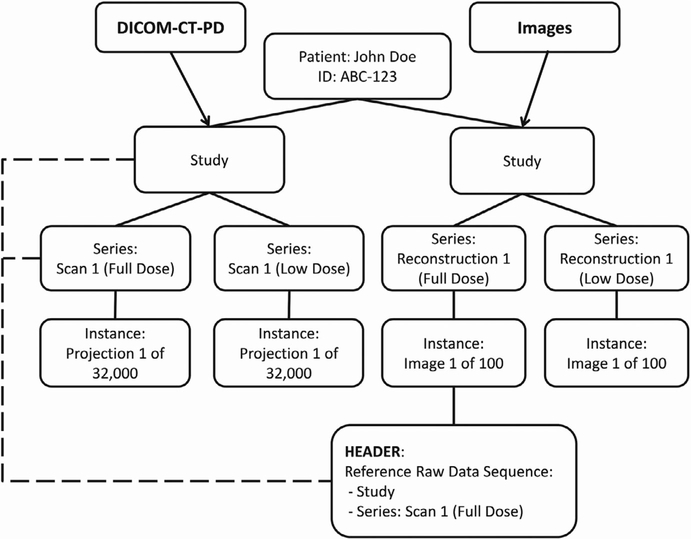
Images included in this dataset follow the standard DICOM image format.21 Figure 3 illustrates the relationship between study, series, and instance UIDs at different dose levels between the DICOM-CT-PD projection data and the associated DICOM images for a given patient. Within each of the image series headers is a tag sequence that helps track and identify the original raw data from which it was derived. This DICOM tag sequence, identified as the Reference Raw Data Sequence (0008,9121), contains the study and series UIDs from the DICOM-CT-PD projection data from which the image series originated.
FIG. 3.
The relationship between DICOM-CT-PD files and the associated image data showing how the relationship between the two is maintained using the reference raw data sequence tags.
An anonymized patient name and identifier is used for each case in the dataset. All patients with a head scan are identified with an N followed by a 3 digit number, chest cases with a C, and abdomen cases with an L. Both the patient name and ID are the same. The series description can help identify if the file is projection data, image data, full dose, or simulated lower dose. Information regarding specific tags and other important details on using the private tags are located in the user manual, accessible with the dataset.
This data collection is named Low Dose CT Image and Projection Data (LDCT-and-Projection-Data) and can be accessed on the TCIA website www.cancerimagingarchive.net or by digital object identifier (DOI) 10.7937/9npb-2637. This dataset is 1.32 TB in size. It is comprised of 299 cases, 13,009,241 files, and 3 clinical reports. The extremely large number of files is a consequence of storing each individual project in its own file.
4. DISCUSSION
The potential value of the described data library has been demonstrated through its use in the 2016 Low Dose CT Grand Challenge sponsored by the Mayo Clinic, American Association of Physicists in Medicine and the National Institute of Biomedical Imaging and Bioengineering.16 The purpose of the challenge was to provide common datasets and evaluation methods to investigators and thereby estimate and compare the diagnostic performance of image-based denoising techniques and iterative reconstruction algorithms for the task of detecting hepatic metastases from simulated low-dose CT data (25% of the full dose). Interest in the challenge was very high, with 90 sites registering to participate from over 20 different countries. Since completion of the challenge, over 500 investigators from over 40 countries have requested access to the abdominal CT studies used in the 2016 Grand Challenge. It thus appears clear that this much larger data library will be very valuable to the research community. The careful annotation of pathology will be of particular value in the training and testing of novel artificial intelligence technologies. [Correction added on January 30, 2021, after first online publication: The 30 abdominal have been deleted.]
Below are examples of research studies utilizing our data:
A deep convolutional neural network using directional wavelets for low-dose x-ray CT reconstruction.25
A residual encoder–decoder convolutional neural network (RED-CNN) for 2D and 3D CT denoising.26,27
A generative adversarial network (GAN) for low-dose CT denoising.28,29
A multiresolution deep learning U-net for sparse-view CT.30
Performance comparison of CNN-based image denoising methods using different loss functions.31
A self-attention CNN for low-dose CT denoising with self-supervised perceptual loss network.32
A cycle-consistent adversarial network (CycleGAN) for low-dose CT image denoising without paired CT images for training.33
A residual CNN for liver extraction from low-dose CT images.34
As additional examples of the value of these data, within our own research program and clinical practice, we have used these and other data to.
determine optimal protocol settings in our large sub-specialty clinical practice,35–38
conduct multireader, multicase (MRMC) studies to discern the impact of different reconstruction algorithms, patient dose levels and other factors on radiologist diagnostic performance and confidence,12–16,20,35–37,39,40
develop, and evaluate using MRMC studies, nonlocal means and deep learning-based image denoising methods,41–43 and
develop model observers and deep learning methods from phantom or patient data to predict human observer performance of radiologists when interpreting patient data to allow rapid optimization of protocols for any scanner model, exam type, or patient characteristics.17–19
This data library, however, does have several limitations.
The DICOM-CT-PD format is an extended DICOM format because its header needed to contain data in private tags beyond those defined in the standard DICOM information object definition. Standard DICOM interfaces will thus not recognize these private tags. To address this, a DICOM-CT-PD data dictionary is available to allow users to read the projection data and associated tags.
The DICOM-CT-PD data provided in this library are based on the projection data right before image reconstruction — after all the data corrections that have been performed by manufacturer (e.g., beam hardening, scattering, nonuniformity). For researchers who would like to develop algorithms to improve upon these corrections, or to take these and other nonidealities into account in the reconstruction algorithm, truly raw projection datasets, without the manufacturer’s corrections, are needed. Because the manufacturers did not provide us with access to these data, they are not provided in this library. We hope that further collaborations with CT manufacturers may provide access to the preprocessed data.
Although the lower-dose simulation method used in the creation of the low-dose data was fully validated, as described above, the simulated lower-dose data are limited in that they may not perfectly reflect what would have occurred had the patient actually been scanned at the lower-dose levels. For example, our confidence in our low-dose simulation approach is diminished when attempting to simulate extremely low-dose data, where the detected number of photons is so low that system electronic noise becomes a major factor. In that situation, manufacturers typically implement nonlinear processes in the data acquisition and processing chain, which are extremely challenging to emulate.
As described previously, we have validated that the simulated reduced dose projection data included in this library are very reasonable simulations of the data that would have been obtained in an actual measurement. Of course, there is always a potential for differences between simulated data and what a real measurement would have produced. We have directly measured this in phantoms and found the differences to be within 5–6%. However, for patient data, it is impossible to determine the magnitude of any such differences, given the difficulty of scanning hundreds of patients at both full and reduced doses. Furthermore, even if such a study were approved by an Institutional Review Board, it would be impossible to exactly match the contrast level enhancement and anatomic positions between two temporally separate scans, which decrease the value of the data for many applications.
We believe that given these limitations on acquiring matching full- and low-dose data, a simulation approach provides the best option for algorithm development and validation, independent of noise mitigation strategies implemented by a specific manufacturer that are unable to be simulated. At the simulated reduced dose levels included in this library, we believe that any differences that may exist between simulated and measured data have negligible impact on algorithms developed using the provided lower-dose data. This belief is supported by the successful use of the data in numerous publications.26–34
The dataset is provided in a paired fashion at both routine dose and simulated lower dose, which is ideal for many techniques involving supervised learning. Many noise reduction techniques trained based on these paired datasets have demonstrated great success in terms of reducing image noise and improving image quality.26–34 However, given the recent success of unsupervised low-dose CT reconstruction, it is desirable also to have unpaired low-dose and full-dose dataset available, which is a topic of interest for future development. Meanwhile, the current paired dataset provided in this work can still be used to meet that purpose using appropriate arrangement of training cases (e.g., organize the cases in an unpaired fashion), which is beneficial as the full-dose images can be used as references even if they are not used in the training and testing.
The number of files associated with each patient exam is extremely large (e.g., tens of thousands). This is because each unique projection is stored in a single file. Hence the number of files is determined by the number of projection views acquired during the scan. In pilot versions of the DICOM-CT-PD format, all projections were in a single file. This required users to import the entire large file (e.g., as large as 4 GB), even if they only wanted to use a few rotations of projection data to quickly perform a reconstruction or other processing action. Thus, based on this and other feedback from a number of colleagues who worked with our early pilot data, we decided to separate each projection view into a unique file. This provides a data structure that is as versatile as possible for current and future research directions.
The library currently contains only data from two scanner manufacturers and three currents, but no longer state-of-the-art, scanner models. As CT technology continues to advance, important new scanner attributes will not be represented in the current library, although with the assistance of scanner manufacturers to convert their projection data into the DICOM-CT-PD format, the size and diversity of the library can be easily expanded.
Although a wide range of anatomy and pathology are contained in the provided patient cases, they represent only a fraction of the clinical uses and findings from CT imaging. Here also, with the assistance of scanner manufacturers, the size and diversity of the library can be expanded to include data from any application or containing any pathology. We look forward to the day when manufacturers will provide tools to allow practices to export projection data from any patient exam or scanner model to a vendor-neutral projection data format, such as DICOM-CT-PD, for use by the research community.
5. CONCLUSION
The Low Dose CT Image and Projection Dataset described herein is publicly available at TCIA’s data repository.44 It comprises full and reduced dose projection data, reconstructed image data, and detailed pixel-based annotation of clinical findings for 299 patient CT exams over the head, chest, and abdomen for commercial scanners from two different CT manufacturers. To the best of our knowledge, no other open source data format or publicly available data repository exists in which projection data, scan geometry, and scan parameters are all accessible for clinical patient CT exams. The lack of such data has limited clinically relevant research in this field to CT scanner manufacturers and their small number of research collaborators. This unique data library will therefore facilitate the development and validation of new CT reconstruction and/or denoising algorithms, including those associated with machine learning or artificial intelligence.
ACKNOWLEDGMENTS
We acknowledge the many individuals that have contributed to this project: Drs. David DeLone, Jeff Fidler, David Levin, Amy Hara, Karl Stierstorfer, Thomas Flohr, Jiang Hsieh, Nobert Pelc, David Clunie, and Gregory Michalak, as well as Tammy Drees, Maria Shiung, Phil Edwards, Jayse Weaver, and Kris Nunez. Research reported in this article was supported by the National Institutes of Health under award numbers R01 EB017095 and U01 EB017185. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. We also express appreciation to the 299 patients who agreed to share their de-identified CT data with the research community and to the Radiology Department at the Mayo Clinic for facilitating collection of these data.
Footnotes
CONFLICT OF INTEREST
Dr. McCollough receives industry funding from Siemens AG, unrelated to this work. The other authors disclose no relevant conflict of interest.
Contributor Information
Taylor R. Moen, Department of Radiology, Mayo Clinic, Rochester, MN, USA
Baiyu Chen, Department of Radiology, Mayo Clinic, Rochester, MN, USA.
David R. Holmes, III, Biomedical Imaging Resource, Mayo Clinic, Rochester, MN, USA.
Xinhui Duan, Department of Radiology, Mayo Clinic, Rochester, MN, USA.
Zhicong Yu, Department of Radiology, Mayo Clinic, Rochester, MN, USA.
Lifeng Yu, Department of Radiology, Mayo Clinic, Rochester, MN, USA.
Shuai Leng, Department of Radiology, Mayo Clinic, Rochester, MN, USA.
Joel G. Fletcher, Department of Radiology, Mayo Clinic, Rochester, MN, USA
Cynthia H. McCollough, Department of Radiology, Mayo Clinic, Rochester, MN, USA.
REFERENCES
- 1.Herman GT, Lent A, Rowland SW. ART: mathematics and applications. A report on the mathematical foundations and on the applicability to real data of the algebraic reconstruction techniques. J Theor Biol. 1973;42:1–32. [DOI] [PubMed] [Google Scholar]
- 2.Hounsfield G A method of and apparatus for examination of a body by radiation such as x or gamma radiation. The Patent Office, London, Patient specification; 1972:1283915. [Google Scholar]
- 3.Bouman CA, Sauer K. A unified approach to statistical tomography using coordinate descent optimization. IEEE Trans Image Process. 1996;5:480–492. [DOI] [PubMed] [Google Scholar]
- 4.Browne JA, Holmes TJ. Developments with maximum likelihood X-ray computed tomography. IEEE Trans Med Imaging. 1992;11:40–52. [DOI] [PubMed] [Google Scholar]
- 5.Erdogan H, Fessler JA. Ordered subsets algorithms for transmission tomography. Phys Med Biol. 1999;44:2835–2851. [DOI] [PubMed] [Google Scholar]
- 6.Sauer K, Bouman C. A local update strategy for iterative reconstruction from projections. IEEE Trans Signal Process. 1993;41:534–548. [Google Scholar]
- 7.Thibault JB, Sauer KD, Bouman CA, Hsieh J. A three-dimensional statistical approach to improved image quality for multislice helical CT. Med Phys. 2007;34:4526–4544. [DOI] [PubMed] [Google Scholar]
- 8.Thibault J-B, Sauer K, Bouman C, Hsieh J. High Quality Iterative Image Reconstruction For Multi-Slice Helical CT, International Conference on Fully 3D Reconstruction in Radiology and Nuclear Medicine,. 2003; available at: https://engineering.purdue.edu/~bouman/publications/origpdf/F3D-2003a.pdf. [Google Scholar]
- 9.Li K, Garrett J, Ge Y, Chen GH. Statistical model based iterative reconstruction (MBIR) in clinical CT systems. Part II. Experimental assessment of spatial resolution performance. Med Phys. 2014;41:071911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li K, Tang J, Chen GH. Statistical model based iterative reconstruction (MBIR) in clinical CT systems: experimental assessment of noise performance. Med Phys. 2014;41:041906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L, Vrieze TJ, Leng S, Fletcher J, McCollough C. Technical Note: Measuring contrast- and noise-dependent spatial resolution of an iterative reconstruction method in CT using ensemble averaging. Med Phys. 2015;42:2261–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter RE, Holmes DR III, Fletcher JG, McCollough CH. Evaluation of pseudo-reader study designs to estimate observer performance results as an alternative to fully crossed, multi-reader, multi-case studies. Acad Radiol. 2020;27:244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher JG, DeLone DR, Kotsenas AL, et al. Evaluation of lower dose spiral head CT for detection of intracranial findings causing neurologic deficit. Am J Roentgenol. 2019;40:1855–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher JG, Fidler JL, Venkatesh SK, et al. Observer performance with varying radiation dose and reconstruction methods for detection of hepatic metastases. Radiology. 2018;289:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher JG, Yu L, Fidler JL, et al. Estimation of observer performance for reduced radiation dose levels in CT: Eliminating reduced dose levels that are too low is the first step. Acad Radiol. 2017;24:876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCollough CH, Bartley A, Carter RE, et al. Low-dose CT for the detection of metastatic liver lesions: Results of the 2016 Low Dose CT Grand Challenge. Med Phys. 2017;44:e339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dilger SK, Yu L, Chen B, et al. Localization of liver lesions in abdominal CT imaging: I. Correlation of human observer performance between anatomical and uniform backgrounds. Phys Med Biol. 2019;64:105011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dilger SK, Yu L, Chen B, et al. Localization of liver lesions in abdominal CT imaging: II. Mathematical model observer performance correlates with human observer performance for localization of liver lesions in abdominal CT imaging. Phys Med Biol. 2019;64:105012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong H, Yu L, Leng S, et al. A deep learning and partial least square regression based model observer for a low-contrast lesion detection task in CT. Med Phys. 2019;46:2052–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Duan X, Yu Z, Leng S, Yu L, McCollough C. Technical Note: development and validation of an open data format for CT projection data. Med Phys. 2015;42:6964–6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Electrical Manufacturers Association, NEMA PS3 / ISO 12052, Digital Imaging and Communications in Medicine (DICOM) Standard, Rosslyn, VA, USA: 2020; Available free at. http://www.dicomstandard.org/. [Google Scholar]
- 22.Yu L, Shiung M, Jondal D, McCollough CH. development and validation of a practical lower-dose-simulation tool for optimizing computed tomography scan protocols. J Comput Assist Tomogr. 2012;36:477–487. [DOI] [PubMed] [Google Scholar]
- 23.Stierstorfer K, Rauscher A, Boese J, Bruder H, Schaller S, Flohr T. Weighted FBP–a simple approximate 3D FBP algorithm for multislice spiral CT with good dose usage for arbitrary pitch. Phys Med Biol. 2004;49:2209–2218. [DOI] [PubMed] [Google Scholar]
- 24.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys. 2017;44:e360–e375. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Zhang Y, Kalra MK, et al. Low-dose CT with a residual encoder-decoder convolutional neural network. IEEE Trans Med Imaging. 2017;36:2524–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan H, Zhang Y, Yang Q, et al. 3-D Convolutional encoder-decoder network for low-dose CT via transfer learning from a 2-D trained network. IEEE Trans Med Imaging. 2018;37:1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, Wei B, Feng P, He P, Guo X, Wang G. Low-dose CT image denoising using a generative adversarial network with a hybrid loss function for noise learning. IEEE Access. 2020;8:67519–67529. [Google Scholar]
- 29.Yang Q, Yan P, Zhang Y, et al. Low-dose CT image denoising using a generative adversarial network with wasserstein distance and perceptual loss. IEEE Trans Med Imaging. 2018;37:1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Y, Ye JC. Framing U-net via deep convolutional framelets: application to sparse-view CT. IEEE Trans Med Imaging. 2018;37:1418–1429. [DOI] [PubMed] [Google Scholar]
- 31.Kim B, Han M, Shim H, Baek J. A performance comparison of convolutional neural network-based image denoising methods: The effect of loss functions on low-dose CT images. Med Phys. 2019;46:3906–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, Hsu W, Xie X, Cong J, Gao W. SACNN: self-attention convolutional neural network for low-dose CT denoising with self-supervised perceptual loss network. IEEE Trans Med Imaging. 2020;39:2289–2301. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Zhou S, Huang J, Yu L, Jin M. Investigation of low-dose ct image denoising using unpaired deep learning methods. IEEE Tran Radiat Plasma Med Sci. 2020:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheema MN, Nazir A, Sheng B, Li P, Qin J, Feng DD. Liver extraction using residual convolution neural networks from low-dose ct images. IEEE Trans Biomed Eng. 2019;66:2641–2650. [DOI] [PubMed] [Google Scholar]
- 35.Fletcher JG, Hara AK, Fidler JL, et al. Observer performance for adaptive, image-based denoising and filtered back projection compared to scanner-based iterative reconstruction for lower dose CT enterography. Abdom Imaging. 2015;40:1050–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fletcher JG, Yu L, Li Z, et al. Observer performance in the detection and classification of malignant hepatic nodules and masses with CT image-space denoising and iterative reconstruction. Radiology. 2015;276:465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Favazza CP, Ferrero A, Yu L, Leng S, McMillan KL, McCollough CH. Use of a channelized Hotelling observer to assess CT image quality and optimize dose reduction for iteratively reconstructed images. J Med Imag. 2017;4:031213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fletcher JG, Levin DL, Sykes A-MG, et al. Observer performance for detection of pulmonary nodules at unenhanced chest CT over a large range of radiation dose levels. Radiology. 2020; under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen B, Leng S, Yu L, Yu Z, Ma C, McCollough CH. Lesion insertion in the projection domain: Methods and initial results. Med Phys. 2015;42:7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen B, Ma C, Leng S, et al. Validation of a projection-domain insertion of liver lesions into CT images. Acad Radiol. 2016;23:1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Yu L, Trzasko JD, et al. Adaptive nonlocal means filtering based on local noise level for CT denoising. Med Phys. 2014;41:011908. [DOI] [PubMed] [Google Scholar]
- 42.Missert AD, Yu L, Leng S, Fletcher JG, McCollough CH. Synthesizing images from multiple kernels using a deep convolutional neural network. Med Phys. 2020;47:422–430. [DOI] [PubMed] [Google Scholar]
- 43.Missert A, Yu L, Leng S, McCollough CH. Simulation of CT images reconstructed with different kernels using a convolutional neural network and its implications for efficient CT workflow. Proc SPIE Med Imag. 2019;10948:109482Y. [Google Scholar]
- 44.McCollough CH, Chen B, Holmes DRI, et al. Data from Low Dose CT Image and Projection Data, The Cancer Imaging Archive; 2020. [Google Scholar]