Abstract
Background
Radiomics refers to the extraction of a large number of image biomarker describing the tumor phenotype displayed in a medical image. Extracted from positron emission tomography (PET) images, radiomics showed diagnostic and prognostic value for several cancer types. However, a large number of radiomic features are nonreproducible or highly correlated with conventional PET metrics. Moreover, radiomic features used in the clinic should yield relevant information about tumor texture. In this study, we propose a framework to identify technical and clinical meaningful features and exemplify our results using a PET non‐small cell lung cancer (NSCLC) dataset.
Materials and methods
The proposed selection procedure consists of several steps. A priori, we only include features that were found to be reproducible in a multicenter setting. Next, we apply a voxel randomization step to identify features that reflect actual textural information, that is, that yield in 90% of the patient scans a value significantly different from random texture. Finally, the remaining features were correlated with standard PET metrics to further remove redundancy with common PET metrics. The selection procedure was performed for different volume ranges, that is, excluding lesions with smaller volumes in order to assess the effect of tumor size on the results. To exemplify our procedure, the selected features were used to predict 1‐yr survival in a dataset of 150 NSCLC patients. A predictive model was built using volume as predictive factor for smaller, and one of the selected features as predictive factor for bigger lesions. The prediction accuracy of the both models were compared with the prediction accuracy of volume.
Results
The number of selected features depended on the lesion size included in the analysis. When including the whole dataset, from 19 features reflecting actual texture only two were found to be not strongly correlated with conventional PET metrics. When excluding lesions smaller than 11.49 and 33.10 mL (25 and 50 percentile of the dataset), four out of 27 features and 13 out of 29 features remained after eliminating features highly correlated with standard PET metrics. When excluding lesions smaller than 103.9 mL (75 percentile), 33 out of 53 features remained. For larger lesions, some of these features outperformed volume in terms of classification accuracy (increase of 4–10%). The combination of using volume as predictor for smaller and one of the selected features for larger lesions also improved the accuracy when compared with volume only (increase from 72% to 76%).
Conclusion
When performing radiomic analysis for smaller lesions, it should be first carefully investigated if a textural feature reflects actual heterogeneity information. Next, verification of the absence of correlation with all conventional PET metrics is essential in order to assess the additional value of radiomic features. Radiomic analysis with lesions larger than 11.4 mL might give additional information to conventional metrics while at the same time reflecting actual tumor texture. Using a combination of volume and one of the selected features for prediction yields promise to increase accuracy and reliability of a radiomic model.
Keywords: clinical value, feature selection, radiomics
1. INTRODUCTION
Quantitative interpretation of positron emission tomography (PET) may be used for diagnosis, prognosis, and treatment response assessment for cancer patients. 1 , 2 To date, the maximum intensity value in the tumor (SUVMAX), the metabolic active tumor volume (MATV), or the total lesion glycolysis (TLG) are often used for this purpose. 3 , 4 Recently, other metrics describing the textural properties of the tumor (i.e., tumor heterogeneity) have gained increasing interest. 5 , 6 These so called radiomic features might yield additional value to conventional metrics and might help to determine prognosis and treatment efficacy more accurately and reliably than conventional metrics. Several studies commented on the promising value of radiomic features for different tumor types. 6 , 7 , 8
While a large number of studies reported on the additional value of radiomics, more and more studies address its challenges and pitfalls. 9 , 10 The sensitivity of radiomic features to all steps in the radiomics workflow (i.e., image acquisition and reconstruction, tumor delineation, image discretization, and image processing) has been discussed in several reports. 8 , 11 , 12 In a clinical workflow, only features that result in comparable values when extracted from a patient scanned several times under the same conditions (repeatable features) and that are yielding only small differences when extracted from scans of the same patient acquired under different conditions (e.g., different scanners) (reproducible features) should be used in a radiomics workflow. 13 Both characteristics are essential in order to guarantee a reliable treatment assessment and an accurate diagnosis independent of, for example, on which PET scanner the patient was examined. Moreover, many radiomic features are highly correlated with conventional metrics such as MATV what makes their additional value questionable. 14 Additionally, a radiomic feature used in the clinic should be relevant and explainable, that is, should reflect the tumor heterogeneity observed in the medical image accurately. 15
It is of utmost importance that all these requirements are fulfilled as it might happen that a feature is found by chance to be predictive for the required task without yielding any meaningful information about tumor heterogeneity. Regarding the large number of radiomic features and the relatively small number of patients included in the majority of studies, this can happen as illustrated in Ref. [16] where the authors used randomly generated feature values for the prediction of overall survival and achieved a maximum cross‐validation accuracy under the curve (AUC) of 0.79. Therefore, each radiomic feature should be carefully checked if it really reflects the tumor heterogeneity observed in the image.
In this study, we propose a procedure to identify and select only those features that may yield technical and clinical meaningful information before using them in a prognostic model in an example dataset. We only include radiomic features which were found to be repeatable and reproducible in a previous multicenter study. Out of these robust features, we further select features using a method by Welch et al. who randomly shuffled tumor intensity values of CT images in order to destroy the underlying tumor texture. By comparing the features extracted from the original image with the features extracted from randomly redistributed voxel value distributions, we can identify features describing tumor heterogeneity beyond randomness. Hereby, we consider a feature as describing actual texture information when the feature value extracted from the original image is significantly different from the feature value of the randomly redistributed voxel intensities. If this is not the case, the feature does not seem to describe actual texture information. Moreover, we eliminate those features which are strongly correlated with conventional PET metrics. Previous studies demonstrated that the correlation with conventional PET metrics depends on the lesion size. 17 To cover this aspect, we apply the proposed feature selection procedure on different tumor volume ranges. The proposed feature selection procedure does not only avoid false‐positive findings and overfitting, but can also be used for feature space reduction allowing to study radiomics performance in more realistically large sample size (typically for PET up to a few hundred at most). We perform a detailed investigation which radiomic features are meeting all described requirements. As an example of the proposed selection procedure, the selected features are used for classifying 1‐yr survival using a logistic regression model. Hereby, we do not aim to build an optimal prognostic model by including, that is, additional clinical parameters. The aim of this study was to illustrate if we can identify features that describe actual tumor texture and to investigate if these features would then also have additional clinical value in the example dataset.
2. MATERIALS AND METHODS
2.1. Dataset
The study was registered at clinical trials.gov (NCT02024113). All patients gave informed consent for study participation and use of their data for scientific research. The dataset consists of 150 patients with Stage I–IV NSCLC. Details about the patient cohort can be found in Table I. Additional clinical parameters such as the smoking status are displayed in Table S1. All patients fasted at least 6 hours before image acquisition. Time between PET scan and tracer injection was around 60 min. All images were corrected for attenuation, scatter, random coincidences, and normalization. Images were acquired on a Gemini TF Big Bore (Philips Healthcare, Cleveland, OH, USA) and reconstructed using the BLOB‐OS‐TOF reconstruction method provided by the vendor. The reconstructed images yielded a cubic voxel size of 4 mm and are compliant with the benchmarks of the European Association of Nuclear Medicine Research Ltd. (EARL).
Table I.
Patient characteristics.
| Cancer stage | IA | 28 patients |
| IB | 13 patients | |
| IIA | 10 patients | |
| IIB | 8 patients | |
| IIIA | 29 patients | |
| IIIB | 15 patients | |
| IV | 47 patients | |
| Age | Mean | 67 yr |
| Std | 9.3 yr | |
| Sex | Men | 93 |
| Women | 57 |
2.2. Tumor segmentation
Image analysis was performed using an in‐house developed tool designed for the analysis of PET images 18 used in previous works. 19 , 20 For the segmentation of the tumor volume of interest (VOI), a semi‐automatic segmentation was performed including all voxels yielding a SUV above 2.5. High‐uptake regions such as the heart or inflammations in the lung were eliminated by the observer. For lesions close to the heart or other high‐uptake regions, a bounding box was manually drawn around the tumor to avoid inclusion of physiologically high‐uptake normal tissues. Every segmentation was manually checked and corrected if necessary. If some voxels of a close high‐uptake region were still included in the VOI, these voxels were excluded manually from the VOI. Only the primary tumor was included in the current analysis, which was defined as the lung lesion with the largest volume.
2.3. Radiomic feature calculation
Radiomic features were calculated using the open‐source software RaCaT which complies with the benchmarks provided by the Image Biomarker Standardization Initiative (IBSI). 21 , 22 Prior to feature calculation, images and corresponding VOIs were resampled to a cubic voxel size of 2 mm using tri‐linear interpolation as recommended by Hatt et al. 10 A resampling to a cubic voxel size leads to a larger number of reproducible radiomic features as some features depend on the number of voxels included in the VOI. 21 After interpolation, all voxels yielding a value above 0.5 in the resampled segmentation mask were included in the VOI. Before textural feature calculation, all images were first converted to standardized uptake values (SUV) and then discretized using a fixed bin size of 0.25 SUV as recommended by various studies. 11 , 23 , 24 , 25 Moreover, these settings were chosen as it has been demonstrated that they lead to the largest number of reproducible features. 26 Exact feature definitions as well as details about the feature calculations are described elsewhere. 21
2.4. Data analysis
All data analysis was performed in Python 3.4 using the packages numpy, scipy, and scikit‐learn.
2.4.1. Feature selection procedure
Only features that were identified to be robust and reproducible in previous work were included in the analysis. 26 In this work, a phantom containing 3D‐printed phantom inserts reflecting realistic tumor heterogeneity was scanned on various PET systems. The inserts were segmented on each scan separately and radiomic features were extracted. The features that only yielded small differences between the different PET systems and delineations were identified to be reproducible and are included in the present study. However, we recommend to make use of semi‐automated segmentation approaches, as much as possible, to avoid observer variability in tumor delineations which in turn will affect the reproducibility of radiomic analysis. Morphological and statistical features were not considered in the feature selection as they remain constant when performing step 1 (randomly shuffling the intensity values within the VOI). This lead in total to 92 radiomic features. A list containing the names of all features included in this study is provided in the supplemental material (Table S1).
The proposed feature selection procedure consists of three steps:
Step 1.) Randomized voxel assignment: Use a voxel randomization method to identify features that are reflecting the tumor heterogeneity observed in the PET image beyond randomness.
Step 2.) Correlation with conventional metrics: Identify the features yielding additional value to conventional PET metrics.
Step 3.) Mutual correlation: From the remaining features, select only the features yielding complementary information to each other.
The described selection steps will be detailed in the next paragraphs.
2.4.2. Step 1: Randomized voxel assignment
In order to use a radiomic feature in the clinic, it is important that the feature reflects the actual heterogeneity displayed in the PET image. This means that a lesion with the same shape but with a different texture should also have a different feature value. To check if a feature is fulfilling this assumption, the intensity values in a VOI were randomly reshuffled similar to a method proposed by Welch et al. 14 In this way, the original texture of a lesion is destroyed. However, instead of shuffling the intensity values of the whole 3D dataset as proposed by Welch et al., we only shuffled the intensity values inside the VOI and preserved in this way the first order statistics. Radiomic features from these randomly generated voxel redistributions were calculated. The procedure was repeated 50 times per image. An example illustrating the original and two randomly generated distributions are displayed in Fig. 1. Features extracted from the randomly generated voxel distributions will be called random features hereafter.
Fig. 1.
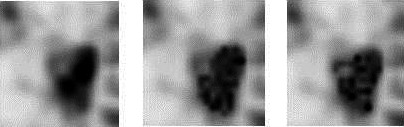
Original tumor (left), two examples (middle and right) of the same tumor after randomly shuffling the intensity values in the VOI.
All features calculated from the original voxel distribution within the tumor VOI were compared with features values from the random distributions. Features yielding different values for the original voxel distributions were regarded as reflecting the actual texture displayed in the image. For this purpose, the mean and standard deviation of the 50 random features were calculated for each tumor separately. A feature calculated from the original voxel distribution was considered to describe random texture when its feature value was within the 95% confidence interval:
With meanrand being the mean value and stdrand being the standard deviation calculated from all 50 random features of one tumor. Every image and tumor was evaluated separately.
Features considered for further analysis needed to yield a value outside the proposed confidence interval for 90% of the patients (note that 90% is used in this paper for illustration purposes, but other thresholds can be applied as deemed appropriate. For example a lower threshold would result in a more liberal selection, thus more features passing this selection step).
2.4.3. Step 2: Correlation with conventional metrics
A radiomic feature should yield additional value to conventional PET metrics. Therefore, all features were checked for their correlation with MATV, SUVPEAK, and SUVMEAN using the Spearman’s rank correlation coefficient. The Spearman’s correlation coefficient is a nonparametric metric describing the relationship between two variables. By comparing the statistical dependence of the rank of the variables, it also captures nonlinear relationships. A correlation above 0.9 was regarded as very strong. 27 Features with a very strong correlation with one conventional metric were regarded as redundant and were therefore discarded from the analysis.
2.4.4. Step 3: Mutual correlation between features
Finally, it is also important that the remaining features are reflecting different tumor characteristics and are therefore not highly correlated among each other. Therefore, all remaining features were checked for a very strong correlation among each other. If two or more features yielded a very strong correlation (correlation coefficient > 0.9), the feature that yielded the lowest correlation with the conventional metrics was kept and tested for its clinical value.
2.5.
2.5.1. Summary of the feature selection procedure
In summary, from the repeatable and reproducible features selected a priori the features that were considered for further analysis were the features that:
described the texture observed in the image beyond randomness: yielded different feature values for the original tumor voxel distribution than for randomly redistributed voxel intensities within the tumor VOI
have additional value to conventional PET metrics: yielded a correlation coefficient below 0.9 with tumor volume, SUVPEAK, and SUVMEAN
yielded complementary information among each other: if features were strongly correlated among each other (0.9 or more), the feature resulting in the lowest correlation with conventional PET metrics was kept
As it has been reported previously that the tumor volume has an impact on the correlation of a feature with conventional metrics, 17 all steps were performed for five volume ranges separately and results were compared:
tumors yielding more than 50 voxels in the original image (>3.2 mL)
tumors larger than the 25% percentile of the dataset (>11.48 mL)
tumors larger than the median of the dataset (>33.04 mL)
tumors larger than 45 mL as indicated by Brooks et al. 28
tumors larger than the 75% percentile (>103.94 mL).
The results of the feature selection procedure will be detailed as function of tumor volume range in the remainder of the paper.
2.6. Comparison with other feature selection methods
The proposed feature selection procedure was compared with two automatic feature selection methods: maximum relevance and minimum redundancy (MRMR) and RELIEF. Both methods aim to select only nonredundant (i.e., noncorrelated) as well as features relevant for the outcome. The MRMR algorithm is selecting the features showing the highest dependence with the outcome variable and at the same time the lowest dependence with other selected features. 29 While the Relief algorithm assigns weights to each feature. 30 The given weight increases when a feature is highly similar between patients in the same group (i.e., alive after 1 year), while the weight decreases when a feature is highly similar to the feature of patients belonging to the inverse group (i.e., dead after 1 year). Features with the highest weights are selected.
Both feature selection methods were performed for each volume range separately. The 10 most important features identified by the two approaches were compared with the features selected by our procedure before eliminating features with a high mutual correlation. The MRMR feature selection was performed in the programming language R using the package mRMRe, 31 while the RELIEF algorithm was implemented in Python 3.6.4.
3. FEATURES FOUND IN PREVIOUS STUDIES
Features found to have clinical value in previous studies were analyzed if they fit the described criteria. This included zone percentage (GLSZM) and entropy (GLCM), 6 , 32 as well as high‐intensity large area emphasis (GLSZM). 32 Moreover, coarseness, contrast, and busyness (NGTDM) were analyzed. 33 , 34
4. CLINICAL VALUE OF SELECTED FEATURES
In order to illustrate the clinical value of features matching the described criteria, three different models were considered:
4.1. Model 1
Each feature was used separately for the prediction of 1‐year survival using stratified cross‐validation and a logistic regression classifier (a detailed description of the classification process is given below). The mean accuracy under the curve (AUC) of all cross‐validation folds was compared with the mean AUC when using the feature volume.
4.2. Model 2
In order to assess their additional value, features found to yield a clinical value in Model 1 were, in a second step, used in combination with MATV in the cross‐validation and the logistic regression model. Also here, the mean AUC of the combined classification was compared with the mean AUC of volume alone.
4.3. Model 3
For features yielding a clinical value in Model 1, receiver operating characteristic curves (ROC) were drawn for the whole dataset. For lesions with volumes below the different thresholds, volume was used as prognostic factor, while for lesions above the thresholds, the selected feature was used. This was done for each selected feature separately. The AUC of the combined prognosis (volume + feature) was compared with the AUC of volume only.
For all models, the clinical value of each feature was determined using the whole dataset and for each volume range separately. Even if a feature was only found to match all criteria for larger lesions, classification was performed for all volume ranges in order to compare the predictive value of the feature for datasets with smaller and larger volume ranges. The distribution of positive/negative outcomes for each volume range is displayed in supplemental Fig. 1.
As the number of patients decreases with the exclusion of smaller lesions, classification was also performed with a subsampled number of patients (=number of patients with volumes > 133.04 mL) for the different volume ranges. This procedure was performed to identify if the number of training samples had an impact on classification accuracy.
5. DESCRIPTION OF CLASSIFICATION PROCESS
As the range of radiomic features differs widely from feature to feature, all features were z‐transformed in order to normalize feature ranges before the start of the classification process. To guarantee that in the training data the number of positives and negatives outcomes (alive/dead after one year) are equally distributed, the underrepresented class was upsampled using the Synthetic Minority Oversampling Technique (SMOTE). 35 The testing was performed on the original data distributions without any over‐ or undersampling. For comparison, the minority class was also randomly oversampled. In order to assess the clinical value of the classifier on independent test datasets, stratified cross‐validation with fivefolds and ten repetitions was performed. Hereby, 80% of the patients were used as training and 20% were used as testing dataset. This procedure was repeated until each data part served once as testing set.
6. RESULTS
6.1. Features remaining after feature selection process
6.1.1. Step 1: Randomized voxel assignment
The number of features describing actual texture (i.e., features outside the random range) depended on the volume range included in the analysis: While 19 features were found to be outside the random range when including the whole dataset, the number of features increased to 27 when excluding lesions smaller than 3.2 mL. Eliminating lesions smaller than 11.48, 33.10, 45, and 103.9 mL led to 29, 45, 45, and 53 features describing actual textural information. All features outside the random range and their correlation coefficients with conventional metrics are listed in Tables S2–S7 for the different volume ranges.
6.1.2. Step 2: Correlation with conventional metrics
The tumor volume had also an effect on the correlation with conventional metrics and therefore on the additional value of the features: The larger the volume range, the more features were found to be highly correlated with standard PET metrics. From the 19 features remaining after Step 1, only two features remained when eliminating features highly correlated with conventional metrics when analyzing the whole dataset. When excluding lesions smaller than 3.2 mL, four of the 27 features remained were not highly correlated with MATV, SUVMEAN, and SUVPEAK. The number of remaining features increased to 13 out of 29, 25 out of 45, 26 out of 45, and 33 out of 53 features.
The behavior of two features as a function of tumor volume and SUVMEAN are displayed in Figs. 2(a) and 2(b). As illustrated, for smaller lesions the original features are inside the random range and therefore not describing actual textural information. With increasing volume the features are found to be outside the proposed random range. The correlation with MATV is very strong for smaller lesions but decreases for larger lesions. While yielding a low correlation with MATV, some features result in a very strong correlation with SUVMEAN as illustrated in Fig. 2(b).
Fig. 2.
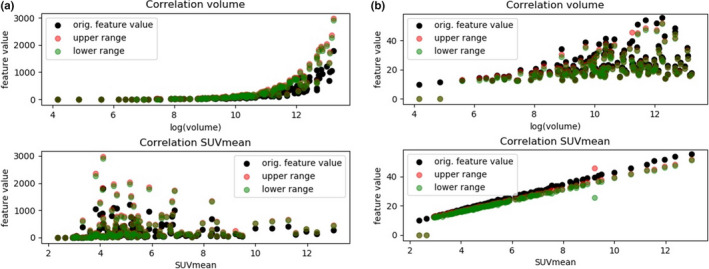
Illustration of the correlation between features and MATV or SUVMEAN: a): the feature Gray‐level nonuniformity GLSZM2D yielding a correlation coefficient with MATV of 0.97 (upper row) and a correlation with SUVMEAN of 0.34 (lower row). b): The feature joint average GLCM 3D avg yielding a correlation of 0.45 with MATV (upper row) and a correlation of 0.99 with SUVMEAN. [Color figure can be viewed at wileyonlinelibrary.com]
6.1.3. Step 3: Mutual correlation between features
When identifying those features yielding complementary information, that is, when eliminating features highly correlated between each other, one feature remained when including the whole dataset as well as lesions larger than 3.2 mL. 2, 9, 11, and 15 features remained when excluding volumes less than 11.48, 33.1, 45, and 103.9 mL, respectively.
The described increase in number of selected features for each step of the feature selection procedure is illustrated in Fig. 3. All features remaining after Step 3 as well as their correlation coefficients with the conventional metrics are displayed in Table II.
Fig. 3.

Number of features outside the random range (criteria 1) and not correlated with conventional metrics (criteria 1 + 2), and number of features after eliminating features highly correlated between each other (after mutual correlation) for the different volume ranges (left: all volumes are included, from left to right: more and more smaller lesions are excluded from the analysis until on the very right only tumors with a MATV > 103.94 mL are left). [Color figure can be viewed at wileyonlinelibrary.com]
Table II.
Features selected for different volume ranges.
| Feature name | Selected by which volume range |
|---|---|
| Gray_level_non_uniformity (GLCM, 2Davg) | All volume ranges |
| Gray_level_non_uniformity (GLCM, 2Dmrg) | All volume ranges >= 3.2 mL |
| long_runs_emphasis (GLRLM, 2Davg) | All volume ranges >= 11.4 mL |
| Run_percentage (GLRLM 2DWmrg) | All volume ranges >= 33.4 mL |
| Run_length_variance (GLRLM 2Dvmrg) | All volume ranges >= 33.4 mL |
| Gray_level_non_uniformity (GLRLM, 3Davg) | All volume ranges >= 33.4 mL |
| Gray_level_non_uniformity (GLDZM 2Davg) | All volume ranges >=33.4 mL |
| Zone_percentage (GLDZM 2Davg) | All volume ranges >= 33.4 mL |
| Gray_level_non_uniformity (GLDZM 3D) | All volume ranges >= 33.4 mL |
| Zone_distance_non_uniformity (GLDZM 2Davg) | All volume ranges >= 45 mL |
| Zone_distance_non_uniformity (GLDZM 2Dmrg) | All volume ranges >= 45 mL |
| coarseness (NGLDM 2Dmrg) | All volume ranges >= 103.9 mL |
| small_distance_emphasis (GLDZM 2Davg) | All volume ranges >= 103.9 mL |
| Dependence_count_non_uniformity (NGTDM 2Dmrg) | ALL volume ranges >= 103.9 mL |
| Dependence_count_entropy (NGTDM 2Davg) | All volume ranges >= 103.9 mL |
Features selected by automatic feature selection methods
The features that were identified by the feature selection method RELIEF are listed in Table III. When all tumor volumes were included in the feature selection, all features selected by RELIEF were either highly correlated with conventional PET metrics or were not describing actual texture.
Table III.
Features selected by the RELIEF algorithm. Features that were also found by our procedure are displayed in bold, while features not reflecting actual texture are marked with (NoTex), features highly correlated with volume or SUVMEAN are marked with (CORR).
| Volume > 0 mL | Volume > 3.2 mL | Volume > 11.48 mL | Volume > 33.04 mL | Volume > 45 mL | Volume > 103.94 mL |
|---|---|---|---|---|---|
| (CORR) Large zone high gray‐level emphasis (GLSZM 3D) | (CORR) Large zone high gray‐level emphasis (GLSZM 3D) | (CORR) Large zone high gray‐level emphasis (GLSZM 3D) | (CORR) Large zone high gray‐level emphasis (GLSZM 3D) | (CORR) Large zone high gray‐level emphasis (GLSZM 3D) | (CORR) Large zone high gray‐level emphasis (GLSZM 3D) |
| (NoTex) coarseness (NGTDM 2Dmrg) | (CORR) Run length variance (GLRLM 2Dvmrg) | (NoTex) Zone distance nonuniformity normalized (GLDZM2Davg) | (CORR) Zone size entropy (GLSZM 2Davg) | (CORR) Zone size entropy2Davg | Zone distance nonuniformity normalized (GLDZM 2Davg) |
| (NoTex) coarseness (NGTDM 3D) | (NoTex) long runs emphasis (GLRLM 2Davg) | difference variance (GLCM 2Dvmrg) | Zone distance nonuniformity normalized GLDZM2Davg | (CORR) Dependence count entropy (NGTDM 2Davg) | (CORR) Zone size entropy (GLSZM 2Davg) |
| (NoTex) difference variance (GLCM 2Dvmrg) | (NoTex) Zone distance nonuniformity normalized (GLDZM2Davg) | (NoTex) difference variance (GLCM 2Dmrg) | (CORR) Dependence count entropy (NGTDM 2Davg) | Zone distance nonuniformity normalized (GLDZM 2Davg) | Run length variance (GLRLM 2Dvmrg) |
| (NoTex) difference variance (GLCM 2Dmrg) | (NoTex) joint maximum (GLCM 2Davg) | (NoTex) joint maximum (GLCM 2Davg) | Run length variance (GLRLM 2Dvmrg) | (NoTex) joint maximum (GLCM 2Davg) | Gray‐level nonuniformity (GLSZM 3D) |
| (NoTex) joint maximum (GLCM 2Dmrg) | Gray‐level nonuniformity (GLCM 2Davg) | SUVPEAK | (NoTex) joint maximum (GLCM )2Davg | (NoTex) joint maximum (GLCM 2Dmrg) | Gray‐level nonuniformity (GLDZM 3D) |
| (NoTex) joint maximum (GLCM 2Davg) | (NoTex) joint maximum (GLCM 2Dmrg) | (NoTex) small distance emphasis (GLDZM 2Davg) | (NoTex) joint maximum (GLCM 2Dmrg) | Run length variance2Dvmrg | Gray‐level nonuniformity (GLCM 3Dmrg) |
| (CORR) contrast (NGTDM 2Dmrg) | Gray‐level nonuniformity (GLCM 3Dmrg) | (NoTex) joint maximum (GLCM 2Dmrg) | (NoTex) Dependence count nonuniformity (NGTDM 2Davg) | (NoTex) small distance emphasis (GLDZM 2Davg) | Gray‐level nonuniformity(GLCM 2Dmrg) |
| (CORR) contrast (NGTDM 2Davg) | Gray‐level nonuniformity (GLCM 2Dmrg) |
(NoTex) Dependence count nonuniformity 2Davg |
difference variance (GLCM 2Dvmrg) | coarseness (NGLDM 2Dmrg) | long runs emphasis (GLRLM 2Davg) |
| (CORR) Low dependence high gray‐level emphasis (NGTDM 2Dmrg) | small distance emphasis (GLDZM 2Davg) (CORR) | Zone size entropy (GLSZM 2Davg) (CORR) | long runs emphasis (GLRLM 2Davg) | long runs emphasis (GLRLM 2Davg) | Run length nonuniformity (2DWmrg) |
When excluding smaller lesions, some features were selected by both RELIEF and our proposed selection procedure. When only including very large lesions (i.e., with a volume > 103.94) eight of 10 features selected by RELIEF were overlapping with the features found by our procedure. The other two features did not reflect actual texture for the majority (more than 95%) of the patients.
Features selected by the MRMR algorithm are listed in Table IV. Similar to the RELIEF feature selection, when including the whole dataset only features eliminated by our procedure were selected (leading to no overlap in selected features). By excluding smaller lesions, more and more features selected by MRMR were also selected by our procedure. A maximum of six overlapping features was found when only including very large lesions. Some features were selected by RELIEF as most representative even though they were identified to not reflect actual texture.
Table IV.
Features selected by the MRMR algorithm for the different volume ranges. Features that were also found by our procedure are displayed in bold, while features not reflecting actual texture are marked with (NoTex), features highly correlated with volume or SUVMEAN are marked with (CORR).
| Volume > 0 mL | Volume > 3.2 mL | Volume > 11.48 mL | Volume > 33.04 mL | Volume > 45 mL | Volume > 103.94 mL |
|---|---|---|---|---|---|
| (NoTex) Gray‐level nonuniformity (GLSZM 2Davg) | Gray‐level nonuniformity (GLSZM2Davg) | (CORR) Short run low gray‐level emphasis (GLRLM 3Davg) | (CORR) Short run low gray‐level emphasis (GLRLM 3Davg) | long runs emphasis (GLRLM 2Davg) | Zone distance nonuniformity (GLDZM 2Davg) |
| (NoTex) long runs emphasis (GLRLM 2Davg) | (NoTex) long runs emphasis (GLRM 2Davg) | (NoTex) Zone distance nonuniformity (GLDZM 2Davg) | Gray‐level nonuniformity (GLSZM 3D) | Zone distance nonuniformity (GLDZM 2Davg) | Gray‐level nonuniformity (2DWmrg) |
| (NoTex) Zone distance nonuniformity normalized (GLDZM 2Davg) | (NoTex) Zone distance nonuniformity normalized (GLDZM 2Davg) | (NoTex) long runs emphasis (GLRLM 2Davg) | long runs emphasis (GLRLM 2Davg) | (NoTex) contrast (NGTDM 2Dmrg) | Gray‐level nonuniformity (GLSZM 3D) |
| (CORR) Gray‐level nonuniformity (3Dmrg) | (CORR) Gray‐level nonuniformity (3Dmr)g | Gray‐level nonuniformity (GLCM 2Dvmrg) | (NoTex) Run length nonuniformity (GLRLM 2Davg) | (NoTex) joint maximum (GLCM 2Dmrg) | Run percentage (GLRLM 2Davg) |
| (NoTex) contrast (NGTDM 2Dmrg) | (NoTex) contrast (NGTDM 2Dmrg) | (NoTex) small distance emphasis (GLDZM 2Davg) | (NoTex) contrast (NGTDM 2Dmrg) | Gray‐level nonuniformity (GLSZM 2Davg) | Gray‐level nonuniformity (GLDZM 2Davg) |
| (NoTex) Morans I | (CORR) Short run low gray‐level emphasis (GLRLM 3Davg) | (NoTex) contrast (NGTDM 2Dmrg) | Gray‐level nonuniformity (GLSZM 2Davg) | (NoTex) Zone size entropy (GLSZM 2Dmrg) | (CORR) Run length nonuniformity (GLRLM 2Davg) |
| (CORR) Short run low gray‐level emphasis (GLRLM 3Davg) | (NoTex) Run length nonuniformity (GLRLM 2Dvmrg) | (NoTex) Small distance emphasis (GLDZM 2Davg) | (NoTex) joint maximum (GLCM 2Dmrg) | (NoTex) Large zone high gray‐level emphasis (GLSZM 3D) | (NoTex) Large zone high gray‐level emphasis (GLSZM 3D) |
| (NoTex) Run length nonuniformity (GLRLM 2Dvmrg) | (NoTex) Large zone high gray‐level emphasis (GLSZM 3D) | (NoTex) Large zone high gray‐level emphasis (GLSZM 3D) | (NoTex) Large zone high gray‐level emphasis (GLSZM 3D) | (CORR) difference variance (GLCM 2Dvmrg) | (CORR) difference variance (GLCM 2Dvmrg) |
| (NoTex) Large zone high gray‐level emphasis (GLSZM 3D) | (CORR) difference variance (GLCM 2Dvmrg) | (CORR) difference variance (GLCM 2Dvmrg) | (CORR) difference variance (GLCM 2Dvmrg) | (CORR) coarseness (NGTDM 3D) | (NoTex) joint maximum (GLCM 2Davg) |
| (CORR) difference variance (GLCM 2Dvmrg) | (NoTex) joint maximum (GLCM 2Davg) | (NoTex) joint maximum (GLCM 2Davg) | Zone distance nonuniformity normalized (GLDZM 2Davg) | Zone distance nonuniformity normalized (GLDZM 2Davg) | Zone distance nonuniformity (GLDZM 2Dmrg) |
6.2. Features found in previous studies
When analyzing features identified to have clinical value in previous studies, the feature busyness (NGTDM) was found to be nonreproducible and was therefore a priori excluded from the analysis. None of the remaining features was passing Step 1 of the feature selection procedure when analyzing the whole dataset. When excluding patients with lesions smaller than 33.4 mL, coarseness (NGTDM) and zone percentage (GLSZM) were fulfilling the requirements of Step 1, that is, yielded actual textural information. Coarseness correlated with MATV showing a correlation coefficient of 0.96 even for bigger lesions. Zone percentage yielded a lower correlation with the conventional metrics for larger volumes (MATV > 33.4 mL) and was the only feature that passed all steps. Therefore, this feature was checked for its possible clinical value in the present dataset.
6.3. Clinical value of selected features
6.3.1. Model 1
The clinical value of the selected features depended on the volume range included in the classification process. Some features were, for example, selected to describe actual texture for lesions with volumes above 33.4 mL but did not yield any clinical value for this volume range (mean AUC around 0.55). However, when including also smaller lesions, that is, when performing the classification process for the whole dataset, their clinical value was comparable to the value of MATV. Nevertheless, in these cases, these features were also highly correlated with MATV (Figure 4).
Fig. 4.
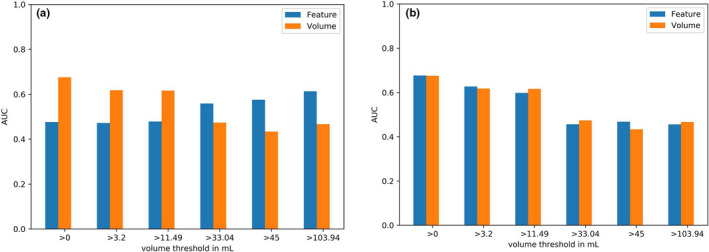
Model 1: Mean cross‐validation AUC of: (a) the feature run percentage which shows an increasing AUC with larger volumes included in the analysis; (b) the feature run length nonuniformity results in a reasonable accuracy when including the whole dataset, while the accuracy is decreasing with decreasing volume range and decreasing correlation with volume. [Color figure can be viewed at wileyonlinelibrary.com]
Features that were found to be highly correlated with SUVMEAN or SUVPEAK did not yield clinical value (mean AUC around 0.5) when used in Model 1. SUVMEAN and SUVPEAK yielded a comparable (low) accuracy when used for classification and yielded therefore no clinical value in the example dataset.
For the rest of the features, the prediction accuracy depended on the feature: The feature contrast (NGTDM 2Dmrg) and zone size entropy (GLSZM 2Davg) were fulfilling all described characteristics but were not yielding a clinical value for 1‐year survival in Model 1, that is, resulted in a mean cross‐validation AUC of around 0.5. The low accuracy was observed for all volume ranges.
The features run length variance (GLRLM 2Dvmrg), run percentage (GLRLM 2Dmrg), difference entropy (GLCM 2Dmrg), Gray‐level nonuniformity (GLRLM 2Davg), long runs emphasis (GLRLM 2Davg), and zone percentage (GLDZM 2Davg) yielded an accuracy around 0.5 when also smaller lesions were included in the classification process. The accuracy increased when only including larger volumes (>33.4 mL) for which they were also found to match all described criteria (Fig. 4). For these larger volumes, they outperformed MATV in terms of accuracy (mean AUC 0.6 vs 0.48).
6.3.2. Model 2
For larger lesions, the features leading to an accuracy improvement in Model 1 led also to an improvement when used together with MATV in the logistic regression model (Fig. 5).
Fig. 5.
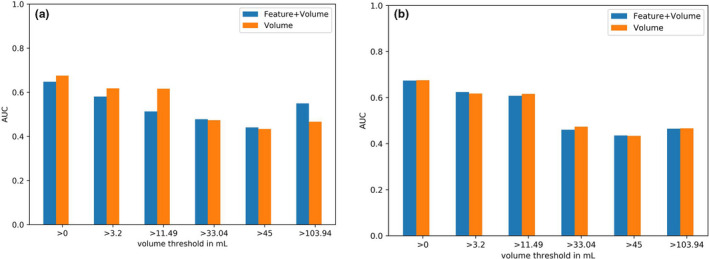
Model 2: Mean cross‐validation AUC of: (a) the feature run percentage, (b) the feature run length nonuniformity used together with MATV in the logistic regression model. [Color figure can be viewed at wileyonlinelibrary.com]
6.3.3. Model 3
For the features outperforming volume for larger lesions, also the combined ROC curves yielded higher AUCs than volume. The combined prediction led to an increase in AUC from 72.5% for volume only to up to 76% for the combined prognosis (Fig. 6). For the feature Gray‐level nonuniformity (GLRLM 2Davg), an increase in AUC was already observed when using the feature for lesions with volumes above 11.4 mL for which the feature also reflected actual heterogeneity. All AUCs of Models 1, 2, and 3 are listed in Table S8.
Fig. 6.
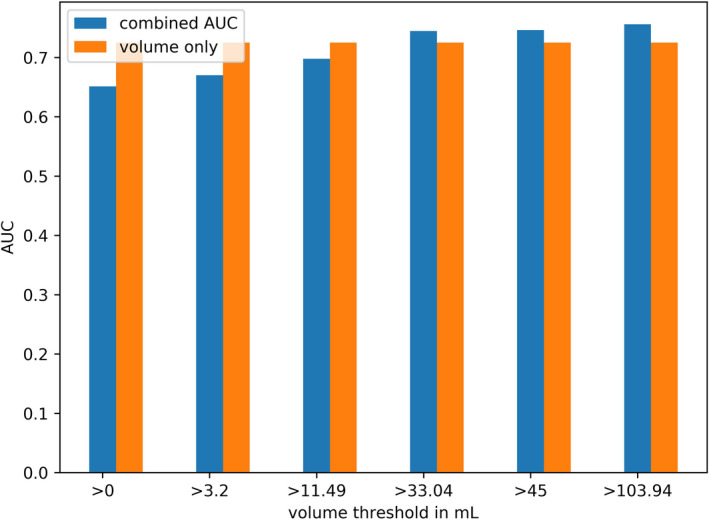
Model 3: Combined AUCs: for lesions above threshold, the feature run percentage is used for prediction, while for features below the threshold, volume is used for prediction. [Color figure can be viewed at wileyonlinelibrary.com]
The oversampling technique had only minimal impact on the accuracy of the classifier as illustrated in the supplemental material. The accuracy when using all patients or a subsample of patients was for all volume ranges comparable, indicating that the difference in accuracy for the different data subsets (by excluding more and more patients) is not due to the different size of training sets.
7. DISCUSSION
In this study, we proposed a feature selection procedure to identify which reproducible radiomic features are at the same time describing the tumor texture beyond randomness and yielding additional value to conventional PET metrics. The impact of lesion size on fulfilling these requirements was assessed.
Our results suggest that the number of plausible radiomic features depends on the lesion size. For smaller lesions, the majority of textural features did not describe actual tumor texture (did not yield feature values significant different from randomness). This result indicates that textural features extracted from small lesions should be handled with care. As small lesions consist only of a few voxels, randomly shuffled and original image are similar and result therefore in similar radiomic features. Hence, it is questionable how accurate a textural feature extracted from a small lesion can reflect underlying lesion heterogeneity. For each textural feature family, a matrix is composed describing, for example, how often a discretized intensity value is appearing consecutively in the VOI or how many connected voxels yield the same intensity value. 36 These textural features might result in a certain value due to the small expansion of the lesion but not due to tumor heterogeneity. The low number of selected radiomic features might be related to the low spatial resolution of PET images which has a high impact on smaller lesions. The low resolution is a general drawback of PET images. Using modern scanners yielding a better spatial resolution might lead to more radiomic features reflecting actual texture. Therefore, if a radiomic model including small lesions is used, it should be carefully checked if a textural feature is describing the underlying tumor characteristics beyond randomness. However, conventional PET metrics such as MATV and SUVmax could be used for the analysis of smaller lesions, while for larger lesions textural features might be used, as was shown with our combined Model 3. Additionally, features extracted from smaller tumors showed also a higher correlation with conventional metrics and might not yield additional value what is in line with the results of Hatt et al. and Brooks et al. 28 , 37
As larger lesions yield more complex anatomical properties, it is natural that for larger lesions more features were selected. However, also for larger lesions, around one third of the features failed to describe texture different from randomness and contain therefore no relevant information about tumor heterogeneity. Especially for small datasets, it can happen that one of these features results by chance in a high correlation with the outcome and results therefore in a good AUC. Thus, a careful check if there is a relationship between a predictive feature and the heterogeneity observed in the medical image is necessary. Moreover, the use of cross‐validation and an external testing set is essential in order to assess the value of a feature in independent test scenarios and lowers the risk of identifying a feature that does not describe relevant information. 13 , 15 , 38
Due to image noise, partial volume effect, and other intrinsic factors, the heterogeneity displayed in a medical image does not reflect the real underlying tumor heterogeneity accurately as was shown previously for MR and PET images. 39 , 40 However, several studies demonstrated that a tumor displayed heterogeneously in a PET image is an indicator for a lower survival chance and higher treatment resistance. 10 , 41 The features identified by the proposed selection procedure are not describing the real tumor heterogeneity, but the heterogeneity observed in the PET image and thus are reflecting the PET tumor phenotype.
The clinical value of the radiomic features selected by our procedure was in our dataset relatively low. Some features resulted in a reasonable mean cross‐validation accuracy when analyzing the whole dataset (i.e., including also smaller volumes), but were in this case also highly correlated with conventional PET metrics. In addition, in this case, no feature yielded additional value to MATV. This fact indicates that the accuracy of these features when applied to the whole dataset is due to the high correlation with volume. With a decrease in correlation with volume (by excluding smaller lesions), the accuracy also decreases what is in line with previous studies reporting a low accuracy when eliminating features that were highly correlated with MATV. 14 , 42
However, when excluding smaller lesions, some features yielded complementary value to MATV, improving the accuracy of around 4–10%. Moreover, using the combination of tumor volume as prognostic factor for smaller lesions and one selected features as prognostic factor for larger lesions led to an increase in AUC up to 76% when compared with using only volume as prognostic factor for the whole dataset (AUC 72.7%). This indicates, that the selected features had additional value only for the larger lesions for which they were also selected. Therefore, it is worthwhile to explore the use of different prognostic imaging biomarkers (e.g., MATV versus a radiomic feature) for different lesion size ranges in future studies.
Brooks et al. indicated that the correlation with MATV decreases with lesions yielding a MATV above 45 mL and radiomic analysis including smaller lesions might be questionable. 28 However, our findings demonstrate that the most adequate threshold is feature dependent what is in line with the findings of Hatt et al. 17 Additionally, Hatt et al. indicated that a more appropriate volume threshold might be 10 mL as this threshold already led to a lower correlation with MATV for some radiomic features. Our results support this finding. Some radiomic features yielded relevant information and were at the same time not highly correlated with conventional metrics when excluding lesions with a volume below 11.4 mL.
One source of uncertainty in the quantitative analysis of PET images results from uncertainties in tumor delineation. As manual segmentations suffer from a high inter‐ and intraobserver variability, a semi‐ or ideally fully automatic segmentation method should preferably be used. Yet, segmentation of tumors in PET images still requires supervision and sometimes manual correction, 43 resulting in intra‐ and inter‐observer variability. It is, however, highly recommended to keep the level of user interaction as low as possible, as we recently showed in Ref. [44]. By using a semi‐automated segmentation approach and workflow, which required delineation adjustment in a few cases (when the tumor was located to another high‐uptake region), we tried to limit the amount of user interaction and we found that this reduces the observer variability as much as possible.
While previous studies concentrated on the correlation of radiomic features with volume, we also investigated the correlation with SUVMEAN and SUVPEAK and found that a large number of radiomic features yields a high correlation with these two parameters as well. Hence, even though some features might yield additional value to MATV, they yield no additional value to other conventional PET metrics. Therefore, the additional value of a feature to all conventional PET metrics (MATV, SUVMEAN, SUVPEAK) should be investigated and features resulting in a very strong correlation with conventional metrics should be discarded from the analysis.
As automatic feature selection methods are data driven, the two automatic feature selection methods used for comparison in our study could not identify features describing nonrelevant or random texture. As our feature selection method does not only check the value of a feature but also its plausibility, it adds an important aspect to automatic feature selection methods. Therefore, it could be used in addition to automatic methods as it can identify features which are found “by chance” to yield clinical value and can therefore help to identify false‐positive findings.
The selected features as well as the volume thresholds used in this study should be validated in a larger, independent patient cohort and are limited to the NSCLC dataset included in this study. Moreover, the used thresholds (i.e., including only features reflecting actual texture for more than 90% of the patients) were chosen as example. In other datasets, these thresholds could be adapted as deemed appropriate or optimal and a more liberal threshold, thus resulting in more selected features, may be feasible when larger datasets are available.
Moreover, the retrospective nature of our study is a clear limitation and our findings should be validated in a larger, prospective cohort. However, the aim of our study was to demonstrate our proposed radiomic feature selection procedure on a clinical dataset. We showed that, especially for smaller lesions, a large number of radiomic features might not reflect actual texture information. Our study demonstrates the need for a more thoughtful performed radiomic analysis and feature selection procedure. The combination of using conventional PET metrics for smaller and textural features for larger lesions might be a solution that should be explored in other datasets. Only with good statistical methods (i.e., using cross‐validation), the use of external testing datasets, as well as a careful check which textural features are repeatable, reproducible, and contain relevant textural information, a radiomic study becomes transferable to other datasets and opens the way for a clinical implementation of radiomics.
8. CONCLUSION
In this study, we proposed a feature selection procedure which identifies reproducible textural that are (a) describing relevant texture and (b) are not highly correlated with conventional PET metrics. Our results show that the larger the lesions the more features are selected. Our results illustrate that when performing textural analysis for small lesions (<11.4 mL), it should be carefully investigated if a textural feature reflects relevant tumor characteristics and yields additional value to conventional metrics. Using tumor volume as prognostic value for smaller lesions and the identified textural features as prognostic value for larger lesions yields promising value for a more accurate and reliable radiomic analysis.
FUNDING
This work is part of the research program STRaTeGy with project number 14929, which is (partly) financed by the Netherlands Organisation for Scientific Research (NWO). This study was financed by the Dutch Cancer Society, POINTING project, grant 10034.
CONFLICT OF INTEREST
The authors have no relevant conflict of interest to disclose.
ETHICAL APPROVAL
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Supporting information
Data S1. Supporting Information.
ACKNOWLEDGMENTS
The authors thank the Center for Information Technology of the University of Groningen for their support and for providing access to the Peregrine high‐performance computing cluster.
REFERENCES
- 1. Vansteenkiste JF, Stroobants SG, Dupont PJ, et al. Prognostic importance of the standardized uptake value on 18 F‐Fluoro‐2‐deoxy‐glucose–positron emission tomography scan in non–small‐cell lung cancer: an analysis of 125 cases. J Clin Oncol. 1999;17:3201–3206. [DOI] [PubMed] [Google Scholar]
- 2. Hammerschmidt S, Wirtz H. Lung Cancer, Dtsch. Aerzteblatt Online; 2009.
- 3. Larson S. Tumor treatment response based on visual and quantitative changes in global tumor glycolysis using PET‐FDG imaging the visual response score and the change in total lesion glycolysis. Clin Positron Imaging. 1999;2:159–171. [DOI] [PubMed] [Google Scholar]
- 4. Vanhove K, Mesotten L, Heylen M, et al. Prognostic value of total lesion glycolysis and metabolic active tumor volume in non‐small cell lung cancer. Cancer Treat Res Commun. 2018;15:7–12. [DOI] [PubMed] [Google Scholar]
- 5. Data TA, Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than. Radiology. 2016;278:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tixier F, Hatt M, Valla C, et al. Visual versus quantitative assessment of intratumor 18F‐FDG PET uptake heterogeneity: prognostic value in non‐small cell lung cancer. J Nucl Med. 2014;55:1235–1241. [DOI] [PubMed] [Google Scholar]
- 7. Kim D‐H, Jung J‐H, Son SH, et al. Prognostic significance of intratumoral metabolic heterogeneity on 18F‐FDG PET/CT in pathological n0 non‐small cell lung cancer. Clin Nucl Med. 2015;40:708–714. [DOI] [PubMed] [Google Scholar]
- 8. Desseroit M‐C, Tixier F, Weber WA, et al. Reliability of PET/CT shape and heterogeneity features in functional and morphologic components of non‐small cell lung cancer tumors: a repeatability analysis in a prospective multicenter cohort. J Nucl Med. 2017;58:406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar V, Gu Y, Basu S, et al. Radiomics : the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hatt M, Tixier F, Pierce L, et al. Characterization of PET/CT images using texture analysis : the past, the present … any future ? Eur J Nucl Med Mol Imaging. 2017;44:151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pfaehler E, Beukinga RJ, de Jong JR, et al. Repeatability of 18 F‐FDG PET radiomic features: a phantom study to explore sensitivity to image reconstruction settings, noise, and delineation method. Med Phys. 2018;46:665–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shiri A, Rahmim P, Ghaffarian P, Geramifar HA, Bitarafan‐Rajabi A. The impact of image reconstruction settings on 18F‐FDG PET radiomic features: multi‐scanner phantom and patient studies. Eur Radiol. 2017;27:4498–4509. [DOI] [PubMed] [Google Scholar]
- 13. Traverso L, Wee AD, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol. 2018;102:1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Welch ML, McIntosh C, Haibe‐Kains B, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother. Oncol. 2019;130:2–9. [DOI] [PubMed] [Google Scholar]
- 15. Buvat I, Orlhac F. The dark side of radiomics: on the paramount importance of publishing negative results. J Nucl Med. 2019;60:1543–1544. [DOI] [PubMed] [Google Scholar]
- 16. Chalkidou MJO, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One. 2015;10:e0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatt M, Majdoub M, Vallieres M, et al. 18F‐FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi‐cancer site patient cohort. J Nucl Med. 2015;56:38–44. [DOI] [PubMed] [Google Scholar]
- 18. Boellaard R. Quantitative oncology molecular analysis suite: ACCURATE. In: SNMMI June 23–26; 2018.
- 19. Kolinger GD, Vállez García D, Kramer GM, et al. Repeatability of [18F]FDG PET/CT total metabolic active tumour volume and total tumour burden in NSCLC patients. EJNMMI Res. 2019;9:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kramer GM, Frings V, Hoetjes N, et al. Repeatability of quantitative whole‐body 18F‐FDG PET/CT uptake measures as function of uptake interval and lesion selection in non‐small cell lung cancer patients. J Nucl Med. 2016;57:1343–1349. [DOI] [PubMed] [Google Scholar]
- 21. Zwanenburg M, Vallières MAA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high‐throughput image‐based phenotyping. Radiology. 2020;295:328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfaehler E, Zwanenburg A, de Jong JR, Boellaard R. RaCaT: an open source and easy to use radiomics calculator tool. PLoS One. 2019;14:e0212223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Velden FHP, Kramer GM, Frings V, et al. Repeatability of radiomic features in non‐small‐cell lung cancer [18F]FDG‐PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol. 2016;18:788–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leijenaar RTH, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG‐PET radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Orlhac F, Soussan M, Chouahnia K, Martinod E, Buvat I. 18F‐FDG PET‐derived textural indices reflect tissue‐specific uptake pattern in non‐small cell lung cancer. PLoS One. 2015;10:e0145063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfaehler E, van Sluis J, Merema BBJ, et al. Experimental multicenter and multivendor evaluation of the performance of PET radiomic features using 3‐dimensionally printed phantom inserts. J Nucl Med. 2020;61:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mukaka MM. Statistics corner: a guide to appropriate use of correlation coefficient in medical research, Malawi. Med J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 28. Brooks FJ, Grigsby PW. The effect of small tumor volumes on studies of intratumoral heterogeneity of tracer uptake. J Nucl Med. 2014;55:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng H, Long F, Ding C. Feature selection based on mutual information criteria of max‐dependency, max‐relevance, and min‐redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27:1226–1238. [DOI] [PubMed] [Google Scholar]
- 30. Kira K, Rendell LA. A Practical Approach to Feature Selection. In: Mach. Learn. Proc. 1992. (Elsevier, 1992), pp. 249–256.
- 31. De Jay N, Papillon‐Cavanagh S, Olsen C, El‐Hachem N, Bontempi G, Haibe‐Kains B. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics. 2013;29:2365–2368. [DOI] [PubMed] [Google Scholar]
- 32. Desseroit M‐C, Visvikis D, Tixier F, et al. Development of a nomogram combining clinical staging with 18F‐FDG PET/CT image features in non‐small‐cell lung cancer stage I‐III. Eur J Nucl Med Mol Imaging. 2016;43:1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cook GJR, Yip C, Siddique M, et al. Are pretreatment 18F‐FDG PET tumor textural features in non‐small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. 2013;54:19–26. [DOI] [PubMed] [Google Scholar]
- 34. Ahn HK, Lee H, Kim SG, Hyun SH. Pre‐treatment 18F‐FDG PET‐based radiomics predict survival in resected non‐small cell lung cancer. Clin Radiol. 2019;74:467–473. [DOI] [PubMed] [Google Scholar]
- 35. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over‐sampling technique. J Artif Intell Res. 2002;16:321–357. [Google Scholar]
- 36. Zwanenburg S, Leger M, Vallières S. Löck, and for the I.B.S. Initiative, Image biomarker standardisation initiative; 2016.
- 37. Hatt M, Tixier F, Cheze Le Rest C, Pradier O, Visvikis D. Robustness of intratumour 18F‐FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur J Nucl Med Mol Imaging. 2013;40:1662–1671. [DOI] [PubMed] [Google Scholar]
- 38. Zwanenburg A, Löck S. Why validation of prognostic models matters? Radiother Oncol. 2018;127:370–373. [DOI] [PubMed] [Google Scholar]
- 39. Yang F, Young LA, Johnson PB. Quantitative radiomics: validating image textural features for oncological PET in lung cancer. Radiother Oncol. 2018;129:209–217. [DOI] [PubMed] [Google Scholar]
- 40. Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging. 2004;22:81–91. [DOI] [PubMed] [Google Scholar]
- 41. Bailly C, Bodet‐Milin C, Bourgeois M, et al. Exploring tumor heterogeneity using PET imaging: the big picture. Cancers (Basel). 2019;11:1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Traverso M, Kazmierski IZ, et al. Machine learning helps identifying volume‐confounding effects in radiomics. Phys Medica. 2020;71:24–30. [DOI] [PubMed] [Google Scholar]
- 43. Hatt M, Laurent B, Ouahabi A, et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal. 2018;44:177–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfaehler E, Burggraaff C, Kramer G, et al. PET segmentation of bulky tumors: strategies and workflows to improve inter‐observer variability. PLoS One. 2020;15:e0230901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information.


