Abstract
Although many impressive metallo‐supramolecular architectures have been reported, they tend towards high symmetry structures and avoid extraneous functionality to ensure high fidelity in the self‐assembly process. This minimalist approach, however, limits the range of accessible structures and thus their potential applications. Herein is described the synthesis of a family of ditopic ligands wherein the ligand scaffolds are both low symmetry and incorporate exohedral functional moieties. Key to this design is the use of CuI‐catalysed azide‐alkyne cycloaddition (CuAAC) chemistry, as the triazole is capable of acting as both a coordinating heterocycle and a tether between the ligand framework and functional unit simultaneously. A common precursor was used to generate ligands with various functionalities, allowing control of electronic properties whilst maintaining the core structure of the resultant cis‐Pd2L4 nanocage assemblies. The isostructural nature of the scaffold frameworks enabled formation of combinatorial libraries from the self‐assembly of ligand mixtures, generating a statistical mixture of multi‐functional, low symmetry architectures.
Keywords: cages, combinatorial library, metallo-supramolecular, self-assembly, asymmetrical
Symmetry is over: A family of functionalised, asymmetric ditopic ligands was synthesised from a common precursor. Each ligand was capable of self‐assembling with PdII ions to selectively form low‐symmetry cis‐Pd2L4 nanocages. Due to the isostructural nature of the ligand framework, multiple ligands were able to be combined to generate combinatorial libraries containing heteroleptic, multi‐functional, low symmetry nanocages.
Introduction
The self‐assembly of metal–organic polyhedra (MOPs) [1] remains a popular tool for supramolecular chemists to generate intricate architectures from minimalist building blocks that have demonstrated myriad functionality. [2] Simplicity of components is advantageous: symmetrical structures mitigate the competing processes of narcissistic (self‐recognition) and integrative self‐sorting (heteromeric‐assembly), [3] whilst minimalist ligands inhibit potentially disruptive effects from functional units. Trading fidelity of assembly for high symmetry and spartan scaffolds, however, limits the scope for developing more sophisticated systems.
Pd2L4 molecular cages [4] have become a common class of MOP and, despite the potential impediments, strategies for accessing lower symmetry variants have been reported. Use of steric [5] and geometric [6] parameters has enabled the self‐assembly of heteroleptic [7] dipalladium nanocages. Exploiting coordination preferences to generate stable metallo‐ligands [8] has allowed mixed‐metal architectures [9] to be realised. Work in this group, [10] and by others, [11] has focused on an alternative, underexplored option: the use of low symmetry ligands. [12] Designed such that steric or geometric parameters, or a combination of the two, direct the self‐assembly process, exclusive formation of specific cage isomers from a potential mixture can be ensured. Although these concepts have been successfully employed in the formation of reduced‐symmetry MOPs, they remain devoid of functionality. Indeed, research into exohedral [13] and endohedral [14] functionalisation of ligand scaffolds for MOPs altogether remains relatively scarce.
The 1,2,3‐triazole, most commonly synthesised using the CuI‐catalysed azide‐alkyne cycloaddition (CuAAC) reaction, [15] has become a ubiquitous unit in supramolecular chemistry. [16] In addition to being the coordinating unit in readily accessible ligands for MOPs, [17] the specificity of the CuAAC reaction makes the triazole ideal as a benign linker between coordinating and functional moieties. [18] Routinely utilised in either of these roles, the function of the triazole is usually determined at an early stage in the design process. Furthermore, the ability of the triazole to fulfil both of these mandates simultaneously is rarely exploited.
This report details the use of asymmetric, mixed‐heterocycle ditopic ligands, with isoquinoline and 1,2,3‐triazole coordinating units, for the formation of functionalised cis‐Pd2L4 architectures. Introduction of the triazole unit as the final step in the ligand synthesis enables derivatization of a common precursor, allowing tuning of ligand properties whilst maintaining the structure of the framework core. This structural consistency was able to be exploited in the assembly of mixed‐ligand combinatorial cage libraries.
Results and Discussion
Based on principles delineated from earlier work, [10] ligand LR (Scheme 1 a), envisaged to be derived from an azide precursor, was designed incorporating both isoquinoline and triazole coordinating units. Optimised models from semi‐empirical calculations (PM6) of the four potential [Pd2 L 4]4+ cage isomers assembled from the hypothetical LMe (where R=CH3) indicated that the cis‐Pd2L4 isomer would be lowest in energy (Scheme 1 b), consistent with previous computational and experimental work, [11] and from a simple visual analysis appeared to be the most favourable structure. Furthermore, the optimised structure of the cis isomer displayed no disquieting distortions of common structural parameters, such as N−Pd−N (≈176°) or alkyne (≈179°) angles, to suggest its formation would be unfavourable.
Scheme 1.
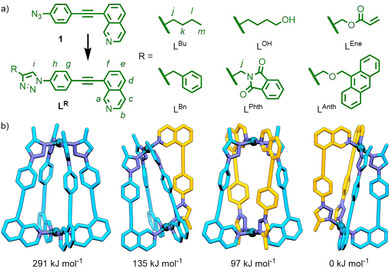
a) Synthesis of ligands LR from azide precursor 1. b) Geometry optimised structures (PM6) of the four potential [Pd2(LMe)4]4+ isomers and their relative energies (from left to right: “all‐up”, “three‐up‐one‐down”, trans and cis). Orange and blue colouring denotes relative ligand orientation.
Buoyed by these rudimentary computational results, the ligand precursor 1 was prepared in 86 % overall yield from commercially available reagents (see Supporting Information for details). Submission of 1 to standard CuAAC reaction conditions (CuSO4⋅5 H2O, sodium ascorbate, DMF, RT) with 1‐hexyne gave LBu in 89 % yield.
Pleasingly, a 2:1 combination of LBu with [Pd(CH3CN)4](BF4)2 in [D6]DMSO resulted in a sharp 1H NMR spectrum composed of a single set of signals (Figure 1 a). The combination of downfield shifts of isoquinoline (Ha and Hb; Δδ=0.50 and 1.25 ppm, respectively) and triazole (Hi; Δδ=0.37 ppm) signals in the 1H NMR spectrum (Figure S42), calculated solvodynamic radius (RS; 9.7 Å) from diffusion‐ordered NMR spectroscopy (DOSY; D=1.03×10−10 m2s−1), and signals observed by mass spectrometry (MS; Figure 1 b) all indicated formation of a species with formula [Pd2(LBu)4]4+. The symmetry of the 1H NMR spectrum and cross‐peaks observed by NOESY (Hb⋅⋅⋅Hj; Figure 1 c) determined that either the cis or trans isomer had been obtained. Diastereotopic splitting of methylene units of the butyl chain (Hj and Hk) suggested that the cis isomer was most likely to have formed. Ultimately, unambiguous confirmation of the cis‐[Pd2(LBu)4]4+ structure came from solid‐state single‐crystal X‐ray diffraction (SCXRD) data (Figure 1 d and e).
Figure 1.
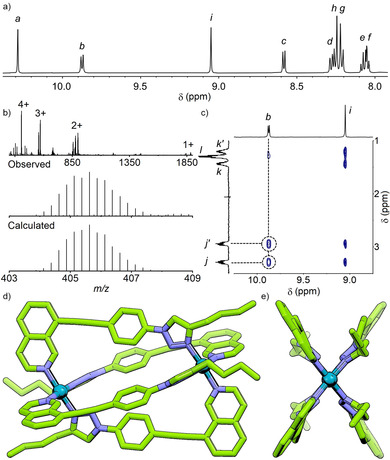
a) Partial 1H NMR spectrum (500 MHz, [D6]DMSO, 298 K) of [Pd2(LBu)4](BF4)4; b) MS with observed and calculated peak for [Pd2(LBu)4]4+; c) partial NOESY spectrum showing through‐space interactions between isoquinoline Hb and butyl chain Hj signals (for signal labelling see Scheme 1); SCXRD structure of [Pd2(LBu)4](BF4)4 shown d) from the side, and e) down the Pd‐Pd axis. N−Pd bond lengths 2.022–2.032 Å; N−Pd−N angles 176.2–176.3°; Pd⋅⋅⋅Pd distance 11.1 Å. Counterions, solvent molecules and hydrogen atoms omitted for clarity.
Having shown that the ligand framework was indeed suitable for the specific formation of cis‐Pd2L4 structures, a number of ligands derived from the common precursor 1 were prepared (Scheme 1 a). In addition to alkyl substituents, self‐assembly of the ligand framework with PdII was found to be compatible with aryl (LBn) and bulkier aromatic (LAnth) and heterocyclic (LPhth) moieties, as well as substituents containing heteroatoms (LOH) and unsaturated units (LEne). Although no SCXRD structures of these cages were obtained, similar MS and NMR spectroscopic details (see Supporting Information) indicated no alteration in the preference for formation of cis‐Pd2L4 cage isomers. With the core cis‐Pd2L4 framework remaining isostructural amongst the ligands examined (Figure 2), the ability to use the triazole substituents to modify the properties of the assemblies was investigated.
Figure 2.
Optimised structures (PM6) of cis‐[Pd2(LR)4]4+ cages assembled from (clockwise from top left) LOH, LEne, LBn, LPhth and LAnth. Hydrogen atoms omitted for clarity.
Although generally considered to be weaker ligands than pyridines, the electronic properties of 1,2,3‐triazoles can be tuned with precision through varying the C‐ and N‐substituents. [19] The potential for fine tuning the ligand strength of triazoles in metallo‐supramolecular systems, however, is largely overlooked. [20] Within the current system, modulation of donor strength was demonstrated through a comparison of the self‐assembly profiles of ligands LBu and LPhth incorporating electron‐donating and ‐withdrawing groups, respectively.
Titration of PdII into a [D6]DMSO solution of LPhth (Figure 3 a) resulted in formation of the expected [Pd2(LPhth)4]4+ cage when a 1:2 metal/ligand ratio was reached (Figure 3 c). At a 1:4 ratio, however, a single species was observed (Figure 3 b) that was spectroscopically distinct from both the free ligand and Pd2L4 cage, although DOSY suggested that it was of a similar size to the latter (R S≈11 Å, compared to 11.3 Å for the cage). The NOESY spectrum (Figure S109) revealed cross‐peaks between Ha and Hb of the isoquinoline, which would only be expected were at least two of these heterocycles to be brought into close proximity through coordination in an anti‐parallel fashion (Figure 3 g); in contrast to the cage, cis‐[Pd2(LPhth)4]4+, no interactions between Hb of the isoquinoline and Hj of the triazole substituent were observed (Figure 3 h). These data are consistent with the formation of a mononuclear complex, [Pd(LPhth)4]2+, with coordination to the PdII ions occurring through the isoquinoline units (a molecular model of one possible conformer of a hypothetical model complex, [Pd(LMe)4]2+, is shown in Figure 3 i).
Figure 3.
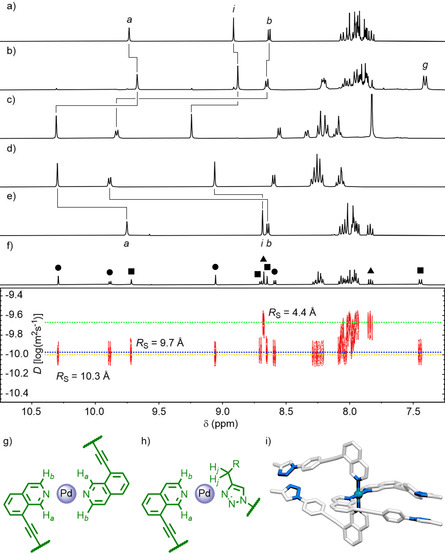
Controlling the self‐assembly profile of LR through tuning the electronic properties of the triazole. Partial 1H NMR spectra (400 MHz, [D6]DMSO, 298 K) of a) LPhth, b) LPhth+0.25 equiv. PdII, c) LPhth+0.50 equiv. PdII, d) LBu+0.50 equiv. PdII, and e) LBu. For signal labelling see Scheme 1. f) Partial DOSY spectrum (500 MHz, [D6]DMSO, 298 K) of LBu+0.25 equiv. PdII. Non‐overlapping peaks have been assigned where possible as [Pd2(LBu)4]4+ (•), [Pd(LBu)4]2+ (▪), and LBu (▴). Chemical structures showing indicative through‐space interactions between protons for g) mononuclear [Pd(LR)4]2+ species and h) [Pd2(LR)4]4+ cage assemblies. i) Molecular model of one possible conformation of a mononuclear [Pd(LMe)4]2+ structure with coordination exclusively through the isoquinoline units.
Additional support for the identity of the mononuclear species came from a model compound, namely the complex formed from the self‐assembly of monodentate ligand precursor 1. A 4:1 mixture of 1 with PdII yielded a sharp set of signals (Figure S113) wherein characteristic peaks—Ha, Hb and Hg—resonated at similar chemical shifts to those observed for the purported [Pd(LPhth)4]2+ complex. Unfortunately, no signals for this latter species were observed by MS (although an isotopic pattern consistent with the formula {[Pd(LPhth)3](X−)}+ (Figure S111) could be seen), presumably due to the instability of the mononuclear complex under the ESI‐MS conditions. SCXRD data was obtained, however, that confirmed the anticipated structure of the model complex, [Pd(1)4]2+ (Figure S130). [21]
The situation with LBu was slightly more complex. The 1H NMR spectrum obtained from a 1:4 mixture of PdII and LBu (Figure 3 f) at first glance appeared to indicate a stoichiometric mixture of [Pd2(LBu)4]4+ (Figure 3 d) and free LBu (Figure 3 e). Closer inspection revealed additional peaks, belonging to neither of these species, that resembled the mononuclear complex seen with LPhth. This suggested that these three species (LBu, [Pd(LBu)4]2+ and [Pd2(LBu)4]4+) might all be present in solution.
2D NMR—in particular NOESY and DOSY—was used to assign non‐overlapping signals to individual components of this mixture. In the NOESY spectrum (Figure S106) cross‐peaks indicative of Hb⋅⋅⋅Hj interactions between the isoquinoline and butyl chain (Figure 3 h) could be seen for the cage assembly but were absent for those signals assigned to the mononuclear complex; additionally, Ha⋅⋅⋅Hb interactions could be seen for the latter (Figure 3 g). DOSY (Figure 3 f) showed peaks assigned to the cage and mononuclear complexes to be diffusing at similar rates (D=10.0 and 9.8×10−11 m2 s−1, respectively) whilst those assumed to be from the free ligand diffused much quicker (D=4.4×10−11 m2s−1), indicative of two similarly sized species and one much smaller (R S=10.3, 9.7 and 4.4 Å, respectively) being present in solution.
Thus, based on the spectral data, it was concluded that a 1:4 mixture of PdII and LBu generated a combination of the dinuclear cage, mononuclear complex (with coordination again presumed to be through the isoquinoline units) and free ligand. It is noted that for free LBu, downfield signals associated with protons Ha and Hb adjacent to the nitrogen atom of the isoquinoline could not be identified. It is assumed that these peaks had broadened significantly, possibly due to transient non‐covalent interactions with the [Pd2(LBu)4]4+ and/or [Pd(LBu)4]2+ complexes.
The proton affinity (PA) of N3 of the triazole, as determined by DFT calculations (B3LYP‐6‐31G(d)), was increased by 16 kJ mol−1 going from the electron‐withdrawing N‐methylenephthalimide substituent in LPhth to the electron‐donating butyl group of LBu (see Supporting Information for details). Using PA as a proxy for ligand strength, these results support the rationalisation of the observed differences in self‐assembly as arising from differences in the σ‐donor ability of the triazoles between the two ligands. The combined experimental and computational results demonstrate effective control of the ligand framework's electronic properties, through tailoring of the triazole substituent, without ultimately affecting the core structure of the cis‐Pd2L4 assembly.
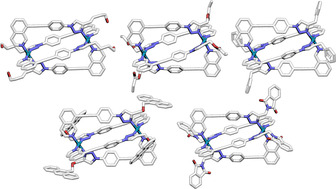
Since the core framework remained constant between the different functionalised ligands, it seemed that it would be possible to generate a statistical combinatorial library of homoleptic and mixed‐ligand cages through self‐assembly of a mixture of more than one ligand. [22] To examine this, PdII and a binary mixture of LBu and either LBn or LPhth were combined in a 1:1:1 ratio in [D6]DMSO. The resultant self‐assembled libraries gave well‐defined 1H NMR spectra (Figures 4 b and S115) that appeared to contain multiple overlapping signals from species of a similar size to the homoleptic assemblies (R S≈10–11 Å). These mixtures were found to be composed of the five possible homoleptic and heteroleptic constitutional assemblies with the formula [Pd2(LBu)x(LBn/LPhth)(4−x)]4+ (Figure 4 d) by MS (Figure 4 e).
Figure 4.
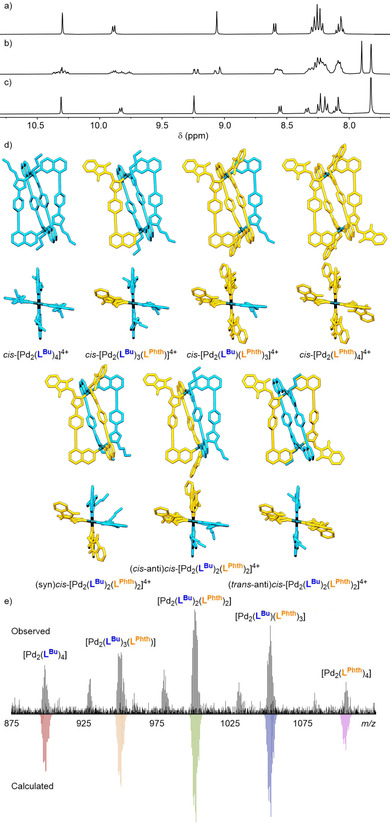
Partial 1H NMR spectra (400 MHz, [D6]DMSO, 298 K) of a) [Pd2(LBu)4](BF4)4, b) combinatorial library [Pd2(LBu)x(LPhth)(4−x)](BF4)4, and c) [Pd2(LPhth)4](BF4)4. d) Optimised structures (PM6) of the members of the combinatorial library [Pd2(LBu)x(LPhth)(4−x)]4+ including the three diastereomeric forms of [Pd2(LBu)2(LPhth)2]4+ (single enantiomers of chiral cages shown). LBu and LPhth ligands have been coloured blue and orange, respectively. e) Observed MS and calculated signals for {[Pd2(LBu)x(LPhth)(4−x)](BF4)2}2+ ions (x=0–4).
It is noted that whilst the homoleptic and cis‐Pd2 L1 3 L2‐type architectures should be present as single diastereomers, cis‐Pd2 L1 2 L2 2 assemblies with asymmetric ligands have the potential to exist as three different diastereomeric forms depending on the relative arrangements and orientations of the ligands (Figure 4 d), although these isomers cannot be distinguished by MS. Additionally, the two Pd2 L1 3 L2 and one of the Pd2 L1 2 L2 2 cages are chiral, resulting in a library of ten cages in total. Herein the three Pd2 L1 2 L2 2 cage isomers have been termed syn, cis‐anti and trans‐anti, where for pairs of the same ligand (in this instance for example, the pair of LBu ligands) syn/anti denotes parallel/antiparallel relative orientations, and cis/trans indicates their relative positions within the cis‐Pd2L4 structure (i.e. adjacent or opposite from each other, respectively).
Having successfully demonstrated statistical mixing of a binary combination of ligands, a ternary system was subsequently examined. LOH, LPhth and LAnth were combined with PdII in a 2:2:2:3 ratio in [D6]DMSO. Unsurprisingly, the resultant 1H NMR spectrum was somewhat complex (Figure S123). The DOSY data, however, were consistent with the formation of appropriately sized assemblies (R S=11.0 Å). The MS data was more complex than for previous systems, likely due to overlapping signals from the multitude of species present in solution, and anion exchange under the MS conditions. However, signals congruent with the three possible tris‐heteroleptic assemblies (i.e., those incorporating at least one of each ligand) were observed (Figure S125). Signals consistent with most of the 15 expected constitutional assemblies of the library could also be seen (Figure 5). As several of the bis‐heteroleptic and all of the tris‐heteroleptic assemblies are expected to exist as a mixture of diastereomers, and a number of these are chiral, the combinatorial library is considered to consist of a total of 45 cis‐Pd2L4 cages (Figure S128).
Figure 5.
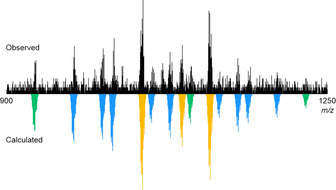
Partial MS of the ternary combinatorial library with calculated signals for {[Pd2L4](BF4)2}2+ ions with one (green), two (blue), and three (orange) different ligands.
Conclusions
A family of asymmetric, ditopic ligands that self‐assemble into exohedrally functionalised, low symmetry cis‐Pd2L4 architectures was realised. CuAAC chemistry was employed to yield these mixed‐heterocycle ligands in which a 1,2,3‐triazole unit served as both a coordinating unit and linker to append functional groups to the assembly scaffold. A variety of functional groups were shown to be tolerated without impacting the core structure of the cage. Furthermore, facile tuning of electronic properties was demonstrated, allowing modification of the ligand self‐assembly profile. Due to the isostructural nature of the core framework of the ligands, and their resultant Pd2L4 assemblies, it was shown possible to generate combinatorial libraries that included mixed‐ligand architectures with multiple functional moieties. With the need to move towards more complex MOPs to advance their utility in various applications, this study has demonstrated an underexplored approach to readily access functional, low symmetry metal–organic assemblies.
Experimental Section
Crystal data for [Pd2(LBu)4](BF4)4: Pd2(C23H20N4)4(BF4)4⋅2(C6H14O)⋅4(C3H7NO), M=2466.48, monoclinic, P21/n, a=13.4683(3), b=18.1293(4), c=24.7113(5) Å, α=90, β=99.400(2), γ=90°, V=5952.8(2) Å3, Z=2, ρ cald=1.376 mg m−3, μ(MoKα)=0.389 mm−1, T=173 K, 12 454 independent measured reflections (R int =0.0322), R 1 =0.0752, wR2=0.2057 [I>2σ(I)], completeness=99.7, 689 parameters, GOF on F 2 = 1.032, max/min residual density 1.859/−0.998 e Å−3. Deposition Number 2043240 contains the supplementary crystallographic data for this paper. These data are provided free of charge by the joint Cambridge Crystallographic Data Centre and Fachinformationszentrum Karlsruhe Access Structures service www.ccdc.cam.ac.uk/structures.
LR were prepared according to the following general procedure: 1 (67.6 mg, 0.25 mmol), CuSO4⋅5 H2O (31.2 mg, 0.125 mmol), sodium ascorbate (49.5 mg, 0.25 mmol) and appropriate alkyne were stirred at RT in DMF (5 mL) for 16 h. The reaction mixture was diluted with EtOAc (40 mL) and washed with 0.1 m EDTA‐Na2 in 1:9 NH4OH(aq)/H2O (10 mL), brine (4×10 mL), dried (MgSO4) and the solvent removed in vacuo, with subsequent purification by column chromatography on silica gel.
[Pd2(LR)4](BF4)4 were prepared by sonicating LR (0.030 mmol) and [Pd(CH3CN)4](BF4)2 (6.7 mg, 0.015 mmol) in [D6]DMSO (0.75 mL) until a homogenous solution was obtained.
For calculated structures in xyz format see DOI: https://doi.org/10.14469/hpc/7366.
Conflict of interest
The author declares no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This work was supported by an Imperial College Research Fellowship. Thanks to Peter Haycock and Corey Fulop for assistance with the collection of NMR data, Dr. Lisa Haigh for MS data, Dr. Andrew White for collection of SCXRD data, Dr. Dan Preston for useful discussions, and Prof. Matthew Fuchter for useful discussions and access to equipment and resources.
J. E. M. Lewis, Chem. Eur. J. 2021, 27, 4454.
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.26434/chemrxiv.13246715.v1).
References
- 1.
- 1a. Fujita M., Chem. Soc. Rev. 1998, 27, 417–425; [Google Scholar]
- 1b. Leininger S., Olenyuk B., Stang P. J., Chem. Rev. 2000, 100, 853–908; [DOI] [PubMed] [Google Scholar]
- 1c. Holliday B. J., Mirkin C. A., Angew. Chem. Int. Ed. 2001, 40, 2022–2043; [PubMed] [Google Scholar]; Angew. Chem. 2001, 113, 2076–2097; [Google Scholar]
- 1d. Ward M. D., Chem. Commun. 2009, 4487–4499; [DOI] [PubMed] [Google Scholar]
- 1e. Chakrabarty R., Mukherjee P. S., Stang P. J., Chem. Rev. 2011, 111, 6810–6918; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1f. Smulders M. M. J., Riddell I. A., Browne C., Nitschke J. R., Chem. Soc. Rev. 2013, 42, 1728–1754; [DOI] [PubMed] [Google Scholar]
- 1g. Ronson T. K., Zarra S., Black S. P., Nitschke J. R., Chem. Commun. 2013, 49, 2476–2490; [DOI] [PubMed] [Google Scholar]
- 1h. Cook T. R., Stang P. J., Chem. Rev. 2015, 115, 7001–7045; [DOI] [PubMed] [Google Scholar]
- 1i. Debata N. B., Tripathy D., Sahoo H. S., Coord. Chem. Rev. 2019, 387, 273–298. [Google Scholar]
- 2.
- 2a. Yoshizawa M., Klosterman J. K., Fujita M., Angew. Chem. Int. Ed. 2009, 48, 3418–3438; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2009, 121, 3470–3490; [Google Scholar]
- 2b. Cook T. R., Vajpayee V., Lee M. H., Stang P. J., Chi K. W., Acc. Chem. Res. 2013, 46, 2464–2474; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2c. Therrien B., Chem. Eur. J. 2013, 19, 8378–8386; [DOI] [PubMed] [Google Scholar]
- 2d. Brown C. J., Toste F. D., Bergman R. G., Raymond K. N., Chem. Rev. 2015, 115, 3012–3035; [DOI] [PubMed] [Google Scholar]
- 2e. Casini A., Woods B., Wenzel M., Inorg. Chem. 2017, 56, 14715–14729; [DOI] [PubMed] [Google Scholar]
- 2f. Kim T. Y., Vasdev R. A. S., Preston D., Crowley J. D., Chem. Eur. J. 2018, 24, 14878–14890; [DOI] [PubMed] [Google Scholar]
- 2g. Sinha I., Mukherjee P. S., Inorg. Chem. 2018, 57, 4205–4221; [DOI] [PubMed] [Google Scholar]
- 2h. Fang Y., Powell J. A., Li E., Wang Q., Perry Z., Kirchon A., Yang X., Xiao Z., Zhu C., Zhang L., Huang F., Zhou H.-C., Chem. Soc. Rev. 2019, 48, 4707–4730; [DOI] [PubMed] [Google Scholar]
- 2i. Sepehrpour H., Fu W., Sun Y., Stang P. J., J. Am. Chem. Soc. 2019, 141, 14005–14020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2j. Rizzuto F. J., von Krbek L. K. S., Nitschke J. R., Nat. Rev. Chem. 2019, 3, 204–222; [Google Scholar]
- 2k. Gosselin E. J., Rowland C. A., Bloch E. D., Chem. Rev. 2020, 120, 8987–9014; [DOI] [PubMed] [Google Scholar]
- 2l. Xue Y., Hang X., Ding J., Li B., Zhu R., Pang H., Xu Q., Coord. Chem. Rev. 2020, 213656. [Google Scholar]
- 3.
- 3a. He Z., Jiang W., Schalley C. A., Chem. Soc. Rev. 2015, 44, 779–789; [DOI] [PubMed] [Google Scholar]
- 3b. Holloway L. R., Bogie P. M., Hooley R. J., Dalton Trans. 2017, 46, 14719–14723; [DOI] [PubMed] [Google Scholar]
- 3c. Bloch W. M., Clever G. H., Chem. Commun. 2017, 53, 8506–8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.For selected examples see:
- 4a. McMorran D. A., Steel P. J., Angew. Chem. Int. Ed. 1998, 37, 3295–3297; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 1998, 110, 3495–3497; [Google Scholar]
- 4b. Chand D. K., Biradha K., Fujita M., Chem. Commun. 2001, 1652–1653; [DOI] [PubMed] [Google Scholar]
- 4c. Clever G. H., Tashiro S., Shionoya M., Angew. Chem. Int. Ed. 2009, 48, 7010–7012; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2009, 121, 7144–7146; [Google Scholar]
- 4d. Crowley J. D., Gavey E. L., Dalton Trans. 2010, 39, 4035–4037; [DOI] [PubMed] [Google Scholar]
- 4e. Liao P., Langloss B. W., Johnson A. M., Knudsen E. R., Tham F. S., Julian R. R., Hooley R. J., Chem. Commun. 2010, 46, 4932–4934; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4f. Kishi N., Li Z., Yoza K., Akita M., Yoshizawa M., J. Am. Chem. Soc. 2011, 133, 11438–11441; [DOI] [PubMed] [Google Scholar]
- 4g. Lewis J. E. M., Gavey E. L., Cameron S. A., Crowley J. D., Chem. Sci. 2012, 3, 778–784; [Google Scholar]
- 4h. Jansze S. M., Wise M. D., Vologzhanina A. V., Scopelliti R., Severin K., Chem. Sci. 2017, 8, 1901–1908; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4i. Preston D., Patil K. M., O'Neil A. T., Vasdev R. A. S., Kitchen J. A., Kruger P. E., Inorg. Chem. Front. 2020, 7, 2990–3001. [Google Scholar]
- 5.
- 5a. Preston D., Barnsley J. E., Gordon K. C., Crowley J. D., J. Am. Chem. Soc. 2016, 138, 10578–10585; [DOI] [PubMed] [Google Scholar]
- 5b. Zhu R., Bloch W. M., Holstein J. J., Mandal S., Schäfer L. V., Clever G. H., Chem. Eur. J. 2018, 24, 12976–12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.
- 6a. Bloch W. M., Abe Y., Holstein J. J., Wandtke C. M., Dittrich B., Clever G. H., J. Am. Chem. Soc. 2016, 138, 13750–13755; [DOI] [PubMed] [Google Scholar]
- 6b. Bloch W. M., Holstein J. J., Hiller W., Clever G. H., Angew. Chem. Int. Ed. 2017, 56, 8285–8289; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 8399–8404; [Google Scholar]
- 6c. Wu K., Zhang B., Drechsler C., Holstein J. J., Clever G. H., Angew. Chem. Int. Ed. 2020, 10.1002/anie.202012425; [DOI] [Google Scholar]; Angew. Chem. 2020, 10.1002/ange.202012425. [DOI] [Google Scholar]
- 7.
- 7a. Pullen S., Clever G. H., Acc. Chem. Res. 2018, 51, 3052–3064; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7b. Bardhan D., Chand D. K., Chem. Eur. J. 2019, 25, 12241–12269. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. Hiraoka S., Sakata Y., Shionoya M., J. Am. Chem. Soc. 2008, 130, 10058–10059; [DOI] [PubMed] [Google Scholar]
- 8b. Wu H. B., Wang Q. M., Angew. Chem. Int. Ed. 2009, 48, 7343–7345; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2009, 121, 7479–7481; [Google Scholar]
- 8c. Smulders M. M. J., Jiménez A., Nitschke J. R., Angew. Chem. Int. Ed. 2012, 51, 6681–6685; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 6785–6789; [Google Scholar]
- 8d. Ramsay W. J., Ronson T. K., Clegg J. K., Nitschke J. R., Angew. Chem. Int. Ed. 2013, 52, 13439–13443; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 13681–13685; [Google Scholar]
- 8e. Li K., Zhang L. Y., Yan C., Wei S. C., Pan M., Zhang L., Su C. Y., J. Am. Chem. Soc. 2014, 136, 4456–4459; [DOI] [PubMed] [Google Scholar]
- 8f. Wise M. D., Holstein J. J., Pattison P., Besnard C., Solari E., Scopelliti R., Bricogne G., Severin K., Chem. Sci. 2015, 6, 1004–1010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8g. Ramsay W. J., Szczypiński F. T., Weissman H., Ronson T. K., Smulders M. M. J., Rybtchinski B., Nitschke J. R., Angew. Chem. Int. Ed. 2015, 54, 5636–5640; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 5728–5732; [Google Scholar]
- 8h. Sanz S., O'Connor H. M., Pineda E. M., Pedersen K. S., Nichol G. S., Mønsted O., Weihe H., Piligkos S., McInnes E. J. L., Lusby P. J., Brechin E. K., Angew. Chem. Int. Ed. 2015, 54, 6761–6764; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 6865–6868; [Google Scholar]
- 8i. Metherell A. J., Ward M. D., Chem. Sci. 2016, 7, 910–915; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8j. Hou Y. J., Wu K., Wei Z. W., Li K., Lu Y. L., Zhu C. Y., Wang J. S., Pan M., Jiang J. J., Li G. Q., Su C. Y., J. Am. Chem. Soc. 2018, 140, 18183–18191; [DOI] [PubMed] [Google Scholar]
- 8k. Preston D., Sutton J. J., Gordon K. C., Crowley J. D., Angew. Chem. Int. Ed. 2018, 57, 8659–8663; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 8795–8799; [Google Scholar]
- 8l. Hardy M., Struch N., Holstein J. J., Schnakenburg G., Wagner N., Engeser M., Beck J., Clever G. H., Lützen A., Angew. Chem. Int. Ed. 2020, 59, 3195–3200; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 3221–3226; [Google Scholar]
- 8m. Lisboa L. S., Findlay J. A., Wright L. J., Hartinger C. G., Crowley J. D., Angew. Chem. Int. Ed. 2020, 59, 11101–11107; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 11194–11200; [Google Scholar]
- 8n. Yang D., Greenfield J. L., Ronson T. K., von Krbek L. K. S., Yu L., Nitschke J. R., J. Am. Chem. Soc. 2020, 142, 19856–19861. [DOI] [PubMed] [Google Scholar]
- 9.
- 9a. Zhang Y. Y., Gao W. X., Lin L., Jin G. X., Coord. Chem. Rev. 2017, 344, 323–344; [Google Scholar]
- 9b. Hardy M., Lützen A., Chem. Eur. J. 2020, 26, 13332–13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lewis J. E. M., Tarzia A., White A. J. P., Jelfs K. E., Chem. Sci. 2020, 11, 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.
- 11a. Hiraoka S., Fujita M., J. Am. Chem. Soc. 1999, 121, 10239–10240; [Google Scholar]
- 11b. Ogata D., Yuasa J., Angew. Chem. Int. Ed. 2019, 58, 18424–18428; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 18595–18599; [Google Scholar]
- 11c. Mishra S. S., Kompella S. V. K., Krishnaswamy S., Balasubramanian S., Chand D. K., Inorg. Chem. 2020, 59, 12884–12894. [DOI] [PubMed] [Google Scholar]
- 12. Lewis J. E. M., Crowley J. D., ChemPlusChem 2020, 85, 815–827. [DOI] [PubMed] [Google Scholar]
- 13.For selected examples see:
- 13a. Kamiya N., Tominaga M., Sato S., Fujita M., J. Am. Chem. Soc. 2007, 129, 3816–3817; [DOI] [PubMed] [Google Scholar]
- 13b. Kikuchi T., Sato S., Fujita M., J. Am. Chem. Soc. 2010, 132, 15930–15932; [DOI] [PubMed] [Google Scholar]
- 13c. Ikemi M., Kikuchi T., Matsumura S., Shiba K., Sato S., Fujita M., Chem. Sci. 2010, 1, 68–71; [DOI] [PubMed] [Google Scholar]
- 13d. Park J., Sun L. B., Chen Y. P., Perry Z., Zhou H. C., Angew. Chem. Int. Ed. 2014, 53, 5842–5846; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 5952–5956; [Google Scholar]
- 13e. Lewis J. E. M., Elliott A. B. S., McAdam C. J., Gordon K. C., Crowley J. D., Chem. Sci. 2014, 5, 1833–1843; [Google Scholar]
- 13f. Wong K. K. G., Hoyas Pérez N., White A. J. P., Lewis J. E. M., Chem. Commun. 2020, 56, 10453–10456. [DOI] [PubMed] [Google Scholar]
- 14.For selected examples see:
- 14a. Tominaga M., Suzuki K., Murase T., Fujita M., J. Am. Chem. Soc. 2005, 127, 11950–11951; [DOI] [PubMed] [Google Scholar]
- 14b. Sato S., Iida J., Suzuki K., Kawano M., Ozeki T., Fujita M., Science 2006, 313, 1273–1276; [DOI] [PubMed] [Google Scholar]
- 14c. Suzuki K., Kawano M., Sato S., Fujita M., J. Am. Chem. Soc. 2007, 129, 10652–10653; [DOI] [PubMed] [Google Scholar]
- 14d. Suzuki K., Takao K., Sato S., Fujita M., J. Am. Chem. Soc. 2010, 132, 2544–2545; [DOI] [PubMed] [Google Scholar]
- 14e. Suzuki K., Sato S., Fujita M., Nat. Chem. 2010, 2, 25–29; [DOI] [PubMed] [Google Scholar]
- 14f. Fujita D., Suzuki K., Sato S., Yagi-Utsumi M., Yamaguchi Y., Mizuno N., Kumasaka T., Takata M., Noda M., Uchiyama S., Kato K., Fujita M., Nat. Commun. 2012, 3, 1093; [DOI] [PubMed] [Google Scholar]
- 14g. Bruns C. J., Fujita D., Hoshino M., Sato S., Stoddart J. F., Fujita M., J. Am. Chem. Soc. 2014, 136, 12027–12034; [DOI] [PubMed] [Google Scholar]
- 14h. Ueda Y., Ito H., Fujita D., Fujita M., J. Am. Chem. Soc. 2017, 139, 6090–6093; [DOI] [PubMed] [Google Scholar]
- 14i. Holloway L. R., Bogie P. M., Lyon Y., Ngai C., Miller T. F., Julian R. R., Hooley R. J., J. Am. Chem. Soc. 2018, 140, 8078–8081. [DOI] [PubMed] [Google Scholar]
- 15.
- 15a. Tornøe C. W., Christensen C., Meldal M., J. Org. Chem. 2002, 67, 3057–3064; [DOI] [PubMed] [Google Scholar]
- 15b. Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B., Angew. Chem. Int. Ed. 2002, 41, 2596–2599; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2002, 114, 2708–2711. [Google Scholar]
- 16. Schulze B., Schubert U. S., Chem. Soc. Rev. 2014, 43, 2522–2571. [DOI] [PubMed] [Google Scholar]
- 17. Vasdev R. A. S., Preston D., Crowley J. D., Dalton Trans. 2017, 46, 2402–2414. [DOI] [PubMed] [Google Scholar]
- 18.
- 18a. Zhao D., Tan S., Yuan D., Lu W., Rezenom Y. H., Jiang H., Wang L. Q., Zhou H. C., Adv. Mater. 2011, 23, 90–93; [DOI] [PubMed] [Google Scholar]
- 18b. Lewis J. E. M., John McAdam C., Gardiner M. G., Crowley J. D., Chem. Commun. 2013, 49, 3398–3400; [DOI] [PubMed] [Google Scholar]
- 18c. Elliott A. B. S., Lewis J. E. M., Van Der Salm H., McAdam C. J., Crowley J. D., Gordon K. C., Inorg. Chem. 2016, 55, 3440–3447; [DOI] [PubMed] [Google Scholar]
- 18d. Samanta S. K., Quigley J., Vinciguerra B., Briken V., Isaacs L., J. Am. Chem. Soc. 2017, 139, 9066–9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suijkerbuijk B. M. J. M., Aerts B. N. H., Dijkstra H. P., Lutz M., Spek A. L., Van Koten G., Klein Gebbink R. J. M., Dalton Trans. 2007, 1273–1276. [DOI] [PubMed] [Google Scholar]
- 20.An exception to this is the work of Crowley and co-workers who observed dramatic differences in the kinetics of self-assembly with triazole-based ligands dependent upon the nature of the N-substituent: Scott S. Ø., Gavey E. L., Lind S. J., Gordon K. C., Crowley J. D., Dalton Trans. 2011, 40, 12117–12124.21792428 [Google Scholar]
- 21.In the SCXRD structure of [Pd(1)4]2+ the complex is in an “all-up” conformation. If [Pd(LPhth)4]2+ were to adopt this conformation exclusively in solution, the observed Ha⋅⋅⋅Hb NOESY cross-peaks would not be expected. It is assumed that in solution both mononuclear species are able to exist as rapidly interconverting conformers through rotation of the ligands about the Pd−N bonds.
- 22.
- 22a. Johnson A. M., Moshe O., Gamboa A. S., Langloss B. W., Limtiaco J. F. K., Larive C. K., Hooley R. J., Inorg. Chem. 2011, 50, 9430–9442; [DOI] [PubMed] [Google Scholar]
- 22b. Frank M., Krause L., Herbst-Irmer R., Stalke D., Clever G. H., Dalton Trans. 2014, 43, 4587–4592; [DOI] [PubMed] [Google Scholar]
- 22c. Frank M., Ahrens J., Bejenke I., Krick M., Schwarzer D., Clever G. H., J. Am. Chem. Soc. 2016, 138, 8279–8287; [DOI] [PubMed] [Google Scholar]
- 22d. Rizzuto F. J., Kieffer M., Nitschke J. R., Chem. Sci. 2018, 9, 1925–1930; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22e. Kieffer M., Bilbeisi R. A., Thoburn J. D., Clegg J. K., Nitschke J. R., Angew. Chem. Int. Ed. 2020, 59, 11369–11373; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 11465–11469. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


