Abstract
The nsP3 macrodomain is a conserved protein interaction module that plays essential regulatory roles in the host immune response by recognizing and removing posttranslational ADP-ribosylation sites during SARS-CoV-2 infection. Thus targeting this protein domain may offer a therapeutic strategy to combat current and future virus pandemics. To assist inhibitor development efforts, we report here a comprehensive set of macrodomain crystal structures complexed with diverse naturally occurring nucleotides, small molecules, and nucleotide analogues including GS-441524 and its phosphorylated analogue, active metabolites of remdesivir. The presented data strengthen our understanding of the SARS-CoV-2 macrodomain structural plasticity and provide chemical starting points for future inhibitor development.
Keywords: Macrodomain, SARS-CoV-2, remdesivir, ADP-ribosylation, structure based design
The recently emerging coronavirus SARS-CoV-2 has become a major global health concern. SARS-Cov-2 is an enveloped (+) single-stranded RNA ((+)ssRNA) coronavirus, encoding several functionally relevant enzymes and protein interaction domains that are necessary for the viral life cycles and deregulation of the innate immune response of the host.1,2 Nonstructural protein 3 (nsP3) is one of the most complex coronavirus proteins comprising several domains and activities.1,3−5 The coding sequence includes a macrodomain (MD) module, which is structurally conserved in coronaviruses and generally binds adenosine diphosphate ribose (ADP-ribose).6 The viral MDs have been shown to play a pivotal role in recognizing and removing posttranslational ADP-ribosylation from specific sites.6−9 This posttranslational modification, which may occur as poly-ADP-ribosylation or mono-ADP-ribosylation (MARylation), is catalyzed by poly-ADP-ribose polymerase/diphtheria toxin-like ADP-ribosyltransferase (PARP/ARTD) enzymes, and it is associated with stress signaling and the innate immune response against pathogens.10,11 Several MARylating PARP enzymes are activated in response to pathogen-associated molecular patterns and interferons,9,11 therefore suggesting that MARylation is a key modification of the innate immune response against viruses.
Several studies have provided strong evidence that viral MDs, including those of several coronaviruses, such as SARS-CoV and MERS-CoV, and different alphaviruses, such as Chikungunya virus and O’nyong’nyong virus, function as hydrolases capable of removing ADP-ribose from proteins.6,9,12−14 Similarly, such de-mono-ADP-ribosylating activity (de-MARylation) has recently been demonstrated for the first nsP3 MD, also known as the Mac1, of SARS-CoV-2,15,16 whereas the other two MDs likely exert distinct activities.6 The MD and its de-MARylation function is important for efficient viral replication, as viruses harboring inactive MDs exhibit reduced replication ability and pathogenicity10,17 and are sensitive to interferon pretreatment.18,19 In addition, the de-MARylation activity of the viral MD has been convincingly linked, directly or indirectly, to the function of viral proteins involved in viral genome replication or in antagonizing the innate immune response.9,10
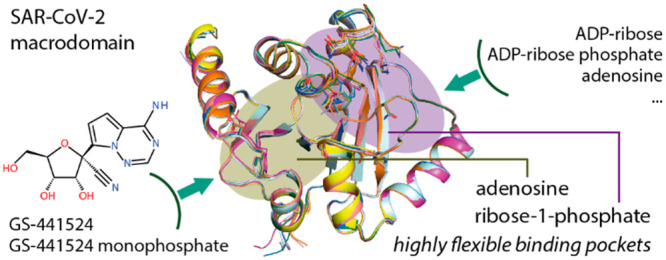
The central role of the SARS-CoV-2 MD in viral replication and in mediating inflammatory responses in the host suggests that targeting this protein module may offer a therapeutic strategy for combating severe respiratory disease. Recent studies have elucidated the crystal structure of this protein module,15,16,20−22 demonstrating that the MD of SARS-CoV-2 exhibits a macro-H2A topology highly similar to homologues of other viruses including SARS-CoV.9,16,23,24 The pocket is lined with a set of highly conserved amino acids and catalytic residues, and thus its interaction with ADP-ribose resembles binding modes reported for other viral MD homologues.16,20 Nevertheless, some subtle structural differences have been observed, suggesting that SARS-CoV-2 MD may possess some degree of plasticity,20 opening a possibility for the binding of diverse small molecules.15
Thus to strengthen our understanding of the nature and druggability of the SARS-CoV-2 MD, we report a collective set of crystal structures, including an apo state as well as high-resolution structures of complexes with diverse naturally occurring nucleosides and nucleotides. A comparison of these structures highlighted the ligand-induced domain plasticity of the ADP-ribose binding site. In addition, we describe the unprecedented discovery of the binding of GS-441524 and its phosphorylated analogue, active metabolites of remdesivir.
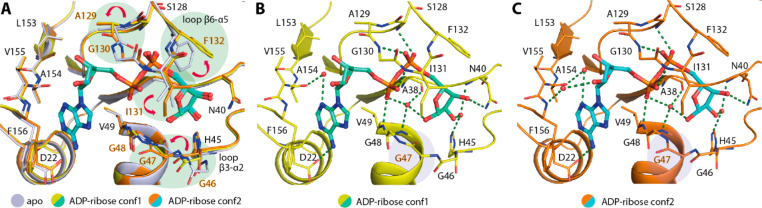
To understand the plasticity of the MD, we first attempted to crystallize an apo state and obtained two orthorhombic crystals that diffracted to 2.16 to 2.20 Å resolution. The presence of five and three molecules in their asymmetric units therefore suggested the potential flexibility of the protein. Indeed, a structural comparison of all chains revealed that whereas the core β-sheet and helices superimposed well, some differences in the conformations of residues lining the ADP-ribose binding pocket were evident (Figure 1A,B). By comparison, the adenosine monophosphate (AMP) binding site was more rigid than the ribose-1-phosphate binding site, in which all three loops connecting β3-α2, β5-α4, and β6-α5 that lined the pocket exhibited various degrees of flexibility. Similar to a previous report,20 we observed diverse conformations of Phe132 and Ile131 located in the phosphate binding site (Figure 1B). The flexibility of the phenylalanine and isoleucine was rather interesting because it potentially determines the availability of the pocket to diverse ligands. This role was supported by conformational changes observed in the complexes with MES and HEPES molecules that were used as buffers in crystallization reagents and occupied the ribose-1-phosphate binding site (Figure 1C,D). Both buffer molecules bound within the phosphate binding site similarly to a previous observation20 and caused conformational changes to the loop β6-α5 residues—in essence, an out-swing of Phe132 and a rotation of the Ile131 side chain. In comparison, a slight difference was observed for the binding of MES that also induced the Ala129-Gly130 main-chain flip, enabling two hydrogen bonds that were absent in the HEPES complex (Figure 1C,D). Overall, the structural changes seen in the apo state and both complexes with these weakly interacting ligands suggested by their high B-factors therefore revealed the potential intrinsic plasticity of the MD, especially within its ribose-phosphate binding pocket, which may form a determinant factor for the binding of ligands.
Figure 1.
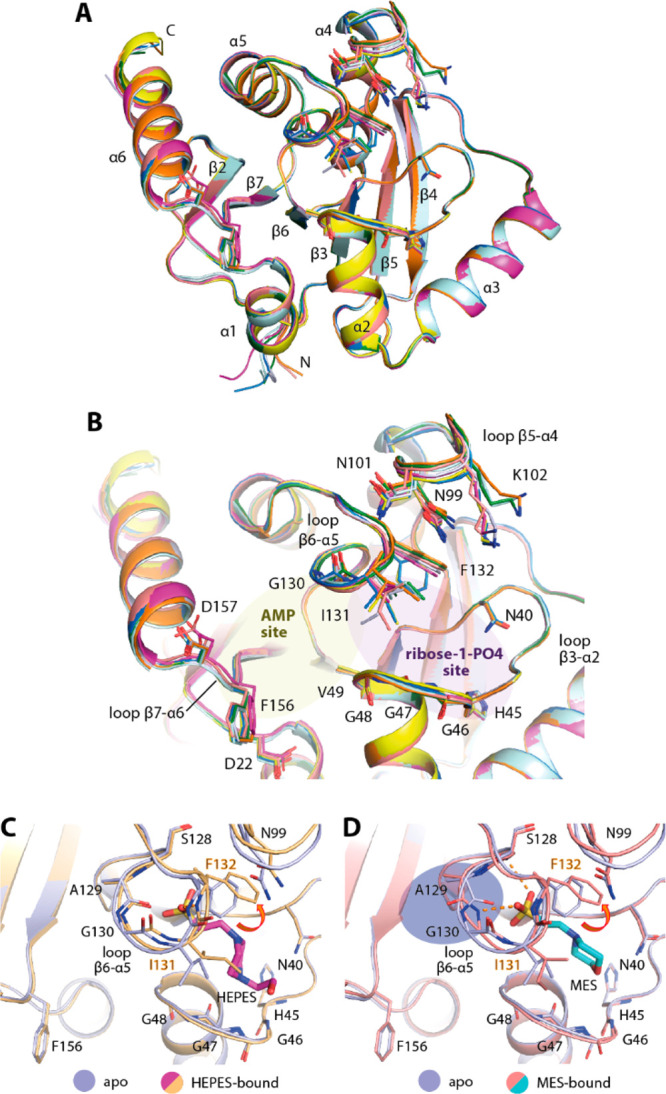
Plasticity of the ADP-ribose binding pocket of the SARS-CoV-2 macrodomain. (A) Superimposition of eight molecules from the asymmetric units of two distinct apo crystal structures (PDB ID: 6YWK and 6YWM). (B) Close up of the ADP-ribose binding pocket of all eight molecules reveals the flexibility of the residues lining the pocket, notably Phe132 and Ile131. (C,D) Binding of HEPES (PDB ID: 6YWK) and MES (PDB ID: 6YWM) used as crystallization reagents within the ribose-phosphate binding site requires conformational changes of Phe132 and Ile131 necessary for an accommodation of the sulfate moieties of the ligands.
To further understand the conformational changes necessary for the binding of ADP-ribose, we performed soaking of the ligand into the apo crystals and compared the ligand-bound form (2.50 Å resolution) with the apo state. Interestingly, a number of significant side-chain alterations were observed, primarily at two regions: loop β6-α5 as well as loop β3-α2, which constructed the ribose-phosphate binding site (Figure 2A). Substantial alterations were noted for the former structural motif, which included an ∼56° outswing of Phe132, vacating the phosphate binding pocket, in addition to a rotation of the Ile131 side chain to pack on top of the terminal ribose and a main-chain carbonyl flip of A129 to enable hydrogen-bond interactions between the Gly130 amine and the phosphate moiety of the ADP-ribose. At the opposing site of the binding pocket, small alterations to the glycine-rich loop β3-α2 were also evident, involving the rearrangement of Gly46–47 main chains and bringing the loop closer to the ligand. Nevertheless, two configurations of this glycine-rich loop were observed for the ADP-ribose complex: one had a Gly47 carbonyl atom flipped “in” (two of five monomers) and the other conformation had an “out”-oriented carbonyl group (in the other three monomers; Figure 2B,C). These structural changes provided further evidence of the high flexibility of this glycine-rich loop, which contains several catalytic residues that are important for enzymatic function.16 Nonetheless, the two conformations of the loop did not alter the contacts between the terminal ribose and the protein, which were also highly conserved compared with similar complexes that have been previously reported.16,20
Figure 2.
Binding of ADP-ribose triggers conformational changes. (A) Superimposition of the apo form and two conformations of the ADP-ribose complex (conf1 and conf2; PDB ID: 6YWL) reveals several conformational changes potentially necessary for the binding of the ligand. (B,C) Two conformations observed for the ADP-ribose complex with a notable difference in the configuration of the Gly47 backbone. Water molecules are shown as red spheres, and green dashed lines indicate hydrogen bonds.
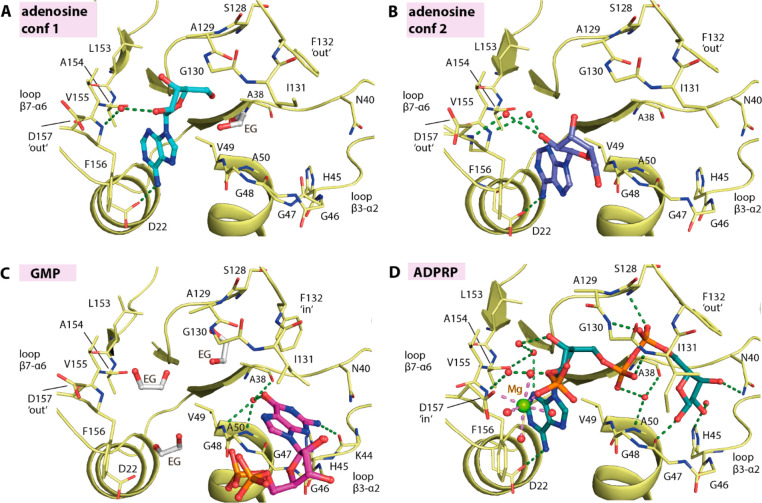
The plasticity of the ADP-ribose binding pocket of the MD prompted us to question whether it might be able to accommodate other nucleosides and nucleotides. Thus we attempted to cocrystallize the protein with selected naturally occurring ligands, including AMP, GDP-glucose, ribose-1-phosphate, β-NAD (nicotinamide adenine dinucleotide), and β-NADP (nicotinamide adenine dinucleotide phosphate), all of which (except for ribose-1-phosphate) led to the successful determination of complexed structures at 1.55 to 2.05 Å resolution. An examination of the electron density suggested, however, the modification of the bound ligands (Figure 3). In the case of AMP, only the adenosine moiety, the product of autohydrolysis lacking the phosphate group, was presented in the structure. Interestingly, the adenosine ligand exerted two binding modes: One resembled that of the adenosine moiety in the ADP-ribose complex, whereas a slight tilt of the adenine ring in the second binding mode positioned the ribose group outside in the solvent-exposed region (Figure 3A,B). Although the hydrogen bond between the adenine amine group and Asp22 was still maintained, different water-mediated contacts between the ribose moiety and the protein were evident.
Figure 3.
Nucleoside and nucleotide binding in the macrodomain. The binding of (A,B) adenosine in two different conformations (PDB ID: 7BF3), (C) GMP (PDB ID: 7BF4), and (D) ADP-ribose-2′-phosphate (ADPRP; PDB ID: 7BF5). The ligands are shown as a stick representation with water molecules shown as red spheres and ethylene glycol molecules labeled as “EG”.
For GDP-glucose, the electron density allowed modeling of only guanosine monophosphate (GMP), a product of hydrolysis. Nonetheless, the binding of GMP in the ribose-1-phosphate binding pocket of the MD was intriguing (Figure 3C). The guanine ring surprisingly occupied the cavity reserved for the terminal ribose moiety of ADP-ribose, not the AMP-binding site, likely due to a poor fit between the guanine 6-hydroxyl and 2-amine groups and Asp22 and the backbone amide of Val155 and Phe156 of loop β7-α6. The bound GMP was stabilized mainly through interactions of the guanine ring, which was sandwiched between Ile131 and the glycine-rich loop β3-α2, forming a network of hydrogen bonds to Lys44, Val49, Ala50, and Ala38. No contribution to binding was observed from the ribose and phosphate groups that were positioned outside the binding pocket in the solvent region.
For β-NAD and β-NADP, the observed electron density suggested that the entities of the bound ligands were likely nicotinamide-cleaved products, resulting in the complex of ADP-ribose and ADP-ribose-2′-phosphate (ADPRP), respectively. Although hydrolase activity catalyzing NAD hydrolysis25 has been documented for some MDs, we assumed that these cleavage products were likely a consequence of autohydrolysis of the compounds because no significant increase in ADP-ribose or ADPRP traces was observed in a high-performance liquid chromatography (HPLC) assay upon an incubation of β-NAD or β-NADP with SARS-CoV-2 MD (data not shown). A structural comparison demonstrated that both the NAD-cleaved product ADP-ribose and the NADP-cleaved product ADPRP assumed a binding mode that was highly identical to that of ADP-ribose (Figures 2 and 3D). Nevertheless, a difference was noted for ADPRP in that the 2′-phosphate group, which was located in the solvent region, could engage additional magnesium-mediated contacts with an inward side chain of Asp157 (Figure 3D).
The flexibility of the ADP-ribose binding pocket with its ability to accept various ligands prompted us to speculate the potential binding of antiviral nucleoside analogues. We therefore selected a set of diverse clinically used antivirals, including abacavir, entecavir, acyclovir, gemcitabine, remdesivir, and remdesivir metabolite GS-441524, and tested the binding of these drugs using cocrystallization. Among the set, an examination of electron density maps revealed that the remdesivir metabolite GS-441524 was the only ligand that showed binding in the crystal structures (2.15 Å resolution; Figure 4A,B). The interaction of this compound was rather unexpected because this metabolite and its active triphosphorylated form have been designed to target the RNA-dependent RNA polymerase (RdRp) of several viruses, including Ebola virus, MERS-CoV, SARS-CoV, and potentially SARS-CoV-2.26−29 In addition, a recent study has further demonstrated that the direct use of GS-441524 can potently inhibit the replication of SAR-CoV-2 in a mouse model.30 On the basis of our crystal structure, a structural comparison demonstrated that the binding mode of GS-441524 in the MD highly resembled that of the adenosine moiety of ADP-ribose. The amine group of the pyrrolotriazine ring, which was sandwiched between Val49 and Phe159, formed a hydrogen bond to Asp22, whereas the hydroxyl groups of the ribose moiety engaged two contacts with the backbone carbonyl atoms of Leu126 and Val155 (Figure 4C). The 1′-nitrile group was accommodated in the space adjacent to loop β7-α6. With a distance of ∼3.3 Å, direct contacts to the backbone amine atoms of Phe156 and Asp157 might be weak, yet charge compatibility with this binding pocket was provided by the negative electrostatic potential of the nitrile group.
Figure 4.
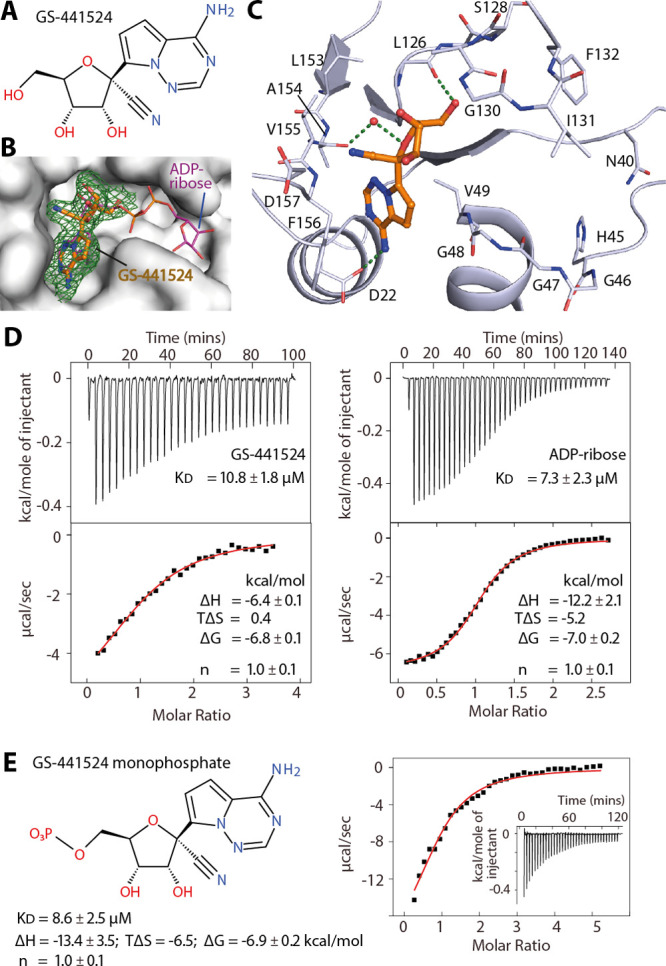
Interaction of the remdesivir metabolite GS-441524 with the SARS-CoV-2 macrodomain. (A) Chemical structure of GS-441524. (B) Surface representation revealing that GS-441524 occupied the binding site of the ADP-ribose adenosine moiety (PDB ID 7BF6). The green mesh represents the |FO| – |FC| omitted electron density map contoured at 3σ for the bound ligand. (C) Detailed interactions between the ligand and the protein. (D) ITC binding isotherm (top) and integrated heat of binding (bottom) for the interactions between either GS-441524 or ADP-ribose and the protein averaged from duplicates. (E) Chemical structure of GS-441524 monophosphate and its ITC binding parameters from a duplicate experiment for the macrodomain.
In comparison with the adenosine substrate, GS-441524 showed improved shape complementarity with the binding pocket (Figures 3A and 4C). To assess this, we performed isothermal titration calorimetry (ITC) to determine the affinities of this ligand, adenosine, AMP, and ADP-ribose. Unfortunately, adenosine and AMP did not yield interpretable ITC binding isotherms. Nonetheless, we observed that the KD values of GS-441524 and ADP-ribose were remarkably comparable (10.8 and 7.3 μM, respectively). This was a surprising result considering that the two phosphate moieties and the terminal ribose are missing in GS-441524. Thus we synthesized the 5′-monophosphate derivative of this compound (GS-441524 monophosphate) and characterized the binding. Interestingly, an installation of 5′-phosphate led to a slightly improved affinity with a similar binding strength compared with ADP-ribose (KD of 8.6 μM), suggesting a contribution of the phosphate group for binding. Nevertheless, the different thermodynamics of the three ligands were of particular note. The presence of the phosphate group in GS-441524 monophosphate and ADP-ribose as well as the additional terminal ribose-1′-phosphate moiety in the latter resulted in large negative binding enthalpy changes, which were counteracted by highly unfavorable entropy changes (TΔS) that were likely attributable to the constrained ligand and potentially the pocket in the bound state.
In conclusion, targeting the MD offers an attractive target for the development of antiviral agents against SARS-CoV-2 and other viruses. To assist the inhibitor discovery efforts, we report a collective set of crystal structures, extending our structural knowledge of this protein. A structural comparison demonstrated that the ADP-ribose binding site, essentially the pocket for ribose-1-phosphate, of the MD possessed high structural plasticity. This was demonstrated by its adaptability to diverse naturally occurring nucleosides and nucleotides, including ADP-ribose-phosphate (ADPRP). This flexibility offers opportunities for the rational targeting of this protein module by small-molecule inhibitors. Indeed, a recent study has reported a number of potential small-molecule binders discovered through crystallographic fragment screening.31 In line with this, our unprecedented discovery of the binding of GS-441524, an active metabolite of remdesivir, supported this hypothesis that the ADP-ribose pocket of the MD indeed represents a druggable site. The low micromolar affinity of GS-441524 and its small molecular weight offer a versatile starting point for ligand design.
Experimental Section
Protein Production
The DNA encoding the SARS-CoV-2 MD was commercially synthesized and subcloned into a pET-28a(+) vector (Genscript, Supplementary Table S1). The expression of the recombinant protein harboring N-terminal His6 tags was carried out in E. coli Rosetta, which was cultured in TB media. The culture was grown at 37 °C until an OD600 of 1.6 was reached prior to cooling to 18 °C. At an OD600 of 2.6 to 2.8, the protein expression was induced by adding 0.5 μM IPTG, and the expression was continued overnight. Cells were harvested by centrifugation and resuspended in buffer containing 50 mM Tris, pH 8.0, 500 mM NaCl, 10 mM imidazole pH 7, 5% glycerol, and 1 mM TCEP. Lysis was performed by sonication, and the supernatant was clarified by centrifugation. The recombinant protein was initially purified using Ni2+-affinity chromatography, and subsequent cleavage of the histidine tag was performed by TEV protease treatment. The cleaved protein was passed through a Ni2+-NTA column and was further purified by size exclusion chromatography using a Superdex S75 column. The pure protein was stored at −80 °C in 25 mM Tris pH 8.0, 150 mM NaCl, and 5% glycerol buffer.
Crystallization and Structure Determination
The MD was buffer-exchanged into 25 mM Tris pH 8.0 and 150 mM NaCl and concentrated to ∼8.5–10 mg/mL. Crystallization was performed using the sitting drop vapor diffusion method at 20 °C, and the conditions are listed in Supplementary Table S2. Microseeding was employed in all crystallization experiments using the initial crystals of the MD, which were stored in 30% broad-molecular-weight PEG smears,32 0.1 M MgCl2, 0.1 M tris pH 7.0. For cocrystallization, the protein was mixed with a 10-fold molar excess of the ligands. In the case of ADP-ribose, soaking was performed overnight using the ligand at 10 mM concentration. Viable crystals were cryoprotected with mother liquor supplemented with 20% ethylene glycol prior to flash cooling in liquid nitrogen.
Diffraction data were collected at the Swiss Light Source and were processed and scaled with XDS33 and aimless,34 respectively. The initial structure solution was obtained with the molecular replacement method using Phaser35 and the coordinate of the MD (PDB ID: 6WEN). Manual model rebuilding alternated with refinement was performed in COOT36 and REFMAC5.37 The geometry of the final models was verified using MOLPROBITY.38 Omitted electron density maps for the bound ligands are shown in Supplementary Figure S1. The summary of the data collection and refinement statistics are in Supplementary Table S2.
Synthesis of GS-441524 Monophosphate
The chemical synthetic procedure is described in the Supporting Information.
Isothermal Calorimetry
The ITC experiment was performed using a NanoITC instrument (TA Instrument) at 25 °C in the buffer containing 20 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM TCEP, and 5% glycerol. For ADP-ribose and GS-441524 monophosphate, the ligand at 500 μM in the syringe was titrated into the reaction cell containing the protein at 35−106 μM, whereas for GS-441524, the protein at 250 μM was titrated into the reaction cell containing the compound at 22 μM. Data analyses were performed with NanoAnalyze software (TA Instrument) using an independent binding model from which the affinities and thermodynamic parameters were calculated.
Glossary
Abbreviations
- nsP3
nonstructural protein 3
- MD
macrodomain
- ADP-ribose
adenosine diphosphate ribose
- AMP
adenosine monophosphate
- GMP
guanosine monophosphate
- NAD
nicotinamide adenine dinucleotide
- NADP
nicotinamide adenine dinucleotide phosphate
- ADPRP
adenosine diphosphate ribose phosphate
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.0c00684.
Supplementary figure and table for the electron density map for the bound ligands, protein sequence, crystallographic data collection, and refinement statistics and chemical synthesis (PDF)
Author Contributions
⊥ X.N. and M.S. contributed equally. All authors have given approval to the final version of the manuscript.
We acknowledge the funding support from Goethe University Frankfurt under the Goethe-Corona-Fonds program and the SGC, a registered charity (no. 1097737) that receives funds from AbbVie, Bayer AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genentech, Genome Canada through Ontario Genomics Institute (OGI-196), EU/EFPIA/OICR/McGill/KTH/Diamond, Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant 875510), Janssen, Merck KGaA (EMD in Canada and U.S.), Merck & Co (MSD outside Canada and U.S.), Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome (106169/ZZ14/Z). This work was also supported by the Fraunhofer Internal Programs under Grant No. Anti-Corona 164-600014. E.P. thanks Deutsche Forschungsgemeinschaft (DFG, Heisenberg-Professur PR1405/7-1) for financial support.
The authors declare no competing financial interest.
This article is made available via the ACS COVID-19 subset for unrestricted RESEARCH re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for the duration of the World Health Organization (WHO) declaration of COVID-19 as a global pandemic.
Supplementary Material
References
- Wu F.; Zhao S.; Yu B.; Chen Y. M.; Wang W.; Song Z. G.; Hu Y.; Tao Z. W.; Tian J. H.; Pei Y. Y.; Yuan M. L.; Zhang Y. L.; Dai F. H.; Liu Y.; Wang Q. M.; Zheng J. J.; Xu L.; Holmes E. C.; Zhang Y. Z. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579 (7798), 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol 2020, 5 (4), 536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.; Kok K. H.; Zhu Z.; Chu H.; To K. K.; Yuan S.; Yuen K. Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes Infect. 2020, 9 (1), 221–236. 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 2004, 303 (5664), 1666–1669. 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Gordon D. E.; Jang G. M.; Bouhaddou M.; Xu J.; Obernier K.; White K. M.; O’Meara M. J.; Rezelj V. V.; Guo J. Z.; Swaney D. L.; Tummino T. A.; Huttenhain R.; Kaake R. M.; Richards A. L.; Tutuncuoglu B.; Foussard H.; Batra J.; Haas K.; Modak M.; Kim M.; Haas P.; Polacco B. J.; Braberg H.; Fabius J. M.; Eckhardt M.; Soucheray M.; Bennett M. J.; Cakir M.; McGregor M. J.; Li Q.; Meyer B.; Roesch F.; Vallet T.; Mac Kain A.; Miorin L.; Moreno E.; Naing Z. Z. C.; Zhou Y.; Peng S.; Shi Y.; Zhang Z.; Shen W.; Kirby I. T.; Melnyk J. E.; Chorba J. S.; Lou K.; Dai S. A.; Barrio-Hernandez I.; Memon D.; Hernandez-Armenta C.; Lyu J.; Mathy C. J. P.; Perica T.; Pilla K. B.; Ganesan S. J.; Saltzberg D. J.; Rakesh R.; Liu X.; Rosenthal S. B.; Calviello L.; Venkataramanan S.; Liboy-Lugo J.; Lin Y.; Huang X. P.; Liu Y.; Wankowicz S. A.; Bohn M.; Safari M.; Ugur F. S.; Koh C.; Savar N. S.; Tran Q. D.; Shengjuler D.; Fletcher S. J.; O’Neal M. C.; Cai Y.; Chang J. C. J.; Broadhurst D. J.; Klippsten S.; Sharp P. P.; Wenzell N. A.; Kuzuoglu-Ozturk D.; Wang H. Y.; Trenker R.; Young J. M.; Cavero D. A.; Hiatt J.; Roth T. L.; Rathore U.; Subramanian A.; Noack J.; Hubert M.; Stroud R. M.; Frankel A. D.; Rosenberg O. S.; Verba K. A.; Agard D. A.; Ott M.; Emerman M.; Jura N.; von Zastrow M.; Verdin E.; Ashworth A.; Schwartz O.; d’Enfert C.; Mukherjee S.; Jacobson M.; Malik H. S.; Fujimori D. G.; Ideker T.; Craik C. S.; Floor S. N.; Fraser J. S.; Gross J. D.; Sali A.; Roth B. L.; Ruggero D.; Taunton J.; Kortemme T.; Beltrao P.; Vignuzzi M.; Garcia-Sastre A.; Shokat K. M.; Shoichet B. K.; Krogan N. J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583 (7816), 459–468. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J.; Kusov Y.; Hilgenfeld R. Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018, 149, 58–74. 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras G. I.; Kustatscher G.; Buhecha H. R.; Allen M. D.; Pugieux C.; Sait F.; Bycroft M.; Ladurner A. G. The macro domain is an ADP-ribose binding module. EMBO J. 2005, 24 (11), 1911–1920. 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack J. G.; Perina D.; Ahel I. Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu. Rev. Biochem. 2016, 85, 431–454. 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]
- Alhammad Y. M. O.; Fehr A. R. The Viral Macrodomain Counters Host Antiviral ADP-Ribosylation. Viruses 2020, 12 (4), 384. 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Singh S. A.; Kerr C. M.; Mukai S.; Higashi H.; Aikawa M. The impact of PARPs and ADP-ribosylation on inflammation and host-pathogen interactions. Genes Dev. 2020, 34 (5–6), 341–359. 10.1101/gad.334425.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B.; Butepage M.; Eckei L.; Krieg S.; Verheugd P.; Shilton B. H. ADP-Ribosylation, a Multifaceted Posttranslational Modification Involved in the Control of Cell Physiology in Health and Disease. Chem. Rev. 2018, 118 (3), 1092–1136. 10.1021/acs.chemrev.7b00122. [DOI] [PubMed] [Google Scholar]
- Li C.; Debing Y.; Jankevicius G.; Neyts J.; Ahel I.; Coutard B.; Canard B. Viral Macro Domains Reverse Protein ADP-Ribosylation. J. Virol. 2016, 90 (19), 8478–8486. 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckei L.; Krieg S.; Butepage M.; Lehmann A.; Gross A.; Lippok B.; Grimm A. R.; Kummerer B. M.; Rossetti G.; Luscher B.; Verheugd P. The conserved macrodomains of the non-structural proteins of Chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci. Rep. 2017, 7, 41746. 10.1038/srep41746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnur D.; Bartlett E.; Mikolcevic P.; Kirby I. T.; Rack J. G. M.; Mikoc A.; Cohen M. S.; Ahel I. Reversible ADP-ribosylation of RNA. Nucleic Acids Res. 2019, 47 (11), 5658–5669. 10.1093/nar/gkz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack J. G. M.; Zorzini V.; Zhu Z.; Schuller M.; Ahel D.; Ahel I. Viral macrodomains: a structural and evolutionary assessment of the pharmacological potential. Open Biol. 2020, 10 (11), 200237. 10.1098/rsob.200237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. H.; Chang S. C.; Chiu Y. C.; Jiang B. C.; Wu T. H.; Hsu C. H. Structural, Biophysical, and Biochemical Elucidation of the SARS-CoV-2 Nonstructural Protein 3 Macro Domain. ACS Infect. Dis. 2020, 6 (11), 2970–2978. 10.1021/acsinfecdis.0c00441. [DOI] [PubMed] [Google Scholar]
- Putics A.; Filipowicz W.; Hall J.; Gorbalenya A. E.; Ziebuhr J. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005, 79 (20), 12721–31. 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Jankevicius G.; Ahel I.; Perlman S. Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis. Trends Microbiol. 2018, 26 (7), 598–610. 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri T.; Eriksson K. K.; Putics A.; Zust R.; Snijder E. J.; Davidson A. D.; Siddell S. G.; Thiel V.; Ziebuhr J.; Weber F. The ADP-ribose-1”-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J. Gen. Virol. 2011, 92 (Pt 8), 1899–1905. 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- Michalska K.; Kim Y.; Jedrzejczak R.; Maltseva N. I.; Stols L.; Endres M.; Joachimiak A. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ 2020, 7 (5), 814–824. 10.1107/S2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad Y. M. O.; Kashipathy M. M.; Roy A.; Gagne J. P.; McDonald P.; Gao P.; Nonfoux L.; Battaile K. P.; Johnson D. K.; Holmstrom E. D.; Poirier G. G.; Lovell S.; Fehr A. R. The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase. J. Virol. 2021, 95 (3), e01969-20. 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick D. N.; Virdi R. S.; Vuksanovic N.; Dahal N.; Silvaggi N. R. Molecular Basis for ADP-Ribose Binding to the Mac1 Domain of SARS-CoV-2 nsp3. Biochemistry 2020, 59 (28), 2608–2615. 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K. S.; Joseph J. S.; Subramanian V.; Clayton T.; Griffith M.; Moy K.; Velasquez J.; Neuman B. W.; Buchmeier M. J.; Stevens R. C.; Kuhn P. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1”-phosphate dephosphorylation by a conserved domain of nsP3. Structure 2005, 13 (11), 1665–1675. 10.1016/j.str.2005.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I.; Pervaiz N.; Khan A.; Saleem S.; Shireen H.; Wei D. Q.; Labrie V.; Bao Y.; Abbasi A. A. Evolutionary and structural analysis of SARS-CoV-2 specific evasion of host immunity. Genes Immun. 2020, 21 (6–8), 409–419. 10.1038/s41435-020-00120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. A.; Kato J.; Kasamatsu A.; Oda H.; Lee D. Y.; Moss J. The ARH and Macrodomain Families of alpha-ADP-ribose-acceptor Hydrolases Catalyze alpha-NAD(+) Hydrolysis. ACS Chem. Biol. 2019, 14 (12), 2576–2584. 10.1021/acschembio.9b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R. T.; Roth J. S.; Brimacombe K. R.; Simeonov A.; Shen M.; Patnaik S.; Hall M. D. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent. Sci. 2020, 6 (5), 672–683. 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J. H.; Tomashek K. M.; Dodd L. E.; Mehta A. K.; Zingman B. S.; Kalil A. C.; Hohmann E.; Chu H. Y.; Luetkemeyer A.; Kline S.; Lopez de Castilla D.; Finberg R. W.; Dierberg K.; Tapson V.; Hsieh L.; Patterson T. F.; Paredes R.; Sweeney D. A.; Short W. R.; Touloumi G.; Lye D. C.; Ohmagari N.; Oh M. D.; Ruiz-Palacios G. M.; Benfield T.; Fatkenheuer G.; Kortepeter M. G.; Atmar R. L.; Creech C. B.; Lundgren J.; Babiker A. G.; Pett S.; Neaton J. D.; Burgess T. H.; Bonnett T.; Green M.; Makowski M.; Osinusi A.; Nayak S.; Lane H. C. Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 2020, 383 (19), 1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T. K.; Jordan R.; Lo M. K.; Ray A. S.; Mackman R. L.; Soloveva V.; Siegel D.; Perron M.; Bannister R.; Hui H. C.; Larson N.; Strickley R.; Wells J.; Stuthman K. S.; Van Tongeren S. A.; Garza N. L.; Donnelly G.; Shurtleff A. C.; Retterer C. J.; Gharaibeh D.; Zamani R.; Kenny T.; Eaton B. P.; Grimes E.; Welch L. S.; Gomba L.; Wilhelmsen C. L.; Nichols D. K.; Nuss J. E.; Nagle E. R.; Kugelman J. R.; Palacios G.; Doerffler E.; Neville S.; Carra E.; Clarke M. O.; Zhang L.; Lew W.; Ross B.; Wang Q.; Chun K.; Wolfe L.; Babusis D.; Park Y.; Stray K. M.; Trancheva I.; Feng J. Y.; Barauskas O.; Xu Y.; Wong P.; Braun M. R.; Flint M.; McMullan L. K.; Chen S. S.; Fearns R.; Swaminathan S.; Mayers D. L.; Spiropoulou C. F.; Lee W. A.; Nichol S. T.; Cihlar T.; Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531 (7594), 381–385. 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Leist S. R.; Schafer A.; Won J.; Brown A. J.; Montgomery S. A.; Hogg A.; Babusis D.; Clarke M. O.; Spahn J. E.; Bauer L.; Sellers S.; Porter D.; Feng J. Y.; Cihlar T.; Jordan R.; Denison M. R.; Baric R. S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11 (1), 222. 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Cao L.; Li G.; Cong F.; Li Y.; Sun J.; Luo Y.; Chen G.; Li G.; Wang P.; Xing F.; Ji Y.; Zhao J.; Zhang Y.; Guo D.; Zhang X. Remdesivir Metabolite GS-441524 Effectively Inhibits SARS-CoV-2 Infection in Mouse Models. J. Med. Chem. 2021, 10.1021/acs.jmedchem.0c01929. [DOI] [PubMed] [Google Scholar]
- Schuller M.; Correy G. J.; Gahbauer S.; Fearon D.; Wu T.; Díaz R. E.; Young I. D.; Martins L. C.; Smith D. H.; Schulze-Gahmen U.; Owens T. W.; Deshpande I.; Merz G. E.; Thwin A. C.; Biel J. T.; Peters J. K.; Moritz M.; Herrera N.; Kratochvil H. T.; Aimon A.; Bennett J. M.; Neto J. B.; Cohen A. E.; Dias A.; Douangamath A.; Dunnett L.; Fedorov O.; Ferla M. P.; Fuchs M.; Gorrie-Stone T. J.; Holton J. M.; Johnson M. G.; Krojer T.; Meigs G.; Powell A. J.; Rack J. G. M.; Rangel V. L.; Russi S.; Skyner R. E.; Smith C. A.; Soares A. S.; Wierman J. L.; Zhu K.; Jura N.; Ashworth A.; Irwin J.; Thompson M. C.; Gestwicki J. E.; von Delft F.; Shoichet B. K.; Fraser J. S.; Ahel I. Fragment Binding to the Nsp3Macrodomain of SARS-CoV-2 Identified Through Crystallographic Screening and Computational Docking. bioRxiv 2020, 2020.11.24.393405. 10.1101/2020.11.24.393405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikuad A.; Knapp S.; von Delft F. Defined PEG smears as an alternative approach to enhance the search for crystallization conditions and crystal-quality improvement in reduced screens. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2015, 71 (Pt 8), 1627–1639. 10.1107/S1399004715007968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66 (2), 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. R.; Murshudov G. N. How good are my data and what is the resolution?. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69 (7), 1204–1214. 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40 (4), 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66 (4), 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalevskiy O.; Nicholls R. A.; Long F.; Carlon A.; Murshudov G. N. Overview of refinement procedures within REFMAC5: utilizing data from different sources. Acta Crystallogr. D Struct Biol. 2018, 74 (3), 215–227. 10.1107/S2059798318000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. J.; Headd J. J.; Moriarty N. W.; Prisant M. G.; Videau L. L.; Deis L. N.; Verma V.; Keedy D. A.; Hintze B. J.; Chen V. B.; Jain S.; Lewis S. M.; Arendall W. B. 3rd; Snoeyink J.; Adams P. D.; Lovell S. C.; Richardson J. S.; Richardson D. C. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27 (1), 293–315. 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.