1. Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and one of the most common diseases diagnosed in the USA among both men and women [1]. Interestingly, the incidence and mortality rates have been declining in the United States over the past decades for those aged over 50, with the pace accelerating to 3% annually from 2003 to 2012. However, there has been an alarming increase (51%) in colorectal cancer incidence in those under 50 since 1994 [2–4]. The reasons for this increase are not fully understood. Known risk factors, such as obesity, alcohol, smoking and a sedentary lifestyle, are likely to underlie early-onset cancers [3].
Genetic predisposition and epigenetic alterations are likely to contribute to this increase. Both play an important role in the disruption of several oncogenic signaling pathways which have been identified in colorectal cancer. Numerous reports have highlighted the importance of early mutations in the adenomatous polyposis coli (APC) tumor suppressor gene and in the CTNNB1 (β-catenin) gene; both of these genes influence the Wnt/β pathway signaling by preventing axin-dependent phosphorylation and degradation of β-catenin [5–8]. In addition to these mutations, a synergistic interaction between the genetic and epigenetic alterations is believed to contribute in the development of CRC [8]. Methylations, such as H3K9 and H3K27, have been found to contribute to the regulation of gene expression in both embryonic stem cells and adult stem cells by silencing genes. On the other hand, H3K4 methylation is critical for gene activation [9]. Another mechanism, which is the subject of intensive research, is the one activated by aldehyde dehydrogenase (ALDH) and that promotes tumor growth. ALDH catalytic activity has been identified as a biomarker of many cancers and cancer stem cells [10]. Our group has recently shown that high ALDH1B1 expression was observed throughout the cells of human colon adenocarcinomas, suggesting a close association between ALDH1B1 presence and activation of Wnt/β-catenin [11, 12]. However, other groups have suggested the ALDH1A1 isozyme to be present in CRC [13, 14]. However, the mechanism by which ALDH1A1 contributes to CRC has not been elucidated.
Athough important signaling pathways and genes have been identified as playing a role in CRC, identification of new prognostic biomarkers continues to be a challenge, because it is a highly heterogeneous cancer, with various molecular alterations taking place throughout the natural course of the disease [15]. Evaluation of the transcriptome allows investigation of alterations in the molecular constituents of the cells and tissues induced by disease. RNA-Seq is one approach that uses deep-sequencing technologies to measure the levels of transcripts and their isoforms, thereby providing a detailed insight into the transcriptome [16]. By identify cellular proteins, proteomics provides additional insights into cellular changes that are downstream of the transcriptome. Metabolite profiling (or metabolomics) provides information about the biochemical consequences of changes in protein expression. This is a powerful approach that measures and identifies small molecules that change during a disease, providing valuable insights into how biochemistry is linked to cell metabolism [17]. Each one of these approaches contributes in a different way in advancing to our understanding of the molecular basis and cellular changes occurring in diseases, such as CRC. They also facilitate the discovery novel biomarkers. For example, transcriptomics can identify the differences in gene expression profiles in key oncogenes and tumor suppressors CRC cells. Proteomics can provide insight into changes in proteins involved in molecular pathways contributing to the development and progression of CRC. Metabolomics can elucidate the biochemical pathways that are altered in the tumor tissue. Each of these approaches enhance our understanding of CRC biology. Recent advances in biocomputational tools make it feasible to (i) analyze large data sets from transcriptomic, proteomic and metabolomic studies, (ii) efficiently integrate them, and (iii) generate a comprehensive systems biology analysis of the disease [18–20]. The use of a multilayer “omics” approach that integrates transcriptomics, proteomics and metabolomics has the potential to greatly increase our understanding of the mechanisms underlying disease. The changes in the metabolome can be linked to alterations in enzymes identified with proteomics, and when combined with transcriptomics data, can enhance the interpretation of the genetic background of thedisease, leading to a more comprehensive molecular view of CRC. The end result of the multi “omics’” approaches to a disease should improve prognosis, and ultimately lead to improved treatment and patient outcomes.
Using a multi-omics approach, we have investigated the impact of genetic suppression (shRNA) of ALDH1A1 expression on transcriptomics, proteomics and untargeted metabolomics analyses in a human colon cancer cell line (COLO320) which has a high constitutive expression of ALDH1A1. The present study (i) generates an integrative omic profile of scramble shRNA vs. ALDH1A1 shRNA COLO320 cells, and (ii) identifies possible alterations in biological pathways caused by suppression of ALDH1A1 expression.
2. Materials and Methods
2.1. Culture of COLO320 cells and puromycin selection
COLO320 cells (American Type Culture Collection; ATCC, VA, USA) were cultured in RPMI 1640 (Life Technologies, Carlsbad, CA) medium supplemented with an antibiotic-antimycotic cocktail (100 U/mL penicillin and 100 μg/mL streptomycin; Life Technologies, Carlsbad, CA) and 10% heat-inactivated fetal bovine serum (FBS; Life Technologies, Carlsbad, CA) under a humidified atmosphere of 5% CO2 in air at 37°C. Cells ( 0.5–1.0×106) were seeded into 25 cm2 culture flasks (Eppendorf AG, Hamburg, Germany) and passaged at 70–80% confluency. To assess the effect of puromycin on cell viability, 0.02–0.1 × 105 cells were seeded in RPMI containing 10% FBS (100 μL/well) in a 96-well cell culture plate (Eppendorf AG, Hamburg, Germany). All cultures were performed in triplicate. After 12 h, the medium was replaced with RPMI 1640 medium with 10% FBS that contained 0–20 μg/mL puromycin (Life Technologies, Carlsbad, CA). Cells were then allowed to incubate for 48 h at 37°C before cell viability was determined using the CellTiter 96® AQueous One Solution Reagent assay (Promega, Madison, WI) per the manufacturer’s protocol.
Cells were passaged using 0.25% trypsin-EDTA followed by counting (ViaStain AOPI Staining Solution,cridine orange/propidium iodide Nexcelom Bioscience, Lawrence, MA) using an automated cellometer (Cellometer K2, Nexcelom Bioscience, Lawrence, MA) and then harvested at 90% confluency.
2.1.1. ALDH1A1 shRNA knockdown
Cellular ALDH1A1 expression was suppressed using MISSION shRNA lentiviral transduction particles containing validated ALDH1A1 shRNA in a pLKO plasmid vector. Scramble control shRNA in pLKO plasmid vector was used to generate control cells (Sigma-Aldrich, St. Louis, MO). The shRNA transfection was carried out as described previously [21] and five individual stable clones were selected using puromycin (10 μg/mL). Western blot analysis was used to verify changes in ALDH1A1 expression using methods described previously [22]. The clone that had the greatest suppression of ALDH1A1 expression was used in the present study.
2.2. Unified sample preparation for RNAseq and proteomic analyses
Knock-down and scramble shRNA-transfected cells were seeded (1×106) into six 150 mm dishes (Corning Inc., Corning, NY, USA) and cultured in RPMI containing 10% heat-inactivated FBS and puromycin (10 μg/mL). Cells were continuously grown in the presence of puromycin until 90% confluency, and then harvested using 5 mL of 0.25% trypsin EDTA (Invitrogen, Thermo Fisher Scientific) for analyses (as described below). The cells were centrifuged at 2,000 rpm for 10 min at room temperature, the supernatant was discarded and the cell pellet was collected. The cell pellet was further rinsed and processed for RNA-seq and proteomics analyses, as described in the following sections, respectively.
2.2.1. RNA-seq analysis
The cell pellet (n=3 for each group) was resuspended in ice-cold Dulbecco’s phosphate-buffered saline (DPBS) and centrifuged at 10,000 rpm for 15 min at 4°C and the supernatant was discarded. This step was repeated three times. Then 600 μL RLT Plus buffer supplemented with 1% β-mercaptoethanol was then added, and the resultant suspension was stored in −80°C. The samples were submitted to the Yale Center for Genome Analysis for RNA-seq analysis.
RNA Seq Library Prep:
mRNA (≈500 ng total) was purified with oligo-dT beads and sheared by incubation at 94°C (first-strand synthesis with random primers). Then, the second strand synthesis was performed using dUTP to generate strand-specific sequencing libraries. The cDNA library was then end-repaired, A-tailed adapters were ligated, and second-strand digestion was performed using uracil-DNA-glycosylase. Indexed libraries (that met appropriate cut-offs for both) were quantified by qRT-PCR using a commercially-available kit (KAPA Biosystems), and insert size distribution was determined using a LabChip GX or Agilent Bioanalyzer. Samples with a yield ≥0.5 ng/μL were used for sequencing.
Flow cell preparation and sequencing:
Sample concentrations were normalized to 10 nM and loaded onto Illumina Rapid or High-output flow cells at a concentration that yielded 150–250 million passing filter clusters per lane. Samples were sequenced using 75 bp single or paired-end sequencing on an Illumina HiSeq 2000 or 2500 according to manufacturer’s protocols. The 6 bp index was read during an additional sequencing read that automatically followed the completion of read 1. Data generated during sequencing runs were simultaneously transferred to the YCGA high-performance computing cluster. A positive control (prepared bacteriophage Phi X library) provided by Illumina was spiked into every lane at a concentration of 0.3% to monitor sequencing quality in real time.
Data analysis and storage:
Signal intensities were converted to individual base calls during a run using the system’s Real Time Analysis (RTA) software. Primary analysis sample de-multiplexing and alignment to the human genome was performed using Illumina’s CASAVA 1.8.2 software suite. The reads were trimmed for quality and aligned with the hg19 reference genome using TopHat2 [23]. The transcripts were assembled using Cufflinks [24]. The assembled transcripts were used to estimate transcript abundance and differential gene-expression using Cuffdiff [24]. The results were visualized using R (CRAN) and CummeRbund [25].
2.2.2. Proteomic analysis
Sample preparation:
The cell pellet was resuspended in ice-cold Dulbecco’s phosphate-buffered saline (DPBS) which was supplemented with complete protease and phosphatase inhibitor cocktails (Roche Diagnostics GmbH, Mannheim, Germany), centrifuged at 10,000 rpm for 15 min at 4°C and the supernatant was discarded. This step was repeated three times. The resultant sample was stored at −80°C and submitted to the Yale Mass Spectrometry (MS) & Proteomics Resource of the W.M. Keck Foundation Biotechnology Resource Laboratory for analysis.
The samples (n=3 for each group) were subjected to three 15 sec bursts of sonication on ice using a Digital Sonifier (Model 450, Branson Ultrasonics Corportation, Danbury, CT). The sonicate was centrifuged at 16,000g for 10 min at 4°C. One hundred μL of the supernatant was added to chloroform:methanol:water (at ratio of 1:4:3 %v/v, respectively) to precipitate proteins, vortexed, and subsequently subjected to centrifugation at 16,000g for 1 min. The top aqueous layer of the supernatant was removed and discarded (with caution not to disturb the formed meniscus) and an additional 400 μL methanol was added to the remaining mixture, and vortexed well. The resultant mixture was centrifuged at 16,000g for 2 min, and the methanol was removed (with caution not to disturb the pellet) and discarded. The protein pellet was dried using a SpeedVac (SPD111V Savant, Fisher Thermo Scientific), and resuspended in 25 μL Rapigest SF surfactant solution (Waters Corporation, Milford, MA) containing 50 mM NH4HCO3 reduced by addition of 5 μL dithiothreitol (45 mM), and subsequently alkylated by the addition of 5 μL iodoacetamide (100 mM). The resultant sample underwent enzymatic digestion by incubation with LysC (at 1:10 enzyme:protein ratio) and trypsin (at 1:10 enzyme:protein ratio) at 37°C overnight. This digestion was quenched with 0.1% formic acid, and subsequently desalted with a C18 UltraMicroSpin columns. The effluents from the de-salting step were dried and then re-dissolved in a mixture of 5 μL 70% formic acid (FA) and 35 μL 0.1% trifluoroacetic acid (TFA). A 2 μL aliquot was reserved to obtain total digested protein amount and 3.2 μL of Pierce Retention Time Calibration Mixture (Cat#88321) was added to each sample resulting in 80 μL total sample volume (containing 0.08 μg/μL sample peptide concentration) prior to injecting 5 μL sample volume on the UPLC Orbitrap Fusion mass spectrometer for normalization of Label-Free Quantitation (LFQ) data.
LC-MS/MS conditions:
Label-Free Quantitation (LFQ) was performed on a Thermo Scientific Orbitrap Fusion mass spectrometer connected to a Waters nano ACQUITY UPLC system equipped with a Waters Symmetry® C18 180 μm × 20 mm trap column and a 1.7-μm, 75 μm × 250 mm nano ACQUITY UPLC column (maintained at 35°C). The digested protein samples were diluted to a final concentration of 0.05 μg protein/μL in 0.1% TFA. Five μL of these solutions were injected in triplicate into the LC MS/MS system in block randomized order. To ensure a high level of identification and quantitation integrity, a resolution of 120,000 and 60,000 was utilized for MS and MS/MS scans, respectively. Trapping was carried out for 3 min at 5 μL/min in 97% Buffer A (0.1% FA in water) and 3% Buffer B (0.075% FA in acetonitrile) prior to eluting with linear gradients that reached 6% B at 5 min, 35% B at 170 min, and 50% B at 175 min, and 97% B at 180 min for 5 min; then dropped down to 3% B at 186 min for 14 min.
Data analysis:
The LC-MS/MS data was processed using Progenesis QI Proteomics software (Nonlinear Dynamics, version 2.0), with protein identification carried out using the Mascot search algorithm. The Mascot search results were exported using a significance cutoff of p<0.05 and FDR of 1% into the Progenesis QI software, where search hits were assigned to corresponding peak features that were extracted from the MS data. Only proteins with two or more unique quantifiable peptides were utilized in downstream analyses. These protein abundances and their accession number were then utilized in Ingenuity Pathway Analysis for networking functions of the identified and quantified proteins.
2.3. Metabolomics
Sample preparation:
Briefly, the cell culture medium was removed from the culture dishes, and the cells were rinsed three times with ice-cold DPBS. For quenching and metabolite extraction, 1 mL ice-cold methanol was added to the plate and the cells were immediately removed from the culture dish by scraping. The cell suspension was collected into 2 mL Protein Lobind Eppendorf tubes that were placed on ice. After cell counting (ViaStain AOPI Staining Solution,cridine orange/propidium iodide Nexcelom Bioscience, Lawrence, MA) using an automated cellometer (Cellometer K2, Nexcelom Bioscience, Lawrence, MA), the cell number was adjusted to 4×106 cells/tube. The cells were lysed by the application of a freeze-thaw cycle (freezing in liquid nitrogen for 5 min, thawing in warm water for 5 min), followed by sonication for 2 min. This process was repeated three times. The samples were then centrifuged at 15,000 rpm for 10 min at 4°C, and the supernatant was transferred to 2 mL tubes and dried using a Speed Vac (SPD111V Savant, Thermo Fisher Scientific). The samples were then reconstituted in isopropanol:acetonitrile:water (2:1:1 %v/v) and 2.5 μL Splash® Lipidomix® (Avanti Polar Lipids Inc.) was added to each sample as an internal standard. A previously published method was utilized [26] to study alterations in the lipid metabolome of the two groups. Samples were randomized and analyzed by UPLC-electrospray ionization quadrupole time-of-flight mass spectrometry in positive mode. A pooled sample containing 10 μL of each individual sample was injected every five injections for quality control.
Data analysis:
Raw data were processed using R-XCMS version 3.0.0 [27]. This workflow involves typical data-processing steps including peak detection, peak filtering, and peak alignment. Final peakpicking parameters were: peak width = c(2,30), snthresh = 5, mzdiff = 0.05, ppm = 15; alignment (bw = 10, mzwid = 0.05) and retention time correction (obiwarp, plottype = c(deviation), profstep = 1). Subsequent data processing was carried out in Matlab R2016b using in-house written routines. More specifically, the data was truncated to peaks with less than 30% missing values. Next, the truncated data was normalized using probabilistic quotient normalization, subsequently missing values were imputed using the k-nearest neighbors method (k=5) and, finally, the data was transformed using a generalized log-transformation (glog) [28]. The glog-transformed data was used as an input to principal component analysis (PCA) for exploratory analysis. PCA analysis revealed tight clustering of the quality control samples, but also two potential outliers (one in the knock-down-group and one in the scramble control-group). The QC-samples and two outliers were not taken into account in subsequent analyses. The kNN-imputed data was used as an input to univariate analysis where the Wilcoxon rank sum test in combination with FDR correction ( =5%) was used to detect significant differences between the knock-down and scramble control cells. Focusing on the significant peaks, network analysis using the Mummichog software was performed, which identified pathways that were significantly (Benjamini–Hochberg False Discovery Rate, FDR adjusted p-value < 0.05) impacted in the studied groups. Mummichog annotates metabolites based on accurate mass m/z and tests significant pathway enrichment within a reference network (human metabolic network) using a Fisher’s exact test [29].
2.4. Bioinformatics and Pathway Analysis of the transcriptome and proteome
Qlucore Omics Explorer v 3.2 (Qlucore AB, Lund, Sweden) was used to identify differentially-expressed transcripts and proteins using a two-tailed Student’s unpaired t-test comparison between the knock-down and scramble control groups. The q-values (q< 0.1) were generated based on the FDR method [30]. Unsupervised hierarchical clustering and heat maps were also generated in the Qlucore program. Pathway analysis of differentially-expressed genes across groups was performed by using the Ingenuity Pathway Analysis Knowledge Base (IPA Build 458397M, version 39408507; Ingenuity Systems, QIAGEN). The top canonical pathways involving those differentially-expressed transcripts and proteins were calculated based on the Fisher’s right-tailed exact test (p < 0.05).
2.4.1. Integrated network analysis of the transcriptome, proteome, and metabolome
Differentially abundant transcripts, proteins (Log2 Fold Change ± 0.5, q < 0.1) and the all the metabolites (tentatively annotated by Mummichog software) were analyzed using MetScape (v3.1.3) [31], a pathway-based networks algorithm in Cytoscape (v3.6.1). The metabolic pathways that were associated with transcript-protein-metabolite interactions were mapped onto each of the networks.
2.5. Raw data availability
The proteomics data from this publication are available at the ProteomeXchange Consortium [32] (dataset identifier PXD012230, http://www.proteomeexchange.org/) via the PRIDE partner repository [33]. For the RNA sequencing data, the FASTQ and processed data are available at Gene Expression Omnibus (GSE121043, https://www.ncbi.nlm.nih.gov/geo/). Metabolomics data are available at the NIH Metabolomics workbench (study ID ST0001093, http://www.metabolomicsworkbench.org/).
3. Results
3.1. Transcriptomic and Proteomic Analyses
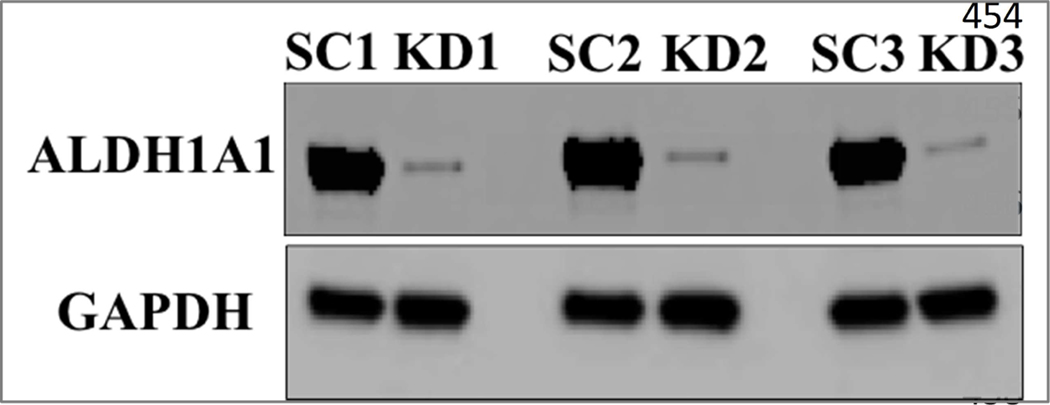
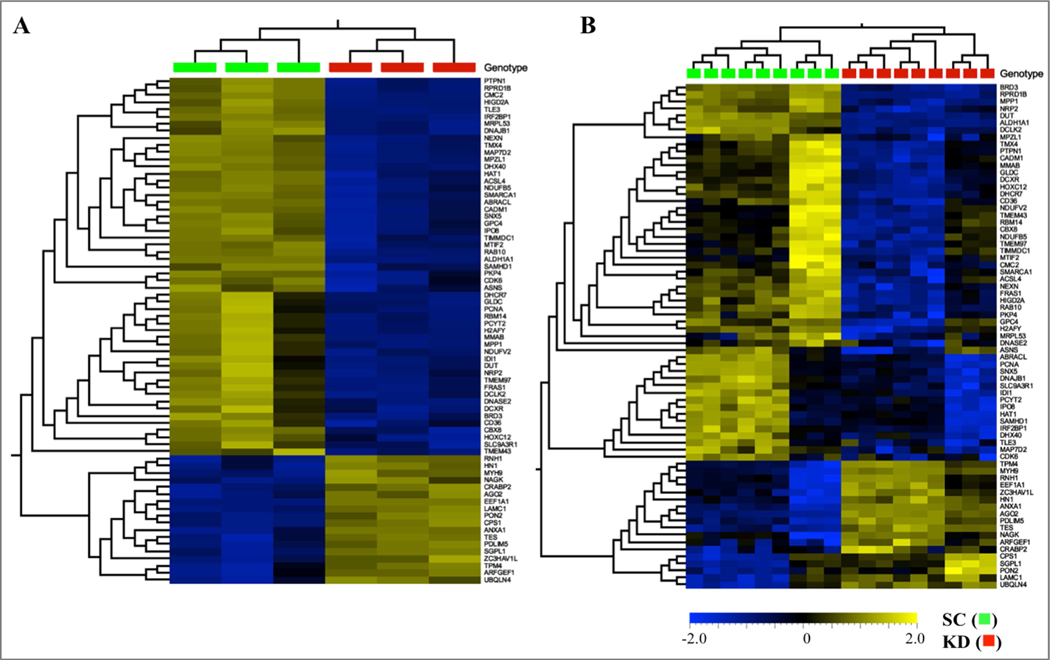
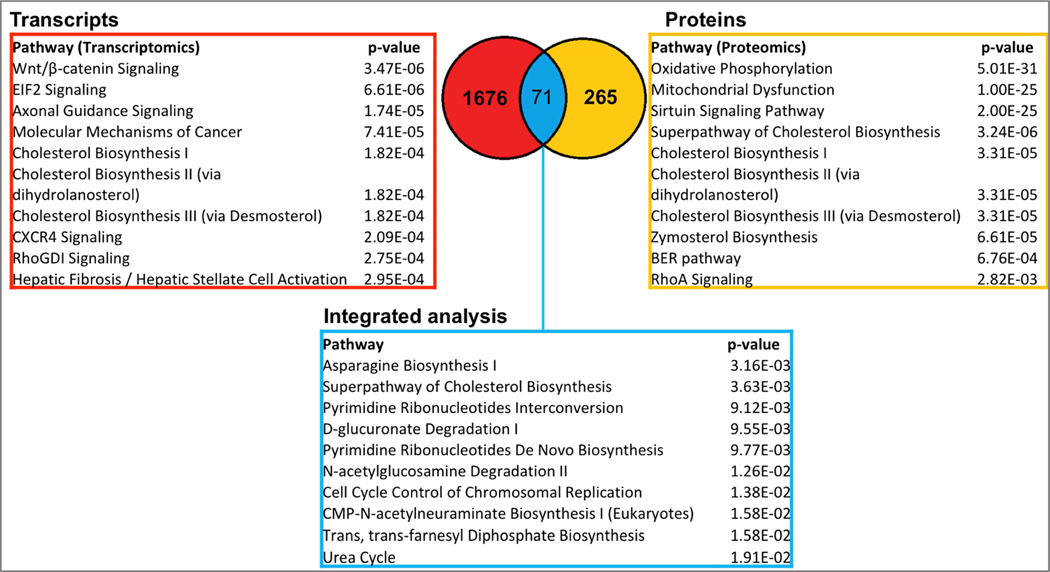
Short hairpin RNA was effective at suppressing expression of ALDH1A1 in COLO320 cells (Fig. 1). RNA-seq and proteomic analyses revealed 1747 differentially-expressed transcripts and 336 differentially-expressed proteins in cells in which ALDH1A1 expression had been suppressed (Fig. 2). Of these, 71 of the differentially-expressed genes related directly to differentially-expressed proteins. Unsupervised hierarchical clustering of the RNA-seq and proteomics data showed that scramble control and ALDH1A1 knock-down samples clustered separately (Fig. 2). Pathway analysis was used to explore the pathways enriched within these differentially-expressed transcripts, proteins and common genes.
Figure 1:
Suppression of ALDH1A1 by shRNA in COLO320 cells. ALDH1A1 and GAPDH expression were determined by Western blot analysis of COLO320 cells that had been transfected with scramble (scramble control, SC) or ALDH1A1 (knock-down, KD) shRNA. Blots are shown for three different samples of the SC or KD clone.
Figure 2:
Hierarchical clustering and heatmaps of 1747 differentially-expressed RNA transcripts (A) and 336 proteins (B) in scramble (scramble control, SC ■) and ALDH1A1 (knock down, KD ■) shRNA-transfected COLO320 cells. Scale bar ranges from −2.0 (light blue) to 2.0 (soft gold) Log2 Fold-Change. RNA transcripts were determined in three separate culture plates of SC and KD cells. Proteomic analyses (B) were determined in triplicate samples obtained from three separate culture plates of SC and KD cells.
In case of transcriptomics results, the most statistically significant pathways were grouped into two categories: those relevant to signaling (such as Wnt/β-catenin (p=3.47E-6, WNT10A and c-MYC genes were down-regulated, with Log2 Fold-Changes=−3.096 and −0.667, q=9.88E-3 and 2.92E-2, respectively)), axonal guidance signaling (1.74E-5), molecular mechanisms of cancer (7.41E-5), and those relevant to lipid metabolism, such as cholesterol biosynthesis I, II and III (p=1.82E-4, for all three pathways) (Fig. 3). Pathway analysis of the differentially-regulated proteins showed oxidative phosphorylation to be the most highly significant pathway (p=5.01E-31), followed by mitochondrial dysfunction (p=1.00E-25) and the sirtuin signaling pathway (p=2.00E-25). For all three pathways, thirty out of the forty proteins that belong to the mitochondrial complex I (NADH:ubiquinone oxidoreductase subunits, NDUF and MT-ND) were all found to be down-regulated (Supplement). In addition, TIMM13 (Log2 Fold-Change=−0.663, q=2.26E-2), PARP1 (Log2 Fold-Change =−0.614, q=1.5E-3), and HIST1H1B (Log2 Fold-Change=−0.734, q=2.89E-3) were all down-regulated in the sirtuin signaling pathway. Furthermore, pathways linked to lipid synthesis, such as the superpathway of cholesterol biosynthesis (p=3.24E-6) and cholesterol biosynthesis (p=3.31E-5) were also significant (Fig. 3).
Figure 3:
Pathway analysis of genes and proteins differentially-expressed in ALDH1A1 knock-down cells. 1747 RNA transcripts (in red) and 336 proteins (in yellow) were mapped to the Ingenuity Knowledge Base. 71 genes and proteins were common between between both lists (blue). Probability (p-values) were calculated using Fisher’s exact test.
Further analysis of the 71 shared differentally-expressed genes and proteins revealed asparagine biosynthesis (p=3.16E-3) as the most enriched pathway. The superpathway of cholesterol biosynthesis was also significant (p=3.36E-3). In both datasets (Supplement), DHCR7 was down-regulated (proteomics: Log2 Fold-Change =−1.488, q=1.55E-4; transcriptomics: Log2 Fold-Change=−1,153, q=0.06), and all of the proteins identified in the proteomics dataset were down-regulated, specifically CYP51A1 (Log2 Fold-Change=−1.465, q=8.54E-5), HMGCS1 (Log2 Fold-Change=−1.396, q=1.55E-5) MSMO1 (Log2 Fold-Change=−1.433, q=2.16E-3), HSD17B7 (Log2 Fold-Change=−0.75, q=3.84E-5), IDI1(Log2 Fold-Change=−1.026, q=6.72E-3) and CYP51A1 (Log2 Fold-Change=−1,026, q=6.72E-3). In addition, pathways, such as pyrimidine ribonucleotides interconversion (p=9.12E-3), N-acetylglucosamine degradation II (p=1.36E-2) and cell cycle control of chromosomal replication (p=1.38E-2) were also significant (Fig. 3).
3.1.2. Metabolomics Analysis
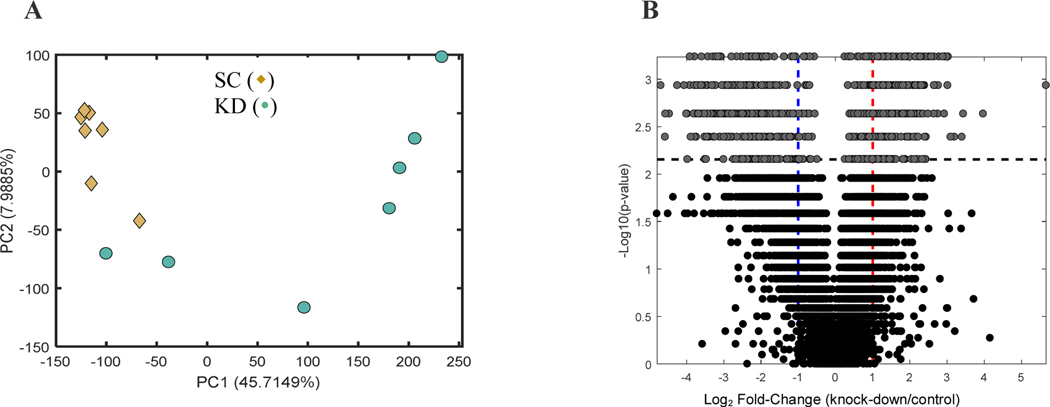
Untargeted metabolomics was performed to determine the variation in metabolic profiles between the scramble control (n=8) vs. ALDH1A1 knock-down cells (n=8). Comprehensive profiles were acquired from the cell extracts and a total of 5,774 features were detected. Unsupervised multivariate analysis was performed for all the samples. Visual inspection of the PCA scores plot revealed two potential outliers: one in the SC group and one in the KO group. This was confirmed by robust PCA (ROBPCA) analysis [34]. Due to the observed skewness in the original PCA scores plot (Supplement), ROBPCA was applied to the glog-transformed data [35]. A scree plot was used to determine how many ROBPCA components should be retained and, subsequently, an outlier detection test was carried out by assessing the score and orthogonal distance of each observation using Hotelling’s T2 and the Q-statistic, respectively. This revealed that the metabolomics profile of the visually-identified outlier in the KO group to be significantly different (p<0.05) from the other KO samples. The outlier in the SC group was not marked as significantly different from the other observations due to its similarity to some of the KO samples (Fig. 4A). Therefore, the ROBPCA analysis was repeated on a reduced data matrix that did not include the KO samples. This resulted in the potential outlier in the SC group being significantly different (p<0.05) from the other SC samples. The principal component analysis that was performed showed separation along the first and the second principal component (Fig. 4A), indicating relevant differences in their basal metabolic activities. There was more variation in the knock-down cells than in the scramble control cells, likely reflective of the variability associated with targeted genetic manipulation by shRNA. Global metabolomics volcano plot analysis revealed 859 ions to be significant (Supplement). The tentative metabolic pathways (shown in Table 1) are mainly linked to lipid metabolism. Of these, the carnitine shuttle pathway (p=5.3E-4) was the most significant pathway. Other pathways linked to energy metabolism, such as pyruvate metabolism and fatty acid oxidation, were also significant (p=1.8E-3 and 2.9E-2, respectively). In addition, both bile acid biosynthesis (p=8.5E-3) and vitamin A (retinol) metabolism (p=2.9E-2) were altered in the ALDH1A1 knock-down cells.
Figure 4:
A) Principal component analysis scores plot of control (scramble shRNA, ◆) and ALDH1A1 (knock-down, ●) COLO320 cells. B) volcano plot (univariate analysis) highlighting 859 significant ions. The horizontal dashed line in the panel shows the FDR-corrected critical p-value (q < 0.05), i.e. all points with p-values smaller tan the critical value (all values above the line) are the 859 ions for which the null hypothesis of no difference was rejected. The blue and red vertical dashed lines correspond to foldchanges of 0.5 and 2, respectively.
Table 1:
Metabolomics pathway analysis using the Mummichog software showing the pathway, the molecules putatively identified (number of metabolites that were identified in the metabolomics dataset and overlap with the metabolites that consist the metabolic pathway), the size of the metabolic pathway (number of metabolites that contribute to the pathway) and the p-value of the pathway (Fisher’s exact test).
| Pathway | Molecules (overlap) | Pathway size | p-value |
|---|---|---|---|
| Carnitine shuttle | 9 | 23 | 5.3 E-4 |
| Pyruvate Metabolism | 4 | 10 | 1.8 E-3 |
| Vitamin E metabolism | 6 | 21 | 2.4 E-3 |
| Propanoate metabolism | 3 | 7 | 3.4 E-3 |
| Glycine, serine, alanine and threonine metabolism | 5 | 18 | 4.1 E-3 |
| Glutathione Metabolism | 2 | 3 | 5.8 E-3 |
| Heparan sulfate degradation | 2 | 3 | 5.8 E-3 |
| Chondroitin sulfate degradation | 2 | 3 | 5.8 E-3 |
| Bile acid biosynthesis | 8 | 38 | 8.5 E-3 |
| Aspartate and asparagine metabolism | 6 | 29 | 1.4 E-3 |
| Arginine and Proline Metabolism | 2 | 5 | 1.8 E-3 |
| Fatty acid oxidation | 2 | 6 | 2.8 E-3 |
| Vitamin A (retinol) metabolism | 5 | 26 | 2.9 E-3 |
| Urea cycle/amino group metabolism | 3 | 14 | 3.9 E-2 |
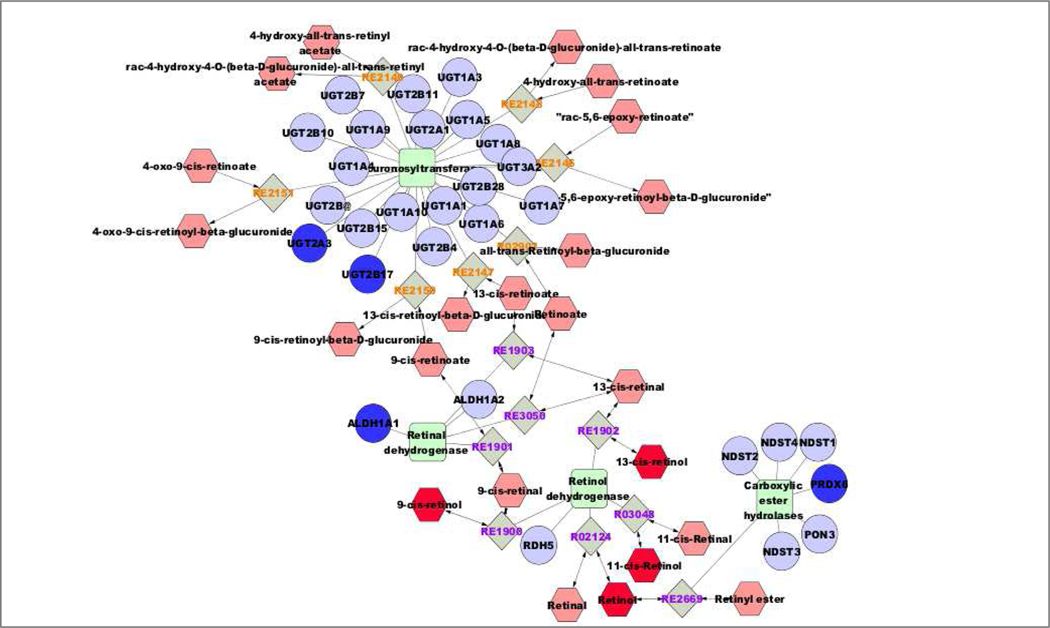
A systems biology approach was used to integrate all three datasets. Pathway visualization on MetScape of the significant genes and proteins (q<0.05) and the putatively-identified metabolites resulted in 61 pathways being identified (Supplement). The generated pathways showed the associations between the transcript-protein-metabolite interactions and how they can be displayed simultaneously on the same map. The identified pathways were related to lipid metabolism (such as omega-3 and omega-6 fatty acid metabolism, de novo fatty acid biosynthesis, glycophospholipid biosynthesis), and to energy metabolism (such as pyrimidine metabolism, TCA cycle, purine metabolism and di-unsaturated fatty acid beta-oxidation). Suppression of the ALDH1A1 gene showed a direct association with the vitamin A (retinol) metabolism pathway (Fig. 5). As shown in Fig. 5, there were three more genes involved in this pathway that were also down-regulated. These included two members of the UDP glucuronosyltransferase 2 family (UGT2B17, Log2 Fold-Change =−8.686, q=0.01, and UGT2A3, Log2 Fold-Change =−2.566, q=0.01) and peroxiredoxin 6 (PRDX6, Log2 Fold-Change =−1.102, q=0.04). The putatively-identified metabolite with m/z 287.2391 (retinol or its isomers 9-, 11- or 13-cis retinol) was down-regulated (Log2 Fold-Change =−2.14, q=0.03) (Supplement).
Figure 5:
Vitamin A (retinol) metabolism visualized in MetScape. Dark blue symbols represent genes and proteins from the transcriptomics and proteomics datasets, and dark red symbols represent putatively annotated metabolites (see Supplement). ALDH1A1 (aldehyde dehydrogenase 1A1, Log2 Fold Change = −5,292, q = 0.001), UGT2B17 (UDP glucuronosyltransferase 2B17, Log2 Fold-Change=−8.686, q=0.01), UGT2A3 (UDP glucuroosyltransferase, Log2 Fold-Change=−2.566, q=0.01) and PRDX6 (peroxiredoxin 6, Log2 Fold-Change=−1.102, q=0.04). The putatively-identified ion was the m/z 287.2391 (retinol or its isomers 9-, 11- or 13-cis retinol, Log2 Fold-Change= −2.142, q=0.03).
4. Discussion
To date, a systems biology integration of transcriptomics, proteomics and metabolomics has not been used to study the role of ALDH genes in CRC. Using this approach, the present study revealed metabolic pathways affected by suppressing the ALDH1A1 gene. New functional links between genes, proteins and metabolites were revealed, providing us with greater understanding of the functional interactions of the disease, and shedding additional insights into the complex biology of CRC.
A large body of research exists that demonstrates metabolites to function as cell signaling molecules involved in many physiological and pathophysiological conditions, including the control of inflammation [36, 37], altered lipid metabolism, [38–40] and CRC [41]. In the present study, the individually analysed datasets uncovered pathways relating to cell signaling and lipid metabolism. Several studies have shown the critical role of Wnt signaling in the proliferation and/or differentiation of stem cells in the intestinal crypts. This role is fundamental and any dysregulation of this pathway is a major contributor to colorectal carcinogenesis, since its activation leads to the accumulation of β-catenin in the nucleus which can be detected in over 80% of CRC tumors [7, 42]. Aberrant Wnt/β-catenin signaling causes unregulated expression of the c-Myc, a proto-oncogene that drives colorectal carcinogenesis. The overexpression of c-Myc is associated with cancer stem cell maintenance and chemoresistance [43–45]. In our work, the transcriptomics dataset showed that both WNT10A and c-MYC were significantly down-regulated, showing that ALDH1A1 may play a role in suppression of these genes in the COLO320 cell line. In addition, the results provide further evidence for targeting ALDHA1 as a drug target for the treatment of CRC.
It is known that metabolic reprogramming is one of the hallmarks of cancer because the energy generated through aerobic glycolysis is considered to be sufficient to offset the energy demands associated with rapid cancer cell division [46]. In a growing tumour, there is enhanced oxidative phosphorylation, mitochondrial biogenesis and oxygen consumption rate [47, 48]. Studies have shown that inhibition of mitochondrial complex I by metformin diminished tumor growth in CRC [49–51]. In our study, thirty out of forty proteins in the mitochondrial complex I (NADH: ubiquinone oxidoreductase subunits, NDUF and MT-ND) were all down-regulated. These changes in a subset of mitochondrial enzymes may underlie the mitochondrial dysfunction pathway identified by the IPA, and lead to the abnormal metabolic and apoptotic processes in CRC. Other studies have shown that alterations in metabolic reprogramming pathways in CRC, such as those involved in pyrimidine biosynthesis and fatty acid oxidation, are caused by aberrant c-Myc expression [52]. Our results showing the down-regulation of c-Myc are consistent with these studies.
In the integrated analysis of both transciptomic and proteomics results, the asparagine biosynthesis I and the superpathway of cholesterol biosynthesis were found to be two most significantly affected pathways by ALDH1A1 gene suppression. It is well-established that cancer cells depend on aerobic glycolysis (a process known as the Warburg effect [46]), and on glutamine to support cell growth and survival [53, 54]. Glutamine is one of the most heavily consumed nutrients by cells in culture [55]. Once imported into the cells, glutamine serves as a carbon source for the tricarboxylic acid (TCA) cycle and as a nitrogen source for the synthesis of nucleotide and non-essential amino acids, such as asparagine [56]. There has been a significant amount of research lately focusing on the role of glutamine and asparagine in cancer. Recent studies have linked increased levels of asparagine to CRC and shown asparagine deprivation to prevent cell proliferation [57–59]. In our study, the down-regulation of asparagine biosynthesis indicates that this pathway is possibly regulated by ALDH1A1. The suppression of this gene could lead to cell growth inhibition, due to asparagine biosynthesis down-regulation, by depriving the cells from essential nutrient supplementation. In addition, studies have shown that alterations in cholesterol biosynthesis play a critical role in colon cancer and a high rate of cholesterol synthesis leads to multidrug resistance (MDR) in this condition [60]. 7-dehydrocholesterol reductase (DHCR7) is the enzyme responsible for converting 7-dehydrocholesterol to cholesterol and the final step in cholesterol synthesis [61, 62]. It was down-regulated in our transcriptomic and proteomic analyses, indicating down-regulation of cholesterol synthesis in cells in which ALDH1A1 expression had been suppressed. Interestingly, lower cholesterol has been associated with increased survival in colon cancer [63, 64]. In addition, many studies have shown that inhibition of cholesterol synthesis using statins can be chemopreventive against CRC; however, the mechanism is unclear [65–67].
The integrated “omics” analysis of all three datasets provided an overview of the possible pathway connections and the crosstalk between the transcripts, the proteins and the metabolites. ALDH1A1 suppression was directly correlated to the vitamin A (retinol) metabolism. Previously published work from our group had shown that retinol binding to fatty acid-binding protein 5 (FABP5) activates orphan nuclear receptor peroxisome proliferator-activated receptor (PPAR)β/δ and acts as procarcinogenic agent [68]. Our integrated omics analysis revealed alterations (down-regulation) in three other genes that are involved in the retinol metabolism, specifically UGT2B17, UGT2A3 and PRDX6. The UGT superfamily is comprised of two major sub-families (UGT1 and UGT2). They are Phase II enzymes that play an important role in the metabolism and biotransformation of exogenous (e.g., drugs, pesticides, components of tobacco smoke) and endogenous (e.g., bilirubin, bile acids, steroid hormones) compounds [69, 70]. Several studies have shown that polymorphisms in phase II enzymes affect CRC risk [70, 71]. Related more specifically to the results of the present study, a UGT2B17 deletion genotype was associated with a decrease in CRC risk [69, 71, 72], and increased levels of UGT2A3 in the primary tumor tissues have been observed in CRC patients with liver metastases [73]. The PRDX6 gene is involved in the control of many cellular physiological functions, including growth, differentiation, apoptosis, embryonic development, and lipid metabolism [74]. PRDX6 binds and reduces phospholipid hydroperoxides, thereby serving a role in the repair of membrane damage caused by oxidative stress. Its expression is regulated mainly? by the redox-active regulators, including c-Myc [75]. Several studies have shown the elevated levels of PRDX6 in breast, bladder lung, ovarian, and pancreatic cancer [76–80]. The present study is the first to show reduced levels of PRDX6 after ALDH1A1 suppression in colon cancer cells and its possible involvement in CRC.
Conclusions
Omics approaches have, in many respects, revolutioned the means by which the molecular mechanisms operating in cells under physiological and pathophysiological conditions can be investigated in an unbiased manner. A systems biology approach not only amplifies the precision of the insights gleaned from each individual omic approach, but also provides even greater insights into the pathways that drive cancer. Using this approach in a CRC cell line, suppression of ALDH1A1 expression was shown to down-regulate oxidative phosphorylation, mitochondrial function, the sirtuin signaling pathway, cholesterol biosynthesis and the vitamin A (retinol) metabolism pathways. How ALDH1A1 causes these changes remains to be established. It may involve already known functions of this protein, such as enzymatic/catalytic activity (i.e., retinoic acid), and/or protein-protein interaction capabilities. In addition, some of the products generated by this protein may be contributors, such as retinoic acid or acetate. This study represents the first effort to integrate multi-omics in the study of ALDHs in the pathophysiology of tumor cells.
Supplementary Material
Highlights.
Role of the ALDH1A1 gene in colorectal cancer cells
Integrated network analysis of transcriptomics, proteomics and metabolomics
Down-regulation of the Vitamin A (retinol) metabolism caused by ALDH1A1 suppression
Down-regulation of the cholesterol pathway caused by ALDH1A1 suppression
Acknowledgements
This work was supported in parts by National Institutes of Health (NIH) Grants AA021724, AA022057 and EY17963. The Orbitrap Fusion mass spectrometer at the Yale MS & Proteomics Resource was funded in part by NIH SIG from the Office Of The Director, National Institutes Of Health of the National Institutes of Health under Award Number (S10OD018034). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Wendy Wang for and Jean Kanyo for preparing the samples and for LC-MS/MS data collection, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A, Colorectal cancer statistics, 2017, CA Cancer J Clin, 67 (2017) 177–193. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2016, CA Cancer J Clin, 66 (2016) 7–30. [DOI] [PubMed] [Google Scholar]
- [3].H. The Lancet Gastroenterology, Colorectal cancer screening: is earlier better?, The lancet. Gastroenterology & hepatology, 3 (2018) 519. [DOI] [PubMed] [Google Scholar]
- [4].Austin H, Henley SJ, King J, Richardson LC, Eheman C, Changes in colorectal cancer incidence rates in young and older adults in the United States: what does it tell us about screening, Cancer Causes Control, 25 (2014) 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Colussi D, Brandi G, Bazzoli F, Ricciardiello L, Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention, Int J Mol Sci, 14 (2013) 16365–16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork K, Ramadoss S, Ding X, Li X, Wang CY, KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/beta-catenin signalling, Nat Commun, 8 (2017) 15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Reya T, Clevers H, Wnt signalling in stem cells and cancer, Nature, 434 (2005) 843–850. [DOI] [PubMed] [Google Scholar]
- [8].Clevers H, The cancer stem cell: premises, promises and challenges, Nat Med, 17 (2011) 313–319. [DOI] [PubMed] [Google Scholar]
- [9].Klose RJ, Zhang Y, Regulation of histone methylation by demethylimination and demethylation, Nat Rev Mol Cell Biol, 8 (2007) 307–318. [DOI] [PubMed] [Google Scholar]
- [10].Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, Vasiliou V, Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress, Free Radic Biol Med, 56 (2013) 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V, Aldehyde dehydrogenase 1B1 (ALDH1B1) is a potential biomarker for human colon cancer, Biochem Biophys Res Commun, 405 (2011) 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Singh S, Arcaroli J, Chen Y, Thompson DC, Messersmith W, Jimeno A, Vasiliou V, ALDH1B1 Is Crucial for Colon Tumorigenesis by Modulating Wnt/beta-Catenin, Notch and PI3K/Akt Signaling Pathways, PLoS One, 10 (2015) e0121648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kozovska Z, Patsalias A, Bajzik V, Durinikova E, Demkova L, Jargasova S, Smolkova B, Plava J, Kucerova L, Matuskova M, ALDH1A inhibition sensitizes colon cancer cells to chemotherapy, BMC Cancer, 18 (2018) 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tomita H, Tanaka K, Tanaka T, Hara A, Aldehyde dehydrogenase 1A1 in stem cells and cancer, Oncotarget, 7 (2016) 11018–11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zarkavelis G, Boussios S, Papadaki A, Katsanos KH, Christodoulou DK, Pentheroudakis G, Current and future biomarkers in colorectal cancer, Annals of gastroenterology, 30 (2017) 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Z, Gerstein M, Snyder M, RNA-Seq: a revolutionary tool for transcriptomics, Nat Rev Genet, 10 (2009) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Patti GJ, Yanes O, Siuzdak G, Innovation: Metabolomics: the apogee of the omics trilogy, Nat Rev Mol Cell Biol, 13 (2012) 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Coman C, Solari FA, Hentschel A, Sickmann A, Zahedi RP, Ahrends R, Simultaneous Metabolite, Protein, Lipid Extraction (SIMPLEX): A Combinatorial Multimolecular Omics Approach for Systems Biology, Mol Cell Proteomics, 15 (2016) 1453–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nakayasu ES, Nicora CD, Sims AC, Burnum-Johnson KE, Kim YM, Kyle JE, Matzke MM, Shukla AK, Chu RK, Schepmoes AA, Jacobs JM, Baric RS, Webb-Robertson BJ, Smith RD, Metz TO, MPLEx: a Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses, mSystems, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hasin Y, Seldin M, Lusis A, Multi-omics approaches to disease, Genome Biol, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guo S, Bai H, Megyola CM, Halene S, Krause DS, Scadden DT, Lu J, Complex oncogene dependence in microRNA-125a-induced myeloproliferative neoplasms, Proc Natl Acad Sci U S A, 109 (2012) 16636–16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y, Thompson DC, Koppaka V, Jester JV, Vasiliou V, Ocular aldehyde dehydrogenases: protection against ultraviolet damage and maintenance of transparency for vision, Prog Retin Eye Res, 33 (2013) 28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Trapnell C, Pachter L, Salzberg SL, TopHat: discovering splice junctions with RNA-Seq, Bioinformatics, 25 (2009) 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L, Differential analysis of gene regulation at transcript resolution with RNA-seq, Nat Biotechnol, 31 (2013) 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L, Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks, Nat Protoc, 7 (2012) 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Charkoftaki G, Jester JV, Thompson DC, Vasiliou V, Nitroen mustard-induced corneal injury involves the sphingomyelin-ceramide pathway, Ocul Surf, 16 (2018) 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G, XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification, Anal Chem, 78 (2006) 779–787. [DOI] [PubMed] [Google Scholar]
- [28].Southam AD, Weber RJ, Engel J, Jones MR, Viant MR, A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics, Nat Protoc, 12 (2016) 310–328. [DOI] [PubMed] [Google Scholar]
- [29].Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B, Predicting network activity from high throughput metabolomics, PLoS Comput Biol, 9 (2013) e1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Benjamini Y, Hochberg Y, Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing, Journal of the Royal Statistical Society. Series B (Methodological), 57 (1995) 289–300. [Google Scholar]
- [31].Karnovsky A, Weymouth T, Hull T, Tarcea VG, Scardoni G, Laudanna C, Sartor MA, Stringer KA, Jagadish HV, Burant C, Athey B, Omenn GS, Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data, Bioinformatics, 28 (2012) 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, Moritz RL, Carver JJ, Wang M, Ishihama Y, Bandeira N, Hermjakob H, Vizcaino JA, The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition, Nucleic Acids Res, 45 (2017) D1100–d1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H, 2016 update of the PRIDE database and its related tools, Nucleic Acids Res, 44 (2016) 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rousseeuw PJ, Vanden Branden K, ROBPCA: A New Approach to Robust Principal Component Analysis AU - Hubert, Mia, Technometrics, 47 (2005) 64–79. [Google Scholar]
- [35].Hubert MR, Peter; Verdonck, Tim, Robust PCA for skewed data and its outlier map, Computational Statistics & Data Analysis, 53 (2009) 2264. [Google Scholar]
- [36].Kasuga K, Suga T, Mano N, Bioanalytical insights into mediator lipidomics, Journal of pharmaceutical and biomedical analysis, 113 (2015) 151–162. [DOI] [PubMed] [Google Scholar]
- [37].Pein H, Ville A, Pace S, Temml V, Garscha U, Raasch M, Alsabil K, Viault G, Dinh CP, Guilet D, Troisi F, Neukirch K, Konig S, Bilancia R, Waltenberger B, Stuppner H, Wallert M, Lorkowski S, Weinigel C, Rummler S, Birringer M, Roviezzo F, Sautebin L, Helesbeux JJ, Seraphin D, Mosig AS, Schuster D, Rossi A, Richomme P, Werz O, Koeberle A, Endogenous metabolites of vitamin E limit inflammation by targeting 5-lipoxygenase, Nat Commun, 9 (2018) 3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nagahashi M, Abe M, Sakimura K, Takabe K, Wakai T, The role of sphingosine-1-phosphate in inflammation and cancer progression, Cancer Sci, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xia D, Wang D, Kim SH, Katoh H, DuBois RN, Prostaglandin E2 promotes intestnal tumor growth via DNA methylation, Nat Med, 18 (2012) 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Tylichova Z, Slavik J, Ciganek M, Ovesna P, Krcmar P, Strakova N, Machala M, Kozubik A, Hofmanova J, Vondracek J, Butyrate and docosahexaenoic acid interact in alterations of specific lipid classes in differentiating colon cancer cells, J Cell Biochem, 119 (2018) 4664–4679. [DOI] [PubMed] [Google Scholar]
- [41].Jain R, Austin Pickens C, Fenton JI, The role of the lipidome in obesity-mediated colon cancer risk, J Nutr Biochem, 59 (2018) 1–9. [DOI] [PubMed] [Google Scholar]
- [42].Sebio A, Kahn M, Lenz HJ, The potential of targeting Wnt/beta-catenin in colon cancer, Expert Opin Ther Targets, 18 (2014) 611–615. [DOI] [PubMed] [Google Scholar]
- [43].Whitfield JR, Soucek L, Tumor microenvironment: becoming sick of Myc, Cell Mol Life Sci, 69 (2012) 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M, The landscape of somatic copy-number alteration across human cancers, Nature, 463 (2010) 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dominguez-Sola D, Gautier J, MYC and the control of DNA replication, Cold Spring Harb Perspect Med, 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vander Heiden MG, Cantley LC, Thompson CB, Understanding the Warburg effect: the metabolic requirements of cell proliferation, Science, 324 (2009) 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, Kalluri R, PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis, Nat Cell Biol, 16 (2014) 992–1003, 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell, 144 (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [49].Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, Chandel NS, Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis, Elife, 3 (2014) e02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ota S, Horigome K, Ishii T, Nakai M, Hayashi K, Kawamura T, Kishino A, Taiji M, Kimura T, Metformin suppresses glucose-6-phosphatase expression by a complex I inhibition and AMPK activation-independent mechanism, Biochem Biophys Res Commun, 388 (2009) 311–316. [DOI] [PubMed] [Google Scholar]
- [51].Owen MR, Doran E, Halestrap AP, Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain, Biochem J, 348 Pt 3 (2000) 607–614. [PMC free article] [PubMed] [Google Scholar]
- [52].Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, Tabata S, Saitoh K, Kato K, Sato S, Igarashi K, Aizawa Y, Kajino-Sakamoto R, Kojima Y, Fujishita T, Enomoto A, Hirayama A, Ishikawa T, Taketo MM, Kushida Y, Haba R, Okano K, Tomita M, Suzuki Y, Fukuda S, Aoki M, Soga T, Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC, Proc Natl Acad Sci U S A, 114 (2017) E7697–e7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wise DR, Thompson CB, Glutamine addiction: a new therapeutic target in cancer, Trends in biochemical sciences, 35 (2010) 427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].DeBerardinis RJ, Cheng T, Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer, Oncogene, 29 (2010) 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kim B, Li J, Jang C, Arany Z, Glutamine fuels proliferation but not migration of endothelial cells, The EMBO journal, 36 (2017) 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang J, Pavlova NN, Thompson CB, Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine, The EMBO journal, 36 (2017) 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Toda K, Kawada K, Iwamoto M, Inamoto S, Sasazuki T, Shirasawa S, Hasegawa S, Sakai Y, Metabolic Alterations Caused by KRAS Mutations in Colorectal Cancer Contribute to Cell Adaptation to Glutamine Depletion by Upregulation of Asparagine Synthetase, Neoplasia, 18 (2016) 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li J, Song P, Zhu L, Aziz N, Zhou Q, Zhang Y, Xu W, Feng L, Chen D, Wang X, Jin H, Synthetic lethality of glutaminolysis inhibition, autophagy inactivation and asparagine depletion in colon cancer, Oncotarget, 8 (2017) 42664–42672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pavlova NN, Hui S, Ghergurovich JM, Fan J, Intlekofer AM, White RM, Rabinowitz JD, Thompson CB, Zhang J, As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid, Cell Metab, 27 (2018) 428–438.e425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gelsomino G, Corsetto PA, Campia I, Montorfano G, Kopecka J, Castella B, Gazzano E, Ghigo D, Rizzo AM, Riganti C, Omega 3 fatty acids chemosensitize multidrug resistant colon cancer cells by down-regulating cholesterol synthesis and altering detergent resistant membranes composition, Mol Cancer, 12 (2013) 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wilcox CB, Feddes GO, Willett-Brozick JE, Hsu LC, DeLoia JA, Baysal BE, Coordinate upregulation of TMEM97 and cholesterol biosynthesis genes in normal ovarian surface epithelial cells treated with progesterone: implications for pathogenesis of ovarian cancer, BMC Cancer, 7 (2007) 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Fitzky BU, Witsch-Baumgartner M, Erdel M, Lee JN, Paik YK, Glossmann H, Utermann G, Moebius FF, Mutations in the Delta7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome, Proc Natl Acad Sci U S A, 95 (1998) 8181–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Dobrzycka M, Spychalski P, Lachinski AJ, Kobiela P, Jedrusik P, Kobiela J, Statins and Colorectal Cancer - A Systematic Review, Exp Clin Endocrinol Diabetes, (2018). [DOI] [PubMed] [Google Scholar]
- [64].Waluga M, Zorniak M, Fichna J, Kukla M, Hartleb M, Pharmacological and dietary factors in prevention of colorectal cancer, J Physiol Pharmacol, 69 (2018). [DOI] [PubMed] [Google Scholar]
- [65].Juneja M, Kobelt D, Walther W, Voss C, Smith J, Specker E, Neuenschwander M, Gohlke BO, Dahlmann M, Radetzki S, Preissner R, von Kries JP, Schlag PM, Stein U, Statin and rottlerin small-molecule inhibitors restrict colon cancer progression and metastasis via MACC1, PLoS Biol, 15 (2017) e2000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chang HL, Chen CY, Hsu YF, Kuo WS, Ou G, Chiu PT, Huang YH, Hsu MJ, Simvastatin induced HCT116 colorectal cancer cell apoptosis through p38MAPK-p53-survivin signaling cascade, Biochim Biophys Acta, 1830 (2013) 4053–4064. [DOI] [PubMed] [Google Scholar]
- [67].Poynter JN, Gruber SB, Higgins PD, Almog R, Bonner JD, Rennert HS, Low M, Greenson JK, Rennert G, Statins and the risk of colorectal cancer, N Engl J Med, 352 (2005) 2184–2192. [DOI] [PubMed] [Google Scholar]
- [68].Singh S, Arcaroli J, Thompson DC, Messersmith W, Vasiliou V, Acetaldehyde and retinaldehyde-metabolizing enzymes in colon and pancreatic cancers, Adv Exp Med Biol, 815 (2015) 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Angstadt AY, Hartman TJ, Lesko SM, Muscat JE, Zhu J, Gallagher CJ, Lazarus P, The effect of UGT1A and UGT2B polymorphisms on colorectal cancer risk: haplotype associations and gene-environment interactions, Genes Chromosomes Cancer, 53 (2014) 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Tukey RH, Strassburg CP, Human UDP-glucuronosyltransferases: metabolism, expression, and disease, Annu Rev Pharmacol Toxicol, 40 (2000) 581–616. [DOI] [PubMed] [Google Scholar]
- [71].Angstadt AY, Berg A, Zhu J, Miller P, Hartman TJ, Lesko SM, Muscat JE, Lazarus P, Gallagher CJ, The effect of copy number variation in the phase II detoxification genes UGT2B17 and UGT2B28 on colorectal cancer risk, Cancer, 119 (2013) 2477–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Scherer D, Koepl LM, Poole EM, Balavarca Y, Xiao L, Baron JA, Hsu L, Coghill AE, Campbell PT, Kleinstein SE, Figueiredo JC, Lampe JW, Buck K, Potter JD, Kulmacz RJ, Jenkins MA, Hopper JL, Win AK, Newcomb PA, Ulrich CM, Makar KW, Genetic variation in UGT genes modify the associations of NSAIDs with risk of colorectal cancer: colon cancer family registry, Genes Chromosomes Cancer, 53 (2014) 568–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wang S, Transcriptome analysis in primary colorectal cancer tissues from patients with and without liver metastases using next-generation sequencing, 6 (2017) 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Nicolussi A, D’Inzeo S, Capalbo C, Giannini G, Coppa A, The role of peroxiredoxins in cancer, Mol Clin Oncol, 6 (2017) 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Fisher AB, Peroxiredoxin 6: a bifunctional enzyme with glutathione peroxidase and phospholipase A(2) activities, Antioxidants & redox signaling, 15 (2011) 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Schremmer B, Manevich Y, Feinstein SI, Fisher AB, Peroxiredoxins in the lung with emphasis on peroxiredoxin VI, Sub-cellular biochemistry, 44 (2007) 317–344. [DOI] [PubMed] [Google Scholar]
- [77].Pylvas M, Puistola U, Kauppila S, Soini Y, Karihtala P, Oxidative stress-induced antioxidant enzyme expression is an early phenomenon in ovarian carcinogenesis, Eur J Cancer, 46 (2010) 1661–1667. [DOI] [PubMed] [Google Scholar]
- [78].Quan C, Cha EJ, Lee HL, Han KH, Lee KM, Kim WJ, Enhanced expression of peroxiredoxin I and VI correlates with development, recurrence and progression of human bladder cancer, J Urol, 175 (2006) 1512–1516. [DOI] [PubMed] [Google Scholar]
- [79].Thongwatchara P, Promwikorn W, Srisomsap C, Chokchaichamnankit D, Boonyaphiphat P, Thongsuksai P, Differential protein expression in primary breast cancer and matched axillary node metastasis, Oncology reports, 26 (2011) 185–191. [DOI] [PubMed] [Google Scholar]
- [80].Park JY, Kim SA, Chung JW, Bang S, Park SW, Paik YK, Song SY, Proteomic analysis of pancreatic juice for the identification of biomarkers of pancreatic cancer, Journal of cancer research and clinical oncology, 137 (2011) 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data from this publication are available at the ProteomeXchange Consortium [32] (dataset identifier PXD012230, http://www.proteomeexchange.org/) via the PRIDE partner repository [33]. For the RNA sequencing data, the FASTQ and processed data are available at Gene Expression Omnibus (GSE121043, https://www.ncbi.nlm.nih.gov/geo/). Metabolomics data are available at the NIH Metabolomics workbench (study ID ST0001093, http://www.metabolomicsworkbench.org/).