Abstract
Background:
Use of new genomic techniques in clinical settings requires that such methods are rigorous and reproducible. Previous studies have shown that quantitation of donor-derived cell-free DNA (%ddcfDNA) by unbiased shotgun sequencing is a sensitive, non-invasive marker of acute rejection after heart transplantation. The primary goal of this study was to assess the reproducibility of %ddcfDNA measurements across technical replicates, manual vs. automated platforms, and rejection phenotypes in distinct patient cohorts.
Methods:
After developing and validating the %ddcfDNA assay, we subjected the method to a rigorous test of its reproducibility: We measured %ddcfDNA in technical replicates performed by two independent laboratories and verified the reproducibility of %ddcfDNA patterns of two rejection phenotypes: acute cellular rejection and antibody-mediated rejection (ACR and AMR, respectively) in distinct patient cohorts.
Results:
We observed strong concordance of technical-replicate %ddcfDNA measurements across two independent laboratories (slope=1.02, R2 >0.99, p<<10−6), as well as across manual and automated platforms (slope=0.80, R2=0.92, p<0.001). The %ddcfDNA measurements in distinct heart-transplant cohorts had similar baselines and error rates. In both patient cohorts, %ddcfDNA temporal patterns associated with rejection phenotypes were similar; however, the quantity of ddcfDNA was significantly higher in samples with severe vs. mild histological rejection grade (2.73% vs. 0.14%, respectively, p<0.001).
Conclusion:
The %ddcfDNA assay is precise and reproducible across laboratories and in samples from two distinct heart-transplant rejection types. These findings pave the way for larger studies to assess the clinical utility of %ddcfDNA as a marker of acute rejection after heart transplantation.
Keywords: reproducibility, cell-free DNA, allograft rejection, heart transplantation, automated
INTRODUCTION
Acute cellular rejection (ACR) and antibody-mediated rejection (AMR) are common after heart transplantation and contribute to poor long-term outcomes. Early diagnosis provides opportunities for interventions to improve outcomes. Currently, however, detection is limited by methodology: endomyocardial biopsy (EMB) shows high inter-observer variability in interpretation and poor performance that may lead to delayed diagnosis, and thus, treatment (1, 2). AlloMap® is an FDA-approved non-invasive diagnostic test with a high negative predictive value for ACR (3), but its utility in predicting other causes of graft injury, such as AMR, remain unknown. Other molecular techniques such as intragraft genomic and proteomic signatures have emerged as reliable markers for graft injury after kidney (4–7), and heart transplantation (8), but both methods require companion EMB analysis and are thus invasive.
Recently, De Vlaminck et al. reported the use of %ddcfDNA as a non-invasive diagnostic tool for detecting heart-transplant rejection (2, 9) by using shotgun sequencing to quantify %ddcfDNA in serial plasma samples from 65 heart transplant recipients (HTRs). That report, employing EMB as a detection standard, demonstrated a diagnostic cutoff of 0.25%ddcfDNA to be sensitive and specific enough to diagnose clinically significant ACR (10), as shown by receiver operator curve (ROC) analysis (area under curve (AUC), ROC = 0.83). Moreover, those data showed that elevations in %ddcfDNA preceded histologic diagnosis by several weeks to months.
In transplantation, cell-free DNA analyses have been used to study infection, microbial diversity, (11, 12), host immunity (13, 14), and to determine the donor tissue of origin (12, 15). These investigations portend much promise in the care of transplant recipients. However, given recent concerns regarding low reproducibility of genomic data (16), we considered it essential to verify the reproducibility of our %ddcfDNA assay. These reproducibility studies are critically important to define test-performance characteristics and to enable rational design of clinical-utility studies toward improving patient outcomes.
MATERIALS AND METHODS
Human subjects and biosample collection:
Rejecting and non-rejecting HTRs were preferentially selected to provide a wide range of %ddcfDNA values and different rejection phenotypes to assess assay reproducibility. We analyzed 127 plasma samples from 21 subjects; 10 subjects from the Stanford Genome Transplant Dynamics (GTD) study (2), 9 HTRs from the NIH-sponsored Acute Cardiac Allograft Cellular Rejection and Cardiac Allograft Vasculopathy Study, and 2 healthy controls. Patients in both cohorts were subjected to a surveillance-biopsy schedule for acute rejection. Additional clinically indicated biopsies were performed when graft dysfunction was suspected. In the NIH cohort, EMBs (n=263) were initially graded using the 1990 International Society of Heart and Lung Transplantation (ISHLT) criteria (17). A second histologic assessment was performed to harmonize grading to the ISHLT 2004 criteria (10) used to grade EMBs within the Stanford cohort. In the NIH cohort, 21 of 263 EMBs showed severe rejection (≥2R grade) on either histologic read; the 8 EMBs that showed concordant results between the two reads were selected for this study. In both cohorts, serial plasma samples were collected prior to EMB. All but 6 samples did not have a corresponding biopsy grade for acute rejection due to poor biopsy sample quality. Plasma from the two healthy control subjects were used to produce standard cell-free DNA mixtures for assay development. Demographic data for both cohorts is summarized in Supplemental Table 2. The Institutional Review Boards of the Stanford University School of Medicine, National Heart Lung and Blood Institute (NHLBI), and INOVA Fairfax Hospital approved this study.
Genotyping:
Donor and recipient genomic DNA was genotyped using Illumina whole-genome array (HumanOmni2.5–8v1.2) at the Cancer Genomics Research Laboratory (National Cancer Institute, NIH) following the manufacturer’s protocol as described previously (2).
Quantification of %ddcfDNA
Cell-free DNA was isolated from plasma (QIAamp Circulating Nucleic Acid Kit, Qiagen) and used to prepare DNA libraries (Mondrain Ovation SP Ultralow Library System). Libraries were sequenced (Illumina HiSeq-2500, 67 ± 25 million, 2 × 75 or 2 × 50 base pair reads/sample).
Sequence reads were analyzed using previously described workflows (2), with minor modifications. Reads were trimmed and filtered to remove PCR duplicates and low-quality reads. The remaining reads were aligned to the human genome build hg19 (www.ucsc.edu). Mapped reads were surveyed for donor and recipient unique SNP, which were used to compute %ddcfDNA.
Statistical analysis
In both cohorts, each %ddcfDNA measurement had a corresponding histologic grading for acute rejection. EMBs were graded as mild (ISHLT grade 0, 1R) or moderate-severe (≥ grade 2R) (10). The %ddcfDNA levels were not normally distributed but were skewed leftward. Thus, the non-parametric Mann-Whitney U test was used to compare groups. Linear regression models were used to determine between-group correlations.
RESULTS
In this study, our goal was to affirm the reproducibility of a new genomic tool (donor-derived cell-free DNA (%ddcfDNA)) for detecting donor-organ rejection in heart-transplant recipients (HTRs). We sought to determine concordance of technical-replicate across laboratories/technicians, a manual vs. automated platform, and to correlate %ddcfDNA patterns with phenotypic data in two rejection types.
Study design and reproducibility of assay workflow
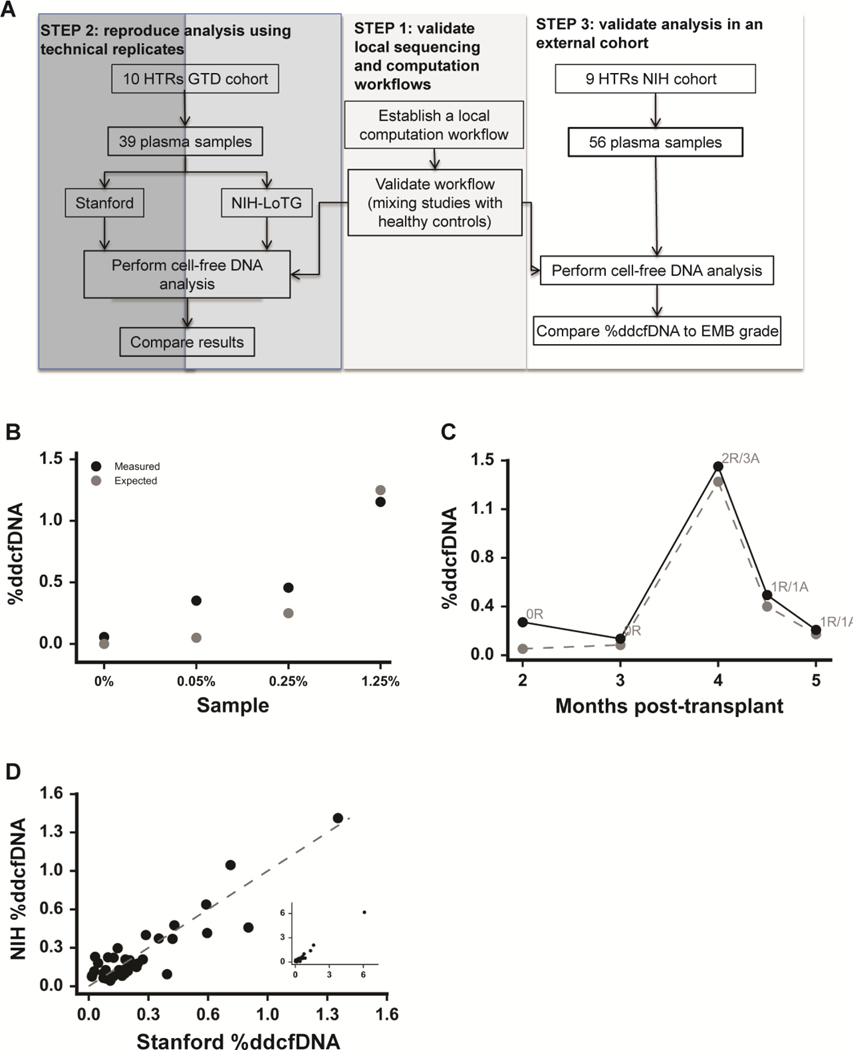
We developed a three-step reproducibility design (Figure 1a) following guidelines from a recent National Institutes of Health (NIH) panel on the reproducibility of genomic data (https://videocast.nih.gov/summary.asp?Live=16381&bhcp=1): (1) establish and validate a local sequencing and computational workflow, (2) confirm internal reproducibility by performing replicate analysis of published data and biologic replicates, and (3) confirm external reproducibility by correlating %ddcfDNA with graft injury (both ACR and AMR) in HTR samples from distinct cohorts.
Figure 1: Development and reproducibility of a local sequencing and computational workflow to determine %ddcfDNA.
a) Experimental design is adapted from NIH panel discussions on reproducing genomic data (www.videocast.nih.gov) suggesting three steps: Step 1 is to develop and validate a local computation workflow. Step 2 is to perform internal validation using sequence files and biosamples previously analyzed. Step 3 (external validation) is to apply the analysis to an external cohort with similar characteristics to the internal validation cohort. (b) Cell-free DNA from two genotyped healthy subjects (one assigned as donor, the other as recipient) were mixed at predefined ratios and sequenced to measure %ddcfDNA. The measured %ddcfDNA was regressed on the expected %ddcfDNA (slope=1.20, r2 =0.97, p-value = 0.032). (c) Serial %ddcfDNA measurements of a GTD subject plotted against months after transplant. Quiescent (0R, 1R) or acute rejection (2R) states are indicated. Solid and dashed lines represent NIH and Stanford results respectively. (d) The linear correlation between %ddcfDNA determined at the NIH (y-axis) and Stanford (x-axis) using replicate plasma samples (n=39). The linear regression line (slope=1.02, r2 = 0.998 and p-value << 10 −6) testing the hypothesis that the slope equals zero is shown.
Step 1 (workflow):
We adopted a sequencing and computation workflow (software, scripts, reference sequence, and parameters) for measuring %ddcfDNA based upon previous reports (2, 9). The method relies on genotyping donor and recipient pairs to identify loci at which the recipient is homozygous but different from the donor (11,401±2,594 of such positions per 10 million reads surveyed). Defining these loci allowed assignment of single-nucleotide polymorphisms (SNPs) unique to donor and recipient; %ddcfDNA represents the ratio of unique donor SNPs to the sum of donor and recipient SNPs.
To correct for any potential errors in computed %ddcfDNA that may have been introduced during the various stages of the assay, we computed (and subtracted) the fraction of incorrect SNPs at positions where donor and recipient were homozygous and identical (24,063±3866 positions per 10 million reads surveyed), as described previously (9). A separate linear-regression analysis of the error rate as described by other investigators (2) did not yield a significant correlation in our analysis (slope =1.56, R2 =0.09, p=0.57, coefficient of variation (CV) = 0.16). We reproduced the local computational workflow and sequencing workflows using mixtures of %ddcfDNA with known ratios from two genotyped healthy controls (one subject assigned as donor, the other as recipient). Computed and expected %ddcfDNA values showed a strong correlation (slope =1.20, R2= 0.97, p=0.032 (Figure 1b)).
Step 2 (internal reproducibility):
Using the established workflow described above in Step 1, we (NIH Laboratory of Transplantation Genomics, NIH-LoTG) re-analyzed %ddcfDNA levels using publicly available sequenced reads deposited by the Stanford Group (www.ncbi.nlm.nih.gov/sra, prjna233479). We obtained nearly identical computed %ddcfDNA values (suppl. Figure 1a) as had been reported by the Stanford group(2). Next, we analyzed 39 technical replicate plasma samples, also analyzed by the Stanford Group. Summary of both assays is shown in Supp Table 1. Within-subject %ddcfDNA measurements showed strong agreement (Figure 1c), as did linear regression of the replicate %ddcfDNA measurements (R2=0.998, p<10−6, Figure 1d). Thus, we have demonstrated reproducibility of both processing and analysis across two independent laboratories (NIH and Stanford).
Step 3 (external reproducibility):
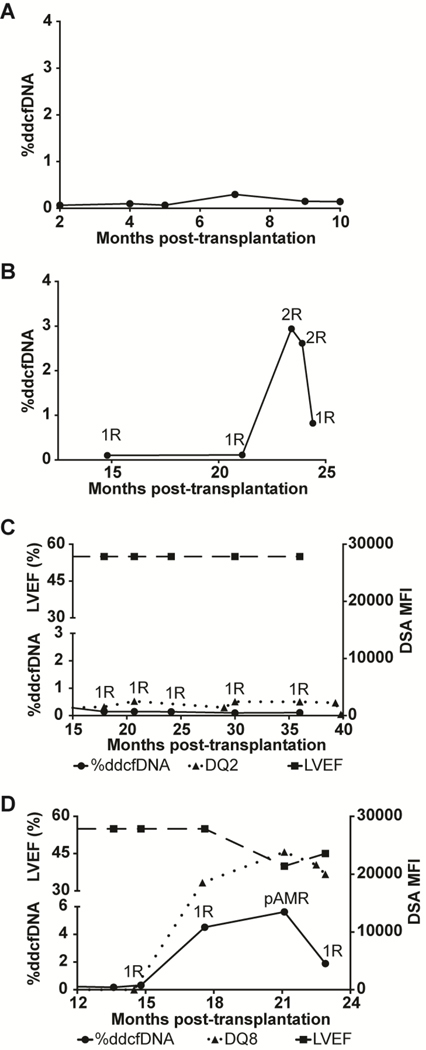
Next, we assessed the reproducibility of %ddcfDNA patterns in distinct patient cohorts. Within both cohorts, most measurements showed %ddcfDNA levels<0.1%, (suppl. Figure 1b). Baseline %ddcfDNA levels (ISHLT grade 0R) were also similar (0.09+0.07 vs. 0.07+0.04 respectively), as were measured error rates (0.04 +0.02 vs. 0.06 +0.01). Replicate %ddcfDNA measurements of 8 samples performed by three technicians showed low variance (coefficient of variance (CV)= 0.04 + 0.0, suppl. Figure 1c). Like the HTRs within the Stanford GTD cohorts, HTR controls (no-rejection) in the NIH cohort (n=2, Figure 2a) showed consistently low levels of %ddcfDNA. Samples from the ACR group had low %ddcfDNA levels when EMB showed non-rejection and elevated %ddcfDNA levels when EMB showed acute rejection (Figure 2b). In the AMR group (n=3), the %ddcfDNA elevations preceded AMR diagnosis by several weeks. All 3 AMR subjects showed DSA >15,000 mean-fluorescence intensity (MFI), a reduction in left ventricular ejection fraction(LVEF) of 10–20% from baseline on echocardiography, clinical signs of congestive heart failure (CHF), and capillaritis on histopathology at the time of diagnosis. One HTR with low DSA MFI and normal histology showed low levels of %ddcfDNA (Figure 2c). Thus, %ddcfDNA levels for individual subjects with ACR and AMR showed similar temporal patterns between the two cohorts, confirming the reproducibility of our approach across rejection types.
Figure 2: Reproducibility of %ddcfDNA patterns of rejection phenotypes in distinct patient cohorts.
Serial %ddcfDNA against time plots of two representative HTRs from the NIH cohort: (a) No rejection; (b) ≥ grade 2R ACR. Corresponding grading of EMB as: No-to-mild rejection (0R or 1R) or moderate to severe rejection (≥2R) are labeled for each %ddcfDNA measurement analyzed. Figures (c) and (d) are representative time plots of two subjects from the NIH cohort with donor-specific antibodies (DSA) against HLA Class II DQ antigens displaced as the mean florescence intensity (MFI-right y-axis) represented in dotted line. Graft function as measured by the left ventricular ejection fraction (upper left y-axis) is shown in the dashed line. The %ddcfDNA (lower left-y-axis) is shown in solid black line. pAMR label in (d) represents time of histopathologic and clinical diagnosis of AMR and initiation of treatment.
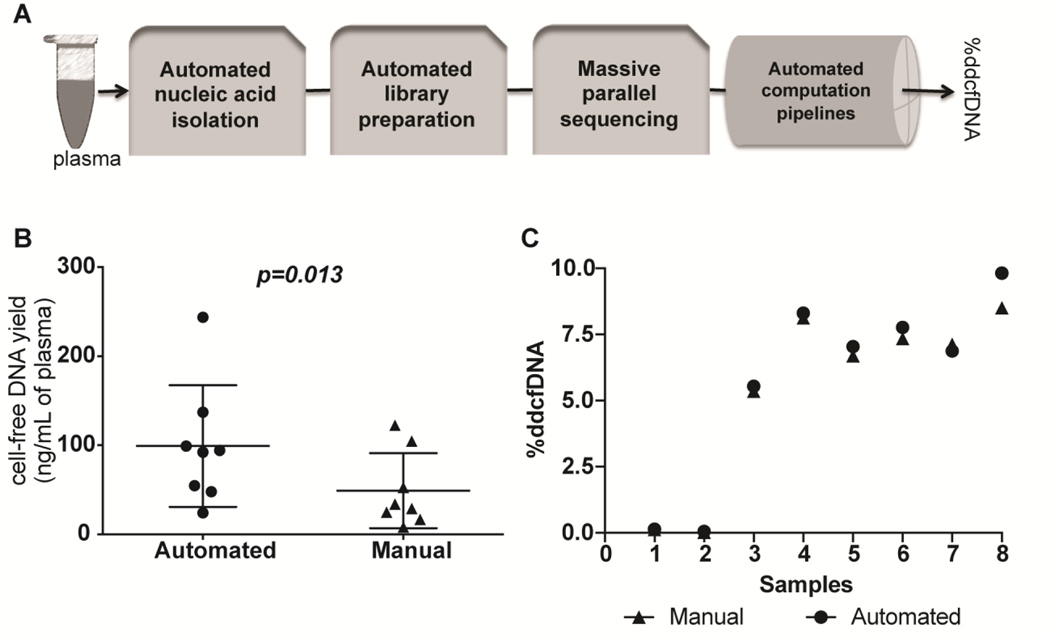
Reproducibility of %ddcfDNA measurements across manual and automated platforms
To enable %ddcfDNA measurement in a future large clinical study, we automated steps for DNA isolation (QiaSymphony, Qiagen) and library preparation (G3 liquid handler, Perkin Elmer) (Figure 3a). Using quantitative PCR, we ascertained that cell-free DNA yields from the automated-platform DNA extraction technique were higher than yields from the manual-platform DNA extraction technique ((99+24 ng/ml vs. 49+15 (mean/SD), p=0.013, Figure 3b)). After filtering non-aligned reads, PCR duplicates, and low-quality reads (18), both platforms produced a similar fraction of high-quality reads ((79+0.5% and 76+8% (p=0.376), respectively)). Calculated %ddcfDNA measurements showed strong correlation between the two platforms (slope=0.80, R2~0.92, p <0.001, (Figure 3c)). The measured error rates were lower with the automated platform than for the manual method (0.02+0.015 vs. 0.05+0.005, p<0.001).
Figure 3: Reproducibility of %ddcfDNA measurement on a high throughput automated workflow.
(a) An automated workflow was assembled with platforms for DNA isolation (QiaSymphony, Qiagen), library preparation (Sciclone G3 NGS workstation, Perkin Elmer), and high throughput sequencing (HiSeq 3000, Illumina). The workflow was coupled to an automated computational workflow with minimal manual manipulation. (b) Cell-free DNA extracted from 8 plasma samples using the semi-automated platform and the traditional manual platform (QIAAmp Circulating Nucleic Acid Kit, Qiagen) were quantified by quantitative PCR and compared using the Student t-test. The horizontal line represents the mean and the error bars represent the standard deviation. (c) The %ddcfDNA for samples analyzed on the automated and manual platforms. Linear regression of the two sets of measurements showed a slope =0.80, r2=0.92 and p-value <0.001. Testing for null hypothesis of slope=1 gives p-value=0.99
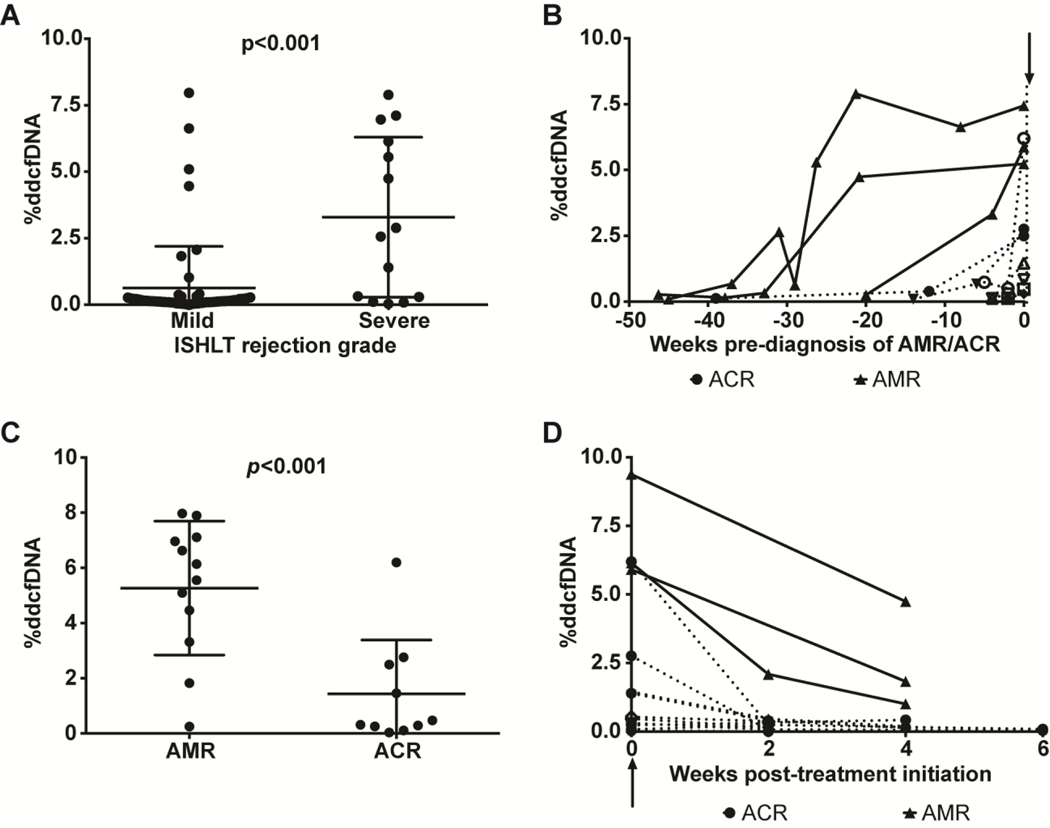
Correlation of %ddcfDNA with rejection severity
In a secondary analysis, we evaluated the relationship between %ddcfDNA and rejection severity. We measured %ddcfDNA from both cohorts (n=105) and compared the 13 measurements that coincided with either ACR or clinical AMR to the 86 measurements that coincided with <2R (mild) rejection (Figure 4a). Six samples without corresponding EMB grading were not included. The 13 ACR/AMR samples (5 Stanford, 8 NIH) had a median 2.72 %ddcfDNA compared to a median 0.14 %ddcfDNA (p<0.001) for mild rejection (Figure 4a). Of the 86 mild-rejection %ddcfDNA measurements, 19 showed %ddcfDNA higher than the previously reported threshold of 0.25% for ≥2R rejection in HTRs (2). Of these, 11 of the %ddcfDNA elevations preceded a severe rejection event (Figure 4b), and 4 %ddcfDNA elevations correlated with treatment for AMR or ACR. The median %ddcfDNA associated with AMR diagnosis (5.8%, n=12) was higher than for ACR (0.39%ddcfDNA, n=10) (p=0.003, Figure 4c). Elevations of %ddcfDNA preceded histopathologic and clinical diagnosis of AMR by 4 to 25 weeks and ACR diagnosis by 0–2 weeks (Figure 4b). These results indicate concordance between rejection severity and %ddcfDNA. The remaining 4 %ddcfDNA elevations showed no clinical or histopathological evidence of graft dysfunction, and %ddcfDNA levels returned to baseline by the time of the subsequent surveillance biopsy. We conjecture that those %ddcfDNA elevations represent subclinical graft injury, that is not detectable by current histopathological methods. For both AMR HTRs and ACR HTRs, %ddcfDNA levels returned to baseline after treatment (Figure 4d).
Figure 4: Correlation between %ddcfDNA and severity of graft injury.
a) The %ddcfDNA values were grouped as <grade 2R (no/mild) or ≥grade 2R rejection based on corresponding biopsy grades and compared (Mann-Whitney U test). (b) Temporal changes of %ddcfDNA preceding a histopathologic diagnosis of ≥grade 2R rejection. Arrow represents time of diagnosis of ≥grade 2R. Interval preceding ≥grade 2R diagnosis (x-axis) was computed by subtracting diagnosis date from sample collection date. Within-subject %ddcfDNA values are linked for subjects diagnosed with AMR (solid lines) or ACR (dashed lines). (c) The %ddcfDNA was compared for AMR vs. ACR diagnoses (Mann Whitney U test), showing significantly higher %ddcfDNA measurements associated with AMR compared to ACR. (d) Temporal changes of %ddcfDNA after initiation of treatment for rejection (arrow). The interval post-treatment was computed as the difference between sample collection date and treatment initiation date.
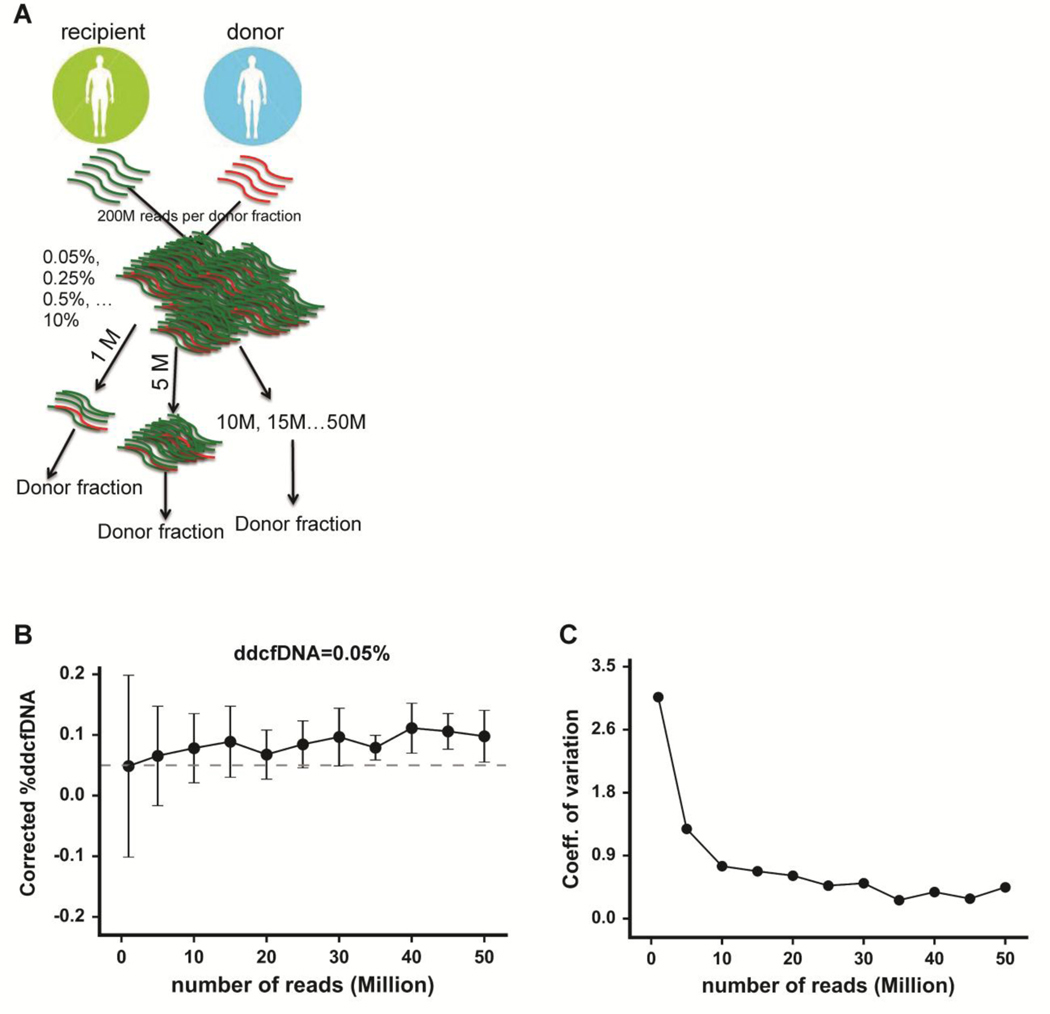
Determining optimal sequencing depth for accurate %ddcfDNA measurements
Genomic tools to clinical management must be both accurate and efficient. Thus, we sought to optimize the threshold sequencing depth to maintain assay precision while minimizing cost and processing time. As shown in Figure 5a, we first constructed two mock genomes using the hg 19 reference genome (www.ucsc.edu) and paired donor/recipient genotype data. The mock genomes were used to create mock master files (19), each with 200 million reads and different donor:recipient ratios (0%−10%). We sampled 1 million reads 10 separate times from each master file, with replacement after each sampling to compute %ddcfDNA. A similar sampling test was repeated at higher depths of 5–50 million reads. We performed two independent simulations, thus yielding 20 %ddcfDNA replicate measurements.
Figure 5: Determining the optimal sequencing depth for precise %ddcfDNA measurement.
(a) A pair of donor and recipient genotypes and the hg19 reference was utilized to simulate two reference genomes. Mock master files of 200 million reads (≈160 bp) per file were generated, each file containing a different fraction of donor reads (0%, 0.05%, 0.25%, 0.5%, 1.25%, 2.5%, 5%). A subset (1, 5, 1o, 15… million) of each master mock file was randomly sampled to determine %ddcfDNA. The analysis was repeated 20 times for each read depth, with replacement after each random sampling. (b) A plot of measured %ddcfDNA against read depth for expected %ddcfDNA of 0.05%. The solid dots represent the mean of the twenty %ddcfDNA values for each read depth. The dashed line shows the expected (true) %ddcfDNA of 0.05%. Error bars represent the standard deviation. (c) A plot of the coefficient of variance (standard deviation/mean) against read depth for expected %ddcfDNA of 0.05%. The arrow points to a read depth of 10 million, beyond which the CV showed less variation.
The 20 replicate %ddcfDNA values fluctuated with respect to expected %ddcfDNA levels, with more fluctuation observed at lower read depths (Figure 5b, suppl. Figure 2a–c). The coefficient of variance (CV) of the %ddcfDNA measurements was initially high at low read depths, then dropped, and remained low thereafter (Figure 5c, suppl. Figures 2d–f). The read depth at which the CV dropped to its lowest point varied inversely with the expected %ddcfDNA (15 million for 0%, 10 million for 0.05%, and 5 million for ≥ 0.25%). We validated these findings using cell-free DNA mixtures made using cell-free DNA from two healthy controls (suppl. Figure 3a–b). From these data, we defined an appropriate threshold read depth of 10–15 million to measure %ddcfDNA levels of 0.05% or higher with minimal variability.
DISCUSSION
With increasingly cheaper technologies, there has been an explosion of genomic analyses that may soon find use in clinical settings. In the absence of rigorously defined standards, concerns have arisen about potential low reproducibility of genomic approaches (16, 20). Having developed a highly sensitive and non-invasive genomic tool (%ddcfDNA) for detecting thoracic-organ rejection, we sought to assess its reproducibility, toward future use in clinical management of HTRs. To achieve this goal, we have developed a three-step process, guided by previous reports (21–24) and a recent NIH reproducibility workshop.
In this study, we established a local (NIH) sequencing and computational workflow to mirror that originally described for %ddcfDNA measurement (2). We then reproduced the workflow using several analytical approaches. These analyses revealed that our %ddcfDNA assay that employs shotgun sequencing is robust, reproducible, and precise as demonstrated by strong agreement between measured %ddcfDNA in technical replicates from independent laboratories and in manual vs. automated platforms.
This study also shows that %ddcfDNA levels correlate precisely with rejection phenotype (ACR, AMR), as well as with severity of graft injury. Our observation that %ddcfDNA elevations precede histopathologic diagnosis of rejection, most strikingly in HTRs with AMR, suggest that elevated %ddcfDNA levels may be used as a marker to initiate treatment earlier than is now feasible, to prevent graft damage and poor health outcomes. The short half-life of %ddcfDNA (15 minutes) and temporal kinetics of %ddcfDNA that decline with rejection treatment also suggest that %ddcfDNA can be an effective tool to monitor treatment efficacy with high temporal resolution. In some HTRs, we observed transient %ddcfDNA elevations without histologic evidence of injury; these may represent self-resolving processes such as ACR. Clinical data on cardiac allograft vasculopathy (CAV) was not collected in this study. Future studies in our group are poised to examine the relationship between %ddcfDNA and CAV. We acknowledge that ischemic reperfusion injury may contribute to early elevations in %ddcfDNA. However, because such initial elevations decay to baseline low-level %ddcfDNA levels within the first week of transplantation (2), we propose that this tool can be used to monitor for rejection earlier than possible with existing tools such as AlloMap®.
Despite these advantages, the performance of the %ddcfDNA assay in conditions that affect cell-free DNA clearance such as kidney and liver disease remains unknown, and require further research. Further, the %ddcfDNA assay may be limited by its labor-intensity and long processing time and the need for genotyping. Our observations that %ddcfDNA was consistently higher (2–5 fold) in AMR compared to ACR may be explored further to determine if %ddcfDNA thresholds can distinguish precisely between the two conditions or used to guide administration of confirmatory tests such as biopsy or DSA measurement. Intriguingly, since T- and B-cell receptor repertoires can be identified by measuring %ddcfDNA levels (14), the method may enable concurrent examination of immunological processes underlying specific rejection phenotypes.
Our results confirm the precision and accuracy of an automated workflow. We also observed that by automating DNA isolation and library preparation, we increased assay capacity 12-fold without compromising DNA yield or assay precision. Coupling the lower sequencing depth (15 million compared to >30 million in our previous assay) and increased automated capacity reduced assay processing time from 10–21 days to 2–3 days and reduced individual assay cost from $330 to $200; the one-time $315 genotype cost for a donor/recipient pair is not included in this price calculation.
The need for genotyping remains a limitation of this %ddcfDNA assay since genotyping requires pre-transplant samples for DNA isolation, which may not be readily available. Recently, novel approaches that measure %ddcfDNA without the need for genotype have been reported (25, 26). If reproduced compared with the shotgun approach, these approaches may obviate the need for genotyping without compromising assay accuracy. With the proposed algorithm used in this study, and the democratization of integrative computational platforms such as Galaxy (27), BioExtract (28), and others (16) to facilitate publication workflows, validation of these genomic tools is likely to become an easier task.
In summary, we show the reproducibility of using %ddcfDNA to quantify allograft injury after heart transplantation, paving the way for larger cohort and clinical utility studies using this tool.
Supplementary Material
ACKNOWLEDGEMENTS
Funding
This study was funded by intramural funds of the National Institutes of Health.
Footnotes
Disclosures
The authors have no conflicts to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Marboe CC, Billingham M, Eisen H, et al. : Nodular endocardial infiltrates (Quilty lesions) cause significant variability in diagnosis of ISHLT Grade 2 and 3A rejection in cardiac allograft recipients. J Heart Lung Transplant 2005;24:S219–26. [DOI] [PubMed] [Google Scholar]
- 2.De Vlaminck I, Valantine HA, Snyder TM, et al. : Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014;6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pham MX, Teuteberg JJ, Kfoury AG, et al. : Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med 2010;362:1890–900. [DOI] [PubMed] [Google Scholar]
- 4.Jelencsics K, Oberbauer R: microRNA and Kidney Transplantation. Adv Exp Med Biol 2015;888:271–90. [DOI] [PubMed] [Google Scholar]
- 5.Halloran PF, de Freitas DG, Einecke G, et al. : The molecular phenotype of kidney transplants. Am J Transplant 2010;10:2215–22. [DOI] [PubMed] [Google Scholar]
- 6.Sellares J, de Freitas DG, Mengel M, et al. : Understanding the causes of kidney transplant failure: the dominant role of antibody-mediated rejection and nonadherence. Am J Transplant 2012;12:388–99. [DOI] [PubMed] [Google Scholar]
- 7.Sis B, Jhangri GS, Bunnag S, Allanach K, Kaplan B, Halloran PF: Endothelial gene expression in kidney transplants with alloantibody indicates antibody-mediated damage despite lack of C4d staining. Am J Transplant 2009;9:2312–23. [DOI] [PubMed] [Google Scholar]
- 8.Holweg CT, Potena L, Luikart H, et al. : Identification and classification of acute cardiac rejection by intragraft transcriptional profiling. Circulation 2011;123:2236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder TM, Khush KK, Valantine HA, Quake SR: Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci U S A 2011;108:6229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart S, Winters GL, Fishbein MC, et al. : Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J Heart Lung Transplant 2005;24:1710–20. [DOI] [PubMed] [Google Scholar]
- 11.De Vlaminck I, Martin L, Kertesz M, et al. : Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A 2015;112:13336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun K, Jiang P, Chan KC, et al. : Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A 2015;112:E5503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vlaminck I, Khush KK, Strehl C, et al. : Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 2013;155:1178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vollmers C, De Vlaminck I, Valantine HA, et al. : Monitoring pharmacologically induced immunosuppression by immune repertoire sequencing to detect acute allograft rejection in heart transplant patients: a proof-of-concept diagnostic accuracy study. PLoS Med 2015;12:e1001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J: Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016;164:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nekrutenko A, Taylor J: Next-generation sequencing data interpretation: enhancing reproducibility and accessibility. Nat Rev Genet 2012;13:667–72. [DOI] [PubMed] [Google Scholar]
- 17.Billingham ME, Cary NR, Hammond ME, et al. : A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: Heart Rejection Study Group. The International Society for Heart Transplantation. J Heart Transplant 1990;9:587–93. [PubMed] [Google Scholar]
- 18.Li H, Durbin R: Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Li L, Myers JR, Marth GT: ART: a next-generation sequencing read simulator. Bioinformatics 2012;28:593–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mack SJ, Milius RP, Gifford BD, et al. : Minimum information for reporting next generation sequence genotyping (MIRING): Guidelines for reporting HLA and KIR genotyping via next generation sequencing. Hum Immunol 2015;76:954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aziz N, Zhao Q, Bry L, et al. : College of American Pathologists’ laboratory standards for next-generation sequencing clinical tests. Arch Pathol Lab Med 2015;139:481–93. [DOI] [PubMed] [Google Scholar]
- 22.Salto-Tellez M, Gonzalez de Castro D: Next-generation sequencing: a change of paradigm in molecular diagnostic validation. J Pathol 2014;234:5–10. [DOI] [PubMed] [Google Scholar]
- 23.Sandve GK, Nekrutenko A, Taylor J, Hovig E: Ten simple rules for reproducible computational research. PLoS Comput Biol 2013;9:e1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studies N-NWGoRiA, Chanock SJ, Manolio T, et al. : Replicating genotype-phenotype associations. Nature 2007;447:655–60. [DOI] [PubMed] [Google Scholar]
- 25.Gordon PM, Khan A, Sajid U, et al. : An Algorithm Measuring Donor Cell-Free DNA in Plasma of Cellular and Solid Organ Transplant Recipients That Does Not Require Donor or Recipient Genotyping. Frontiers in cardiovascular medicine 2016;3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grskovic M, Hiller DJ, Eubank LA, et al. : Validation of a Clinical-Grade Assay to Measure Donor-Derived Cell-Free DNA in Solid Organ Transplant Recipients. J Mol Diagn 2016;18:890–902. [DOI] [PubMed] [Google Scholar]
- 27.Giardine B, Riemer C, Hardison RC, et al. : Galaxy: a platform for interactive large-scale genome analysis. Genome Res 2005;15:1451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lushbough CM, Brendel VP: An overview of the BioExtract Server: a distributed, Web-based system for genomic analysis. Adv Exp Med Biol 2010;680:361–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.