Abstract
In recent years, undifferentiated small round cell sarcoma (USRCS) have been divided into a variety of new, rare, sarcoma subtypes including the group of Ewing-like sarcoma that has the morphological appearance of Ewing sarcoma but carries CIC-DUX4, BCOR-CCNB3 and other gene fusions different from the classical EWSR1-ETS. Using high-throughput RNA-seq analyses we identified a novel recurrent gene fusion, CRTC1-SS18, in two cases of USRCS that lacked any known translocation. RNA-seq results were confirmed by RT-PCR, long-range PCR and FISH. In vitro, we show that the cells expressing the gene fusion were morphologically distinct and demonstrated enhanced oncogenic potential compared to control cells. Expression profile comparisons with tumours of other sarcoma subtypes demonstrated that both cases clustered close to EWSR1-CREB1-positive tumours. Moreover, these analyses indicated an enhanced NTRK1 expression in CRTC1-SS18-positive tumours. We conclude that the novel gene fusion identified in this study adds a new subtype to the USRCS with unique gene signatures and maybe of therapeutic relevance.
Keywords: Undifferentiated small round cell sarcoma, Ewing Sarcoma, RNA-seq, gene fusion
Introduction
Diagnosing small round blue cell tumours on biopsy has been challenging because of their lack of specific features in small specimens. Amongst undifferentiated small round cell sarcoma (USRCS), Ewing-like Sarcoma (ELS) shares some of the morphological features of Ewing Sarcoma (ES), but lacks the classical EWSR1-ETS gene fusion [1–2]. ES are mainly characterised by chromosomal translocations at chromosome 22q12 that fuse the EWSR1 gene with one of the ETS gene family of transcription factors, such as FLI1 or ERG, in 90–95% of ES cases. The classical ES gene fusion protein acts as an oncogene and plays an essential role in tumorigenesis and proliferation of ES cells [2]. Recent studies have identified recurrent gene fusions in ELS; namely; CIC-DUX4 and BCOR-CCNB3 [3–6]. The identification of these gene fusions suggests that other, yet to be identified, gene fusions could be associated with this type of tumour. Furthermore, novel gene fusions in ELS have been reported recently, in case reports, including CIC-FOXO4 BCOR-MAML3 and ZC3H7B-BCOR [7–9]. Identifying these genetically defined entities may contribute towards understanding the pathogenesis and the behaviour of these tumours.
By applying RNA sequencing technology to investigate USRCS, we discovered a novel CRTC1-SS18 gene fusion in two samples from two different cancer centres. Combining both samples we were able to find similarities at clinical, pathological and molecular levels. Moreover, by cloning the fusion gene we were able to demonstrate its oncogenic properties, adding the CRTC1-SS18 fusion gene to the increasing number of described oncogenes.
Materials and Methods
One patient sample (Case 1) was obtained from the Royal Orthopaedic Hospital NHS Foundation Trust Tumour Bank (with permission; REC 12/EM/0048). The other patient sample (Case 2) was obtained from resection material sent to the Centre Léon Bérard for molecular diagnosis. Both samples were acquired with informed consent from the patient and or/next of kin and ethical approval from institutional and local research committee boards. Patient samples were anonymised and used in accordance with the principles expressed in the Declaration of Helsinki.
Immunohistochemistry
Case1:
For immunohistochemistry, 2 μm thick sections were cut and antigens retrieved in an epitope retrieval solution of pH 8 (RE7116; Novocastra, Newcastle, UK) at 68 °C for 17 h in a stirred water bath. The antibody clones, dilutions and sources are as follows: CD99 (12E7, 1:25; Dako, Ely, UK), vimentin (V9, 1:100, Novocastra), CD31 (JC70, 1:100, Dako), CD34 (QBend10, 1:50, Dako), cytokeratin AE1/AE3 (1:100, Dako), CD45 (2B11+PD7/26, Dako), cytokeratin MNF116 (1:50, Dako), desmin (D33, 1:100, Dako), α-smooth muscle actin (SMA) (1A4, 1:200, Dako), epithelial membrane antigen (EMA) (E29, 1:100, Dako), HMB-45 (1:200, Dako), S100 (NCL-L-S100p, 1:1000, Novocastra), WT1 (C-19, 1:500, Santa Cruz, Insight Biotechnology Limited, PO Box 520, Wembley, Middlesex,UK), TLE1 (M-101, 1:50, Santa Cruz), ERG (Erg-1/2/3 C-1; 1:50, Santa Cruz), INI1 (1:25, BD Transduction Laboratories (BD Biosciences), Becton Dickinson UK, Oxford, UK), BCOR (C-10, 1:50, Santa Cruz), ETV4 (16, 1:50, Santa Cruz) and Ki-67 (MIB1, 1:200, Dako).
Case 2:
Sections were cut at 4 μm thick from formalin-fixed-paraffin-embedded (FFPE) tissue and immunostained using a VentanaBenchmark XT automatic stainer (Ventana, Tuscon, AZ, USA). Signals were revealed using the ultraView Universal Dab Detection kit (Ventana). The following antibodies were used: CD99 (12E7, Dako), EMA (E29, 1:50, Dako), desmin (D33, 1:80, Dako), cytokeratin AE1/AE3 (AE1/AE3, 1:50, Dako), caldesmon (h-CD, 1:100, Dako), myogenin (F5D, 1:100, Dako), S100 protein (Z0311, 1:800, Dako), CD34 (QBend-10, 1:25, Dako), INI1 (25, 1:50, BD Transduction Laboratories), BCOR (C-10, 1:50, Santa Cruz), ETV4 (16, 1:50, Santa Cruz). Immunohistochemistry for NTRK1 was performed using a 32 min incubation with the NTRK1 antibody (ab76291, clot EP1058Y, Abcam, dilution 1:200) on a Ventana ULTRA machine with Cell Conditioning Solution 1 pre-treatment for 64 min.
Fluorescence in situ hybridization (FISH) analyses
FISH analyses were performed on formalin-fixed paraffin-embedded (FFPE) tissue sections using the ZytoLight SS18 Dual Color Break Apart Probe (#Z-2097–200, Zytovision, Bremerhaven, Germany) by assessing at least 100 non-overlapping intact nuclei by two independent operators. The positive threshold to call the FISH assay positive was 15%.
Comparative Genomic Hybridization on array (CGH-array) analyses
Genomic DNA was extracted from FFPE tissue using a QIAamp, DNA micro kit (Qiagen, Hilden, Germany). Genomic DNA and human reference DNA (Promega) were labeled with cyanine 5 (Cy5) and cyanine 3 (Cy3), respectively, using the Genomic DNA High-Throughput ULS Labeling Kit (Agilent Technologies, Santa Clara, CA, USA) and co-hybridized onto a Sureprint G3 Human CGH microarray 4×180K (Agilent Technologies) following manufacturer’s recommendations. Data were analysed using Agilent Genomic Workbench software v7.0, or by Cytogenomics software (v2.9.2.4, Agilent), and expressed according to the human reference genome hg19 (GRCh37, Genome Reference Consortium Human Reference 37). The identification of aberrant copy number segments was based on ADM-2 segmentation algorithm with a threshold of 6.0.
Fresh frozen tissue RNA sequencing
Total RNA was extracted from fresh frozen tissue (Case 1) using a Qiagen RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. RNA quality and quantity was measured with Agilent 2100 bioanalyzer (Agilent Technologies). An RNA sample was sent to Oxford Gene Technology (OGT; Begbroke, Oxfordshire, UK) to perform RNA sequencing (RNA-Seq) using the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA). In brief, cDNA libraries were prepared from 1 μg total RNA using the Illumina TruSeq RNA Sample Prep Kit v2. All sequencing was paired-end (100 bp) and performed over 100 cycles and the read files (Fastq) were generated from the sequencing platform via the manufacturer’s software. Mapping and alignment procedure were processed through the Tuxedo suit. The human sequence genome (hg19) was used as a reference and aligned to the sequence reads. FusionCatcher software was used to identify gene fusions from RNA-seq data [10]. RNA-seq data has been deposited (SRA accession: SRP131744).
FFPE RNA sequencing
RNA was extracted from FFPE tissue sections (Case 1 and Case 2) using Trizol reagent (Thermo Fisher Scientific, Courtaboeuf, France) and extracted by subsequent phenol/chloroform. RNase-free DNase Set (Qiagen, Courtabouef, France) was used to remove DNA. The DNase was eliminated by a further Trizol extraction. All RNA were quantified by spectrophotometry (NanoDrop; Thermo Fisher Scientif) and quality was controlled (DV200 value cutoff > 13%) using a TapeStation with Hs RNA ScreenTape (Agilent Technologies). One hundred nanograms of total RNA was used to prepare a library using a TruSeq RNA Access Library Prep Kit (Illumina). Fourteen libraries were pooled at 4 nM with 1% PhiX as an internal control. Sequencing was performed (75 cycles, paired end) using a NextSeq 500/550 High Output V2 kit and an Illumina NextSeq 500 (Illumina). Alignments were performed using STAR algorithm [11] against the GRCh38 reference genome and fusion gene assessments were made using STAR-Fusion [12], FusionCatcher [10] and FusionMap [13] tools. Expression profiles were extracted from fastq files using Kallisto [14], and transformed as log2(TPM+2) prior to quantile normalization using the Limma package v 3.32.2 performed in the R environment v3.4.1 [15]. Only genes with a coding sequence annotation (based on Ensembl GRCh38p5 annotation) and with a maximum expression value across all samples above 2 were considered for the clustering analysis, which was performed using Ward’s distance on the 10% most variant genes based on their interquartile range. RNA-seq data has been deposited (SRA accession: SRP131744).
RT-PCR and Sanger Sequencing
cDNA was generated from total RNA using SuperScript III (Invitrogen) and random primers (Promega). The RT-PCR reactions were performed using 2.5 μl 10X buffer, 2.5 μl dNTPs (2.5 mM), 5 μl 5X GC rich solution, 1 μl Forward Primer (20 pmol), 1 μl Reverse Primer (20 pmol), 0.1 μl Fast Start DNA polymerase (Roche, Burgess Hill, UK). Primer sets used for PCR amplification were CRTC1-F (TCGAACAATCCGCGGAAATT) and SS18-R (GTGCTGGTAAAAGAGACTGCA) and PCR products were visualized using 2% (w/v) agarose gel (Bioline). The PCR products were extracted from the gel and purified using a QIAquick Gel Extraction Kit (Qiagen). A BigDye Terminator V3.1 kit (Applied Biosystems) was used for the cycle sequencing reaction, and PCR products of CRTC1-SS18 gene fusions were directly Sanger sequenced using an ABI 3730 DNA analyser (Applied Biosystems).
Long-Range-PCR (LR-PCR)
LR-PCR was carried out using PrimeSTAR GXL DNA polymerase (Takara Bio, Shiga, Japan). Each LR-PCR reaction was set up using 50 ng DNA, 1 × 5X PrimeSTAR GXL buffer, 200 μM each dNTPs, 0.2 μM of forward primer, 0.2 μM of reverse primer, 1.25 U PrimeSTAR GXL DNA polymerase enzyme, made up to a final volume of 50 μl using sterilized distilled water. The PCR was carried out using the following conditions; 30 cycles of [10 s at 98 °C and 10 min at 60 °C]. The LR-PCR primers used in this study are listed in supplementary materials, Table S1. The size of the PCR product from the gene fusion was unknown; therefore genome walking through both genes and a rough estimation of the product size was carried out. The genome walking covered the exonic and intronic regions of both genes involved in the fusion. LR-PCR was performed on the genomic DNA of the tumour sample. One forward primer (F3) was anchored on exon 1 of the CRTC1 gene and different reverse primers spanning ~2.5 kbp of intron 1 were used to amplify this region and to identify the breakpoint. The samples were electrophoresed on 0.9% agarose gels to determine the size of the PCR product. After confirming the breakpoint of CRTC1-SS18 gene fusion at the genomic level, LR-PCR was performed on both the tumour sample and the corresponding normal tissue to confirm that this fusion was somatic. The PCR-product was then extracted from the gel and sequenced.
Plasmid construction.
The CRTC1-SS18 expression construct was made by PCR amplification of the entire fusion construct using the cDNA generated from Case 1 tumour RNA. This amplicon was sub-cloned into the expression vector, pFlag-CMV-4 (Sigma-Aldrich) using EcoRI and XbaI restriction sites. The primers used were Forward: 5’-cg g aat tcg aag atg gcg act tcg aac aat c-3’ and, Reverse: 5’- cg tctaga t tca ctg ctg gta att tcc ata c-3’. Plasmid constructs were verified by sequencing. Expression of this plasmid generates an N-terminal FLAG-tagged protein. The construct plasmid and associated empty vector were transfected into HEK293 cells (ATCC, Manassas VA, USA), clones were isolated and expression was validated by western blotting using an anti-FLAG antibody (Cat.No. F1804, clone M2, 1:1000 Sigma-Aldrich). Clones were maintained in DMEM and 10% fetal bovine serum supplemented with 1 mg/ml G418 (Life Technologies).
Cell migration assays
Suspensions containing 25,000 cells of a stable CRTC1-SS18 expressing HEK293 clone, or a clone containing the empty vector, suspended in serum free DMEM media were seeded into a 24-well format Boyden chamber cell culture insert (8 μm pore size, PET membrane) (BD Falcon, Bedford MA, USA). The lower chamber contained DMEM, 10% FCS as an attractant. Chambers were incubated for 16 h. Cells were fixed in methanol; cells on the upper side of the chamber were removed and those remaining on the underside stained with crystal violet. Migrated cells were photographed then the crystal violet was solubilized in 500 μl of 33% acetic acid and the optical density measured at 540 nm (n=20).
Cell invasion assays
A modified migration assay was carried out in which 50,000 cells were seeded into Boyden chambers (8 μm pore size, PET membrane) (BD Falcon) pre-coated with 100μl of Geltrex basement membrane matrix (Thermo Scientific). Chambers were incubated for 16 h, Invaded cells were counted by microscopy with the observer unaware of the cell type (n=22).
Soft agar, anchorage-independent growth assay
Anchorage independent growth in soft agar was assessed using the CytoSelect 96-Well Cell Transformation Assay kit (Cell Biolabs San Diego CA, USA) following the manufacturer’s instructions. In brief, 2,500 cells were seeded, in agar, supplemented with DMEM, 10% FCS, per well. Following incubation for 8 d, the agar was solubilized and viable cells were lysed, stained and quantified by fluorometry (excitation 492 nm, emission 520 nm; n=8). ).
Results
Clinical presentation and pathological findings of the index case, Case 1.
A 35 year old man presented to The Royal Orthopaedic Hospital with a rapidly growing lump on his right thigh. Magnetic resonance imaging revealed a heterogeneous mass within the sartorius muscle and the findings were in keeping with a soft tissue sarcoma. A diagnosis of USRCS was made on the biopsy material of the tumor which did not carry the chimeric gene fusions associated with Ewing sarcoma (EWSR1-FLI1 or ERG), mesenchymal chondrosarcoma (EWS–NR4A3 and TAF2N-NR4A3), DSRCT (EWS-WT1) or synovial sarcoma (SS18-SSX). The tumour was also negative for CIC-DUX4 and BCOR-CCNB3. In view of this diagnosis the patient received four cycles of vincristine, ifosfamide, doxorubicin and etoposide (VIDE). Radiologically there was no response to chemotherapy and the mass was excised. The patient developed bilateral lung metastases 18 months after diagnosis and died of disease 92 months later.
Pathological Findings: grossly, an intramuscular, well circumscribed grayish white fleshy tumour with haemorrhagic and necrotic foci measuring 70×65×53 mm was observed (Figure 1A). Histologically, the tumour consisted of solid sheets and nests of small round cells surrounded by desmoplastic stroma reminiscent of desmoplastic small round cell tumour. The tumour cells had scant amounts of eosinophilic cytoplasm and small irregularly shaped round nuclei with stippled chromatin. Some had prominent grooves and small nucleoli. Focal areas of necrosis and mitotic figures were identified (7 per 10 high power field (HPF)). Rosette formation and glandular differentiation were not identified (Figure 1B, C). Immunohistochemically, the tumour cells were diffusely positive for vimentin and CD99 (Figure 1D). The tumour cells did not stain for CD31, CD34, AE1/AE3, CD45, CK(MNF116), desmin, EMA, HMB45, SMA, S100, HMB45, WT1, TLE-1, ERG, ETV4, BCOR or CCNB3. INI1 was retained. The Ki-67 labelling index was up to 20%.
Figure 1. Macroscopy and microscopy images of the Case 1.
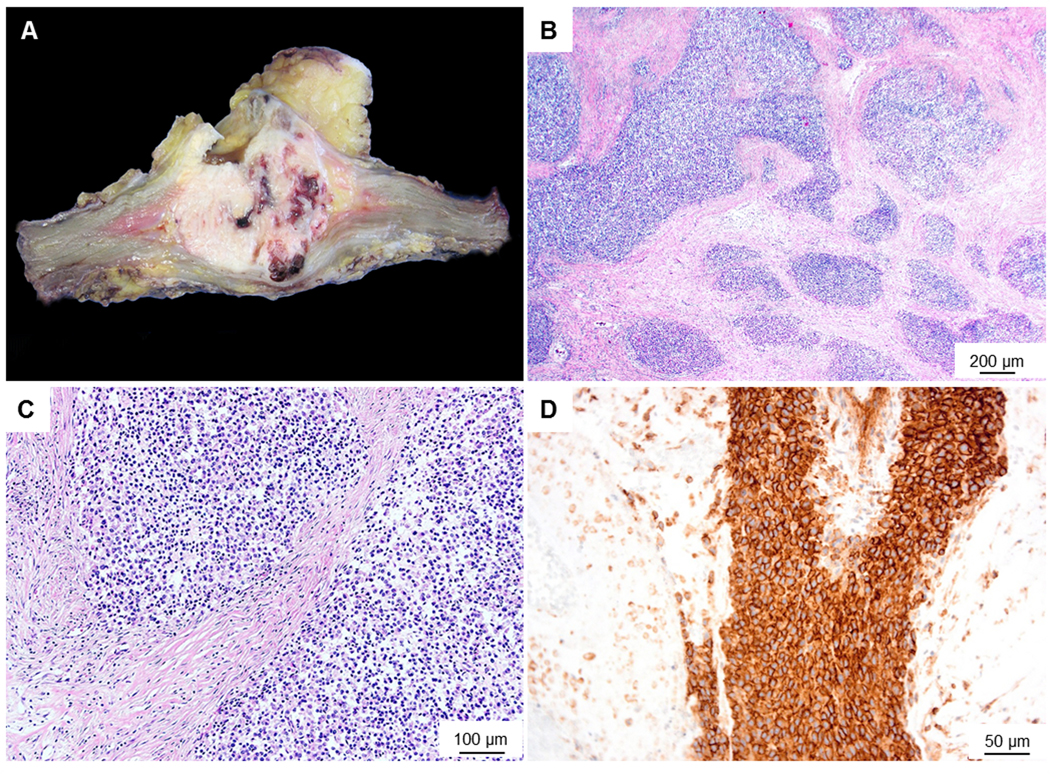
(A) Macroscopic image shows a fairly well circumscribed fleshy tumour with foci of necrosis and haemorrhage. (B) Microscopic image (low-power) shows the tumour to be composed of solid sheets and nests surrounded by a desmoplastic stroma. (C) High power view of the tumour shows small sized cells with scant cytoplasm. (D) Membranous staining with CD99- mimicking Ewing’s sarcoma.
RNA-seq analysis and confirmation of gene fusion
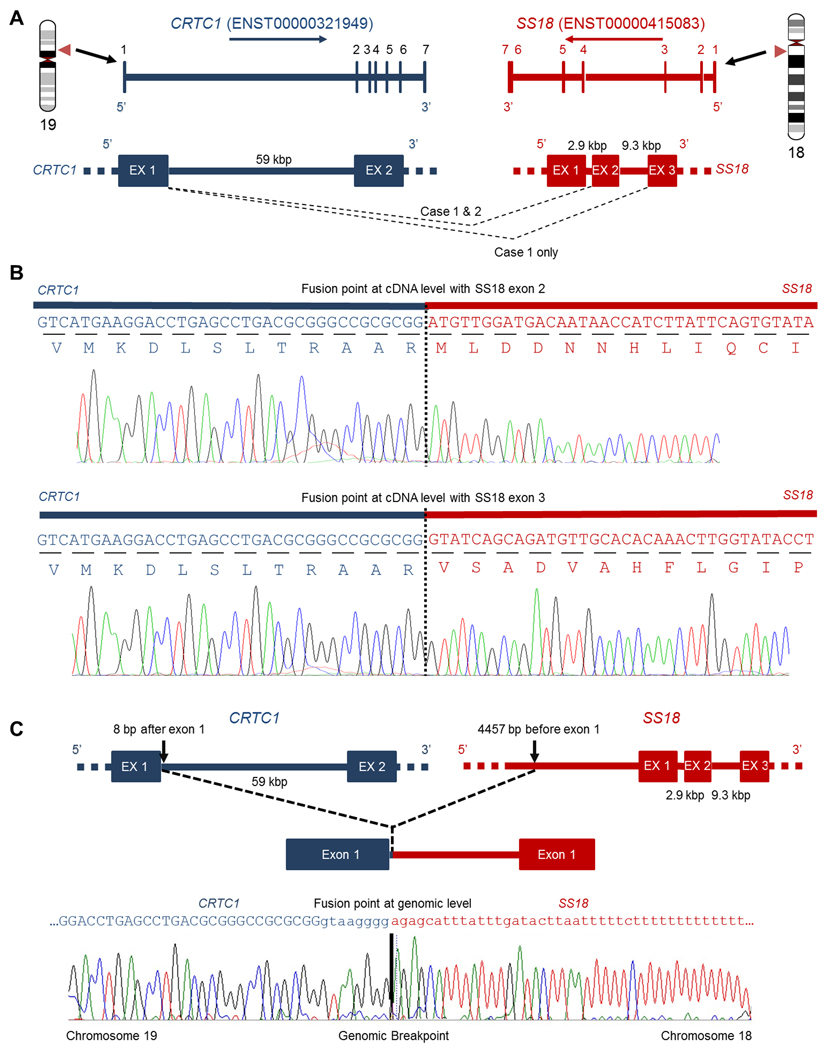
RNA-seq analysis of the index case (Case 1) revealed a novel gene fusion involving CRTC1 and SS18 genes in the tumour sample. Two alternative splicing fusion transcripts were detected that linked exon 1 of the CRTC1 gene with exon 2 or exon 3 of the SS18 gene (Figure 2A,B; supplementary material, Figure S1). A balanced translocation resulted in the SS18 gene being fused with exon 1 of the CRTC1 gene, generating an in-frame fusion protein (Figure 2A,B and Figure 4A). The CRTC1-SS18 fusion transcripts were confirmed by RT-PCR and Sanger sequencing (Figure 2B and supplementary material, Figure S2A). In order to map the fusion breakpoints at the genomic level, Long Range-PCR was carried out to reveal the genomic sequence around the breakpoints (supplementary material, Figure S2B–D and Table S1). The CRTC1 breakpoint was found to be 8 bp from the 3’end of exon 1 (cDNA fusion point) and the SS18 breakpoint was 4457 bp before the 5’end of exon 1 (Figure 2C).
Figure 2. RNA-Seq identification of the CRTC1-SS18 gene fusion.
(A) The genomic intron/exon structure of the CRTC1 gene (blue) and SS18 (red). CRTC1(Ex1)-SS18(Ex2) gene fusion was found in Case 1 and Case 2, whilst CRTC1(Ex1)-SS18(Ex3) alternative splicing gene fusion was found only in Case 1. (B) Sanger sequencing chromatogram of the RT-PCR product is shown and confirmed the CRTC1(Ex1)-SS18(Ex2) fusion junction and the CRTC1(Ex1)-SS18(Ex3) fusion junction in Case 1. (C) Schematic of the Exon-intron structure of the CRTC1-SS18 gene fusion at the DNA level. The intergenic breakpoint for both genes is shown and was confirmed by Sanger sequencing of the LR-PCR product in Case 1.
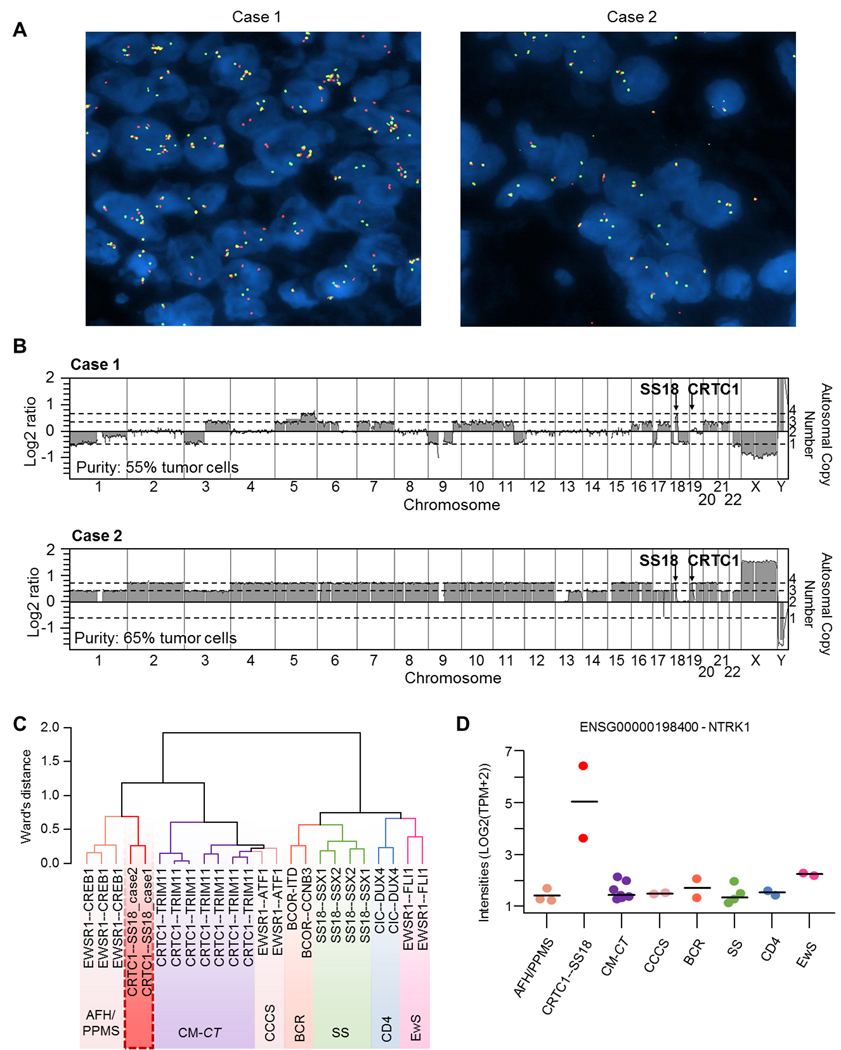
Figure 4.
Genomic and transcript profiles of CRTC1-SS18 positive sarcomas. (A) FISH analysis using an SS18 break-apart probe, showing multiple copies of chromosome 18 carrying the SS18 gene. Most cells were tetrasomic or greater for the SS18 locus on chromosome 19. In case 1, the translocation was balanced (presence of red dots) whereas in case 2 the translocation was unbalanced (the red signal is lost while the green signal remains). (B) Array-comparative genomic hybridization (aCGH) profiles. Case 1 presented a diploid genome with the loss of chromosome 1, 3p, 9, part of 11q, 17p, 18 and 22, together with gains on chromosomes 3q, 5, 6, 7, 9, 10, 11, 16, 17, 20 and 21. The region around the SS18 gene on chromosome 18 was focally gained while the CRTC1 gene locus was normal, which is in accordance with a balanced translocation. Case 2 presented a tetraploid genome with the loss of one copy of chromosomes 1, 3, 13, 14, 17, 21 and 22 and with homozygous deletion of chromosome region 17q22, impacting the MBTD1, UTP18 and CA10 genes. The 5’ region of SS18 (on the minus strand) and 3’ region of CRTC1 (on the plus strand) were lost, in accordance with an unbalanced translocation. (C) Hierarchical clustering of RNAseq data placed the two CRTC1-SS18 cases close to EWSR1-CREB1-positive tumours. (D) NTRK1 expression from RNAseq data. AFH/ PPMS: Angiomatoid fibrous histiocytoma / Primary pulmonary myxoid sarcoma; CCCS: cutaneous clear cell sarcomas; CM-CT: Cutaneous Melanocytomas with CRTC1-TRIM11 fusion; BCR: BCOR-Rearranged sarcomas; CD4: CIC-DUX4 sarcomas; EwS: Ewing Sarcomas; SS: synovial sarcomas.
Clinical presentation and pathological findings of Case 2.
RNA sequencing revealed a second case of a CRTC1-SS18 positive USRCS (Case 2): a 42 year old woman who presented to the CHU Gui de Chauliac (Montpellier, France) with a mass from the popliteal fossa, later diagnosed as an undifferentiated small cell sarcoma. The patient underwent radio- and chemo-therapy prior to the surgical removal of the tumour. A tumour fragment was sent to the Centre Léon Bérard’s department of pathology for a second opinion and molecular diagnosis.
Pathological findings: An intramuscular mass, well circumscribed by a diffuse calcified matrix, measuring 110×75×70 mm was observed. The tumour consisted of bundles and nests of cells embedded in a fibrous stroma focally myxoid (Figure 3A). There were areas of necrosis. Cytologically, the tumour was composed of oval to epithelioid cells (Figure 3B). The cells were medium size with abundant eosinophilic cytoplasm and ovoid and vesicular nuclei with nucleoli (Figure 3C). Immunohistochemically, the tumour cells were positive for CD99 staining and negative for keratins, EMA, desmin, caldesmon, myogenin, S100 protein, CD34, ETV4 and BCOR. INI1 expression was retained. NTRK1 was diffusely positive.
Figure 3.
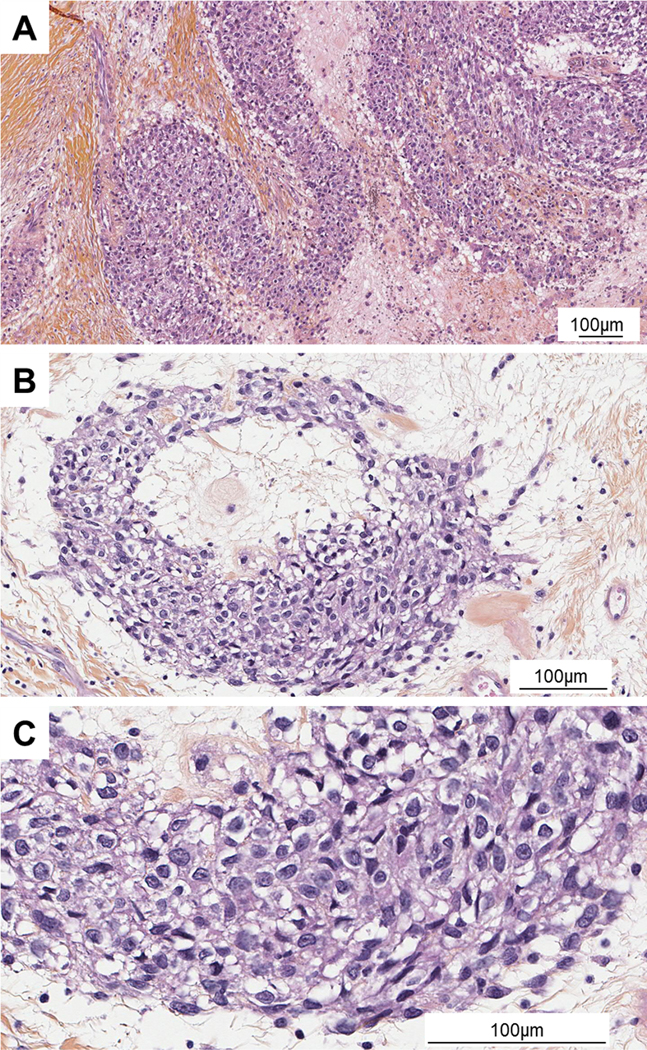
Microscopy images of Case 2. (A) HE (low power objective x10): Sheets and nests cells in desmoplastic stroma. (B) HE (x20) Nest of epithelioid cells. (C) HE (objective x40) Cells were of medium size with abundant eosinophilic cytoplasm and ovoid and vesicular nuclei with nucleoli.
RNA-seq performed on the FFPE material evidenced an in-frame fusion between exon 1 of CRTC1 to exon 2 of SS18. In this case the translocation was unbalanced, as demonstrated by FISH (Figure 2A, 4A). Of note, the cell morphology was similar in the tumour biopsy acquired before treatment, which also harboured the CRTC1-SS18 fusion gene.
Genomic and transcript profiles
Array-comparative genomic hybridization analyses revealed that Case 1 had a diploid genome and a balanced CRTC1-SS18 translocation, whereas Case 2 had a tetraploid genome with an unbalanced CRTC1-SS18 translocation (see Figure 4B for specific chromosomal gains and losses for each case). To be able to compare both samples, RNA-sequencing of Case 1 was also performed on an FFPE sample, in the same pipeline as Case 2. Hierarchical clustering analysis (of RNA-seq data) demonstrated that both samples clustered together and close to the EWSR1-CREB1 positive tumors but not with Ewing or Ewing-like sarcomas (Figure 4C). Also, CRCT1-SS18 samples did not cluster with the recently described cutaneous melanocytomas harbouring a CRTC1-TRIM11 fusion gene [16]. Furthermore, RNA-seq data revealed enhanced NTRK1 expression in the two cases with the CRTC1-SS18 gene fusion compared to other sarcomas with known translocations such as EWSR1-CREB1, BCOR-CCNB3, CIC-DUX4, EWSR1-FLI1, etc (Figure 4D) and this was confirmed at the protein level by immunohistochemistry (supplementary material, Figure S3).
Functional analysis of the CRTC1-SS18 gene fusion product
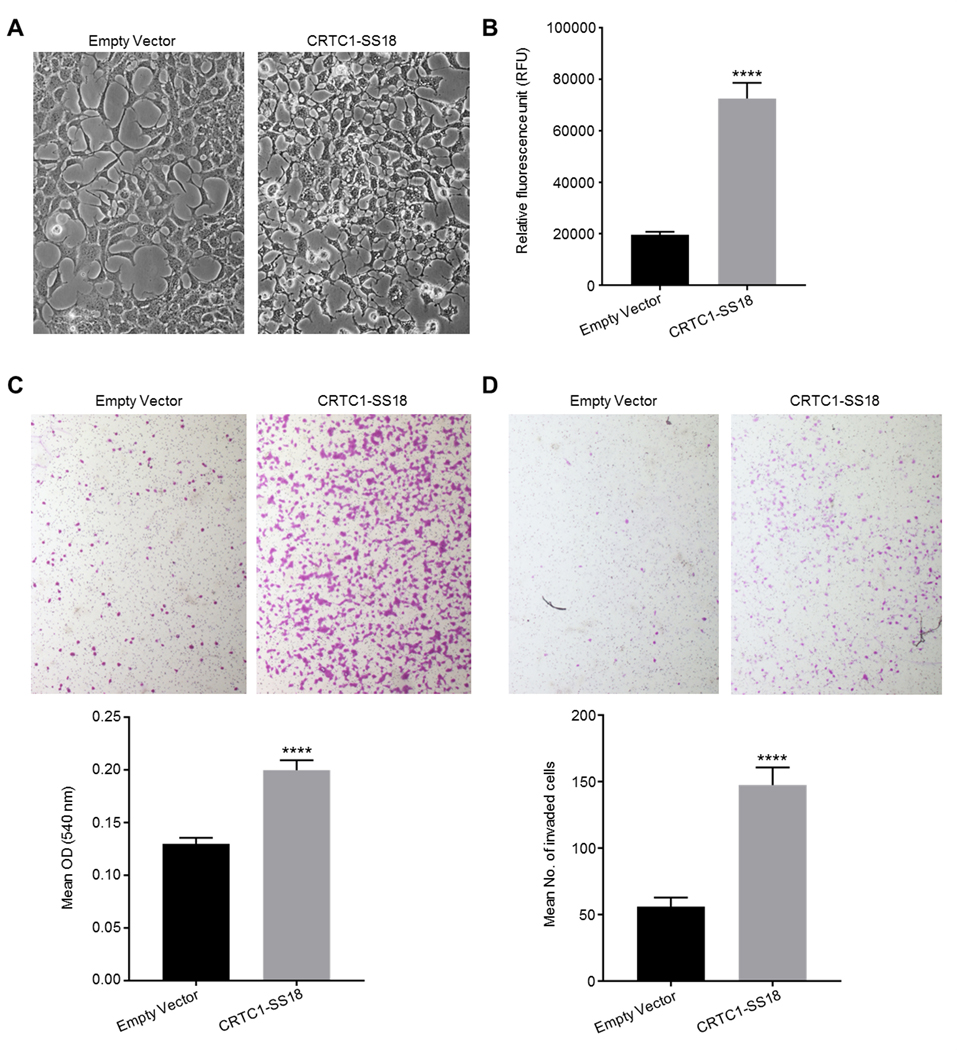
To determine if the product of the CRTC1-SS18 gene fusion identified had any potential oncogenic activity, the fusion gene from Case 1 was cloned into a tagged mammalian expression vector and human HEK293 clones expressing the construct were generated; a fusion protein of the predicted size (~57 kDa) was observed (supplementary material, Figure S4). The clones expressing the CRTC1-SS18 fusion were morphologically distinct from control clones, showing extended pseudopodia and pronounced intracytoplasmic vacuoles (Figure 5A). We proceeded to assess the anchorage-independent growth potential of these cells (hallmark of transformation). CRTC1-SS18-expressing HEK293 cells were seeded into semi-solid agar, in 96-well microtiter plates, and incubated for 8 d. The number of viable cells was then determined using a commercial fluorescence assay. Following this relatively short period of incubation the assay indicated that there were 3.7-times the number of viable cells expressing CRTC1-SS18 compared to control HEK293s, transfected with an empty plasmid. This increase in viable cells was statistically significant (Figure 5B). As expression of this fusion protein appeared to increase anchorage-independent growth, we continued to assess these cells for other hallmarks of malignancy. We carried out assays to determine the migratory and invasive potential of the CRTC1-SS18 expressing cells. In a Boyden chamber assay, where cells are encouraged to pass through 8 μm pores, significantly more HEK293 cells expressing CRTC1-SS18 migrated than control cells, this was determined both visually and by a colorimetric assay (Figure 5C). In a similar assay, where the membrane of the chambers are coated in a basement membrane matrix to model the invasive potential of these cells, 2.6-times more CRTC1-SS18 expressing cells invaded through the membrane than control cells in a 16 h period (Figure 5D).
Figure 5.
Biological analysis of the CRTC1-SS18 gene fusion. (A) Morphological analysis of CRTC1-SS18 expressing cells. CRTC1-SS18 fusion-expressing clones were morphologically distinct from control HEK293s; the cytoplasmic component was small, with pronounced vacuoles and extended, thin pseudopodia (both images taken under phase contrast at 200X magnification). (B) Expression of the CRTC1-SS18 fusion protein in HEK293 cells significantly increased anchorage-independent growth potential. The number of viable, colony-forming cells present, following incubation in soft agar was increased 2.1-fold in HEK293 cells expressing CRTC1-SS18 compared to HEK293 cells transfected with a control plasmid. Viable cells were determined by a fluorometric assay. p<0.0001, t=8.61, df=14. Error bars represent SEM (n=8). (C) Expression of the CRTC1-SS18 fusion protein in HEK293 cells significantly increased cell migration. The number of HEK293 cells expressing CRTC1-SS18 that migrated through 8 μm pores in a Boyden chamber assay in 16 h was significantly increased compared to HEK293 cells transfected with a control plasmid. This was apparent by inspection by microscopy (at 4X objective magnification) and by a colorimetric assay. p<0.0001, t=6.2.2, df=38. Error bars show SEM (n=20). (D) Expression of the CRTC1-SS18 fusion protein in HEK293 cells significantly increased cell invasive potential. The number of HEK293 cells expressing CRTC1-SS18 that invaded through an 8 μm pores coated in a basement membrane matrix in 16 h was significantly increased compared to HEK293 cells transfected with a control plasmid. Invaded cells were counted at 40X objective magnification, the mean number of invaded cells was 2.6-times greater for CRTC1-SS18 expressing cells than for control cells. p<0.0001, t=6.108, df=42. Error bars show SEM (n=22).
Discussion
Ewing-like sarcoma (ELS) or undifferentiated small round cell sarcoma (USRCS) is a subtype of SBRCT that has a morphological appearance close to that of Ewing sarcoma but lacks the characteristic EWSR1-ETS gene fusion. Recently, some of these USRCS sarcomas have been shown to carry gene fusions involving CIC-DUX4, CIC-FOXO4, BCOR-CCNB3, BCOR-MAML3 and ZC3H7B-BCOR [3–8]. In this report we have identified by RNA sequencing a novel recurrent CRTC1-SS18 gene fusion in two USRCS that were negative for known gene fusions in this sarcoma type. The CREB Regulated Transcription Coactivator 1 (CRTC1) gene belongs to a family of highly conserved CREB (cAMP response element-binding protein) coactivators [17–18]. CRTC1 has already been involved in other translocations such as the CRTC1-MAML2 fusion in mucoepidermoid carcinoma (MEC) of salivary, bronchial and thyroid glands [19–22] and the CRTC1-TRIM1 fusion in cutaneous melanocytomas [16]. The SS18 protein (NBAF Chromatin Remodelling Complex Subunit SS18) functions as a transcriptional coactivator and interacts directly with members of the SWI/SNF chromatin remodelling complex [23]. The SS18-SSX fusion is a result of the chromosomal translocation t(X;18)(p11;q11) in virtually all cases of synovial sarcoma (SS), which accounts for approximately 10–20% of all soft tissue sarcomas [24–26].
These two cases of CRTC1-SS18 fusion sarcomas are regarded as a distinct entity from all other ELS, DRSCT and poorly differentiated synovial sarcoma described in the literature. Poorly differentiated synovial sarcoma is characterised by high cellularity, polygonal to small round cell morphology, frequent mitoses and necrosis. These poorly differentiated SS may be distinguished by expression of high molecular weight cytokeratins, CD99 and demonstrating the characteristic t(X;18)(p11:2;q11:2). However these two cases of CRTC1-SS18 sarcoma had a distinct morphology in contrast to those seen in poorly differentiated synovial sarcoma and lacked the characteristic t(X;18)(p11:2;q11:2) translocation. Only one other ELS tumour with a CIC-FOXO4 gene fusion has been described with desmoplastic stroma which showed immunoreactivity with CD99 and focal WT1. This tumour occurred in the neck of an elderly male. DSRCT commonly arises in the abdominal cavity in children and young adults and are characterised histologically by nests of small round cells surrounded desmoplastic stroma, immunohistochemically by positive staining for keratins, desmin and WT1, and genetically by the presence of a recurrent translocation t(11;22)(p13;q12). However the tumours we describe here occurred in a lower limb of adult patients, tumour cells were larger than cells of DSRCT with more cytoplasm and showed immunoreactivity with CD99 but did not carry the EWS-WT1 gene fusion.
Morphologically the two tumours we present here shared features: large fibrous stroma, small to medium cells with eosinophilic cytoplasm and vesicular nuclei. Expression profile analyses confirmed that our CRTC1-SS18 positive sarcomas were not related to Ewing or Ewing-like sarcomas, but rather to EWSR1-CREB1 positive tumours. Of note, CRTC1-TRIM11 cutaneous melanocytomas were also found resembling EWSR1-CREB1 clear cell sarcomas [16] but hierarchical clustering clearly separated both types of CRTC1 fused tumours. Finally, we also present here evidence at both RNA and protein level, that NTRK1 is expressed at higher levels in the CRTC1-SS18 sarcomas than in other related tumours. We could not find any fusion involving NTRK1 in these tumours explaining its elevated level Nevertheless, NTRK1 expression may therefore be useful as a marker for differential diagnosis but most importantly may be used as a therapeutic target. In addition, we demonstrate that the cells expressing the CRTC1-SS18 gene fusion were morphologically distinctive and had enhanced oncogenic potential compared to control cells.
In summary, we have presented two cases of USRCS with a novel CRTC1-SS18 gene fusion. It would be beneficial to screen more samples to determine the frequency of CRTC1-SS18 gene fusion in other USRCS. The severe clinical phenotype (lung metastasis at an early age) of case 1 (Case 2 has 6 months of follow-up), the novel CRTC1-SS18 gene fusion and the expression profile data indicate that these tumours may be classified as a new type of USRCS. Except sporadic and unique cases, to our knowledge, the two USRCS in this study are the only cases of another sarcoma type other than SS involving SS18 as a recurrent gene fusion partner. The discovery of this new fusion should enable better classification and study of these rare sarcomas. CRTC1-SS18 gene fusion sarcoma should be considered in the differential diagnosis of USRCS, DSRCT and in poorly differentiated tumours which show SS18 split signals with FISH. Elevated levels of NTRK1 in CRTC1-SS18 positive sarcomas may be of therapeutic importance and amiable to treatment with tyrosine-kinase inhibitors [27]. Considered all together, for the field of rare to ultra-rare sarcomas, this study offers a nice example of the need to assess samples from different cancer centres to identify recurrent fusions and to be able to characterize new sarcoma subtypes. Further collaborations between groups is therefore required to depict the whole landscape of small round cell sarcomas.
Supplementary Material
Table S1. The sequence of primers used for long range PCR
Figure S1. Read coverage for CRTC1(Ex1)-SS18(Ex2) and CRTC1-SS18(Ex3) identified in Case 1 using RNA-seq data
Figure S2. RT-PCR validation of CRTC1-SS18 fusions and Long-Range PCR to determine genomic break points
Figure S3. Immunostaining of NTRK1 (Case 2), showing strong positivity; scale bar=100 μm
Figure S4. Validation of CRTC1-SS18 expression in stably-transfected HEK293 clones
Acknowledgements
This research was funded in part by the Kuwait Medical Genetics Centre (KMGC), Ministry of Health, Kuwait, the Royal Orthopaedic Hospital NHS Foundation Trust, Birmingham, England, the Centre Léon Bérard, Lyon, France, the Institut National de la Santé et de la Recherche Médicale, the Italian Ministry of Health (Ricerca Corrente RC L2029, IRCCS Galeazzi Orthopaedic Institute) and by the Department of Biomedical, Surgical and Dental Sciences (University of Milan, grant number 15–63017000-700).
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
References:
- 1.Machado I, Navarro S, Llombart-Bosch A. Ewing sarcoma and the new emerging Ewing-like sarcomas: (CIC and BCOR-rearranged-sarcomas). A systematic review. Histol Histopathol 2016; 31: 1169–1181. [DOI] [PubMed] [Google Scholar]
- 2.Antonescu C. Round cell sarcomas beyond Ewing: emerging entities. Histopathology 2014; 64: 26–37. [DOI] [PubMed] [Google Scholar]
- 3.Pierron G, Tirode F, Lucchesi C, et al. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet 2012; 44: 461–466. [DOI] [PubMed] [Google Scholar]
- 4.Kawamura-Saito M, Yamazaki Y, Kaneko K, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet 2006; 15: 2125–2137. [DOI] [PubMed] [Google Scholar]
- 5.Antonescu CR, Owosho AA, Zhang L, et al. Sarcomas With CIC-rearrangements Are a Distinct Pathologic Entity With Aggressive Outcome: A Clinicopathologic and Molecular Study of 115 Cases. Am J Surg Pathol 2017; 41: 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puls F, Niblett A, Marland G, et al. BCOR-CCNB3 (Ewing-like) sarcoma: a clinicopathologic analysis of 10 cases, in comparison with conventional Ewing sarcoma. Am J Surg Pathol 2014; 38: 1307–1318. [DOI] [PubMed] [Google Scholar]
- 7.Sugita S, Arai Y, Tonooka A, et al. A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol 2014; 38: 1571–1576. [DOI] [PubMed] [Google Scholar]
- 8.Specht K, Zhang L, Sung YS, et al. Novel BCOR-MAML3 and ZC3H7B-BCOR Gene Fusions in Undifferentiated Small Blue Round Cell Sarcomas. Am J Surg Pathol 2016; 40: 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brohl AS, Solomon DA, Chang W, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014; 10: e1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicorici D, Satalan M, Edgren H, et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data bioRxiv 2014. [Google Scholar]
- 11.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas B, Dobin A, Stransky N, et al. STAR-Fusion: Fast and Accurate Fusion Transcript Detection from RNA-Seq. Aviv Regev. bioRxiv 2017; DOI: 10.1101/120295. [DOI] [Google Scholar]
- 13.Ge H, Liu K, Juan T, et al. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics 2011; 27: 1922–1928. [DOI] [PubMed] [Google Scholar]
- 14.Bray NL, Pimentel H, Melsted P, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016; 34: 525–527. [DOI] [PubMed] [Google Scholar]
- 15.R Development Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 16.Cellier L, Perron E, Pissaloux D, et al. Cutaneous Melanocytoma With CRTC1-TRIM11 Fusion: Report of 5 Cases Resembling Clear Cell Sarcoma. Am J Surg Pathol 2017; DOI: 10.1097/PAS.0000000000000996. [DOI] [PubMed] [Google Scholar]
- 17.Conkright MD, Canettieri G, Screaton R, et al. TORCs: transducers of regulated CREB activity. Mol Cell 2003; 12: 413–423. [DOI] [PubMed] [Google Scholar]
- 18.Iourgenko V, Zhang W, Mickanin C, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci USA 2003; 100: 12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordkvist A, Gustafsson H, Juberg-Ode M, et al. Recurrent rearrangements of 11q14–22 in mucoepidermoid carcinoma. Cancer Genet Cytogenet 1994; 74: 77–83. [DOI] [PubMed] [Google Scholar]
- 20.Tonon G, Modi S, Wu L, et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003; 33: 208–213. [DOI] [PubMed] [Google Scholar]
- 21.Enlund F, Behboudi A, Andren Y, et al. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin’s tumors. Exp Cell Res 2004; 292: 21–28. [DOI] [PubMed] [Google Scholar]
- 22.Tirado Y, Williams MD, Hanna EY, et al. CRTC1/MAML2 fusion transcript in high-grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin′s tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer. 2007; 46: 708–715. [DOI] [PubMed] [Google Scholar]
- 23.Middeljans E, Wan X, Jansen PW, et al. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS One 2012; 7(3):e33834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark J, Rocques PJ, Crew AJ, et al. Identification of novel genes,and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994; 7: 502–8. [DOI] [PubMed] [Google Scholar]
- 25.Skytting B, Nilsson G, Brodin B, et al. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst 1999; 91: 974–5. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen TO, Poulin NM, Ladanyi M. Synovial sarcoma: recent discoveries as a roadmap to new avenues for therapy. Cancer Discov 2015; 5: 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schram AM, Chang MT, Jonsson P, et al. Fusions in solid tumours: diagnostic strategies, targeted therapy, and acquired resistance. Nat Rev Clin Oncol 2017; 14(12): 735–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The sequence of primers used for long range PCR
Figure S1. Read coverage for CRTC1(Ex1)-SS18(Ex2) and CRTC1-SS18(Ex3) identified in Case 1 using RNA-seq data
Figure S2. RT-PCR validation of CRTC1-SS18 fusions and Long-Range PCR to determine genomic break points
Figure S3. Immunostaining of NTRK1 (Case 2), showing strong positivity; scale bar=100 μm
Figure S4. Validation of CRTC1-SS18 expression in stably-transfected HEK293 clones