Abstract
Background and Aims
Crassulacean acid metabolism (CAM) is often considered to be a complex trait, requiring orchestration of leaf anatomy and physiology for optimal performance. However, the observation of trait correlations is based largely on comparisons between C3 and strong CAM species, resulting in a lack of understanding as to how such traits evolve and the level of intraspecific variability for CAM and associated traits.
Methods
To understand intraspecific variation for traits underlying CAM and how these traits might assemble over evolutionary time, we conducted detailed time course physiological screens and measured aspects of leaf anatomy in 24 genotypes of a C3+CAM hybrid species, Yucca gloriosa (Asparagaceae). Comparisons were made to Y. gloriosa’s progenitor species, Y. filamentosa (C3) and Y. aloifolia (CAM).
Key Results
Based on gas exchange and measurement of leaf acids, Y. gloriosa appears to use both C3 and CAM, and varies across genotypes in the degree to which CAM can be upregulated under drought stress. While correlations between leaf anatomy and physiology exist when testing across all three Yucca species, such correlations break down at the species level in Y. gloriosa.
Conclusions
The variation in CAM upregulation in Y. gloriosa is a result of its relatively recent hybrid origin. The lack of trait correlations between anatomy and physiology within Y. gloriosa indicate that the evolution of CAM, at least initially, can proceed through a wide combination of anatomical traits, and more favourable combinations are eventually selected for in strong CAM plants.
Keywords: Yucca gloriosa, Yucca aloifolia, Yucca filamentosa, Asparagaceae, Agavoideae, CAM photosynthesis, leaf anatomy, hybrid
INTRODUCTION
A fundamental aim of comparative biology is to elucidate how, when, and why traits evolve, and the biological consequences of trait evolution. Some traits have simple genetic architecture: changes may be induced by mutations to single genes or regulatory elements, as in the case of hair colour in mice (Hoekstra et al., 2006), flower colour and pollinator shifts in Erythranthe guttata (Bradshaw and Schemske, 2003; Yuan et al., 2013), and herbicide resistance in barley (Lee et al., 2011). Other traits are more complex, in that they are actually a sum of phenotypic states orchestrated across an organism. For example, the evolution of C4 photosynthesis requires both altered biochemical pathways as well as changes to leaf anatomy (Hatch, 1987; Christin et al., 2013; Sage et al., 2014), and burrowing behaviour in field mice relies on changes to separate genetic modules (Weber et al., 2013). Because complex traits are unlikely to evolve via a single mutation (Lenski et al., 2003), one might expect various intermediate phenotypes to exist through the evolutionary progression from ancestral to derived character states. Species exhibiting intermediate phenotypes could be instrumental to ordering the sequence of events that led to the evolution of a complex trait. Intermediate phenotypes also lend insight into the genetic landscape of a complex trait: for example, genetic linkage can restrict which trait combinations are possible and can affect how quickly natural selection can act upon them (Gerrish et al., 2007; Barton, 2010).
Crassulacean acid metabolism (CAM) is an example of a complex plant trait involving biochemistry, anatomy and physiology that works tandemly with the C3 Calvin Benson cycle to increase the water use efficiency of plants. The C3 pathway uses Rubisco, an enzyme that has both carboxylating and oxygenating functions. Under high temperatures or conditions that promote stomatal closure, such as drought stress, rates of Rubisco oxygenation increase and force C3 plants to undergo photorespiration, an energetically costly process. CAM concentrates CO2 in an effort to reduce oxygenation via Rubisco and consequently photorespiratory stress. CAM species open their stomata at night, when lower temperatures and higher relative humidity reduce evapotranspiration. Incoming CO2 is initially converted to malate and stored in the vacuole. During the day the stomata largely remain closed, and the stored CO2 is decarboxylated from malate, surrounding Rubisco and the C3 machinery with elevated CO2 concentrations. The CAM carbon-concentrating mechanism reduces levels of photorespiration while simultaneously increasing overall water use efficiency. As a result, CAM plants are often found in hot, arid or seasonally dry habitats – often, but not always, where water is limiting.
Because all CAM plants retain and use the entire C3 machinery, many species can fix carbon through a mixture of both pathways (Winter, 2019). Strong CAM plants use CAM for the vast majority of their carbon uptake, while C3+CAM species use a mix of both pathways to fix CO2. For example, CAM cycling plants fix nocturnally respired CO2 through the CAM pathway but otherwise have C3 physiology. Moreover, plants can vary not only in their ability to use CAM, but also the degree to which CAM can be modulated under abiotic stress. Both strong CAM and C3+CAM can alter the relative contribution of CAM to CO2 fixation as a response to abiotic stressors. C3+CAM species can upregulate the CAM pathway (‘facultative CAM’) or downregulate the contribution of the C3 pathway, whereas strong CAM species may increase the degree of C3 carbon fixation when exceptionally well-watered (Hartsock and Nobel, 1976). It is unclear how intermediate phenotypes fit into the evolutionary trajectory of CAM, but the prevalence of intermediate CAM species (Winter, 2019) suggests that such a dynamic phenotype can be advantageous under certain situations (i.e. seasonal drought) (Winter et al., 2008; Herrera, 2009). CAM photosynthesis has evolved at least 60 times independently (Edwards and Ogburn, 2012), although this is probably an inaccurate count due to the difficulties associated with surveying intermediate CAM, particularly facultative forms involving induction of CAM only under specific conditions. Additionally, attempts to delineate when CAM evolved within extant CAM lineages are made difficult by a lack of phylogenetic resolution, particularly in very diverse lineages.
Specific anatomical traits have long thought to be required for maximum CAM function (Nelson et al., 2005; Nelson and Sage, 2008). To be able to store large amounts of CO2 as malate, CAM plants require larger vacuoles; indeed, CAM plants typically have larger mesophyll cells than their C3 counterparts (Heyduk et al., 2016a; Males, 2018). Intercellular airspace (IAS) is often reduced in CAM species (Nelson and Sage, 2008; Barrera Zambrano et al., 2014). One theory is that tight packing of cells reduces the amount of CO2 leakage that can occur during the day, when malate is decarboxylated and results in high concentrations of CO2 in the cells (Nelson and Sage, 2008). Alternatively, reduced IAS may just be a result of larger cells packed into a leaf whose size may be limited by other factors, including the need to maintain hydraulic connectivity (Maxwell et al., 1997). Finally, CAM plants are often described as having thick, succulent leaves (Gibson, 1982). The importance and timing of these anatomical changes remains unclear: in some systems, species that are C3+CAM look anatomically like their C3 relatives (Silvera et al., 2005; Males, 2018), whereas other lineages evolved succulent leaf anatomy prior to CAM (Heyduk et al., 2016b) or coincident with the origin of CAM (Barrera Zambrano et al., 2014). As a result, our understanding of the importance of leaf anatomy on CAM function remains unclear.
One of the greatest challenges in understanding the concerted evolution between CAM biochemistry and anatomy is a lack of systems in which genetic segregation produces variation within and among these traits. While comparisons between C3 and strong CAM species have helped us define a suite of traits that seem to segregate with photosynthetic pathway (e.g. Luttge et al., 1986; Gravatt and Martin, 1992; Heyduk et al., 2016a), these comparisons conflate trait differences with evolutionary distance and yield little insight into how suites of CAM traits have been assembled repeatedly in plant evolutionary history. Are traits assembled sequentially, such that a certain phenotype must arise (e.g. large cells) before a secondary phenotype can evolve (e.g. accumulation of malate)? Or are there a number of trait combinations that can arise in any order and span phenotypic space, but selection repeatedly favours certain combinations to maximize the efficiency of CAM?
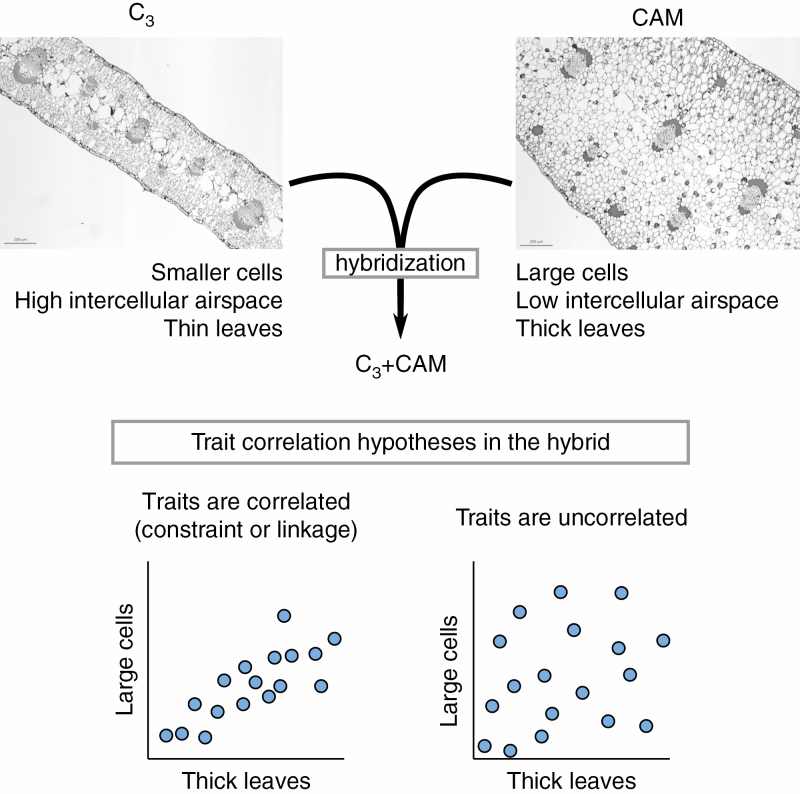
To understand whether anatomy and physiology are correlated we measured anatomical and photosynthetic traits in a C3+CAM species, Yucca gloriosa, a naturally occurring homoploid hybrid species resulting from a wild cross between Y. aloifolia (CAM) and Y. filamentosa (C3) (Rentsch and Leebens-Mack, 2012; Heyduk et al., 2016a). All three Yucca species overlap in the southeastern United States, with Y. filamentosa having the broadest range that extends into the northeast and midwest, Y. aloifolia being more restricted to the southeast, and Y. gloriosa inhabiting the narrowest natural range, occurring only in the coastal regions of the Atlantic seaboard between Florida and Virginia. Previous work has demonstrated contrasting photosynthetic pathways in the parental species and intermediate physiology and anatomy in the hybrid (Heyduk et al., 2016a, b). Genetic screens based on microsatellite data have suggested that while Y. gloriosa still retains a mixture of both parental genomes, genotypes are not identical and thus not likely to be F1 hybrids (Rentsch and Leebens-Mack, 2012; Heyduk et al., 2016a). Here we examine the extent of intraspecific variation in photosynthetic and anatomical traits across 24 genotypes of the C3+CAM hybrid, Y. gloriosa. Specifically, we assess the relationship between anatomy and photosynthetic phenotype (Fig. 1) and the extent of environmentally driven genotypic variation. We show that genotypes vary in the degree of CAM used, as well as the level of upregulation of CAM under drought stress; we further find that there is little correlation between anatomical traits and photosynthetic phenotypes.
Fig. 1.
Anatomical traits typically associated with C3 and strong CAM plants, showing possible resulting trait associations in a hybrid between a C3 and a CAM species.
MATERIAL AND METHODS
Genotypes of Y. gloriosa were collected from across its geographical range (Virginia to Florida) (Supplementary Data Table S1) as ramets, then transplanted to the University of Georgia Department of Plant Biology glasshouses. Plants were grown in 60 : 40 soil/sand mix with once-weekly watering and fertilizer as needed, and maintained until proper rooting was established and significant new growth was noticeable (minimum 6 months). All plants were grown in the same glasshouse, with no additional growth light beyond sunlight, and generally experiencing day/night temperatures of roughly 28 °C/21 °C, although temperatures varied throughout the year. Beginning in spring 2016, 24 genotypes were randomly assigned to growth chamber experimental runs in sets of four genotypes. For each experimental run, 34 clonal replicates for each of the four genotypes were acclimated in the growth chamber for 4 d before manipulation. Growth chambers had 12-h days (beginning at 0700 h), with 30 °C/17 °C day/night temperatures and a relative humidity of 30–40 %. Light intensity at leaf level was ~400 µmol m−2 s−2 and plants were kept well-watered during the acclimation phase. On the first experimental day (‘day 1’), gas exchange measurements were collected every 2 h for a 24-h period, beginning 1 h lights turned on, using a LiCor 6400XT (Lincoln, NB, USA). After day 1, plants were allowed to dry down, soil moisture information was collected with an ML2 soil moisture probe (Delta-T Devices, London, UK) on experimental days (Supplementary Data Table S2 and Fig. S1), and on day 7 plants were re-measured for gas exchange identically to day 1. At the end of day 7, plants were re-watered, then measured a final time for gas exchange on day 9. Experimental runs were conducted in April, September and November 2016, and February, March, April and August 2017 on a total of 24 genotypes. To compare net carbon gain between genotypes, the area beneath the curve formed from plotting time course LiCor data was calculated. Area under the curve (AUC) was estimated per genotype per treatment using the auc() function in the DescTools (Signorell, 2019) package in R v.3.5.1 (R Core Team, 2019).
Leaf samples for titratable acidity measurements were collected on days 1, 7 and 9 2 h before lights off and 2 h before lights were turned back on. Fully expanded leaves that were un-shaded by others in the rosette were preferentially sampled; different leaves were sampled on days 1, 7 and 9, taking care to avoid sampling from old, dying or partially expanded leaves. Leaf punches were taken in triplicate at both time points from all individual plants, then were immediately flash frozen and stored at −80 °C. Leaves were quickly weighed once removed from the freezer and placed in 60 ml of 20 % EtOH. Samples were boiled until the volume was reduced by half, at which point the total volume was returned to 60 ml by adding water of pH 7.0. Samples were boiled to half volume once more, refilled to 60 ml with water, then allowed to cool to room temperature. The room temperature liquid was cleared of leaf debris and titrated with 0.002 m NaOH to a final pH of 7.0. Total µmol of H+ was calculated as (ml of 0.002 m NaOH × 2 mm)/grams of frozen tissue.
Leaf cross-sections were collected in April 2018 and April 2019 from clonal replicates of the same genotypes measured for gas exchange, with two samples collected per genotype from separate biological replicates when available. Cross-sections were sampled from plants growing in the University of Georgia glasshouses under a once-weekly watering regime and fertilizer addition as needed. Leaves were cut, fixed in formalin, embedded in paraffin, then sectioned at the University of Georgia Veterinary Hospital Histology Lab. Sections were stained with Toluidine Blue and mounted on glass slides. For each of the separate plants sectioned per genotype, two images were taken on a Zeiss (Oberkochen, Germany) microscope at 5× and 10× magnification, taking care to avoid imaging edges or damaged sections. Images were analysed in ImageJ (NIH, Bethesda, MD, USA) to collect measurements of cell size and IAS, as well as leaf thickness. For cell size, the areas of five adaxial and abaxial mesophyll cells were measured per image. IAS was measured as a fraction of intercellular air per total cell area and is reported as a per cent of mesophyll. Leaf thickness was measured in triplicate across each image analysed for cell size and IAS. Stomatal density was measured by painting both adaxial and abaxial leaf surfaces with clear nail polish (collected March 2019), then removing with tape and imaging stomatal impressions with a Zeiss microscope. Stomatal measurements were taken from two biological replicates per genotype, when available. Previously collected data on adaxial and abaxial cell sizes from additional genotypes of Y. gloriosa was also included (Heyduk et al., 2016a); although IAS was measured on this previous dataset, due to image quality, we suspect IAS may have been overestimated in the data previously published. IAS was therefore re-analysed for all data published in 2016. ANOVAs or ANCOVAs were performed, as appropriate, to determine the effect of Y. gloriosa genotype on phenotypic traits; in the case of CO2 uptake and acid accumulation, treatment (watered and drought) was included as a factor (Supplementary Data Table S3).
To compare the hybrid to the parental species, previously published data on Yucca filamentosa and Yucca aloifolia were included as well (Heyduk et al., 2016a, b). The parental datasets are smaller, in that a total of five and seven genotypes were measured for various traits in Y. aloifolia and Y. filamentosa, respectively, with two to four replicates per genotype of each species. The parental species were grown in the same conditions as Y. gloriosa: plants were collected as rhizome cuttings from the wild, grown in the University of Georgia glasshouses for at least 6 months (where they were sampled for leaf anatomical traits), then placed in the same growth chamber with identical conditions for gas exchange and titratable acidity measurements conducted using largely the same methods as described above for Y. gloriosa. Similarly, leaf anatomical traits were collected and measured in the parental species using the same methods as for Y. gloriosa (and are fully described in Heyduk et al., 2016a). All statistical analyses were conducted in R v.3.5.1, and raw data and genotypic means can be found at www.github.com/kheyduk/Yucca_physiology. ANOVAs were calculated across traits to determine differences between species (Supplementary Data Table S4). We correlated both raw data (i.e. individual plant traits) as well as genotypic means using cor.test() in R and adjusted the resulting P-values for multiple testing with the Benjamini–Hochberg correction. Correlations were conducted pairwise on all traits, except in cases where one trait was a subset of another (e.g. nocturnal CO2 total assimilation is a subset of total daily CO2 assimilation). Correlations were conducted on individual values and genotypic means of all three species together, then separately for just Y. gloriosa. No correlations were tested within the parental species, as the data pulled from earlier work did not have enough within-genotype replication. For a trait combination to be reported as significant, it had to be significant when correlated across both individuals and genotypic means; for significant correlations, only the across-individual statistics are reported, while genotypic mean statistics can be found in Supplementary Data Tables S5 and S6.
RESULTS
Gas exchange and titratable acidity
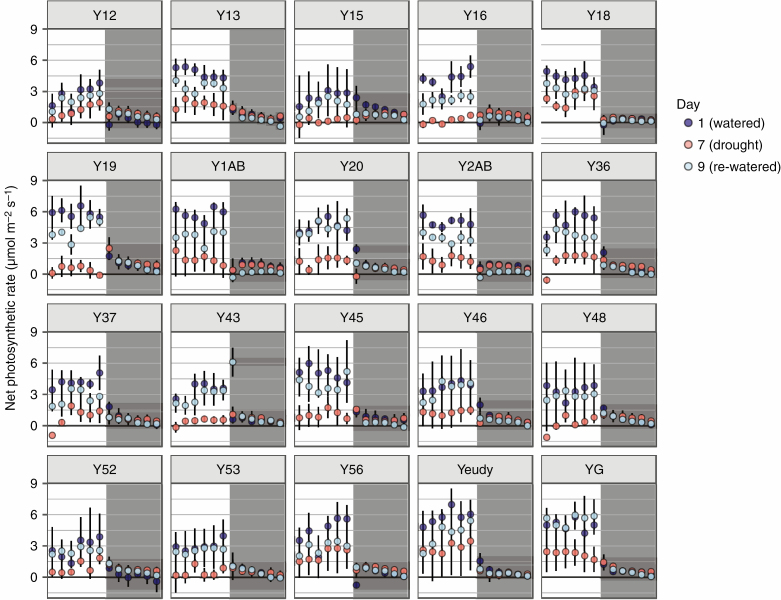
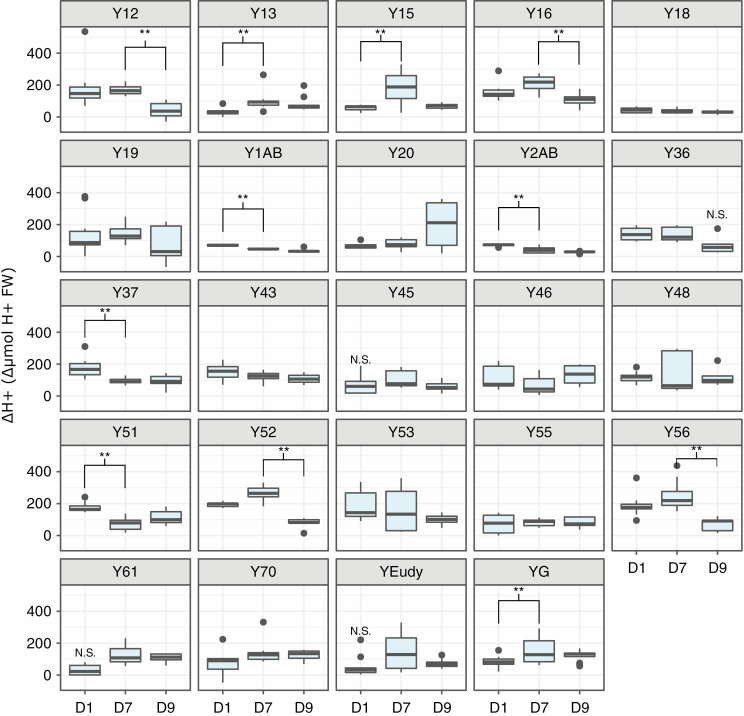
Genotypes of Yucca gloriosa varied in their gas exchange patterns over a diel cycle (Fig. 2; Supplementary Data Fig. S2). The majority of genotypes had some level of C3 daytime CO2 fixation as well as slight nocturnal CO2 assimilation under well-watered conditions (Fig. 2). Under drought stress, overall responses varied. Some genotypes had a nearly total shutdown of gas exchange during the day under drought stress, whereas others maintained positive levels. Because plants dried down at slightly variable rates (Table S2 and Fig. S1), we examined the effect of genotype and soil moisture on CO2 uptake: well-watered plants had only a slightly significant effect of genotype (F19,51 = 2.16, P < 0.05) on CO2 uptake, while drought-stressed plants had a significant effect of soil moisture (F1,48 = 15.02, P < 0.001). However, nocturnal CO2 assimilation (and thus the level of CAM performed) was not significantly related to soil moisture under either well-watered (F1,51 = 0.03, P = 0.87) or drought stress (F1,50 = 0.64, P = 0.43). Instead, a clear genotype × environment (G×E) signal was observed via an ANOVA (Type III) assessing the interaction of genotype and treatment (well-watered vs. drought) on nocturnal CO2 uptake (interaction: F21,131 = 2.36, P < 0.01, excluding re-watered measurements) (Table S3). At night, certain genotypes (e.g. 16 and 1AB, Fig. 2) were able to increase CO2 assimilation under drought stress relative to well-watered conditions. No hybrid genotype fully replicated the levels of nocturnal CO2 assimilated by Y. aloifolia, and many had the ability to use CAM even when well-watered, indicating Y. gloriosa does not have strictly facultative CAM but rather weak CAM with drought inducibility. Nocturnal acid accumulation in the hybrid Y. gloriosa, like gas exchange, had a significant interaction effect between genotype and treatment (F23,120 = 3.73, P < 0.001, excluding re-watered measurements) (Table S3). In general, the majority of genotypes showed an increase in leaf acidity over the night period, indicative of CAM; most genotypes displayed some level of acid accumulation on all days of the experiment, regardless of water status (Fig. 3). Four genotypes had gas exchange removed from the analysis (Y51, Y55, Y61, and Y70) because of a malfunction with the LiCor during the experimental run; however, titratable acidity and leaf anatomy were unaffected and are still reported.
Fig. 2.
(A) Gas exchange of Yucca gloriosa genotypes measured every 2 h over a 24-h period beginning at 1 h after lights on (0800 h). White and grey backgrounds specify daytime and night-time measurements, respectively. Mean and standard deviation are shown for days 1 (well-watered), 7 (drought stress) and 9 (re-watered). Four samples were omitted due to potentially inaccurate LiCOR measurements (genotypes 51, 55, 61 and 70).
Fig. 3.
Levels of leaf titratable acidity (AM H+ equivalents − PM H+ equivalents to pH 7.0) across well-watered (D1), drought (D7) and re-watered (D9) time points in 24 genotypes of Yucca gloriosa. If any given value is not significantly different from 0 (no acid accumulation), N.S. is shown above the box. Significant differences between time points within a genotype are indicated by brackets above the boxes.
Inter- and intraspecific response to drought
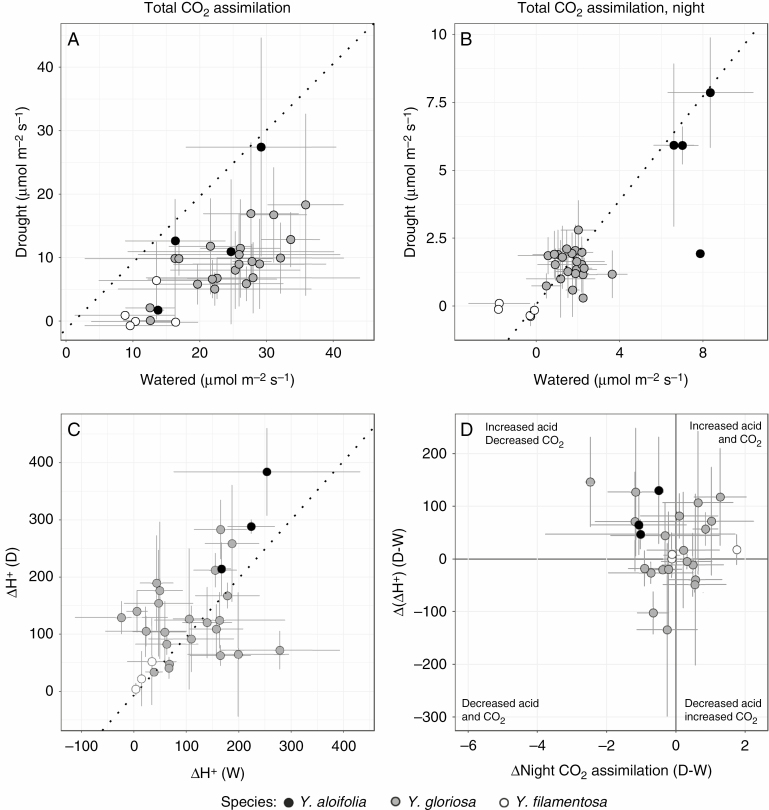
When compared to the parental genotypes for which gas exchange measurements are available, many Y. gloriosa genotypes had some of the highest net CO2 assimilation values (as calculated by the area under the gas exchange curves) during both well-watered and drought conditions (Fig. 4A). However, night-time net CO2 assimilation was intermediate in Y. gloriosa compared to parental species and tended toward the C3 parent Y. filamentosa (Fig. 4B). When drought-stressed, Y. filamentosa genotypes showed a decrease in overall CO2 assimilation (Fig. 4B) under drought stress. Yucca aloifolia showed on average a decrease in night-time CO2 assimilation under drought stress (Fig. 4A, B) (Heyduk et al., 2016a). Yucca gloriosa genotypes varied in their gas exchange drought response; certain genotypes increased the amount of CO2 acquired at night, whereas others decreased net night-time CO2 acquisition, similar to Y. aloifolia.
Fig. 4.
Physiological effect of drought stress on genotypic means (and standard deviation) across all three species. (A) Estimated total CO2 assimilation, based on the area under the LiCOR curves across the entire diel cycle, for both hybrid and parental genotypes under well-watered and drought-stressed conditions. (B) Estimated total CO2 assimilation based on the area under the LiCOR curves at night only, for both hybrid and parental genotypes under well-watered and drought-stressed conditions. (C) Leaf acid accumulation under well-watered conditions (W) vs. drought-stressed conditions (D), with genotypic mean and one standard deviation. Dashed line indicates equal values under both conditions; genotypes that fall above or below indicate that greater or lower amounts of acid, respectively, were accumulated under drought stress than under well-watered conditions. (D) Comparison of the change in night-time CO2 assimilation induced by drought stress (x-axis) to the change in leaf acid accumulation induced by drought (y-axis). Quadrants are labelled with the phenotype observed.
Drought-induced response in leaf acid accumulation varied across hybrid genotypes as well and spanned a larger range of values than either parent. While Y. filamentosa never accumulated significant levels of leaf acid (well-watered: t5 = 1.71, P = 0.07; drought-stressed: t5 = 1.40, P = 0.11), Y. aloifolia had appreciable levels of acid accumulation over the night period under well-watered conditions and increased the amount of acid accumulated under drought (Fig. 4B). Yucca gloriosa genotypes spanned the range from low levels of acid accumulation to CAM-like levels under well-watered conditions, and genotypes varied in their ability to increase the amount of acid stored under drought. A few genotypes responded to drought with significant and positive increases in leaf acidity on day 7 relative to day 1 (e.g. Y13 and YG). Genotype Y18 was a notable exception in its lack of acid accumulation and lack of response to drought stress, corresponding to its negligible rates of CO2 assimilation during the dark period (Fig. 2). Genotypes that had high levels of acid accumulation under well-watered conditions tended to decrease acid under drought, while those that had lower levels of acid accumulation under well-watered conditions tended to increase the amount of acid stored in leaves under drought stress (Fig. 4B).
Because CAM can be defined by both acid accumulation and night-time CO2 assimilation, comparing the response of genotypes through both phenotypes can indicate the mode of CAM employed under drought stress. For example, Y. aloifolia reduced the amount of CO2 assimilated at night, but typically increased leaf acid accumulation (Fig. 4D), indicating more reliance on recycling CO2 when drought-stressed. Some genotypes of Y. gloriosa decreased reliance on atmospheric CO2 and increased acid accumulation with drought stress (upper left quadrant, Fig. 4D). Others responded to drought by increasing both night-time CO2 assimilation and leaf acid accumulation (upper right quadrant, Fig. 4D). A few genotypes were negatively impacted by drought in that they reduced both leaf acid accumulation and night-time CO2 uptake, such that stress appears to have diminished their CAM capacity (lower left quadrant, Fig. 4D). Finally, a few genotypes appeared to increase the amount of night-time CO2 assimilation but decrease the level of acid stored in the leaves (lower right quadrant, Fig. 4D); however in many of these latter cases the error bars overlap zero, and therefore we cannot reject the expectation that night-time CO2 uptake is coupled with acid accumulation in these genotypes. Regardless, the general diversity of drought responses in the hybrid Y. gloriosa is clear.
Leaf anatomy
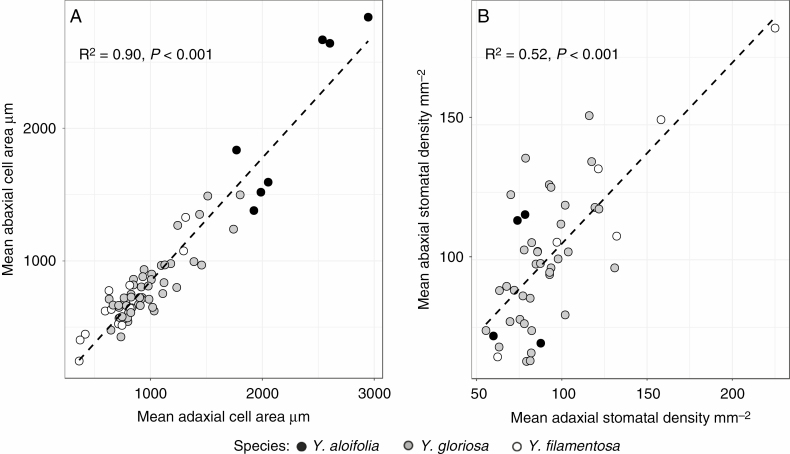
All anatomical traits were significantly different between species, based on ANOVA (Benjamini–Hochberg corrected P-values) (Supplementary Data Table S4), with the exception of abaxial stomatal density. Within Y. gloriosa, the only anatomical traits significantly different between genotypes were IAS (F25,21 = 3.36, P < 0.001) and mean stomatal density (averaged abaxial and adaxial values) (F4,13 = 3.73, P < 0.01) (Table S3). As with physiological traits, anatomical differences between Y. aloifolia and Y. filamentosa were stark, while the hybrid largely filled the phenotypic space between. Cell sizes on both adaxial and abaxial sides of the leaf were larger in Y. aloifolia than in either of the other two species (Fig. 4A). Stomatal density, conversely, was lowest on average in Y. aloifolia and greatest in Y. filamentosa (Fig. 5B). Both cell sizes and stomatal densities on adaxial and abaxial sides of the leaf were highly positively correlated (Fig. 5A, B) across all individuals (cell size: t67 = 25.19, R2 = 0.90, P < 0.001; stomata: t46 = 7.12, R2 = 0.52, P < 0.001).
Fig. 5.
Mean cell sizes (A) and stomatal densities (B) on adaxial and abaxial sides of the leaf per individual plant. In both cases, the dashed line is the regression line from the lm() function in R. R2 and P-values are reported based on correlation tests in R.
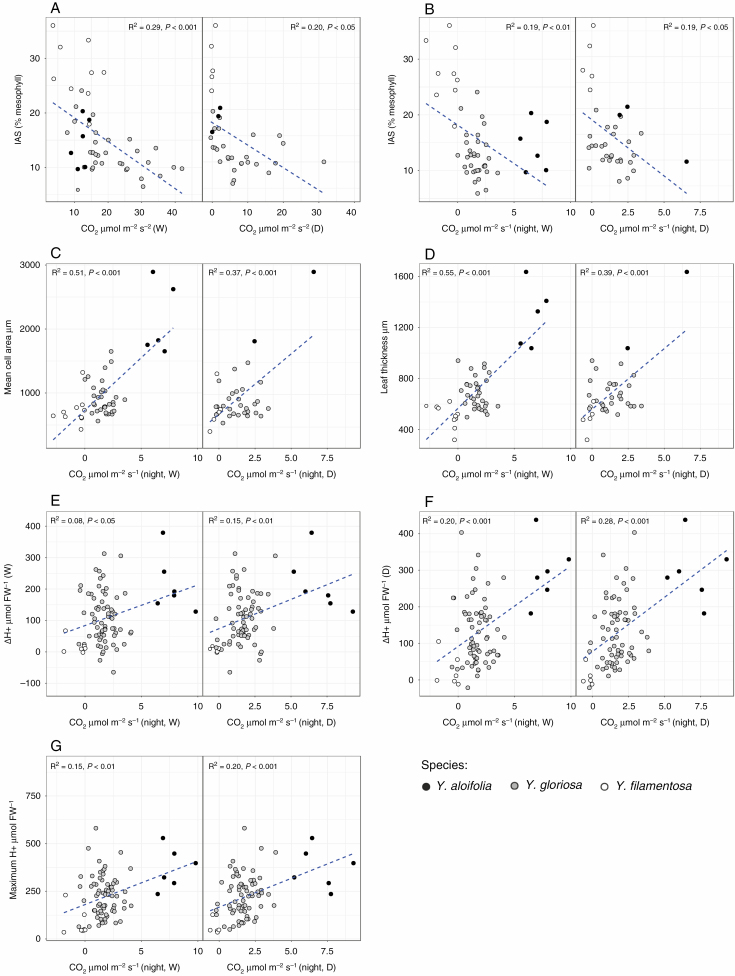
Many anatomical traits were correlated to nocturnal CO2 assimilation, but not total CO2 uptake or leaf acid accumulation (Fig. 6) (Supplementary Data Table S5 and Fig. S3). Total CO2 assimilation across the whole day under both well-watered and drought stress was negatively correlated to IAS, albeit with a relatively low R2 in both cases (Fig. 6A). Nocturnal CO2 uptake was negatively correlated to IAS, and positively correlated to levels of acid accumulation (under both watered and drought conditions), the maximum amount of acid held within a leaf at any time point, mean cell size and leaf thickness (Fig. 6B–G). The amount of leaf acid accumulated under drought stress was correlated to total nocturnal CO2 assimilation under drought stress (R2 = 0.14, P < 0.01). Within Y. gloriosa, nearly all the trait correlations were not significant (Table S6). The only significant correlations for traits in Y. gloriosa were between mean cell size and leaf thickness (R2 = 0.57, P < 0.001) and between total CO2 assimilation under water and drought conditions (R2 = 0.24, P < 0.001).
Fig. 6.
Scatterplots and regression lines with R2 and P-values for a subset of traits (Supplementary Data Fig. S4). Individual data per plant are plotted, and correlations are shown for individual plant data across all three species together, rather than genotypic means (Table S5). (A) Total CO2 assimilation under watered (W) and drought (D) plotted against intercellular airspace (IAS). (B–G) Nocturnal CO2 assimilation under watered and drought plotted against IAS (B), mean mesophyll cell area (C), leaf thickness (D), leaf acid accumulation under watered (E) and drought (F) treatments, and against the maximum amount of acid present at any time point (G). Leaf acidity measurements are per gram of frozen weight (f. wt).
DISCUSSION
Detailed physiological and anatomical measurements in Y. gloriosa have revealed among-genotype variation in CAM phenotypes, and that anatomical and physiological traits show a lack of correlation within Y. gloriosa. Under drought stress, the levels of daytime CO2 assimilation were largely driven by environment (i.e. soil moisture content) whereas nocturnal CO2 assimilation rates and acid accumulation were influenced by a combination of genotype and environmental effects. Our results reveal a continuum of photosynthetic traits across Y. gloriosa genotypes, including variation in drought response. Anatomical measurements were largely not predictive of physiological traits within Y. gloriosa. In contrast, cell size, IAS and leaf thickness were predictive of nocturnal CO2 uptake in cross species comparisons. These observations suggest that anatomical characteristics can be decoupled from photosynthetic physiology of CAM within the homoploid hybrid species Y. gloriosa.
Interspecific correlations of anatomy and physiology
The few studies that have linked leaf anatomy to CAM photosynthetic capacity have provided often contrasting results on how important various anatomical traits are for CAM. In a study that compared phylogenetically unrelated strong CAM and C3+CAM species, cell size was found to be unrelated to CAM (Nelson and Sage, 2008). In contrast, a study of Clusia species that ranged from C3 to CAM with intermediates showed palisade mesophyll cell size was significantly correlated to the proportion of CO2 uptake at night (Barrera Zambrano et al., 2014). Across the three Yucca species examined here, cell size (area) was related to nocturnal CO2 uptake under both watered and drought conditions, although such a relationship did not exist at the intraspecific level within Y. gloriosa. All studies use cell size as a proxy for vacuolar size, which in theory would limit the storage capacity of malate. It is possible that vacuolar size is not linearly related to cell size (but see Chan and Marshall, 2014), and that inconsistent results on the importance of cell size between studies is related to using anatomical proxies for the true trait of interest. Alternatively, and more probably, studies that control for phylogenetic distance, such as the present one and those conducted across Clusia species, reduce noise introduced by sampling across evolutionarily distant lineages and may provide a more accurate assessment of anatomical importance.
In addition to cell size, IAS is often cited as a critical trait for CAM, although whether it evolves as a byproduct of tight cell packing (Maxwell et al., 1997) or as a way to reduce CO2 efflux remains unclear (Borland et al., 2018). IAS was strongly correlated to strength of CAM when measured across unrelated CAM and C3+CAM species (Nelson and Sage, 2008), but had little role in determining the strength of CAM when assessed within the genus Clusia (Barrera Zambrano et al., 2014). IAS was correlated to nocturnal CO2 assimilation when tested across all three species of Yucca here, but was not correlated to leaf acid accumulation, and showed no relationship to any other traits within Y. gloriosa. That IAS is not predictive of physiology in Y. gloriosa (neither nocturnal CO2 uptake nor the amount of leaf acids that accumulate) is surprising, given that contrasts between C3 and CAM species have repeatedly shown the latter have significantly reduced IAS (Heyduk et al., 2016a, b; Males, 2018). Together, the IAS trends across and within Yucca species show that while IAS may be required for constitutive CAM, there exists a large intermediate space where IAS predicts very little about photosynthetic functionality.
For many anatomical and physiological traits, Y. gloriosa has intermediate values compared to the two parental species and often occupies a much broader range of trait values (Supplementary Data Fig. S3). It is possible that our limited sampling of the parental species, drawn from previous work, reduces our ability to accurately assess trait space in Y. aloifolia and Y. filamentosa. However, multiple genotypes were sampled across the ranges of both parental species, and thus the greater variation found within the hybrid is likely to be due to genomic mixing, rather than sampling bias. While trait values in Y. gloriosa were typically intermediate, one notable exception was the transgressive values of total CO2 assimilation under both watered and drought-stressed conditions. Due to Y. gloriosa’s nearly C3-level of daytime CO2 fixation coupled with the ability to use low-level CAM, total CO2 uptake rates far exceed that of either parent, at least in certain genotypes. Such a mixed photosynthetic strategy may be particularly valuable on the coastal dunes that Y. gloriosa is restricted to, because although rainfall in the southeastern USA is not particularly limiting, any water that does fall probably percolates through the sandy substrate quickly.
Despite the potentially novel phenotypes that Y. gloriosa exhibits relative to its parental species, they are unlikely to underlie the speciation of the hybrid from its progenitors. All three Yucca species are found across the southeastern US coast, although only Y. aloifolia and Y. gloriosa grow with any frequency on the coastal dunes. Yucca filamentosa is typically further away from the ocean in the coastal scrub, although it can be found in exposed sand near brackish inlets (K. Heyduk, unpubl. res.). Homoploid hybrid species can be formed and maintained either through chromosomal structural rearrangements that form a reproductive barrier between the new species and its progenitors, or via ecological differentiation, whereby the new combination of traits in the hybrid allows for habitation of a novel niche relative to the parental species (Gross and Rieseberg, 2005). As the habitats of the Yucca species studied here largely overlap, particularly Y. gloriosa and Y. aloifolia, the latter at first seems unlikely, despite Y. gloriosa being clearly distinct in total CO2 assimilation rates. However, flowering time of the three species is markedly different: Y. filamentosa typically flowers earliest, in late May and June, followed by Y. aloifolia. Yucca gloriosa has been noted to flower at the end of the summer and into autumn (Trelease, 1902); whether the later flowering time was selected for in order to reduce backcrossing, or was instead a byproduct of the initial hybridization events, remains unknown. Additionally, other biotic interactions (e.g. below-ground mutualisms or pollinators) or microhabitat variation are largely untested as potential drivers of Yucca speciation (but see Rentsch and Leebens-Mack, 2014). Chromosomal structural rearrangements may explain an apparent lack of backcrossed individuals, but we do not currently have the genomic data to test this hypothesis.
Intraspecific variation for CAM upregulation
Genotypes of Y. gloriosa used variable levels of CAM, and differentially upregulated CAM under drought stress. The differential drought response was a result of two separate axes of the CAM phenotype: both leaf acid accumulation and nocturnal CO2 uptake varied by environment, and could do so in a de-coupled manner (Fig. 4). That is, certain genotypes increased the amount of leaf acids accumulated based not on increasing atmospheric CO2 uptake but instead by presumably re-fixing respired CO2. Such a response indicates that many of the required enzymes are present and regulated correctly, but that stomatal aperture responded negatively to drought at night. Reducing net CO2 uptake but increasing leaf acid accumulation is the typical response of Y. aloifolia to drought stress as well. In general, the response to drought stress in Y. gloriosa was transgressive relative to parental phenotypes, in that genotypes of Y. gloriosa were able to respond to drought stress in ways that neither parent could. For example, certain genotypes could increase both CO2 uptake and leaf acid accumulation under drought stress; this response was not seen in any of the parental genotypes measured here. Other genotypes occupied a part of trait space where nocturnal CO2 uptake increased under drought stress, but leaf acids decreased (Fig. 4D). How incoming CO2 is processed in these genotypes remains unclear and warrants additional exploration in these genotypes, especially through metabolomic and genomic analyses to help pinpoint alternative pathways.
The segregation of CAM drought response in Y. gloriosa also presents an ideal system with which to interrogate the molecular components of drought response in facultative CAM species. CAM has been touted as a potential trait for increasing food and biofuel crop drought tolerance through bioengineering (Borland et al., 2014, 2015), and early efforts to transform C3 species to CAM were instrumental in generating an abundance of genomics data for CAM species. Yet fully committing a C3 plant to CAM may result in costs that outweigh any gains in drought tolerance; larger leaves and cells will require more energy and time to produce, and constitutive CAM usage is not ideal when drought may be intermittent. Instead, drought tolerance engineering efforts should look to facultative CAM or C3+CAM, as in Y. gloriosa, which outperforms its parental species in terms of total CO2 uptake, and may result in faster biomass gains, although this remains to be tested. The natural variation for CAM induction and upregulation in Y. gloriosa, as well as the uncoupling of various CAM traits, including anatomy, acid accumulation and CO2 uptake, make Y. gloriosa ideal for investigating the molecular basis of particular CAM traits and their regulation via abiotic signalling.
Future work should continue to examine intraspecific variation in plant anatomical and photosynthetic traits, particularly in intermediate species, as well as variation under different environmental conditions. There is likely to be significant variation even in non-hybrid species. The grass Alloteropsis semialata has both C3, C4 and intermediate individuals, and is a model system for understanding how C4 evolved in this species (Ueno and Sentoku, 2006; Lundgren et al., 2016). Moreover, photosynthetic traits are likely to vary across the geographical range of a species, especially if that range has variation in environmental cues. For example, photorespiration rates vary across populations of Flaveria linearis, an intermediate C3–C4 species (Teese, 1995). Finally, photosynthetically intermediate species are not the only ones capable of showing intraspecific variation in leaf anatomical and photosynthetic traits. Accessions of the C4Gynandropsis gynandra have high enough intraspecific variation for C4 traits that crosses between phenotypically distinct genotypes could allow for genetic mapping of traits of interest (Reeves et al., 2018). While it has not been examined extensively, strong CAM species have the potential to exhibit intraspecific variation, and understanding that variation can help us better understand the overall plasticity of complex traits such as CAM photosynthesis.
Implications for the evolution of CAM
While Y. gloriosa is a hybrid and therefore represents a somewhat atypical avenue for trait evolution, it still allows us a glimpse into how a trait such as CAM might be assembled. The homoploid nature of Y. gloriosa means that the genomic content of the two parental species is not highly divergent, and that the mix of traits found in Y. gloriosa genotypes are not a result of a highly perturbed genome but more like what may be expected of an intraspecific cross between phenotypically divergent parents. The mixture of traits within Y. gloriosa allows us to speculate on the genomic architecture underlying the CAM phenotype. It seems unlikely that many of the traits are genetically linked – that is, the few relationships between traits within Y. gloriosa mean the underlying genes are dispersed in physical genomic location and that they are not necessarily expressed in or regulated by similar pathways. For example, there is nothing in the genome of Y. gloriosa that requires large cells to develop low IAS (or vice versa), or that CAM activity is in any way linked genetically to leaf thickness. The variation in and lack of association between traits in Y. gloriosa also implies, unsurprisingly, that the CAM phenotype is highly quantitative, and that recombination can break up many of the underlying traits. The genetic architecture of CAM does not fully explain why such a mix of traits has remained in Y. gloriosa. Perhaps not enough generations have passed for the traits to sort into parental types, or perhaps the environment in some way promotes the maintenance of Y. gloriosa’s interemediate phenotypes. Alternatively, Y. gloriosa is not a particularly rare species in its native habitat, but its populations are small and relatively isolated. In such small populations, selection has a weaker effect than drift, which can lead to less advantageous combinations of traits persisting in a population (Ohta, 1992). Additional research using reciprocal transplants could facilitate our understanding of whether intermediate traits like those found in Y. gloriosa can confer a fitness advantage in some circumstances.
The variation and lack of correlation between traits underlying the CAM phenotype in Y. gloriosa also give insight into how CAM is assembled over evolutionary time. While certain traits appear fixed when we examine strong C3 and CAM species, intermediate species are important for understanding the processes that may have led to trait fixation and correlation across traits. After all, selection acts not on the species level, but on individuals, and indeed there is a broad phenotypic space within Y. gloriosa for the traits examined here that selection could act upon. That selection seems to recurrently end up on a particular anatomical phenotype in CAM species (i.e. larger cells, thicker leaves) despite no genetic constraint for such a correlation suggests there is an optimal combination of traits for CAM efficiency. The pattern of convergent evolution of trait combinations, paired with intermediate species showing highly variable trait combinations, implies a funnel shape to the evolutionary trajectory of CAM. Species can use a degree of CAM without committing to any particular leaf anatomy (Edwards, 2019), meaning that initial transitions to using C3+CAM can follow broad and varied routes. This is in contrast to the evolution of C4 photosynthesis, which, like CAM, requires specific anatomical characteristics. In C4 lineages, anatomical changes occur prior to the evolution of C4 biochemistry (McKown and Dengler, 2007; Lundgren et al., 2019); in some cases, like the PACMAD clade of grasses, these anatomical changes happen early enough in evolutionary time that they are thought to have facilitated repeated origins of C4 (Christin et al., 2013). In contrast, ‘weak’ CAM or C3+CAM has no anatomical constraints in Yucca. There is, however, an upper bound where further investment in CO2 fixation by the CAM pathway requires dedicated anatomy, although the exact threshold of that transition point remains unclear. The funnel shape to the evolution of CAM, whereby no anatomical constraints impact low levels of CAM function, means that ordering of events on the evolutionary trajectory from C3 to CAM will be exceedingly difficult, as lineages can take various routes through the intermediate zone.
While the lack of correlated traits in an intermediate C3+CAM hybrid species has implications for broader questions on the evolution of CAM, future work can elaborate upon the patterns seen here and help to assess how generalizable these results are. Sampling of parental genotypes and overall range was limited, and thus there may exist greater variation among traits in the parental C3 and CAM species as well. Indeed, most studies that examine the correlation of anatomy to photosynthetic physiology do not sample intraspecific variation, and therefore it remains a largely unexplored area of CAM. The growth conditions used in this study were based on previous work in the Yucca system, but modulating those conditions may reveal deeper levels of variation across environmental gradients. Finally, the Yucca hybrid system is a single example of intermediacy between C3 and CAM, and other C3+CAM species should continue to be examined via detailed physiology and anatomy to advance fundamental understanding of how CAM evolves. Investigations within and between species exhibiting a mix of CAM, C3 and intermediate species will continue to provide insights into whether the decoupling of CAM traits we observe in a hybrid species holds more generally.
CONCLUSIONS
Comparisons between C3 and CAM species have suggested suites of traits are correlated to maximize the efficiency of each photosynthetic pathway. CAM species have large cells for storing malate, and the cells are often packed together densely in large, thick leaves to minimize CO2 leakage back into the atmosphere; C3 plants have large amounts of airspace between significantly smaller cells to facilitate the diffusion of CO2 to the sites of Rubisco carboxylation. These trends have been seen repeatedly in independent CAM lineages, but few studies have examined intermediate C3+CAM plants, and even fewer have assessed intraspecific variation for traits. The C3+CAM hybrid Y. gloriosa examined here not only has a greater range of traits than either of its parental species, but it also lacks many of the trait correlations commonly associated with the ability to use CAM. Indeed, no single leaf anatomical trait could predict the amount of CO2 acquired via CAM in the hybrid species. The lack of correlation within the intermediate Y. gloriosa suggests that the evolutionary trajectory to CAM from C3 passes through a stage where many combinations of anatomical and photosynthetic physiology traits are viable. Furthermore, in Yucca at least, anatomical and physiological traits are not genetically linked, supporting existing hypotheses that suites of leaf traits found repeatedly in CAM species have been selected for in order to maximize photosynthetic efficiency. Finally, we find that there is extensive intraspecific variation in the ability to upregulate CAM under drought stress in Y. gloriosa. Using the variation for CAM in this hybrid, we can begin to interrogate the genetic mechanisms that link environmental cues to CAM photosynthesis.
SUPPLEMENTARY DATA
Figure S1: Soil moisture measurements. Figure S2: Stomatal conductance measurements. Figure S3: PCA of physiological and anatomical traits. Figure S4: Pairwise correlations between traits. Table S1: GPS coordinate information for all accessions used in this study.Table S2: Soil dry down information for Yucca gloriosa genotypes measured for gas exchange and titratable acidity.Table S3: ANOVA/ANCOVA of trait differences among genotypes of Y. gloriosa and treatments.Table S4: ANOVA of trait differences across the three species of Yucca.Table S5: Correlations (raw and genotypic means) across traits in all three species.Table S6: Correlations (raw and genotypic means) across traits in Y. gloriosa.
ACKNOWLEDGEMENTS
The authors would like to thank the state parks of Georgia, Florida, South Carolina, North Carolina and Virginia, as well as the National Park Service, for their assistance in collection and permissions. Special thanks to Nida Moledina, Dan Tepstov, Grace Manning, Rushi Patel and Charmi Patel for measuring thousands of titration samples, the glasshouse staff at UGA (particularly Mike Boyd and Greg Cousins) for granting us growth chamber access, Rick Field and Amanda Cummings for assistance in collecting anatomical samples, and Cody Howard and Ed McAssey for comments on the manuscript.
FUNDING
This work was supported by the National Science Foundation [no. DEB 1442199 to J.L-M.] and the Donnelley Postdoctoral Fellowship at Yale University [to K.H.].
LITERATURE CITED
- Barton NH. 2010. Genetic linkage and natural selection. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 365: 2559–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera Zambrano VA, Lawson T, Olmos E, Fernández-García N, Borland AM. 2014. Leaf anatomical traits which accommodate the facultative engagement of crassulacean acid metabolism in tropical trees of the genus Clusia. Journal of Experimental Botany 65: 3513–3523. [DOI] [PubMed] [Google Scholar]
- Borland AM, Hartwell J, Weston DJ, et al. 2014. Engineering crassulacean acid metabolism to improve water-use efficiency. Trends in Plant Science 19: 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland AM, Leverett A, Hurtado-Castano N, Hu R, Yang X. 2018. Functional anatomical traits of the photosynthetic organs of plants with crassulacean acid metabolism. In: Adams WW III, Terashima I, eds. The leaf: a platform for performing photosynthesis. Cham: Springer International Publishing, 281–305. [Google Scholar]
- Borland AM, Wullschleger SD, Weston DJ, et al. 2015. Climate-resilient agroforestry: physiological responses to climate change and engineering of crassulacean acid metabolism (CAM) as a mitigation strategy. Plant, Cell & Environment 38: 1833–1849. [DOI] [PubMed] [Google Scholar]
- Bradshaw HD, Schemske DW. 2003. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426: 176–178. [DOI] [PubMed] [Google Scholar]
- Chan YH, Marshall WF. 2014. Organelle size scaling of the budding yeast vacuole is tuned by membrane trafficking rates. Biophysical Journal 106: 1986–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Osborne CP, Chatelet DS, et al. 2013. Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proceedings of the National Academy of Sciences of the United States of America 110: 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ. 2019. Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. The New Phytologist 223: 1742–1755. [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Ogburn RM. 2012. Angiosperm responses to a low-CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories. International Journal of Plant Sciences 173: 724–733. [Google Scholar]
- Gerrish PJ, Colato A, Perelson AS, Sniegowski PD. 2007. Complete genetic linkage can subvert natural selection. Proceedings of the National Academy of Sciences of the United States of America 104: 6266–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AC. 1982. The anatomy of succulence. In: Ting IP, Gibbs M, eds. Crassulacean acid metabolism: Proceedings of the fifth annual symposium in botany. Rockville, MD: American Society of Plant Physiologists, 1–17. [Google Scholar]
- Gravatt DA, Martin CE. 1992. Comparative ecophysiology of five species of Sedum (Crassulaceae) under well-watered and drought-stressed conditions. Oecologia 92: 532–541. [DOI] [PubMed] [Google Scholar]
- Gross BL, Rieseberg LH. 2005. The ecological genetics of homoploid hybrid speciation. The Journal of Heredity 96: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsock TL, Nobel PS. 1976. Watering converts a CAM plant to daytime CO2 uptake. Nature 262: 574–576. [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895: 81–106. [Google Scholar]
- Herrera A. 2009. Crassulacean acid metabolism and fitness under water deficit stress: if not for carbon gain, what is facultative CAM good for? Annals of Botany 103: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, Burrell N, Lalani F, Leebens-Mack J. 2016a. Gas exchange and leaf anatomy of a C3-CAM hybrid, Yucca gloriosa (Asparagaceae). Journal of Experimental Botany 67: 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, McKain MR, Lalani F, Leebens-Mack J. 2016b. Evolution of a CAM anatomy predates the origins of Crassulacean acid metabolism in the Agavoideae (Asparagaceae). Molecular Phylogenetics and Evolution 105: 102–113. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Moreno-Villena JJ, Gilman IS, Christin PA, Edwards EJ. 2019a. The genetics of convergent evolution: insights from plant photosynthesis. Nature Reviews. Genetics 20: 485–493. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Ray JN, Ayyampalayam S, et al. 2019. b. Shared expression of crassulacean acid metabolism (CAM) genes pre-dates the origin of CAM in the genus Yucca. Journal of Experimental Botany 70: 6597–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313: 101–104. [DOI] [PubMed] [Google Scholar]
- Lee H, Rustgi S, Kumar N, et al. 2011. Single nucleotide mutation in the barley acetohydroxy acid synthase (AHAS) gene confers resistance to imidazolinone herbicides. Proceedings of the National Academy of Sciences of the United States of America 108: 8909–8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Ofria C, Pennock RT, Adami C. 2003. The evolutionary origin of complex features. Nature 423: 139–144. [DOI] [PubMed] [Google Scholar]
- Lundgren MR, Christin PA, Escobar EG, et al. 2016. Evolutionary implications of C3–C4 intermediates in the grass Alloteropsis semialata. Plant, Cell & Environment 39: 1874–1885. [DOI] [PubMed] [Google Scholar]
- Lundgren MR, Dunning LT, Olofsson JK, et al. 2019. C4 anatomy can evolve via a single developmental change. Ecology Letters 22: 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttge U, Stimmel KH, Smith JAC, Griffiths H. 1986. Comparative ecophysiology of CAM and C3 bromeliads II: field measurements of gas exchange of CAM bromeliads in the humid tropics. Plant, Cell and Environment 19: 377–383. [Google Scholar]
- Males J. 2018. Concerted anatomical change associated with crassulacean acid metabolism in the Bromeliaceae. Functional Plant Biology 45: 681–695. [DOI] [PubMed] [Google Scholar]
- Maxwell K, von Caemmerer S, Evans JR. 1997. Is a low internal conductance to CO2 diffusion a consequence of succulence in plants with crassulacean acid metabolism? Australian Journal of Plant Physiology 24: 777. [Google Scholar]
- McKown AD, Dengler NG. 2007. Key innovations in the evolution of Kranz anatomy and C4 vein pattern in Flaveria (Asteraceae). American Journal of Botany 94: 382–399. [DOI] [PubMed] [Google Scholar]
- Nelson EA, Sage RF. 2008. Functional constraints of CAM leaf anatomy: tight cell packing is associated with increased CAM function across a gradient of CAM expression. Journal of Experimental Botany 59: 1841–1850. [DOI] [PubMed] [Google Scholar]
- Nelson EA, Sage TL, Sage RF. 2005. Functional leaf anatomy of plants with crassulacean acid metabolism. Functional Plant Biology 32: 409. [DOI] [PubMed] [Google Scholar]
- Ohta T. 1992. The nearly neutral theory of molecular evolution. Annual Review of Ecology and Systematics 23: 263–286. [Google Scholar]
- R Core Team . 2019. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Reeves G, Singh P, Rossberg TA, Sogbohossou EOD, Schranz ME, Hibberd JM. 2018. Natural variation within a species for traits underpinning C4 photosynthesis. Plant Physiology 177: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch JD, Leebens-Mack J. 2012. Homoploid hybrid origin of Yucca gloriosa: intersectional hybrid speciation in Yucca (Agavoideae, Asparagaceae). Ecology and Evolution 2: 2213–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch JD, Leebens-Mack J. 2014. Yucca aloifolia (Asparagaceae) opts out of an obligate pollination mutualism. American Journal of Botany 101: 2062–2067. [DOI] [PubMed] [Google Scholar]
- Sage RF, Khoshravesh R, Sage TL. 2014. From proto-Kranz to C4 Kranz: building the bridge to C4 photosynthesis. Journal of Experimental Botany 65: 3341–3356. [DOI] [PubMed] [Google Scholar]
- Signorell A. 2019. DescTools: tools for descriptive statistics. R package version 0.99.34. https://cran.r-project.org/package=DescTools. [Google Scholar]
- Silvera K, Santiago LS, Winter K. 2005. Distribution of crassulacean acid metabolism in orchids of Panama: evidence of selection for weak and strong modes. Functional Plant Biology 32: 397. [DOI] [PubMed] [Google Scholar]
- Teese P. 1995. Intraspecific variation for CO2 compensation point and differential growth among variants in a C3–C4 intermediate plant. Oecologia 102: 371–376. [DOI] [PubMed] [Google Scholar]
- Trelease W. 1902. The Yucceae. Missouri Botanical Garden Annual Report 1902: 27–133. [Google Scholar]
- Ueno O, Sentoku N. 2006. Comparison of leaf structure and photosynthetic characteristics of C3 and C4 Alloteropsis semialata subspecies. Plant, Cell & Environment 29: 257–268. [DOI] [PubMed] [Google Scholar]
- Weber JN, Peterson BK, Hoekstra HE. 2013. Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature 493: 402–405. [DOI] [PubMed] [Google Scholar]
- Winter K. 2019. Ecophysiology of constitutive and facultative CAM photosynthesis. Journal of Experimental Botany 70: 6495–6508. [DOI] [PubMed] [Google Scholar]
- Winter K, Garcia M, Holtum JA. 2008. On the nature of facultative and constitutive CAM: environmental and developmental control of CAM expression during early growth of Clusia, Kalanchöe, and Opuntia. Journal of Experimental Botany 59: 1829–1840. [DOI] [PubMed] [Google Scholar]
- Yuan YW, Sagawa JM, Young RC, Christensen BJ, Bradshaw HD Jr. 2013. Genetic dissection of a major anthocyanin QTL contributing to pollinator-mediated reproductive isolation between sister species of Mimulus. Genetics 194: 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.