Abstract
Background
Flagella‐mediated motility is both a crucial virulence determinant of Helicobacter pylori and a factor associated with gastrointestinal diseases. Flagellar formation requires flagellins to be glycosylated with pseudaminic acid (Pse), a process that has been extensively studied. However, the transfer of Pse to flagellins remains poorly understood. Therefore, the aim of this study is to characterize a putative glycosyltransferase jhp0106 in flagellar formation.
Materials and Methods
Western blotting and chemical deglycosylation were performed to examine FlaA glycosylation. Protein structural analyses were executed to identify the active site residues of Jhp0106, while the Jhp0106‐FlaA interaction was examined using a bacterial two‐hybrid assay. Lastly, site‐directed mutants with mutated active site residues in the jhp0106 gene were generated and investigated using a motility assay, Western blotting, cDNA‐qPCR analysis, and electron microscopic examination.
Results
Loss of flagellar formation in the Δjhp0106 mutant was confirmed to be associated with non‐glycosylated FlaA. Furthermore, three active site residues of Jhp0106 (S350, F376, and E415) were identified within a potential substrate‐binding region. The interaction between FlaA and Jhp0106, Jhp0106::S350A, Jhp0106::F376A, or Jhp0106::E415A was determined to be significant. As well, the substitution of S350A, F376A, or E415A in the site‐directed Δjhp0106 mutants resulted in impaired motility, deficient FlaA glycosylation, and lacking flagella. However, these phenotypic changes were regardless of flaA expression, implying an indefinite proteolytic degradation of FlaA occurred.
Conclusions
This study demonstrated that Jhp0106 (PseE) binds to FlaA mediating FlaA glycosylation and flagellar formation. Our discovery of PseE has revealed a new glycosyltransferase family responsible for flagellin glycosylation in pathogens.
Keywords: flagellin, glycosyltransferase, Helicobacterpylori, Jhp0106, motility
1. INTRODUCTION
The flagellum is a crucial virulence factor of Helicobacter pylori and is essential for the colonization of gastric mucosa. 1 , 2 , 3 , 4 Flagella‐mediated motility facilitates H.pylori pathogenesis during the initial colonization. It also helps H.pylori achieve dense colonization and is associated with the pathological outcomes of gastrointestinal diseases such as chronic gastritis, gastric and duodenal ulcer, mucosa‐associated tissue lymphoma, and gastric adenocarcinoma. 2 , 5 , 6 , 7 , 8 Most H.pylori cells harbor three to five polar‐sheathed flagella, which are comprised of the basal body, hook, and filament. 9 , 10 More than forty different proteins are involved in flagellar formation, rotation, and chemo‐response, making a flagellum one of the most complex structures in H.pylori cells. In H.pylori, the main components of the filament are FlaA (major flagellin) and FlaB (minor flagellin), whose gene expression is under a hierarchical regulation by the three transcriptional sigma factors RpoD/σ80, RpoN/σ54, and FliA/σ28. 11 , 12 This RpoD/RpoN/FliA regulatory system ensures that the synthesis of early flagellar regulators and structural genes is established before the expression of flaA begins. Furthermore, the synthesis and assembly of mature FlaA is also modulated by glycosylation via a post‐translational modification. 13 , 14
Glycosylation on flagellins enables flagella assembly and bacterial motility. 15 H.pylori utilizes a sialic acid‐like nonulosonic acid (pseudaminic acid, Pse) for its flagellin glycosylation, similar to the O‐linked glycosylation in Aeromonas, Campylobacter, and Magnetospirillum. 16 , 17 , 18 Six enzymes, PseB, PseC, PseH, PseG, PseI, and PseF, are involved in the biosynthesis of Pse in A.caviae, A.hydrophila, C. jejuni, and H. pylori. 13 , 16 , 17 , 19 , 20 , 21 , 22 Mutation of the genes coding for Pse biosynthesis results in non‐motile aflagellate cells, indicating the importance of O‐linked glycosylation in virulent pathogens. Although the biosynthesis of O‐linked glycosylation has been extensively studied, how a synthesized Pse element is transferred onto flagellins remains an unsolved puzzle. It has been suggested that the glycosyltransferase PseE in C.jejuni and other members of a motility‐associated factor (Maf) family play a role in Pse transfer. With the nature of low protein sequence homology and structural complexity in glycosyltransferases, conducting a molecular study to investigate Pse glycosyltransferase has been challenging. 13 , 17 , 19 , 23 , 24 , 25 Recently, Sulzenbacher et al. solved the first crystal structure of Maf in M. magneticum AMB‐1. 18 Their structural insights provide an excellent fundament for future characterizations of Pse glycosyltransferases.
In the genome of H.pylori strain J99, we have identified a homolog of an maf gene, named jhp0106. We hypothesize that Jhp0106 is a Pse glycosyltransferase for flagellin glycosylation. Applying our findings from the previous study, 26 the expression of jhp0106 is under the control of the CsrA/RpoN regulatory system and essential for flagellar formation and, consequently, bacterial motility. In this study, we aimed to bridge the gap between Pse biosynthesis and flagellin glycosylation. We conducted a combination of computational, phenotypic, and molecular analyses to gain further information on the characteristics of glycosyltransferase Jhp0106, and to understand the role of Jhp0106 in flagellin FlaA glycosylation and flagella‐associated motility in H.pylori.
2. MATERIALS AND METHODS
2.1. Bacterial cultivation
The bacterial strains and plasmids used in this study are described in Table 1. H.pylori was grown on CDC anaerobic blood agar (BBL, Microbiology Systems, Cockeysville, MD, USA) or in Brucella broth containing 10% (v/v) horse serum (Gibco BRL, Life Technologies, Rockville, MD, USA) at 37°C in microaerophilic conditions (5% O2, 10% CO2 and 85% N2). E.coli was grown on Luria‐Bertani (LB) (BD Biosciences, San Jose, CA, USA) agar or in broth. Bacteria harboring antibiotic resistance determinants were grown in the presence of the appropriate antibiotics at the following concentrations: ampicillin (Amp, 100 μg/mL); chloramphenicol (Cm, 25 μg/mL); and kanamycin (Km, 50 μg/mL for E.coli, 10 μg/mL for H.pylori).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference |
|---|---|---|
| E.coli strain | ||
| DH5α | F− Ψ 80dlacZ∆M15 ∆(lacZYA‐argF) U169 hsdR17 recA1 thi−1 relA1 | Laboratory stock |
| XL1‐Blue MRF’ |
∆(mcrA)183 ∆(mcrCB‐hsdSMR‐mrr)173 endA1 supE44 thi−1 recA1gyrA96 relA1 lac [F’ proAB lacIqZ ∆M15 Tn5 (Kmr)] |
Stratagene |
| H.pylori strain | ||
| J99 | Wild‐type strain isolated from patient with duodenal ulcer; motile | 8 |
| SW862 | ∆jhp0106 revertant, derived from SW863; motile | 26 |
| SW863 | ∆jhp0106 insertion mutant; non‐motile; J99 strain having a truncated form of Jhp0106; Kmr | 26 |
| SW866 | ∆flaA mutant; non‐motile; Kmr | 26 |
| SW867 | ∆flaA revertant, derived from SW866; motile | 26 |
| SW871 | ∆jhp0106::S350A mutant; non‐motile. J99 strain having Jhp0106 with amino acid residue 350 substitution from serine to alanine | This study |
| SW872 | ∆jhp0106::F376A mutant; non‐motile. J99 strain having Jhp0106 with amino acid residue 376 from phenylalanine to alanine | This study |
| SW869 | ∆jhp0106::E415A mutant; non‐motile. J99 strain having Jhp0106 with amino acid residue 415 substitution from glutamate to alanine | This study |
| Plasmid | ||
| yT&A | E.coli cloning vector; Ampr | RBC Bioscience |
| yT&A‐pTRG‐FlaA | yT&A derivative plasmid containing full‐length FlaA; Ampr | This study |
| yT&A‐pBT‐Jhp0106 | yT&A derivative plasmid containing full‐length JHP0106; Ampr | This study |
| pBT | Bacterial two‐hybrid vector for N‐terminal RGS‐λcI fusion; p15A ori; Cmr | Stratagene |
| pBT‐Jhp0106 | pBT expressing full‐length Jhp0106; Cmr | This study |
| pBT‐GrlA | pBT expressing full‐length GrlA; Cmr | Laboratory stock |
| pBT‐Jhp0106 S350A | pBT expressing full‐length Jhp0106 containing a point mutation: amino acid residue serine altered to alanine on position 350; Cmr | This study |
| pBT‐Jhp0106 F376A | pBT expressing full‐length Jhp0106 containing a point mutation: amino acid residue phenylalanine altered to alanine on position 376; Cmr | This study |
| pBT‐Jhp0106 E415A | pBT expressing full‐length Jhp0106 containing a point mutation: amino acid residue glutamate altered to alanine on position 415; Cmr | This study |
| pTRG | Bacterial two‐hybrid vector for N‐terminal RGS‐RNAPα fusion; ColE1 ori; Tcr | Stratagene |
| pTRG‐Jhp0106 | pTRG expressing full‐length Jhp0106; Tcr | This study |
| pTRG‐FlaA | pTRG expressing full‐length FlaA; Tcr | This study |
| pTRG‐GrlR | pTRG expressing full‐length GrlR; Tcr | Laboratory stock |
Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant.
2.2. SDS‐PAGE and Western blotting analysis
Bacterial total proteins were subjected to 8% SDS‐PAGE, and the proteins were transferred to PVDF membranes, followed by blocking in 5% skimmed milk. Membranes were probed overnight at 4°C with diluted 1:5000 mouse anti‐FlaA polyclonal antibody (in‐house) or 1:5000 mouse anti‐Hsp60 monoclonal antibody (Sigma‐Aldrich) in TBS buffer containing 0.1% Tween 20. The washing and visualization of the probed membrane were carried out as described in our previous study. 27
2.3. Deglycosylation of flagellin FlaA proteins
To remove O‐linked glycosaminoglycan chains, trifluoromethanesulphonic acid (TfOH) treatment was conducted with modifications. 28 In short, we used French press to extract the total proteins in the supernatant fraction of H.pylori cells, freeze‐dried the collected supernatant, and then dissolved proteins in an anisole:TfOH mixture (ratio of 1:2, v:v) in an ice‐water bath for 3 hours. The TfOH‐treated proteins were extracted and precipitated in a 50 × ether:n‐hexane mixture (ratio of 9:1, v:v) at −40°C for 3 hours, to recover deglycosylated proteins. The proteins were then pelleted and washed again before resuspended in the 50 × ether:n‐hexane mixture. The resultant suspension was stored at −40°C overnight to allow a sufficient precipitation. After centrifugation, the pelleted deglycosylated proteins were vacuum‐dried and dissolved in distilled water. We also used 5 mol/L guanidine hydrochloride to dissolve the insoluble deglycosylated proteins and the TfOH‐untreated samples. All samples were subjected to 8% SDS‐PAGE for Western blotting as described above.
2.4. In silico simulation of Jhp0106
Homologous protein modeling of Jhp0106 was generated on the SWISS‐MODEL server (https://swissmodel.expasy.org/). Three‐dimensional structural analysis and superimposition were performed using the program PyMOL Molecular Graphics System (http://www.pymol.org). The coordinates and structure factors for the Maf of M.magneticum AMB‐1 retrieved from the Protein Data Bank with accession numbers 5MU5. We selected twenty‐two homologous protein sequences of Jhp0106 (protein ID: WP_079993006.1; NCBI genome accession number: NC_000921.1) in Epsilonproteobacteria retrieved from the UniProtKB Database (https://www.uniprot.org/uniprot/) using the BLASTp program (Table S1). Alignment of Jhp0106 and its homologous protein sequences was generated using the CLC Sequence Viewer version 8.0 (CLC Bio, Cambridge, MA). The secondary structural prediction of Jhp0106 was carried out using Jpred4, a protein secondary structure prediction server (http://www.compbio.dundee.ac.uk/jpred/). The coiled‐coil domain was predicted using the DeepCoil software. 29
2.5. Bacterial two‐hybrid assay
Plasmid construction: the 1,533 bp full‐length flaA fragment obtained from the H.pylori J99 genomic DNA was amplified using pTRG‐FlaA‐BamHI‐F and pTRG‐FlaA‐XhoI‐R primers (Table 2). The resultant flaA amplicon was ligated into plasmid yT&A (RBC Bioscience, Taiwan) to generate plasmid yT&A‐pTRG‐FlaA. After BamHI/XhoI digestion, a BamHI/XhoI‐cleaved 1,539 bp flaA fragment was inserted into BamHI/XhoI‐digested pTRG (Stratagene, La Jolla, CA). This plasmid was designated “plasmid pTRG‐FlaA.” Using the same strategy, we constructed yT&A‐pBT‐Jhp0106 and pBT‐Jhp0106. The strategic detail of creating a bait plasmid (pBT‐Jhp0106‐S350A) carrying a point mutation is given. We first performed site‐directed mutagenesis PCR to replace the targeted amino acids of Jhp0106 using Herculase II fusion DNA polymerase (Agilent, Waldbronn, Germany) with primers 0106‐Ser350toAla‐F and 0106‐Ser350toAla‐R (Table 2). This inverse PCR amplicon was self‐ligated to form pBT‐Jhp0106‐S350A, followed by Sanger sequencing validation. The same approach was applied to construct pBT‐Jhp0106‐F376A and pBT‐Jhp0106‐E415A.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5’−3’) |
|---|---|
| pBT−0106‐EcoRI‐F | GAATTCTATGAATATTTATCAAAAAAACTTGCAAGCTCT |
| pBT−0106‐BglII‐R | AGATCTTTACCATTCTTTCAAAGCCATTTTGATC |
| pTRG‐FlaA‐BamHI‐F | GGATCCATGGCTTTTCAGGTCAATACAAATATCAAT |
| pTRG‐FlaA‐XhoI‐R | CTCGAGCTAAGTTAAAAGCCTTAAGATATTTTGTTGAACG |
| 0106‐Ser350toAla‐F | GGCATAGGCATGGCCGCAGCGAACATG |
| 0106‐Ser350toAla‐R | CATGTTCGCTGCGGCCATGCCTATGCC |
| 0106‐Phe376toAla‐F | GGGCAAGATTTGAGCGCTTCACAAAGCGGTAACA |
| 0106‐Phe376toAla‐R | TGTTACCGCTTTGTGAAGCGCTCAAATCTTGCCC |
| 0106‐Glu415toAla‐F | GGGGGTAATGGGAAAGTAGCAACCACT TTAGTGTGGAAA |
| 0106‐Glu415toAla‐R | TTTCCACACTAAAGTGGTTGCTACTTTCCCATTACCCCC |
| jhp0106‐Mut−1 | CCGGAATTCCGCTTCTTCAACTTGCGTTT |
| jhp0106‐Mut−2 | CCGGAATTCAAATCAACCAGCATTTCACG |
| gyrA‐RealT−1 | GACACCGCAGTTTATGATGC |
| gyrA‐RealT−1 | TTCTGGCTTCAGTGTAACGC |
| jhp0548(flaA)‐qPCR−1 | AATGGCGGTCAGGATTTAAC |
| jhp0548(flaA)‐qPCR−2 | TGTGAGTCAGAAGCCGAAAC |
Assay: a BacterioMatch two‐hybrid system (Stratagene) was applied to determine the interaction between Jhp0106 and FlaA. Briefly, the two types of plasmids, pBT and pTRG, containing the respective bait (Jhp0106 or its mutants) and target (FlaA) genes were used to simultaneously transform to E.coli XL1‐Blue MRF’ cells. Protein‐protein interaction was determined by measuring the expression of the reporter gene (β‐galactosidase). The activity of β‐galactosidase was quantified using Miller units and then applied for a comparison between different clones to identify protein‐protein interaction. 30
2.6. Construction of the jhp0106 site‐directed mutant strains
For generating the jhp0106 mutants carrying point mutation on amino acid residues S350, F376, and E415, the individual 1004‐bp jhp0106 mutant fragments were amplified from pBT‐Jhp0106‐S350A, pBT‐Jhp0106‐F376A, and pBT‐Jhp0106‐E415A (Table 1) using the primers jhp0106‐Mut‐1 and jhp0106‐Mut‐2 (Table 2). The generated amplicons were EcoRI‐cleaved, ligated into pUC18 to designate as KO‐plasmids (Table 1). These KO‐plasmids were naturally transformed into the H.pylori Δjhp0106 mutant to create jhp0106 mutants with amino acid substitutions (S350A, F376A, or E415A). The kanamycin‐sensitive transformants of H.pylori were chosen for validating specific point mutations by Sanger sequencing analysis.
2.7. Spreading motility assay and quantitation of motility
Spreading motility was assessed by measuring the diameter of the migration of the bacteria through the soft agar (0.3%) from the inoculated center toward the periphery of the plate as described in our previous study. 27 After seven‐day inoculation, the motility of each strain was recorded and calculated as the mean ±SD of three independent experiments.
2.8. RNA isolation and cDNA preparation
H.pylori was grown on Brucella/10% horse serum agar plates for 32–36 hours, transferred to 50 mL Brucella broth/10% horse serum at an initial OD600 of 0.2, and inoculated at 37°C with shaking. Once bacterial OD600 reached 1.2, we preserved RNA profile by adding 200 μL of iced‐cold 5% acidic phenol in ethanol to one mL of bacterial culture, and stored the cells at −80°C until use. RNA isolation was performed using GENEzol TriRNA Bacteria kit (Geneaid, Taiwan) according to the manufacturer's recommendations, followed by RQ1 RNase‐free DNase treatment (Promega) at 37°C for 30 minutes. The DNase‐treated RNAs were immediately purified using RNA Cleanup kit (Geneaid) and eluted in nuclease‐free water (Ambion) with addition of RNasin ribonuclease inhibitor (Promega). We validated the quality and quantity of the purified DNase‐treated RNA samples by agarose gel electrophoresis and Nanodrop measurement. For cDNA conversion, DNase‐treated RNA (1.5 µg), random hexamer primers (Thermo Scientific), dNTPs, and M‐MLV reverse transcriptase (Promega) were used to generate cDNA via reverse transcription at 37°C for 1 hour. The cDNA samples were store at −20°C until testing.
2.9. Relative quantitation of gene expression by cDNA‐qPCR analysis
Relative quantitation of gene expression was determined by qPCR analysis using Applied Biosystems Real‐Time PCR StepOnePlus platform (Grand Island, NY). The 10‐fold diluted cDNA, qPCR primers (Table 2), and 2 × Fast SYBR Green Green Master mix (Applied Biosystems) were prepared according to the manufacturer's instructions. Cycling conditions were as follows: 95°C for 2 minutes; 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data were collected at 60°C, followed by a dissociation curve. The relative gene expression was calculated based on ΔΔCt methods, with subtraction to gyrA Ct value as endogenous control.
2.10. Transmission electron microscope (TEM) analysis of bacterial flagella
The H.pylori wild‐type and Δjhp0106 mutant strains (SW863, SW871, SW872, and SW869) were harvested from broth after 24–28 hours of inoculation under microaerophilic condition at 37°C. The bacterial cells were pelleted by centrifugation (100 × g, 10 minutes) and resuspended by finger flicking in distilled water (OD600 = 8–12). 31 Two µL of the suspension was deposited directly onto carbon‐coated Formvar copper grids (300 mesh) (Electron Microscopy Sciences, PA, USA) for 1 minute. Excess liquid was removed by capillary absorption using the sharp edge of Watman filter paper. The grids were then stained with 1% (w/v) phosphotungstic acid (Sigma‐Aldrich) in distilled water for 25 seconds, washed once with distilled water, and vacuumed dried for 8 hours. The stained grids were examined at 2000 magnifications (Electron Microscopy Facility, National Yang Ming University) using a JEM‐1400PLUS transmission microscope (JEOL Ltd, Japan) at 100 kV. The micrographs were taken from two independent experiments.
2.11. Statistical analysis
The Student t test and paired t test were applied for the parametric differences, whereas one‐way ANOVA (Tukey's multiple comparison test) (GraphPad Prism 5) was used to compare groups of variances obtained from more than two datasets. All statistical significance was two‐tailed with a p value<0.05.
3. RESULTS
3.1. Loss of motility in the Δjhp0106 mutant was associated with non‐glycosylated FlaA proteins
Previous observation on non‐motile aflagellate H.pylori cells suggested the involvement of jhp0106 in flagellar formation. 26 In this study, we aimed to discover key factors that influence flagellar formation in the background of jhp0106 disruption. We first examined the expression of FlaA, the major constituent of the flagellar filament, in the Δjhp0106 insertion strain (SW863). In comparison with FlaA (57 kDa) in the whole‐cell proteins of the WT strain, we found the FlaA signals with lower molecular mass (≈ 53 kDa) in the SW863 strain (Figure 1A). This lower molecular mass of FlaA was similar to that deduced from putative FlaA sequence, suggesting the higher molecular mass of FlaA detected in the WT strain was probably a consequence of post‐translational modification. The same phenomenon was seen when FlaA signals (57 kDa) were recovered in the Δjhp0106 revertant (SW862) and ΔflaA revertant (SW867) strains. In the ΔflaA mutant strain (SW866), the absence of FlaA signals confirmed the specificity of anti‐FlaA sera that were used. This observant post‐translational modification of FlaA in the WT, SW862, and SW867 strains was postulated to flagellin glycosylation via Jhp0106. To test this hypothesis, we chose a chemical deglycosylation method using TfOH 28 to remove carbohydrate elements from glycoproteins while the protein backbone remains intact. As shown in Figure 1B, FlaA with molecular mass (≈ 53 kDa) was detected in the TfOH‐treated whole‐cell proteins of the WT and SW863 strains, and that of the SW863 strain without TfOH treatment. This result supported our aforementioned hypothesis that Jhp0106 mediates FlaA glycosylation.
FIGURE 1.
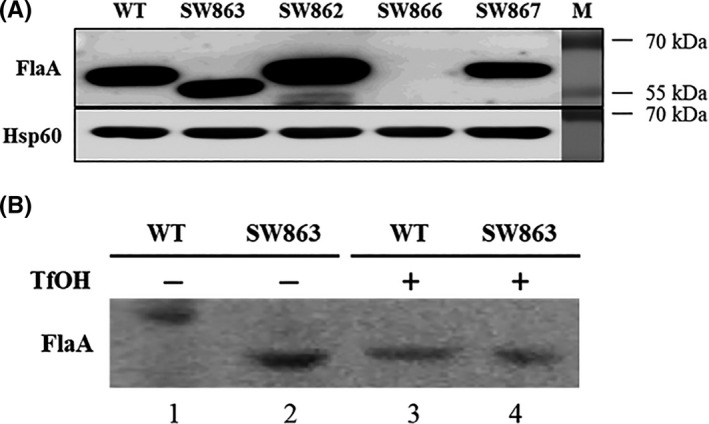
Western blot analysis of FlaA in H.pylori. (A) Detection of FlaA in the wild‐type and mutant strains. The whole‐cell proteins of the wild type (WT), the ∆jhp0106 (SW863), ∆jhp0106 revertant (SW862), ∆flaA mutant (SW866), and ∆flaA revertant (SW867) were probed with anti‐FlaA mouse polyclonal antibody. Anti‐Hsp60 mouse monoclonal antibody was used as internal control. (B) Glycosylation of FlaA verified by trifluoromethanesulphonic acid (TfOH) treatment. The total proteins extracted from the WT and SW863 strains were treated with or without TfOH and analyzed by Western blotting. TfOH chemically removed conjugated carbohydrates that were glycosylated on FlaAs (represented in the form of higher molecular mass ≈ 57 kDa) (lane 1), resulting in non‐glycosylated FlaAs (represented in the form of lower molecular mass ≈ 53 kDa) (lanes 2, 3, and 4)
3.2. Jhp0106 as a glycosyltransferase of the Maf family by in silico prediction
To explore the role of Jhp0106 as a glycosyltransferase, we applied computational analysis to elucidate the structural domains of Jhp0106. A fine‐scale homologous modeling of Jhp0106 (Figure 2A, left) was performed and compared to the glycosyltransferase Maf of M.magneticum AMB‐1 (Figure 2A, right), a reported Maf associated with flagellin glycosylation (PDB entry 5MU5). 18 The central α/β domain of the Jhp0106 formed a putative substrate‐binding region with high similarity to that of the Maf. By this modeling, the glycosylation of FlaA by the Maf homolog Jhp0106 might be achieved via discovered three potential yet conserved active site residues of Jhp0106, serine (S) 350, phenylalanine (F) 376, and glutamate (E) 415 (Figure 2B). Furthermore, we defined the N‐terminal, central α/β domain, and C‐ terminal domains of Jhp0106 (Figure 2C). Of which, a MAF_flag10 motif structure (pfam 01973) of the Maf family and a coiled‐coil domain for protein interaction were present in the central α/β and C‐terminal domains, respectively. We further assessed and confirmed the conservation of these uncharacterized active site residues (S350, F376, and E415) of Jhp0106 in Epsilonproteobacteria (Figure S1). These in silico predictions collectively gave us the confidence to postulate that Jhp0106 is a glycosyltransferase of the Maf family, and the three uncharacterized active site residues (S350, F376, and E415) may be involved in the glycosylation onto FlaA.
FIGURE 2.
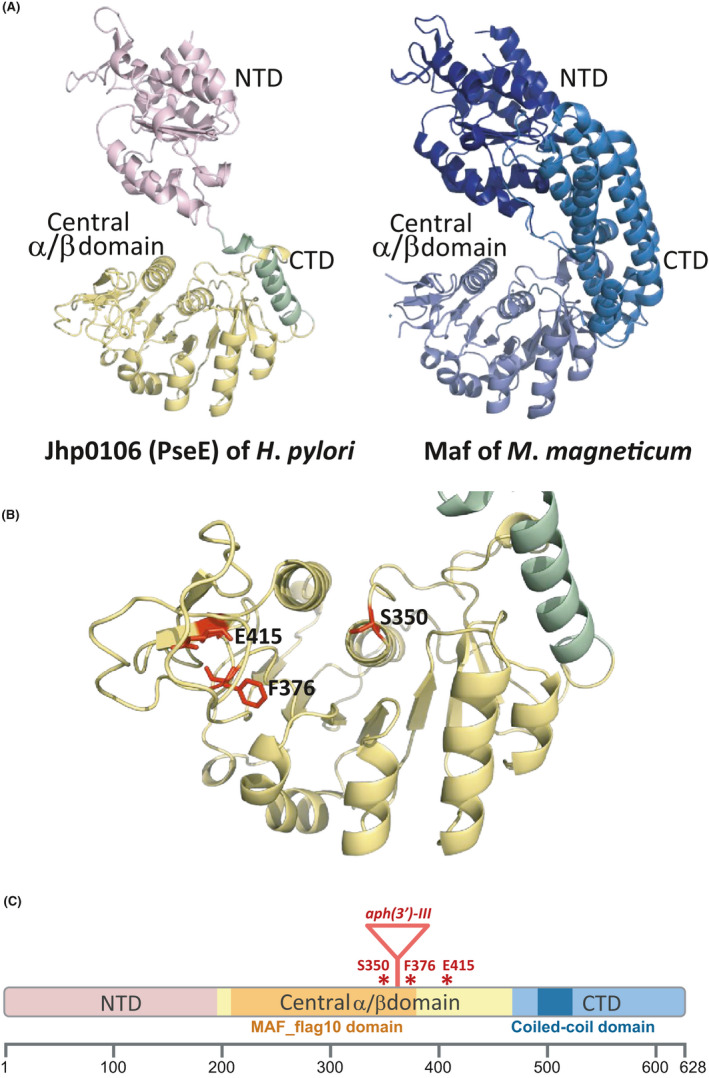
In silico analysis of Jhp0106 structural homology and the location of functional domains. (A) Domain arrangement of Jhp0106 (PseE) (from residue N3 to K464) (left) and Maf of Magnetospirillum magneticum AMB‐1 (from residue I7 to D664) (PDB entry 5MU5) (right). The N‐terminal domain (NTD) was oriented according to structural homology, and the positioning and structure of the central α/β domain and the C‐terminal domain (CTD) also revealed high coordination. (B) Structural modeling on the central α/β domain of the Jhp0106. The central α/β domain revealed a putative substrate‐binding pocket where three potential active site residues S350, F376, and E415 of Jhp0106 were discovered (marked in highlighted). (C) Functional domain prediction of Jhp0106. Jhp0106 was comprised of the N‐terminal domain (residues 1–192), central α/β domain (residues 193–464), and C‐terminal domain (residues 465–628). Notably, two distinct functional domains were found as the MAF_flag 10 domain (pfam 01973) (residues 206–376) (colored in orange), as well as the coiled‐coil domain (residues 495–526) (colored in dark blue). The putative active site residues (S350, F376, and E415) were marked as asterisks, while the insertion of a kanamycin‐resistance cassette (aph(3’)‐III) at residue 360 was indicated as a triangle
3.3. Corroboration of FlaA‐Jhp0106 interaction
To demonstrate that Jhp0106 can glycosylate FlaA, we conducted a bacterial two‐hybrid assay to determine protein interaction between Jhp0106 and FlaA. In principle, an interaction between two proteins is represented by the expression of lacZ in a reporter E.coli strain. A weak yet significant interaction between Jhp0106 and FlaA was identified (Figure 3, sample 4) when compared to our negative controls (sample 1), background signal controls (samples 2, 3, 5, 7, and 9), and positive control (sample 11). Nonetheless, recombinant Jhp0106 with amino acid substitutions (S350A, F376A, or E415A) retained prominent interaction with FlaA (samples 6, 8, and 10), indicating the point mutations of Jhp0106 did not abolish FlaA binding. In conclusion, our results proved the presence of interaction between Jhp0106 and FlaA.
FIGURE 3.
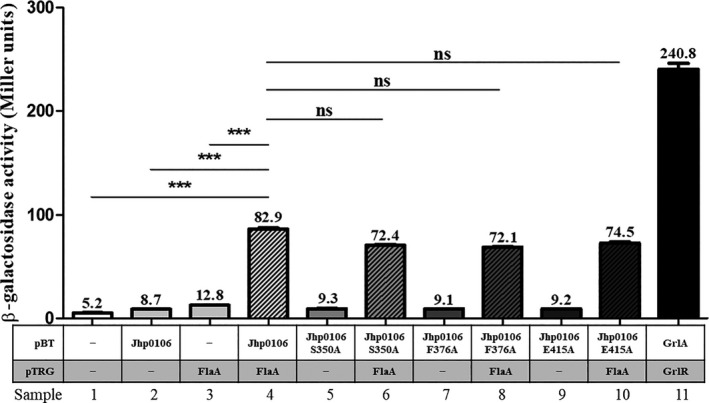
The Jhp0106‐FlaA interaction assessed by a bacterial two‐hybrid system. Jhp0106‐FlaA protein interaction was investigated using a bacterial two‐hybrid system in E.coli. The gene flaA was cloned into pTRG to determine whether FlaA interacts with Jhp0106 and the Jhp0106‐derivatives carrying S350A, F376A, or E415A mutation that were cloned in pBT. E.coli cells harboring empty vectors pBT (bait) and pTRG (target) (sample 1) served as negative control. In parallel, E.coli cells harboring only pBT‐Jhp0106 (sample 2), pTRG‐FlaA (sample 3), pBT‐Jhp0106::S350A (sample 5), pBT‐Jhp0106::F376A (sample 7), or pBT‐Jhp0106::E415A (sample 9) served as background signal controls. For positive control, E.coli cells obtained both pBT‐GrlA and pTRG‐GrlR (sample 11). An increase of measured β‐galactosidase activity is an indication of positive interaction between the tested target and bait proteins (samples 4, 6, 8, and 10). The results were the representative of three independent experiments (means ±SEM). *** denotes statistical significance with p < 0.001; ns stands for “not significant.”
3.4. Characterization of Jhp0106’s active site in the site‐directed Δjhp0106 mutants
To investigate which amino acid residues contribute to Jhp0106 activity, we experimentally constructed Δjhp0106 mutants carrying the respective point mutation of the putative active site residues (S350, F376, and E415) (Table 1). The accuracy of the mutated nucleosides in the Δjhp0106 site‐directed mutants was verified by Sanger sequencing analysis. Interestingly, we only observed a diffuse spreading growth pattern in the WT and Δjhp0106 revertant strain SW862 (motility diameter ≈ 1.5 cm) (Figure 4A,B). On the contrary, in the Δjhp0106 mutants SW863, Δjhp0106::S350A (SW871), Δjhp0106::F376A (SW872), and Δjhp0106::E415A (SW869), colonies displayed small condensed spots (≈ 0.5 – 0.7 cm) as the indicative of a typical pattern for non‐motile cells (Figure 4A,B). A possibility of growth difference resulting in varied motility was ruled out since comparable growth rate was recorded in the all tested strains (Figure 4C). Notably, the expression of FlaA in the SW871, SW872, and SW869 strains exhibited lower mass ≈ 53 kDa, a non‐glycosylated form of FlaA (Figure 4D). However, the great reduction of FlaA in the SW871, SW872, and SW869 strains was unexpected, leading us to examine the expression of flaA (Figure 4E). A significant increase of the flaA expression (2.78‐fold) was merely observed in the SW863 strain, whereas the comparable levels of flaA transcripts were shown in the WT, SW871, SW872, and SW869 strains. These results indicated the decrease of FlaA in the SW871, SW872, and SW869 strains was not due to the change of flaA expression. Taken together, our phenotypic analyses demonstrated that Jhp0106 functions as a glycosyltransferase for FlaA, and the three active site residues (S350, F376, and E415) of Jhp0106 contributes to glycosylation.
FIGURE 4.
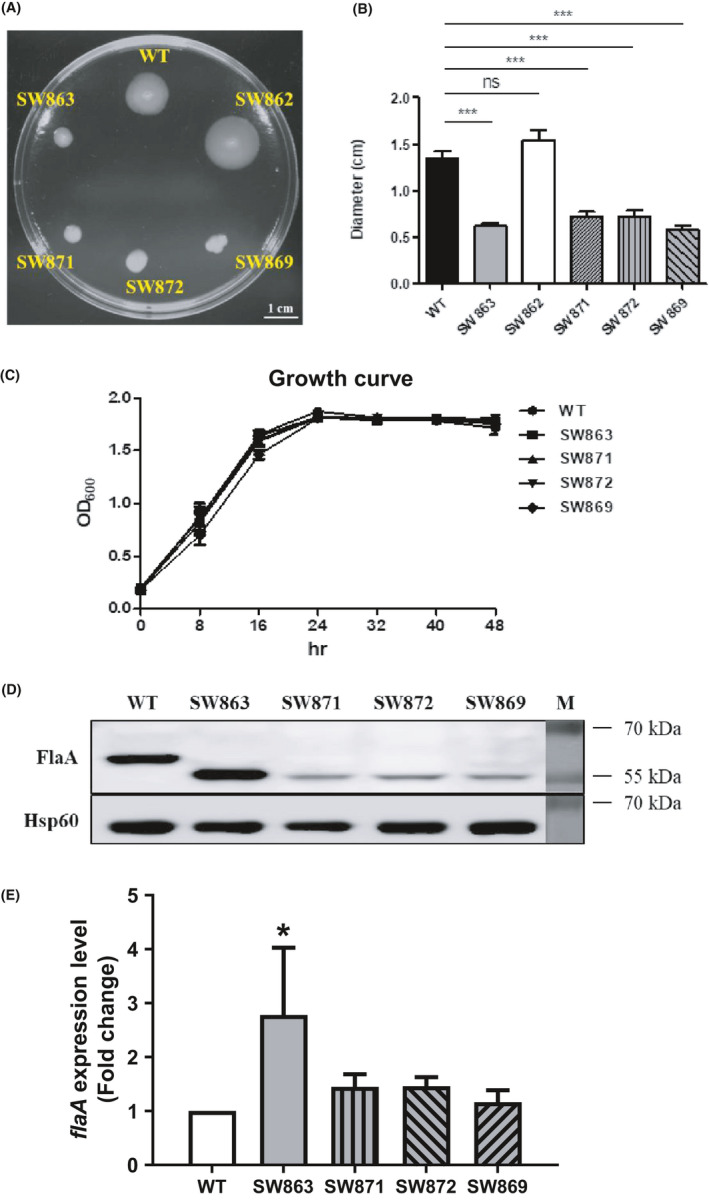
Phenotypic analysis of the ∆jhp0106 insertion mutant and site‐directed mutants. (A) The motility of the SW863, SW862, ∆jhp0106::S350A (SW871), ∆jhp0106::F376A (SW872), ∆jhp0106::E415A (SW869), and the WT strains was evaluated by a soft‐agar motility assay. (B) Motility diameters were quantified and calculated as the mean ±SD of three independent experiments. *** represents statistical significance (p < 0.001). ns stands for “not significant.” (C) Growth of the WT, SW863, SW871, SW872, and SW869 strains showed a comparable trend over a course of 48‐hour inoculation. The results were calculated as the mean ±SD of three independent experiments. (D) Expression of FlaA detected in the whole‐cell proteins of the site‐directed ∆jhp0106 mutants by Western blotting. To be noted, the molecular mass of FlaA in the SW871, and SW872, and SW869 strains were similar to that in the SW863 strain, but lower than the WT strain. Surprisingly, FlaA yield was reduced in the SW871, and SW872, and SW869 strains. (E) Expression of flaA mRNA examined using cDNA‐qPCR analysis. An increase of flaA transcripts (2.78‐fold) was detected in the SW863 strain, whereas no significant change of flaA expression observed in the SW871, SW872, and SW869 strains. The results were measured as the mean ±SD of three independent biological repeats
3.5. Lack of flagellation in the site‐directed Δjhp0106 mutants
To demonstrate the relation between FlaA glycosylation and flagellar formation, we examined the formation of flagellar filament using negative staining by a transmission electron microscope. In Figure 5A, long flagella were observed in the wild‐type strain, whereas no flagellar filament was detected in the SW863 strain (Figure 5B). This result was consistent with the aflagellate SW863 cells shown in our previous study. 26 Interestingly, none of cells from the site‐directed Δjhp0106 mutant strains (SW871, SW872, and SW869) displayed flagella (Figure 5C,D,E), implying that non‐glycosylated FlaAs were unable or less efficient to form flagellar filaments.
FIGURE 5.
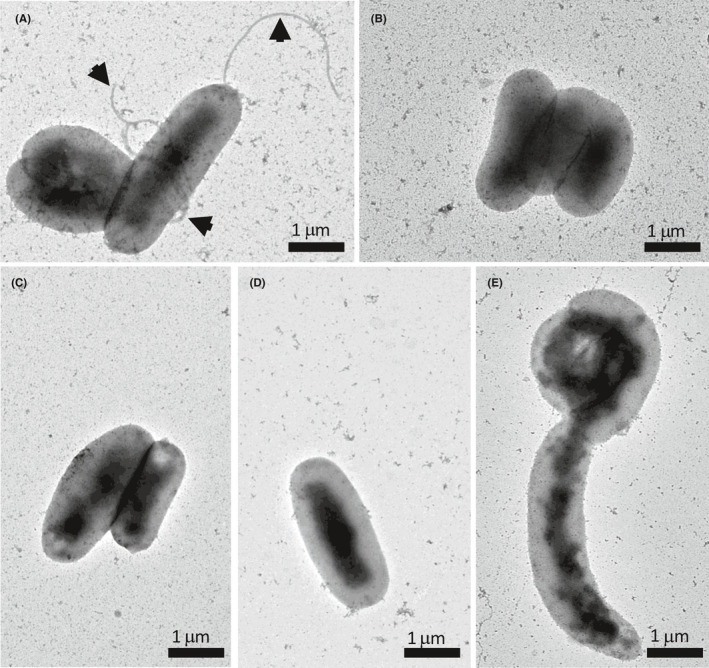
Transmission electron micrographs of negative‐stained H.pylori. Flagellar formation in the (A) wild‐type, (B) SW863 (C) SW871, (D) SW872, and (E) SW869 strains was examined by phosphotungstic acid staining and transmission electron microscope. The arrowheads point to the presence of bacterial flagella. Scale bars represent one micrometer
4. DISCUSSION
In this study, we successfully characterized the function of jhp0106 in flagellar formation in H.pylori. We first verified the impaired motility in the Δjhp0106 insertion mutant SW863 was a result of non‐glycosylated FlaA. By performing computational analyses, three uncharacterized yet conserved active site residues (S350, F376, and E415) at the central α/β domain of Jhp0106 were identified for potential substrate‐binding region. We further investigated the impact of Jhp0106’s active site on FlaA glycosylation and flagellar formation by examining FlaA‐Jhp0106 interaction via a bacterial two‐hybrid assay, and by phenotypic and molecular analyses in the site‐directed Δjhp0106 isogenic strains. Collectively, we demonstrated that Jhp0106 binds to FlaA and mediates FlaA glycosylation, together with a discovery of the active site of glycosyltransferase Jhp0106. Hence, the glycosyltransferase Jhp0106 is suggestively named “PseE,” as a member of the pseudaminic acid biosynthesis for flagellin glycosylation.
Disruption of jhp0106 and its active site residues (S350, F376, and E415) severely affected the motility of H.pylori with a remarkably differential abundance of FlaA in the Δjhp0106 insertion and sited‐directed mutants (Figures 1, 4). Furthermore, our electron microscopic analysis showed no flagella on the bacterial surface of the Δjhp0106 insertion and sited‐directed mutants (Figure 5B‐E). This provided an evident support that the PseE‐mediated glycosylation on FlaA was critical for flagellar formation. However, the phenomenon of non‐glycosylated FlaA variation in different Δjhp0106 mutants was found to be uncorrelated to flaA transcription in the mutants (Figure 4E), leading us to hypothesize that protein degradation on non‐glycosylated FlaAs may occur in the sited‐directed Δjhp0106 mutants.
The reduction of FlaA abundance in the sited‐directed Δjhp0106 mutants drew our attention and questioned why the non‐glycosylated FlaAs in the SW863 strain were not degraded. Based on the findings from our study and the flagellin glycosylation events investigated in H.pylori and other pathogens, 16 , 17 , 32 , 33 , 34 , 35 , 36 , 37 it is likely that in the wild‐type strain (Figure 6, the left panel), PseE would first bind to FlaA, followed by sugar transfer to glycosylate FlaA. The glycosylated FlaA within this FlaA‐PseE complex would possibly be recognized by a flagellin‐specific chaperone FliS. 33 , 34 , 35 With the chaperone‐mediated protection, the unfolded glycosylated FlaA would be transported efficiently via a flagellar type three secretion system (fT3SS) to extracellular surface. Thus, the flagellar filaments are formed conferring to H.pylori's motility for invasion. In an Aeromonas caviae study, 16 flagellin glycosylation was found to occur earlier than chaperone binding and flagellar secretion via the T3SS. However, the export process of glycosylated and non‐glycosylated FlaA was not tightly coupled with FliS chaperone binding, which was reported in a recent study in H.pylori G27 strain. 35 Their observation on the formation of short flagellar filaments in the ΔfliS mutant suggested a less efficient secretion of FlaA and impaired flagellar assembly. Differing from the situation in the wild‐type, in the SW863 strain (Figure 6, the middle panel), which generated truncated PseEs without a putative coiled‐coil domain by the aph(3’)‐III insertion in the jhp0106 gene (Figure 2C), the non‐glycosylated FlaAs would be stably present in the cytosol and not bound to the truncated PseEs. The transportation of non‐glycosylated FlaA by FliS is less efficient; hence, no apparent flagella formed on the surface of non‐motile H. pylori SW863 cells (Figure 5B). In contrast, the expressed PseE with the respective mutated active site residue in the SW871, SW872, and SW859 strains (Figure 6, the right panel) would still likely bind to FlaA, as inferred from our examination on FlaA‐PseE interaction in the bacterial two‐hybrid assay (Figure 3). Although these mutated PseEs bound to FlaA, they would be unable to transfer sugar to FlaA. Therefore, this particular PseE‐bound non‐glycosylated FlaA would probably undergo proteolytic degradation, resulting in aflagellate non‐motile cells (Figure 5C‐E). This mechanism of proteolytic degradation is presumed to avoid transportation of incorrect or immature form of FlaA proteins.
FIGURE 6.
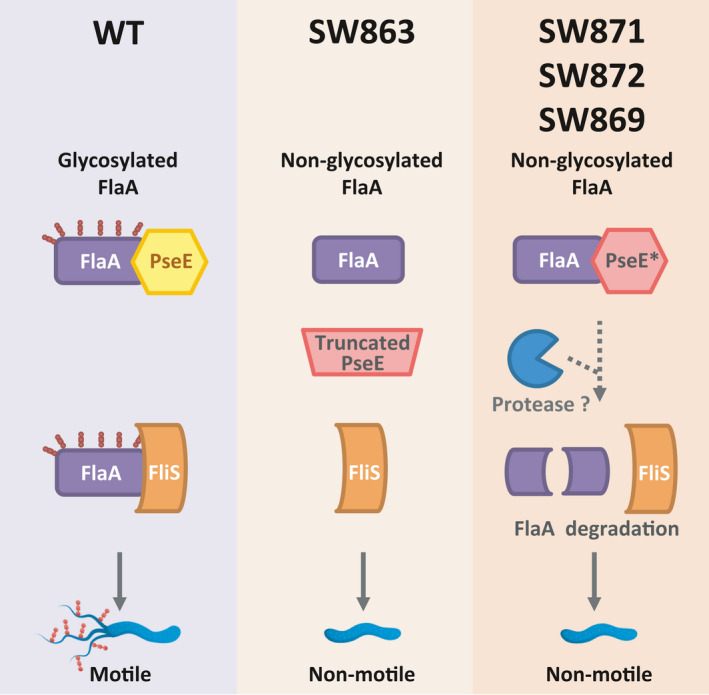
Process of flagellin (FlaA) glycosylated by Jhp0106 (PseE) in H.pylori. Based on our findings and the previous studies on flagellin chaperones, we proposed that Jhp0106 (PseE) performs a role as a glycosyltransferase responsible for the glycosylation of flagellin FlaA. In the wild‐type strain (the left panel), PseE would bind to newly synthesized FlaA, then transfer sugar analog to modify FlaA. Flagellin chaperone FliS would possibly recognize this glycosylated FlaA bound‐PseE complex. Sequentially, the glycosylated FlaA, which were bound and protected by FliS, would be transported via the flagellar type three secretion system (fT3SS). Upon this, the mature form of glycosylated FlaA would be assembled extracellularly into a flagellar filament, conferring H. pylori's motility for invasion. However, in the insertion mutant strain SW863, whose PseE becomes truncated due to the aph(3’)‐III insertion, the mutant cells lack not only the ability of glycosylation but also the capability of protein binding (the middle panel). Therefore, the synthesized FlaA might not bind to the truncated PseE and would be waiting to be glycosylated in the cytosol. These non‐glycosylated FlaAs might not be transported efficiently by FliS and the fT3SS, resulting in aflagellate non‐motile cells. In contrast to the SW863 strain, the non‐glycosylated FlaA encounters differently in the SW871, 872, and 869 strains (the right panel). As the site‐directed PseE proteins only carry one point mutation in the potential substrate‐binding region, they would still be synthesized and bound to FlaA. Adversely, this non‐glycosylated FlaA‐PseE complex might not be favored and protected by FliS, resulting in degradation of the non‐glycosylated FlaA by unknown proteases. This probably makes the SW871, 872, and 869 strains are aflagellate and non‐motile. The dotted arrow‐line represents an inferred biological process
In conclusion, our study used computational and molecular characterization to demonstrate that Jhp0106 (PseE) contributes to FlaA glycosylation in H.pylori. The motility of H.pylori is a crucial virulence determinant in bacterial pathogenesis. Therefore, understanding the function of Jhp0106 in flagellar formation not only expanded our current knowledge of flagellin glycosylation, but also revealed a previously unknown glycosyltransferase family responsible for flagellin glycosylation in clinically important pathogens. This new finding will also assist in the development of an innovative non‐antibiotic treatment using small molecules (sugar analogs) to antagonize FlaA maturation and prevent H.pylori colonization.
CONFLICT OF INTEREST
The authors have no disclosure or other conflicts of interests to report.
AUTHOR CONTRIBUTIONS
Kai‐Yuan Yang, Cheng‐Yen Kao, and Marcia Shu‐Wei Su contributed equally to this work. MSWS, STH, CHT, PJT, and JJW designed research. KYY, YLC, MSWS, and JWC performed research. SW and MSWS conducted protein structural analyses. MSWS, CYK, and JJW wrote the manuscript. All authors read and approved the final manuscript.
5. ETHICS APPROVAL AND CONSENT TO PARTICIPATE
All the mice were taken care and performed according to the guideline given by the Institutional Animal Care and Use Committee (IACUC) of the National Cheng Kung University with the approval number 109127.
CONSENT FOR PUBLICATION
Not applicable.
Supporting information
Figure S1
Table S1
ACKNOWLEDGEMENTS
We would like to thank Drs. Ryan Holroyd and Xiaoxi Brook Lin for their helpful comments and editing on this manuscript. We also thank Wei‐Jiun Tsai for his great assistance with the PyMOL software and Chia‐Hsuan Su for graphic design. We appreciate the technical support from Yui‐Ying Yu and Shu‐Li Ho at the Electron Microscopy Facility in the National Yang‐Ming University. This study was supported by the Ministry of Science and Technology, Taiwan (the MOST grants 107‐2320‐B‐010‐012 and 108‐2320‐B‐010‐002).
Kai‐Yuan Yang, Cheng‐Yen Kao, and Marcia Shu‐Wei Su contributed equally to this work.
DATA AVAILABILITY STATEMENT
The dataset supporting the conclusions of this article is included in the article.
REFERENCES
- 1. Ottemann KM, Lowenthal AC. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun. 2002;70(4):1984‐1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eaton KA, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64(7):2445‐2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kao CY, Sheu BS, Sheu SM, et al. Higher motility enhances bacterial density and inflammatory response in dyspeptic patients infected with Helicobacter pylori . Helicobacter. 2012;17(6):411‐416. [DOI] [PubMed] [Google Scholar]
- 4. Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria. Int J Med Microbiol. 2002;291(8):605‐614. [DOI] [PubMed] [Google Scholar]
- 5. Diaconu S, Predescu A, Moldoveanu A, Pop CS, Fierbinteanu‐Braticevici C. Helicobacter pylori infection: old and new. J Med Life. 2017;10(2):112‐117. [PMC free article] [PubMed] [Google Scholar]
- 6. Parsonnet J, Friedman GD, Vandersteen DP, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325(16):1127‐1131. [DOI] [PubMed] [Google Scholar]
- 7. Graham DY, Hepps KS, Ramirez FC, Lew GM, Saeed ZA. Treatment of Helicobacter pylori reduces the rate of rebleeding in peptic ulcer disease. Scand J Gastroenterol. 1993;28(11):939‐942. [DOI] [PubMed] [Google Scholar]
- 8. Alm RA, Ling L‐S, Moir DT, et al. Genomic‐sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori . Nature. 1999;397(6715):176‐180. [DOI] [PubMed] [Google Scholar]
- 9. Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter . Annu Rev Microbiol. 2011;65:389‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsang J, Hoover TR. Basal body structures differentially affect transcription of RpoN‐ and FliA‐dependent flagellar genes in Helicobacter pylori . J Bacteriol. 2015;197(11):1921‐1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39(1):14‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niehus E, Gressmann H, Ye F, et al. Genome‐wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori . Mol Microbiol. 2004;52(4):947‐961. [DOI] [PubMed] [Google Scholar]
- 13. Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori . Mol Microbiol. 2003;48(6):1579‐1592. [DOI] [PubMed] [Google Scholar]
- 14. Logan SM. Flagellar glycosylation ‐ a new component of the motility repertoire? Microbiology (Reading). 2006;152(Pt 5):1249‐1262. [DOI] [PubMed] [Google Scholar]
- 15. Valguarnera E, Kinsella RL, Feldman MF. Sugar and spice make bacteria not nice: protein glycosylation and its influence in pathogenesis. J Mol Biol. 2016;428(16):3206‐3220. [DOI] [PubMed] [Google Scholar]
- 16. Parker JL, Lowry RC, Couto NA, Wright PC, Stafford GP, Shaw JG. Maf‐dependent bacterial flagellin glycosylation occurs before chaperone binding and flagellar T3SS export. Mol Microbiol. 2014;92(2):258‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McNally DJ, Hui JPM, Aubry AJ, et al. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81–176 using a focused metabolomics approach. J Biol Chem. 2006;281(27):18489‐18498. [DOI] [PubMed] [Google Scholar]
- 18. Sulzenbacher G, Roig‐Zamboni V, Lebrun R, et al. Glycosylate and move! The glycosyltransferase Maf is involved in bacterial flagella formation. Environ Microbiol. 2018;20(1):228‐240. [DOI] [PubMed] [Google Scholar]
- 19. Canals R, Vilches S, Wilhelms M, Shaw JG, Merino S, Tomas JM. Non‐structural flagella genes affecting both polar and lateral flagella‐mediated motility in Aeromonas hydrophila . Microbiology (Reading). 2007;153(Pt 4):1165‐1175. [DOI] [PubMed] [Google Scholar]
- 20. Schoenhofen IC, Lunin VV, Julien J‐P, et al. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori . J Biol Chem. 2006;281(13):8907‐8916. [DOI] [PubMed] [Google Scholar]
- 21. Tabei SMB, Hitchen PG, Day‐Williams MJ, et al. An Aeromonas caviae genomic island is required for both O‐antigen lipopolysaccharide biosynthesis and flagellin glycosylation. J Bacteriol. 2009;191(8):2851‐2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thibault P, Logan SM, Kelly JF, et al. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem. 2001;276(37):34862‐34870. [DOI] [PubMed] [Google Scholar]
- 23. Schoenhofen IC, McNally DJ, Brisson JR, Logan SM. Elucidation of the CMP‐pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP‐N‐acetylglucosamine by a single enzymatic reaction. Glycobiology. 2006;16(9):8C‐14C. [DOI] [PubMed] [Google Scholar]
- 24. Karlyshev AV, Linton D, Gregson NA, Wren BW. A novel paralogous gene family involved in phase‐variable flagella‐mediated motility in Campylobacter jejuni . Microbiology (Reading). 2002;148(Pt 2):473‐480. [DOI] [PubMed] [Google Scholar]
- 25. Parker JL, Day‐Williams MJ, Tomas JM, Stafford GP, Shaw JG. Identification of a putative glycosyltransferase responsible for the transfer of pseudaminic acid onto the polar flagellin of Aeromonas caviae Sch3N. Microbiologyopen. 2012;1(2):149‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kao CY, Chen JW, Wang S, Sheu BS, Wu JJ. The Helicobacter pylori J99 jhp0106 gene, under the control of the CsrA/RpoN regulatory system, modulates flagella formation and motility. Front Microbiol. 2017;8:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kao CY, Sheu BS, Wu JJ. CsrA regulates Helicobacter pylori J99 motility and adhesion by controlling flagella formation. Helicobacter. 2014;19(6):443‐454. [DOI] [PubMed] [Google Scholar]
- 28. Edge AS. Deglycosylation of glycoproteins with trifluoromethanesulphonic acid: elucidation of molecular structure and function. Biochem J. 2003;376(Pt 2):339‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ludwiczak J, Winski A, Szczepaniak K, Alva V, Dunin‐Horkawicz S. DeepCoil‐a fast and accurate prediction of coiled‐coil domains in protein sequences. Bioinformatics. 2019;35(16):2790‐2795. [DOI] [PubMed] [Google Scholar]
- 30. Dove SL, Hochschild A. A bacterial two‐hybrid system based on transcription activation. Methods Mol Biol. 2004;261:231‐246. [DOI] [PubMed] [Google Scholar]
- 31. Kim KW, Jung WK, Park YH. Comparison of pre‐stain suspension liquids in the contrasting ability of neutralized potassium phosphotungstate for negative staining of bacteria. J Microbiol Biotechnol. 2008;18(11):1762‐1767. [DOI] [PubMed] [Google Scholar]
- 32. Radomska KA, Wosten M, Ordonez SR, Wagenaar JA, van Putten JPM. Importance of Campylobacter jejuni FliS and FliW in flagella biogenesis and flagellin secretion. Front Microbiol. 2017;8:1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Altegoer F, Mukherjee S, Steinchen W, et al. FliS/flagellin/FliW heterotrimer couples type III secretion and flagellin homeostasis. Sci Rep. 2018;8(1):11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mukherjee S, Babitzke P, Kearns DB. FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis . J Bacteriol. 2013;195(2):297‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lam WW, Sun K, Zhang H, Au SW. Crystal structure of flagellar export chaperone FliS in complex with flagellin and HP1076 of Helicobacter pylori . Front Microbiol. 2020;11:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schirm M, Schoenhofen IC, Logan SM, Waldron KC, Thibault P. Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal Chem. 2005;77(23):7774‐7782. [DOI] [PubMed] [Google Scholar]
- 37. Josenhans C, Vossebein L, Friedrich S, Suerbaum S. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol Lett. 2002;210(2):165‐172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Data Availability Statement
The dataset supporting the conclusions of this article is included in the article.


