Abstract
Air pollution has been documented to contribute to severe respiratory diseases like asthma and chronic obstructive pulmonary disorder (COPD). Although these diseases demonstrate a shift in the lung microbiota towards Proteobacteria, the effects of traffic generated emissions on lung microbiota profiles have not been well-characterized. Thus, we investigated the hypothesis that exposure to traffic-generated emissions can alter lung microbiota and immune defenses. Since a large population of the Western world consumes a diet rich in fats, we sought to investigate the synergistic effects of mixed vehicle emissions and high-fat diet consumption. We exposed 3-month-old male C57Bl/6 mice placed either on regular chow (LF) or a high-fat (HF: 45% kcal fat) diet to mixed emissions (ME: 30 μg PM/m3 gasoline engine emissions + 70 μg PM/m3 diesel engine emissions) or filtered air (FA) for 6 h/d, 7 d/wk for 30 days. Levels of pulmonary immunoglobulins IgA, IgG, and IgM were analyzed by ELISA, and lung microbial profiling was done using qPCR and Illumina 16 S sequencing. We observed a significant decrease in lung IgA in the ME-exposed animals, compared to the FA-exposed animals, both fed a HF diet. Our results also revealed a significant decrease in lung IgG in the ME-exposed animals both on the LF diet and HF diet, in comparison to the FA-exposed animals. We also observed an expansion of Enterobacteriaceae belonging to the Proteobacteria phylum in the ME-exposed groups on the HF diet. Collectively, we show that the combined effects of ME and HF diet result in decreased immune surveillance and lung bacterial dysbiosis, which is of significance in lung diseases.
Keywords: Air pollution, Lung microbiome, Immunoglobulins, Proteobacteria
1. Introduction
The lungs are among the first organs to be exposed to the harmful effects of inhaled air pollution. Air pollutants including particulate matter (PM), polycyclic aromatic hydrocarbons, gaseous mixtures of nitrogen dioxide, carbon monoxide, and volatile organic compounds have all been implicated in causing severe lung damage (Marino et al., 2015; Moorthy et al., 2015; Xing et al., 2016). Exposure to air pollutants is associated with the exacerbation of several respiratory diseases such as asthma, bronchitis, and chronic obstructive pulmonary disorder (COPD) (Andersen et al., 2011; Faustini et al., 2013). Air pollution exposures result in around 7 million premature deaths in a year, of which 43% are deaths due to COPD, and 26% due to respiratory infections (WHO Global Health Observatory, 2016). The incidence of the occurrence of these diseases is also higher in heavily polluted regions suggesting that air pollutants play a role in either development or exacerbation of underlying lung conditions (Kim et al., 2018). Air pollutants have been documented to affect immune response at the mucosal surfaces by altering immunoglobulin production and releasing inflammatory mediators (Hiraiwa and van Eeden, 2013; Li et al., 2017). Although there has been a lot of interest in the immunological consequences of air pollution, we are only beginning to explore the impact of these pollutants on the newly identified lung microbiome.
Lungs were historically considered to be sterile, but recent advances in sampling techniques and 16 S rRNA sequencing have demonstrated that the lower respiratory tract is replete with a wide variety of microorganisms– both in health and disease (Dickson et al., 2016). The healthy lung microbiome is variable due to the dynamic responses of inhalation, exhalation, mucociliary clearance, host-immune responses, etc., that occur continuously within the lungs. Despite these fluxes, most of the bacteria in the healthy lungs belong to 4 major phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Mathieu et al., 2018). The commensal microbial diversity is crucial in maintaining several homeostatic functions such as immune system development and regulation. With advances in lung microbiome studies, we now understand that these microorganisms are not mere bystanders, but they play a significant role in modulating the immune environment on the lungs. Studies in germ-free mice have demonstrated that IgA production is significantly reduced within their airway lumen, making them vulnerable to antigen challenges (Ruane et al., 2016). In the absence of microbial stimulation in germ-free mice, they were also found to have decreased mucus production, which severely impedes their mucociliary defense mechanism (Yun et al., 2014).
In disease states, it is observed that the diversity of commensal bacteria is often affected when certain bacteria with selective advantages proliferate and outcompete the others. In many inflammatory diseases of the lung, a shift in the microbiota profile towards Proteobacteria is observed (Hilty et al., 2010; Molyneaux et al., 2013), primarily because these microorganisms have unique abilities to thrive in inflammatory environments (Rizzatti et al., 2017). The human microbiome is understood to be influenced by several factors, including diet and environmental exposures (Tasnim et al., 2017). There are a few emerging studies that show inhaled air pollutants from both anthropogenic and natural sources can also induce alteration in the Firmicutes: Bacteroidetes ratio in the GI tract, which increases susceptibility to inflammation (Fitch et al., 2020; Mutlu et al., 2018).
A large percentage of the Western world consumes a diet rich in fats, which has contributed to the epidemic of obesity, characterized by low-grade inflammation (Duan et al., 2018). High-fat (HF) diet consumption alone has been documented to cause microbial shifts with an increase in Firmicutes in the gastrointestinal tract (Murphy et al., 2015). However, to date, the synergistic effects of traffic-generated air pollutant mixtures and HF diet on the lung microbiota have not been characterized. To address this gap in knowledge, we investigated the hypothesis that exposure to a mixture of gasoline and diesel emissions can alter the lung microbiota and immune defenses in wildtype mice placed on an HF diet. The interactions between environmental exposures, diet, microbiome, and the immune system are vital in understanding the development of diseases. In the following experiments, we exposed C57Bl/6 wildtype mice to a mixture of gasoline and diesel emissions and placed them on either a standard mouse chow or HF diet, and analyzed immunoglobulin levels and lung microbiota profiles.
2. Materials and methods
2.1. Animals and inhalational exposure
Three-month-old C57Bl/6 male mice (C57BL/6NTac, Taconic, Germantown, NY) were placed either on a standard mouse chow (LF, containing 12% fat, n = 12) or a high-fat (HF, n = 12) diet (TD88137 Custom Research Diet, Harlan Teklad, Madison, WI; 21.2% fat content by weight, 45% kcal from fat, 1.5 g/kg cholesterol content) for 30 days prior to exposures. Mice were then exposed to whole-body inhalation to either filtered air (FA, n = 6 per diet) or a mixture of gasoline and diesel engine exhaust (ME: 30 μg PM/m3 gasoline engine emissions + 70 μg PM/m3 diesel engine; n = 6 per diet) for 6 h/d, 7 d/wk, for 30 days. ME was created by combining exhaust from a 1996 GM gasoline engine and a Yanmar diesel generator system, and exposures chemistries and PM characterized, as previously reported (Lucero et al., 2017; Lund et al., 2011; McDonald et al., 2004, 2008; Mumaw et al., 2016; Oppenheim et al., 2013). Particle size distribution, composition, and mass concentration were determined, as previously described (Suwannasual et al., 2018). The particle mass size distribution had a median of 1 μm (range: < 0.5–20 μm), particle number size distribution for this exposure had a median size of approximately 60 nm; with total particle mass for the mixture measured at 102.5 ± 20.9 μg/m3 over the 30 d study. Particle mass concentration by gravimetric analysis of Teflon membrane filters at the inlet of the chamber and inside the exposure chamber was conducted once/wk throughout the exposure protocol. The FA exposure consisted of HEPA-filtered ambient air. Mice were kept, 4 to a cage, in standard shoebox cages within AAALAC International-approved rodent housing facility (2 m3 exposure chambers) for the entirety of the study, which maintained a constant temperature (20–24 °C) and humidity (30–60% relative humidity). Chow and water were provided ad libitum, except during daily exposures when chow was removed. All procedures were approved by the Animal Care and Use Committee at the Lovelace Respiratory Research Institute and conform to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996).
Tissue Collection: Animals were sacrificed 14–16 h after their last exposure. Mice were anesthetized with Euthasol (0.1 ml per 30 g mouse) and euthanized by exsanguination. The lungs were dissected and immediately snap-frozen in liquid nitrogen.
The nomenclature used are as follows: (a) LF FA: C57Bl/6 mice placed on LF diet and exposed to FA, (b) LF ME: C57Bl/6 mice placed on LF diet and exposed to ME, (c) HF FA: C57Bl/6 mice placed on HF diet and exposed to FA, (d) HF ME: C57Bl/6 mice placed on HF diet and exposed to ME.
2.2. ELISA
Lung tissues (n = 6 per group) were homogenized in a beat beater with sterile saline and the supernatants were used for Immunoglobulin ELISAs. The concentration of IgA (Fisher Scientific, EMIGA), IgG (Fisher Scientific, 88-50400-22), and IgM (Fisher Scientific, 88-50470-22) were measured in 10-fold diluted lung tissue homogenates using ELISA according to the manufacturer’s recommendations. The samples were processed in triplicates, and values were determined from a known value standard curve, using a sigmoidal four-parameter logistic (4-PL) curve-fit.
2.3. qPCR
DNA from homogenized lung tissues (n = 6 per group) was extracted using ZR Fecal DNA miniprep (Zymo Research). qPCR was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and the CFX96 Real-Time system (Bio-Rad). For bacterial 16 S rRNA analysis, samples were normalized to Eubacteria utilizing known-concentration standards. Bacterial primers used are described in Table 1.
Table 1.
Primer sequences used for qPCR analysis.
| Primer | Sequence | Tm |
|---|---|---|
| Eubacteria FP | 5′-ACTCCTACGGGAGGCAGCAGT-3′ | 68.4 °C |
| Eubacteria RP | 5′-ATTACCGCGGCTGCTGGC-3′ | 70.7 °C |
| Enterobacteriaceae FP | 5′-GTGCCAGCMGCCGCGGTAA-3′ | 76.3 °C |
| Enterobacteriaceae RP | 5′-GCCTCAAGGGCACAACCTCCAAG-3′ | 73.6 °C |
| Bacteroidetes FP | 5′-GGTTCTGAGAGGAGGTCCC-3′ | 62.7 °C |
| Bacteroidetes RP | 5′-GCTGCCTCCCGTAGGAGT-3′ | 64.4 °C |
| Firmicutes FP | 5′-GGAGYATGTGGTTTAATTCGAAGCA-3′ | 63.9 °C |
| Firmicutes RP | 5′-AGCTGACGACAACCATGCAC-3′ | 66.5 °C |
| Actinobacteria FP | 5′-CGCGGCCTATCAGCTTGTTG-3′ | 69.9 °C |
| Actinobacteria RP | 5′-CCGTACTCCCCAGGCGGGG-3′ | 74.9 °C |
FP, forward primer; RP, reverse primer; Tm, melting temperature.
2.4. Illumina MiSeq sequencing
Genomic DNA was isolated using ZR Fecal DNA miniprep (Zymo Research) from lung homogenates (n = 6 per group). 16 S rRNA genes (variable region 4, V4) were amplified using a composite forward primer and a reverse primer with a unique 10-base barcode used to tag PCR products from respective samples, as described in Fan et al. (2015).
2.5. 16 S Bioinformatics and statistical analyses
Sequence reads obtained from the 16 S rRNA sequencing were analyzed by Mothur Software (version: 1.39.5) from the pipeline of MiSeq SOP (Schloss et al., 2009); standard procedure of this pipeline was followed. In this pipeline (https://rpubs.com/maddieSC/mo-thur_SOP_May_2018), the paired-end reads, that is, the forward and the corresponding reverse reads obtained from paired-end sequencing, were combined to form “contigs.” Furthermore, these sequence reads are from the V4 region (~ 250–300 bp) of the 16 S rRNA sequences; however, due to the PCR errors, sequence reads longer than 300 bp, and sequences with ambiguous base calls could be generated (Kozich et al., 2013). These low-quality sequences were removed from further analysis. Next, following the standard protocol of Mothur, the duplicate sequence reads were merged as it is not computationally useful to align the same sequence multiple times. These processed sequences were then aligned to the reference database, SILVA, containing the 16 S rRNA gene sequences. The database was first customized to our regions of interest (V4 regions) using command pcr.seqs; this was performed for the improvement of overall alignment. Following the alignment, any sequences that did not align to V4 region sequences in the customized database were removed. After the alignment, parts of sequences overhanging at both ends were removed. Gap characters (“−”) inserted during the alignment were removed. Further, these sequences were de-noised via a pre-clustering step (sequences were sorted based on the abundance and clustered based on their nucleotide difference less than 2) and removal of chimeras. These sequences were clustered based on species-level (97% or more) similarity to form Operational Taxonomic Units (OTUs), hence giving the absolute abundance matrix. Once the OTUs were established, taxonomic identity was assigned to each OTU. We used the Greengenes database for the taxonomic classification as the usage of this database is known to provide lower taxonomic level assignments and leaves fewer sequences unclassified. Additionally, a consensus confidence threshold was set at the 80% classification cut-off (default in Mothur) to specify the taxonomic identities. Finally, normalized taxonomic abundance for OTUs in each sample was obtained by dividing the abundance values by the total number of sequences in the sample, using the “normalized.shared” function. Normalized values were then approximated to the nearest integer. Note that the samples with very low sequence count (< 3) were eliminated from the further statistical analysis.
These data were further used as the input for the downstream statistical analyses performed. Alpha diversity quantifying the diversity of microbial species within a sample was estimated using the Chao1 index (for species richness) and Shannon index (for species diversity; the more the richness of the species and the more the species evenly distributed in a sample, the greater the Shannon diversity) (Chao, 1984; Chazdon et al., 1998; Shannon and Weaver, n.d.). Beta diversity representing the diversity in microbial species between different samples was calculated in the form of UniFrac weighted and unweighted principal coordinate analysis (PCoA) plot (Lozupone and Knight, 2005). Unweighted UniFrac is a qualitative measure that estimates the distance between two microbial communities based on the fraction of the branch length in a phylogenetic tree leading to descendant taxa in exclusively one or the other community (Lozupone et al., 2007). Weighted UniFrac, an extension of the unweighted Unifrac, also takes into account the relative abundance of taxa represented in the communities. Alpha diversity analysis was performed and the UniFrac (weighted and unweighted) PCoA plots were generated in the R environment using the Phyloseq package. The diversity estimators including AMOVA and ANOVA were implemented with mothur.
2.6. Statistics
Data were analyzed by two-way ANOVA with a Sidak-Holm multiple pairwise comparison post-hoc test using GraphPad Prism 7 for Fig. 1 and Fig. 2A-D. Data are expressed as mean ± SEM, and a p < 0.05 was considered statistically significant.
Fig. 1.
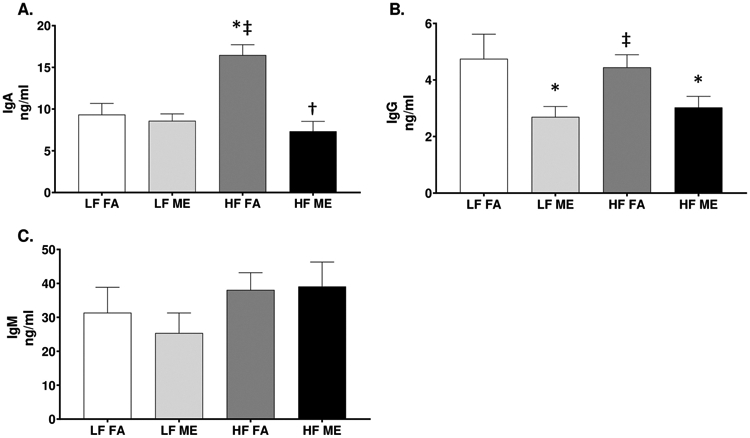
A decrease in lung IgG and IgA is observed in mice exposed to ME and HF diet. ELISA of (A) IgA, (B) IgG, and (C) IgM in lung tissue homogenates of C57Bl/6 mice placed either on regular chow (LF) or a high-fat (HF) diet and exposed to either filtered air (FA) or whole-body inhalation to a mixture of gasoline and diesel engine exhaust (ME: 30 μg PM/m3 gasoline engine emissions + 70 μg PM/m3 diesel engine) for 6 h/d, 7 d/wk for a period of 30 days. Data are depicted as the mean ± SEM with *p < 0.05 compared to LF FA, †p < 0.05 compared to HF FA, ‡p < 0.05 compared to LF ME by two way ANOVA.
Fig. 2.
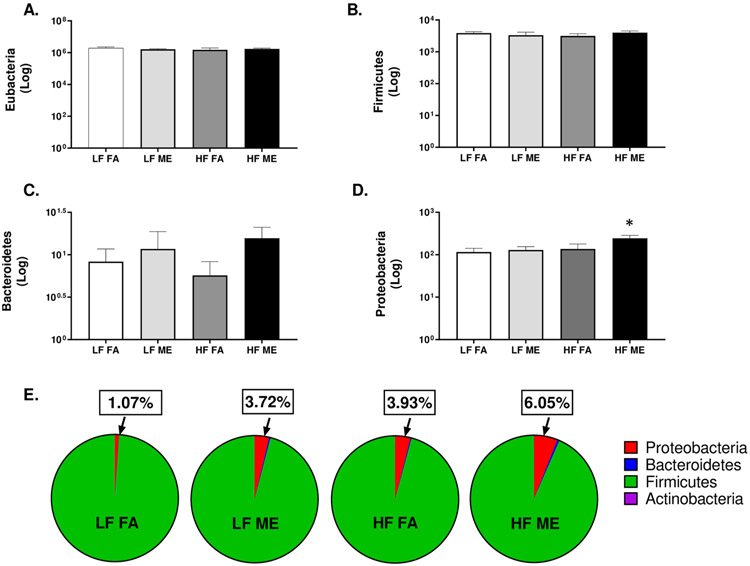
Exposure to ME results in an increase in the abundance of Proteobacteria. qPCR of lung tissue homogenates for (A) total bacteria (Eubacteria) and phyla - (B) Firmicutes, (C) Bacteroidetes, (D) Proteobacteria, (E) pie charts representing all major phyla in C57Bl/6 mice placed either on regular chow (LF) or a high-fat (HF) diet and exposed to either filtered air (FA) or whole-body inhalation to a mixture of gasoline and diesel engine exhaust (ME: 30 μg PM/m3 gasoline engine emissions + 70 μg PM/m3 diesel engine) for 6 h/d, 7 d/wk for a period of 30 days. *p < 0.05 compared to LF FA by two way ANOVA.
3. Results
3.1. Exposure to ME alters immunoglobulin levels within the lungs of C57Bl/6 wildtype mice
Although IgA is the predominant immunoglobulin in mucosal secretions, IgG and IgM are also locally present within the lungs and aid in the exclusion of invading antigens. To investigate whether exposure to inhaled vehicle emissions can alter airway defenses, we quantified the levels of IgA, IgG, and IgM within lung homogenates by ELISA. When compared to LF FA and LF ME groups, we observed IgA to significantly increase in the HF FA group (Fig. 1A, p < 0.001). Interestingly, when compared to the HF FA, we observed a significant decrease in IgA in the HF ME group (Fig. 1A, p < 0.001). The respective F values for IgA levels are: exposure = 17.020, diet = 6.021, exposure x diet interaction = 12.27. We also observed a significant decrease in IgG in the LF ME and HF ME groups (Fig. 1B, p = 0.023, F = 9.140 for exposure), compared to the LF FA group. IgM levels were found to be unaltered across all groups (Fig. 1C).
3.2. Exposure to ME results in an increased abundance of Proteobacteria in C57Bl/6 mice on the HF diet
To determine whether the bacterial abundance within the lungs was altered with our exposures, we quantified the total bacterial load using qPCR. We obtained a total of 6 log copies/ml of bacterial DNA (Fig. 2A). However, there were no statistical differences observed in the total bacterial abundance across all groups. There were also no significant alterations observed within Firmicutes and Bacteroidetes between the groups (Fig. 2B, C). Interestingly, we observed that the abundance of Proteobacteria was significantly elevated only in the HF ME groups (Fig. 2D, p = 0.031). The respective F values for Proteobacteria are: exposure = 2.499, diet = 3.492, exposure x diet interaction = 1.165. Actinobacteria was barely detected by qPCR (data not shown). When we measured the percentages of the individual phyla, contributing to the total bacterial abundance, for each of the study groups, we observed that the overall percentage of Proteobacteria was much higher in the lungs of the HF ME group (Fig. 2E).
3.3. Exposure to ME results in the increased relative abundance of Enterobacteriaceae in C57Bl/6 mice on the HF diet
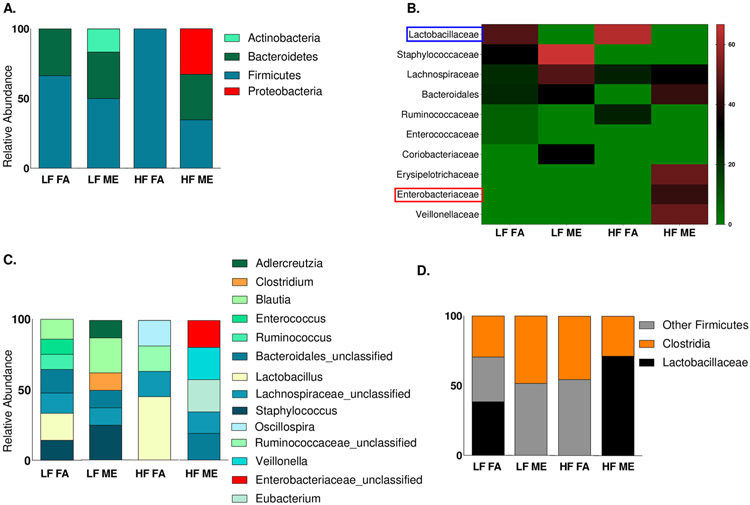
To confirm the expansion of Proteobacteria observed by qPCR, we performed Illumina MiSeq sequencing analysis of the 16 S rRNA region. We observed a similar increase in abundance in the Proteobacteria phylum in the HF ME groups alone (Fig. 3A). Although we observed Proteobacteria in the LF ME groups in the absolute abundance file, they had very low reads and were removed during the normalization of this dataset. Further classification revealed that most of the bacteria within the Proteobacteria phylum belonged to the Enterobacteriaceae family (Fig. 3B, C). Interestingly, we observed that Lactobacillus predominantly present in both the LF and HF control groups was absent in the ME exposed groups (Fig. 3B, D). We found that bacterial alterations are occurring within the Firmicutes phyla as well. Clostridia were found to be expanding in the ME exposed groups on both LF and HF diets (Fig. 3D). Although most of these bacteria within the Clostridia class belong to the Lachnospiraceae family, only the LF ME group showed the presence of Clostridium species (Fig. 3C).
Fig. 3.
Exposure to ME and HF diet increases the relative abundance of Enterobacteriaceae belonging to the Proteobacteria phylum. 16 S Illumina sequencing of the lung bacterial DNA at the (A) phylum and (C) genus level, (B) heatmap showing the relative abundance at the family level and (D) relative abundance of major bacteria in the Firmicutes phlya in lung tissues of C57Bl/6 mice placed either on regular chow (LF) or a high-fat (HF) diet and exposed to either filtered air (FA) or whole-body inhalation to a mixture of gasoline and diesel engine exhaust (ME: 30 μg PM/m3 gasoline engine emissions + 70 μg PM/m3 diesel engine) for 6 h/d, 7 d/wk for a period of 30 days.
3.4. Bacterial diversity is altered with exposure to ME in C57Bl/6 mice
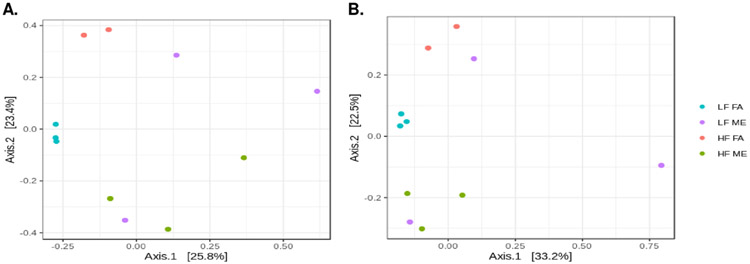
To investigate exposure mediated shifts in lung microbial diversity, we performed α-diversity and β-diversity calculations. The α-diversity was estimated using the Shannon (diversity) index (Fig. 4A) and Chao1 (richness) index (Fig. 4B), both of which showed a decrease in the bacterial diversity in the HF ME group. However, these results were not statistically significant by ANOVA due to the low abundance of these microorganisms. We observed significant differences in β-diversity in lung microbiota profiles in the exposed and control groups. Fig. 5A is an unweighted PCoA plot constructed using unweighted UniFrac distances representing the total variance in bacterial communities. Each dot is representative of the lung microbiome of one animal in each of the four groups. Although we had six animals in each group, we were unable to obtain reads for all the samples due to the low abundance of microorganisms within the lungs. Despite these challenges, we still observe a clear separation between the ME exposed and FA control groups. Both the unweighted and weighted PCoA plots show LF FA and HF FA samples are dispersed away from the LF ME and HF ME groups (Fig. 5A, B). Analysis of Molecular Variance (AMOVA) was performed to assess the variations among different groups. AMOVA showed UniFrac distances with a significant p-value of 0.006 between LF FA and LF ME and a p-value of 0.007 between the LF FA and HF ME groups (Table 2). A significant variation was also observed between the control groups on LF or HF diets (Table 2, p = 0.014).
Fig. 4.
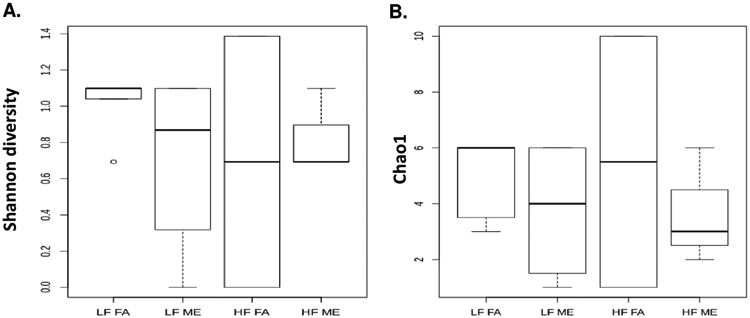
Bacterial alpha diversity analysis of exposure and diet groups. Alpha diversity analysis using (A) Shannon index (diversity) and (B) Chao1 index (richness) compared in C57Bl/6 mice placed either on regular chow (LF) or a high-fat (HF) diet and exposed to either filtered air (FA) or whole-body inhalation to a mixture of gasoline and diesel engine exhaust (ME: 30 μg PM/m3 gasoline engine emissions + 70 μg PM/m3 diesel engine) for 6 h/d, 7 d/wk for a period of 30 days.
Fig. 5.
Differential clustering of bacterial groups is observed with ME exposures. β-diversity calculations using (A) unweighted and (B) weighted analyses. Each circle is representative of one animal in each of the groups of C57Bl/6 mice placed either on regular chow (LF) or a high-fat (HF) diet and exposed to either filtered air (FA) or whole-body inhalation to a mixture of gasoline and diesel engine exhaust (ME: 30 μg PM/m3 gasoline engine emissions + 70 μg PM/m3 diesel engine) for 6 h/d, 7 d/wk for a period of 30 days.
Table 2.
AMOVA analysis of microbiota profiles from lungs of C57Bl/6 mice exposed via inhalation to mixed emissions.
| Exposure groups | Sum Sq | Df | Mean Sq | P-value |
|---|---|---|---|---|
| LF FA vs. LF ME | 0.006 | |||
| Among groups | 0.970628 | 1 | 0.970628 | |
| Within groups | 3.84797 | 10 | 0.384797 | |
| Total | 4.81859 | 11 | ||
| HF FA vs. HF ME | 0.106 | |||
| Among groups | 0.58214 | 1 | 0.58214 | |
| Within groups | 2.72784 | 6 | 0.45464 | |
| Total | 3.30998 | 7 | ||
| LF FA vs. HF ME | 0.007 | |||
| Among groups | 0.778862 | 1 | 0.778862 | |
| Within groups | 3.14873 | 9 | 0.349858 | |
| Total | 3.92759 | 10 | ||
| HF FA vs. LF FA | 0.014 | |||
| Among groups | 0.766791 | 1 | 0.766791 | |
| Within groups | 2.42088 | 7 | 0.345841 | |
| Total | 3.18767 | 8 | ||
| HF FA vs. LF ME | 0.504 | |||
| Among groups | 0.510557 | 1 | 0.510557 | |
| Within groups | 3.42708 | 7 | 0.489583 | |
| Total | 3.93764 | 8 | ||
| HF ME vs. LF ME | 0.088 | |||
| Among groups | 0.603311 | 1 | 0.603311 | |
| Within groups | 4.15492 | 9 | 0.461658 | |
| Total | 4.75824 | 10 |
FA, filtered air; ME, mixed vehicle emissions; LF, standard mouse chow; HF, high-fat diet; Df, degrees of freedom; Sum Sq, sum of squares; Mean Sq, mean of squares.
4. Discussion
Recent studies point to an increase in the incidence of lung diseases in heavily polluted regions, with mounting evidence implicating air pollution to be a primary risk factor for increased hospitalizations of individuals with respiratory diseases, such as asthma and COPD (Kurt et al., 2016; Moore et al., 2016; Raji et al., 2020). Many of these lung diseases have been associated with an increase in bacteria belonging to the Proteobacteria phylum (Dickson et al., 2013; Rizzatti et al., 2017). However, to date, an association between air pollution-mediated lung microbial alterations or their effects on the onset or progression of lung diseases has not been made. In this study, we report for the first time to our knowledge that exposure to a mixture of gasoline and diesel emissions in combination with the HF diet affects immune defenses and causes lung bacterial dysbiosis with an expansion of Proteobacteria. Both the gaseous and the PM component of traffic generated air pollutants were incorporated in our study to determine the overall impacts of vehicle derived pollutants. We utilized the HF diet component alongside our exposures since the standard Western diet that constitutes > 30% fat is present in much of the human population. Using this model, we sought to analyze the synergistic effects of inhaled air pollutants and the HF diet.
The results obtained indicate that exposures to mixed emissions cause a decrease or degradation of immunoglobulins that play crucial roles in protecting the airways. ME-exposure resulted in decreased pulmonary IgG in both the LF and HF-diet animals, while pulmonary IgA was only decreased with ME exposure within the HF-diet group and was not statistically different from the LF FA or LF ME groups. In addition to viral neutralization, IgA functions to prevent bacterial adherence on the surface of airway epithelial cells, thereby preventing unwarranted activation of immune responses (Mantis and Forbes, 2010). It is possible that with concurrent consumption of a HF diet, ME exposure mediates a reduction in pulmonary IgA that promotes decreased immune surveillance leading to an unwarranted bacterial outgrowth of Proteobacteria within the lungs of those animals. The decrease in IgA may be associated with degradation caused by toxic proteases released by neutrophils that rapidly infiltrate the airway lumen when exposed to pollutants (Pilette et al., 2003). Importantly, IgA was found to be significantly elevated in the HF FA groups. An increase in intestinal IgA in response to HF diets has been documented (Kunisawa et al., 2014); however, diet outcomes on IgA expression in the lung are not as well characterized. It is plausible that a gut-lung axis mediated response may contribute to alterations in IgA expression in the lungs of mice on the HF diet (Enaud et al., 2020). A decrease in IgG in ME exposed groups on both LF and HF diet suggests that the decrease is exposure mediated. A previous study of ambient particulate matter exposure on Sprague–Dawley rats increased secretory IgA levels but decreased IgG (Li et al., 2017). A decrease in salivary IgA has been observed in children living in polluted regions (Mehrbani et al., 2016). These discrepancies observed between different studies can be explained by several reasons, including different study models, variations in the components of exposed particulates and gaseous emissions, as well as the duration and intensity of exposures. Both immune suppression and initiation of pronounced inflammation have been observed in response to different particulates in air pollution, all of which have detrimental consequences on the host.
In response to air pollutant exposures, immune cells have been documented to generate reactive oxygen and nitrogen species (ROS, RNS) to mediate the challenge, and it is a well-understood mechanism of air-pollution mediated toxicity (Laskin et al., 2010; Lodovici and Bigagli, 2011). Although inflammation was traditionally understood to expel potential antigens, recent evidence suggests that specific microbial communities can exploit byproducts of inflammation to proliferate and worsen inflammatory responses (Scales et al., 2016). For example, macrophages and neutrophils release ROS in the form of and RNS in the form of as antimicrobial effectors that decompose to generate nitrates in the process of eliminating antigens. These nitrates can be utilized selectively by bacteria belonging to the Proteobacteria phylum for anaerobic respiration and growth (Winter and Bäumler, 2014). We suspect that air pollution-induced oxidative stress, along with the degradation of IgA, may be responsible for the observed increase in Proteobacteria in the HF ME group of study animals. Baseline inflammation may be higher in the HF ME groups as the HF diet alone are reported to increase inflammation; however, this was not assessed in the current study (Duan et al., 2018).
The bacterial composition was found to be altered in the HF FA groups with an abundance of Firmicutes. HF diet has been shown to increase Firmicutes in the gastrointestinal tract. Thus, a possible gut-lung interplay may contribute to an increased predominance of Firmicutes within the HF FA groups (Zhang and Yang, 2016). Beneficial bacteria such as Lactobacillus that has anti-inflammatory properties (Mortaz et al., 2013) were found to decrease in the ME exposed groups. This was accompanied by an increase in Clostridia in the ME exposed groups, with LF ME groups exhibiting an increase in the Clostridium species. Many bacteria in the Clostridia class are beneficial commensals, but Clostridium species and their outgrowth have been associated with having pathogenic outcomes in several lung diseases (Palmacci et al., 2009; Shu et al., 2008). Actinobacteria was also found to be elevated only in the LF ME group by sequencing. However, we are unaware if an increase in Actinobacteria has pathogenic outcomes within the lungs. The roles of these commensal lung bacteria are not well-known, considering our rudimentary knowledge of the lung microbiome.
The shifts observed in the Firmicutes and Proteobacteria phylum may be causing the reduced bacterial diversity observed in our results in the HF ME groups as there is an outgrowth of bacteria with selective advantages. Although not statistically significant, the Shannon diversity index shows reduced bacterial diversity in the HF ME group. The Chao1 index that measures richness of the microbial profile also revealed a decrease in the abundance of bacteria in the HF ME groups, suggesting opportunistic pathogens may be replacing commensals. The PCoA plots show a distinct separation of each group, reinstating our observations that exposures cause shifts in the microbial profile.
We also notice the expansion of Enterobacteriaceae within the Proteobacteria phylum. Bacteria belonging to the Proteobacteria phylum have been associated with many mucosal diseases (Yang and Jobin, 2014). It is plausible that the expansion of these bacteria could potentially enhance the inflammatory response, possibly due to a lack of IgA responses to safeguard against unwarranted bacterial adherence. In the absence of IgA, Proteobacteria can proliferate and adhere to epithelial linings and induce inflammatory signaling by activating Toll-like receptors (Tana et al., 2003). Thus, with decreasing IgA observed in the lungs of the HF ME-exposed animals in our study, there could be unwarranted expansion and adherence of Proteobacteria on epithelial linings that could enhance inflammation (Huffnagle et al., 2017).
With this study, we hope to shed light on the effects that air pollutants can have on the lungs and how they may exacerbate lung diseases; however, there are a few limitations to note. First, the concentration of ME used in the current study (100 μg/m3 PM) would be considered a high environmental exposure scenario. However, this PM concentration is within (or below) the range of PM observed in near roadway, occupational, and heavily populated urban area exposure scenarios (Costa et al., 2017; IQAir, 2021; Pronk et al., 2009). Moreover, a high degree of variability is observed across all the groups, which we believe is characteristic of the lung microbiome profile owing to dynamic responses within the lungs. The microbial composition is constantly renewing, and our observations at a certain time point within a group may not necessarily be identical at another time point. These exposures were done for a total of 30 days, and the reported findings are taken from only the one-time point, which is a limitation to this study. We were unable to detect bacteria in many of our samples by 16 S sequencing that affected our n-value. However, the qPCR results from the same samples helped to confirm the expansion of Proteobacteria. We were also unable to confirm inflammation and generation of ROS-RNS with this study due to the lack of available tissues; however, subsequent studies are currently underway to investigate these mechanisms.
5. Conclusion
Considering the increasing number of lung disorders that occur in urban populations exposed to high air pollution levels, we sought to determine whether traffic-generated air pollutants can affect lung immunoglobulin levels and microbiota with or without concurrent consumption of an HF diet. Our study demonstrates that high-fat diet and/or exposure to traffic-generated air pollution can affect pulmonary immunoglobulin levels and alter the lung microbial profile. Understanding the lung microbiota shifts in response to the environment and diet as a possible contributor(s) to the pathophysiology of lung diseases is paramount to identifying mechanistic pathways involved in air-pollutant mediated effects on pulmonary disorders and overall human health.
Acknowledgments
We would like to acknowledge the Chemistry and Inhalation Exposure group, in the Environmental Respiratory Health Program, at Lovelace Biomedical and Environmental Research Institute for the characterization and monitoring of the animal exposures. We would like to thank Xinying Niu from Mirpuri lab at UT Southwestern for her assistance in generating bacterial standards. We also thank Dr. Jay Raadt from the Office of Research Consulting at UNT for his assistance with the 16 S bioinformatics data.
Funding
This work was supported by the National Institute of Environmental Health Sciences at the National Institute of Health grants [R00 ES016586] and [R15 ES026795], as well as internal funding from the University of North Texas to A.K.L.
Abbreviations:
- 4-PL
Four-parameter logistic
- AMOVA
Analysis of molecular variance
- COPD
Chronic obstructive pulmonary disorder
- ELISA
Enzyme linked immunosorbent assay
- FA
Filtered air
- HF diet
High-fat diet
- IgA
Immunoglobulin A
- IgG
Immunoglobulin G
- IgM
Immunoglobulin M
- LF
Regular chow
- ME
Mixture of gasoline and diesel engine exhaust
- OTUs
Operational Taxonomic Units
- PCoA
Principal coordinate analysis
- PM
Particulate matter
- qPCR
Quantitative PCR
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Andersen ZJ, Hvidberg M, Jensen SS, Ketzel M, Loft S, Sørensen M, Tjøneland A, Overvad K, Raaschou-Nielsen O, 2011. Chronic obstructive pulmonary disease and long-term exposure to traffic-related air pollution: a cohort study. Am. J. Respir. Crit. Care Med 183 (4), 455–461. 10.1164/rccm.201006-0937OC. [DOI] [PubMed] [Google Scholar]
- Chao A, 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat 11 (4), 265–270. [Google Scholar]
- Chazdon RL, Colwell RK, Denslow JS, Guariguata MR, 1998. Statistical methods for estimating species richness of woody regeneration in primary and secondary rain forests of Northeastern Costa Rica. Man and the Biosphere Series: Forest biodiversity research, monitoring and modeling: conceptual background and old-world case studies, vol. 20, 285–309. https://cgspace.cgiar.org/handle/10568/17965. [Google Scholar]
- Costa LG, Cole TB, Coburn J, Chang YC, Dao K, Roque' PJ, 2017. Neurotoxicity of traffic-related air pollution. NeuroToxicology 59, 133–139. 10.1016/j.neuro.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Huffnagle GB, 2013. The role of the bacterial microbiome in lung disease. Expert Rev. Respir. Med 7 (3), 245–257. 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB, 2016. The microbiome and the respiratory tract. Annu. Rev. Physiol 78, 481–504. 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Zeng L, Zheng C, Song B, Li F, Kong X, Xu K, 2018. Inflammatory links between high fat diets and diseases. Front. Immunol 9, 2649. 10.3389/fimmu.2018.02649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enaud R, Prevel R, Ciarlo E, Beaufils F, Wiëers G, Guery B, Delhaes L, 2020. The gut-lung axis in health and respiratory diseases: a place for inter-organ and interkingdom crosstalks. Front. Cell. Infect. Microbiol 10 (9) 10.3389/fcimb.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim M, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY, 2015. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat. Med 21 (7), 808–814. 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustini A, Stafoggia M, Colais P, Berti G, Bisanti L, Cadum E, Cernigliaro A, Mallone S, Scarnato C, Forastiere F, EpiAir Collaborative Group, 2013. Air pollution and multiple acute respiratory outcomes. Eur. Respir. J 42 (2), 304–313. 10.1183/09031936.00128712. [DOI] [PubMed] [Google Scholar]
- Fitch MN, Phillippi D, Zhang Y, Lucero J, Pandey RS, Liu J, Brower J, Allen MS, Campen MJ, McDonald JD, Lund AK, 2020. Effects of inhaled air pollution on markers of integrity, inflammation, and microbiota profiles of the intestines in Apolipoprotein E knockout mice. Environ. Res 181, 108913 10.1016/j.envres.2019.108913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WOC, 2010. Disordered microbial communities in asthmatic airways. PLoS One 5 (1), e8578. 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraiwa K, van Eeden SF, 2013. Contribution of lung macrophages to the inflammatory responses induced by exposure to air pollutants. Mediat. Inflamm. 2013, 1–10. 10.1155/2013/619523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G, Dickson R, Lukacs N, 2017. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 10 (2), 299–306. 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IQAir, World’s most polluted cities 2019 (PM2.5). ⟨https://www.iqair.com/world-most-polluted-cities?continent=&country=&state=&page=1&perPage=50&cities=. Accessed January 7, 2021⟩.
- Kim D, Chen Z, Zhou L-F, Huang S-X, 2018. Air pollutants and early origins of respiratory diseases. Chronic Dis. Transl. Med 4 (2), 75–94. 10.1016/j.cdtm.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD, 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol 79 (17), 5112–5120. 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisawa J, Hashimoto E, Inoue A, Nagasawa R, Suzuki Y, Ishikawa I, Shikata S, Arita M, Aoki J, Kiyono H, 2014. Regulation of intestinal IgA responses by dietary palmitic acid and its metabolism. J. Immunol 193 (4), 1666–1671. 10.4049/jimmunol.1302944. [DOI] [PubMed] [Google Scholar]
- Kurt OK, Zhang J, Pinkerton KE, 2016. Pulmonary health effects of air pollution. Curr. Opin. Pulm. Med 22 (2), 138–143. 10.1097/MCP.0000000000000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskin DL, Sunil VR, Fakhrzadeh L, Groves A, Gow AJ, Laskin JD, 2010. Macrophages, reactive nitrogen species, and lung injury. Ann. N. Y. Acad. Sci 1203, 60–65. 10.1111/j.1749-6632.2010.05607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, He F, Liao B, Zhou Y, Li B, Ran P, 2017. Exposure to ambient particulate matter alters the microbial composition and induces immune changes in rat lung. Respir. Res 18 (1), 143. 10.1186/s12931-017-0626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodovici M, Bigagli E, 2011. Oxidative stress and air pollution exposure. J. Toxicol 2011, 1–9. 10.1155/2011/487074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R, 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol 73 (5), 1576–1585. 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R, 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol 71 (12), 8228–8235. 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero J, Suwannasual U, Herbert LM, McDonald JD, Lund AK, 2017. The role of the lectin-like oxLDL receptor (LOX-1) in traffic-generated air pollution exposure-mediated alteration of the brain microvasculature in Apolipoprotein (Apo) E knockout mice. Inhal. Toxicol 29 (6), 266–281. 10.1080/08958378.2017.1357774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AK, Lucero J, Harman M, Madden MC, McDonald JD, Seagrave JC, Campen MJ, 2011. The oxidized low-density lipoprotein receptor mediates vascular effects of inhaled vehicle emissions. Am. J. Respir. Crit. Care Med 184 (1), 82–91. 10.1164/rccm.201012-1967OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantis NJ, Forbes SJ, 2010. Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol. Investig 39 (0), 383–406. 10.3109/08820131003622635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino E, Caruso M, Campagna D, Polosa R, 2015. Impact of air quality on lung health: myth or reality? Ther. Adv. Chronic Dis 6 (5), 286–298. 10.1177/2040622315587256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu E, Escribano-Vazquez U, Descamps D, Cherbuy C, Langella P, Riffault S, Remot A, Thomas M, 2018. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front. Physiol 9, 1168. 10.3389/fphys.2018.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JD, Barr EB, White RK, Chow JC, Schauer JJ, Zielinska B, Grosjean E, 2004. Generation and characterization of four dilutions of diesel engine exhaust for a subchronic inhalation study. Environ. Sci. Technol 38 (9), 2513–2522. 10.1021/es035024v. [DOI] [PubMed] [Google Scholar]
- McDonald JD, Barr EB, White RK, Kracko D, Chow JC, Zielinska B, Grosjean E, 2008. Generation and characterization of gasoline engine exhaust inhalation exposure atmospheres. Inhal. Toxicol 20 (13), 1157–1168. 10.1080/08958370802449696. [DOI] [PubMed] [Google Scholar]
- Mehrbani SP, Babaloo Z, Eslami H, Abdollahian T, Tabatabai V. motameni, 2016. The effects of air pollution on the salivary iga levels in children. Biomed. Pharmacol. J 9 (2), 659–662. [Google Scholar]
- Molyneaux PL, Mallia P, Cox MJ, Footitt J, Willis-Owen SAG, Homola D, Trujillo-Torralbo M-B, Elkin S, Kon OM, Cookson WOC, Moffatt MF, Johnston SL, 2013. Outgrowth of the bacterial airway microbiome after rhinovirus exacerbation of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med 188 (10), 1224–1231. 10.1164/rccm.201302-0341OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore E, Chatzidiakou L, Kuku M-O, Jones RL, Smeeth L, Beevers S, Kelly FJ, Barratt B, Quint JK, 2016. Global associations between air pollutants and chronic obstructive pulmonary disease hospitalizations. A systematic review. Ann. Am. Thorac. Soc. 13 (10), 1814–1827. 10.1513/AnnalsATS.201601-064OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy B, Chu C, Carlin DJ, 2015. Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol. Sci. 145 (1), 5–15. 10.1093/toxsci/kfv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaz E, Adcock IM, Folkerts G, Barnes PJ, Paul Vos A, Garssen J, 2013. Probiotics in the management of lung diseases. Mediat. Inflamm 2013, 1–10. 10.1155/2013/751068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumaw CL, Levesque S, McGraw C, Robertson S, Lucas S, Stafflinger JE, Campen MJ, Hall P, Norenberg JP, Anderson T, Lund AK, McDonald JD, Ottens AK, Block ML, 2016. Microglial priming through the lung-brain axis: the role of air pollution-induced circulating factors. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 30 (5), 1880–1891. 10.1096/fj.201500047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EA, Velazquez KT, Herbert KM, 2015. Influence of high-fat-diet on gut microbiota: a driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metab. Care 18 (5), 515–520. 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Comba IY, Cho T, Engen PA, Yazıcı C, Soberanes S, Hamanaka RB, Niğdelioğlu R, Meliton AY, Ghio AJ, Budinger GRS, Mutlu GM, 2018. Inhalational exposure to particulate matter air pollution alters the composition of the gut microbiome. Environ. Pollut. 240, 817–830. 10.1016/j.envpol.2018.04.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim HA, Lucero J, Guyot A-C, Herbert LM, McDonald JD, Mabondzo A, Lund AK, 2013. Exposure to vehicle emissions results in altered blood brain barrier permeability and expression of matrix metalloproteinases and tight junction proteins in mice. Part. Fibre Toxicol 10, 62. 10.1186/1743-8977-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmacci C, Antocicco M, Bonomo L, Maggi F, Cocchi A, Onder G, 2009. Necrotizing pneumonia and sepsis due to Clostridium perfringens: a case report. Cases J. 2, 50. 10.1186/1757-1626-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilette C, Ouadrhiri Y, Dimanche F, Vaerman J-P, Sibille Y, 2003. Secretory component is cleaved by neutrophil serine proteinases but its epithelial production is increased by neutrophils through NF-κB– and p38 mitogen-activated protein kinase–dependent mechanisms. Am. J. Respir. Cell Mol. Biol 28 (4), 485–498. 10.1165/rcmb.4913. [DOI] [PubMed] [Google Scholar]
- Pronk A, Coble J, Stewart P, 2009. Occupational exposure to diesel engine exhaust: a literature review. J. Expo. Sci. Environ. Epidemiol 19, 443–457. 10.1038/jes.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji H, Riahi A, Borsi SH, Masoumi K, Khanjani N, AhmadiAngali K, Goudarzi G, Dastoorpoor M, 2020. Acute effects of air pollution on hospital admissions for asthma, COPD, and bronchiectasis in Ahvaz, Iran. Int. J. Chronic Obstr. Pulm. Dis 15, 501–514. 10.2147/COPD.S231317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A, 2017. Proteobacteria: a common factor in human diseases. BioMed. Res. Int 2017, 1–7. 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruane D, Chorny A, Lee H, Faith J, Pandey G, Shan M, Simchoni N, Rahman A, Garg A, Weinstein EG, Oropallo M, Gaylord M, Ungaro R, Cunningham-Rundles C, Alexandropoulos K, Mucida D, Merad M, Cerutti A, Mehandru S, 2016. Microbiota regulate the ability of lung dendritic cells to induce IgA class-switch recombination and generate protective gastrointestinal immune responses. J. Exp. Med 213 (1), 53–73. 10.1084/jem.20150567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales BS, Dickson RP, Huffnagle GB, 2016. A tale of two sites: how inflammation can reshape the microbiomes of the gut and lungs. J. Leukoc. Biol 100 (5), 943–950. 10.1189/jlb.3MR0316-106R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF, 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75 (23), 7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C, Weaver W, (n.d.). The Mathematical Theory of Communication, 131. [Google Scholar]
- Shu C-C, Yao M, Hung C-C, Ku S-C, Yu C-J, Chang Y-L, 2008. Lung abscess due to Clostridium baratii infection in a patient with invasive pulmonary Aspergillosis. J. Clin. Microbiol 46 (3), 1153–1154. 10.1128/JCM.02446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwannasual U, Lucero J, McDonald JD, Lund AK, 2018. Exposure to traffic-generated air pollutants mediates alterations in brain microvascular integrity in wildtype mice on a high-fat diet. Environ. Res. 160, 449–461. 10.1016/j.envres.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tana, Watarai S, Isogai E, Oguma K, 2003. Induction of intestinal IgA and IgG antibodies preventing adhesion of verotoxin-producing Escherichia coli to Caco-2 cells by oral immunization with liposomes. Lett. Appl. Microbiol. 36 (3), 135–139. 10.1046/j.1472-765x.2003.01278.x. [DOI] [PubMed] [Google Scholar]
- Tasnim N, Abulizi N, Pither J, Hart MM, Gibson DL, 2017. Linking the gut microbial ecosystem with the environment: does gut health depend on where we live? Front. Microbiol 8, 1935. 10.3389/fmicb.2017.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Bäumler AJ, 2014. Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes 5 (1), 71–73. 10.4161/gmic.27129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y-F, Xu Y-H, Shi M-H, Lian Y-X, 2016. The impact of PM2.5 on the human respiratory system. J. Thorac. Dis 8 (1), E69–E74. 10.3978/j.issn.2072-1439.2016.01.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jobin C, 2014. Microbial imbalance and intestinal pathologies: connections and contributions. Dis. Model. Mech 7, 1131–1142. 10.1242/dmm.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y, Srinivas G, Kuenzel S, Linnenbrink M, Alnahas S, Bruce KD, Steinhoff U, Baines JF, Schaible UE, 2014. Environmentally determined differences in the murine lung microbiota and their relation to alveolar architecture. PLoS One 9 (12), e113466. 10.1371/journal.pone.0113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Yang X-J, 2016. Effects of a high fat diet on intestinal microbiota and gastrointestinal diseases. World J. Gastroenterol 22 (40), 8905–8909. 10.3748/wjg.v22.i40.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Global Health Observatory, 2016. https://apps.who.int/gho/data/view.main.BODAMBIENTAIRDTHS. (Accessed 10 August 2020). [Google Scholar]