Abstract
Background
Autophagy of alveolar macrophages is a crucial process in ischemia/reperfusion injury-induced acute lung injury (ALI). Bone marrow-derived mesenchymal stem cells (BM-MSCs) are multipotent cells with the potential for repairing injured sites and regulating autophagy. This study was to investigate the influence of BM-MSCs on autophagy of macrophages in the oxygen-glucose deprivation/restoration (OGD/R) microenvironment and to explore the potential mechanism.
Methods
We established a co-culture system of macrophages (RAW264.7) with BM-MSCs under OGD/R conditions in vitro. RAW264.7 cells were transfected with recombinant adenovirus (Ad-mCherry-GFP-LC3B) and autophagic status of RAW264.7 cells was observed under a fluorescence microscope. Autophagy-related proteins light chain 3 (LC3)-I, LC3-II, and p62 in RAW264.7 cells were detected by Western blotting. We used microarray expression analysis to identify the differently expressed genes between OGD/R treated macrophages and macrophages co-culture with BM-MSCs. We investigated the gene heme oxygenase-1 (HO-1), which is downstream of the phosphoinositide 3-kinase/protein kinase B (PI3K/Akt) signaling pathway.
Results
The ratio of LC3-II/LC3-I of OGD/R treated RAW264.7 cells was increased (1.27 ± 0.20 vs. 0.44 ± 0.08, t = 6.67, P < 0.05), while the expression of p62 was decreased (0.77 ± 0.04 vs. 0.95 ± 0.10, t = 2.90, P < 0.05), and PI3K (0.40 ± 0.06 vs. 0.63 ± 0.10, t = 3.42, P < 0.05) and p-Akt/Akt ratio was also decreased (0.39 ± 0.02 vs. 0.58 ± 0.03, t = 9.13, P < 0.05). BM-MSCs reduced the LC3-II/LC3-I ratio of OGD/R treated RAW264.7 cells (0.68 ± 0.14 vs. 1.27 ± 0.20, t = 4.12, P < 0.05), up-regulated p62 expression (1.10 ± 0.20 vs. 0.77 ± 0.04, t = 2.80, P < 0.05), and up-regulated PI3K (0.54 ± 0.05 vs. 0.40 ± 0.06, t = 3.11, P < 0.05) and p-Akt/Akt ratios (0.52 ± 0.05 vs. 0.39 ± 0.02, t = 9.13, P < 0.05). A whole-genome microarray assay screened the differentially expressed gene HO-1, which is downstream of the PI3K/Akt signaling pathway, and the alteration of HO-1 mRNA and protein expression was consistent with the data on PI3K/Akt pathway.
Conclusions
Our results suggest the existence of the PI3K/Akt/HO-1 signaling pathway in RAW264.7 cells under OGD/R circumstances in vitro, revealing the mechanism underlying BM-MSC-mediated regulation of autophagy and enriching the understanding of potential therapeutic targets for the treatment of ALI.
Keywords: Bone marrow mesenchymal stem cells, Oxygen-glucose deprivation/restoration, Phosphoinositide 3-kinase/protein kinase B signaling pathway, Macrophages, Autophagy, Whole-genome microarray assay
Introduction
The pathological processes of acute lung injury (ALI) are normally accompanied by broad infiltration, increased autophagy, decreased phagocytosis, and excessive production of inflammatory mediators of pulmonary macrophages, which largely contribute to severe pulmonary edema.[1–3] One of the biological characteristics of macrophages is their role in autophagy. Generally, a regulated amount of autophagy is considered to be a survival mechanism to protect cells from hypoxia, starvation, and infection,[1,2] but overactivated autophagy causes apoptosis or necrosis.[4,5] To date, an increasing number of studies have shown that the level of autophagy mediated by alveolar macrophages (AMs) is crucial in ALI induced by ischemia/reperfusion injury (IRI).[4,6,7]
Bone marrow-derived mesenchymal stem cells (BM-MSCs) are multipotent cells[8] with strong potential to travel to injured sites and produce anti-apoptosis, depression of inflammatory factors as well as participate in the regulation of autophagy.[9–13] For example, studies have demonstrated that BM-MSCs appear to be effective for therapy[14,15] in restoration[9–11] and treatment[16–18] of lung injuries, such as pulmonary hypertension and chronic obstructive pulmonary disease. However, the regulation of autophagy by BM-MSCs in macrophages and the related signaling pathways under IRI-induced ALI remain unclear.
The phosphoinositide 3-kinase/protein kinase B (PI3K/Akt) signaling pathway is involved in the regulation of a wide range of cellular processes,[19] including nutrition, metabolism, proliferation, and apoptosis.[20,21] Furthermore, the PI3K/Akt pathway also regulates autophagy through downstream molecules.[20,21] It has been recognized that heme oxygenase-1 (HO-1) is a molecule that is downstream of the PI3K/Akt pathway, and the up-regulation or overexpression of HO-1 might play a protective role under the stimulation of hypoxia, oxidative stress, and IRI.[22–24] Previous studies have shown that reactive oxygen species or transforming growth factor-beta 1 up-regulate the expression of HO-1 via the PI3K/Akt pathway,[25] and activated PI3K/Akt up-regulates the expression of HO-1 via Nrf2.[25–27] Interestingly, HO-1 is also involved in the modulation of cellular autophagy. In liver IRI, HO-1 was found to induce autophagy to protect hepatic cells from injury.[28] Conversely, in acute renal injury, knocking out HO-1 increased oxidative stress and autophagy, causing cellular death. However, knocking down HO-1 decreased autophagy and reduced oxidative stress.[29] However, it is still a question whether HO-1 is expressed in macrophages under IRI circumstances.
Thus, in the present study, we established a co-culture system of macrophages (RAW264.7) with BM-MSCs under oxygen-glucose deprivation/restoration (OGD/R) circumstances in vitro to determine whether BM-MSC-mediated regulation of autophagy in RAW264.7 cells occurs via the PI3K/Akt signaling pathway and to screen the downstream gene of HO-1 based on a whole-genome microarray assay.
Methods
Cell culture
Human BM-MSCs were purchased from ScienCell Company (Catalogue No. 7500, Carlsbad, CA, USA). Vials of 5 × 106 cells (passage 1) were thawed, plated into culture chambers (Corning, Lowell, MA, USA) in mesenchymal stem cell medium (catalog No.7501, ScienCell) supplemented with 5% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) and 1% penicillin/streptomycin (P/S); then, they were incubated at 37°C in 5% CO2 for 6 to 7 days until 85% to 90% confluence was reached. BM-MSCs (passages 3–6) were used for experiments.
Human embryonic lung fibroblasts (HELFs) (catalog No. GNHu5) and RAW264.7 mouse macrophages (catalog No. TCM13/SCSP5036) were purchased from the cell bank of the Chinese Academy of Sciences. The culture conditions for HELFs and RAW264.7 cells were the same; they were cultured in high glucose Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS and 1% P/S and were incubated at 37°C in 5% CO2 for 6 to 7 days until 70% to 80% confluence was reached. HELFs and RAW264.7 cells (passages 3–6) were used for experiments. All procedures were performed according to the manufacturer's instructions.
In vitro co-culture of RAW264.7 cells with BM-MSCs or HELFs under OGD/R conditions
A six-well Transwell co-culture system (0.4-μm pore polycarbonate, Corning) was used to establish co-culture of RAW264.7 cells with BM-MSCs or HELFs. RAW264.7 cells were grown (50%–70% cell density) in six-well plates. The 3rd to 6th passages of BM-MSCs (1 × 105 cells/well) were seeded in the upper wells of a six-well Transwell system and then were cultured overnight. Then, the upper wells of BM-MSCs were transferred to the lower wells of the six-well plate that contained RAW264.7 cells, producing a co-culture system. The co-culture system of RAW264.7 cells with HELFs was the same as that of BM-MSCs.
The OGD/R model was established as previously reported.[7,30,31] Briefly, the standard DMEM culture medium was replaced with an equal volume of glucose-free DMEM and incubated at 37°C in 1% CO2 for 8 h (to mimic ischemia). Then, the glucose-free DMEM was replaced with standard medium and incubated at 37°C in 5% CO2 for 12 h (to mimic reperfusion).
Experimental groups were randomly divided as follows: (1) Control (RAW264.7); (2) Control + BM-MSCs (RAW264.7 + BM-MSCs); (3) OGD/R (OGD/R-induced RAW264.7); (4) OGD/R + BM-MSCs (OGD/R-induced RAW264.7 + BM-MSCs); and (5) OGD/R + HELF (OGD/R-induced RAW264.7 + HELF). Among those groups, the control, OGD/R, and OGD/R + BM-MSC groups were also used for whole-gene microarray analysis, and each experiment was performed in triplicate and marked as Con-1,2,3; OGD/R-1,2,3; or OGD/R + BM-MSC-1,2,3.
Recombinant adenovirus (Ad-mCherry-GFP-LC3B) for tracking autophagy in RAW264.7 cells
RAW264.7 cells infected with Ad-mCherry-GFP-LC3B (Beyotime Biotechnology, Shanghai, China) express intracellular fluorescent fusion proteins of mCherry (red fluorescence), green fluorescence fusion protein (GFP, green fluorescence), and light chain 3B (LC3B, yellow fluorescence, merged red and green, representing the autophagic intensity), which indicate the autophagic status of RAW264.7 cells observed under a fluorescence microscope.
Detailed procedures were performed according to the manufacturer's instructions. Briefly, the infection of RAW264.7 cells with Ad-mCherry-GFP-LC3B occurred under optimized infection conditions (multiplicity of infection [MOI]), which was based on the formula: plaque-forming unit = cell number × MOI. RAW264.7 cells were cultured overnight in high glucose DMEM at 37°C in 5% CO2 to reach 50% confluence. Based on the variable MOI values being tested, a certain proportion of Ad-mCherry-GFP-LC3B virus was added into wells containing RAW264.7 cells, and then were incubated at 37°C in 5% CO2 away from light. The best infection conditions were determined, including the best MOI, best observation time point, and 20% to 70% infection efficiency of adenovirus, which enabled the autophagic vitality of RAW264.7 cells could be investigated.
Western blotting
Proteins were extracted from cultured RAW264.7 cells using radioimmunoprecipitation assay buffer (Beyotime Biotechnology). The Western blotting analysis was performed according to standard procedures as described in earlier studies. Briefly, equal amounts of protein from cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Bio-Rad, Hercules, California, USA). The membranes were blocked with blocking solution (Beyotime Biotechnology) for 1 h and subsequently were incubated with primary and secondary antibodies. Immunoreactive bands were visualized with a chemiluminescence detection system (Thermo Scientific Pierce, Rockford, IL, USA). All antibodies used in these studies were purchased from Cell Signaling Technology (Danvers, MA, USA).
Inhibition of the PI3K/Akt pathway with LY294002 in RAW264.7 cells
LY294002 (20 μmol/L Selleck Chemicals, Houston, TX, USA), a PI3K inhibitor, was applied to selectively inhibit the PI3K/Akt pathway in RAW264.7 cells. The procedures were performed according to the manufacturer's instructions.
Extraction and clean-up of total RNA from RAW264.7 cells
RAW264.7 cells were lysed, and mRNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. The concentration and integrity of the RNA in each group were measured at optical density (OD) 260/280 nm and OD260/230 nm ratios with an ultra-microspectrophotometer (NanoDrop ND-1000; Agilent, Santa Clara, CA, USA). Total RNA clean-up was performed using a QIAGEN RNeasy Mini Kit (Qiagen 74104, Germany) according to the manufacturer's protocol. Purified RNA should be between 1.8 and 2.1 in the OD260/280 nm ratio and have an OD260/230 nm ratio >1.8. Clear bands representing ribosomal RNAs 5S, 18S, and 28S were visualized without degradation on formaldehyde denaturing gel electrophoresis.
Cy3-uridine triphosphate (UTP)-labelled RNA and labeled cRNA quality control
Purified RNA was labeled and amplified with Cy3-UTP using a Quick Amp Labeling Kit, One-Color (p/n 5190-0442; Agilent), according to the manufacturer's instructions. Cy3-UTP-labeled cRNA was purified using a QIAGEN RNeasy Mini Kit (74104; Qiagen) according to the manufacturer's protocol.
PCR analysis of mRNA expression of differential genes
After extraction and measurement of RNA from RAW264.7 cells, quantitative real-time PCR (qRT-PCR) was performed. The primer sequences were designed by BioTNT Corporation (Shanghai, China), and β-actin served as an internal reference. A ReverTra Ace qPCR RT Kit (Hitachi, Toyobo, Japan) was used for RNA reverse transcription, which was performed according to the manufacturer's instructions.
Microarray expression analysis
One microgram of total RNA from each sample was used to generate amplified and biotinylated sense-strand complementary DNA (cDNA) from the entire expressed genome, which was performed according to the manufacturer's instructions (Quick Amp Labeling Kit, p/n 5190-0442; Agilent; QIAGEN RNeasy Mini Kit, 74104; Qiagen). Agilent Mouse 4 × 44K Gene Expression Microarrays (v2), which include nearly 40,000 mouse genome informatic points and transcripts based on RefSeq, GenBank, and RIKEN databank, were hybridized with the cDNA samples for 17 h in a 65°C incubator. Then, they were washed and stained and finally scanned using the Agilent Microarray Scanner (p/n G2565BA; Agilent) with Feature Extraction software (Agilent). Scanned fluorescence values were transferred into Genespring GX (ver.12.1; Agilent). The extracted data were used to compare the signal ratio (fold change) between groups.
The fold change reflects genome changes. Differentially expressed genes (DEGs) were defined as fold change ≥2.0 and P ≤ 0.05 as determined by Student's t test. Based on the R package,[32] gene ontology (GO) analysis was used for locating and identifying DEGs within cells[33]; Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis[34] was used to identify the various pathway distributions of differential genes.
Statistical analysis
The values of all of the measurements are presented as the mean ± standard deviation. Comparisons between groups were performed with Student's t test. The analysis was performed with GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA). Each experiment was performed in triplicate. Values of P < 0.05 were considered statistically significant.
Results
Co-cultured BM-MSCs down-regulated the elevated levels of autophagy in RAW264.7 cells under OGD/R conditions
Recombinant adenovirus (Ad-mCherry-GFP-LC3B) was used to infect RAW264.7 cells to confirm the BM-MSC-mediated modulation of autophagy in RAW264.7 cells, and intracellular expression of the mCherry-GFP-LC3B fusion proteins was observed under a fluorescence microscope. The fluorescence images displayed a decrease in yellow fluorescence in RAW264.7 cells in the OGD/R group, suggesting increased autophagic intensity. However, the yellow fluorescence of RAW264.7 cells in the OGD/R + BM-MSC group was higher than that in the OGD/R group, indicating the down-regulation of BM-MSCs resulting in elevated autophagy in RAW264.7 cells was induced by the OGD/R conditions [Figure 1]. For comparison, there was dispersed intracellular yellow fluorescence in RAW264.7 cells in the control group, representing relatively low autophagy.
Figure 1.
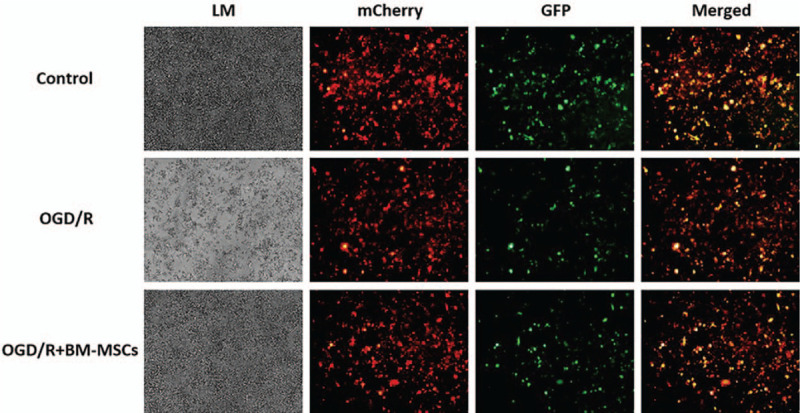
Morphological observation of the growth of RAW264.7 cells under light microscopy and intracellular expression of the mCherry-GFP-LC3B fusion protein under a fluorescence microscope. Original magnification, ×200. BM-MSCs: Bone marrow-derived mesenchymal stem cells; GFP: Green fluorescence fusion protein; LM: Morphological growth of RAW264.7 cells under light microscopy; mCherry: Red fluorescence fusion protein; OGD/R: Oxygen-glucose deprivation/restoration.
Co-cultured BM-MSCs regulated autophagy-related proteins in RAW264.7 cells under OGD/R conditions
To further elucidate the effect of BM-MSCs on autophagy in RAW264.7 cells under OGD/R conditions, the autophagy-related proteins LC3-I, LC3-II, and p62 in RAW264.7 cells were detected by Western blotting. Quantitative statistical analysis showed that there was a significant change in the LC3-II/LC3-I ratio (1.27 ± 0.20 vs. 0.44 ± 0.08, t = 6.67, P < 0.05) and reduced expression of p62 in the OGD/R group compared to those levels in the control group (0.77 ± 0.04 vs. 0.95 ± 0.10, t = 2.90, P < 0.05), and co-culture with BM-MSCs significantly decreased the LC3-II/LC3-I ratio (0.68 ± 0.14 vs. 1.27 ± 0.20, t = 4.12, P < 0.05) and up-regulated the expression of p62 compared to the levels in the OGD/R group (1.10 ± 0.20 vs. 0.77 ± 0.04, t = 2.80, P < 0.05) [Figure 2].
Figure 2.
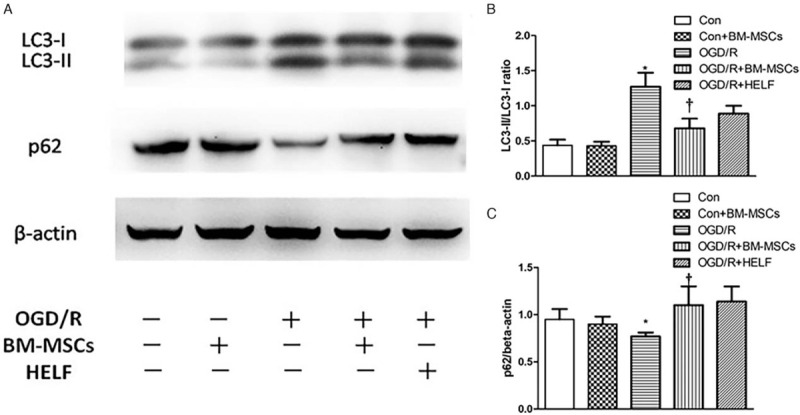
Autophagy-related proteins LC3-I, LC3-II, and p62 in RAW264.7 cells were detected by Western blotting. (A) The protein band visualization for each group. (B) Statistical analysis of the LC3-II/LC3-I ratio among the five groups. ∗P < 0.05 vs. control group, †P < 0.05 vs. OGD/R group. (C) Statistical analysis of the expression of p62 among the five groups. ∗P < 0.05 vs. control group, †P < 0.05 vs. OGD/R group. Data are expressed as mean ± standard deviation. All the data were based on three dependent experiments. BM-MSCs: Bone marrow-derived mesenchymal stem cells; Con: Control; HELF: Human embryonic lung fibroblasts; LC3: Light chain 3; OGD/R: Oxygen-glucose deprivation/restoration.
Co-cultured BM-MSCs down-regulated autophagic levels in RAW264.7 cells through activation of the PI3K/Akt signaling pathway under OGD/R conditions
To elucidate the signaling pathway related to autophagy in RAW264.7 cells, the autophagy-related pathway proteins PI3K, p-Akt, and Akt were detected by Western blotting. The proteins visualized after the blot and the quantitative statistical analysis showed that there were significantly lower expression levels of PI3K (0.40 ± 0.06 vs. 0.63 ± 0.10, t = 3.42, P < 0.05) and the p-Akt/Akt (0.39 ± 0.02 vs. 0.58 ± 0.03, t = 9.13, P < 0.05) ratio in the OGD/R than in the control group. However, the expression of PI3K (0.54 ± 0.05 vs. 0.40 ± 0.06, t = 3.11, P < 0.05) and the p-Akt/Akt ratio (0.52 ± 0.05 vs. 0.39 ± 0.02, t = 9.13, P < 0.05) were significantly higher in the OGD/R + BM-MSC group than they were in the OGD/R group, suggesting that the OGD/R-induced inhibition of the PI3K/Akt signaling pathway caused the elevation of autophagic levels in RAW264.7 cells, whereas co-cultured BM-MSCs down-regulated autophagic levels in RAW264.7 cells through activation of the PI3K/Akt signaling pathway [Figure 3].
Figure 3.
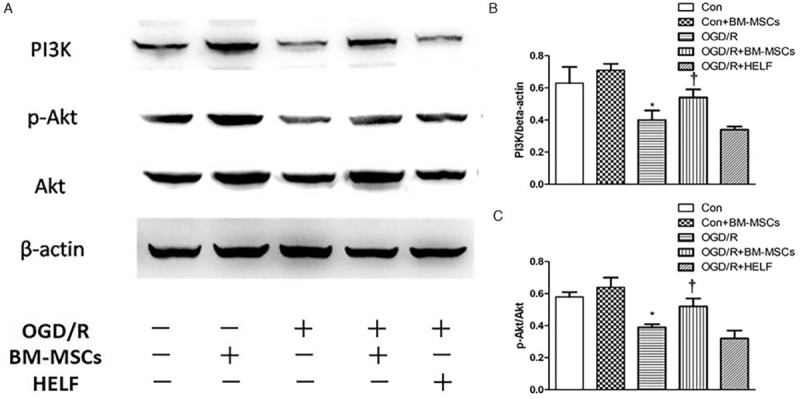
Western blotting analysis for the detection of the autophagy-related pathway proteins PI3K, p-Akt, and Akt in RAW264.7 cells in each group. (A) The protein band visualization for PI3K, p-Akt, and Akt in each group. (B) Statistical analysis of PI3K expression among the five groups. ∗P < 0.05 vs. control group, †P < 0.05 vs. OGD/R group. (C) Statistical analysis of the p-Akt/Akt ratio among the five groups. ∗P < 0.05 vs. control and control + BM-MSCs groups, †P < 0.05 vs. OGD/R and OGD/R + HELF groups. Data are expressed as mean ± standard deviation. All the data were based on three dependent experiments. Akt: Protein kinase B; BM-MSCs: Bone marrow-derived mesenchymal stem cells; Con: Control; HELF: Human embryonic lung fibroblasts; LC3: Light chain 3; OGD/R: Oxygen-glucose deprivation/restoration; PI3K: Phosphoinositide 3-kinase.
LY294002 blocked the BM-MSC-mediated modulation of autophagy in RAW264.7 cells through the PI3K/Akt pathway
To further verify the role of BM-MSCs in regulating autophagy in RAW264.7 cells through the PI3K/Akt pathway, RAW264.7 cells were pre-treated with LY294002 (a PI3K inhibitor), and the autophagy-related protein LC3-I/LC3-II was detected under OGD/R conditions. The proteins visualized after the blot and the quantitative statistical analysis showed that with pre-treatment of RAW264.7 cells with LY294002, there was a significantly increased LC3-II/LC3-I ratio in the OGD/R + BM-MSCs + LY294002 group compared to the OGD/R + BM-MSCs group (1.29 ± 0.15 vs. 0.89 ± 0.08, t = 4.08, P < 0.05) [Figure 4A and 4B], further indicating that the down-regulation of the autophagic level of RAW264.7 cells by BM-MSCs occurred through the PI3K/Akt signaling pathway under OGD/R conditions.
Figure 4.
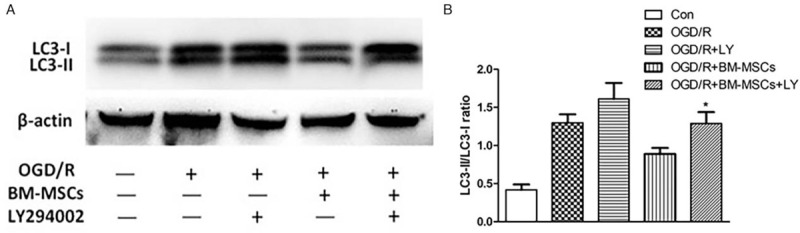
Western blotting analysis for the detection of the autophagy-related proteins LC3-I and LC3-II and statistical analysis of the LC3-II/LC3-I ratio in the five groups of RAW264.7 cells pre-treated with LY294002. (A) The protein band visualization of Western blotting in each group. (B) Statistical analysis of the LC3-II/LC3-I ratio among the five groups; ∗P < 0.05 vs. OGD/R + BM-MSCs group. Data are expressed as mean ± standard deviation. All the data were based on three dependent experiments. BM-MSCs: Bone marrow-derived mesenchymal stem cells; Con: Control; HELF: Human embryonic lung fibroblasts; LY: LY294002; LC3: Light chain 3; OGD/R: Oxygen-glucose deprivation/restoration.
Whole-genome expression profile and summary of DEGs
Based on the patterns of the genes identified by the microarray, the fluorescence intensity in each genetic locus was recorded as original data and treated with Agilent Genespring GX software to obtain the level of transcription for each gene in RAW264.7 cells. Cluster detection was performed along with the level of gene transcription in each gene. A clustering diagram showed the overall difference in all gene transcription between the OGD/R and control groups, as well as the OGD/R + BM-MSC and OGD/R groups [Figure 5A].
Figure 5.
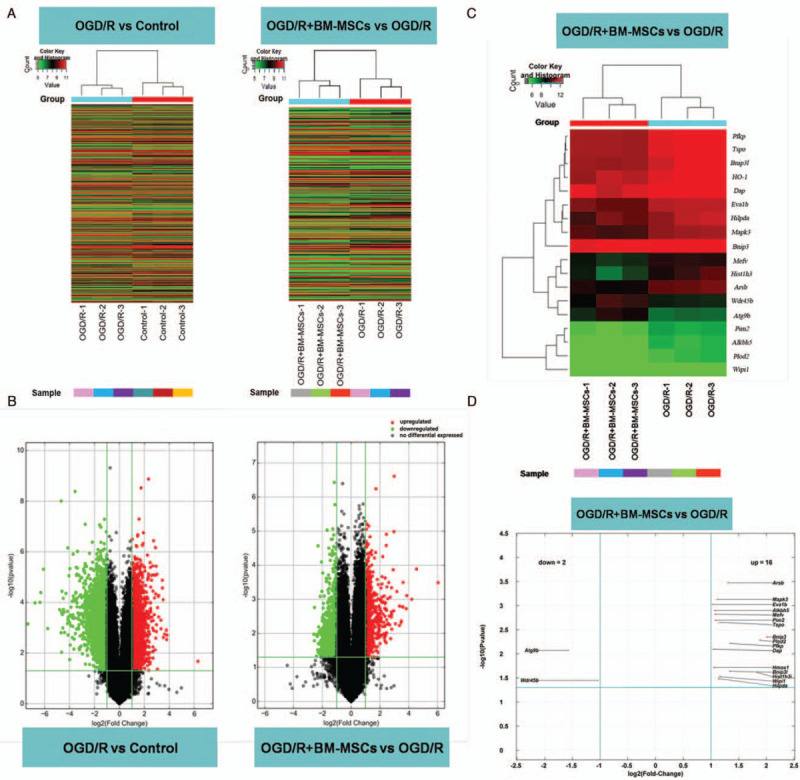
Whole-genome expression profile and summary of DEGs. (A) Clustering diagram showing the overall difference in transcription in RAW264.7 cells between the OGD/R and control groups as well as between the OGD/R + BM-MSCs and OGD/R groups. Fluorescence intensity represents the level of gene transcription (red: high; blue: low). (B) Illustration of changes with a volcano plot. The DEGs between compared groups (fold change ≥2.0 and P ≤ 0.05). Colors represent the level of changes in transcription (red: up-regulation; blue: down-regulation). (C) Clustering diagram of DEGs related to autophagy between the OGD/R+BM-MSC group and the OGD/R group. Fluorescence intensity represents the level of transcription (red: high; blue: low). (D) Volcano plot illustrates DEGs related to autophagy. There were 16 up-regulated genes (including HO-1 and Mapk3) and two down-regulated genes (Atg9b and Wdr45b) between the OGD/R + BM-MSC group and the OGD/R group. BM-MSCs: Bone marrow-derived mesenchymal stem cells; DEGs: Differentially expressed genes; HO-1: Heme oxygenase-1; OGD/R: Oxygen-glucose deprivation/restoration.
Volcano plots were used to visualize genome-wide gene expression. Through the detection of the whole-genome expression profile, more than 26,000 genes and transcripts were screened. Afterward, the filtration and comparison of DEGs were performed by analyzing the fold change and P-value, as illustrated with volcano plots. The criteria for DEGs were based on the level of change in gene transcription (fold change ≥2.0 and P ≤ 0.05) between compared groups [Figure 5B]. The results showed that 1785 genes were up-regulated and 1856 genes were down-regulated in the OGD/R group compared to the control group. However, 699 genes were up-regulated and 735 genes were down-regulated in the OGD/R + BM-MSC group compared to the OGD/R group.
Statistical analysis of the expression of DEGs related to autophagy
The selected DEGs between the OGD/R + BM-MSC and OGD/R groups were subjected to GO and BP analysis to confirm the up- and down-regulation of autophagy-related genes. There were a total of 18 genes with significantly different expression between the OGD/R + BM-MSC group and the OGD/R group (fold change ≥2.0 and P ≤ 0.05), including 16 up-regulated genes (such as HO-1 and Mapk3) and two down-regulated genes (Atg9b and Wdr45b). The results are shown in Supplementary Table 1 and are visualized in a cluster diagram [Figure 5C] and a volcano plot [Figure 5D].
Identification of the DEG HO-1
In our previous study and increased research, the target gene HO-1 might refer to IRI. We selected the HO-1 gene for further confirmation of its differential expression through pathway analysis with the KEGG databank to identify a link between HO-1 expression and PI3K/Akt under OGD/R conditions. Fluorescent qPCR and Western blotting were used to determine the mRNA expression and protein level of the HO-1 gene. The results showed significant up-regulation of both mRNA (0.89 ± 0.17 vs. 0.40 ± 0.07, t = 4.62, P < 0.05) and protein expression in the HO-1 gene in RAW264.7 cells when they were co-cultured with BM-MSCs under OGD/R conditions (0.70 ± 0.09 vs. 0.48 ± 0.02, t = 4.13, P < 0.05) [Figure 6A and 6B], which was in accordance with the data from the whole-genome expression profile.
Figure 6.
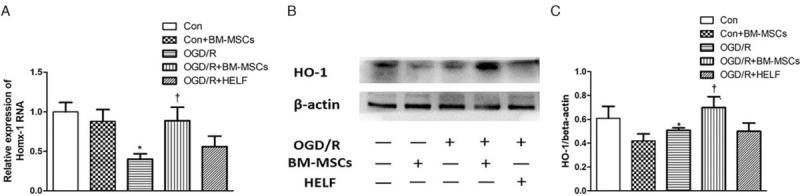
Analysis of the differentially expressed gene HO-1 in RAW264.7 cells among groups. (A) Statistical analysis of fluorescent quantitative polymerase chain reaction for expression of HO-1 mRNA; ∗P < 0.05 vs. Control group, +P < 0.05 vs. OGD/R group. (B) Western blotting and statistical analysis for protein expression of HO-1. ∗P < 0.05 vs. control group, +P < 0.05 vs. OGD/R group. Data are expressed as mean ± standard deviation. BM-MSCs: Bone marrow-derived mesenchymal stem cells; Con: Control; HELF: Human embryonic lung fibroblasts; HO-1: Heme oxygenase-1; OGD/R: Oxygen-glucose deprivation/restoration.
Discussion
There are complex mechanisms in IRI-induced ALI. Clinically, a major problem in ALI is excessive inflammation. In particular, broad infiltration, increased autophagy, and the increased release of inflammatory mediators by pulmonary macrophages are the leading causes of ALI, and these factors largely contribute to severe pulmonary edema.[1–3] Therefore, the reduction of excessive inflammation, and more specifically, modulating the regulation of the autophagic level in macrophages, could be the central point in the alleviation of IRI-induced ALI. In the present study, based on a co-culture system of RAW264.7 cells with BM-MSCs under OGD/R conditions in vitro, we verified that BM-MSCs modulate autophagy in RAW264.7 cells via the PI3K/Akt signaling pathway and tentatively identified the downstream gene expression of HO-1 in the PI3K/Akt signaling pathway with a whole-genome microarray assay. Following pre-treatment with LY294002, the effect of BM-MSCs on autophagy in RAW264.7 cells was blocked. The results confirmed the existence of the PI3K/Akt/HO-1 signaling pathway, and BM-MSCs down-regulated the autophagic level in RAW264.7 cells in this way. This is interesting because this is the first study to uncover the mechanism underlying the BM-MSC-mediated modulation of autophagy in RAW264.7 cells, and, more broadly, this study enriches our understanding of the autophagy of macrophages in ALI as well as areas to target in BM-MSCs for ALI therapy.
Autophagy of macrophages is involved in multiple lung diseases.[3,35–37] Regulated moderate amount autophagy is considered to be a survival mechanism that protects cells from hypoxia, starvation, and infection,[1,2] but overactivated autophagy causes apoptosis or necrosis.[4,5] Thus, precise modulation of autophagy is a key link for the maintenance of cellular homeostasis.[38–40] Increasing studies have shown that the modulation of the autophagic level of AMs is a crucial process in IRI-induced ALI[4,6,7]; therefore, trying to find an effective therapeutic strategy related to this process has recently become a focal point, but few studies have examined the effect of BM-MSCs on the autophagy of macrophages under OGD/R circumstances. In the present study, with stimulation of OGD/R-induced hypoxia in RAW264.7 cells, the LC3-II/LC3-I ratio was found to significantly increase, and p62 expression decreased, which was accompanied by the up-regulation of autophagy; however, co-culture with BM-MSCs significantly reduced the LC3-II/LC3-I ratio and increased p62 expression in RAW264.7 cells. The alterations in autophagy induced by IRI were similar to what was observed in previous studies.[40–42] In our recent work, pre-treatment with BM-MSCs down-regulated autophagy in AMs and alleviated the inflammatory response in IRI-induced ALI mice; therefore, the present research provided further evidence for an effective therapeutic strategy based on BM-MSCs in IRI-induced ALI.[14,15]
The PI3K/Akt signaling pathway is recognized as a typical pathway involved in the regulation of a wide range of cellular processes,[22,43,44] including nutrition, metabolism, proliferation, and apoptosis.[20,21] However, it remains unknown which signaling pathway underlying BM-MSCs regulates autophagy of macrophages under IRI conditions. In the present study, the autophagy-related PI3K/Akt signaling pathway as well as the regulatory role of BM-MSCs on autophagy was demonstrated. The results showed lower expression of PI3K and the p-Akt/Akt ratio in the OGD/R and OGD/R + HELF groups; with co-culture of BM-MSCs, there was significantly higher expression of PI3K and an increase in the p-Akt/Akt ratio in the OGD/R + BM-MSC group; with pre-treatment with LY294002, there was a significantly higher LC3-II/LC3-I ratio in both the OGD/R + LY294002 and OGD/R + BM-MSC + LY294002 groups compared to both the OGD/R and OGD/R + BM-MSC groups. These findings are interesting because, on the one hand, they confirmed the existence of the autophagy-related PI3K/Akt signaling pathway in RAW264.7 cells. On the other hand, BM-MSCs modulated autophagy in RAW264.7 cells via the PI3K/Akt signaling pathway. The results provide a further understanding of the mechanisms guiding autophagy regulation.
Moreover, through screening downstream molecules in the PI3K/Akt pathway with a whole-genome microarray assay, the DEG HO-1, which is a downstream molecule of the PI3K/Akt signaling pathway, was identified. Therefore, the existence of a PI3K/Akt/HO-1 signaling pathway was confirmed in RAW264.7 cells based on KEGG database analysis. The final results confirmed that the expression of mRNA and protein of HO-1 were also up-regulated, and the alteration of HO-1 was consistent with involvement in the PI3K/Akt pathway. This finding was interesting because as a downstream molecule of the PI3K/Akt pathway, HO-1 is involved in the modulation of cellular autophagy, and the up-regulation or overexpression of HO-1 might play a protective role following stimulation by hypoxia, oxidative stress, and IRI.[22–24] However, in the present study, some other DEGs related to autophagy, such as Mapk3 (extracellular regulated protein kinases 1/2, ERK1/2) and Bnip3/Bnip3l (Nip3-like protein X, Nix), were also identified, which might implicate the mitogen-activated protein kinase/ERK and hypoxia-inducible factor-1α/Bnip3/Beclin-1 pathways, respectively, so future investigations on different genes related to different signaling pathways are needed.
There were some limitations in this study, and further investigations are needed to achieve the following: (1) understanding the communication between BM-MSCs and RAW264.7 cells in a co-culture system; (2) identification of the role of HO-1 in the regulation of autophagy; and (3) identification of the regulatory effects of BM-MSCs on other possible pathways.
Overall, in this study, we verified that BM-MSCs modulate the autophagy of RAW264.7 cells via the PI3K/Akt pathway and downstream signaling molecule HO-1 under OGD/R circumstances in vitro. These findings provide evidence for BM-MSC therapy as a clinical option for IRI-induced lung injury.
Funding
The work was supported by a grant from the National Natural Science Foundation of China (No. 81490533).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Wang NF, Bai CX. Bone marrow-derived mesenchymal stem cells modulate autophagy in RAW264.7 macrophages via the phosphoinositide 3-kinase/protein kinase B/heme oxygenase-1 signaling pathway under oxygen-glucose deprivation/restoration conditions. Chin Med J 2021;134:699–707. doi: 10.1097/CM9.0000000000001133
Supplemental digital content is available for this article.
References
- 1.Su CF, Kao SJ, Chen HI. Acute respiratory distress syndrome and lung injury: pathogenetic mechanism and therapeutic implication. World J Crit Care Med 2012; 1:50–60. doi: 10.5492/wjccm.v1.i2.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubenfeld GD, Herridge MS. Epidemiology and outcomes of acute lung injury. Chest 2007; 131:554–562. doi: 10.1378/chest.06-1976. [DOI] [PubMed] [Google Scholar]
- 3.Matthay MA, Zemans RL. The acute respiratory distress syndrome: pathogenesis and treatment. Annu Rev Pathol 2011; 6:147–163. doi: 10.1146/annurev-pathol-011110-130158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol 2011; 21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009; 137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Cao H, Li J. Autophagy induced by DAMPs facilitates the inflammation response in lungs undergoing ischemia-reperfusion injury through promoting TRAF6 ubiquitination. Cell Death Differ 2017; 24:683–693. doi: 10.1038/cdd.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan T, Chen L, Huang Z. Autophagy decreases alveolar macrophage apoptosis by attenuating endoplasmic reticulum stress and oxidative stress. Oncotarget 2016; 7:87206–87218. doi: 10.18632/oncotarget.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding DC, Shyu WC, Lin SZ. Mesenchymal stem cells. Cell Transplant 2011; 20:5–14. doi: 10.3727/096368910X. [DOI] [PubMed] [Google Scholar]
- 9.Teixeira FG, Carvalho MM, Sousa N. Mesenchymal stem cells secretome: a new paradigm for central nervous system regeneration? Cell Mol Life Sci 2013; 70:3871–3882. doi: 10.1007/s00018-013-1290-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sueblinvong V, Loi R, Eisenhauer PL. Derivation of lung epithelium from human cord blood-derived mesenchymal stem cells. Am J Respir Crit Care Med 2008; 177:701–711. doi: 10.1164/rccm.200706-859OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merino-Gonzalez C, Zuniga FA, Escudero C. Mesenchymal stem cell-derived extracellular vesicles promote angiogenesis: potencial clinical application. Front Physiol 2016; 7:24.doi: 10.3389/fphys.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimczak A, Kozlowska U. Mesenchymal stromal cells and tissue-specific progenitor cells: their role in tissue homeostasis. Stem Cells Int 2016; 2016:4285215.doi: 10.1155/2016/4285215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong X, Wang P, Wu Q. Human umbilical cord blood derived mesenchymal stem cells improve cardiac function in cTnT(R141W) transgenic mouse of dilated cardiomyopathy. Eur J Cell Biol 2016; 95:57–67. doi: 10.1016/j.ejcb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Frausin S, Viventi S, Verga FL. Wharton's jelly derived mesenchymal stromal cells: biological properties, induction of neuronal phenotype and current applications in neurodegeneration research. Acta Histochem 2015; 117:329–338. doi: 10.1016/j.acthis.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Caseiro AR, Pereira T, Ivanova G. Neuromuscular regeneration: perspective on the application of mesenchymal stem cells and their secretion products. Stem Cells Int 2016; 2016:9756973.doi: 10.1155/2016/9756973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Zhang H, Zeng M. Bone marrow mesenchymal stem cells protect alveolar macrophages from lipopolysaccharide-induced apoptosis partially by inhibiting the Wnt/beta-catenin pathway. Cell Biol Int 2015; 39:192–200. doi: 10.1002/cbin.10359. [DOI] [PubMed] [Google Scholar]
- 17.Horie S, Laffey JG. Recent insights: mesenchymal stromal/stem cell therapy for acute respiratory distress syndrome. F1000Res 2016; 5: doi: 10.12688/f1000research.8217.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devaney J, Horie S, Masterson C. Human mesenchymal stromal cells decrease the severity of acute lung injury induced by E. coli in the rat. Thorax 2015; 70:625–635. doi: 10.1136/thoraxjnl-2015-206813. [DOI] [PubMed] [Google Scholar]
- 19.Niesler U, Palmer A, Radermacher P. Role of alveolar macrophages in the inflammatory response after trauma. Shock 2014; 42:3–10. doi: 10.1097/SHK.0000000000000167. [DOI] [PubMed] [Google Scholar]
- 20.Li GM, Liang CJ, Zhang DX, Zhang LJ, Wu JX, Xu YC. XB130 knockdown inhibits the proliferation, invasiveness, and metastasis of hepatocellular carcinoma cells and sensitizes them to TRAIL-induced apoptosis. Chin Med J 2018; 131:2320–2331. doi: 10.4103/0366-6999.241800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L, Liu Y, Wang M. Celastrus orbiculatus extract triggers apoptosis and autophagy via PI3K/Akt/mTOR inhibition in human colorectal cancer cells. Oncol Lett 2016; 12:3771–3778. doi: 10.3892/ol.2016.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y, Zhang Z, Li Q. Hyperbaric oxygen preconditioning protects lung against hyperoxic acute lung injury in rats via heme oxygenase-1 induction. Biochem Biophys Res Commun 2015; 456:549–554. doi: 10.1016/j.bbrc.2014.09.074. [DOI] [PubMed] [Google Scholar]
- 23.Jones AW, Durante W, Korthuis RJ. Heme oxygenase-1 deficiency leads to alteration of soluble guanylate cyclase redox regulation. J Pharmacol Exp Ther 2010; 335:85–91. doi: 10.1124/jpet.110.169755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang WZ, Jones AW, Wang M. Preconditioning with soluble guanylate cyclase activation prevents postischemic inflammation and reduces nitrate tolerance in heme oxygenase-1 knockout mice. Am J Physiol Heart Circ Physiol 2013; 305:H521–H532. doi: 10.1152/ajpheart.00810.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Ma Q, Li J. Hyperglycemia enhances the invasive and migratory activity of pancreatic cancer cells via hydrogen peroxide. Oncol Rep 2011; 25:1279–1287. doi: 10.3892/or.2011.1150. [DOI] [PubMed] [Google Scholar]
- 26.Kang JS, Choi IW, Han MH. The cytoprotective effects of 7,8-dihydroxyflavone against oxidative stress are mediated by the upregulation of Nrf2-dependent HO-1 expression through the activation of the PI3K/Akt and ERK pathways in C2C12 myoblasts. Int J Mol Med 2015; 36:501–510. doi: 10.3892/ijmm.2015.2256. [DOI] [PubMed] [Google Scholar]
- 27.Li W, Ma F, Zhang L. S-Propargyl-cysteine exerts a novel protective effect on methionine and choline deficient diet-induced fatty liver via Akt/Nrf2/HO-1 pathway. Oxid Med Cell Longev 2016; 2016:4690857.doi: 10.1155/2016/4690857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carchman EH, Rao J, Loughran PA. Heme oxygenase-1-mediated autophagy protects against hepatocyte cell death and hepatic injury from infection/sepsis in mice. Hepatology 2011; 53:2053–2062. doi: 10.1002/hep.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolisetty S, Traylor AM, Kim J. Heme oxygenase-1 inhibits renal tubular macroautophagy in acute kidney injury. J Am Soc Nephrol 2010; 21:1702–1712. doi: 10.1681/ASN.2010030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Zhou J, Zhang D. Bone marrow-derived mesenchymal stem cells enhance autophagy via PI3K/AKT signalling to reduce the severity of ischaemia/reperfusion-induced lung injury. J Cell Mol Med 2015; 19:2341–2351. doi: 10.1111/jcmm.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narasimhan P, Liu J, Song YS. VEGF stimulates the ERK 1/2 signaling pathway and apoptosis in cerebral endothelial cells after ischemic conditions. Stroke 2009; 40:1467–1473. doi: 10.1161/STROKEAHA.108.534644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogojeska J, Alexa A, Altmann A. Rtreemix: an R package for estimating evolutionary pathways and genetic progression scores. Bioinformatics 2008; 24:2391–2392. doi: 10.1093/bioinformatics/btn410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torto-Alalibo T, Purwantini E, Lomax J. Genetic resources for advanced biofuel production described with the Gene Ontology. Front Microbiol 2014; 5:528.doi: 10.3389/fmicb.2014.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y, Morishima K. Blast KOALA and ghost KOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 2016; 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Confalonieri M, Salton F, Fabiano F. Acute respiratory distress syndrome. Eur Respir Rev 2017; 26:160116.doi:10.1183/16000617.0116-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeki AA, Yeganeh B, Kenyon NJ. Autophagy in airway diseases: a new frontier in human asthma? Allergy 2016; 71:5–14. doi: 10.1111/all.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fetterman JL, Holbrook M, Flint N. Restoration of autophagy in endothelial cells from patients with diabetes mellitus improves nitric oxide signaling. Atherosclerosis 2016; 247:207–217. doi: 10.1016/j.atherosclerosis.2016.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eltzschig HK, Bratton DL, Colgan SP. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat Rev Drug Discov 2014; 13:852–869. doi: 10.1038/nrd4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Wang JS, Zheng ZK. Participation of autophagy in lung ischemia-reperfusion injury in vivo. J Surg Res 2013; 182:e79–e87. doi: 10.1016/j.jss.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Cao H, Li J. Autophagy induced by DAMPs facilitates the inflammation response in lungs undergoing ischemia-reperfusion injury through promoting TRAF6 ubiquitination. Cell Death Differ 2017; 424:683–693. doi: 10.1038/cdd.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu R, Chen ZF, Yan J. Complement C5a exacerbates acute lung injury induced through autophagy-mediated alveolar macrophage apoptosis. Cell Death Dis 2014; 5:e1330.doi: 10.1038/cddis.2014.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P, Guan YF, Du H. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy 2012; 8:77–87. doi: 10.4161/auto.8.1.18274. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Lopez N, Singh R. ATGs: scaffolds for MAPK/ERK signaling. Autophagy 2014; 10:535–537. doi: 10.4161/auto.27642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ou L, Lin S, Song B. The mechanisms of graphene-based materials-induced programmed cell death: a review of apoptosis, autophagy, and programmed necrosis. Int J Nanomedicine 2017; 12:6633–6646. doi: 10.2147/IJN.S140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


