SUMMARY
In mammalian cells, specialized DNA polymerase ζ (pol ζ) contributes to genomic stability during normal DNA replication. Disruption of the catalytic subunit Rev3l is toxic and results in constitutive chromosome damage· including micronuclei. As manifestations of this genomic stress are unknown· we examined the transcriptome of pol ζ-defective cells by RNA sequencing (RNA-seq). Expression of 1,117 transcripts is altered by > 4-fold in Rev3l-disrupted cells· with a pattern consistent with an induction of an innate immune response. Increased expression of interferon-stimulated genes at the mRNA and protein levels in pol ζ-defective cells is driven by the cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS)-signaling partner stimulator of interferon genes (STING) pathway. Expression of key interferon-stimulated chemokines is elevated in basal epithelial mouse skin cells with a disruption of Rev3l. These results indicate that the disruption of pol ζ may simultaneously increase sensitivity to genotoxins and potentially engage parts of the innate immune response· which could add an additional benefit to targeting pol ζ in cancer therapies.
In brief
Martin et al. show that disruption of the catalytic subunit of DNA polymerase ζ, Rev3l, results in the accumulation of DNA damage and increased expression of interferon-stimulated genes. The cGAS-STING pathway, part of the innate immune system that responds to genomic stress, drives this expression signature in pol ζ-deficient cells.
Graphical Abstract
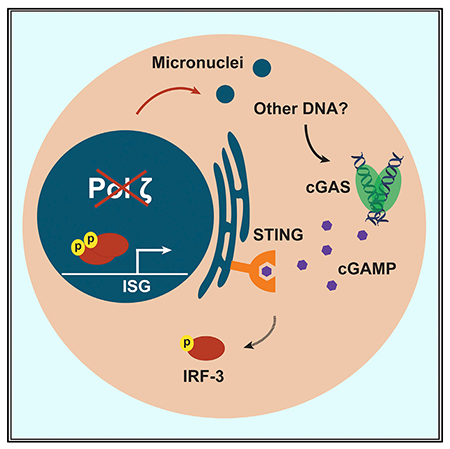
INTRODUCTION
Mammalian genomes encode an array of translesion DNA polymerases, which provide a diverse tool kit to tolerate assorted genomic lesions (Lange et al., 2011). While most translesion DNA polymerases are required for cells to survive various exogenous genotoxic assaults, they are not essential for mammalian development or unchallenged cellular survival (Lange et al., 2011). An exception is DNA polymerase ζ (pol ζ). The pol ζ catalytic subunit is encoded by the Rev3l gene (Martin and Wood, 2019). Germline disruption of Rev3l results in embryonic lethality in mice (Bemark et al., 2000; Esposito et al., 2000; Wittschieben et al., 2000). The indispensable nature of pol ζ reflects genome protective functions that are inadequately understood. Disruption of Rev3l in B cells or keratinocytes in mice leads to acute genomic stress in the target tissues (Daly et al., 2012; Lange et al., 2013, 2018; Schenten et al., 2009; Wittschieben et al., 2010). Primary mouse embryonic fibroblasts (MEFs) rapidly accumulate chromosome breaks at the first metaphase following Rev3l disruption (Lange et al., 2012). In addition, chromosome rearrangements are observed in Rev3l-deficient MEFs in a p53 null background (Wittschieben et al., 2006). Loss of pol ζ in primary MEFs cripples cell proliferation, with the cells failing to replicate past roughly two rounds of cell division following Rev3l disruption (Lange et al., 2012). If cells can survive Rev3l disruption, this promotes spontaneous tumorigenesis in mouse models (Lange et al., 2013; Wittschieben et al., 2010). Given that the loss of pol ζ function results in chromosome breaks and structural abnormalities (Lange et al., 2012; Wittschieben et al., 2006), this could generate genetic variety to promote oncogenic growth, but only if the cells can overcome the disruption of pol ζ. This makes it of great interest to understand the cellular consequences of the loss of Rev3l.
The dramatic growth suppression in Rev3l knockout MEFs appears to be ameliorated by blunting DNA damage quality control responses, for example, by p53 deletion (Wittschieben et al., 2006) or large T-antigen expression (Lange et al., 2012) (which inhibits p53 and other targets). However, p53 deletion utterly fails to rescue the embryonic lethality of Rev3l disruption in mice (O-Wang et al., 2002; Van Sloun et al., 2002; Wittschieben et al., 2006). This implies that there are additional, p53-independent, growth-suppressive responses to the genomic strain caused by Rev3l disruption. We set out to clarify the consequences of the sustained genomic stress induced by the loss of pol ζ in mammalian cells. Starting with an unbiased transcriptome-wide approach, we discovered that the loss of pol ζ induces the constitutive expression of immune system-related genes, in particular, interferon-stimulated genes (ISGs). Furthermore, we found that the cytosolic nucleic acid sensor cyclic guanosine monophosphate-AMP synthase (cGAS) and its downstream signaling partner stimulator of interferon genes (STING) drive this response in absence of functional pol ζ. Given that the cGAS-STING axis can inhibit cell growth, this provides a further potential explanation for why the loss of pol ζ function is remarkably toxic to proliferating cells.
RESULTS AND DISCUSSION
A shortened REV3L construct rescues known phenotypes of pol ζ disruption
To dissect the consequences of the poorly resolved genome protection function of pol ζ, we set up a complementation system using large T-antigen immortalized MEFs (Lange et al., 2012), either with a pol ζ-proficient background, Rev3l heterozygous (HET), or a pol ζ deficient background, Rev3l knockout (KO). The KO cell lines (Rev3l−/Δ) were generated by adenovirus-Cre disruption of a single allele in Rev3l−/lox MEFs (Lange et al., 2012). HETs (Rev3l+/D), which retain pol ζ activity, provides a control for this process since they also had deletion of a single Rev3l allele using adenovirus-Cre with Rev3l+/lox MEFs as the parental cell line. We have not observed phenotypic differences between Rev3l HETs and WT cells or mice (Lange et al., 2012).
A biochemically active shortened human REV3L construct, TR4-2 (Lee et al., 2014) (Figure 1A), with an N-terminal FLAG-hemagglutinin (HA) tag, was stably expressed in Rev3l KO MEFs (Figures 1B and S1A). To test that our complementation system was in fact addressing both external and endogenous genome-protective functions of pol ζ, we tested both cellular sensitivity to cisplatin and the formation of micronuclei in unchallenged cells. Micronuclei are a marker of genomic instability and arise when a whole chromosome or fragment of a chromosome missegregates and forms its own discrete nuclear compartment. DNA breaks that result in chromosome fragmentation can lead to acentric chromosomes that form micronuclei (Fenech et al., 2011).
Figure 1. Shortened REV3L construct rescues phenotypes of pol ζ disruption.
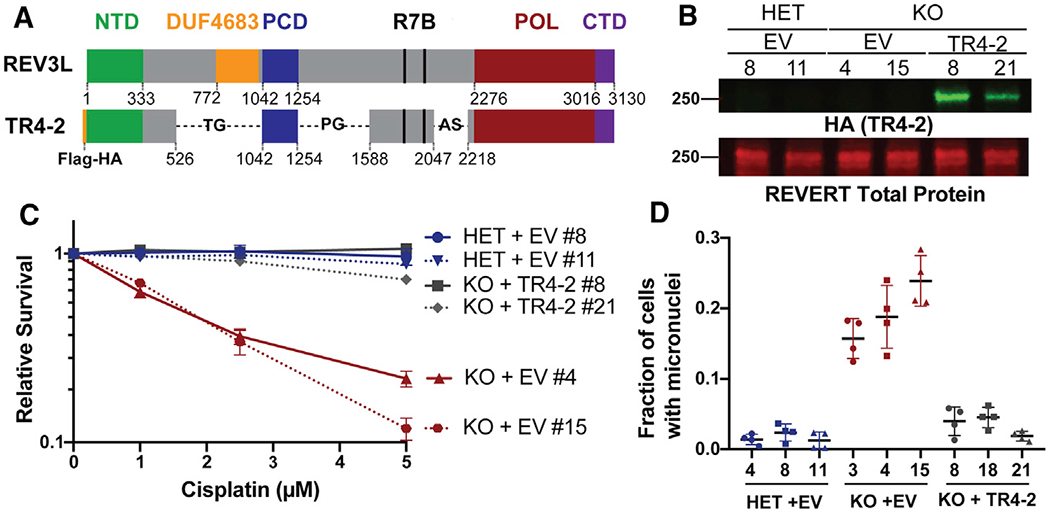
(A) Schematic of full-length human REV3L and human REV3L construct TR4-2. TR4-2 retains most conserved domains and binding sites of REV3L, including the regions that coordinate interactions with the accessory subunits of pol ζ, the C-terminal domain (CTD, purple), the 2 REV7 binding sites (R7B, black), and a positively charged domain (PCD) of uncertain function. The B-family catalytic core is formed by folding of the N-terminal domain (NTD, green) and the polymerase domain (POL, red) (Martin and Wood, 2019).
(B) Immunoblot with HA antibody showing stable expression of TR4-2 with an N-terminal FLAG-HA tag in Rev3l KO MEF clones.
(C) Stable expression of TR4-2 in Rev3l KO MEF clones reverses hypersensitivity to cisplatin. MEFs were exposed to the indicated cisplatin concentrations for 48 h, and relative survival was quantified with the ATPlite assay. The error bars represent the standard deviation of 3 replicates.
(D) Stable expression of TR4-2 decreases micronuclei formation in unchallenged Rev3l KO MEF clones. The error bars represent the standard deviation of four replicates. See also Figure S1.
As expected, Rev3l KO + empty vector (EV) MEFs were hypersensitive to cisplatin relative to the Rev3l HET MEFs + EV (Figures 1C and S1B). Stable expression of TR4–2 in Rev3l KO MEFs reversed the hypersensitivity to cisplatin (Figures 1C and S1B). This indicates that in this context, TR4–2 can restore the function of REV3L in DNA damage tolerance. Consistent with large-scale genomic stress, ~15%-23% of unchallenged Rev3l KO MEFs had at least 1 micronucleus, relative to Rev3l HET MEFs, which had a micronucleus frequency of ~1%-3% (Figure 1D). Stable TR4–2 expression in Rev3l KO MEFs restored micronucleus frequency to near normal, showing that TR4–2 can restore some Rev3l genome-protective functions. We used this isogenic system as a tool to probe the unknown consequence of the loss of pol ζ.
Loss of polymerase ζ alters the transcriptome
To uncover the type of stress occurring in cells lacking pol ζ, we performed genome-wide mRNA sequencing on a controlled set of immortalized clones: Rev3l HET + EV, Rev3l KO + EV, and Rev3l KO + TR4-2. To focus on major changes, we set a strict threshold (>|2| log2 fold change and false discovery rate [FDR] < 0.05). Expression analysis of 17,346 mapped transcripts revealed that 1,117 transcripts were either upregulated or downregulated in the Rev3l KO + EV relative to the Rev3l HET + EV MEFs (Figure 2A). The majority (~68%) were upregulated (Figure 2A). These upregulated or downregulated genes displayed no statistically significant enrichment or depletion for DNA replication or canonical DNA damage-sensing pathways. This is not completely unexpected, given that p53 promotes much of the transcriptional response to DNA damage, and our MEFs have inactivated p53 due to large T-antigen immortalization. In these immortalized high-passage cells, we are likely to observe stable transcriptional alterations, rather than an acute response.
Figure 2. Cells deficient in DNA polymerase ζ have an altered transcriptome.
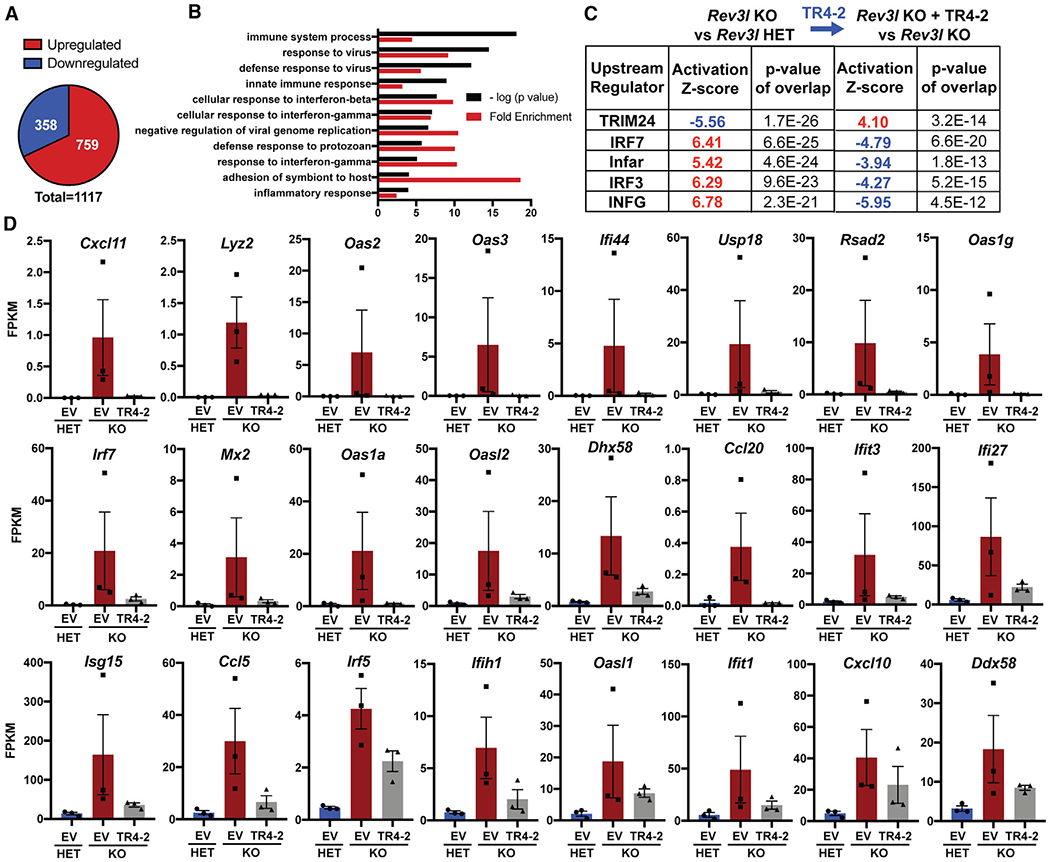
(A) Differentially expressed genes in Rev3l KO + empty vector (EV) relative to Rev3l HET + EV using the threshold of a fold change >4 and a false discovery rate of <0.05.
(B) Top 10 GO (Gene Ontology) terms reveal enrichment of immune system-related genes in upregulated genes in Rev3l KO MEFs.
(C) Upstream regulator analysis reveals the dataset is consistent with the predicted activation of positive regulators of an interferon response in Rev3l KO MEFs.
(D) Expression of individual interferon-stimulated genes (ISGs) is elevated in Rev3l KO MEFs and is suppressed by TR4-2 expression. The fragments per kilobase of transcript per million mapped reads (FPKM) of 3 clones for each condition, HET + EV (ID nos. 4, 8, 11), KO + EV (ID nos. 3, 4, 15), and KO +TR4-2 (ID nos. 8, 18, 21) is graphed. The error bars represent SEMs. See also Figure S2 and Table S1.
Instead, the upregulated genes displayed an enrichment in immune system-related pathways, as revealed by Gene Ontology (GO) analysis (Figure 2B). Upstream regulator analysis was used to analyze all differentially expressed genes. This revealed that the alterations in the transcriptome are consistent with the activation of positive regulators of the interferon response, including key transcription factors in this pathway, IRF3 and IRF7 (Figure 2C). Importantly, the predicted activation of all five of the identified upstream regulators was reversed by expression of the Rev3l TR4–2 cDNA (Figure 2C). This shows that the changes in differential gene expression stem from a function of Rev3l. The same trends were also observed by applying a substantially lower threshold (log2 FC > |0.5|) for differentially expressed genes. This increased the dataset to 2,071 differentially expressed transcripts. Pathway analysis showed a negative correlation with predicted TRIM24 activation. TRIM24 suppresses ISG expression (Tisserand et al., 2011), confirming that our data are consistent with the expression of ISGs.
To explore whether our dataset is in fact consistent with an interferon-like response, we analyzed a curated set of 24 known ISGs and observed increased expression in the Rev3l KO relative to the Rev3l HET MEFs, which was largely reduced by the stable expression of the Rev3l cDNA TR4-2 (Figure 2D). It is notable that while the expression of TR4-2 conferred nearly complete rescue of cisplatin sensitivity (Figure 1C) and micronuclei formation (Figure 1D), the expression of some individual ISGs was only partially reversed (Figures 2D and S2; Table S1). It is possible that the expression of some genes in the MEF clones may have been stabilized by epigenetic changes, not reversible by the reintroduction of Rev3l. It is also possible that segments of REV3L not present in TR4-2 (e.g., the DUF4683 domain) (Figure 1A) may contribute to a specific gene expression function of REV3L but not to the DNA damage bypass function. These results reveal that the disruption of pol ζ promotes the induction of ISGs.
The cGAS-STING pathway drives the expression of ISG expression in Rev3l-disrupted cells
Given that our complemented cell lines were generated using lentivirus constructs and grown continually under selection, we moved to validate the results in the parental Rev3l KO and Rev3l HET MEF cell lines and 1 additional set of cell lines to limit extraneous variables. We confirmed an increase in the mRNA expression of specific ISGs in the Rev3l KO MEFs relative to the control cell lines, including key chemokines (Figure 3A). To extend these findings to the protein level, we examined ISG products by immunoblotting. Corresponding to an increase in mRNA levels, we also observed an increase in the protein levels of known ISGs MDA5 (encoded by the Ifih1 gene), ISG15, and viperin (encoded by the Rsad2 gene) (Figure 3B).
Figure 3. Disruption of Rev3l results in increased expression of ISGs and proteins.
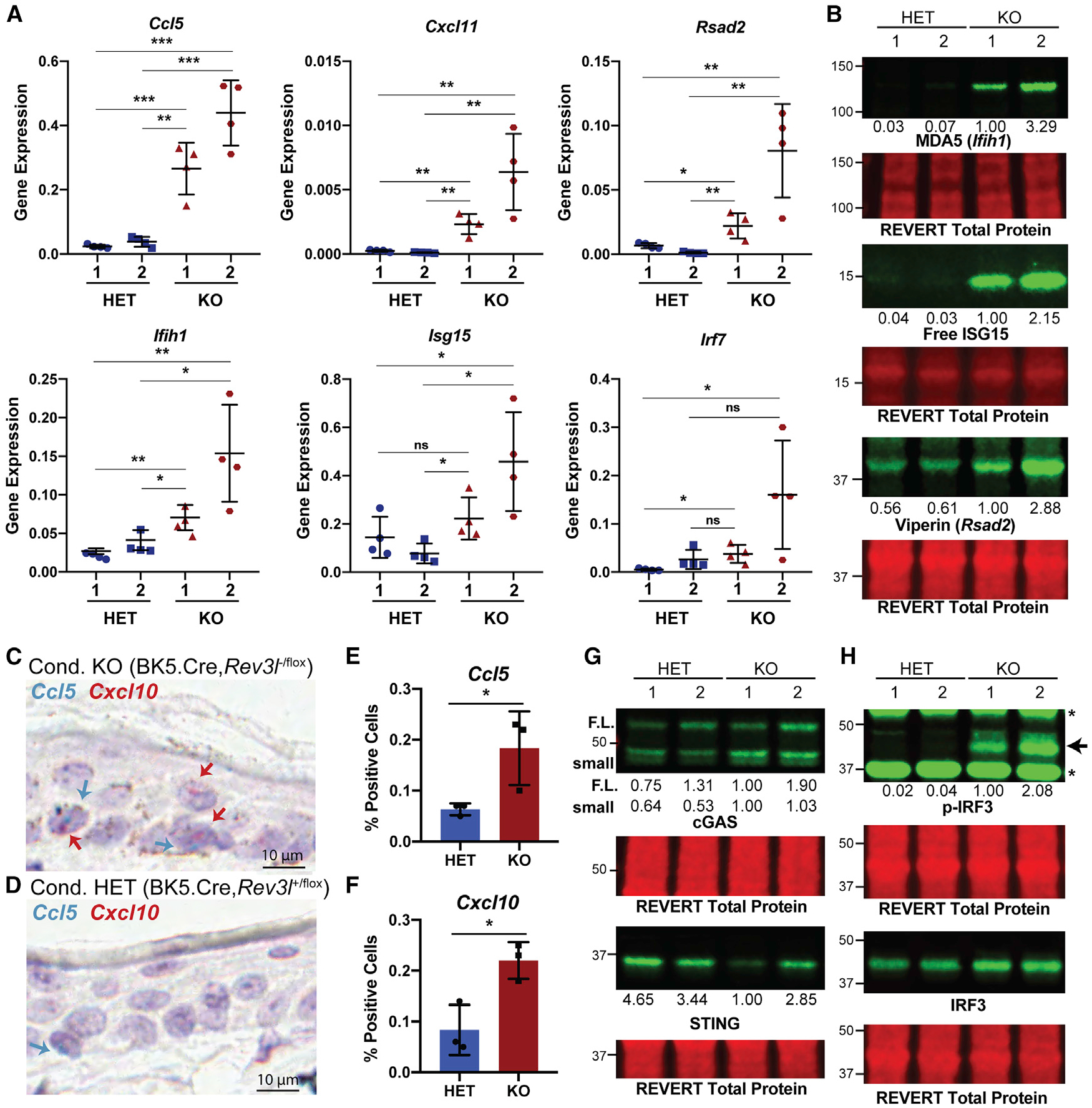
(A) Gene expression (2−ΔCt) of selected ISGs normalized to hypoxanthine phosphoribosyltransferase (HPRT) detected by qRT-PCR. Error bars represent standard deviation. Unpaired 2-tailed Student’s t test, *p < 0.05, **p < 0.01, and ***p < 0.001.
(B) Immunoblots showing increased protein levels of ISGs MDA5, ISG15, and viperin. The quantification of each protein relative to KO 1 is shown underneath the blot. The signals were first normalized to total protein.
(C) Representative image of CCL5 (blue) and CXCL10 (red) signals in the epithelia of conditional Rev3l KO mice (BK5.Cre,Rev3l−/flox) positive cells are shown with blue and red arrows, respectively. A 10 μm scale bar is shown. The brown staining is epithelial melanin present in the KO model.
(D) Same as (C), except for conditional Rev3l HET mice (BK5.Cre,Rev3l+/flox). A 10 μm scale bar is shown.
(E) Quantification for percentage of cells positive for CCL5 in the epithelia for 3 mice of each genotype.
(F) Same as (E), except for CXCL 10.
(G) Immunoblot showing the presence of components of the innate immune system, cGAS and STING, in MEFs, with reduced STING in pol ζ KO cells. Quantification of the signal intensity of cGAS and STING relative to KO 1 is displayed underneath the blot. The signal intensity of each band was first normalized to total protein. For cGAS, we detect 2 bands: 1 at full-length molecular weight (F.L.) and 1 (small) that we confirm is another cGAS species in Figure 4C.
(H) Enhanced phosphorylation of S888 of IRF3 (corresponding to S396 in humans) in Rev3l KO MEFs. The relative quantification of p-IRF3 signal intensity normalized to IRF3 is displayed underneath the p-IRF3 blot. The signal intensities were first normalized to total protein signal. See also Figure S3.
These results were obtained by analysis of isogenically matched pairs of Rev3l-proficient and Rev3l-deficient immortalized cell lines. An important question is whether the disruption of REV3L function drives the induction of ISGs in primary cells. This is experimentally challenging, because complete disruption in primary cells rapidly promotes senescence in primary MEFs (Lange et al., 2012). We instead took advantage of a mouse model system in which Rev3l is specifically disrupted only in keratin-5-expressing cells, principally the epithelial cells of the skin and hair follicles. These mice survive, but they have a constitutively lower cell density in the basal epithelium (Lange et al., 2013). The skin is extremely sensitive to physical insults, including UV irradiation and wounding (Lange et al., 2013, 2018). Rev3l-defective epithelial cells struggle with re-proliferation because of ongoing DNA replication stress, which accounts for the fragility of the epithelia (Lange et al., 2013, 2018).
We hypothesized that the expression of ISGs in the immortalized MEFs may depend on DNA damage incurred during replication. Thus, we examined primary cells arising from proliferation in the skin. In normal mice, stem cells in the hair follicles undergo division and migrate to continuously repopulate the epithelium. Using skin sections from mice previously reported (Lange et al., 2018), we tested the expression of the mRNA of CXCL10 and CCL5 by in situ hybridization (Figures 3C–3F and S3). We found that with 3 independent mice in Rev3l-defective cells, CXCL10 and CCL5 levels were significantly upregulated by 2.6-fold and 2.9-fold, respectively (Figures 3E and 3F).
These data indicate that an interferon-like branch of the innate immune system may be activated due to the disruption of pol ζ function. Since it seems unlikely that pol ζ plays a direct role in transcriptional regulation, the next obvious question is how and why loss of pol ζ induces the expression of ISGs.
The major consequence of pol ζ disruption in unchallenged mammalian cells is increased genomic instability as evidenced by multiple markers, including chromosome fragmentation and aberrations and micronuclei (Lange et al., 2012, 2013, 2016; Wittschieben et al., 2006) (Figure 1D). Therefore, it seems likely that this transcriptional response ultimately stems from the vast genomic damage induced by the loss of pol ζ function. Consistent with this hypothesis, the innate immune system can respond to endogenous DNA that has escaped from the nucleus, in addition to its more canonical role in recognizing and mounting an interferon response to foreign DNA. In some instances, this response can halt cell growth to shut down propagations of virally infected cells and cells with dangerously fragmented genomes.
Mammalian cells have a host of cytosolic nucleic acid sensors that patrol the cytosol for DNA. One of these, cGAS, is increasingly recognized to be of paramount importance in the induction of an interferon response to both exogenous and endogenous cytosolic DNA (Ablasser and Chen, 2019). Binding of double-stranded DNA to cGAS activates the enzymatic activity of cGAS and leads to the production of 2′,3′ cyclic GMP-AMP (cGAMP). cGAMP binds to the STING receptor on the membrane of the endoplasmic reticulum, resulting in the activation of kinases, including TBK1, which can in turn phosphorylate and activate interferon regulatory factor 3 (IRF3), a central transcription factor in the interferon response.
Given that the cGAS-STING axis has been implicated specifically in responding to endogenous DNA damage and has been correlated with micronuclei formation (a potential source for cGAS activation), we asked whether cGAS and STING promote the induction of expression of ISGs due to the loss of function of pol ζ. Consistent with most MEFs having a functional innate immune system, both Rev3l KO and HET MEF cell lines expressed both cGAS and STING (Figure 3G). In the Rev3l KO cells, the steady-state levels of STING (but not cGAS) are somewhat lower (decreased by ~3- to 4-fold for KO 1 and decreased by ~20%– 60% in KO 2). The decreased STING protein level is consistent with a constitutive activation of the cGAS-STING pathway, as cGAS activation leads to a negative feedback loop resulting in STING degradation (Gonugunta et al., 2017; Prabakaran et al., 2018). Importantly, we detected an increase in IRF3 phosphorylated at S888 (corresponding to S396 in humans), indicative of IRF3 activation in Rev3l KO MEFs (Panne et al., 2007; Yoneyama et al., 2002) (Figure 3F).
This led us to investigate whether cGAS-STING drives the expression of ISGs in cells with disrupted pol ζ. Knockdown of either cGAS or STING significantly reduces the mRNA expression of selected ISGs and dramatically decreases corresponding protein levels (Figures 4A–4D). In addition, the depletion of cGAS or STING in Rev3l KO MEFs markedly reduced S888 phosphorylation of IRF3 (Figure 4E). This indicates that the disruption of pol ζ function promotes the upregulation of ISGs driven by the cGAS-STING pathway.
Figure 4. The cGAS-STING axis promotes expression of ISGs due to loss of pol ζ function.
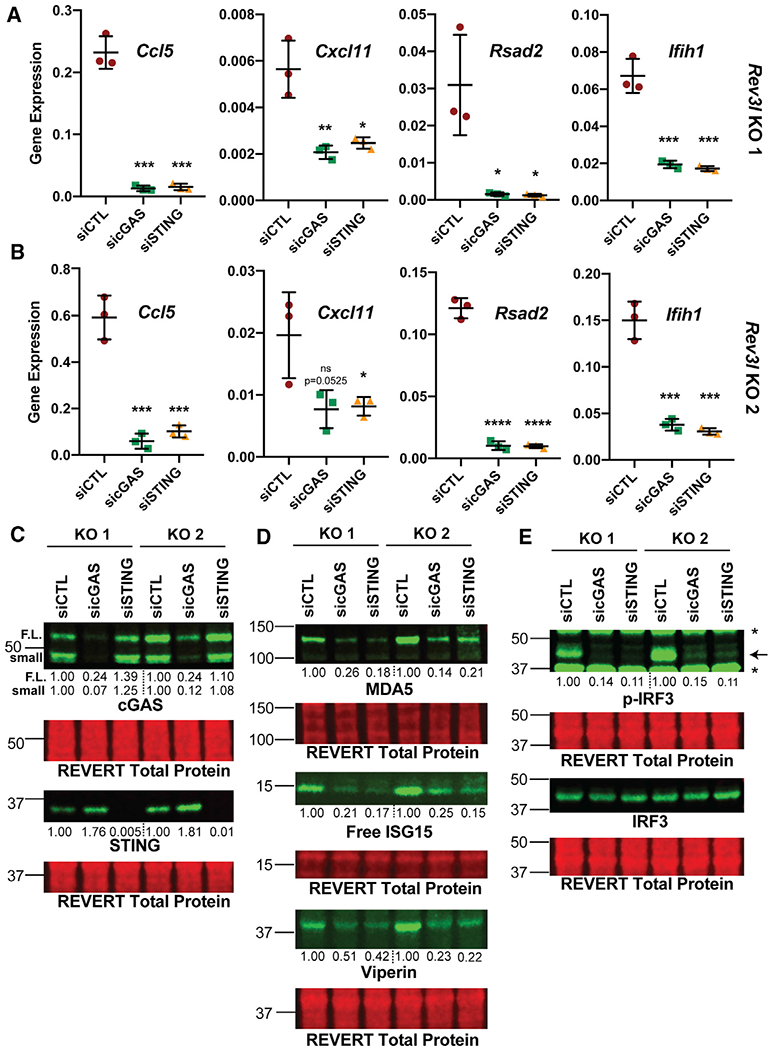
(A) Knockdown of cGAS or STING reduces mRNA expression of CCL5, CXCL11, RSAD2 (which encodes viperin), and IFIH1 (which encodes MDA5) as detected by qRT-PCR in Rev3l KO 1. Gene expression (2−ΔCt) of selected ISGs normalized to HPRT detected by qRT-PCR. Error bars represent standard deviation. Unpaired 2-tailed Student’s t test, *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
(B) Same as in (A), except with Rev3l KO 2.
(C) Efficient knockdown of cGAS or STING protein levels. The relative quantification of the full-length cGAS (F.L.), a cGAS smaller species (small), and STING are displayed underneath each blot. The signal intensities were first normalized to total protein.
(D) MDA5, ISG15, and viperin protein levels decrease with cGAS and STING knockdown. The quantification of each protein relative to negative control dicer-substrate short interfering RNA duplex (siCTL) is shown underneath the blot. The signal intensities were first normalized to total protein signal.
(E) Phosphorylation of S888 in mouse (analogous to the human S396) of IRF3 in Rev3l KO MEFs decreases with knockdown of cGAS and STING. The relative quantification of p-IRF3 signal intensity-normalized IRF3 is displayed underneath the p-IRF3 blot. The signal intensities were first normalized to total protein signal.
Pol ζ stands apart from the other translesion polymerases in that it is required for mammalian development and proliferation of primary cells. Now we can say that in addition to activating p53-dependent responses, disruption of pol ζ function promotes the activation of the cGAS-STING pathway.
It is remarkable that the disruption of an enzyme commonly thought of as a specialized translesion synthesis polymerase can lead to the constitutive activation of the cGAS-STING pathway. Recently, cells with a loss of function of key DNA repair enzymes RNaseH2, BRCA2, and BLM have been shown to have an elevated cGAS-STING response that correlates with an increase in micronuclei (Gratia et al., 2019; Mackenzie et al., 2016, 2017; Reisländer et al., 2019). There are several sources of DNA damage that may give rise to a sustained response, including cytosolic mitochondrial DNA (West et al., 2015) and cytosolic DNA arising from stalled and processed replication forks (Coquel et al., 2018). DNA stress may continually arise from DNA replication fork collapse in the absence of pol ζ, which could promote the formation of micronuclei and also release small fragments of DNA. Furthermore, some nuclear genes, including pol ζ, control mitochondrial DNA integrity. There is evidence that mitochondrial function is compromised without pol ζ (Singh et al., 2015). It remains to be seen whether micronuclei are the primary instigator of cGAS activation in cells lacking pol ζ or whether they are more of an indicator of DNA degradation.
Our experiments were performed with MEFs immortalized by large T-antigen, which allowed cellular survival in the absence of pol ζ. In addition to blunting p53 activity and inactivating Rb, large T-antigen has been implicated in impairing an interferon response to nucleic acids in primary MEFs and suppressing cGAS-STING activation in human 293 cells through an unknown mechanism or mechanisms (Lau etal., 2015; Reus et al., 2020). Which functions of the large T-antigen enable cells to tolerate Rev3l deletion is an interesting but unexplored question. It could involve the poorly understood role of large T-antigen in inhibiting an interferon response or be tied to the blockage of p53 and Rb functions, or both.
An interferon response can result in shutting down cell growth. Specifically, cGAS has been tied to promoting senescence in primary cells (Glück et al., 2017; Yang et al., 2017). Primary MEFs lacking pol ζ only make it approximately two cell divisions before cell growth completely halts, which is accompanied by an increase in senescent cells (Lange et al., 2012). Further demonstrating the essential nature of pol ζ, Rev3l-defective primary keratinocytes are unable to proliferate in a mouse model, and germline disruption of Rev3l blocks embryo development (Lange et al., 2012, 2013; Martin and Wood, 2019). It is possible that cGAS-STING drives this severe growth arrest in primary cells due to the loss of pol ζ function.
In addition to widening our understanding of the lengths that cells go to protect themselves from the genomic damage induced by the impairment of pol ζ, these results could have an impact on translational approaches. A potential approach, suggested by experiments in laboratory settings, has been to disrupt pol ζ function to enhance chemotherapeutic effectiveness (Berdis, 2008; Sail et al., 2017; Xu et al., 2013). An inhibitor that impairs the interaction of pol ζ with its regulator REV1 has been developed that sensitizes cancer cells and xenograft tumors to cisplatin treatment (Wojtaszek et al., 2019). This approach would have multiple advantages for therapy by enhancing DNA damage sensitivity and eliminating pol ζ-dependent point mutations during lesion bypass. Our work suggests that such inhibitors may also induce a cGAS-STING response due to the increased chromosomal instability. Promoting the activation of the cGAS-STING pathway as a way to engage the immune system is being widely explored as a new tool in cancer therapy (Hoong et al., 2020). Given our results, it would be exciting to investigate whether pol ζ disruption could also engage the innate immune system in a way that could be exploited for cancer treatment.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Additional information and requests for resources and reagents can be directed to and fulfilled by Lead Contact, Dr. Richard Wood (rwood@mdanderson.org).
Materials availability
This study did not generate unique reagents.
Data and code availability
The datasets produced in this study have been uploaded to Gene Expression Omnibus. Accession number for the RNA sequencing data is GEO: GSE163313. The full immunoblot images in Figures 1,3,4, and S1 have been uploaded to Mendeley Data: https://dx.doi.org/10.17632/5348dspthz.1
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The immortalized Rev3l knockout (KO) and heterozygous (HET) mouse embryonic fibroblast (MEF) parental cell lines in this study were as described (Lange et al., 2012). In brief, the Rev3l KO MEFs were generated by large T-antigen immortalization of primary MEFs isolated from mouse embryos with one null Rev3l and one floxed allele, followed by ex vivo Ad-Cre deletion of the floxed allele, and clonal selection to ensure a homogeneous population with the genotype Rev3l ID. The Rev3l HET MEFs were isolated in the same manner expect starting MEFs from embryos with one wild-type and one floxed allele, resulting in MEFs with the genotype Rev3l+/D. The floxed allele was generated by placing loxP sites flanking two conserved exons, corresponding to residues 2776–2860, that contain the three conserved catalytic aspartate residues of REV3L (Wittschieben et al., 2010). The null allele replaced these two exons with a lacZ-neoR cassette (Wittschieben et al., 2000). In this study two sets of clones were analyzed. Cells were grown in DMEM (Sigma #5796), 10% fetal bovine serum (FBS) and 1 X penicillin/streptomycin (GIBCO #15140122). Cell lines were genotyped for Rev3l allele status as described (Lange et al., 2012), and karyotyping confirmed the presence of mouse chromosomes only. The RNA sequencing data show that the Rev3l+/A MEF cell line expresses characteristic Y chromosome genes (Kdm5d, Uty, Eif2s3y, Ddx3y) indicating that it was derived from a male embryo (Mizukami et al., 2019). The Rev3l ID MEF does not express these genes but expresses Xist, indicating derivation from a female embryo. All cell lines were negative for mycoplasma infection as shown by regular checks with a MycoAlert Mycoplasma Detection Kit (Lonza Bioscience).
METHOD DETAILS
Generation of TR4-2 expressing Rev3l KO MEFs
TheTR4–2 construct, a gift from Dr. Wei Yang (Lee et al., 2014), was cloned into the pCDH backbone with an N-terminal Flag-HAtag as the same previously described for the full length Rev3l cDNA (Tomida et al., 2015). The pCDH-Flag-HA-TR4–2 or pCDH-Flag-HA empty vector was stably inserted into the MEF cell lines using lentiviral infection as previously described (Lange et al., 2016). Three single clones were isolated for analysis and continually grown in 1 mg/mL of puromycin to ensure stable integration of the construct.
RNA isolation
RNA was isolated from 1.5 x 106 (Figure 2, RNA seq) or 2.5 x 105 cells (Figure 3, gene expression) or 1 x 106 cells (Figure 4, gene expression), using the RNeasy Kit (QIAGEN # 74104) following the manufacturer’s instructions including the on-column DNase I digestion (QIAGEN #79254).
Genome-wide mRNA sequencing
mRNA libraries were prepared using the Illumina TruSeq Stranded mRNA kit following manufacturer’s instructions and 75 base paired end sequencing was run on the Illumina HiSeq 3000. Three biological replicates were prepared for each condition. 41-46 million pairs of reads were generated per sample. Each pair of reads represents a cDNA fragment from the library.
Sequence mapping
The reads were mapped to the mouse genome mm10 by TopHat (version 2.0.10) (Kim et al., 2013). By reads, the overall mapping rate is 96.0%-97.1%. 93.9%-95.3% fragments have both ends mapped to the mouse genome.
Identifying differential expression
GENCODE Release M19 (Mudge and Harrow, 2015) using htseq-count from HTSeq package (version 0.6.0) (Anders et al., 2015) was used to determine the number of fragments in identified genes. A normalized estimate of gene expression was calculated as fragments per kilobase of exon model per million reads mapped (FPKM). If genes had less than 10 fragments in all the samples, they were excluded from differential expression analysis. R/Bioconductor package edgeR (version 3.8.6) (Robinson et al., 2010) was used to statistically assess differentially expressed genes. Genes with FDR (false discovery rate) < 0.05, fold change > |0.5| and length > 200 bp were called as differentially expressed. More stringent cut-offs for fold change were used for various analyses as described below.
Analysis of differentially expressed genes
For gene ontology analysis, genes upregulated more than 4-fold with an FDR < 0.05 were entered into DAVID 6.8 on 02/22/20 and the top 10 GOTERMS_BP_Direct were plotted based on −log(p value) (Huang etal., 2009a, 2009b). For upstream regulator analysis, differential expressed genes both upregulated and downregulated more than 4-fold with an FDR < 0.05. were entered into IPA (QIAGEN Inc., https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/). Given that upregulated genes are overrepresented in the Rev3l KO differentially expressed genes, this results in an expected bias for upstream regulators that also predominately result in the upregulation of targeted genes which we see in our dataset. Since, bias-corrected z-scores are not reported for upstream regulators with a |bias term| > 0.5, we’ve reported the uncorrected z-score here. Given that the majority of differentially expressed genes are upregulated in our dataset, bias is to be expected for transcription regulators that primarily induces expression of genes.
Gene expression analysis by quantitative PCR
High Capacity cDNA Reverse Transcription (Applied Biosciences #4368814) was used to prepare cDNA from 1000 ng of total RNA from each sample. qPCR was run iTaq Universal SYBR Green Supermix (Biorad #1725121) on the Applied Biosystems 7900HT Fast Real-Time PCR System. Gene expression of calculated using the 2▵DCt method normalizing to the Hprt gene. See STAR methods table for the primers used for mouse Hprt and target genes (Mackenzie et al., 2016; West et al., 2015; Yang et al., 2007).
Immunoblotting
Cells were lysed (3 million cells/100 mL) in lysis buffer (Tris-HCl: 50 mM, pH 7.5, NaCl 250 mM, EDTA 1 mM, Triton X-100 0.1%, 1 X Protease/Phosphatase Inhibitor Cocktail CST #5872) for 30 min on ice with mixing every 10 min. Debris was pelleted by centrifugation at 15,000 x g for 20 min at 4°C. Protein amounts were quantified using Biorad Protein Assay (Biorad #500-0006) and a bovine serum albumin standard curve (Biorad #500-0007) following manufacturer’s instructions. The samples were denatured using 4 x loading buffer (LI-COR #928-40004). 25 μg of protein / well and Precision Plus Protein All Blue Standards (Biorad #161-0373) were run on 4%-20% polyacrylamide gels (Biorad #4561096) in 1 x Tris/Glycine/SDS buffer (Biorad #161-0772). Protein was transferred to Immobilon-FL PVDF Membrane (Millipore #IPFL00010) in 1 x Tris/Glycine buffer (Biorad #161-0772), 20% methanol. After transfer, membranes were dried. Total protein was measured using REVERT total protein stain kit (LI-COR #926-11010) following the manufacturer’s instructions. Membranes were blocked for 1 h in 0.5X Odyssey Blocking Buffer(OBB, LI-COR #927-50000) in Tris Buffered Saline (TBS) and then incubated in primary antibody overnight at 4°C. The primary antibodies were used at the following dilutions in 0.5XOBB/TBS/0.2% Tween-20. Rabbit anti-HA-Tag (C29F4) (CST #3724,1:1000), Rabbit anti-cGAS (Mouse specific) (D3O8O) (CST #31659, 1:1000), rabbit anti-STING (D2P2F) (CST # 13647, 1:1000), rabbit anti-MDA-5 (D74E4) (CST # 5321, 1:1000), rabbit anti-ISG15 (CST # 2743, 1:500), rabbit anti-Phospho-IRF-3 (Ser396) (4D4G) (CST #4947, 1:1000), rabbit anti-IRF-3 (D83B9) (CST #4302, 1:1000), mouse anti-viperin (Millipore #MAB106, 1:250), rabbit anti-STAT1 (D1K9Y) (CST: #14994, 1:1,000), rabbit anti-TBK1 (D1B4) (CST: #3504, 1:1,000), and rabbit anti-Phospho-TBK1 (S172) (D52C2) (CST: #5483, 1:1,000).
After primary incubation membranes were washed three times in TBS/0.1% Tween-20, and incubated for 1 h in secondary antibody either goat anti-Rabbit 800CW (LI-COR #926-32211) or goat anti-mouse 800CW (Li-Cor #827-08364) diluted 1:20,000 in 0.5X OBB /TBS /0.2% Tween-20/0.01% SDS. After primary incubation, membranes were washed three times in TBS/0.1% Tween-20, rinsed with TBS, then dried and imaged on the LI-COR Odyssey FC.
Knockdown of cGAS and STING protein levels
600.0 cells were seeded into 10 cm plates. The next day, cells were transfected with 1 nM of the appropriated siRNA using Lipofectamine RNAiMAX (ThermoFisher #13778150) following the manufacturer’s protocols. The following dicer-substrate short interfering RNAs were used: siCTL (IDT: Negative Control DsiRNA # 51-01-14-03), sicGAS (IDT: DsiRNA Duplex mm.Ri.Mb21d1.13.1), and siSTING (IDT: DsiRNA Duplex mmRi.Tmem173.13.2). After 48 h, cells were harvested in paired pellets for RNA and protein analysis and flash frozen in liquid nitrogen.
Cisplatin sensitivity
10,000 cells were seeded in triplicate into 96 well plates. The following day cells were treated with the appropriate concentration of cisplatin or vehicle control. After 48 h, the relative survival of the cells was calculated by using the ATPlite assay (Perkin Elmer #606016943) following the manufacturer’s instructions.
Micronuclei frequency
20.0 cells per chamber were seeded on four-chamber slides. After 48 h, cells were fixed in 100% methanol and slides were stained with DAPI. The slides were mounted and imaged. Micronuclei were counted as discrete units distinct from the nucleus.
Single-molecule RNA in situ hybridization
Skin tissues were from previously described 7-10 wk old mice of genotypes BK5.Cre; Rev3l+/lox'°x and BK5.Cre; Rev3r/lox. In these mice, the floxed allele of Rev3l is deleted specifically in epithelial cells where the keratin 5 promoter (BK5) is active (Lange et al., 2018). Sections of formalin fixed, paraffin-embedded archival blocks were stained with hematoxylin and eosin. RNA in situ hybridization experiments were performed using RNAscope® (Wang et al., 2012). Paired double-Z oligonucleotide probes were designed against target RNA using custom software. The following probes were used Mm-Ccl5, cat no. 469601, NM_013653.3,11 pairs, nt 4527 and Mm-Cxcl10-C2, cat no. 408921-C2, NM_021274.2, 16 pairs, nt 11-1012. The RNAscope® 2.5 HD Duplex Reagent Kit (Advanced Cell Diagnostics, Newark, CA) was used according to the manufacturer’s instructions. Each sample was quality controlled for RNA integrity with probes specific to the housekeeping genes PPIB and POLR2A. Negative control background staining was evaluated using a probe specific to the Bacillus subtilis dapB gene. Brightfield images were acquired using an Aperio ScanScope CS microscope using a 40x objective. Fields of interfollicular basal epithelial cells with defined nuclei were scored. Positive cells usually contained only one hybridization focus. Images from three individual mice of each genotype were visualized with QuPath and scored (15 fields per mouse, total > 800 epithelial cells/mouse).
QUANTIFICATION AND STATISTICAL ANALYSIS
Unpaired two-tailed Student’s t tests were applied to qPCR, micronuclei frequency, and in situ hybridization results using Prism 8. The significance threshold was set at a <0.05. On all graphs the mean is graphed and the error bars represent ± SD. For the graph of gene expression (FPKM) from RNA sequencing data (Figure 2D), the error bars represent SEM.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-HA-Tag (C29F4) | Cell Signaling Technologies | Cat#3724; RRID:AB_1549585 |
| Rabbit anti-cGAS (Mouse specific) (D3O8O) | Cell Signaling Technologies | Cat#31659; RRID:AB_2799008 |
| Rabbit anti-STING (D2P2F) | Cell Signaling Technologies | Cat#13647; RRID:AB_2732796 |
| Rabbit anti-MDA-5 (D74E4) | Cell Signaling Technologies | Cat#5321; RRID:AB_10694490 |
| Rabbit anti-Phospho-IRF-3 (Ser396) (4D4G) | Cell Signaling Technologies | Cat#4947; RRID:AB_823547 |
| Rabbit anti-IRF-3 (D83B9) | Cell Signaling Technologies | Cat#4302; RRID:AB_1904036) |
| Rabbit anti-STAT1 (D1K9Y) | Cell Signaling Technologies | Cat#14994; RRID:AB_2737027 |
| Rabbit anti-TBK1 (D1B4) | Cell Signaling Technologies | Cat#3504; RRID:AB_2255663 |
| rabbit anti-Phospho-TBK1 (S172) (D52C2) | Cell Signaling Technologies | Cat#5483; RRID:AB_10693472 |
| Mouse anti-Viperin | Millipore | Cat#MABF106 RRID:AB_11203644 |
| Goat anti-Rabbit 800CW | LI-COR | Cat#926-32211; RRID:AB_621843 |
| Goat anti-Mouse 800CW | LI-COR | Cat#827-08364; RRID:AB_10793856 |
| Critical commercial assays | ||
| REVERT Total Protein Stain Kit | LI-COR | Cat#926-11010 |
| ATPlite Luminescence Assay System | Perkin Elmer | Cat#6016941 |
| MycoAlert Mycoplasma Detection Kit | Lonza Bioscience | Cat#LT07-218 |
| Deposited data | ||
| Raw and analyzed RNA sequencing data | This paper | GEO: GSE163313 |
| Full immunoblots for the cropped immunoblots in Figures 1, 3, 4, and S1. | This paper | Mendeley Data: https://dx.doi.org/10.17632/5348dspthz.1 |
| Experimental models: cell lines | ||
| Rev3l HET 1 MEFs: Rev3L(+/Δ) mT/mG (+/−) MEFs from mouse embryo 3, TAg immortalized clone 6 (or 3(+)6) | Lange et al., 2012, 2016 | N/A |
| Rev3l HET 2 MEFs: same as above except clone 10 (or 3(+)10) | Lange et al., 2012, 2016 | N/A |
| Rev3l KO 1 MEFs: Rev3L(−/Δ) mT/mG (+/−) MEFs from mouse embryo 4, TAg immortalized clone 5 (or 4(-)5) | Lange et al., 2012, 2016 | N/A |
| Rev3l KO 2 MEFs: same as above except clone 10 (or 3(+)10) | Lange et al., 2012, 2016 | N/A |
| Rev3l HET + EV #Clone: HET 1 + pCDH-EF1-Flag-HA-Empty, clones: 4, 8 and 11 | This study | N/A |
| Rev3l KO + EV #Clone: KO 1 + pCDH-EF1-Flag-HA-Empty, clones: 3, 4, and 15 | This study | N/A |
| Rev3l KO + TR4-2 #Clone: KO 1 + pCDH-EF1-Flag-HA-TR4-2, clones: 8, 18, and 21 | This study | N/A |
| Oligonucleotides | ||
| “siCTL”: Negative Control DsiRNA Duplex | Integrated DNA Technologies | Cat#51-01-14-03 |
| “sicGAS”: cGAS (MB21D1) DsiRNA Duplex | Integrated DNA Technologies | Design ID: mm.Ri.Mb21d1.13.1 |
| “siSTING”: STING (TMEM173) DsiRNA Duplex | Integrated DNA Technologies | Design ID: mmRi.Tmem173.13.2 |
| For primers used for qPCR in this study, see Table S2. | See Table S2 | N/A |
| Recombinant DNA | ||
| pLEXm-His8-MBP-TR4-2 | Lee et al., 2014 | N/A |
| pCDH-EF1-Flag-HA-TR4-2 | This study | N/A |
| pCDH-EF1-Flag-HA-Empty | Tomida et al., 2015 | N/A |
| Software and algorithms | ||
| TopHat (version 2.0.10) | Kim et al., 2013 | N/A |
| HTSeq package (version 0.6.0) | Anders et al., 2015 | N/A |
| R/Bioconductor package edgeR (version 3.8.6) | Robinson et al., 2010 | N/A |
| Prism (version 8) | GraphPad | N/A |
Highlights.
Loss of pol ζ (Rev3l) leads to chromosome damage marked by increased micronuclei
Rev3l loss increases expression of interferon-stimulated genes (ISGs) and proteins
Expression of ISG chemokines is elevated in Rev3l-disrupted primary epithelial cells
The cGAS-STING pathway drives this ISG expression signature in Rev3l-disrupted cells
ACKNOWLEDGMENTS
We thank Sarita Bhetawal and Megan Lowery for assistance with cell culture and advice. We thank Dr. Yue Lu, Dr. Bin Liu, and Kevin Lin for invaluable assistance with the RNA expression data analysis, and Dr. Manu Sebastian, Jimi Lynn Young, and Carlos Perez for in situ hybridization. We are grateful to Winnie Cheng for assistance with the micronuclei experiments. We thank Dr. Wei Yang (NIH) for the gift of the TR4-2 cDNA. Funding was provided by National Institutes of Health grant no. CA193124, Department of Defense grant no. W81XWH-17-10239, and the J. Ralph Meadows Chair in Carcinogenesis Research to R.D.W. S.K.M. was supported by a CPRIT Research Training Grant award (RP170067). We thank the cores at The University of Texas MD Anderson Cancer Center in the Department of Epigenetics and Molecular Carcinogenesis that contributed to our data collection. RNA sequencing (Next Generation Sequencing Core) and quantitative PCR were supported by CPRIT grants RP120348 and RP170002. In situ hybridization performed by the Research Histology, Pathology, and Imaging Core was supported by P30 CA16672-39 DHHS/NCI Cancer Center Support Grant.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108775.
DECLARATION OF INTERESTS
R.D.W. is a scientific advisor for Repare Therapeutics and a shareholder.
REFERENCES
- Ablasser A, and Chen ZJ (2019). cGAS in action: expanding roles in immunity and inflammation. Science 363, eaat8657. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Khamlichi AA, Davies SL, and Neuberger MS (2000). Disruption of mouse polymerase zeta (Rev3) leads to embryonic lethality and impairs blastocyst development in vitro. Curr. Biol. 10, 1213–1216. [DOI] [PubMed] [Google Scholar]
- Berdis AJ (2008). DNA polymerases as therapeutic targets. Biochemistry 47, 8253–8260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel F, Silva MJ, Técher H, Zadorozhny K, Sharma S, Nieminuszczy J, Mettling C, Dardillac E, Barthe A, Schmitz AL, et al. (2018). SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61. [DOI] [PubMed] [Google Scholar]
- Daly J, Bebenek K, Watt DL, Richter K, Jiang C, Zhao ML, Ray M, McGregor WG, Kunkel TA, and Diaz M (2012). Altered Ig hypermutation pattern and frequency in complementary mouse models of DNA polymerase ζ activity. J. Immunol. 188, 5528–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Godindagger I, Klein U, Yaspo ML, Cumano A, and Rajewsky K (2000). Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr. Biol. 10, 1221–1224. [DOI] [PubMed] [Google Scholar]
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, and Thomas P (2011). Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26, 125–132. [DOI] [PubMed] [Google Scholar]
- Glück S, Guey B, Gulen MF, Wolter K, Kang T-W, Schmacke NA, Bridgeman A, Rehwinkel J, Zender L, and Ablasser A (2017). Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonugunta VK, Sakai T, Pokatayev V, Yang K, Wu J, Dobbs N, and Yan N (2017). Trafficking-mediated sting degradation requires sorting to acidified endolysosomes and can be targeted to enhance anti-tumor response. Cell Rep. 21, 3234–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratia M, Rodero MP, Conrad C, Bou Samra E, Maurin M, Rice GI, Duffy D, Revy P, Petit F, Dale RC, et al. (2019). Bloom syndrome protein restrains innate immune sensing of micronuclei by cGAS. J. Exp. Med 216, 1199–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoong BYD, Gan YH, Liu H, and Chen ES (2020). cGAS-STING pathway in oncogenesis and cancer therapeutics. Oncotarget 11, 2930–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Takata K, and Wood RD (2011). DNA polymerases and cancer. Nat. Rev. Cancer 11, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Wittschieben JP, and Wood RD (2012). DNA polymerase ζ is required for proliferation of normal mammalian cells. Nucleic Acids Res. 40, 4473–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Bedford E, Reh S, Wittschieben JP, Carbajal S, Kusewitt DF, DiGiovanni J, and Wood RD (2013). Dual role for mammalian DNA polymerase ζ in maintaining genome stability and proliferative responses. Proc. Natl. Acad. Sci. USA 110, E687–E696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Tomida J, Boulware KS, Bhetawal S, and Wood RD (2016). The Polymerase Activity of Mammalian DNA Pol ζ Is Specifically Required for Cell and Embryonic Viability. PLoS Genet. 12, e1005759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Bhetawal S, Reh S, Powell KL, Kusewitt DF, and Wood RD (2018). DNA polymerase ζ deficiency causes impaired wound healing and stress-induced skin pigmentation. Life Sci. Alliance 1, e201800048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Gray EE, Brunette RL, and Stetson DB (2015). DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 350, 568–571. [DOI] [PubMed] [Google Scholar]
- Lee YS, Gregory MT, and Yang W (2014). Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc. Natl. Acad. Sci. USA 111, 2954–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Lettice L, Tarnauskaitė Ž, Reddy K, Dix F, Revuelta A, Abbondati E, Rigby RE, Rabe B, et al. (2016). Ribonuclease H2 mutations induce a cGAS/STING-dependent innate immune response. EMBO J. 35, 831–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, et al. (2017). cGAS surveillance of 11 micronuclei links genome instability to innate immunity. Nature 548, 461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SK, and Wood RD (2019). DNA polymerase ζ in DNA replication and repair. Nucleic Acids Res. 47, 8348–8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami H, Kim JD, Tabara S, Lu W, Kwon C, Nakashima M, and Fukamizu A (2019). KDM5D-mediated H3K4 demethylation is required for sexually dimorphic gene expression in mouse embryonic fibroblasts. J. Biochem 165, 335–342. [DOI] [PubMed] [Google Scholar]
- Mudge JM, and Harrow J (2015). Creating reference gene annotation for the mouse C57BL6/J genome assembly. Mamm. Genome 26, 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O-Wang J, Kajiwara K, Kawamura K, Kimura M, Miyagishima H, Koseki H, and Tagawa M (2002). An essential role for REV3 in mammalian cell survival: absence of REV3 induces p53-independent embryonic death. Biochem. Biophys. Res. Commun 293, 1132–1137. [DOI] [PubMed] [Google Scholar]
- Panne D, McWhirter SM, Maniatis T, and Harrison SC (2007). Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J. Biol. Chem 282, 22816–22822. [DOI] [PubMed] [Google Scholar]
- Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, Reinert L, Cai Y, Jensen SB, Skouboe MK, et al. (2018). Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 37, e97858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisländer T, Lombardi EP, Groelly FJ, Miar A, Porru M, Di Vito S, Wright B, Lockstone H, Biroccio A, Harris A, et al. (2019). BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat. Commun 10, 3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus JB, Trivino-Soto GS, Wu LI, Kokott K, and Lim ES (2020). SV40 Large T Antigen Is Not Responsible for the Loss of STING in 293T Cells but Can Inhibit cGAS-STING Interferon Induction. Viruses 12, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sail V, Rizzo AA, Chatterjee N, Dash RC, Ozen Z, Walker GC, Korzhnev DM, and Hadden MK (2017). Identification of Small Molecule Translesion Synthesis Inhibitors That Target the Rev1-CT/RIR Protein-Protein Interaction. ACS Chem. Biol 12, 1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, Alt FW, and Rajewsky K (2009). Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J. Exp. Med 206, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Li X, Owens KM, Vanniarajan A, Liang P, and Singh KK (2015). Human REV3 DNA Polymerase ζ Localizes to Mitochondria and Protects the Mitochondrial Genome. PLoS ONE 10, e0140409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserand J, Khetchoumian K, Thibault C, Dembélé D, Chambon P, and Losson R (2011). Tripartite motif 24 (Trim24/Tif1α) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor α (Rarα) inhibition. J. Biol. Chem 286, 33369–33379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida J, Takata K, Lange SS, Schibler AC, Yousefzadeh MJ, Bhetawal S, Dent SY, and Wood RD (2015). REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 43, 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sloun PP, Varlet I, Sonneveld E, Boei JJ, Romeijn RJ, Eeken JC, and De Wind N (2002). Involvement of mouse Rev3 in tolerance of endogenous and exogenous DNA damage. Mol. Cell. Biol 22, 2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, and Luo Y (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn 14, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. (2015). Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben J, Shivji MK, Lalani E, Jacobs MA, Marini F, Gearhart PJ, Rosewell I, Stamp G, and Wood RD (2000). Disruption of the developmentally regulated Rev3l gene causes embryonic lethality. Curr. Biol 10, 1217–1220. [DOI] [PubMed] [Google Scholar]
- Wittschieben JP, Reshmi SC, Gollin SM, and Wood RD (2006). Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 66, 134–142. [DOI] [PubMed] [Google Scholar]
- Wittschieben JP, Patil V, Glushets V, Robinson LJ, Kusewitt DF, and Wood RD (2010). Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 70, 2770–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek JL, Chatterjee N, Najeeb J, Ramos A, Lee M, Bian K, Xue JY, Fenton BA, Park H, Li D, et al. (2019). A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy. Cell 178, 152–159.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xie K, Zhang XQ, Pridgen EM, Park GY, Cui DS, Shi J, Wu J, Kantoff PW, Lippard SJ, et al. (2013). Enhancing tumor cell response to chemotherapy through nanoparticle-mediated codelivery of siRNA and cisplatin prodrug. Proc. Natl. Acad. Sci. USA 110, 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Wei L, Pfeffer SR, Du Z, Murti A, Valentine WJ, Zheng Y, and Pfeffer LM (2007). Identification of CXCL11 as a STAT3-dependent gene induced by IFN. J. Immunol 178, 986–992. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Ren J, Chen Q, and Chen ZJ (2017). cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 114, E4612–E4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, and Fujita T (2002). Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res 22, 73–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets produced in this study have been uploaded to Gene Expression Omnibus. Accession number for the RNA sequencing data is GEO: GSE163313. The full immunoblot images in Figures 1,3,4, and S1 have been uploaded to Mendeley Data: https://dx.doi.org/10.17632/5348dspthz.1


