Abstract
Momilactones from rice have allelopathic activity, the ability to inhibit growth of competing plants. Transferring momilactone production to other crops is a potential approach to combat weeds, yet a complete biosynthetic pathway remains elusive. Here, we address this challenge through rapid gene screening in N. benthamiana, a heterologous plant host. To do so, we solved a central problem with this technique: diminishing yields remain a bottleneck for multi-step pathways. We increased intermediate and product titers by re-routing diterpene biosynthesis from the chloroplast to the cytosolic, high-flux mevalonate pathway. This enabled the discovery and reconstitution of a complete route to momilactones (>10-fold yield improvement versus rice). Pure momilactone B isolated from N. benthamiana inhibits germination and root growth in Arabidopsis, validating allelopathic activity. We demonstrate the broad utility of this approach by applying it to forskolin, a hedgehog inhibitor, and taxadiene, an intermediate in taxol biosynthesis (~10-fold improvement versus chloroplast expression).
Introduction
Herbicides are one of the largest chemical inputs to agriculture, and herbicide resistance is among the most commonly engineered traits in crops1. An alternative to the exogenous addition of chemical herbicides is to utilize mechanisms that plants have evolved themselves to limit the growth of competitive plant species, a process termed allelopathy2. Although this phenomenon has been described for numerous plant pairs, few molecules have been directly shown to impart allelopathic activity. A lead example are the momilactones, a group of diterpenes found predominantly in rice that enable seedlings to limit growth of nearby competing plants3. The most potent momilactone, momilactone B, has been attributed to inhibit the growth of common rice field monocot weeds, Echinochloa crus-galli and Echinochloa colonum, at IC50 of 6.1 and 5.0 μM for shoots and 6.3 and 12.5 μM for roots, respectively4. Momilactone B also inhibits growth and germination of numerous monocot and dicot plant species5–8.
The association of rice allelopathy with a single metabolic pathway along with the high demand for herbicides raises the intriguing possibility of engineering momilactone biosynthesis as a type of in situ-produced herbicide in crops3,9. However, the potential utility of momilactones is restricted by the lack of a complete set of biosynthetic genes which precludes pathway engineering for in planta function and/or production in a heterologous host. Sourcing diterpenes from the native plant is hindered by complex extraction processes from plant matrix, high production costs, and endemic and environmental constraints related to growth and harvesting. For instance, current methods of momilactone production are limited to the laboratory scale: access requires either hydroponic growth of thousands of rice seedlings to collect liters of root exudates or direct extraction from kilograms of the plant source to obtain yields of 0.75–37.8 μg/g in dried husk and 1.06–12.73 μg/g of dry weight in other rice organs5,10–12. Even with reliable access to pure compounds, the utility of many diterpenoids with agricultural relevance is limited without engineering tools for transferring these biosynthetic pathways from one plant to another for in situ production.
Metabolic engineering of a biosynthetic pathway in a heterologous host can alleviate sourcing problems. Engineering in microbial hosts have resulted in strategies for the manufacture of a handful of plant-derived natural products13–17. Furthermore, stable engineering of pathways in a plant host using tissue-specific and synthetic temporal control of pathway induction opens the possibility for the introduction of new functional traits. Engineering of transgenic plant lines has resulted in the tissue-specific production of β-carotene in golden rice18. Despite the promise of metabolic engineering for production of difficult-to-access compounds, these approaches are limited by the lack of required biosynthetic genes for the majority of plant pathways. These approaches become even more critical for terpenes such as the momilactones, where the native plant source is not a viable route for production.
Transient expression of pathways in the model plant Nicotiana benthamiana (a relative of tobacco) has accelerated biosynthetic gene discovery. Three major bottlenecks are alleviated, at least partially, in N. benthamiana: First is the need for rapid combinatorial testing of difficult-to-express plant enzyme candidates without knowledge of the sequence of biochemical steps. Second, assay of enzyme candidates requires access to complex intermediates that often do not accumulate in the native plant producer or heterologous expression host and are challenging synthetic targets. Third, the reconstitution of multi-step linear plant pathways suffers from diminishing yields that come with increasing pathway length. For example, the reconstitution of the pathway steps in momilactone biosynthesis to an unknown, oxidized syn-pimaradiene intermediate, necessary to study the formation of a lactone and hemiacetal ring, has not been accomplished.
In recent years, transient expression in N. benthamiana resulted in the reconstitution of partial and complete pathways from major classes of natural products19–24. Despite the utility of tobacco, diterpene pathways remain challenging targets. These pathways typically include extensive oxidations requiring numerous enzymes acting in sequence, and are especially prone to compounding yield effects that limit precursor accumulation. For instance, access to momilactone precursors could lead to the discovery of variants with stronger allelopathic bioactivity and provide insights into which moieties are associated with allelopathic effects. More importantly, reconstitution of a complete pathway would provide the first step towards transferring the allelopathic properties to another crop.
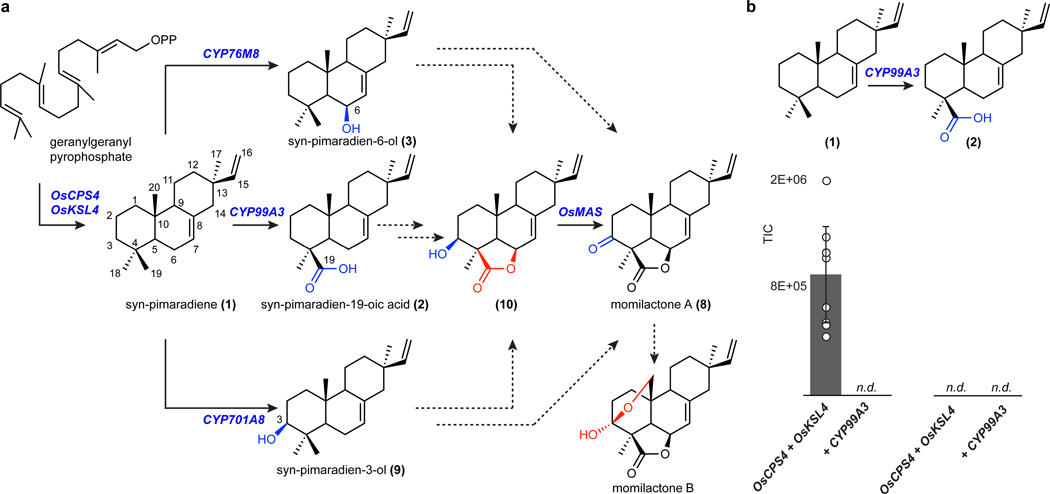
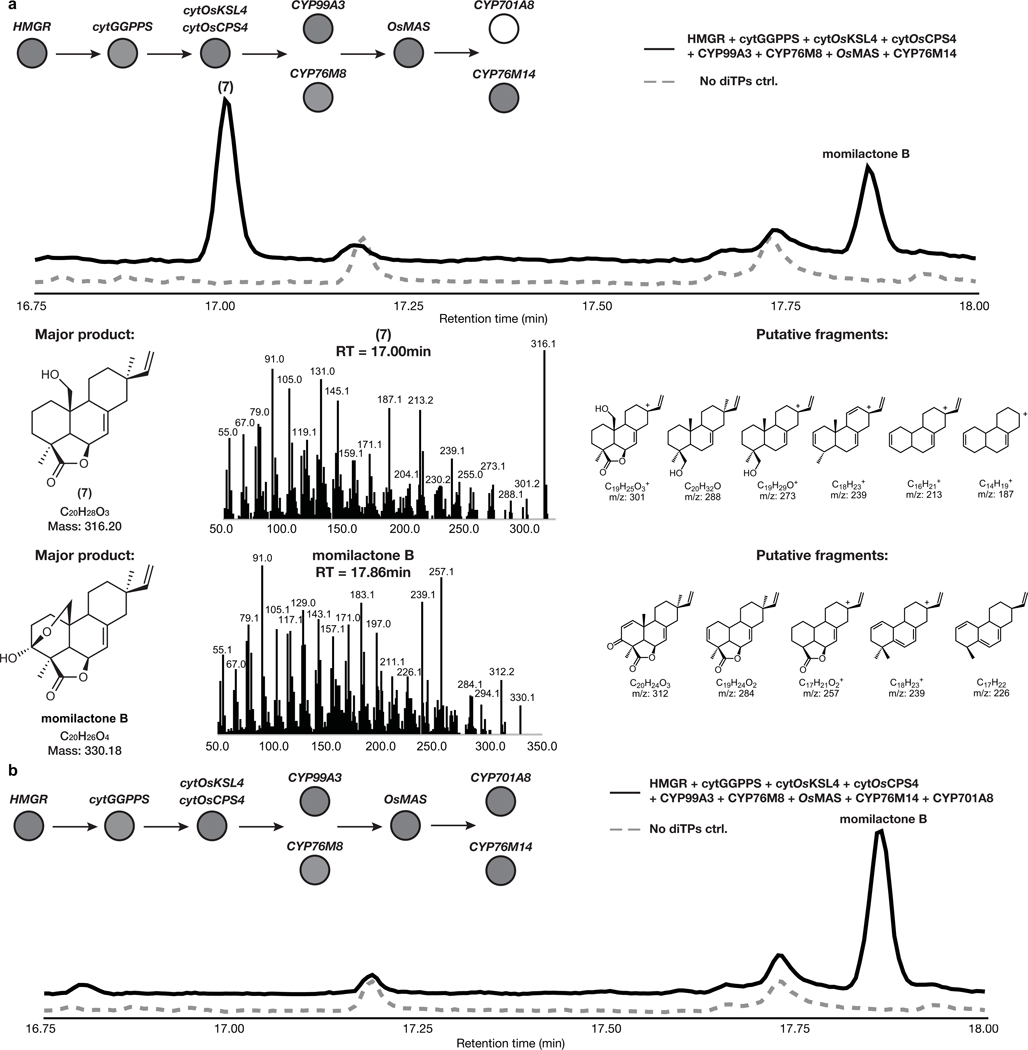
Despite the identification of several enzymes involved in momilactone biosynthesis, the order of known steps and the reconstitution of multiple characterized enzymes in vivo remains elusive3 (Fig. 1a). Most notably, enzymes involved in the formation of the characteristic lactone and hemiacetal rings, important moieties presumably required for allelopathic activity, remain to be characterized. To date, the required diterpene synthases to generate the tricyclic momilactone scaffold, OsCPS4 and OsKSL4, have been identified25–28. The genes encoding these diterpene synthases are clustered with genes encoding cytochrome P450 enzymes (CYPs), CYP99A3 and CYP99A2, and a short chain dehydrogenase reductase (SDR), OsMAS, in a gene cluster on rice chromosome 4. CYP99A3 along with two additional non-clustered cytochrome P450 enzymes, CYP76M8 and CYP701A8, oxidize the diterpene scaffold at different carbons, yet sequential activity of these CYPs to form a scaffold oxidized at multiple positions has not be shown 29–31. The clustered OsMAS encodes a SDR capable of oxidation of the surrogate substrate 3β-hydroxy-9β-pimara-7,15-dien-19,6β-olide (10) to momilactone A (8) through in vitro assays32.
Fig. 1: Proposed momilactone biosynthetic pathway including previously characterized steps.
(a) Chemical functionalities in blue indicate previously characterized reactions. Missing steps are shown in dotted arrows and predicted functionalities are highlighted in red. (b) Transient expression of CYP99A3 with requisite diterpene synthases, OsCPS4 and OsKSL4, cytGGPPS and DXS in N. benthamiana. Values and error bars represent the mean and the standard deviation, n = 9 biological independent leaf samples examined over three independent experiments.
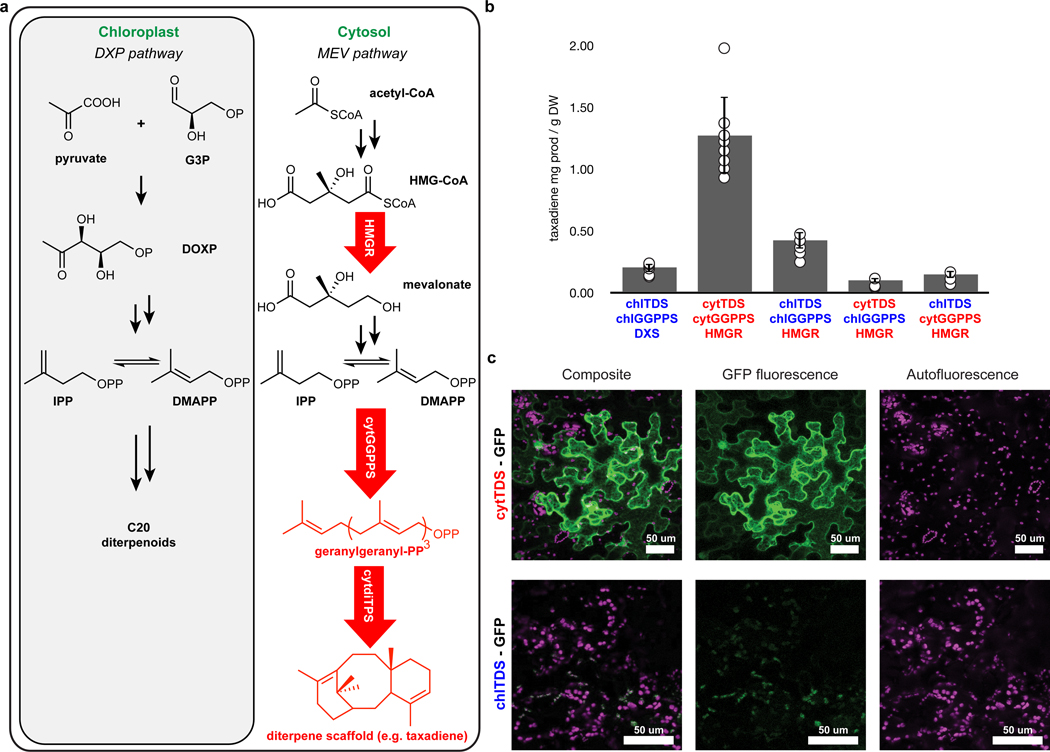
Diminishing yields that make discovery of full diterpenoid pathways difficult can be alleviated with an ample supply of isoprene precursor building blocks. Plants synthesize terpenoids via two distinctly compartmentalized, endogenous pathways, the cytosol-localized mevalonate (MEV) pathway and the chloroplast-localized deoxyxylulose phosphate (DXP) pathway. Although both pathways operate simultaneously and produce the same 5-carbon isoprene equivalents that feed terpene biosynthesis generally, the condensation and subsequent cyclization to a terpene scaffold is catalyzed by distinctly localized prenyltransferases and terpene synthases (Supplementary Figure 1). Sesqui- and triterpenoid biosynthesis typically is fed by the cytosolic MEV pathway involving C15 and C30 pyrophosphate intermediates, while mono-, di-, and tetraterpenoid biosynthesis typically is fed by the plastidial DXP pathway involving C10, C20 and C40 intermediates. Engineering approaches to increase the flux of both the MEV and DXP pathways in N. benthamiana involve overexpression of rate-limiting enzymes: HMGR (3-hydroxy-3-methylglutaryl-CoA reductase) for the MEV pathway, and DXS (1-deoxy-d-xylulose-5-phosphate synthase) in the DXP pathway33–36. These approaches increased yields of both di- and triterpene products produced through multi-step biosynthetic routes; however, additional yield improvements would facilitate pathway reconstitution and discovery and simplify access to desired products.
Here we develop an engineering strategy for improved diterpene production in N. benthamiana to enable discovery of the missing genes required to complete the full 7-step momilactone biosynthetic pathway from rice. Our approach involves redirecting diterpene production from the plastids to an engineered high-flux cytosolic pathway by modifying the subcellular localization of the requisite diterpene synthases. We show that this approach made pathway discovery possible, and facilitated the large-scale isolation of the potent allelopathic compound momilactone B from a heterologous plant host. As proof of concept, we demonstrate that momilactone B, isolated and purified from N. benthamiana leaves, exhibits in vitro germination and growth inhibition in assays using the model plant Arabidopsis thaliana. We also demonstrate the versatility of this approach for increasing yield in diterpenoid pathways using the clinically relevant anticancer agent taxol and the adenylate cyclase activator forskolin as key examples. Taken together, our work constitutes a general diterpenoid engineering strategy for rapid pathway discovery and access to valuable and difficult-to-access diterpenes with potent biological activity.
Results
Establishing a method for diterpene yield improvement
In our initial experiments towards discovery of the missing genes in the momilactone pathway, we tested the activity of full-length, native, chloroplast-localized versions of the two requisite diterpene synthases OsCPS4 and OsKSL4 in N. benthamiana leaves using Agrobacterium-mediated transient protein expression. In an effort to obtain high yields of the diterpene scaffold, we co-expressed with the established rate-limiting enzymes from the DXP pathway, geranylgeranyl pyrophosphate synthase GGPPS (chlGGPPS) and DXS (Supplementary Figure 2). Although we observed production of syn-pimaradiene (1) in N. benthamiana with this set, we did not observe the subsequent metabolite in the pathway, syn-pimaradien-19-oic acid (2), after co-expressing the additional downstream pathway gene CYP99A329 (Fig. 1b). The role of CYP99A3 has previously been confirmed after expression in E. coli, so we considered that, in N. benthamiana, the required substrate, 1, does not accumulate to high enough levels due to limited flux through the plastidial DXP pathway. Given that di- and triterpene pathways both require the common isoprene precursors and the capacity for higher flux has been established for triterpenoid engineering in N. benthamiana, we reasoned that rewiring diterpene biosynthesis to be fed through the cytosolic MEV pathway might increase levels of 1 and enable pathway reconstitution (Fig. 2a). This approach would require re-routing both the momilactone diterpene synthases, OsCPS4 and OsKSL4, and the native chlGGPPS, the enzyme that generates the required C20 geranylgeranyl pyrophosphate substrate, from the chloroplast to the cytosol.
Fig. 2: High-titer production of diterpenoid-derived products N. benthamiana through cytosolic diterpenoid engineering.
(a) Redirecting diterpenoid biosynthesis from plastids to the cytosol requires changing subcellular localization of the required prenyltransferase and diterpene synthase(s). Yield improvement via the MEV pathway requires the overexpression of the rate-limiting gene HMGR. Arrows in red represent cytosolic-localized and overexpressed genes. G3P, glyceraldehyde 3-phosphate; DOXP, 1-deoxy-d-xylulose-5-phosphate synthase; IPP, isopentyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; cytGGPPS, cytosolic geranylgeranyl pyrophosphate synthase; cytdiTPS, cytosolic diterpene synthase. (b) Taxadiene biosynthesis via cytosolic engineering leads to a 6-fold yield improvement compared to plastid engineering. Overexpression of both cytTDS and cytGGPPS is required for titer improvement. Values and error bars represent the mean and the standard deviation, n = 9 biological independent leaf samples examined over three independent experiments. DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DW, dry weight. (c) Subcellular localization of overexpressed cytTDS-GFP and chlTDS-GFP fusion proteins suggests that removal of chloroplast localization peptides is sufficient to disrupt targeting. Images using confocal microscopy show z-stacks of maximum intensity projections scaled to the same intensity range for GFP fluorescence (green; 488nm excitation, 500–550 emission), chlorophyll autofluorescence (magenta; 488nm excitation, 650–700 emission) and the two-channel composite. Images are representative of three biological replicates repeated over two independent experiments.
We first tested this approach for improving yields using the well-studied taxadiene synthase (TDS) as a model system (Supplementary Figure 3). Terpene synthases are classified according to their reaction-initiation mechanism. Class I terpene synthases initiate reactions through ionization of an isoprenyl pyrophosphate and Class II synthases initiate reactions through protonation of C-C double bonds or epoxides37. TDS is a monofunctional Class I diterpene synthase, and this single enzyme is capable of generating a cyclic diterpene scaffold directly from GGPP38. We reasoned that TDS would be simpler to initially test, in comparison to the momilactone diterpene synthase pair, the class II OsCPS4 and class I OsKSL4, which must both be expressed together to generate the cyclic diterpene scaffold39,40. To re-route chlTDS and chlGGPPS to the cytosol, we designed modified versions bearing N-terminal truncation of the native chloroplast localization sequences. We anticipated the encoded products of cytTDS and cytGGPPS would reside in the cytosol and co-expressed them with the rate-limiting MEV pathway enzyme HMGR (Fig. 2a). Analysis of leaf extracts revealed up to a ten-fold yield increase in taxadiene (12) production levels compared to expression of native chlTDS in combination with overexpression of rate-limiting steps of the DXP pathway (which is reported to be on the order of 3.5–10-fold higher than expression of native chlTDS alone) (Fig. 2b)35,41. We found that N-terminal truncation of both cytTDS and cytGGPPS was necessary to boost yields efficiently: expression of either enzyme with its native chloroplast transit sequence intact did not result in a yield increase (e.g. compare cytTDS and chlGGPPS to chlTDS and chlGGPPS, Fig. 2b). This strategy enabled detection of four minor products resulting from expression of cytTDS that were not previously observed after plastidial taxadiene reconstitution (Supplementary Figure 4). One of these products likely corresponds to a confirmed isomer of 12. Given that TDS has not been shown to generate oxidized taxadiene products, the two minor products with masses corresponding to oxidized taxadiene scaffolds possibly originate from oxidation with endogenous N. benthamiana enzymes. Expression of cytGGPPS and HMGR also altered endogenous triterpene metabolism (Supplementary Figure 5).
Although the increased diterpene yield after truncation of these synthases suggests integration with the cytosolic pathway, we directly examined altered localization of the modified proteins. We designed C-terminally fused GFP variants of TDS and GGPPS with and without their N-terminal chloroplast signal sequence and examined their localization in N. benthamiana epidermal cells using confocal microscopy. This analysis confirmed that deletion of chloroplast localization sequences disturbed plastidial targeting for TDS (RuBisCo and GFP alone were used as controls) (Fig. 2c). Furthermore, partial transfer was observed for GGPPS, even without optimization of the site for N-terminal sequence truncation (Supplementary Figure 6). Taken together, these data support our hypothesis that cytosolic re-routing is responsible for the observed increases in diterpene scaffold yield.
Cytosolic re-routing for reconstitution of multistep pathways
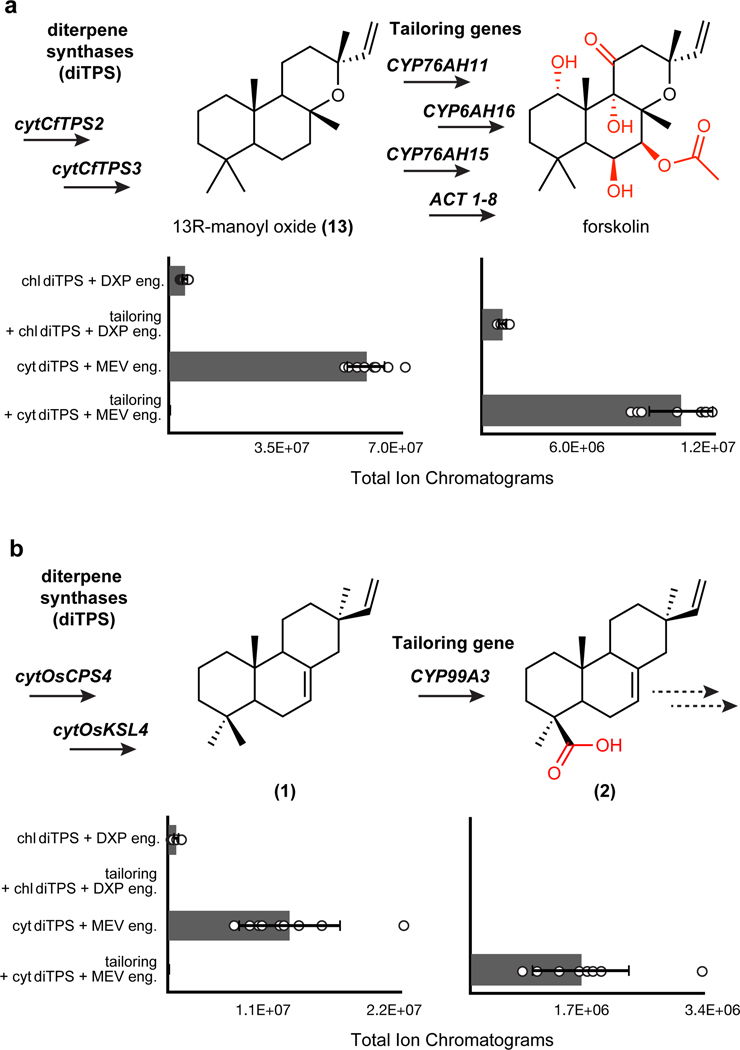
With a strategy for yield improvement in the context of TDS, we next used an established, complete terpene pathway to test whether similar gains would be observed for products derived from diterpene synthases pairs and, more importantly, whether this strategy would be compatible with expression of multiple endoplasmic reticulum membrane-bound CYPs that typically constitute part of full diterpenoid pathways. We chose to examine the production of forskolin (14), a hedgehog signal inhibitor, given that the full set of genes had previously been characterized in N. benthamiana20,42 (Supplementary Figure 7). To generate the labdane ring system of forskolin, we transiently expressed the terpene synthase pair CfTPS2 and CfTPS3 from Coleus forskohlii (coleus) in N. benthamiana leaves to produce 13R-manoyl oxide (13). Co-expression of the cytosolic-localized Class I and Class II diterpene synthases cytCfTPS2 and cytCfTPS3 with HMGR and cytGGPPS resulted in a 12-fold yield improvement in 13 over co-expression of the respective chloroplast-localized synthases with DXS and chlGGPPS (Fig. 3a). Given the success of increasing diterpenoid supply with cytosolic-localized diterpene synthases, we co-expressed the additional tailoring genes for reconstitution of the complete pathway. Cytosolic expression of the full 6-step forskolin pathway resulted in a 7-fold yield improvement compared to plastidial expression. Our results indicated that our cytosolic engineering strategy appears general to different classes of terpene synthases, and that yield improvements are propagated through a multistep pathway including series of tailoring enzymes (Fig. 3a and Supplementary Figure 8).
Fig. 3: Biosynthesis of tailored diterpenoids is enhanced via cytosolic expression in momilactone and forskolin pathways.
a,b, Overexpression of genes for forskolin (a) and momilactone (b) biosynthesis via cytosolic (MEV) and chloroplast (DXP) engineering strategies. Values and error bars represent the mean and the standard deviation, n=9 biological independent leaf samples examined over three independent experiments.
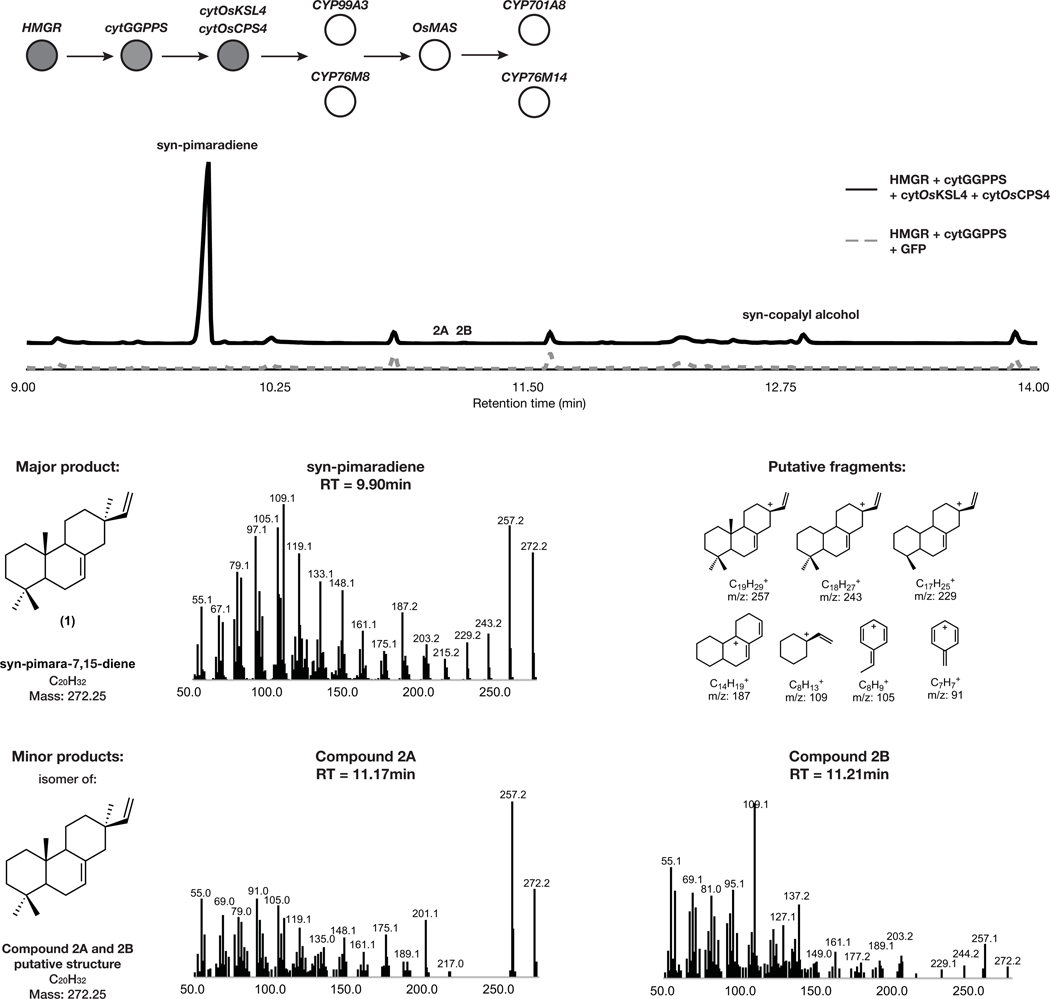
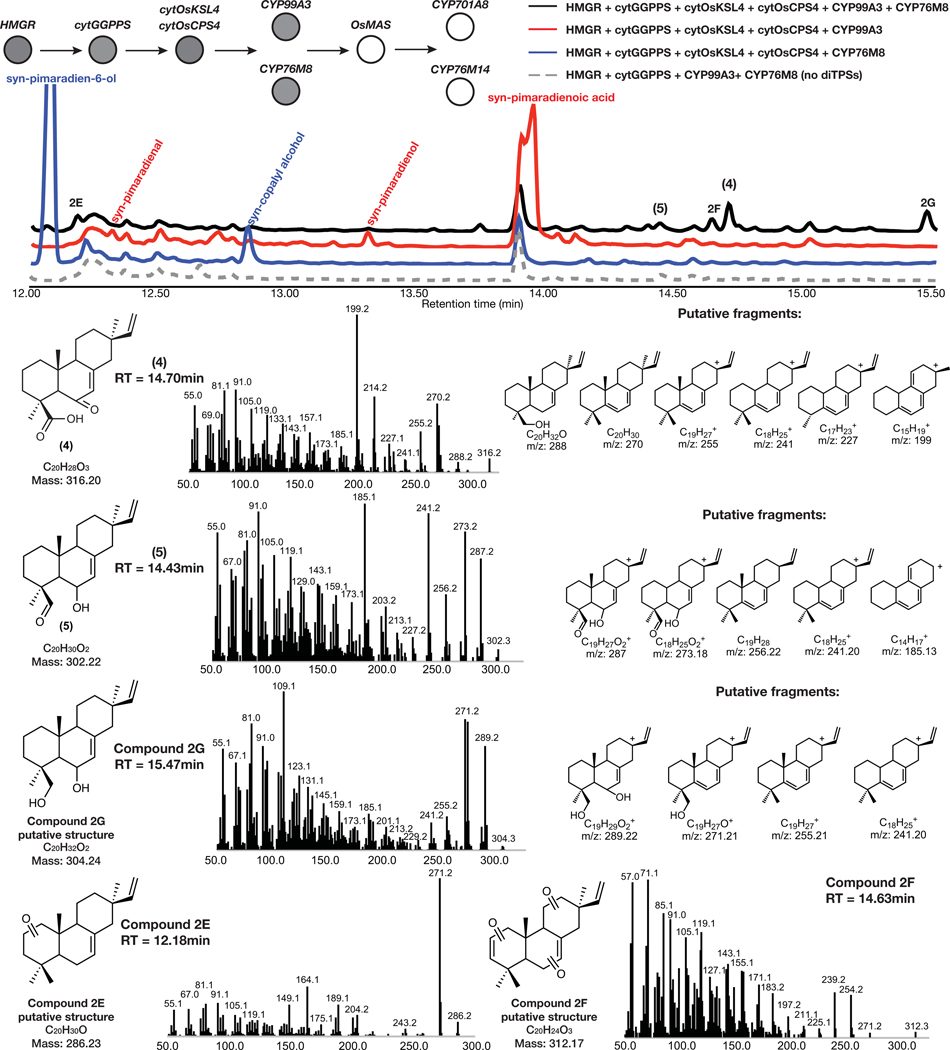
With an approach established for improved yields of complex diterpene products derived from selective oxidation, we tested this strategy for expediting the discovery of a complete, multi-step momilactone biosynthetic pathway. Similarly to what we observed in forskolin and taxadiene biosynthesis, co-expression of known momilactone pathway genes cytOsCPS4, cytOsKSL4, cytGGPPS and HMGR resulted in a 10-fold yield improvement in 1 over co-expression of the respective chloroplast-localized diterpene synthase genes with DXS and chlGGPPS (Fig. 3b). Furthermore, co-expression of these modified enzymes allowed for the detection of minor products that were not previously observed via chloroplast expression (Extended Data Fig. 1) as well as products related to putative intermediates formed en route to the diterpene scaffold (Supplementary Figure 9).
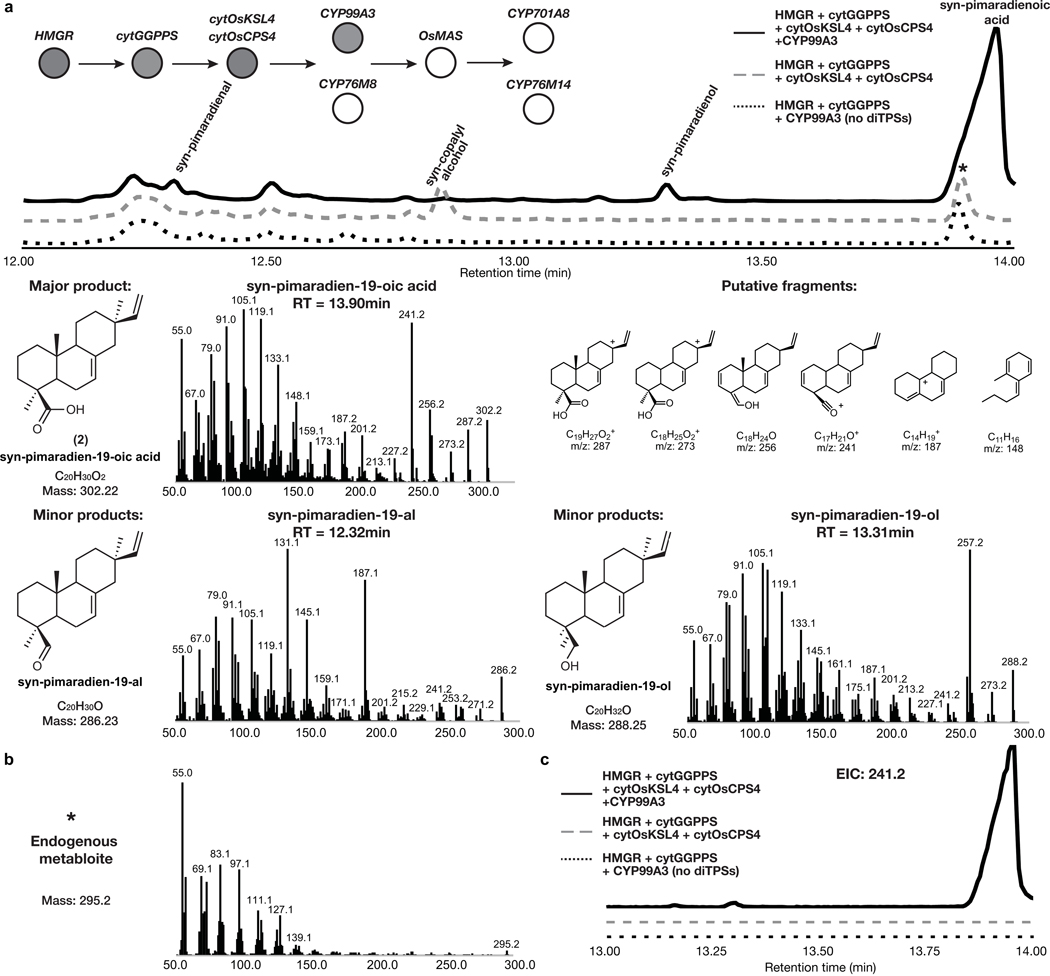
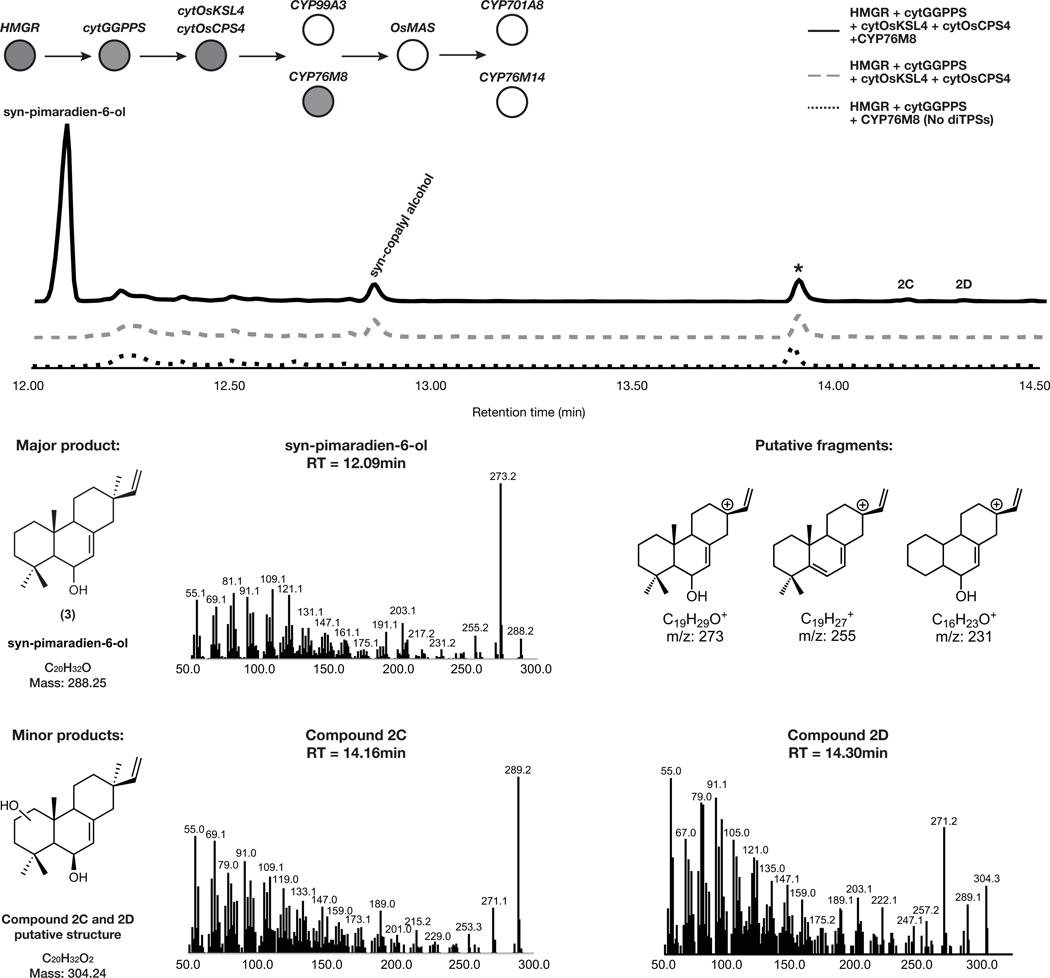
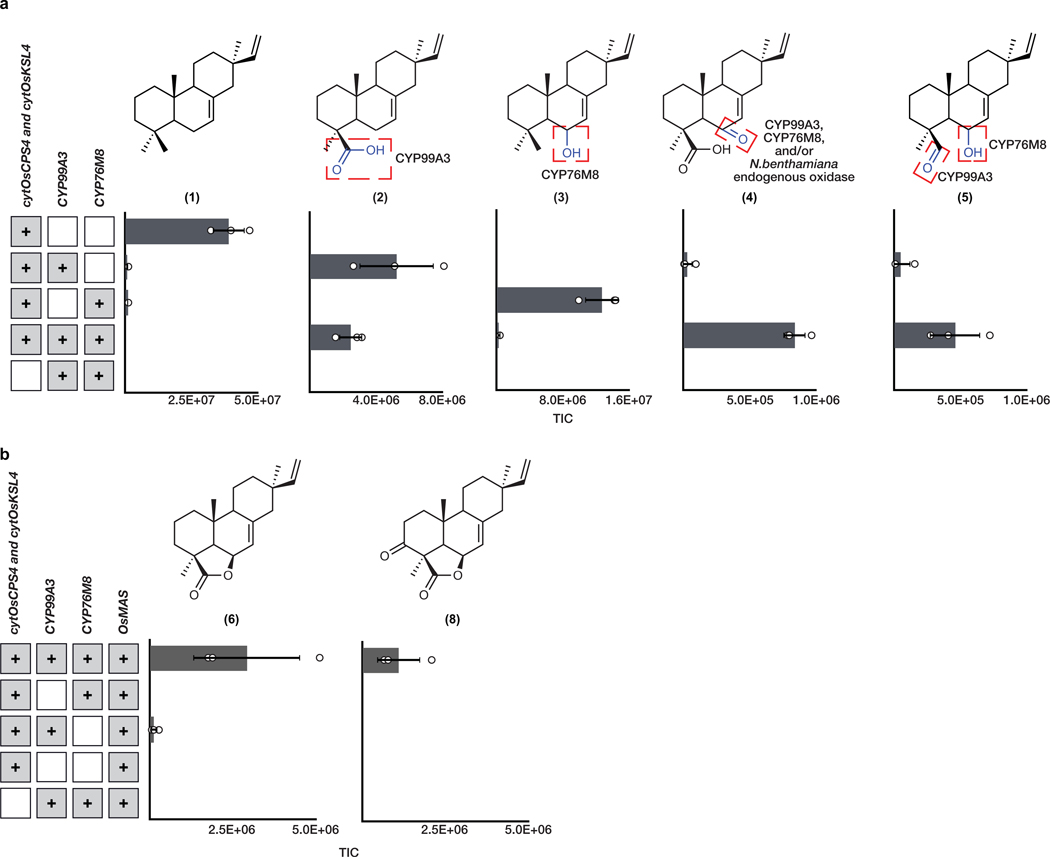
Downstream of the momilactone diterpene synthase are two pathway-associated enzymes, cytochromes P450 CYP99A3 and CYP76M8 (Fig. 1a). These have previously been assayed individually with isolated substrates and shown to produce two related momilactone intermediates, 2 and syn-pimaradien-6-ol (3)29,30. We found similar accumulation of these intermediates after transient expression of either of these P450s in combination with our established engineered momilactone synthase system (Fig. 3b, Extended Data Fig. 2 and Extended Data Fig. 3).
Elucidation of missing steps in momilactone pathway
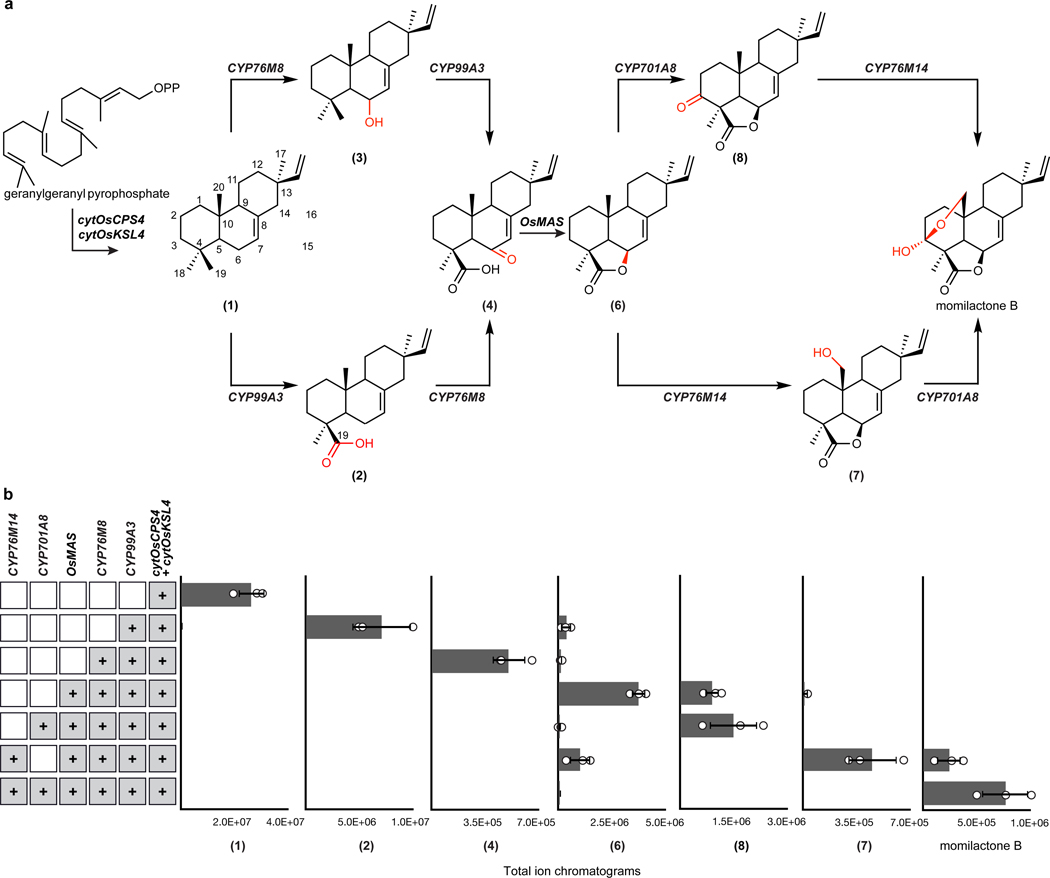
We predicted that the remaining steps to complete the biosynthesis of momilactone would be catalyzed by CYPs, SDRs and 2-oxoglutarate dependent dehydrogenases (2-ODDs) (Fig. 1a). We took two strategies to identify candidate genes encoding these enzymes. First, we selected genes associated with the established momilactone biosynthetic gene cluster (Supplementary Figure 10). Second, we looked for gene candidates co-expressed with known pathway enzymes using a publicly available gene expression dataset. This approach revealed 15 biosynthetic candidates co-expressed with rice diterpene synthase genes cytOsCPS4 and cytOsKSL4 and the remaining clustered genes CYP99A2, CYP99A3 and OsMAS (Supplementary Table 1). This list contained the genes encoding previously characterized CYP enzymes CYP76M8 and CYP701A8, which act on 1, along with other closely related enzymes found in the same CYP subfamilies30,43. Interestingly, the candidate CYP76M14 is closely related to CYP76M8 (65% amino acid identity). The evolutionary history of the CYP76M subfamily in rice suggests that CYP76M14 could be involved in specialized diterpenoid biosynthesis30.
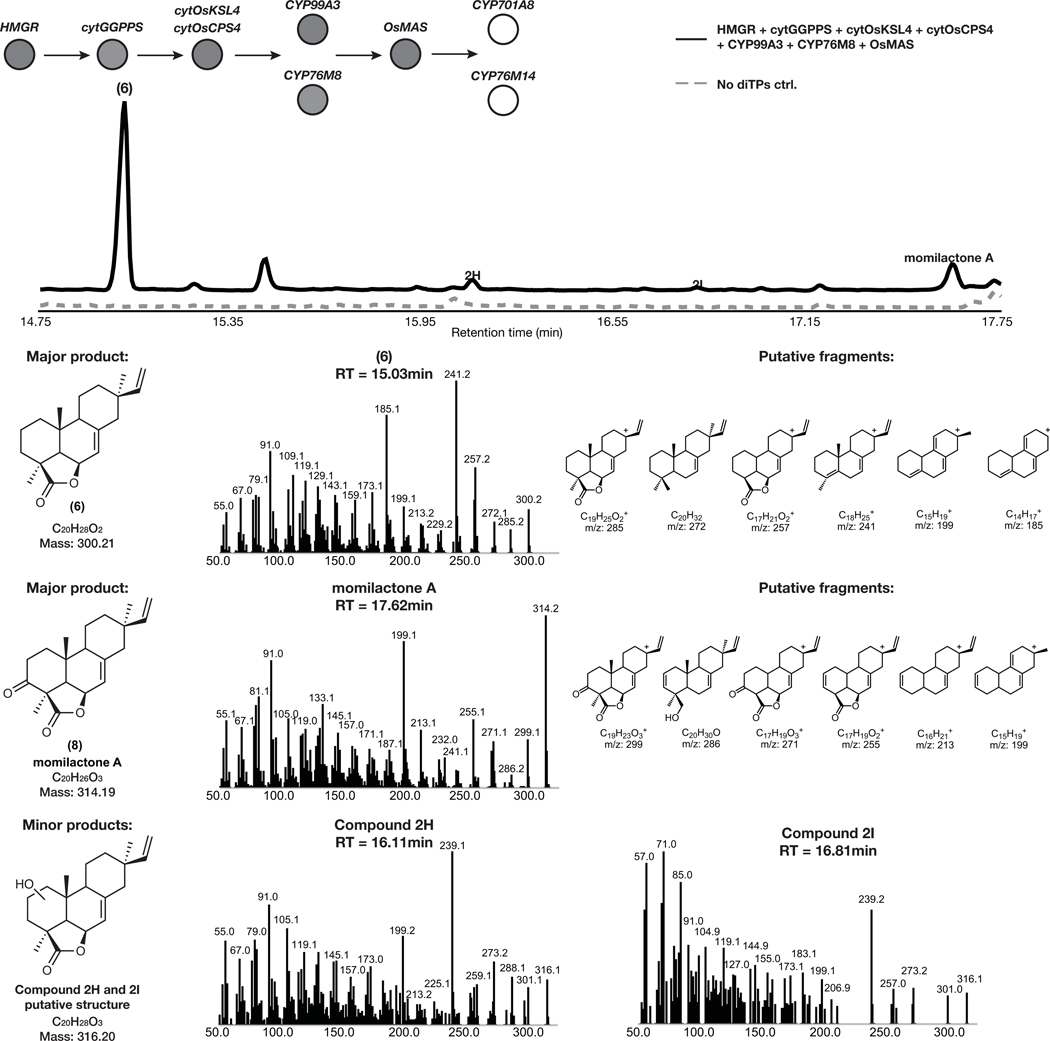
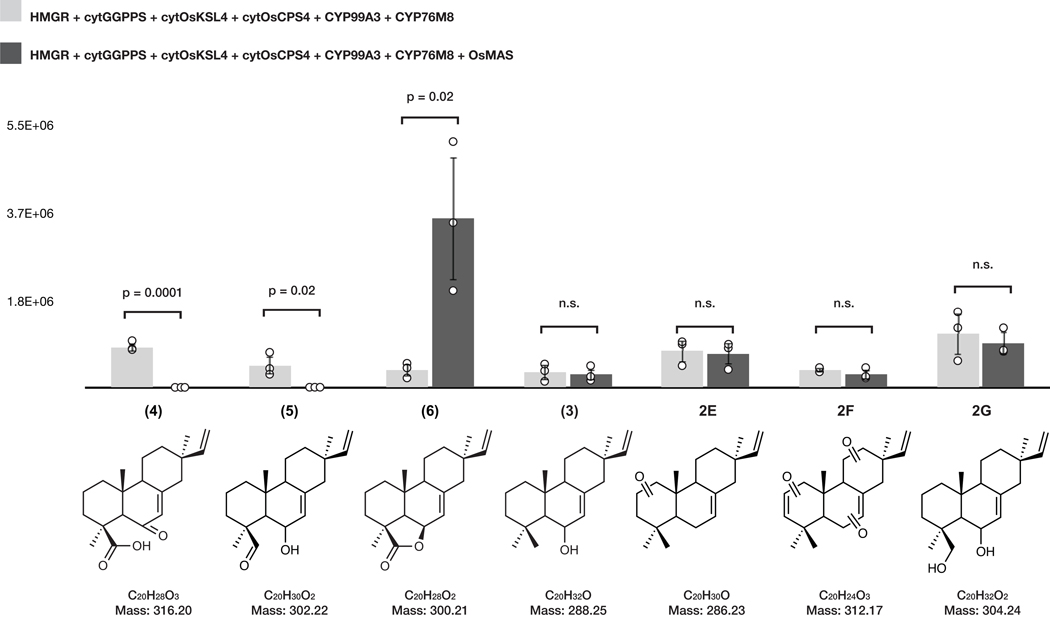
Given that 2 is the most-oxidized pathway intermediate, we first screened candidates for conversion of 2 to a novel metabolite. Each candidate gene was tested by transient expression in N. benthamiana leaves in combination with cytOsCPS4 and cytOsKSL4, HMGR, cytGGPPS, and CYP99A3. Leaf extracts were analyzed using GC-MS for consumption of 2 and the production of any new oxidized product. Our screen revealed that the enzyme CYP76M8, previously shown to hydroxylate 1 to 3, resulted in the reduction of the levels of 2 and accumulation of multiple diterpene products with varying degrees of oxidation (4 and 5) (Extended Data Fig. 4 and Extended Data Fig. 5a)30. The accumulation of multiple oxidized intermediates may represent overoxidation of accumulating reactive intermediates by endogenous N. benthamiana in the absence of a complete biosynthetic pathway. Further candidate testing revealed that transient expression of OsMAS in combination with cytOsCPS4 and cytOsKSL4, HMGR, cytGGPPS, CYP99A3, and CYP76M8 in N. benthamiana leaves was necessary for the production of the lactone 6. (Fig. 4a,b and Extended Data Figs. 5b and 6). Addition of OsMAS resulted in decreased levels of 4 and 5, suggesting that these scaffolds could be either true pathway intermediates or derivatives of pathway intermediates (Extended Data Fig. 7). Finally, additional screening in combination with cytOsCPS4 and cytOsKSL4, HMGR, cytGGPPS, CYP99A3, CYP76M8, and OsMAS revealed that two additional CYP genes, CYP701A8 and CYP76M14, encode enzymes that could respectively oxidize 6 to the C3 ketone 8, and to the C20 hydroxy 7 (Fig. 4a,b and Extended Data Fig. 8a). CYP701A8 has previously been shown to oxidize 1 to syn-pimaradien-3-ol (9)31. Momilactone B (15) was produced upon co-expression of both CYP701A8 and CYP76M14 (Fig. 4a,b and Extended Data Fig. 8b). Formation of the hemiacetal ring characteristic of momilactone B could occur through nucleophilic attack of the C20 alcohol at C3 to form the ring. These results demonstrate complete reconstitution of momilactone biosynthesis in N. benthamiana and the complete transfer of an allelopathic pathway from one plant to another.
Fig. 4: Complete reconstitution of momilactone biosynthesis in Nicotiana benthamiana.
(a) Proposed momilactone biosynthetic pathway. (b) Average GC-MS ion abundances of major intermediates of the momilactone pathway after overexpression of indicated genes via cytosolic engineering. Values and error bars represent the mean and the standard deviation, n=3 biological independent leaf samples examined over three independent experiments.
Isolation and characterization of momilactone B
With a pathway established in N. benthamiana, we scaled up production and isolated substantial quantities of momilactone B for structural characterization. Our scale-up allowed for the recovery of 3.6 mg of momilactone B from 80 infiltrated tobacco plants for an isolated yield of 16.7 μg/g of fresh weight (167 μg/g of dry weight) of leaves (Supplementary Table 1). We confirmed that the isolated product matched previous reports via NMR (Supplementary Figures 11, 12, and 13). Compared to the native host, where yields of momilactone B range between 0.75–37.8 μg/g in dry weight, access of momilactone B via N. benthamiana is a robust and improved method for product isolation given a 5–220-fold yield improvement10–12 (Supplementary Table 2).
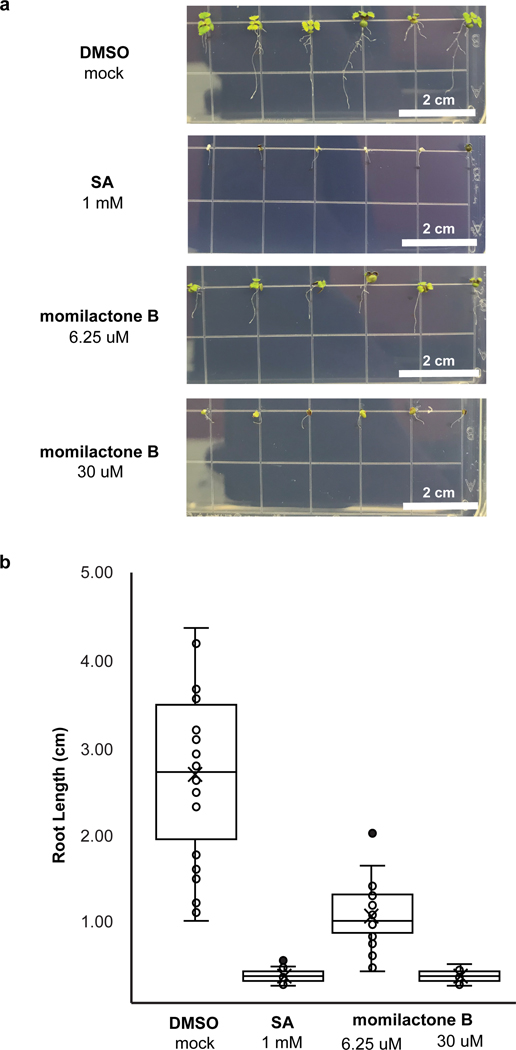
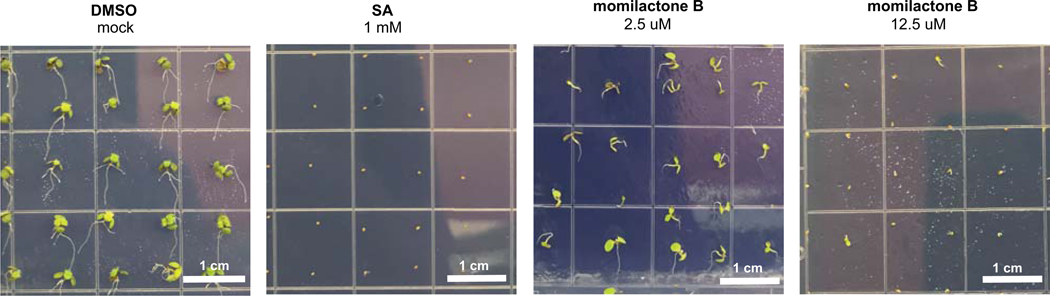
Although the allelopathic activity of momilactone B has been reported, as a proof of concept for the transfer of allelopathic activity from one plant to another, we confirmed that the compound produced in N. benthamiana could inhibit seed germination and plant growth4–8. We monitored germination efficiency of Arabidopsis thaliana seeds in the presence of momilactone B over 10 days, and observed delayed germination of seeds at 2.5 μM and 12.5 μM and complete inhibition at 60 μM compared to a mock DMSO control (Extended Data Fig. 9 and Supplementary Table 3). We then monitored root growth inhibition of Arabidopsis seedlings in the presence of momilactone B. Root growth was impaired at 6.25 μM and growth was completely stunned at 30 μM (Fig. 5). Our results demonstrate that momilactone B can be accessed through this general in planta engineering strategy, and suggest that other complex diterpenoid intermediates, many which do not accumulate in the native plant, could be accessed for rapid structural characterization and assay of biological activities.
Fig. 5. Momilactone B inhibits A. thaliana seed root growth.
(a) Phenotype of 10-day old A. thaliana seedlings grown on MS medium plates with different momilactone B concentrations. A. thaliana seeds were first germinated on MS medium plates. At day 3, seedlings were transferred to MS medium plates with varying momilactone B concentration. (b) Root length of A. thaliana seedlings. Box plot values correspond to n = 28 biological independent samples examined over two independent experiments. Middle line is the median and the mean is marked with an x. Lower and upper hinges correspond to the first (Q1) and third (Q3) quartiles, The lower whisker extends to Q1 – 1.5 x IQR (where IQR is the interquartile range) and the upper whisker extends to Q3 + 1.5 x IQR. Outliers are shown as solid data points.
Discussion
Our results demonstrate that production of substantial amounts of momilactones which, to date, has not been accessible using microbial hosts, is possible through Agrobacterium-mediated transient expression in N. benthamiana. Compared to microbial hosts, metabolic engineering in N. benthamiana provides an optimal environment for plant mRNA processing and for protein folding, sorting, splicing and localization. Precursor supply, intermediates from primary metabolism, and enzyme cofactors required to support plant biosynthetic pathways are also plentiful in a plant chassis. Furthermore, tobacco expresses reductase partners required by CYPs, enzymes that catalyze the oxidation of the vast majority of diterpenoid scaffolds. Engineering and discovery of pathways is also facilitated by combinatorial expression of candidate enzymes where multiple Agrobacterium strains, each containing a different construct, can be co-infiltrated in bulk to rapidly screen candidate genes and build metabolic pathways44. Our work highlights the utility of a plant host for rapid reconstitution of long, complex metabolic pathways.
Despite the advantages of a plant host for pathway discovery, background metabolism can interfere with engineered pathways due to increased genetic and metabolic complexity relative to microbial hosts. For instance, N. benthamiana contains approximately 90 times more CYPs than S. cerevisiae. Specialized plant CYPs involved in native diterpenoid metabolism in N. benthamiana could interfere in pathway reconstitution and shunt intermediates into off-pathway products. Likewise, given the modularity of diterpene biosynthesis, undesired diterpenoid scaffolds could be generated by a native Class I synthase with a non-native Class II synthase or vice-versa41. This possibility is highlighted by the low but detectable production of 8 and momilactone B we observe without co-expression of CYP701A8 (Fig. 4b). This suggests that a N. benthamiana CYP701A8 homolog or related enzyme could oxidize the momilactone scaffold at the C3 position; however, activity from other overexpressed pathway CYPs cannot be ruled out.
Similar to other in vivo platforms, pathway reconstitution using N. benthamiana does not reveal the precise order of biosynthetic steps when multiple tailoring reactions occur upon addition of an enzyme to a multi-step pathway. In the context of our work, addition of CYP701A8 suggests that this enzyme is responsible for the two-step oxidation of 6 to the C3 carbonyl 8. However, previously reported activities of CYP701A8 and OsMAS would in fact support that CYP701A8 only catalyzes the one-step oxidation to the hydroxyl, and subsequently OsMAS oxidizes this product to the carbonyl 8 (refs. 31,32). Likewise, the order of events involved in the biosynthesis of 4 and 5 formed by CYP99A3 and CYP76M8 is unclear given that CYP99A3 and CYP76M8 have only been shown to produce either 2 or 3 (refs. 29,30). Although we provide a direct route for biosynthesis and isolation of momilactones, our platform cannot differentiate whether CYP99A3 or CYP76M8 is involved in the presumed conversion of the C6 alcohol to the C6 carbonyl, or if possible interference of native N. benthamiana metabolism is responsible for said oxidation. In vitro enzyme assays and metabolic profiling of rice KO lines with this set of enzymes would ultimately provide more concrete proof of the main pathway towards momilactones.
Plants synthesize terpenoids via two distinctly compartmentalized, endogenous pathways. Thus, the capability of metabolic engineering in plants is rich. Numerous approaches have been established to increase terpene yields using transient expression in N. benthamiana. Strategies include expression of rate-limiting enzymes, localization of enzymes to various plant compartments, and addition of terpene synthase transcription factors45–47. To date, the highest levels of a terpenoid engineered in planta have been reported via transient overexpression of a truncated HMGR and a β-amyrin synthase34. However, metabolic flux analysis in plant glandular trichomes, specialized cells involved in the biosynthesis and secretion of volatile terpenes, suggest that the limit of terpene biosynthetic capacity in both the MEV and DXP pathways remains unexploited48,49. Accumulation of high titers of steviol glycosides in Stevia rebaudiana, which are reported to constitute up to 20% of the plant’s mass, suggest untapped potential for further enhancements in plant hosts50.
The lack of elucidated plant biosynthetic pathways hinders our capabilities to combat agricultural losses to insects and weeds. Nonetheless, advances in pathway discovery and metabolic engineering are small, positive steps towards mitigating pest problems. Engineering tools focusing on transferring herbicidal diterpene pathways for in situ production will be needed given our current limitations to transfer only a few genes from one plant to a few number of plant hosts. Better tools will be required to expand host range, to increase the length of genes transferred, and for spatial and temporal tuning. The stable engineering of a long pathway in another crop will also require understanding of a natural product’s biological target and mechanism of action. Likely, non-biosynthetic genes, such as transporters, transcription factors and/or resistance genes, may need to be engineered to diminish allelopathic susceptibility in the case of a transgenic plant producing momilactones. If present, gene editing and protein engineering may be required to remodel a native biological target in the host plant to confer resistance. Despite these difficult challenges, the transferring of allelopathic pathways, and generally, multi-step biosynthetic pathways between plants will open exciting frontiers in agriculture.
Online methods
Cloning of biosynthetic genes
mRNA from Arabidopsis thaliana 7-day old seedlings was isolated using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) according to the manufacturer’s instructions. cDNA was generated using Super Script IV First Strand Synthesis System (Invitrogen). Gene sequences for HMGR, DXS, and RuBisCo were amplified by PCR using Q5® High-Fidelity DNA Polymerase and gene specific primers form Integrated DNA Technologies (Supplementary Table 4)51,52. All other gene sequences were synthesized by Gen9, Inc and PCR amplified as described above (Supplementary Table 5). Gibson assembly was used to insert PCR amplicons into AgeI and XhoI (New England Biolabs) linearized pEAQ-HT vector53. All synthesized genes were designed with 5’ pEAQ-HT vector overlaps necessary for Gibson assembly. Constructs were transformed into either E. coli Top10 chemically competent cells (Invitrogen) or E. coli 5-alpha chemically competent cells (New England Biolabs). Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit (Qiagen). Sequence confirmation was carried using Sanger DNA sequencing (Elim Biopharm).
Cloning of fusion proteins
All diterpene synthases and GGPPS sequences were analyzed using Localizer and ChloroP 1.1 Predictor Server and LOCALIZER for the presence of a chloroplast transit peptide54,55. N-terminal truncation variants were designed according to the predictor algorithm’s results (Supplementary Table 6). GFP was C-terminally fused to the sequence of interest through Gibson assembly and constructs were transformed into pEAQ-HT vector as described above. A peptide linker consisting of Gly-Ser-Gly-Ser was used to join the protein of interest to GFP.
Plant growth conditions
Nicotiana benthamiana plants were grown at room temperature under a 16 h light cycle and watered bi-weekly using 2 g/L of fertilizer (Peters Excel® 15–5-15). Small-scale experiments were carried using 5 week old plants. Large-scale experiments for product isolation were carried using 5 – 7 week old plants.
Heterologous expression of genes of interest in N. benthamiana via agroinfiltration
pEAQ-HT plasmids containing a gene of interest were independently transformed into chemically competent Agrobacterium tumefaciens cells (strain GV3101) using the freeze-thaw method. Selection was carried at 30°C for 2–3 days using LB agar plates containing 50 and 30 μg/mL of kanamycin and gentamicin, respectively. Single colonies were then picked, sequenced, and re-streaked on a fresh plate for 2 days. Patches of cells were then scraped off, resuspended in 500 μL of LB, and centrifuged at 5,000 g for 5 min. The supernatant was discarded, the pellet was resuspended in 1 mL of Agrobacterium induction media (10 mM MES pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone; Acros Organics) and incubated for 2 hours at room temperature. A. tumefaciens suspensions at OD600 = 0.15 (per strain) were prepared by diluting strains with Agrobacterium induction media. For small scale experiments, the underside of N. benthamiana leaves was infiltrated using a needleless 1 mL syringe. For large scale experiments involved in product isolation, whole plants were infiltrated under vacuum. Leaves from different plants (n=3 or n=9) were collected either 5 days post-infiltration (small scale) or 8 days post-infiltration (large scale) and immediately extracted. All small scale experiments were repeated three times with similar results.
Metabolite extraction of N. benthamiana leaves
Leaf tissue was collected using a 1 cm DIA leaf disc cutter. Each biological replicate consisted of 4 leaf discs from the same leaf (approx. 0.04 g FW leaves). Discs were placed inside a 2 mL safe-lock microcentrifuge. 500 μL of ethyl acetate (EtOAc) (Fisher Scientific, ACS & HPLC grade) was added to each sample and these were then homogenized in a ball mill using 5 mm stainless steel beads a ball mill at 25 Hz for 2 min (Retsch MM 400). Samples were placed on a shaker and incubated at room temperature for 2 hours at 300 rpm. After incubation, the samples were briefly centrifuged. Without disturbing the pellet, 450 μL of extract were transferred to a new microcentrifuge tube and evaporated to dryness under N2. The samples were reconstituted in 45 μL of EtOAc and transferred to appropriate vials before GC-MS analysis.
GC-MS analysis of leaf extracts and standards
Samples were analyzed using an Agilent 7820A gas chromatogram coupled to an Agilent 5977B mass spectrometer. The samples were analyzed on an Agilent VF-5HT column (30 m x 0.25 mm x 0.1 μm) using a constant flow of 1 mL/min of helium. The inlet was set at 280°C in split mode with a 10:1 split ratio. The injection volume was set to 1 μL. Oven conditions were as follows: 130°C for 2 min, ramp to 250°C at 8°C/min, ramp to 310°C at 10°C/min and hold for 5 min. The MSD transfer line was set to 250°C, the MS Source was set to 230°C, and the MS Quad was set to 150°C. MS data was collected using Scan mode from 50 −550 m/z with a 4 min solvent delay. Geranylgeraniol was purchased from Santa Cruz Biotechnology (CAS 24034–73-9). Forskolin was purchased from Fisher Scientific (CAS 66575–29-9).
Large scale extraction and isolation of momilactone B
216 g of leaves from 80 infiltrated plants were cut into small pieces of approx. 0.25 cm2 in area. Leaves were placed in a 4 L flask with a magnetic stir bar and extracted using 2.7 L of EtOAc for 48 h at room temperature with constant stirring (1 g FW leaves per 12.5 mL). Extracts were filtered using vacuum filtration and dried using rotary evaporation. Flash chromatography was performed using a 7 cm DIA column loaded with silica (SiliaFlash® P60). Hexanes (Fisher Scientific, ACS & HPLC grade) and EtOAc were used as the running solvents. 500 mL fractions were collected via isocratic elution (83% hexanes, 17% EtOAc). Fractions were analyzed via GC-MS, and those containing the compound of interest, were pooled and dried using rotary evaporation. Further flash chromatography followed using a 5.9 cm DIA column loaded with silica. 14mL fractions were collected via isocratic elution (71% hexanes, 29% EtOAc). GC-MS was used to identify the fractions containing the compound of interest. These were then pooled and dried. Additional flash chromatography was performed using a 3 cm DIA column loaded with silica. 8mL fractions were collected using 90% DCM (Fisher Scientific, ACS & HPLC grade) and 10% EtOAc. Fractions containing the compound of interest were identified via GC-MS. Fractions were then pooled, dried, and characterized using 1H NMR and 13C NMR. A standard curve based on GC-MS ionization was prepared using the isolated material for yield quantification. The amount of momilactone B recovered was 3.6 mg.
NMR analysis
1H NMR and 13C NMR spectra were acquired using a Varian Inova 600 MHz spectrometer at room temperature. Shifts are referenced to the residual solvent peak (CDCl3, Acros Organics) and reported downfield in ppm using Me4Si as the 0.0 ppm internal reference standard.
Image acquisition for subcellular localization analysis using confocal microscopy
Subcellular localization of fusion proteins in N. benthamiana was determined using a Leica TCS SP8 laser scanning confocal microscope in resonant scanning mode using LASX software. Cells were imaged at room temperature using a 63 × 1.4 oil objective. GFP fluorescence was imaged by exciting with a white laser set at 488 nm. Emission was collected from 500 – 550 nm on a HyD SMD hybrid detector. A lifetime gate filter of 1 – 8 ns was applied to reduce background chloroplast autofluorescence. Chloroplast spectra were simultaneously imaged using the same emission excitation. Emission was collected from 650 – 700 nm on a HyD SMD hybrid detector. Images were collected using 8-line averaging. Images shown are representative of three biological replicates. Experiments were repeated two times with similar results. Images were analyzed using Fiji – ImageJ (Version 1.0).
Transcriptome data mining and analysis of Oryza sativa dataset
Publicly available gene expression data from Oryza sativa roots subjected to fungal (Magnaporthe oryzae) elicitation was downloaded from the Gene Expression Omnibus (GEO) from NCBI with GEO Accession: GSE1836156. The dataset consisted of expression data collected from roots infected with pathogen and a mock at 2, 4 and 6 days after elicitation. All expression data for replicates was averaged and log2-scaled. Fold changes between mock at 2 dpi and the five other conditions were calculated. Linear regression analysis to calculate Pearson’s R coefficient on the fold changes was preformed using OsCPS4, OsKSL4, CYP99A3, CYP99A2 and OsMAS as bait genes. The obtained list was ranked by decreasing Pearson’s R coefficient. The top 100 microarray probes (per bait gene list) were then mapped to the respective O. sativa genes. Candidate genes were then annotated both via Pfam assignment and via the best blastx hit using the Arabidopsis thaliana proteome as a reference. The final list of candidates was further refined to only include candidates with Pfam assignments belonging to CYP, 2-ODD, SDR, and terpene synthase families (Supplementary Table 1, Supplementary Table 5 and Supplementary Table 7).
Arabidopsis thaliana seed sterilization and stratification
Approximately 100 μL of Arabidopsis thaliana ecotype Col 0 seeds were placed in a 1.5 mL Eppendorf tube and sterilized using 1 mL of 70% (v/v) EtOH. The seeds were gently inverted by hand for 2 min. The remaining EtOH was discarded, and the seeds were then sterilized using 1 mL of 50% (v/v) bleach, 0.025% Tween 20 (v/v) solution. The tube was gently inverted by hand for 12 min. The remaining bleach solutions was discarded and the seeds were washed 8 times using 1 mL of sterile H2O. The seeds were kept in the dark at 4°C for 2 days for stratification. Sterilization steps were conducted in a laminar flow hood.
Arabidopsis thaliana germination inhibition assay
All Arabidopsis inhibition assays were carried using 1x Murashige and Skoog (MS) medium with vitamins plates (PhytoTechnology Laboratories) containing 1.5% (w/v) agar at pH 5.7. Momilactone B was added to the media after autoclaving and subsequent cooling to approximately 45°C and added at the desired final concentration (2.5 μM, 12.5 μM or 60 μM) using a 12.5 mM stock solution in DMSO. Salicylic acid (Sigma-Aldrich CAS 69–72-7) was added at a final concentration of 1 mM. When necessary, equivalent amounts of DMSO were added to plates to control for differences in DMSO concentration. 100 seeds were transferred to each plate and arranged in a 10 by 10 grid with equal spacing between seeds. Plates were grown vertically in a growth chamber at 50% humidity, 22°C and photon flux (100 μmol/m2 per s) under a 16 h light / 8 h dark cycle. Seed germination was monitored for 10 days. Germination was recorded when the radicle and cotyledon were visible. Experiments were conducted as biological duplicates.
Arabidopsis thaliana root growth inhibition assay
A. thaliana (Col 0) seeds were germinated and grown vertically for 3 days in a growth chamber at 50% humidity, 22°C and photon flux (100 μmol/m2 per s) under a 16 h light / 8 h dark cycle in 1x MS media plates (1.5% (w/v) agar, pH 5.7). Seedlings were transferred to 1x MS media plates (1.5% (w/v) agar, pH 5.7) containing the desired amount of DMSO, momilactone B (6.25 μM and 30 μM) or salicylic acid (1 mM) under sterile conditions. 14 seedlings were aligned into two equally distributed rows of seven seedlings. Plates were grown vertically under the same growth conditions previously described. Seedlings were monitored for 10 days. Changes in root length were analyzed using Fiji – ImageJ (Version 1.0). Experiments were conducted as biological duplicates.
Statistical analysis
Statistical analysis was performed in Microsoft Excel 2018.
Data availability statement
Data supporting the findings of this study are presented in the published article (including its Supplementary Information) or are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Fig. 1. Characterization of pathway intermediates from overexpression of cytOsCPS4 + cytOsKSL4 via MEV engineering.
GC-MS total ion chromatogram (TIC) and MS spectra of major and minor pathway products from overexpression of HMGR, cytGGPPS, cytOsCPS4, and cytOsKSL4 in N. benthamiana. Representative TICs are shown for the experimental sample and GFP control. Three total products with m/z corresponding to diterpene derived scaffolds were observed only upon expression of pathway genes. The major product identified corresponded to 1 and putative structures for two minor products were hypothesized. For m/z corresponding to syn-copalyl alcohol see Supplemental Figure 9. Putative ion structures for the major product were assigned as reference. Putative structures are preliminary and based on predicted chemical formulas combined with analysis of the fragment ions observed in the MS spectra.
Extended Data Fig. 2. Characterization of pathway intermediates from overexpression of cytOsCPS4 + cytOsKSL4 + CYP99A3 via MEV engineering.
GC-MS total ion chromatogram (TIC) and MS spectra of products from overexpression of HMGR, cytGGPPS, cytOsCPS4, cytOsKSL4, and CYP99A3 in N. benthamiana. Representative TICs are shown for the experimental sample and controls. Three products with m/z corresponding to diterpene derived scaffolds were observed only upon expression of pathway genes. The major product identified corresponded to 2 and the two minor products corresponded to syn-pimaradien-19-al and syn-pimaradien-19-ol29. For m/z corresponding to syn-copalyl alcohol see Supplementary Figure 9. Putative ion structures for 2 were assigned as reference. Putative structures are preliminary and based on predicted chemical formulas combined with analysis of the fragment ions observed in the MS spectra. An endogenous metabolite co-eluting with 2 was identified (* in the TIC). (b) MS spectra of endogenous metabolite. (c) GC-MS extracted ion chromatogram (EIC) using m/z = 241.2, corresponding to an abundant fragment found in the MS of 2, confirmed that 2 is only present upon expression of the requisite diTPSs and CYP99A3.
Extended Data Fig. 3. Characterization of pathway intermediates from overexpression of cytOsCPS4 + cytOsKSL4 + CYP76M8 via MEV engineering.
GC-MS total ion chromatogram (TIC) and MS spectra of major and minor pathway products from overexpression of HMGR, cytGGPPS, cytOsCPS4, cytOsKSL4, and CYP76M8 in N. benthamiana. Representative TICs are shown for the experimental sample and the no diterpene synthase control. Three total products with m/z corresponding to diterpene derived scaffolds were observed only upon expression of pathway genes. The major product identified corresponded to syn-pimaradien-6-ol and putative structures for two minor products were hypothesized (For syn-copalyl alcohol see Supplemental Figure 9, for *, corresponding to an endogenous metabolite, see Extended Data Figure 2). Putative ion structures for the major product were assigned as reference. Putative structures are preliminary and based on predicted chemical formulas combined with analysis of the fragment ions observed in the MS spectra.
Extended Data Fig. 4. Characterization of pathway intermediates from overexpression of cytOsCPS4 + cytOsKSL4 + CYP99A3 + CYP76M8 via MEV engineering.
GC-MS total ion chromatogram (TIC) and MS spectra of products from overexpression of HMGR, cytGGPPS, cytOsCPS4, cytOsKSL4, CYP99A3, and CYP76M8 in N. benthamiana. Representative TICs are shown for the experimental sample and controls. Products with m/z corresponding to diterpene derived scaffolds were observed only upon expression of pathway genes. Putative ion structures for 4, 5, and 2G were assigned as reference. Putative structures are preliminary and based on predicted formulas combined with analysis of the fragment ions observed in the MS spectra.
Extended Data Fig. 5. Oxidation of syn-pimaradiene scaffold via overexpression of various tailoring oxidases.
(a) Oxidation of syn-pimaradiene can proceed via overexpression of either CYP99A3 or CYP76M8. n=3 biological independent leaf samples examined over three independent experiments. (b) Overexpression of OsMAS, CYP99A3 and CYP76M8 is required for biosynthesis of the lactone ring in 6 and 8. Average GC-MS ion abundances of oxidized syn-pimaradiene scaffolds after overexpression of indicated genes via cytosolic engineering. Values and error bars represent the mean and the standard deviation of biological triplicates. n=3 biological independent leaf samples examined over three independent experiments.
Extended Data Fig. 6. Characterization of pathway intermediates from overexpression of cytOsCPS4 + cytOsKSL4 + CYP99A3 + CYP76M8 + OsMAS via MEV engineering.
GC-MS total ion chromatogram (TIC) and MS spectra of major and minor pathway products from overexpression of HMGR, cytGGPPS, cytOsCPS4, cytOsKSL4, CYP99A3, CYP76M8, and OsMAS in N. benthamiana. Representative TICs are shown for the experimental sample and the no diterpene synthase control. Four total products with m/z corresponding to diterpene derived scaffolds were observed only upon expression of pathway genes. Two major products identified corresponded to 6 and 8, and putative structures for two minor products were hypothesized. Putative ion structures for the major product were assigned as reference. Putative structures are preliminary and based on predicted chemical formulas combined with analysis of the fragment ions observed in the MS spectra.
Extended Data Fig. 7. Expression of OsMAS results in the reduction of levels of several oxidized syn-pimaradiene scaffolds.
Average GC-MS ion abundances of oxidized syn-pimaradiene intermediates after overexpression of indicated genes via cytosolic engineering. Co-expression with OsMAS results in reduction of levels of 4, 5, and increase in levels of 6. Values and error bars represent the mean and the standard deviation of biological triplicates. n=3 biological independent leaf samples examined over three independent experiments. P values were calculated using unpaired, two-tailed t-tests.
Extended Data Fig. 8. Characterization of pathway intermediates from overexpression of cytOsCPS4 + cytOsKSL4 + CYP99A3 + CYP76M8 + OsMAS + CYP76M14 + CYP701A8 via MEV engineering.
GC-MS total ion chromatogram (TIC) and MS spectra of products from overexpression of HMGR, cytGGPPS, cytOsCPS4, cytOsKSL4, CYP99A3, CYP76M8, OsMAS, CYP76M14, and CYP701A8 in N. benthamiana. Representative TICs are shown for the experimental sample and control. (a) Two products with m/z corresponding to diterpene derived scaffolds were observed only upon expression of pathway genes. The products identified corresponded to momilactone B and 7. Putative ion structures were assigned as reference. Structures of 7 is preliminary and based on a predicted chemical formula combined with analysis of the fragment ions observed in the MS spectra. (b) The major product identified corresponded to momilactone B (See Supplementary Figure 11 and 12 for structural characterization).
Extended Data Fig. 9. Momilactone B inhibits A. thaliana seed germination.
(a) Phenotype of A. thaliana seeds after 10 days of incubation in MS medium plates with different momilactone B concentrations. SA, salicylic acid.
Supplementary Material
Acknowledgements
This work was supported by a National Institutes of Health (NIH) U01 Grant 110699 and NIH R01 Grant 121527. We thank C. Liou and K. Smith for discussions and feedback with the manuscript. We thank E. Holmes, Y. Chen and J. Kim for help in developing Arabidopsis inhibition assays, H. Cartwright at the Advanced Imaging Facility in the Carnegie Institution for Science – Department of Plant Biology for training and assistance with confocal microscopy imaging, C. Miller for help with confocal imagining analysis and K. Smith and S. Kim for help with flash column purification and NMR analysis.
Footnotes
Competing financial interest statement
Authors declare no competing financial interests.
References
- 1.Benbrook CM Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 28, 3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farooq M, Jabran K, Cheema ZA, Wahid A. & Siddique KHM The role of allelopathy in agricultural pest management. Pest Manag Sci 67, 493–506 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Kato-Noguchi H. & Peters RJ The role of momilactones in rice allelopathy. J. Chem. Ecol. 39, 175–185 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Kato-Noguchi H, Hasegawa M, Ino T, Ota K. & Kujime H. Contribution of momilactone A and B to rice allelopathy. J. Plant Physiol. 167, 787–791 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Kato-Noguchi H, Ino T, Sata N. & Yamamura S. Isolation and identification of a potent allelopathic substance in rice root exudates. Physiol. Plant. 115, 401–405 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Kato-Noguchi H, Ota K. & Kujime H. Absorption of momilactone A and B by Arabidopsis thaliana L. and the growth inhibitory effects. J. Plant Physiol. 169, 1471–1476 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Toyomasu T. et al. Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Biosci. Biotechnol. Biochem. 72, 562–567 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Chung I-M, Hahn S-J & Ahmad A. Confirmation of potential herbicidal agents in hulls of rice, Oryza sativa. J. Chem. Ecol. 31, 1339–1352 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Jabran K. & Farooq M. in Allelopathy (eds. Cheema ZA, Farooq M. & Wahid A) 349–385 (Springer Berlin Heidelberg, 2013). doi: 10.1007/978-3-642-30595-5_15 [DOI] [Google Scholar]
- 10.Kato T. et al. Momilactones, growth inhibitors from rice, oryza sativa L. Tetrahedron Lett. 14, 3861–3864 (1973). [Google Scholar]
- 11.Chung IM, Kim JT & Kim S-H Evaluation of allelopathic potential and quantification of momilactone A,B from rice hull extracts and assessment of inhibitory bioactivity on paddy field weeds. J. Agric. Food Chem. 54, 2527–2536 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Quan NV et al. Momilactones A and B Are α-Amylase and α-Glucosidase Inhibitors. Molecules 24, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paddon CJ et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Li Y. et al. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. USA 115, E3922–E3931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown S, Clastre M, Courdavault V. & O’Connor SE De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. USA 112, 3205–3210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Y. et al. Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. USA 112, 6224–6229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo X. et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567, 123–126 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Paine JA et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 23, 482–487 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Lau W. & Sattely ES Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349, 1224–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pateraki I. et al. Total biosynthesis of the cyclic AMP booster forskolin from Coleus forskohlii. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caputi L. et al. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science 360, 1235–1239 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Hodgson H. et al. Identification of key enzymes responsible for protolimonoid biosynthesis in plants: Opening the door to azadirachtin production. Proc. Natl. Acad. Sci. USA 116, 17096–17104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polturak G. et al. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 210, 269–283 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Crocoll C, Mirza N, Reichelt M, Gershenzon J. & Halkier BA Optimization of Engineered Production of the Glucoraphanin Precursor Dihomomethionine in Nicotiana benthamiana. Front. Bioeng. Biotechnol. 4, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu M, Hillwig ML, Prisic S, Coates RM & Peters RJ Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39, 309–318 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Otomo K. et al. Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J. 39, 886–893 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Otomo K. et al. Diterpene cyclases responsible for the biosynthesis of phytoalexins, momilactones A, B, and oryzalexins A-F in rice. Biosci. Biotechnol. Biochem. 68, 2001–2006 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Wilderman PR, Xu M, Jin Y, Coates RM & Peters RJ Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135, 2098–2105 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Hillwig ML & Peters RJ CYP99A3: functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 65, 87–95 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q. et al. Characterization of CYP76M5–8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287, 6159–6168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitaoka N, Wu Y, Xu M. & Peters RJ Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl. Microbiol. Biotechnol. 99, 7549–7558 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimura K. et al. Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Henry LK et al. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 4, 721–729 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Reed J. et al. A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab. Eng. 42, 185–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brückner K. & Tissier A. High-level diterpene production by transient expression in Nicotiana benthamiana. Plant Methods 9, 46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Herpen TWJM et al. Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS One 5, e14222 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wendt KU & Schulz GE Isoprenoid biosynthesis: manifold chemistry catalyzed by similar enzymes. Structure 6, 127–133 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Wildung MR & Croteau R. A cDNA clone for taxadiene synthase, the diterpene cyclase that catalyzes the committed step of taxol biosynthesis. J. Biol. Chem. 271, 9201–9204 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Bohlmann J, Meyer-Gauen G. & Croteau R. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95, 4126–4133 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerbe P. & Bohlmann J. Plant diterpene synthases: exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 33, 419–428 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Andersen-Ranberg J. et al. Expanding the Landscape of Diterpene Structural Diversity through Stereochemically Controlled Combinatorial Biosynthesis. Angew. Chem. Int. Ed. Engl. 55, 2142–2146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pateraki I. et al. Manoyl oxide (13R), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii. Plant Physiol. 164, 1222–1236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Hillwig ML, Wu Y. & Peters RJ CYP701A8: a rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 158, 1418–1425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christ B. et al. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 10, 3206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Q. et al. Reconstitution of the costunolide biosynthetic pathway in yeast and Nicotiana benthamiana. PLoS One 6, e23255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickschat JS, Rinkel J, Rabe P, Beyraghdar Kashkooli A. & Bouwmeester HJ 18-Hydroxydolabella-3,7-diene synthase - a diterpene synthase from Chitinophaga pinensis. Beilstein J Org Chem 13, 1770–1780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong L, Jongedijk E, Bouwmeester H. & Van Der Krol A. Monoterpene biosynthesis potential of plant subcellular compartments. New Phytol. 209, 679–690 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Lange BM & Turner GW Terpenoid biosynthesis in trichomes--current status and future opportunities. Plant Biotechnol. J. 11, 2–22 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Tissier A. Trichome Specific Expression: Promoters and Their Applications in Transgenic Plants - Advances and Limitations. (InTech, 2012). doi: 10.5772/1409 [DOI] [Google Scholar]
- 50.Ceunen S. & Geuns JMC Steviol glycosides: chemical diversity, metabolism, and function. J. Nat. Prod. 76, 1201–1228 (2013). [DOI] [PubMed] [Google Scholar]
Methods references
- 51.Learned RM & Fink GR 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Arabidopsis thaliana is structurally distinct from the yeast and animal enzymes. Proc. Natl. Acad. Sci. USA 86, 2779–2783 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Estévez JM, Cantero A, Reindl A, Reichler S. & León P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 276, 22901–22909 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Sainsbury F, Thuenemann EC & Lomonossoff GP. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Emanuelsson O, Nielsen H. & von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sperschneider J. et al. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 7, 44598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marcel S, Sawers R, Oakeley E, Angliker H. & Paszkowski U. Tissue-adapted invasion strategies of the rice blast fungus Magnaporthe oryzae. Plant Cell 22, 3177–3187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar S. et al. Mechanistic Insights into Taxadiene Epoxidation by Taxadiene-5α-Hydroxylase. ACS Chem. Biol. 11, 460–469 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Abidi SL Chromatographic analysis of plant sterols in foods and vegetable oils. J. Chromatogr. A 935, 173–201 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Kim JH, Lee YJ, Kim BG, Lim Y. & Ahn J-H Flavanone 3beta-hydroxylases from rice: key enzymes for favonol and anthocyanin biosynthesis. Mol. Cells 25, 312–316 (2008). [PubMed] [Google Scholar]
- 60.Hefner J, Ketchum RE & Croteau R. Cloning and functional expression of a cDNA encoding geranylgeranyl diphosphate synthase from Taxus canadensis and assessment of the role of this prenyltransferase in cells induced for taxol production. Arch. Biochem. Biophys. 360, 62–74 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are presented in the published article (including its Supplementary Information) or are available from the corresponding author upon reasonable request.