Abstract
G-protein-coupled receptors (GPCRs) are the most important signal transducers in higher eukaryotes. Despite considerable progress, the molecular basis of subtype-specific ligand selectivity, especially for peptide receptors, remains unknown. Here, by integrating DNP-enhanced solid-state NMR spectroscopy with advanced molecular modeling and docking, the mechanism of the subtype selectivity of human bradykinin receptors for their peptide agonists has been resolved. The conserved middle segments of the bound peptides show distinct conformations that result in different presentations of their N and C termini toward their receptors. Analysis of the peptide–receptor interfaces reveals that the charged N-terminal residues of the peptides are mainly selected through electrostatic interactions, whereas the C-terminal segments are recognized via both conformations and interactions. The detailed molecular picture obtained by this approach opens a new gateway for exploring the complex conformational and chemical space of peptides and peptide analogs for designing GPCR subtype-selective biochemical tools and drugs.
GPCRs respond to a wide variety of stimuli, for example photons, amines, ions, peptides, as well as small proteins, and trigger downstream signaling pathways by activating heterotrimeric G proteins1. They form the most important class of signal transducers in higher eukaryotes. In recent years, the structural characterization of GPCRs by X-ray crystallography has contributed to an unparalleled understanding of their molecular architecture and the structural aspects of ligand binding, receptor activation and allosteric modulation2-4. The wealth of newly obtained structural data has created a strong demand for advanced spectroscopy such as solution and solid-state nuclear magnetic resonance (ssNMR) to gain insights into the mechanism of signaling bias, structural plasticity5-7, ligand binding and ligand–receptor interactions8-12.
Despite these major advances in understanding the molecular basis of GPCR signaling, the foundations of subtype selectivity, especially for peptide ligand GPCRs, remains poorly understood, which hampers mechanistic understanding and rational drug design for peptide receptors. GPCR subtypes are closely related receptors with high sequence similarity, but they can differentiate between sets of ligands that are highly similar in structure or sequence by binding to them with substantially different affinities13,14. Recently, subtype selectivity of rhodopsin-like GPCRs has been studied with non-native, small-molecule ligands, revealing rearrangements of the seven transmembrane bundles to confer binding specificity15,16. In the case of peptide ligands, however, this situation becomes more challenging because of their size and inherent complexity.
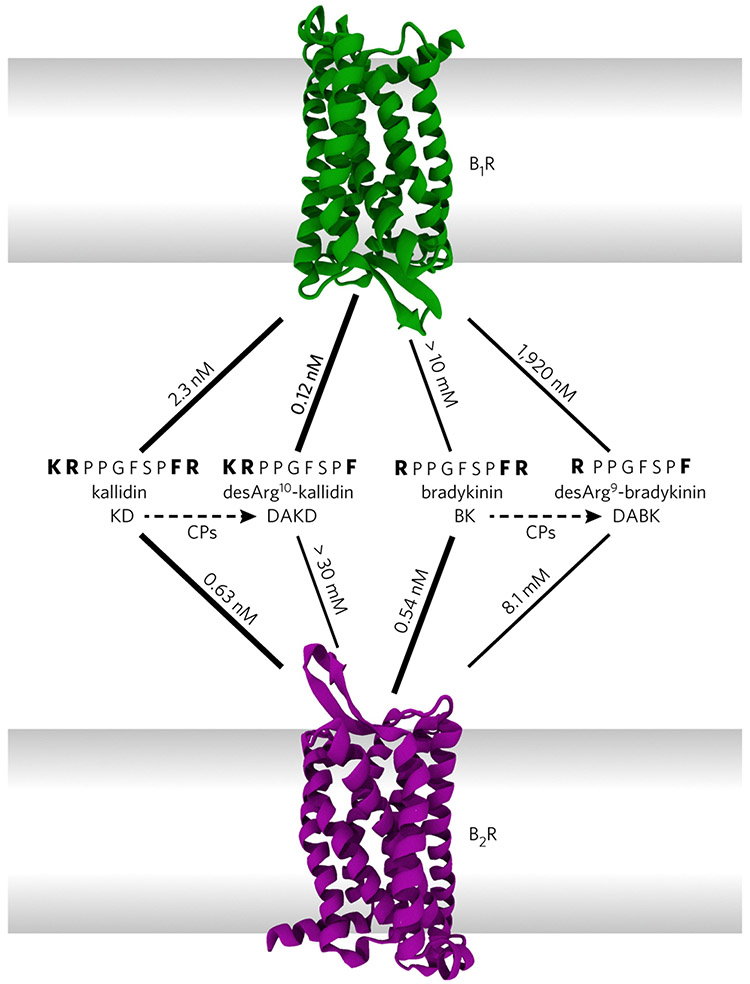
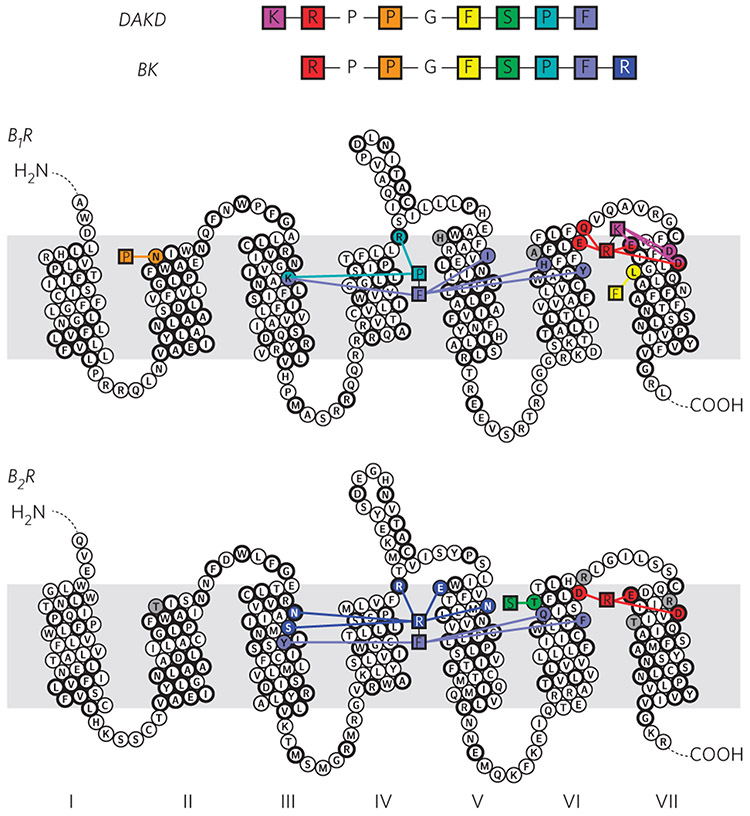
Here, we address the molecular basis of subtype selectivity for kinin peptides by human bradykinin receptors (BRs). The peptides kallidin (KD) and bradykinin (BK) are derived from different kininogen isoforms. KD differs from BK only in the presence of one additional N-terminal lysine residue17 (Fig. 1). Both are high-affinity agonists for the human bradykinin 2 receptor (B2R), which regulates vasodilation, and thereby blood pressure, as well as other cardiovascular functions18. In vivo, carboxypeptidases convert KD and BK into desArg10-kallidin (DAKD) and desArg9-bradykinin (DABK) by removing their C-terminal arginine residues. The resulting peptides display only weak binding affinity to the B2R. However, KD and DAKD bind to the human bradykinin 1 receptor (B1R) as high affinity-agonists and trigger downstream signaling related to inflammation and pain19. In contrast, BK and DABK, which lack the additional N-terminal lysine residue, exhibit rather low affinity to the B1R (Fig. 1). Both receptors share a high overall sequence identity (41%), and it is assumed that the residues forming the peptide-binding pocket of the BRs are highly conserved14. It is therefore puzzling how these receptors differentiate between peptides with high sequence similarity in such a selective manner.
Figure 1 ∣. Affinities of kinin peptides for their respective human bradykinin receptors, B1R and B2R.
Kallidin (KD) and bradykinin (BK) derive from kininogen by proteolytic cascades and differ only by an additional N-terminal lysine residue in KD. Both peptides are high-affinity ligands for B2R. Removal of the C-terminal arginine (dashed lines) by carboxypeptidases (CPs) yields desArg10-kallidin (DAKD) and desArg9-bradykinin (DABK). Despite their similarity, only DAKD, but not DABK, binds with high affinity to B1R13.
In the absence of 3D structures for B1R and B2R, we address this question by comparing structures of bound peptide agonists determined by ssNMR and combining these data with advanced molecular modeling and docking. Because wild-type, non-engineered human B1R can only be prepared in small quantities that are insufficient for conventional NMR studies, we made use of dynamic nuclear polarization (DNP) for enhancing the detection sensitivity of our ssNMR experiments by approximately 100-fold. DNP makes use of unpaired electrons in the form of stable radicals added to the sample as a polarization source to increase the NMR signal (Fig. 2a). DNP-enhanced ssNMR with magic-angle sample spinning (MAS) has just recently emerged as a tool in membrane protein research. The signal enhancements enabled challenging applications suffering from small spin numbers. Examples include the analysis of trapped photointermediate states20,21, visualizing cross-protomer interactions22, ligand-binding studies on mammalian transporter complexes23 or even studies on proteins directly within the cellular context24-26.
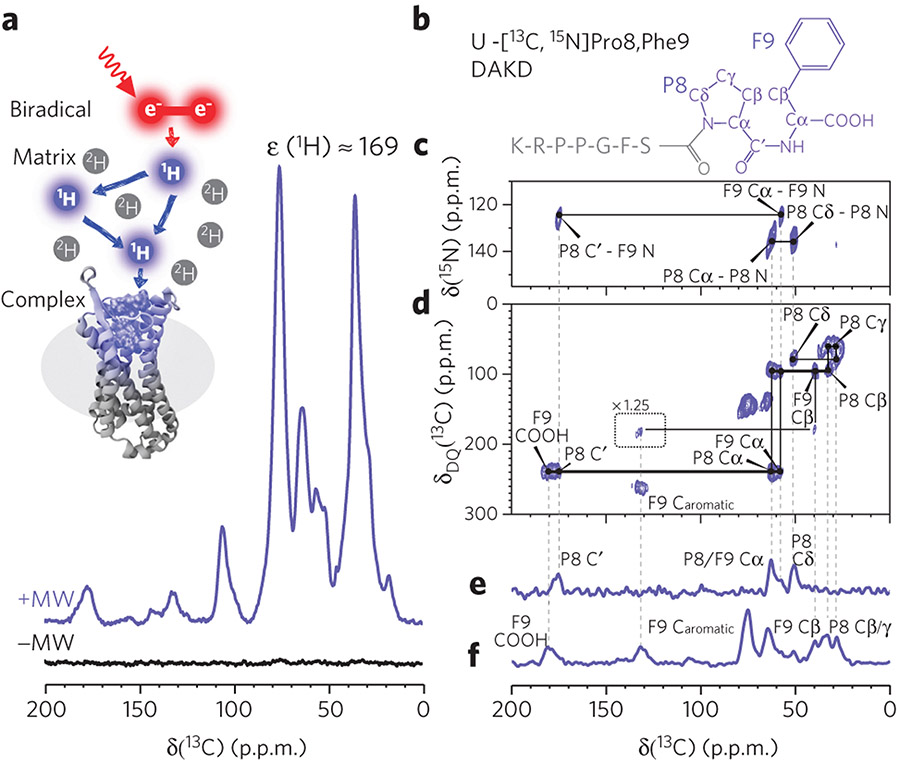
Figure 2 ∣. Experimental setup and exemplary spectra of B1R in complex with DAKD.
(a) A sample containing the DAKD–B1R complex doped with the biradical AMUPol is subjected to magic-angle sample spinning under continuous wave microwave irradiation, resulting in polarization transfer from electrons via protons to the sites of interest. As a result, a large signal enhancement of the B1R–DAKD complex (purple) is observed in comparison to conventional NMR (black) (see Online Methods). (b) One of the DAKD labeling schemes used here: U-[13C,15N]P8F9 DAKD. The chemical shift assignment was accomplished by following the connection of the signals on TEDOR 15N-13C spectra (c), DQ-SQ 13C-13C (d), TEDOR-filtered 13C-spectra (e), and DQ/REDOR doubly filtered 13C-spectra (f). The pulse program used for f is presented in Supplementary Figure 6. Caromatic, carbons on a phenyl ring. Dashed lines guide the chemical shift connectivity among different spectra.
Here, DNP-enhanced ssNMR reveals a substantially different fold of DAKD bound to human B1R in comparison to earlier reported BK bound to human B2R10. The combination of NMR data with advanced docking and modeling enabled a comparative analysis of peptide-binding GPCR interactions. Overall, selectivity is controlled by peptide interactions with nonconserved residues in the binding pockets of B1R and B2R. Our findings show that subtype selectivity in peptide receptors is distinct from that in small ligand receptors and indicate that the subtype selectivity of BRs is the result of multiple chemical and conformational factors, which act together in a complex and synergistic manner.
RESULTS
DNP-enhanced solid-state NMR on DAKD with B1R
Human wild-type BRs, B1R in particular, remain at low expression levels in eukaryotic hosts after intensive optimizations and have limited stability. Our initial attempts to reconstitute the B1R into proteoliposomes or lipid cubic phase failed to yield samples suitable for DNP ssNMR studies because of either the difficulty of controlling protein orientation or phase destruction while cooling the samples to the DNP operating temperature (ca. 100 K). As B1R in 1% n-dodecyl β-d-maltoside (DDM) and 0.1% cholesteryl hemisuccinate (CHS) mixed detergent micelles (Supplementary Fig. 1) shows a high DAKD binding affinity close to that in native membranes, we decided to directly characterize the DAKD–B1R complex in homogeneous solution. This approach allows accessibility of all binding sites and also leads to high DNP signal enhancements (over 100 times; Fig. 2a), substantially better than those achieved on inhomogeneous liposome samples. The cryogenic conditions needed for DNP-enhanced ssNMR experiments also extend the sample lifetime remarkably, permitting time-consuming NMR experiments to be performed on a GPCR-peptide complex of low stability.
To alleviate signal overlap, the complete DAKD sequence was covered by six nonoverlapping isotope labeling schemes, which were designed for the optimized separation of peptide 13C signals based on the characteristic chemical shift dispersions of each site (Supplementary Table 1). To resolve the majority of the DAKD signals, we chose DQ-SQ (double-quantum single-quantum) 13C-13C and TEDOR (transferred-echo double resonance) 15N-13C 2D correlation spectroscopy as the main NMR methods. These experiments have been shown to be suitable for studying membrane proteins under DNP conditions, and both serve as efficient filters for selectively removing the natural abundance 13C signals from the receptor and detergent23. As an example, 13C-13C DQ-SQ and 15N-13C TEDOR spectra of U-[13C,15N]P8F9-DAKD (Fig. 2b) in complex with B1R are shown in Figure 2c,d. Following the characteristic spectral patterns, most of the 13C and 15N signals on these 2D spectra could be assigned unambiguously. The same approach was applied to five other labeled peptide–B1R complexes (Supplementary Figs. 2-5). Remaining overlapping signals were resolved by additional spectroscopic editing and filtering experiments. As shown in Figure 2e,f, the NMR signal of 13C in P8, which is close to and therefore strongly dipolar-coupled to 15N of P8, could be selectively detected in TEDOR-type experiments. On the contrary, the NMR signals of 13C nuclei that are further away from 15N, including the C-terminal carboxylate group, were resolved by applying a REDOR-type filter, which dephases the 13C magnetization that would be built up by TEDOR. Eventually, we unambiguously assigned almost all backbone and side chain 13C and 15N resonances of DAKD in complex with or in the absence of B1R (Supplementary Tables 2 and 3) in DNP-enhanced ssNMR spectra.
By comparing the chemical shifts (Supplementary Table 4) of B1R-bound and free DAKD peptides recorded under the same conditions, we could already identify some interaction areas. The N-terminal residues reveal the most pronounced 15N chemical shift perturbations (Supplementary Fig. 7a-c). The side chain and the N-terminal amine group of K1DAKD show significant up-field shifts for their 15N resonances upon binding to B1R, which would be in line with electrostatic interactions between this group and the receptor. Similarly, the guanidine group of R2DAKD also exhibits a small observable shift in its 15N resonance (Supplementary Fig. 7b), which is also an indicator of similar interactions as for K1DAKD. The binding-induced chemical shift perturbations exhibit a different pattern at the C-terminal residues. The presence of B1R causes a considerable shift of the F9DAKD Cα-N cross-peak in the TEDOR spectrum, which points to a defined conformational change of the C-terminal backbone structure (Supplementary Fig. 7d). We have also monitored this signal in a DAKD analog (DALK, KRPPGKSPL), which differs only at its C-terminal residue and acts as a high-affinity antagonist for B1R. Furthermore, a significant shift of the L9DALK Cα-N peak was detected in the presence of receptor (Supplementary Fig. 8). This finding resembles the observations for DAKD, and therefore suggests that the C-terminal part of these peptides forms a common motif for interaction with the B1R.
The receptor-bound structure of DAKD
Despite the significant signal enhancement provided by DNP, all attempts to record long-range distance restraints were unsuccessful because of the limited coherence lifetime under our experimental conditions. Therefore, the DAKD backbone structure in the B1R–DAKD complex had to be calculated based on the backbone 13C and 15N chemical shifts.
To obtain a converging and verified solution using these sparse data, we calculated the DAKD backbone structure from torsion angle restraints directly predicted from chemical shifts (‘backward’ approach). The obtained solution was examined by an extensive ‘forward’ protocol based on random structural libraries from which chemical shifts were predicted and statistically compared to experimental values. The best matching set was finally clustered. Combining both approaches allowed us to assess the robustness of this methodology.
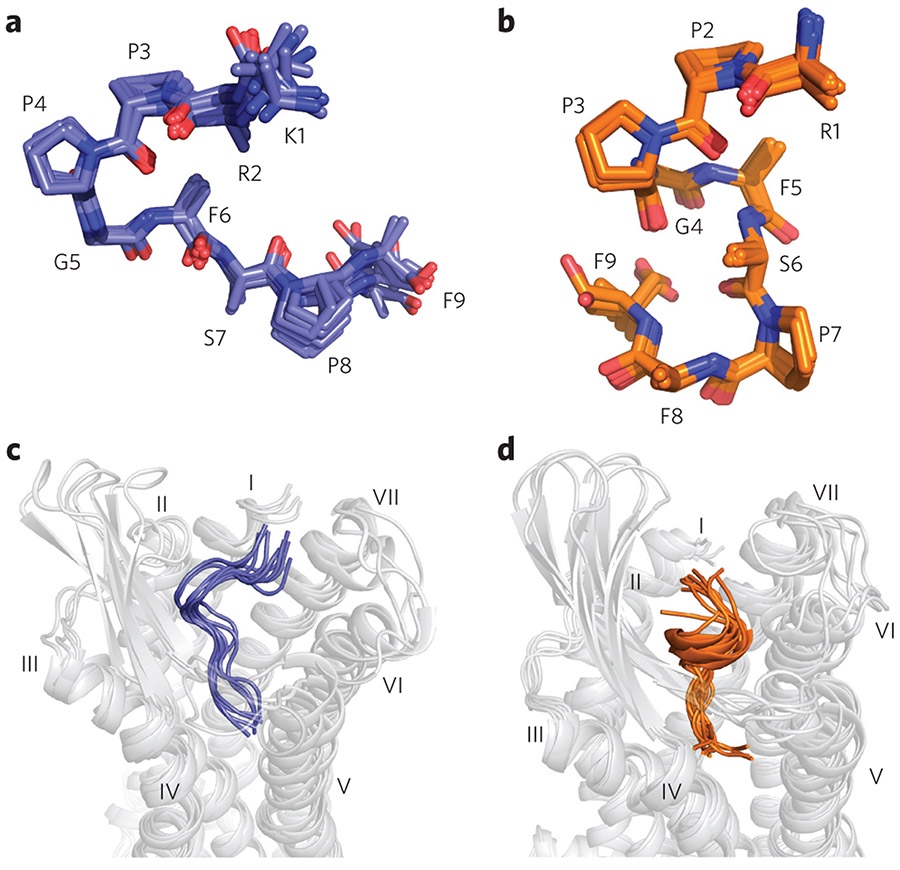
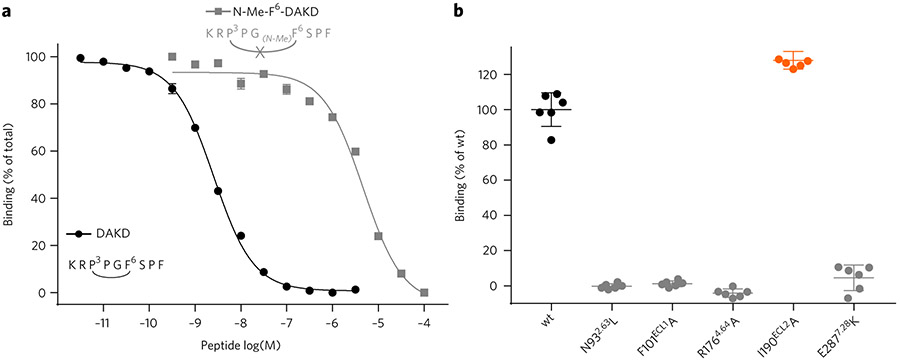
The determined DAKD backbone conformation is depicted in Figure 3a, displaying a V-shaped fold bearing a characteristic β-turn-like structure in the middle part of the peptide (P3DAKD–F6DAKD). This structural motif was verified biochemically using an engineered DAKD analog in which the amide of F6DAKD was methylated. This modification disrupts the β-turn-like structure and should therefore stretch the peptide conformation. Indeed, our binding assay shows that this DAKD analog has a 1,000-fold lower binding affinity for B1R than the native peptide (Fig. 4a).
Figure 3 ∣. Backbone structures of DAKD in complex with human B1R in comparison to BK bound to human B2R.
Only backbone and Cβ atoms are shown. (a) The backbone structure of DAKD calculated from NMR data features a V-shaped fold with a β-turn-like structure around P3–F6. (b) The NMR-based backbone structure of BK is characterized by an overall S-shape with a 310-helix-like segment (P2–F5) in the middle. (c,d) Rosetta modeling of DAKD in B1R (c) and BK in B2R (d) reproduces the characteristic V-shape of DAKD and the S-shape fold of BK (see text and Supplementary Fig. 10 for further details).
Figure 4 ∣. Functional characterization of peptide variants and B1R mutants.
(a) Disruption of the central β-turn in DAKD results in a strong decrease in affinity for the B1R. The methylation of the amide nitrogen of F6DAKD disrupts the central β-turn and results in a 1,000-fold decrease of DAKD binding affinity (KI DAKD: 1.11 ± 0.04 nM (circles); DAKD linearized: 2.03 ± 0.17 μM (rectangles)). (b) Verification of key peptide interaction sites predicted from B1R–DAKD models by site-directed mutagenesis (maximal binding of DAKD normalized to wild type; n = 6). I190ECL2A shows an increased binding activity (129%, orange scatter), whereas all other mutations result in a complete loss of DAKD binding. Expression levels of B1R mutants were assessed by western blot (Supplementary Fig. 13).
To elucidate the molecular origins of subtype selectivity between B1R and B2R, we calculated the backbone structure of BK bound to B2R from our previously reported NMR data10 using the method described above. B2R-bound BK shows a conformation strikingly distinct from that of B1R-bound DAKD. It features an S-shaped structure with a 310-helix-like segment in the middle (Fig. 3b). Moreover, the C-terminal part in BK is folded in a turn-like structure, whereas an extended open conformation is observed for DAKD. Interestingly, binding of BK to the B2R causes a major conformational rearrangement of the peptide, whereas the structure of DAKD is essentially the same both when in solution and when bound to the B1R (see Supplementary Fig. 9 for a comparison of all structures). All chemical shift values and the structure refinement statistics are summarized in Supplementary Tables 2-8.
Docking and modeling
To understand their subtype-specific binding behavior, models of DAKD in complex with B1R and of BK in complex with B2R were generated using Rosetta multiple-template comparative modeling and flexible peptide docking. The aim was to identify the binding interface of DAKD and BK at B1R and B2R, respectively. B1R and B2R models were created on the basis of 24 experimentally determined class A GPCR structures (Supplementary Tables 9 and 10). Both models show common structural features, as observed in the crystal structures of the other peptide-binding GPCRs, such as an α-bulge in helix V and proline-kinks in helices IV and VI. The extracellular loops (ECL) exhibit valid conformations with ECL2, adopting a β-sheet, which is found in many peptide-binding GPCRs. The obtained models were found to be independent of the activation state of the used GPCR templates (see Online Methods).
DAKD and BK were simultaneously folded and docked into the B1R and B2R models, respectively, using the Rosetta FlexPepDock application27. In both cases, ligand docking converged to a single solution. This approach was chosen over a direct docking of the NMR-derived structure into the receptor models, as it allows a better sampling of the conformational space by Rosetta. The Rosettaderived models of docked DAKD and BK have the same distinct conformations as described above, i.e., a V-shaped backbone structure with a type-II β-turn for B1R-bound DAKD and an S-shaped fold with a central 310-helix for B2R-bound BK (see Fig. 3c,d and Supplementary Fig. 10).
These distinct peptide structures suggest some major differences within their respective binding pockets, which have been believed to be similar overall. Approximately 3/4 of the B1R binding pocket residues (defined here for a region within 5 Å distance to any ligand atom) are sequence counterparts of B2R. Within the group of counterpart residues, the sequence identity and similarity between B1R and B2R are 45% and 67%, respectively, which is slightly higher than those for the complete receptor (41% and 59%). Furthermore, DAKD and BK were found to occupy a similar sized region within the binding pocket. The change in the solvent-accessible surface area of B1R and B2R upon binding was quantified to be 1,713 ± 45 Å2 and 1,814 ± 129 Å2, respectively.
B1R–DAKD and the B2R–BK receptor–peptide interfaces
To identify possible ligand-binding sites in B1R and B2R, we collected statistics of the contact frequency of peptide–receptor residue pairs. The number of pairwise residue interactions with a Rosetta score less than −1.0 Rosetta energy units (REU) across the 1,000 top-scoring models was counted (Supplementary Figs. 11 and 12), providing a likelihood of residue–residue interactions. For DAKD, we observed a binding mode that agrees well with previously reported mutagenesis28,29 and modeling studies30 and fulfills most of the predicted contacts from which we had derived a set of upper distance restraints to guide ligand docking.
The N terminus of DAKD is facing transmembrane helices (TMH) VI and VII, with the N-terminal amine group and the side chains of K1DAKD and R2DAKD located next to a cluster of polar, acidic residues (Fig. 5a,c). K1DAKD is coupled via electrostatic interactions with E2877.28, D2887.29 and D2917.32, whereas the side chain of R2DAKD is involved in contacts primarily with E2736.58, Q2776.62, E2877.28 and D2917.32 (Supplementary Fig. 11). A role of E2736.58 and D2917.32 in DAKD binding was reported previously30,31, and replacement of these residues by alanine decreased ligand-binding affinity. Similarly, the charge-inverting mutation E2877.28K fully abolishes DAKD binding to the receptor (Fig. 4b).
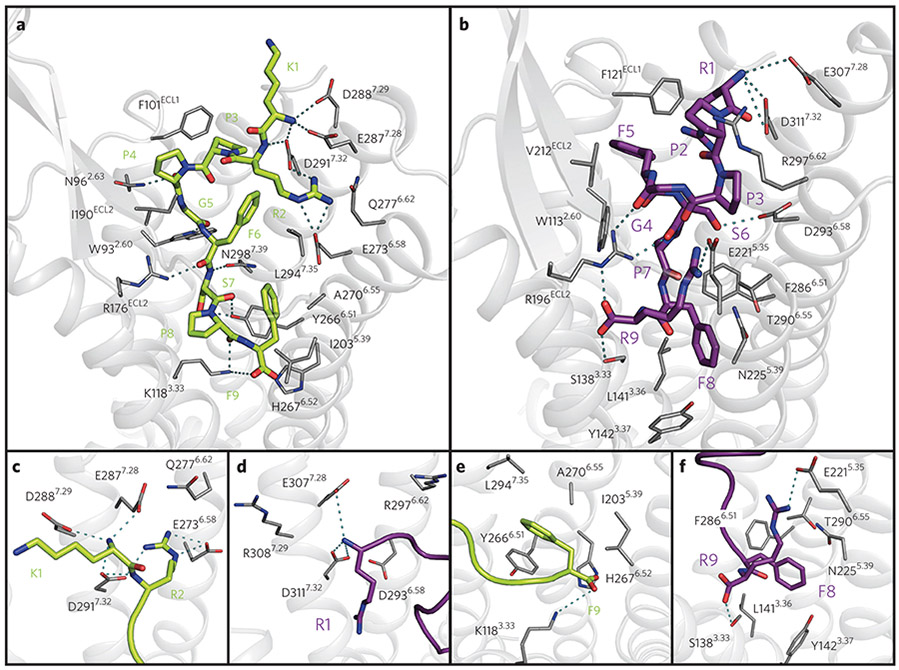
Figure 5 ∣. Structural characterization of the B1R–DAKD (green) and B2R–BK (purple) binding pocket.
(a) Top view of DAKD docked to a comparative model of B1R. (b) Top view of BK docked to a comparative model of B2R. (c) Side view of the DAKD and (d) BK N-terminal binding site at TMH VI and VII. (e) Side view of the DAKD and BK (f) C-terminal binding site between TMH3, 5 and 6. The ligand is shown as thick sticks. Receptor residues predicted to be involved in ligand binding are labeled and are shown as thin sticks. Predicted interactions are indicated by dotted cyan lines. Atoms are colored by type (oxygen, red; nitrogen, blue; sulfur, yellow).
The C terminus of the peptide is facing TMH III, V and VI, which are known to be involved in receptor activation2. The C terminus of DAKD interacts electrostatically with the side chain of K1183.33, and the phenyl ring of F9DAKD is surrounded by a group of hydrophobic residues (I2035.39, Y2666.51, A2706.55 and L2947.35) (Fig. 5a,e). These observations match previous experimental data29 identifying K1183.33 as the B1R key residue that discriminates between DAKD and BK binding.
In the middle part of the peptide, the interaction between the side chain of F6DAKD and L2947.35 (Fig. 5a) occurs in 89% of our models (Supplementary Fig. 11). Meanwhile, the hydrogen bond interaction involving the backbone carbonyl of the neighboring residue, G5DAKD, and the Nε atom of W932.60 is the most frequently observed contact (94%) in our B1R–DAKD models (Supplementary Fig. 11). The indole ring of W932.60 is fixed in a preferred orientation for interaction with G5DAKD by a π–π stacking with the F3027.43 phenyl ring, suggesting an indirect role for F3027.43 in ligand binding through concerted interactions involving residues in the binding pocket. This picture is compatible with previous observations that the F302A mutation in B1R negatively modulates DAKD binding30.
Another frequently detected interaction in our B1R models is that between the hydroxyl groups of receptor residue Y2666.51 and S7DAKD (Fig. 5a; Supplementary Fig. 11). When Y2666.51 is replaced with alanine or phenylalanine, no change in binding affinity is observed30. We found that another polar residue, N2987.32, adjacent to Y2666.51, can possibly substitute a hydrogen bond to the OH group of S7DAKD, which would require only a flipping of the S7DAKD side chain, and this could compensate for the loss of a hydroxyl group of Y2666.51.
In addition to reproducing previous experimentally determined interactions, our B1R–DAKD models predict a series of new binding contacts, which we tested experimentally via site-directed mutagenesis. Proline residues P3DAKD and P4DAKD make hydrophobic contacts with F101 and I190 in ECL1 and ECL2, respectively (Fig. 5a; Supplementary Fig. 11), whereas the backbone at P4DAKD is stabilized by a hydrogen bond with N962.63. Furthermore, R176 in ECL2 provides several hydrogen bond contacts to the backbone of F6DAKD and P8DAKD, although the exact geometry differs slightly between the different receptor models. It is conceivable that these residues have not been considered part of the receptor binding site in previous modeling studies30-33 that were based solely on the formerly available structures of bacteriorhodopsin or bovine rhodopsin. Our modeling approach using multiple genetically and functionally more closely related GPCR templates provides more confident receptor models, especially for their extracellular loop regions, supported by the biochemical validation of newly predicted receptor–peptide contacts. Indeed, replacing F101(ECL1) with an alanine residue fully abolishes DAKD binding to the B1R. Polar contacts between N962.63–P4DAKD and R176(ECL2)–F6DAKD and R176(ECL2)–P8DAKD are crucial for DAKD binding, as the introduction of hydrophobic residues in these sites (N962.63L and R176(ECL2)A) also results in a complete loss of binding affinity of DAKD (Fig. 4b). By contrast, introduction of an alanine at position I190(ECL2) maintains DAKD binding with an IC50 comparable to that of wild-type B1R (Fig. 4b), which is in agreement with the predicted frequency of the I190–P4DAKD interaction (28%; Supplementary Fig. 11) and/or the ability of alanine to compensate the hydrophobic contacts provided by a flexible Ile side chain in a loop.
A similar analysis was also carried out for the B2R–BK receptor–peptide interface (Fig. 5b,d,f and Supplementary Fig. 12). Compared to DAKD, BK adopts a similar overall pose within the B2R binding pocket, with the N terminus facing TMH VI and VII and the C terminus bound to TMH III, V and VI. The observed B2R–BK contacts, as obtained by a per-residue breakdown of the interface energy of our models (Supplementary Fig. 12), match well with residues found in a previous site-directed mutagenesis study of the B2R binding pocket31. Furthermore, our B2R binding model predicts several interactions that have not been described before, such as hydrophobic contacts between F5BK and the aromatic side chains of W1132.60 and F121 in ECL1, as well as several hydrogen bonds between the central portion of the BK backbone and R196 in ECL2.
DISCUSSION
Our data indicate that the subtype selectivity of BRs is the result of multiple chemical and conformational factors, which act in a complex and synergistic manner.
BRs discriminate between the N-terminal parts of their respective peptides, BK and DAKD, mainly via their binding chemistry. Our work reveals that a cluster of acidic residues located at the extracellular side of TMH VI/VII and ECL3 are crucial for the binding of positively charged N-terminal residues. The location of such a cluster is supported by the previous work on B1R/B2R chimeric receptors30,31. In B1R, K1DAKD and R2DAKD interact with E2877.28, D2887.29 and D2917.32 and E2736.58, Q2776.62, E2877.28 and D2917.32, respectively (Figs. 5 and 6; Supplementary Figs. 11 and 12). These residues are all conserved between the B1R and the B2R except D2887.29 and Q2776.62, which are replaced by R2976.62 and R3087.29 in the B2R (Supplementary Fig. 14). The B2R agonist BK carries only one charged N-terminal residue, R1BK, which interacts mainly with residues D2936.58, E3077.28 and D3117.32. These residues coincide with those in B1R that are responsible for binding to R2DAKD. This means that R2DAKD and R1BK have a similar protein interaction interface, but the N-terminal residues K1DAKD and R1BK of both agonists have a different position within the binding pockets of both receptors.
Figure 6 ∣. Representation of key interactions responsible for high affinity binding of DAKD to B1R and of BK to B2R.
B1R discriminates between DAKD and BK mainly via electrostatic interactions at the N terminus, whereas B2R selects via a complex interaction network as a result of different C-terminal structures of the BK and DAKD. The residues conserved among B1R and B2R are shown in bold circles.
Due to the replacement of Q2776.62 and D2887.29 in B1R by R2976.62 and R3087.29 in B2R, the negative charge density, and therefore the electrostatic binding capacity, of B2R for peptide ligands with a positively charged N terminus is strongly reduced. This conclusion is consistent with the observation that replacement of these residues by alanine or positively charged amino acids substantially impairs BK binding in B2R31. Both the altered charge density and different N-terminal position in the binding pocket together could explain why B1R selects peptides that contain two charged N-terminal residues K1 and R2 (DAKD or KD) over those starting with only positively charged N-terminal residue R1 (BK, DABK). The N-terminal peptide selectivity of B1Rs in other species further supports this argument. In dog B1R, for example, D7.29 is replaced by a neutral Asn residue (Supplementary Table 11), and the receptor shows no significant selectivity between DAKD and DABK34. Furthermore, in some rodents, residue E7.28 is replaced by a Lys residue, which inverts the local charge and can impose a strong perturbation on the chemical architecture of the N-terminal binding cluster. Indeed B1Rs of some of these rodents are known to have a diminished, or even reversed, N-terminal selectivity for human kinin peptides34. Our mutagenesis study also validates the strong impact of this residue in species-specific subtype selectivity.
The presented data suggest that B1R and B2R discriminate between the C-terminal parts of their respective peptide ligands via specific peptide conformations and peptide–receptor interactions. The C-terminal segments of DAKD and BK fold into distinct conformations in B1R and B2R, respectively. The four C-terminal residues of BK form a turn-like structure in which the R9BK residue flips back toward the extracellular surface. The formation of such a turn-like structure in DAKD is hampered by the lack of an additional C-terminal Arg residue, as present in BK. The drastic differences in the C-terminal folding are connected to the distinct receptor–-peptide interactions. In B1R, residues K1183.33, I2035.39, Y2666.51 and H2676.52 show contacts with F9DAKD, and residues K1183.33 and R176ECL2 show contacts with P8DAKD (Figs. 5 and 6; Supplementary Fig. 11). In B2R, residues N1343.29, S1383.33, M1924.60, R196ECL2, E2215.35 and N2255.39 interact with R9BK, and Y1423.37, F2886.51 and Q2876.52 with F8BK (Figs. 5 and 6; Supplementary Fig. 12). Residue H2215.35 in the B1R is replaced by E2215.35 in the B2R, which provides an electrostatic binding site in the B2R for peptides with a C-terminal Arg. In addition, residue I2255.39 in the B1R is replaced by N2255.39 in B2R, which offers a better matching in binding pocket polarity for peptides with polar side chains at the C terminus. In agreement with our model, the previously reported N2255.39A mutation decreases BK binding by a factor of 5 (ref. 31).
Moreover, the positively charged residue K1183.33 in B1R is substituted by a neutral S1383.33 in B2R, which is more compatible with the positively charged and flipped C-terminal Arg residue R9BK. This finding is in line with earlier suggestions that residue 3.33 is key for triggering receptor subtype specificity29. In addition, other sites with conserved differences between mammalian B1Rs and B2Rs also contribute to the binding of the peptide C-terminal region. Such a complex pattern indicates a highly integrated network for accommodating the distinct C-terminal folding of DAKD and BK. It can be envisaged that such a complex interaction network could also serve as a subtle regulator for functional switching between agonism and antagonism while not compromising the binding affinity, which has been indeed suggested for the DALK peptide30.
The reconfiguration of the N- and C-terminal binding networks leads to different positioning of the N terminus of the peptide, as well as different orientation of the C-terminal residues in the receptor (Fig. 6). Together with these changes, residues in the middle part of the peptide switch to distinct binding sites on the receptor, allowing polarity matching as shown in Figure 6. The middle parts of the peptides, which share the common sequence, serve as linkers, allowing correct presentation of the peptide N and C termini with respect to the receptor. The importance of the conformation of this central segment is highlighted by the drastic loss of affinity of DAKD upon methylation of the backbone amide of F6, which is expected to impair the formation of the central β-turn (Fig. 4a).
It may be peculiar that free and bound structures of DAKD show high similarity, but one could speculate about potential biological reasons: B1R shows high basal activity comparable to that of agonized B2R13,35. Therefore, B1R could already adopt or sample a ‘partially activated’ conformation, which is ready to bind a prestructured DAKD without the need of major structural reorganizations. In contrast, B2R shows larger activity differences between its ground and activated states.
In summary, although the human B1R and B2R show high sequence identity, many nonconserved residues in the peptide-binding pocket reshape the binding landscape by casting distinct interactions to selectively accommodate peptides that have similar sequences but distinct structures. Whereas the mechanism of subtype selectivity of some GPCRs is evident from major structural divergence of the orthosteric binding pockets, for example the opioid receptors36, other receptor subtypes such as orexin and muscarinic receptors only display minor rearrangements of specific residues and subtle changes of the size and shape of the binding pocket15,16. Recent crystallographic studies on the orexin15 and endothelin B receptors37 furthermore attempted to attribute the subtype selectivity of peptide ligand GPCRs to both the N-terminal region of these receptors15 and their TMH cores37. However, the mechanism of human BR subtype selectivity of peptide ligands is strikingly more complex than previously assumed, which is caused by the intrinsic complexity of the conformational and chemical space of peptides. The diversity in the receptor sequence, the polymorphism of peptide conformation and the distinct binding chemistry are all required to create the unique subtype selectivity in these peptide receptors. The requirements of both N- and C-terminal binding at distal sites on the receptor also justify the tremendous difficulty of efficiently developing small-molecule regulators of BRs. Small sized ligands, which lack the capacity to establish distal interactions within the receptors emulating the peptide N- and C-terminal binding modes, are less likely to be promising antagonists.
As demonstrated in this work, the integration of DNP-enhanced ssNMR with advanced molecular modeling and docking techniques offers a powerful and novel way to obtain structural and mechanistic insights into challenging GPCR targets.
ONLINE METHODS
Peptide synthesis and labeling schemes.
Uniformly 15N-13C-labeled variants of DAKD (KRPPGFSPF) and DALK (KRPPGFSPL) were ordered from Eurogentec, Cologne, Germany and Thermo Fisher Scientific, Ulm, Germany. Linearized DAKD (KRPPGFSPF, methylation of the amide nitrogen at F6) was from Thermo Scientific. Radiolabeled DAKD (3,4-PROLYL-3,4-3H(N)) was from PerkinElmer, Rodgau, Germany. Unlabeled DAKD for binding assays was obtained from Eurogentec. Peptides used in this study are summarized in Supplementary Table 1.
GPCR production in Sf9 cells.
The cDNA encoding the full-length human B1R was codon optimized for expression in insect cells and cloned into the pOET1 transfer vector (Oxford Expression Technologies, Oxford, UK) via 5′ BamHI, 3′ HindIII restriction sites. The receptor was flanked with an N-terminal decahistidine and FLAG tag as well as a C-terminal StrepII tag. Recombinant baculoviruses were generated using the flashBAC kit according to manufacturer’s instructions (Oxford Expression Technologies). High titer baculoviruses were used to infect Sf9 cells at a cell density of 1.75 to 2 × 106 cells/ml cultured in TMN-FH medium (c.c.pro GmbH, Oberdorla, Germany) supplemented with 2 mM glutamine (PAA Laboratories, GE Healthcare, Munich, Germany), 5% (v/v) FCS (BioWest, Nuaillé, France), 7.5 nM vitamin B12 (Sigma-Aldrich), 50 μg/ml gentamicin (Thermo Fisher Scientific), 0.1% pluronic F-68 (Applichem, Darmstadt, Germany). Cells were harvested 96 h past infection and stored at −80 °C until further use. Cell pellets were resuspended in 50 mM HEPES-NaOH (pH 7.6), 100 mM NaCl, 10 mM EDTA (supplemented with protease inhibitors: 5 μg/ml leupeptin, 1 mM EDTA, 1 μM E64, 2 μg/ml pepstatin A, 10 μg/ml aprotinin, 1 mM PMSF) and lysed by nitrogen decompression. Nonlysed cells and debris were collected at 1,000g for 10 min at 4 °C and membranes were pelleted by ultracentrifugation (210,000g for 90 min at 4 °C). Membranes were resuspended in 50 mM HEPES-NaOH (pH 7.6), 100 mM NaCl, 1 mM EDTA, and 5% (w/v) glycerol, flash frozen in liquid nitrogen and stored at −80 °C until purification. The receptor was solubilized in 1% n-dodecyl β-d-maltoside (DDM) and 0.1% cholesteryl hemisuccinate (CHS) for 3 h at 4 °C after dilution of the membranes in buffer A (50 mM HEPES-NaOH (pH 7.6), 150 mM NaCl, 5% (w/v) glycerol, 200 nM 15N-13C-labeled DAKD or DALK). Nonsolubilized material was removed by ultracentrifugation (210,000g for 45 min at 4 °C) and cleared solubilizates were loaded onto HisTrap HP columns (GE Healthcare, Munich, Germany) using Äkta systems (GE Healthcare). Receptors were washed with buffer A1 (buffer A supplemented with 20 mM imidazole, 200 nM 15N-13C-labeled DAKD or DALK, 0.07% DDM, 0.007% CHS) and buffer A2 (buffer A supplemented with 50 mM imidazole, 200 nM 15N-13C-labeled DAKD or DALK, 0.07% DDM, 0.007% CHS) and eluted in buffer B (buffer A supplemented with 400 mM imidazole, 0.07% DDM, 0.007% CHS). Receptors were concentrated in 50 kDa molecular weight cut-off concentrators (Amicon, Merck Millipore, Darmstadt, Germany), and buffer was exchanged to 50 mM HEPES-NaOD (pD 7.6), 150 mM NaCl, 5% (w/v) [12C-2H]glycerol (Euriso-Top, Saint-Aubin, France) in 76% D2O/18% H2O. The labeled peptide was added in molar excess during concentration and omitted in the final concentration steps to reduce nonspecific binding. 300–400 μg receptor–ligand complex (equivalent to 285–340 μM) were used per sample and were supplemented with 10 mM AMUPol, mixed in a 1:1 ratio [12C-2H]glycerol, and transferred to 3.2 mm sapphire or zirconium oxide rotors. Samples were frozen in situ in the cryo-gas flow of the spectrometer or in liquid nitrogen.
Reference samples contained 10 μg 15N-13C-labeled DAKD or DALK in 50 mM HEPES-NaOD (pD 7.6), 150 mM NaCl, 5% (w/v) [12C-2H]glycerol (76% D2O/18% H2O), 10 mM AMUPol and 4% DDM/0.4% CHS to mimic detergent increase during receptor–ligand complex concentration.
Heterologous expression of B1Rs in HEK293T cells.
Wild-type and mutant B1R sequences were synthesized (GenScript) and cloned into pcDNA3.1 for mammalian expression. Sequence integrity was verified by sequencing. HEK293T cells were cultured in DMEM medium (Sigma-Aldrich Chemie GmbH, Steinheim, Germany) supplemented with 10% FBS and 1% penicillin/streptomycin in a 5% CO2 incubator at 37 °C. Cells were passaged once in 3 d until they reached 100% confluence. At 24 h before transfection cells were seeded into a 10-cm culture dish (Thermo Fisher Scientific Nunc, Waltham, MA, USA) at a cell density of 2.2 × 106 cells per dish (five dishes per construct). Transfection was performed with 10 μg DNA using Lipofectamine 2000 (Thermo Fisher Scientific Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. At 48 h post transfection, growth medium was removed, cells were washed with PBS and detached using a cell scraper. Cells were collected by centrifuging at 130g for 5 min and snap frozen in liquid nitrogen until further use.
Cells were resuspended in breaking buffer (25 mM HEPES-NaOH pH 7.5; 1 mM EDTA) and protease inhibitors (Complete EDTA free, Roche Applied Science; 1 mM PMSF) at a cell density of 4 × 106 cells/ml. Cells were disrupted by nitrogen decompression in a pressurized vessel (Parr, Moline, USA). Intact cells and cell debris were removed by centrifugation (1000g for 10 min at 4 °C). Membranes were then pelleted using an ultracentrifuge (100,000g for 60 min at 4 °C) and resuspended in membrane buffer (25 mM HEPES-NaOH pH 7.5; 150 mM NaCl; 10% glycerol) using a glass dounce homogenizer. Total membrane protein was quantified using the bicinchoninic acid (BCA) method (Pierce Biotechnology, Rockford, USA), with bovine serum albumin as a standard. Membranes were aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C until further use. After resuspension in lysis buffer (25 mM HEPES-NaOH, pH 7.5, 10 mM EDTA and protease inhibitors (Complete EDTA free, Roche Applied Science); 1 mM PMSF (Carl Roth)) cells were lysed by nitrogen decompression (Parr Instrument Company, Moline, IL, USA). Nonlysed cells and debris were removed by centrifugation (1,000g for 10 min at 4 °C), and membranes were subsequently pelleted by ultracentrifugation (180,000g for 60 min at 4 °C). Membranes were homogenized in membrane buffer (25 mM HEPES-NaOH pH 7.5, 150 mM NaCl,5% (w/v) glycerol), aliquoted, flash frozen and stored at −80 °C until use. Total membrane protein content was determined by the bicinchoninic acid (BCA) method using bovine serum albumin as the standard.
For western blotting, membrane suspensions equivalent to 150 μg total membrane protein were treated with 4 U benzonase endonuclease in benzonase buffer (50 mM Tris–HCl pH 8.0, 1 mM MgCl2) and protease inhibitors (Complete EDTA free, Roche Applied Science; 1 mM PMSF) for 20 min at 4 °C, resolved on a 4–12% Bis–Tris NuPAGE gel (Thermo Fisher Waltham, USA), and transferred onto PVDF membranes. The PVDF membrane was blocked with 5% nonfat milk at room temperature (20–22 °C) for 1 h in TBST buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl; 0.05% Tween-20) and incubated with a monoclonal alkaline phosphatase-coupled anti-FLAG M2 antibody (Sigma-Aldrich, A8592 Steinheim, Germany) for 1 h at room temperature. The PVDF membrane was washed five times for 5 min with TBST and developed in alkaline phosphate buffer (100 mM Tris–HCl, pH 9.5, 100 mM NaCl, 5 mM MgCl2) containing 0.33 mg/ml 5-bromo-4-chloro-3-indolyl phosphate p-toluidinium (BCIP) and 0.165 mg/ml nitro-blue tetrazolium chloride (NBT).
Binding assays.
B1R binding assays in Sf9 membranes were executed by incubation of 10–50 μg total membrane protein with increasing concentrations of radiolabeled DAKD ([3H]Lys[Des-Arg9]bradykinin ([3H]DAKD, PerkinElmer, Boston, MA, USA)) (for determination of dissociation constants (KD)) or unlabeled DAKD variants (for determination of inhibition constants) in 50 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 1× SyntheChol (Sigma-Aldrich Steinheim, Germany) for 60 min at room temperature. Binding was terminated by rapid filtration over GF/B glass fiber filters, wash steps were executed with 50 mM HEPES-NaOH (pH 7.6), and remaining radioactivity was analyzed by liquid scintillation counting. Competition binding assays using receptors expressed in HEK293T cells were performed accordingly, using 100 μg total membrane protein in 50 mM Tris–HCl (pH 7.4), 5 mM MgCl2 and 1 nM [3H]DAKD. GF/B filters were washed with ice-cold water. The affinity state of solubilized B1R was analyzed by immobilizing the receptor via the C-terminal StrepII tag on StrepTactin beads (Qiagen, Hilden, Germany) and incubation with increasing concentrations of radiolabeled DAKD in buffer A. Beads were washed with buffer A and analyzed by liquid scintillation counting. Data were evaluated with Prism6 (GraphPad Software, La Jolla, USA) and determined dissociation constants were used to convert half maximal inhibitory concentrations (IC50) to inhibition constants (KI) via the Cheng–Prusoff equation. Data are represented as means ± s.e.m. from two or three independent experiments each performed in triplicate (n = 3).
DNP-enhanced solid-state NMR experiments.
All DNP-enhanced MAS ssNMR experiments were carried out on a Bruker Avance II DNP ssNMR spectrometer operating at 400.197 MHz (9.40 T). The spectrometer is equipped with a Bruker 3.2 mm HCN cryo-MAS probehead. The dry nitrogen gas for cryo-MAS and temperature control is pre-cooled in a low temperature heat exchanger maintained by continuous liquid nitrogen supply. High-power continuous-wave (CW) microwave irradiation was generated in a CPI gyrotron (Communications and Power Industries) and transmitted to sample location in the probehead via corrugated waveguides. For approaching the optimized DNP enhancement, the microwave frequency was adjusted to 263.580 GHz by setting the cavity temperature. The microwave output power was about 35 W as calibrated using an external water load. The microwave power attenuation of the corrugated waveguide was about 4.8 dB, which permits about 1/3 of the gyrotron output power to reach the sample. The stability of the microwave power was monitored by an external thermometer coupled to a mirror-load device.
All the samples were loaded into the 3.2 mm rotors (sapphire or ZrO2 material) and sealed with Vespel caps. The rotors were snap-frozen in liquid nitrogen before NMR experiments or directly in the low temperature cryo-MAS gas flow. All samples were transferred on dry ice/in liquid nitrogen and preserved at −80 °C between measurements. The MAS frequency was stabilized at 8,000 ± 6 Hz at about 110 K in all measurements.
1H-13C cross-polarization (CP) was achieved using a ramped spin-lock (SL) pulse (80–100) for protons and a constant amplitude SL pulse for carbons. The average SL power for 1H was about 48 kHz, and the field strength for 13C SL was about 40 kHz. The contact time was set to 800 μs. 13C-13C and 15N-13C 2D spectra were constructed using DQ-SQ and out-and-back TEDOR approaches. These two methods were chosen based on their good efficiency and robustness under DNP conditions. DQ-SQ 13C-13C 2D spectra were recorded using the largest possible F1 window38. The double quantum coherence was excited by POST-C7 pulses39 at 56 kHz with 100 kHz CW heterodecoupling. Both DQ excitation and reconvention times were set to 500 μs (four rotor periods), corresponding to 14 POST-C7 units at 8,000 kHz MAS. The DQ efficiency was about 20–25%. Typically, 512 to 1,024 scans were accumulated for each t1 point of DQ-SQ spectra of peptide-GPCR samples. The DQ-SQ 13C-13C 2D spectra were acquired with 1,536 (F2) by 32 or 48 (F1) points for spectral windows of 296 (SQ) and 560 (DQ) p.p.m., respectively, and were processed with a 4,094 (F2) by 1,024 (F1) matrix. An exponential window function with Lorentzian broadening factor of 100 Hz was applied on both direct and indirect dimension. The TEDOR 15N-13C 2D experiments were conducted using a scheme shown in ref. 40. Briefly, each train of recoupling pulses before and after the central 13C π pulse is composed of four 15N π pulses (10 μs). This corresponds to 1,000 μs (eight rotor period) total recoupling time in whole pulse sequence. CW heterodecoupling at 100 kHz were applied during TEDOR recoupling. Typically, 2,048 scans were accumulated for each t1 point of TEDOR experiments on peptide–GPCR samples. The TEDOR 15N-13C 2D spectra were acquired with 1,024 (F2) by 32 (F1) points for spectral windows of 296 (13C) and 49 (15N) p.p.m., respectively, and were processed with a 4,094 (F2) by 1,024 (F1) matrix. A Gaussian window function with Lorentzian broadening factor of 20 Hz and a Gaussian broadening factor 0.05 was applied on direct dimension. Indirect dimension was processed using a pure cosine window function.
The DQ and REDOR doubly filtered 1D 13C spectrum (Fig. 2f) was acquired using the pulse sequence shown in Supplementary Figure 6. The DQF step was set as mentioned above. The “REDOR” dephasing is achieved by two sets of dipolar recoupling pulses, each containing four 15N π pulses (10 μs), tethered symmetrically by a rotor synchronized 13C π pulse (8 μs). CW heterodecoupling at 100 kHz were applied during the DQF and REDOR dephasing periods. For all experiments 100 kHz decoupling using SPINAL64 (ref. 41) was applied during acquisition.
All 13C chemical shifts reported in Supplementary Tables 2 and 3 were referenced indirectly to TSP via alanine (LT) and adamantine (RT) signals. 15N chemical shifts were referenced indirectly via the gyromagnetic ratios to liquid ammonia.
Peptide structure calculation.
Backbone structures were calculated with torsion angle restraints derived by a ‘backward’ approach based on chemical shifts. These structures were examined using a ‘forward’ method based on chemical shifts predicted from a library of random conformations. In addition, the robustness of this procedure was tested on the structure of free DAKD in solution, determined by liquid-state NMR based on additional distance restraints.
(a). B1R-bound DAKD backbone structure calculation from chemical shifts (‘backward’ approach).
Backbone torsion angle restraints were generated from chemical shifts (Supplementary Table 2) using TALOS+42, TALOS-N43 or PREDITOR44. They were used by CYANA2.1 (ref. 45) for structure calculations in torsion angle space. 500 initial models were annealed through 20,000 steps under torsion angle restraints with a weight set to 0.10. A bundle size of ten output structures was chosen. The structure refinement statistics was analyzed by CYANA and iCING46 (Supplementary Table 7).
(b). B1R-bound DAKD backbone structure calculation via a ‘forward’ approach based on Flexible-Meccano/SHIFTX predictions.
As a control, an unrestrained backbone ensemble containing 300,000 conformations was generated by Flexible-Meccano47. The backbone chemical shifts of each conformation in this collection were predicted using SHIFTX48, and the deviations between the predicted and experimental values were calculated. A simple search for the best matching sets of chemical shifts returned ambiguous conformational ensemble, and additional conditions for selecting the correct structures had to be introduced.
Therefore, a conformational test ensemble containing 500 DAKD structures was calculated by CYANA starting from 12,500 initial models using torsion angle restraints generated from our experimental chemical shifts by TALOS+ ‘backward’ calculations. For each structure in this ensemble, 13C chemical shifts were recalculated by SHIFTX. These predicted chemical shifts were compared to the experimental input values for each site. Those showing significantly large deviations (R2DAKD Cα, F6DAKD C, S7DAKD C) were excluded from the following analysis to reduce a biasing introduced by the intrinsic deviation between the SHIFTX and TALOS+ algorithms. The intrinsic deviation of SHIFTX-predicted chemical shifts within the CYANA test ensemble (‘cutoff’) is represented by the sum of the absolute values of the largest backbone 13C chemical shift differences.
where and are the predicted 13C chemical shifts of a certain nuclei in the ith and jth peptide structure in the CYANA ensemble, respectively.
For each of the 300,000 members of the Flexible-Meccano ensemble, absolute values of the differences between SHIFTX-predicted and experimental backbone 13C chemical shifts were calculated and summed up
where is the predicted 13C chemical shift of certain nuclei in the ith peptide structure in the Flexible-Meccan ensemble, and is the experimental 13C chemical shift of this nuclei.
Within the Flexible-Meccano ensemble, ΣΔCS assumes a Gaussian-like distribution with a minimum value at 6.61 p.p.m., a maximum at 13 p.p.m. and a width of 12 p.p.m. The value of the cutoff parameter was now used to select the best candidates with minimal ΣΔCS. A set of 1,490 structures was found between the global minimal ΣΔCS of 6.61 p.p.m. and the cutoff value. The use of such a “large” set of conformations selected by defined cutoff improves structural clustering compared to previous protocols9.
To derive restraints for further structure refinements, the backbone torsion angles (ϕ, ψ) from the selected set of structures were extracted and fitted by a Gaussian distribution (Supplementary Fig. 15a,b). Its mean and s.d. values were taken as the mean torsion angle and the allowed deviation. Three of the torsion angles (G5 ϕ, F6 ψ, and S7 ψ) showed a double distribution. The independence of these ambiguous torsion angles was examined using a ‘Ramachandran’ plot correlating these pairs of torsion angles extracted from each individual in the top 1,490 structures (Supplementary Fig. 15c). Eventually, we obtained eight (2×(G5) × 2× (F6) × 2× (S7)) sets of backbone torsion angle restraints extracted from the Flexible-Meccano/SHIFTX ensemble. These eight sets of restraints were used as inputs in CYANA calculations. Eight distinct clusters (labeled A-H) with ten structures each were generated.
(c). Comparison of DAKD structures calculated from forward, backward and Rosetta/docking approaches.
Using approaches (a) and (b), 11 structural ensembles were generated in total. To elucidate the relationships of these ensembles, a systematic analysis was conducted. In addition, the structural ensemble derived from the Rosetta/Docking approach was included for comparison. The 120 structures within the 12 clusters were compared by computing 6,600 pairwise backbone heavy atom r.m.s. deviation values. They were used to visualize the structural similarities in a string plot (Supplementary Fig. 16): TALOS+/TALOS-N/PREDITOR ensembles and the cluster A and E from the forward approaches are closely related to each other and agree well with the Rosetta/docking solution. The clusters B, C, D, F, G, and H could be immediately excluded. The clusters A and E mainly differ from the ‘backward’ solutions in their S7 ψ angle but only A matches the other solutions.
Based on the string plot in Supplementary Figure 16, the ensemble calculated by CYANA using TALOS+ restraints agrees best with the Rosetta/Docking solution. In addition, as shown in Supplementary Table 7, the TALOS+/CYANA approach yielded better structures compared to TALOSN/CYANA and PREDITOR/CYANA in terms of violations of restraints and backbone torsion angle distributions on Ramachandran plots. Therefore, the combination of TALOS+/CYANA was selected as the method of choice for all further calculations.
(d). Determination of DAKD (free) and B2R-bound and free BK peptide conformations using TALOS+/CYANA.
The torsion angle restraints of DAKD without receptor and BK peptide with and without receptor were generated by TALOS+, and the structures were calculated using the same backward CYANA protocol as described in section (a) above. The chemical shifts of DAKD without receptor are listed in Supplementary Table 3. The chemical shift values used for BK structure determination were previously reported in Lopez et al.10 The statistics of structure determination of DAKD and BK peptides in receptor-bound and free states were summarized in Supplementary Table 8.
For further validation, the structure of free DAKD was determined by solution-state NMR based on chemical shift as well as distance restraints (see Section (e) below and Supplementary Tables 5 and 6). For comparison, the same procedure as described in (a) was applied using the experimental solution-state NMR chemical shifts to derive backbone torsion angle restraints. In addition, a ‘forward’ calculation approach as described in section (b) was applied. Briefly, we started from the same random conformation library of DAKD (300,000 conformations) and re-ranked all the individuals according to the level of matching with experimental solution NMR chemical shifts of free DAKD. The P2 Cα and P8 C chemical shifts were excluded due to the large deviations among different sets of programs. The first round of selection with a ΣΔCS cutoff at 7.10 p.p.m. yielded a sublibrary containing 4,745 conformations (1.58% of full library). The backbone torsion angle distributions within this sublibrary were extracted and used to build the representative bundles as CYANA restraints. The final result contained four bundles as a result of two independent ambiguities in G5 ϕ and S7 ψ angles.
We then ran a comparative analysis of all the backward and forward solutions of free DAKD based on backbone r.m.s. deviation. As shown in the string plot (Supplementary Fig. 17), the TALOS-based backward calculations, as well as two of the forward clusters, resemble the solution NMR structure in terms of backbone r.m.s. deviation. A second round of selection based on S7 ψ angle led to the convergence via identification of a unique solution from all forward bundles. In general, the bundles passing the selection steps correctly depicted the V-shaped folding of DAKD in solution, which appears similar to the bound state. These analyses demonstrate that our computational pipeline could reliably determine the conformations of the peptides.
(e). Determination of solution NMR structure of free DAKD using CS and distance-restraints.
As further validation, the structure of free DAKD was determined by solution-state NMR based on chemical shift as well as distance restraints. The NMR experiments of the free peptide (3 mM sample in 50 mM MES buffer, pH 5.6, 100 mM NaCl and 10% D2O) were conducted at a temperature of 295 K on a Bruker Avance III HD 600 MHz spectrometer, equipped with Prodigy cryogenic triple-resonance probes. NMR spectra were acquired and processed using TopSpin version 3.5 (Bruker BioSpin 2017). For the chemical shift assignment, the following experiments were conducted: homonuclear 2D 1H1H-ROESY (100, 200, 300, and 500 ms mixing time) and 2D 1H1H-TOCSY (20 and 80 ms mixing time), and heteronuclear 2D 1H13C-edited-HSQC, 2D 1H13C-HMBC and 2D 1H15N-sofast-HMQC at natural abundance. The solution-NMR structure calculations of free DAKD were performed using ARIA 1.2 with CNS 1.1 (refs. 49,50). The standard simulated annealing (SA) protocols were used, including ROE distance calibration and spin-diffusion correction. The three 2D 1H,1H-ROESY spectra (100, 200, and 300 ms mixing time) used for the structure calculation were manually peak picked and assigned using Sparky 3.114 (T.D. Goddard and D.G. Kneller, University of California, San Francisco). Backbone dihedral angle restraints have been included based on TALOS-N43 predictions when in agreement with prior calculations for which only ROEs were used. Fifty starting structures were generated based on a linear template molecule. For each iteration (0–7), in which 50 structures were calculated, the ROE distance restraints were recalibrated by ARIA based on the 20 lowest energy structures. The violation tolerance was progressively reduced to 0.1 Å in the last iteration (8) in which 200 structures were calculated. For the structure calculations, a four-stage SA protocol was used using torsion angle dynamics. The high temperature stage consisted of 10,000 steps at 10,000 K. This step was followed by refinement and cooling down stages: 8,000 steps at 2,000 K, 5,000 steps to 1,000 K and 10,000 steps to 50 K. During the SA protocol the force constant for the distance restraints was set to 0, 10, 10, and 50 kcalmol−1Å−2 for the successive stages. The final 20 lowest energy structures were further refined in explicit water. The solution-state NMR chemical shifts are provided in Supplementary Table 5. The structure calculation refinement statistics are summarized in Supplementary Table 6. The DAKD structure in solution is displayed in Supplementary Figure 9c.
Homology modeling and docking.
The overall workflow for receptor modeling and peptide docking is summarized in Supplementary Figure 18. Structural models of the B1R–DAKD and B2R–bradykinin complexes were generated using the protein structure prediction software package Rosetta, version 3.5 (ref. 51). Comparative models of B1R and B2R were built based on 24 experimentally determined class A GPCR structures as possible templates (Supplementary Table 9). These structures were aligned with MUSTANG52, and the resulting multiple-sequence alignment was aligned with the B1R and B2R sequence using ClustalW53. The N- and C-terminal sequence termini were truncated by 31 and 22 residues for B1R and 51 and 39 residues for B2R because of a lack of coordinates for the aligned residues in most of the template structures. The sequence alignment was manually adjusted to remove gaps within transmembrane helix regions and to ensure that highly conserved residues and helix endings remain aligned (Supplementary Table 10). Transmembrane helix regions were predicted by programs PSIPRED54 and OCTOPUS55. The B1R or B2R residues were threaded onto the atomic coordinates of the aligned residues within each of the 24 template GPCRs. 4,000 B1R and B2R models were assembled by Rosetta comparative modeling56 using segments of the threaded structures and sequence-based fragments. All models underwent all-atom refinement in internal and Cartesian coordinate space with gradient minimization. The models were clustered based on backbone r.m.s. deviation with automatic radius detection, and the top-scoring models from the ten largest clusters were selected and checked for incorrect structural features such as helix–helix clashes or unlikely helix kinks. To sample loop conformations more thoroughly, these receptor models were subjected to another round of comparative modeling in which the extracellular loop regions were individually reconstructed using only seven peptide-binding GPCRs as templates: angiotensin II type-1 receptor (PDB code 4YAY), C–C chemokine receptor type 5 (PDB code 4MBS), κ-opioid receptor (PDB code 4DJH), δ-opioid receptor (PDB code 4N6H), μ-opioid receptor (PDB code 4DKL) and orexin receptor type 2 (PDB code 4S0V). An atom pair constraint between B1R residues C1103.25 and C189ECL2 and between B2R residues C1303.25 and C211ECL2 was included to account for the expected, highly conserved disulfide bond. Furthermore, a β-strand pairing constraint was applied to B1R residues R176ECL2 – L192ECL2 and B2R residues R196ECL2 – S214ECL2. A β-sheet secondary structure formation of that region was predicted by PSIPRED54 and observed in all template GPCR structures. In each step, 4,000 receptor models were created and 10–20 models with a high score and valid loop conformations were selected from the 10 largest clusters and used as input for the next round of loop modeling.
To avoid a collapse of the receptor loops into the receptor binding pocket during all-atom relaxation, and thus an occlusion of the ligand binding site, the peptide ligand was placed within the receptor pocket before and during loop modeling and re-docked afterwards. A set of 10 B1R and 20 B2R comparative models were used as input structures for the final ligand docking step. DAKD and bradykinin were simultaneously folded and docked into B1R and B2R using the Rosetta FlexPepDock application27. This protocol combines Monte Carlo Metropolis–based rigid body moves and peptide backbone conformational sampling in Rosetta’s low-resolution centroid mode with subsequent full-atom refinement and side chain optimization. Experimental information about putative ligand binding residues was used to derive restraints to guide ligand docking. Restraints were implemented as a set of ambiguous distance restraints between the Cα atom of the proposed binding residue of the receptor and the Cα atom of each ligand residue with a 10 Å distance cutoff. Only the distance giving the lowest energy was used to calculate the restraint energy of a specific receptor residue. For B1R, restraints were derived for residues K1183.33 (ref. 29) and A2706.55, E2736.58, D2917.32, L2947.35 and F3027.43 (all ref. 30). For B2R, residues W1132.60 (ref. 57), S1383.33 (refs. 28,29), F2866.51, T2906.5, D2936.58, D3117.32 and Q3157.36 (all ref. 31) were used to construct distance restraints for ligand docking. A total of 50,000 B1R–DAKD and 72,000 B2R–bradykinin models were generated. Models were selected by clustering of the 1,000 best models by combined Rosetta total, peptide and interface score (Supplementary Fig. 19). Putative ligand-binding residues of B1R and B2R were identified by a per-residue breakdown of the Rosetta interface energy and counting all interactions with a score lower than −1.0 Rosetta energy units within the 1,000 best models. The compliance of the structural models of DAKD and bradykinin with the experimental chemical shift data was checked by back-calculating chemical shifts from structure using the programs SPARTA+58 and SHIFTX2 (ref. 59) (Supplementary Fig. 20). A final set of 10 B1R–DAKD and B2R–bradykinin models that showed the smallest chemical shift r.m.s. deviation relative to the experimental data were selected as representative models.
Our used set of templates contained GPCR structures both in the active and the inactive states. It has previously been suggested60 that the Rosetta comparative modeling protocol is insensitive to the state of the GPCR templates so that it would not affect modeling and docking. To further validate this assumption, we compared the similarity of active and inactive structures, as well as the similarity of our receptor models with each of the two subgroups by calculating r.m.s. deviations with the structure-based alignment tool MAMMOTH. The average r.m.s. deviation value of active structures (3.0 ± 0.5 Å) is not considerably different from inactive structures (3.3 ± 0.6 Å) and comparable to the average r.m.s. deviation when all templates were combined (3.4 ± 0.5 Å). No significant differences are found when calculating pairwise r.m.s. deviations of B1R or B2R with active (3.8 ± 0.1 Å / 3.5 ± 0.4 Å) and inactive structures (3.8 ± 0.1 Å / 3.6 ± 0.2 Å) which shows that the receptor models are indistinguishable with respect to the activation state and suggests that their modeling is indeed insensitive to the state of the template GPCRs.
Life sciences reporting summary.
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
We would like to thank T. Mosler and M. Radloff for excellent technical assistance. The German Research Foundation has supported this work through an equipment grant (GL 307/8-1). Funding by DFG project G-NMR and by SFB 807 “Transport and communication across membranes,” the Cluster of Excellence Frankfurt Macromolecular Complexes and the Max Planck Society is acknowledged. The work was also supported by BMRZ through infrastructure support by the State of Hesse. Work in the Meiler laboratory is supported through NIH (R01 GM080403, R01 GM099842 and R01 GM073151) and NSF (CHE 1305874).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
Data availability.
The authors declare that all data supporting the findings of this study are available within the article, its Supplementary Information file and from the corresponding authors upon reasonable request.
References
- 1.Kobilka BK G protein coupled receptor structure and activation. Biochim. Biophys. Acta 1768, 794–807 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen SGF et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasmussen SGF et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White JF et al. Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isogai S et al. Backbone NMR reveals allosteric signal transduction networks in the β1-adrenergic receptor. Nature 530, 237–241 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Nygaard R et al. The dynamic process of β2-adrenergic receptor activation. Cell 152, 532–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu JJ, Horst R, Katritch V, Stevens RC & Wüthrich K Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkamp S et al. Structure of monomeric interleukin-8 and its interactions with the N-terminal binding site-I of CXCR1 by solution NMR spectroscopy. J. Biomol. NMR 69, 111–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaiser A et al. Unwinding of the C-terminal residues of neuropeptide Y is critical for Y2 receptor binding and activation. Angew. Chem. Int. Ed. Engl 54, 7446–7449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez JJ et al. The structure of the neuropeptide bradykinin bound to the human G-protein coupled receptor bradykinin B2 as determined by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl 47, 1668–1671 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Luca S et al. The conformation of neurotensin bound to its G protein-coupled receptor. Proc. Natl. Acad. Sci. USA 100, 10706–10711 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connor C et al. NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc. Natl. Acad. Sci. USA 112, 11852–11857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leeb-Lundberg LM, Marceau F, Müller-Esterl W, Pettibone DJ & Zuraw BL International union of pharmacology. XLV. Classification of the kinin receptor family: from molecular mechanisms to pathophysiological consequences. Pharmacol. Rev. 57, 27–77 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Surgand J-S, Rodrigo J, Kellenberger E & Rognan D A chemogenomic analysis of the transmembrane binding cavity of human G-protein-coupled receptors. Proteins 62, 509–538 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Yin J et al. Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors. Nat. Struct. Mol. Biol 23, 293–299 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Thal DM et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature 531, 335–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillmeister P & Persson PB The Kallikrein-Kinin system. Acta Physiol. (Oxf.) 206, 215–219 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Hess JF, Borkowski JA, Young GS, Strader CD & Ransom RW Cloning and pharmacological characterization of a human bradykinin (BK-2) receptor. Biochem. Biophys. Res. Commun 184, 260–268 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Menke JG et al. Expression cloning of a human B1 bradykinin receptor. J. Biol. Chem 269, 21583–21586 (1994). [PubMed] [Google Scholar]
- 20.Becker-Baldus J et al. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA 112, 9896–9901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mak-Jurkauskas ML et al. Energy transformations early in the bacteriorhodopsin photocycle revealed by DNP-enhanced solid-state NMR. Proc. Natl. Acad. Sci. USA 105, 883–888 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maciejko J et al. Visualizing specific cross-protomer interactions in the homo-oligomeric membrane protein proteorhodopsin by dynamic-nuclear-polarization-enhanced solid-state NMR. J. Am. Chem. Soc 137, 9032–9043 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lehnert E et al. Antigenic peptide recognition on the human ABC transporter TAP resolved by DNP-enhanced solid-state NMR Spectroscopy. J. Am. Chem. Soc 138, 13967–13974 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Kaplan M et al. Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR. Nat. Methods 12, 649–652 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Jacso T et al. Characterization of membrane proteins in isolated native cellular membranes by dynamic nuclear polarization solid-state NMR spectroscopy without purification and reconstitution. Angew. Chem. Int. Ed. Engl 51, 432–435 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Frederick KK et al. Sensitivity-enhanced NMR reveals alterations in protein structure by cellular milieus. Cell 163, 620–628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raveh B, London N, Zimmerman L & Schueler-Furman O Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One 6, e18934 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fathy DB, Kyle DJ & Leeb-Lundberg LM High-affinity binding of peptide agonists to the human B1 bradykinin receptor depends on interaction between the peptide N-terminal L-lysine and the fourth extracellular domain of the receptor. Mol. Pharmacol 57, 171–179 (2000). [PubMed] [Google Scholar]
- 29.Fathy DB, Mathis SA, Leeb T & Leeb-Lundberg LM A single position in the third transmembrane domains of the human B1 and B2 bradykinin receptors is adjacent to and discriminates between the C-terminal residues of subtype-selective ligands. J. Biol. Chem 273, 12210–12218 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Ha SN et al. Identification of the critical residues of bradykinin receptor B1 for interaction with the kinins guided by site-directed mutagenesis and molecular modeling. Biochemistry 45, 14355–14361 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Jarnagin K et al. Mutations in the B2 bradykinin receptor reveal a different pattern of contacts for peptidic agonists and peptidic antagonists. J. Biol. Chem 271, 28277–28286 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Kyle DJ Structural features of the bradykinin receptor as determined by computer simulations, mutagenesis experiments, and conformationally constrained ligands: establishing the framework for the design of new antagonists. Braz. J. Med. Biol. Res 27, 1757–1779 (1994). [PubMed] [Google Scholar]
- 33.Gieldon A, Lopez JJ, Glaubitz C & Schwalbe H Theoretical study of the human bradykinin-bradykinin B2 receptor complex. ChemBioChem 9, 2487–2497 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Regoli D, Rizzi A, Perron SI & Gobeil F Jr. Classification of kinin receptors. Biol. Chem 382, 31–35 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Leeb-Lundberg LM, Kang DS, Lamb ME & Fathy DB The human B1 bradykinin receptor exhibits high ligand-independent, constitutive activity. Roles of residues in the fourth intracellular and third transmembrane domains. J. Biol. Chem 276, 8785–8792 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Granier S et al. Structure of the δ-opioid receptor bound to naltrindole. Nature 485, 400–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shihoya W et al. Activation mechanism of endothelin ETB receptor by endothelin-1. Nature 537, 363–368 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Hong M Solid-state dipolar INADEQUATE NMR spectroscopy with a large double-quantum spectral width. J. Magn. Reson 136, 86–91 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Hohwy M, Jakobsen HJ, Eden M, Levitt MH & Nielsen NC Broadband dipolar recoupling in the nuclear magnetic resonance of rotating solids: A compensated C7 pulse sequence. J. Chem. Phys 108, 2686–2694 (1998). [Google Scholar]
- 40.Jaroniec CP, Filip C & Griffin RG 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. J. Am. Chem. Soc 124, 10728–10742 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Fung BM, Khitrin AK & Ermolaev K An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson 142, 97–101 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Delaglio F, Cornilescu G & Bax A TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Y & Bax A Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berjanskii MV, Neal S & Wishart DS PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 34, W63–W69 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Güntert P, Mumenthaler C & Wüthrich K Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol 273, 283–298 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Doreleijers JF et al. CING: an integrated residue-based structure validation program suite. J. Biomol. NMR 54, 267–283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozenne V et al. Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics 28, 1463–1470 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Neal S, Nip AM, Zhang H & Wishart DS Rapid and accurate calculation of protein 1H, 13C and 15N chemical shifts. J. Biomol. NMR 26, 215–240 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Brünger AT et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr 54, 905–921 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Linge JP, O’Donoghue SI & Nilges M Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 339, 71–90 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Leaver-Fay A et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konagurthu AS, Whisstock JC, Stuckey PJ & Lesk AM MUSTANG: a multiple structural alignment algorithm. Proteins 64, 559–574 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Larkin MA et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Jones DT Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol 292, 195–202 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Viklund H & Elofsson A OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24, 1662–1668 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Song Y et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bellucci F et al. A different molecular interaction of bradykinin and the synthetic agonist FR190997 with the human B2 receptor: evidence from mutational analysis. Br. J. Pharmacol 140, 500–506 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen Y & Bax A SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR 48, 13–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han B, Liu Y, Ginzinger SW & Wishart DS SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR 50, 43–57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen ED, Norn C, Frimurer TM & Meiler J Assessment and challenges of ligand docking into comparative models of G-protein coupled receptors. PLoS One 8, e67302 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article, its Supplementary Information file and from the corresponding authors upon reasonable request.