Abstract
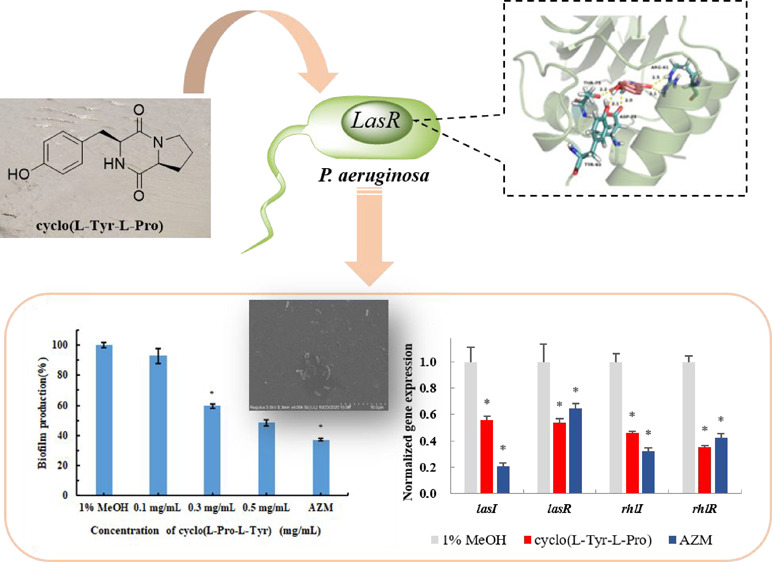
Bacterial quorum sensing (QS) is anticipated as a new potential target for the development of antimicrobial drugs. An anti-QS substance against Chromobacterium violaceum CV026 and Pseudomonas aeruginosa PA01 has been isolated and purified from the crude extracts of the marine fungus Penicillium chrysogenum DXY-1, and the accurate structure was identified as cyclo(l-Tyr-l-Pro). This cyclic dipeptide at sub-minimum inhibitory concentration can decrease the QS-regulated violacein production of C. violaceum CV026 by 79% and QS-mediated pyocyanin production, proteases, and elastase activity of P. aeruginosa PA01 by 41%, 20%, and 32%, respectively. In addition, it can also destroy the biofilm formation and decrease QS gene expression of P. aeruginosa PA01. Molecular docking was further performed, and the obtained data indicated that this dipeptide blocks the effect of QS autoinducers through competitive binding to the same pocket of the receptor proteins. We expect this anti-QS cyclic dipeptide to be a potential pro-drug treating drug-resistant P. aeruginosa infections, and these findings could relieve the alarming problem of microbial resistance to antimicrobial drugs.
Introduction
Antimicrobial resistance1 is one of the greatest health threats that humans face now and will face in the future.2 Tackling this increasingly serious antimicrobial resistance is a primary global concern.3,4 Currently used antibiotics can prevent infection by directly killing or inhibiting microorganisms. However, they may stimulate bacteria quickly developing resistance via changing the targets of antibiotics. Due to the self-evolution of bacteria, employing the conventional antibacterial screening strategies to solve the problem of drug-resistant bacterial infection is challenging.5 Therefore, it is exceptionally urgent to discover new targets and develop novel antimicrobial agents using nonconventional antibacterial screening approaches.6 In recent decades, the quorum sensing (QS) system, able to regulate the survivability and pathogenicity of microorganism, has been an appealing target for the development of new antimicrobial drugs7 and has attracted interest of pharmacologists and chemists. In contrast to conventional antibiotics that kill or inhibit bacterial growth, QS inhibitors (QSIs) can prevent bacterial infection by manipulating the pathogenic processes regulated by QS,8,9 such as virulence factor expression, biofilm formation, bacterial migration, and secretion regulation. Since these inhibitors put less selective pressure on bacteria, thus barely inducing drug resistance mutations, QSIs have been proposed as novel antibacterial compounds.10
Ocean-dwelling organisms can secrete marine natural products representing a highly diverse reserve of bio-actives because of their adaptation to extreme conditions characterized by high pressure and salt concentration, low temperatures, low nutrient, and low dissolved oxygen.11,12 A great deal of QSIs has been found in many kinds of marine microbes, including bacteria, fungi, algae, mosses, corals, and sponges. These QSIs have been reported to be active against various pathogens of Gram-negative and Gram-positive bacteria. Among the natural and artificial synthetic QSIs, the most intensively studied group is analogs and derivatives of N-acyl homoserine lactones, a kind of QS autoinducer in Gram-negative bacteria.13,14 In our previous work, a QSI against both Chromobacterium violaceum CV026 and Pseudomonas aeruginosa PA01 was isolated from a marine bacterium named Rheinheimera aquimaris QSI02, and its structure was finally identified as a cyclic dipeptide cyclo(l-Ser-l-Trp).15 Our recent work found that crude extracts of a marine fungus Penicillium chrysogenum DXY-116 discovered from marine sediments surrounding the Taiwan Strait (East Sea, Fuzhou, China) can inhibit the violacein production of the C. violaceum CV026, and a tyrosol has been identified as a QSI suppressing the QS-mediated virulence factor production and biofilm formation of P. aeruginosa PA01.
In this work, further tests and purification of the crude extracts of P. chrysogenum DXY-1 were accomplished and a cyclic dipeptide cyclo(l-Tyr-l-Pro) was identified as another active content. As reported, this cyclic dipeptide has been isolated from various microbial metabolites17−19 and was proven as an extracellular QS signal molecule produced by bacteria that can compete with the autoinducer C6HSL.20,21 However, a detailed study of this cyclic dipeptide suppressing the QS system of P. aeruginosa PA01 has not been reported. We first discovered this cyclic dipeptide from P. chrysogenum and evaluated its inhibitory ability against the QS system of C. violaceum CV026 and P. aeruginosa PA01. Besides inhibition of virulence factors and biofilm formation, its effect on QS gene (lasI, lasR, rhlI, and rhlR) expression of P. aeruginosa PA01 was also investigated. Also, further molecular docking data indicated that this cyclic dipeptide can bind the receptor protein competitively with the autoinducer, resulting in the inhibition of the QS system.
Results and Discussion
Cyclic dipeptides, widely biosynthesized by nonribosomal peptide synthetases and cyclic dipeptide synthetases in kinds of organisms, have been reported exhibiting various biological activities and regarded as one of the promising building blocks of drug candidates.22−24 Since they consist of endogenous amino acids, peptide drugs always show some advantages over nonpeptide drugs for their high activity and low toxicity. However, there are also some disadvantages like short half-life and low oral bioavailability. The superiority of the cyclic dipeptides to linear oligopeptides is their stability and oral bioavailability due to the lack of target bounds for exopeptidases.
Among the cyclic dipeptides isolated from secondary metabolites, prolyl-containing peptides make up a large proportion22,25 because of their bis-ring forming favor. Aromatic residues such as Phe and Tyr are also common in natural bioactive cyclic dipeptides.26 Rowley and co-workers reported the isolation of cyclo(l-Tyr-l-Pro) from Bacillus cereus D28 and its activity as an antagonist of bioluminescence against Vibrio harveyi.(17) McCormick and colleagues disclosed that cyclo(l-Tyr-l-Pro), cyclo(l-Tyr-d-Pro), and cyclo(l-Phe-l-Pro) from human vaginal isolate Lactobacillus reuteri RC-14 were able to interfere with the agr-mediated QS system of Gram-positive bacteria Staphylococcus.(27)
Isolation and Structure Confirmation of Cyclo(l-Tyr-l-Pro)
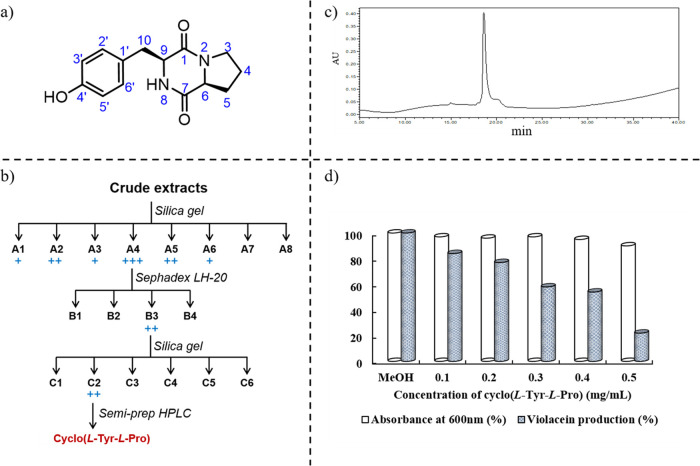
P. chrysogenum DXY-1 was cultured and treated as a previous protocol,16 and the crude extracts were separated with a silica gel column in different eluents, Sephadex LH-20, and preparative HPLC successively (Figure 1b). QSI activity of constituents during purification was monitored with C. violaceum CV026. As a result, cyclo(l-Tyr-l-Pro) (Figure 1a) was identified as a principle active compound. In combination with a comparison to the previously published literature,28 the structure was confirmed by various analytical methods, such as UV, ESI-MS, and NMR. Detailed NMR data are shown in the Supporting Information.
Figure 1.
(a) Structure of QS inhibitor cyclo(l-Tyr-l-Pro). (b) Isolation route of the crude extracts. (c) HPLC analysis of the obtained compound (gradient 5%–100% methanol in H2O for 40 min). (d) Effects of different concentrations of cyclo(l-Tyr-l-Pro) on the growth of C. violaceum CV026 and the production of violacein. MeOH (1% v/v) was used as a negative control.
Minimum Inhibitory Concentrations of Cyclo(l-Tyr-l-Pro)
Minimum inhibitory concentrations (MICs) of isolated cyclo(l-Tyr-l-Pro) against C. violaceum CV026 and P. aeruginosa PA01 were 3.0 and 6.2 mg/mL, respectively. To investigate whether the trace impurity generated during the separation process interfere with the bioactivity and if the stereochemistry of this cyclic dipeptide influences its activity, we tested the antimicrobial activity of chemically synthesized cyclo(l-Tyr-l-Pro) and its isomers against P. aeruginosa PA01. The MICs of different cyclic dipeptides are listed in Table 1; the MIC of synthetic cyclo(l-Tyr-l-Pro) was similar to that of the isolated one. However, the other synthetic isomers all showed less activity than the natural one, which only contains natural amino acid residues.
Table 1. MICs of Cyclic Dipeptides against P. aeruginosa PA01.
| cyclic dipeptide | MIC (mg/mL) |
|---|---|
| cyclo(l-Tyr-l-Pro)i | 6.2 |
| cyclo(l-Tyr-l-Pro)s | 6.4 |
| cyclo(l-Tyr-d-Pro) | 12.8 |
| cyclo(d-Tyr-l-Pro) | 12.8 |
| cyclo(d-Tyr-d-Pro) | 12.8 |
Anti-QS Activity of Cyclo(l-Tyr-l-Pro)
The violacein production of C. violaceum CV026 was only activated by adding the exogenous C6HSL as an inducer into the medium because of a lack of signal molecule synthase gene cviI. Spectrophotometric analysis data indicated that the violacein production decreased after adding cyclo(l-Tyr-l-Pro) to this medium. Furthermore, the inhibitory efficacy correlated positively with the concentration of cyclo(l-Tyr-l-Pro) (Figure 1d). Also, at a concentration of 0.5 mg/mL, it can inhibit the production of violacein of C. violaceum CV026 by 79%.
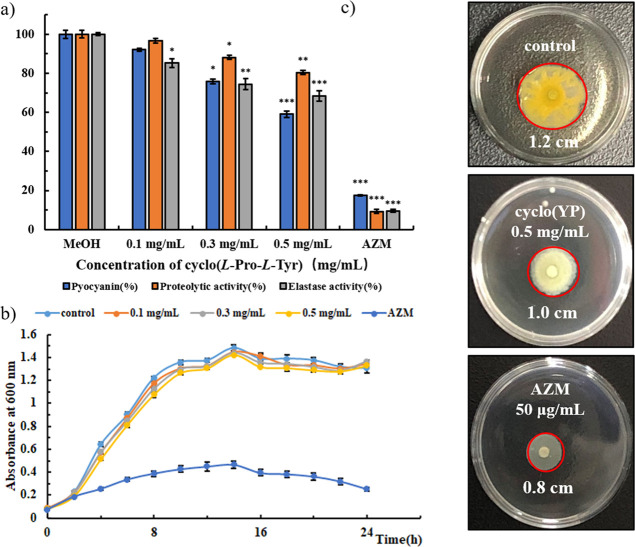
P. aeruginosa can cause disease in plants, animals, and human beings and exhibit multidrug resistance.29 The QS system in P. aeruginosa regulates the biofilm formation and production of virulence factors such as pyocyanin, elastase, and proteases.30 Inhibition of QS-controlled virulence in P. aeruginosa may be a novel strategy in treating infection caused by this bacterium.31 After treatment with different concentrations of cyclo(l-Tyr-l-Pro), the changes in the production of virulence factors of P. aeruginosa PA01 were analyzed by a quantitative chemical assay. Statistical analysis was performed using a one-way ANOVA test. As shown in Figure 2a, after being treated with this cyclic dipeptide, the production of pyocyanin and activities of proteases and elastase were inhibited dose dependently. Synthetic cyclo(l-Tyr-l-Pro) showed the same anti-QS tendency as the isolated one (Supporting Information, Figure S4). Also, at a concentration of 0.5 mg/mL cyclo(l-Tyr-l-Pro), which is lower than one-tenth MIC, the production of pyocyanin was inhibited by 41%, and activities of proteases and elastase were decreased by 20% and 32%, respectively. Also, significance analysis results indicated that they are significantly different versus control. Although the positive control 50 μg/mL AZM exhibited excellent inhibitory activity on the virulence factor production, it also severely affected the bacterial survival (Figure 2b), which further increased the antimicrobial resistance of P. aeruginosa. We also investigate the anti-QS activity of synthetic cyclo(Tyr-Pro) isomers and find that at the concentration of sub-MIC, they can also inhibit the production of QS-regulated virulence factor and biofilm formation dose dependently (Supporting Information, Figure S5). Meanwhile, the three isomers were less active than cyclo(l-Tyr-l-Pro), which were consistent with the MIC results.
Figure 2.
(a) Inhibitory effects of cyclo(l-Tyr-l-Pro) on the virulence factor production in P. aeruginosa PA01 at different concentrations. (b) Growth curve of P. aeruginosa PA01 under treatment with cyclo(l-Tyr-l-Pro). (c) Swarming motility of P. aeruginosa PA01. AZM (50 μg/mL) and MeOH (1% v/v) were used as positive and negative controls, respectively. Values are presented as mean ± SD (n = 3). Statistical analysis was performed using a one-way ANOVA test. *P < 0.05, * *P < 0.005, ***P < 0.0005 (vs control).
Then, the isolated dipeptide was tested for its reduction of P. aeruginosa PA01 swarming. As shown in Figure 2c, after being treated with 0.5 mg/mL cyclo(l-Tyr-l-Pro), the swarming motility migration diameter was ∼1.0 cm, which is smaller than the colony diameter recorded in the control group (∼1.2 cm). Also, the color of the colonies treated with the isolated cyclic dipeptide was light compared with that of the untreated one.
Inhibitory Effects on Biofilm Formation
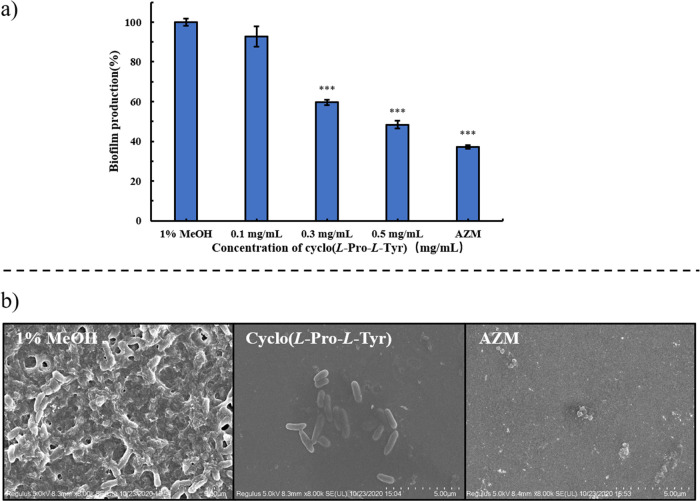
The impact of cyclo(l-Tyr-l-Pro) on P. aeruginosa PA01 biofilm formation was detected through a crystal violet assay method. The data showed that the treatment with 0.5 mg/mL cyclo(l-Tyr-l-Pro) resulted in the biofilm formation reducing to 48% compared with negative control (Figure 3a). SEM results indicated that, under the treatment with cyclo(l-Tyr-l-Pro), the bacterial cells were entirely in the form of short rods, but the biofilm was destroyed, leaving the bacteria naked and exposed (Figure 3b). These results supported the fact that cyclo(l-Tyr-l-Pro) did not damage bacterial cells, only inhibited the QS-regulated pathogenic behavior of P. aeruginosa.
Figure 3.
(a) Inhibition of biofilm formation under treatment of cyclo(l-Tyr-l-Pro) assessed by crystal violet staining. Values are presented as mean ± SD (n = 3). Statistical analysis was performed using a one-way ANOVA test. *P < 0.05, **P < 0.005, * **P < 0.0005 (vs control). (b) SEM images of P. aeruginosa PA01 biofilms treated with 1% MeOH, 0.5 mg/mL cyclo(l-Tyr-l-Pro), and 50 μg/mL AZM.
Inhibitory Effects of Cyclo(l-Tyr-l-Pro) on QS Gene Expression
Fluorescence real-time quantitative PCR was performed to investigate the effects of cyclo(l-Tyr-l-Pro) on QS gene expression of P. aeruginosa PA01. As shown in Figure 4, 0.5 mg/mL cyclo(l-Tyr-l-Pro) reduced expression of lasI, lasR, rhlI, and rhlR significantly by 43%, 46%, 54%, and 64%, respectively. For lasR and rhlR, the inhibition ratio was superior to AZM (50 μg/mL).
Figure 4.
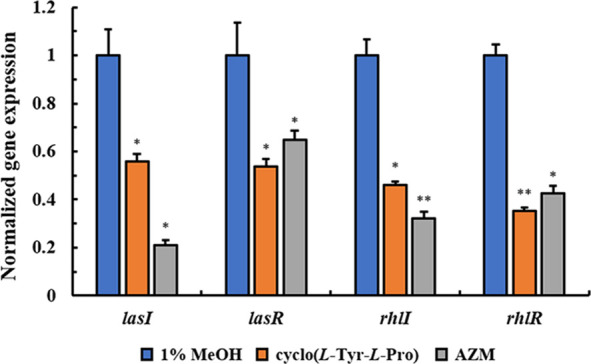
Effects of cyclo(l-Tyr-l-Pro) on P. aeruginosa PA01 QS gene expression. Values are presented as mean ± SD (n = 3). Statistical analysis was performed using a one-way ANOVA test. *P < 0.05, * *P < 0.005 (vs control).
Molecular Docking Analysis
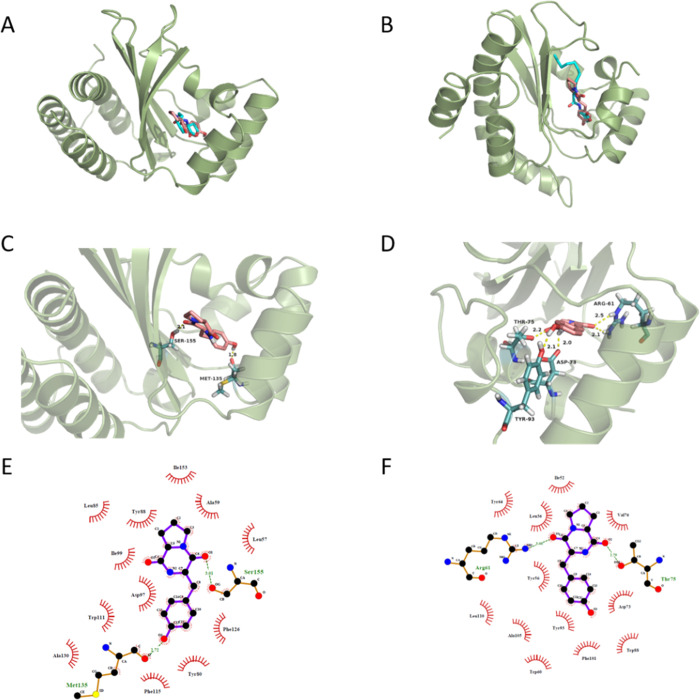
To investigate the possible mechanism of how cyclo(l-Tyr-l-Pro) quenches the QS system in P. aeruginosa PA01, the interaction between cyclo(l-Tyr-l-Pro) and the QS receptor CviR of C. violaceum CV026 and LasR of P. aeruginosa PA01 was evaluated by molecular docking. Crystal structures of CviR (PDB ID, 3QP1) and LasR (PDB ID, 2UV0) were used for the docking calculation (Figure 5). The natural ligands C6HSL and 3-oxo-C12HSL were docked back into their corresponding protein structures as a control, and the credible docking methods were established simultaneously. The control docking results were consistent with the reported X-ray structures.32,33 The docking results suggested that cyclo(l-Tyr-l-Pro) interacted with receptor proteins at the same pockets as natural ligands (Figure 5A,B). The binding energy for cyclo(l-Tyr-l-Pro) with CviR was −8.47 kcal/mol, which was lower than that for the autoinducer C6HSL (−6.65 kcal/mol). Hydrogen bonds were formed between cyclo(l-Tyr-l-Pro) and Ser155 and Met135 in CviR (Figure 5C). According to the data, the carbonyl oxygen of the Tyr moiety of the cyclic dipeptide interacted with the H species of the hydroxyl group of Ser 155, forming a hydrogen bond with the bond length ∼2.1 Å. At the same time, the H species on phenolic hydroxyl interacted with the carbonyl oxygen of Met135, forming a hydrogen bond with the bond length ∼1.8 Å. The key hydrophobic interactions with nearby residues are shown in Figure 5E. For LasR protein, there are four similar chains in 2UV0. Therefore, we chose the H chain as the target in docking, the binding energy for this cyclic dipeptide (−8.22 kcal/mol) was proximate with the natural ligand (−8.20 kcal/mol). Hydrogen bonds formed at Tyr93 (with phenolic hydroxyl, ∼2.1 Å), Thr75 (with carbonyl oxygen of Tyr moiety, ∼2.2 Å), Asp73 (with hydrogen of amide, ∼2.0 Å), and Arg61 (with carbonyl oxygen of Pro moiety, ∼2.1 and 2.5 Å) (Figure 5D). The key hydrophobic interactions between cyclo(l-Tyr-l-Pro) and surrounding residues are illustrated in Figure 5F. It is concluded that cyclo(l-Tyr-l-Pro) achieved the inhibition of the QS system in P. aeruginosa PA01 by competing with the natural ligand of binding to the same amino acid sites in LasR; such a competitive binding results in a possible change in the conformation of receptor protein that led to the loss of the corresponding physiological functions.
Figure 5.
(A, B) Docked conformation of natural ligand C6HSL (left) and 3-oxo-C12HSL (right) with cyan sticks and cyclo(l-Tyr-l-Pro) with pink sticks into the active site of 3QP1 and 2UV0 receptor proteins respectively. The hydrogen bonds between cyclo(l-Tyr-l-Pro) and (C) 3QP1 or (D) 2UV0 are shown with dotted yellow lines. (E, F) Ligplot of cyclo(l-Tyr-l-Pro) bound to the 3QP1 (left) and H chain of 2UV0 (right) showing the key hydrophobic interactions.
However, we must point out that cyclo(l-Tyr-l-Pro) seems less active against both model bacteria in comparison with cyclo(l-Ser-l-Trp)15 involved in our previous work. We speculated that less bindable groups and the rigid structure of the N-bridged heterocycle accounted for this. To elucidate the structure and activity relationship inside, further study on the interaction between receptor proteins and dipeptides as well as the structural modification is undergoing.
Conclusions
A cyclo(l-Tyr-l-Pro) was isolated and identified from the crude extracts of marine fungus P. chrysogenum DXY-1 and had shown a prominent QS-regulated virulence inhibiting effect on model bacteria C. violaceum CV026 and P. aeruginosa PA01. Furthermore, this cyclic dipeptide inhibited the QS-mediated pathogenicity via competitively binding to the active pocket of receptor protein to block the activation of downstream genes required for virulence factor expression in these two indicator strains. The results obtained in this study will provide a theoretical basis for the molecular design of QS oriented multitarget antimicrobial peptides and the development of novel short peptide antibiotics in order to alleviate bacterial multidrug resistance and virulence. This study will lead to better antimicrobial drug design and provide a theoretical basis for the synthesis of QS inhibitors, offering promising antipathogenic drugs.
Materials and Methods
Bacterial Strains
C. violaceum CV026, (mini-Tn5 mutant of C. violaceum ATCC 31532), and P. aeruginosa PA01 were used as QS indicator strains in this study. Each strain was cultured for 12 h with Luria Bertani (LB) broth containing yeast extract (0.5% w/v), peptone (1% w/v), and NaCl (1% w/v) in 100 mL of distilled water. The medium was solidified with 1.2% agar and supplemented with appropriate antibiotics (C. violaceum CV026, 50 μg/mL kanamycin) when required.
Chemical Agents and Instruments
All commercial organic solvents and salts (Sigma Aldrich, TCI, Adamas, J&K, Energy, etc.) were used without further purification. Cyclic dipeptides were purchased from GL Biochem (Shanghai) Ltd. Azithromycin was purchased from Macklin, and kanamycin sulfate was purchased from Sangon Biotech. Flash chromatography was performed using a 200–300 or 100–200 mesh silica gel (Qingdao Haiyang Chemical Co., Ltd.). Preparative HPLC separations were performed using Shimadzu LC-20AR semipreparative solvent delivery units and a Shimadzu SPD-20A UV detector equipped with a Shimadzu C18 column (5.0 μm, 20 × 250 mm) at a flow rate of 10 mL/min. NMR spectra were acquired using a Bruker 400 MHz, and d6-DMSO (J&K) was used as the solvent during NMR analysis. Optical density (OD) was recorded on a microplate reader SpectraMax Plus 384 (Molecular Devices, USA). Fluorescence real-time quantitative PCR was done on a real-time system (Bio-Rad Laboratories, Hercules, CA).
Isolation and Structural Elucidation
A 30 L fermentation broth of marine-derived fungus P. chrysogenum DXY-1 was treated with a homogenizer to disrupt the mycelia and then extracted three times with EtOAc as previous work. Subsequently, the crude extract was separated on a silica gel with CH2Cl2/MeOH (v/v, 10:1) as an eluent, yielding eight fractions (A1–A8). Using the agar well diffusion method, each fraction was tested using C. violaceum CV026 as a monitor strain to identify the target fraction with an anti-QS activity. The target fraction was purified by ordinal chromatographic methods including column chromatography over Sephadex LH-20 and a silica gel and finally semipreparative RP-HPLC using a C18 column to obtain the pure QSI (gradient 5%–100% of MeOH in H2O in 40 min, >95% purity by HPLC). The pure QSI was further identified employing different analytical techniques like UV–vis, MS, 1H NMR, 13C NMR, and 2D NMR.
Violacein Assay
The effect of separated fractions and pure QSI compound on the production of violacein was quantified via the following procedure. In summary, 400 μL of C. violaceum CV026 was grown in 20 mL of LB broth containing different concentrations of components, 20 μL (50 μg/mL) of kanamycin sulfate, and 100 μL of C6HSL, and the broth was allowed shaking at 150 rpm at 28 °C for 12 h. Then, from each flask, 1 mL of culture was removed, placed to a 1.5 mL Eppendorf tube, and centrifuged at 12000 rpm for 10 min to obtain insoluble violacein. Afterward, the supernatant was discarded, and 1 mL of DMSO was added to the deposit. The solution was centrifuged again at 12,000 rpm for 10 min, and the supernatant was collected to measure the absorbance at a wavelength of 585 nm using a microplate reader (SpectraMax Plus 384, Molecular Devices, USA). The harvested bacterial cells were resuspended in ddH2O (1 mL) to determine the cell growth through measurement of the optical density at a wavelength of 600 nm (OD600).
Pyocyanin Assay
P. aeruginosa PA01 was grown to the logarithmic phase and inoculated into LB medium, containing different concentrations of QSI compound, 1% MeOH, and 50 μg/mL azithromycin (AZM), at an initial OD600 of 0.05, and the medium was allowed shaking for 12 h at 37 °C. After centrifugation, the broth (5 mL) was extracted with CHCl3 (3 mL), and the organic layer was acidized with 0.2 M HCl (1 mL). The resulting solution was centrifuged, and the aqueous layer was collected to measure its absorbance at 520 nm. The decrease in pyocyanin production was calculated based on the percent change in absorbance. P. aeruginosa ATCC 27853 was used as the quality control strain, which required MICs that approximated therapeutic concentrations, and was stable and reproducible.
Proteolytic Activity Assay
Proteolytic activity was investigated using a modified protocol previously reported.15 First, 0.1% activated P. aeruginosa PA01 was cultured in LB media containing different concentrations of QSI compound, 1% MeOH, or AZM (50 μg/mL) at 37 °C and 150 rpm for 12 h. After centrifugation, the supernatant (100 μL) was filtered using a 0.22 μm nylon filter and then diluted with 400 μL of buffer (50 mM K2HPO4, 0.8% azocasein, pH 7.0). After incubation at 30 °C for 3 h, HCl (1.5 M) was added to the solution and the mixture was reacted at 0 °C for 10 min. Immediately after centrifugation, NaOH solution (0.5 M) was added to the liquid supernatant. The sample was centrifuged again to collect the top layer, and its optical density at 440 nm was measured. The percent change in absorbance was then calculated to determine the proteolytic activity.
Elastase Activity Assay
P. aeruginosa PA01 was activated and cultured in the conditions as described above in Proteolytic Activity Assay. After centrifugation, the top layer (100 μL) was filtered using a 0.22 μm nylon filter and added to 900 μL of Na2HPO4 buffer (pH 7.0) with 10 mg elastin-Congo red (Elastin Products). After incubation at 37 °C for 2 h, the samples were centrifuged at 10,000g for 10 min. The absorbance of the top layer was measured at 495 nm to calculate the elastase activity.
Swarming Motility of P. aeruginosa PA01
Swarming motility of P. aeruginosa PA01 was investigated in 35 × 10 mm Petri dishes containing 2 mL of agar medium [glucose (10 mg/mL), agar (5 mg/mL), tryptone (5 mg/mL), and yeast extract powder (2 mg/mL)] with control solution, cyclo-dipeptide (0.5 mg/mL), and AZM (50 μg/mL). A filter paper (diameter 2.0 mm) soaked in 2 μL of bacterial suspension (OD600 = 0.05) was centered on each dish and incubated at 37 °C for 48 h. After incubation, colony diameters were measured to test the swarming motility of bacteria.34
Biofilm Assay
The biofilm assay was conducted in 96-well plates as a previous protocol.35 In brief, P. aeruginosa PA01 was inoculated into LB medium at an initial OD600 of 0.05 and cultured with the QSI compound, 1% MeOH, or AZM at 37 °C for 24 h. The plates were washed with PBS three times to remove the broth, then dried at room temperature for 20 min, and stained with 1% crystal violet for 5 min. The stained biofilms were washed with PBS followed by the addition of 200 μL of 95% ethanol. The absorbance was read at a wavelength of 545 nm (Spectra Max Plus384, Molecular Devices, USA). To further confirm the QS inhibitory effect of the extract compound, the suspension of P. aeruginosa PA01 (OD600 = 0.05) with appropriate concentration of extracted QSI and control agents were cultured in a six-well plate containing sterile slide glasses placed at the bottom of the wells, and the plates were then incubated at 37 °C for 2 days. After incubation, the slide glasses were fixed with 2.5% glutaraldehyde and sputter-coated with gold for SEM observations and photomicrography.
Fluorescence Real-Time Quantitative PCR
Expression of P. aeruginosa PA01 QS genes was investigated following the reported protocol36 using a real-time system (Bio-Rad Laboratories, Hercules, CA). The reaction conditions in this study were as follows: 95 °C for 3 min followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. The relative fold changes of mRNA levels were measured using the comparative cycle threshold (ΔΔCq) method for gene expression. The experiments were repeated independently three times using different RNA samples.
Docking Studies
Molecular docking studies were performed to identify the conformational changes in the protein structure due to the interaction of the QSI compound with the receptor proteins CviR and LasR. The energies of the isolated QSI and autoinducers were minimized with ChemBio3D Ultra 14.0, and the structures were saved as PDB form. The crystal structures of C. violaceum CviR (Protein Data Bank (PDB) ID: 3QP1, resolution of 2.0 Å) and P. aeruginosa LasR (PDB ID: 2UV0, resolution of 1.8 Å) were downloaded from the RCSB PDB website, and the co-crystal organic moiety was removed. Docking was accomplished using AutoDockTools-1.5.6 following a general protocol that we previously reported15 The visual graphics of the interaction between small molecules were output via PyMol and LigPlus.
Acknowledgments
This work was financially supported by the Natural Science Foundation of China (U1805234 and 22007013), Natural Science Foundation of Fujian Province of China (2019J01264), Program for Innovative Research Team in Science and Technology in Fujian Province University, 100 Talents Program of Fujian Province, Special Funds of the Central Government Guiding Local Science and Technology Development (2020L3008), and Scientific Research Start-up Fund for High-Level Talents in Fujian Normal University (004828).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00020.
NMR spectroscopic data, UV–vis spectrum of isolated cyclo(l-Tyr-l-Pro), inhibitory effect of isolated and synthesized cyclo(l-Tyr-l-Pro) on QS-regulated virulence factor expression of P. aeruginosa PA01, and anti-QS activity of different isomers of cyclo(l-Tyr-l-Pro) (PDF)
Author Contributions
∥ X.Y. and L.L. contributed equally to this work and are joint first authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Song D.; Meng J.; Cheng J.; Fan Z.; Chen P.; Ruan H.; Tu Z.; Kang N.; Li N.; Xu Y.; Wang X.; Shu F.; Mu L.; Li T.; Ren W.; Lin X.; Zhu J.; Fang X.; Amrein M. W.; Wu W.; Yan L. T.; Lü J.; Xia T.; Shi Y. Pseudomonas aeruginosa quorum-sensing metabolite induces host immune cell death through cell surface lipid domain dissolution. Nat. Microbiol. 2019, 4, 97. 10.1038/s41564-018-0290-8. [DOI] [PubMed] [Google Scholar]
- Mullard A. Momentum builds around new antibiotic business models. Nat. Rev. Drug Discov. 2014, 13, 711–713. 10.1038/nrd4455. [DOI] [PubMed] [Google Scholar]
- Woolhouse M.; Farrar J. Policy: An intergovernmental panel on antimicrobial resistance. Nature 2014, 509, 555–557. 10.1038/509555a. [DOI] [PubMed] [Google Scholar]
- Woolhouse M. E. J.; Ward M. J. Microbiology Sources of antimicrobial resistance. Science 2013, 341, 1460–1461. 10.1126/science.1243444. [DOI] [PubMed] [Google Scholar]
- Sprenger M.; Fukuda K. ANTIMICROBIAL RESISTANCE. New mechanisms, new worries. Science 2016, 351, 1263–1264. 10.1126/science.aad9450. [DOI] [PubMed] [Google Scholar]
- Laxminarayan R.; Sridhar D.; Blaser M.; Wang M.; Woolhouse M. Achieving global targets for antimicrobial resistance. Science 2016, 353, 874–875. 10.1126/science.aaf9286. [DOI] [PubMed] [Google Scholar]
- Whiteley M.; Diggle S. P.; Greenberg E. P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D.; Wezeman R. J.; Siegel A. P.; Blackwell H. E. Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J. Am. Chem. Soc. 2005, 127, 12762–12763. 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- O’Loughlin C. T.; Miller L. C.; Siryaporn A.; Drescher K.; Semmelhack M. F.; Bassler B. L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 17981–17986. 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Cell communication: stop the microbial chatter. Nature 2014, 511, 493–497. 10.1038/511493a. [DOI] [PubMed] [Google Scholar]
- de la Calle F. Marine microbiome as source of natural products. Microb. Biotechnol. 2017, 10, 1293–1296. 10.1111/1751-7915.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marris E. Marine natural products: drugs from the deep. Nature 2006, 443, 904–905. 10.1038/443904a. [DOI] [PubMed] [Google Scholar]
- Fuqua C.; Parsek M. R.; Greenberg E. P. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 2001, 35, 439–468. 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Welch M.; Mikkelsen H.; Swatton J. E.; Smith D.; Thomas G. L.; Glansdorp F. G.; Spring D. R. Cell-cell communication in Gram-negative bacteria. Mol. BioSyst. 2005, 1, 196–202. 10.1039/B505796P. [DOI] [PubMed] [Google Scholar]
- Sun S.; Dai X.; Sun J.; Bu X.; Weng C.; Li H.; Zhu H. A diketopiperazine factor from Rheinheimera aquimaris QSI02 exhibits anti-quorum sensing activity. Sci. Rep. 2016, 6, 39637. 10.1038/srep39637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.; Sun S.; Li L.; Dai X.; Li H.; He Q.; Zhu H. Tyrosol from marine Fungi, a novel Quorum sensing inhibitor against Chromobacterium violaceum and Pseudomonas aeruginosa. Bioorg. Chem. 2019, 91, 103140. 10.1016/j.bioorg.2019.103140. [DOI] [PubMed] [Google Scholar]
- Teasdale M. E.; Donovan K. A.; Forschner-Dancause S. R.; Rowley D. C. Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar. Biotechnol. 2011, 13, 722–732. 10.1007/s10126-010-9334-7. [DOI] [PubMed] [Google Scholar]
- Capon R. J.; Stewart M.; Ratnayake R.; Lacey E.; Gill J. H. Citromycetins and bilains A-C: new aromatic polyketides and diketopiperazines from Australian marine-derived and terrestrial Penicillium spp. J. Nat. Prod. 2007, 70, 1746–1752. 10.1021/np0702483. [DOI] [PubMed] [Google Scholar]
- Mitova M.; Tommonaro G.; Hentschel U.; Müller W. E.; De Rosa S. Exocellular cyclic dipeptides from a Ruegeria strain associated with cell cultures of Suberites domuncula. Mar. Biotechnol. 2004, 6, 95–103. 10.1007/s10126-003-0018-4. [DOI] [PubMed] [Google Scholar]
- Degrassi G.; Aguilar C.; Bosco M.; Zahariev S.; Pongor S.; Venturi V. Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: cross-talk with quorum sensing bacterial sensors. Curr. Microbiol. 2002, 45, 250–254. 10.1007/s00284-002-3704-y. [DOI] [PubMed] [Google Scholar]
- Holden M. T.; Ram Chhabra S.; de Nys R.; Stead P.; Bainton N. J.; Hill P. J.; Manefield M.; Kumar N.; Labatte M.; England D.; Rice S.; Givskov M.; Salmond G. P.; Stewart G. S.; Bycroft B. W.; Kjelleberg S.; Williams P. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other gram-negative bacteria. Mol. Microbiol. 1999, 33, 1254–1266. 10.1046/j.1365-2958.1999.01577.x. [DOI] [PubMed] [Google Scholar]
- de Carvalho M. P.; Abraham W.-R. Antimicrobial and biofilm inhibiting diketopiperazines. Curr. Med. Chem. 2012, 19, 3564–3577. 10.2174/092986712801323243. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A.; Ivanov V. T. Conformational states and biological activity of cyclic peptides. Tetrahedron 1975, 31, 2177–2209. 10.1016/0040-4020(75)80216-X. [DOI] [Google Scholar]
- Borthwick A. D. 2,5-Diketopiperazines: synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem. Rev. 2012, 112, 3641–3716. 10.1021/cr200398y. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Mohandas C.; Nambisan B.; Kumar D. R. S.; Lankalapalli R. S. Isolation of proline-based cyclic dipeptides from Bacillus sp. N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties. World J. Microbiol. Biotechnol. 2013, 29, 355–364. 10.1007/s11274-012-1189-9. [DOI] [PubMed] [Google Scholar]
- Brockmeyer K.; Li S. M. Mutations of Residues in Pocket P1 of a Cyclodipeptide Synthase Strongly Increase Product Formation. J. Nat. Prod. 2017, 80, 2917–2922. 10.1021/acs.jnatprod.7b00430. [DOI] [PubMed] [Google Scholar]
- Li J.; Wang W.; Xu S. X.; Magarvey N. A.; McCormick J. K. Lactobacillus reuteri-produced cyclic dipeptides quench agr-mediated expression of toxic shock syndrome toxin-1 in staphylococci. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3360–3365. 10.1073/pnas.1017431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buedenbender L.; Robertson L. P.; Lucantoni L.; Avery V. M.; Kurtböke D. I.; Carroll A. R. HSQC-TOCSY Fingerprinting-Directed Discovery of Antiplasmodial Polyketides from the Marine Ascidian-Derived Streptomyces sp. (USC-16018). Mar. Drugs 2018, 16, 189. 10.3390/md16060189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strateva T.; Yordanov D. Pseudomonas aeruginosa - a phenomenon of bacterial resistance. J. Med. Microbiol. 2009, 58, 1133–1148. 10.1099/jmm.0.009142-0. [DOI] [PubMed] [Google Scholar]
- Papaioannou E.; Utari P. D.; Quax W. J. Choosing an appropriate infection model to study quorum sensing inhibition in Pseudomonas infections. Int. J. Mol. Sci. 2013, 14, 19309–19340. 10.3390/ijms140919309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukarieh F.; Williams P.; Stocks M. J.; Cámara M. Pseudomonas aeruginosa Quorum Sensing Systems as Drug Discovery Targets: Current Position and Future Perspectives. J. Med. Chem. 2018, 61, 10385–10402. 10.1021/acs.jmedchem.8b00540. [DOI] [PubMed] [Google Scholar]
- Bottomley M. J.; Muraglia E.; Bazzo R.; Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- Chen G.; Swem L. R.; Swem D. L.; Stauff D. L.; O’Loughlin C. T.; Jeffrey P. D.; Bassler B. L.; Hughson F. M. A strategy for antagonizing quorum sensing. Mol. Cell 2011, 42, 199–209. 10.1016/j.molcel.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M. C.; Paul S.; Gupta P.; Tribedi P.; Sarkar S.; Manna D.; Bhattacharjee S. 3-Amino-4-aminoximidofurazan derivatives: small molecules possessing antimicrobial and antibiofilm activity against Staphylococcus aureus and Pseudomonas aeruginosa. J. Appl. Microbiol. 2016, 120, 842–859. 10.1111/jam.13063. [DOI] [PubMed] [Google Scholar]
- Kim H.-S.; Lee S.-H.; Byun Y.; Park H.-D. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci. Rep. 2015, 5, 8656. 10.1038/srep08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J.; Sun F.; Feng W.; Sun Y.; Qiu X.; Xiong L.; Liu Y.; Chen Y. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J. Appl. Microbiol. 2016, 120, 966–974. 10.1111/jam.13073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.