This study demonstrates that nuclear DEK governs quiescence and metabolic homeostasis of HSC and preserves HSC potential. Mechanistically, DEK dampens chromatin accessibility in HSC by inducing deacetylation of H3K27 and governs gene expression, including Akt1/2.
Abstract
The oncogene DEK is found fused with the NUP214 gene creating oncoprotein DEK-NUP214 that induces acute myeloid leukemia (AML) in patients, and secreted DEK protein functions as a hematopoietic cytokine to regulate hematopoiesis; however, the intrinsic role of nuclear DEK in hematopoietic stem cells (HSCs) remains largely unknown. Here, we show that HSCs lacking DEK display defects in long-term self-renew capacity, eventually resulting in impaired hematopoiesis. DEK deficiency reduces quiescence and accelerates mitochondrial metabolism in HSCs, in part, dependent upon activating mTOR signaling. At the molecular level, DEK recruits the corepressor NCoR1 to repress acetylation of histone 3 at lysine 27 (H3K27ac) and restricts the chromatin accessibility of HSCs, governing the expression of quiescence-associated genes (e.g., Akt1/2, Ccnb2, and p21). Inhibition of mTOR activity largely restores the maintenance and potential of Dek-cKO HSCs. These findings highlight the crucial role of nuclear DEK in preserving HSC potential, uncovering a new link between chromatin remodelers and HSC homeostasis, and have clinical implications.
Introduction
Hematopoietic stem cells (HSCs) can differentiate into all blood cell lineages and are essential for the continuous replenishment of the hematopoietic system (Dzierzak and Bigas, 2018; Eaves, 2015; Bigas and Waskow, 2016). Quiescence is a fundamental characteristic of HSCs that ensures longevity and protects HSCs against genetic damage and stem cell exhaustion. HSC homeostasis requires a coordinated cell-cycle balance between quiescence and activation (Nakamura-Ishizu et al., 2014; Calvi and Link, 2015), and requires regulated metabolic activity (Ito et al., 2019; Ito and Ito, 2018; Signer et al., 2014; Liang et al., 2020; Ho et al., 2017), to preserve the self-renewal capacity of HSCs and meet the demands of hematopoiesis. Thus, elucidating the molecular networks that regulate HSC quiescence and metabolism will expand the understanding of HSC homeostasis and be beneficial for therapeutic uses of HSCs.
DEK was initially identified in a fusion with the NUP214 nucleoporin in a subtype of acute myeloid leukemia (Ho et al., 2017; Soekarman et al., 1992), and the oncoprotein DEK-NUP214 targets a population of long-term HSC (LT-HSC) for leukemogenic transformation (Oancea et al., 2010). The DEK protein is conserved in mammals and is the only member of its protein class (Riveiro-Falkenbach and Soengas, 2010; Broxmeyer et al., 2013), which has no known enzymatic activity but contains a central DNA binding domain of the SAF (scaffold attachment factor) or SAP (after SAF-A/B; acinus; Pias) box (Kappes et al., 2004; Böhm et al., 2005). DEK has been revealed to promote transcription through direct promoter and enhancer binding (Sandén et al., 2014; Fu et al., 1997), or by acting as a coactivator of the nuclear splicing factor U2AF to regulate intron removal (Soares et al., 2006; Yue et al., 2020). Importantly, in vitro studies have revealed chromatin and histone binding activities for DEK, and DEK interacts with heterochromatin protein 1α, indicating DEK as a chromatin architectural protein (Kappes et al., 2011). Besides intracellular DEK, DEK is one of two known nuclear proteins that can be secreted (Mor-Vaknin et al., 2006; Mor-Vaknin et al., 2011). Extracellular DEK may act as a hematopoietic cytokine, which increases LT-HSC number and decreases hematopoietic progenitor cell (HPC) number via the CXC chemokine receptor CXCR2 (Capitano et al., 2019). The above evidence implies that DEK has crucial roles in hematopoiesis and leukemogenesis; thus, illuminating the exact functions of DEK in HSCs is essential if we are to use DEK as a hematopoietic cytokine or target DEK in HSCs. Importantly, this will also help us to understand the roles of the oncoprotein DEK-NUP214 in acute myeloid leukemia.
However, the most important and fundamental roles of nuclear DEK in HSC remain largely unknown. Serrano-Lopez et al. (2018) reported that loss of nuclear DEK has no obvious effect on hematopoiesis in mice at 8–10 wk of age. Controversially, Broxmeyer et al. (2012) found that DEK-deficient HPCs show enhanced CFU-granulocyte by promoting HPC proliferation in vitro, suggesting a role for DEK in cell-cycling regulation of hematopoietic cells. Here, we aimed to explore the precise role of DEK in HSCs and to elucidate the molecular mechanisms involved. Unexpectedly, we found that DEK deficiency reduces hematopoiesis in mice under steady state or stress. Notably, DEK is essential for quiescence maintenance of HSCs and governs mitochondrial metabolism by restricting mammalian target of rapamycin (mTOR) activity in HSCs. We next conducted a series of functional and mechanistic studies, which revealed that DEK recruits the nuclear receptor corepressor 1 (NCoR1)/histone deacetylase 3 (HDAC3) complex to induce deacetylation of histone 3 at lysine 27 (H3K27), thus playing as a chromatin accessibility regulator and transcriptionally regulating its critical target genes (e.g., Akt1/2). Furthermore, knockdown of DEK in human CD34+ primitive hematopoietic cells also promotes cell cycling, suggesting that DEK is essential for the quiescence maintenance of HSCs. Our studies uncover an essential role of DEK in regulating HSC potential and provide fundamental knowledge of nuclear DEK, thus offering insights for future clinical applications.
Results
DEK deficiency decreases the HSC pool
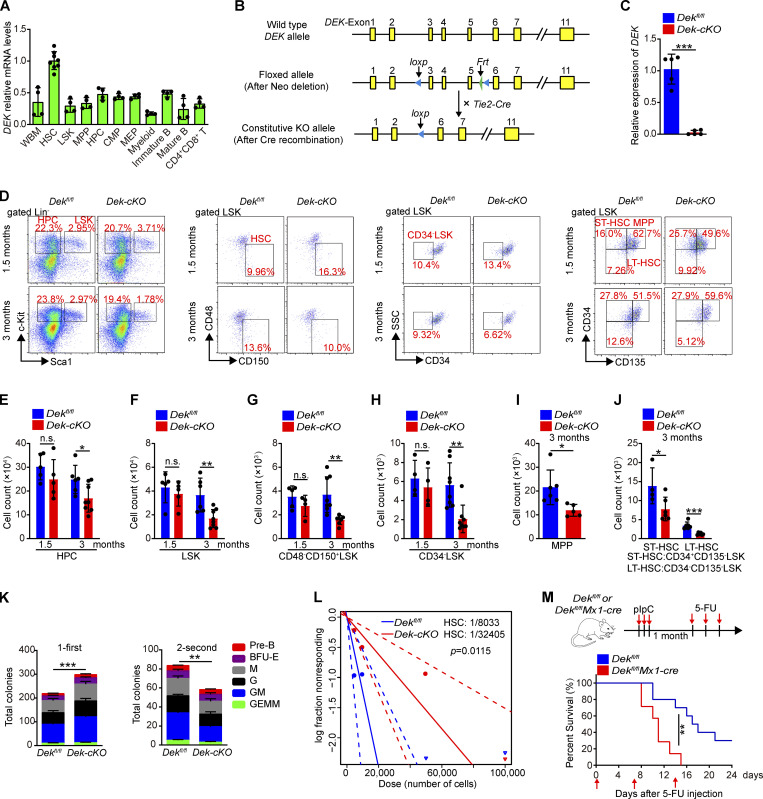
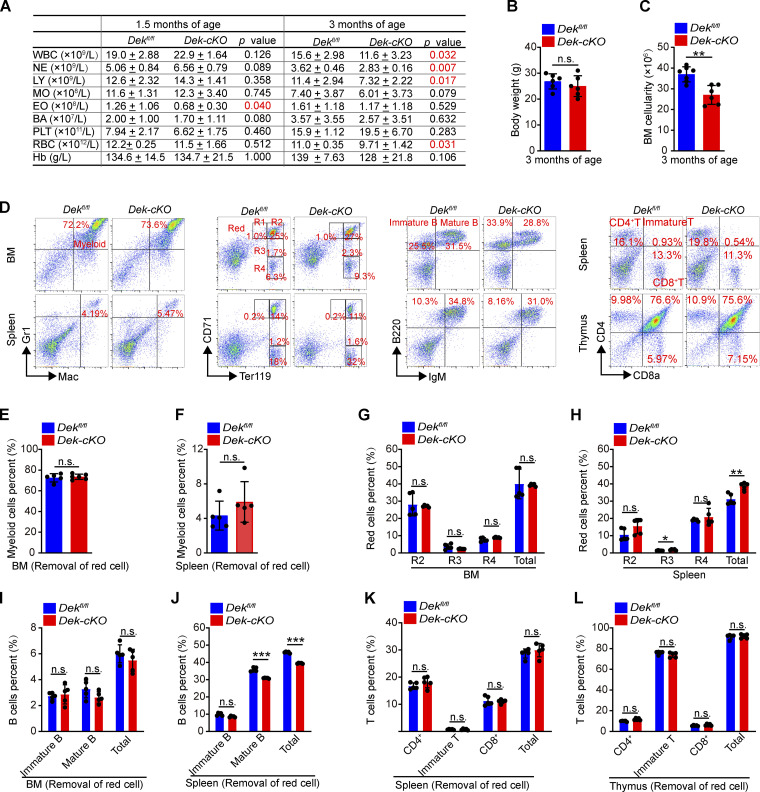
To determine the role of DEK in hematopoiesis, we characterized the expression of DEK in subsets of primitive and mature bone marrow (BM) cells. DEK exhibited higher expression in LT-HSCs than in LSK (Lin−Sca-1+c-Kit+) cells, HPCs, or mature cells (Fig. 1 A), indicating an important role for DEK in HSCs. Dek−/− knockout mice showed no significant changes in HSC or HPC number at 8–10 wk of age (Serrano-Lopez et al., 2018). Here, we first generated a conditional knockout allele of the DEK gene (Dekfl/+), in which exon 3 to exon 5 is flanked with loxp sites. Then, we crossed Dekfl/+mice with Tie2-Cre mice, in which a transgene encoding Cre recombinase is expressed in endothelial cells and HSCs under the control of the Tie2 promoter, to generate control Dekfl/fl mice and Dekfl/flTie2-Cre mice, with conditional deletion of Dek (called Dek-cKO mice here; Fig. 1, B and C). We found no obvious changes in hematological parameters in Dek-cKO mice at 1.5 mo of age (Fig. S1 A), but these mice exhibited considerably fewer blood cells compared with Dekfl/fl mice at 3 mo of age (Fig. S1 A). DEK deficiency also resulted in decreased BM cellularity of mice at 3 mo of age (Fig. S1, B and C); however, the frequency of myeloid, red, B, and T cells were comparable between Dek-cKO mice and Dekfl/fl mice (Fig. S1, D–L). Thus, our data indicate that deletion of DEK results in impaired hematopoiesis but has slight effects on the differentiation of mature blood cells.
Figure 1.
DEK is highly expressed in HSCs and maintains HSC pool. (A) qRT-PCR analysis of DEK in different hematopoietic cell subsets in WT mice (n = 4–8), including WBM (whole BM cells), HSCs (Lin−c-Kit+Sca-1+CD48−CD150+), LSK cells (Lin−c-Kit+Sca-1+), MPPs (Lin−c-Kit+Sca-1+CD48+CD150−), HPCs (Lin−c-Kit+Sca-1−), CMPs (CD34+CD16/32medHPC), MEPs (CD34−CD16/32−HPC), myeloid cells (Gr1+Mac+), immature B cells (B220+IgM−), mature B cells (B220+IgM+), and CD4+CD8+ T cells. (B) Gene targeting strategies for hematopoietic lineage-specific KO of DEK in mice. (C) Relative mRNA expression of DEK in freshly sorted Dekfl/fl and Dek-cKO HSCs (n = 5). (D) FACS analysis of HPCs, LSK cells, HSCs, MPPs (CD34+CD135+LSK), ST-HSCs (CD34+CD135−LSK), and LT-HSCs (CD34−CD135−LSK) in BM cells of Dekfl/fl and Dek-cKO mice at 1.5 or 3 mo of age. (E–H) Count of HPCs, LSK cells, and HSCs (CD48−CD150+LSK or CD34−LSK) in BM cells of Dekfl/fl and Dek-cKO mice at 1.5 or 3 mo of age (n = 5–8). (I and J) Count of MPPs, ST-HSCs, and LT-HSCs in BM cells of Dekfl/fl and Dek-cKO mice at 3 mo of age (n = 6). (K) In vitro assay of the CFUs of megakaryocyte colonies (GEMM), granulocyte-macrophage colonies (GM), granulocyte colonies (G), macrophage colonies (M), and burst-forming unit-erythroid colonies (BFU-E) at 10–12 d after plating Dekfl/fl and Dek-cKO BM cells (n = 8). For the second plating, live cells from the colonies obtained during the first plating were plated as before and cultured for 10–12 d (n = 4). (L) The CRU frequency is determined by extreme limiting dilution analysis, showing the estimated HSC frequency in the BM of Dekfl/fl and Dek-cKO mice (n = 3 donor mice, five recipients at each cell concentration). (M) Survival curve of Dekfl/fl and Dekfl/flMx1-Cre mice following sequential 5-FU treatment (n = 10). Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test or Mantel-Cox test. Data are representative of three independent experiments.
Figure S1.
DEK deletion impairs hematopoiesis in mice. (A) PB complete blood cell counts of Dekfl/fl and Dek-cKO mice at 1.5 and 3 mo of age (n = 5–7). WBC, white blood cells; NE, neutrophils; LY, lymphocytes; MO, monocytes; EO, eosinophils; BA, basophils; PLT, platelets; Hb, hemoglobin. (B and C) Weight analysis and BM cells count in Dekfl/fl and Dek-cKO mice at 3 mo of age (n = 6). (D) FACS analysis of myeloid cells (Mac+Gr1+), red cells (R1: Ter119medCD71high; R2: Ter119highCD71high; R3: Ter119highCD71med; R4: Ter119highCD71low), B cells (immature B: IgM−B220+; mature B: IgM+B220+), and T cells (immature T: CD8a+CD4+; CD4+ T; CD8+ T) in BM, spleen, and thymus of Dekfl/fl and Dek-cKO mice at 3 mo of age. (E and F) Percent analysis of myeloid cells in BM and spleen cells (n = 5). (G and H) Percent analysis of red cells in BM and spleen cells (n = 5). (I and J) Percent analysis of B cells in BM and spleen cells (n = 5). (K and L) Percent analysis of T cells in spleen and thymus cells (n = 5). Error bars represent means ± SD. **, P < 0.01; ***, P < 0.001; Student’s t test. Data are representative of three independent experiments.
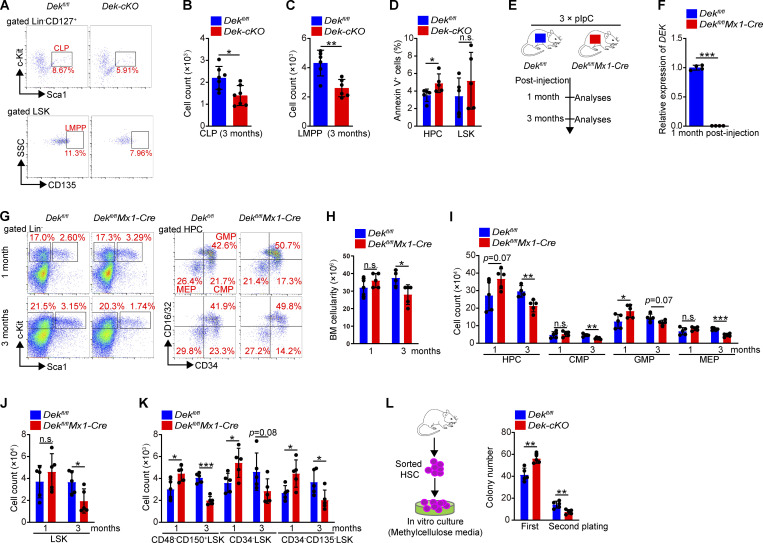
We found that neither the frequency, nor the total number of HPCs, LSKs, and HSCs exhibited considerable changes in Dek-cKO mice at 1.5 mo of age, but these mice had a strikingly lower number of stem-enriched cells compared with control mice at 3 mo of age (Fig. 1, D–H). The total number of multipotent progenitors (MPPs), short-time HSCs (ST-HSCs), and LT-HSCs were also considerably decreased in Dek-cKO mice compared with Dekfl/fl mice at 3 mo of age (Fig. 1, D, I, and J), as well as the number of common lymphoid progenitors (CLPs) and lymphoid-primed multipotent progenitors (LMPPs; Fig. S2, A–C). We found that DEK deletion resulted in a slight increase in the apoptosis of hematopoietic stem/progenitor cells (HSPCs) under homeostatic state (Fig. S2 D). To further elucidate whether DEK regulated adult hematopoiesis, we generated Dekfl/flMx1-Cre (Fig. S2, E and F). The number of HPCs was elevated at 1 mo after induction of deletion of DEK, but the number of BM cells and HPCs were reduced at 3 mo after induction of DEK deletion (Fig. S2, G–I). Moreover, Dekfl/flMx1-Cre mice showed an increased number of HSCs at 1 mo after induction of DEK deletion, but these mice exhibited a considerably decreased number of LSK cells and HSCs at 3 mo after polyinosinic-polycytidylic acid (pIpC) injection (Fig. S2, J and K), indicating a transient expansion but eventual reduction of the HSC pool upon DEK deletion.
Figure S2.
DEK deletion decreases the HSPC pool in mice. (A) FACS analysis of CLPs (Lin−CD127+Sca-1loc-Kitlo) and LMPPs (Lin− Sca-1+c-Kit+CD135+) in BM cells of Dekfl/fl and Dek-cKO mice. (B and C) Count of CLPs and LMPPs in BM cells of Dekfl/fl and Dek-cKO at 3 mo of age (n = 6–7). (D) Analysis of apoptotic HPCs and LSK cells in BM cells of Dekfl/fl and Dek-cKO at 3 mo of age. Annexin V+ cells represent early and later apoptotic cells (n = 5). (E) Experimental schematic for the generation of mice with inducible deletion of DEK, Dekfl/flMx1-Cre. Mice were treated with an i.p. injection of 10 µg pIpC per gram of body weight every second day for a total of three injections. The mice were sacrificed for analyses at 1 or 3 mo after pIpC injection. (F) Relative mRNA expression of DEK in freshly sorted HSCs of Dekfl/fl and Dekfl/flMx1-Cre mice at 1 mo after pIpC injection (n = 4). (G) FACS analysis of LSK cells and HPCs in Dekfl/fl and Dekfl/flMx1-Cre BM cells at 1 or 3 mo after pIpC injection. (H) BM cells count of Dekfl/fl and Dekfl/flMx1-Cre mice after pIpC injection (n = 5). (I) Count of HPC, CMP, GMP, and MEP in BM cells of Dekfl/fl and Dekfl/flMx1-Cre mice after pIpC injection (n = 5). (J) LSK cells count in Dekfl/fl and Dekfl/flMx1-Cre BM cells (n = 5). (K) HSC (CD48−CD150+LSK, CD34−LSK, or CD34−CD135−LSK) count in Dekfl/fl and Dekfl/flMx1-Cre BM cells (n = 5). (L) In vitro assay of the CFUs at 10–12 d after plating Dekfl/fl and Dek-cKO HSC cells. For the second plating, live cells from the colonies obtained during the first plating were plated as before and cultured for 10–12 d (n = 5). Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test. Data are representative of three independent experiments.
The CFU assay showed that DEK-deficient BM cells and HSC (from Dek-cKO mice at 1.5 mo of age) gave rise to a considerably increased number of total colonies (Fig. 1 K and Fig. S2 L); however, the replated CFU assay showed strikingly decreased colonies (Fig. 1 K and Fig. S2 L), suggesting that the expansion of DEK-deficient HSPCs is at the expense of self-renewal. To further determine the abundance of functional HSCs in Dek-cKO BM cells, we performed a limiting-dilution assay (Fig. 1 L). Donor engraftment assessment showed that Dek-cKO mice displayed a fourfold reduction in the frequency of functional HSCs at 3 mo of age compared with Dekfl/fl mice (1/32405 vs. 1/8033, P = 0.012; Fig. 1 L). We also injected the cell cycle–dependent myelotoxic agent 5-fluorouracil (5-FU) into Dekfl/flMx1-Cre mice weekly. Dekfl/flMx1-Cre mice died markedly earlier than Dekfl/fl mice (Fig. 1 M). Altogether, the above data suggested that DEK was essential for HSC maintenance and hematopoiesis.
DEK deficiency compromises HSC self-renewal in a cell-intrinsic manner
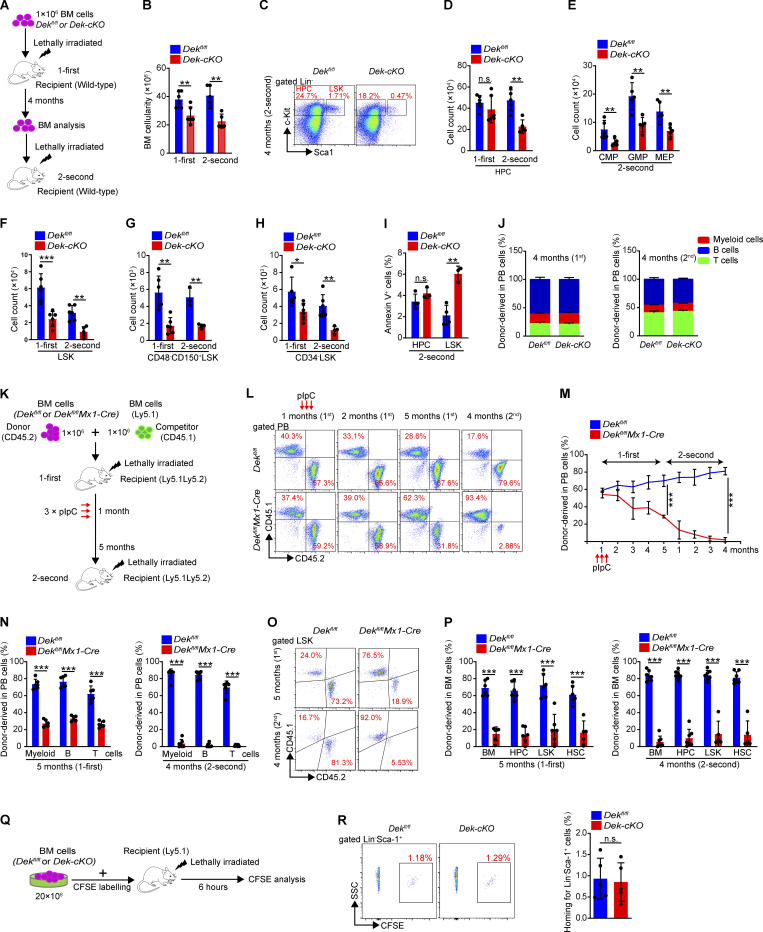
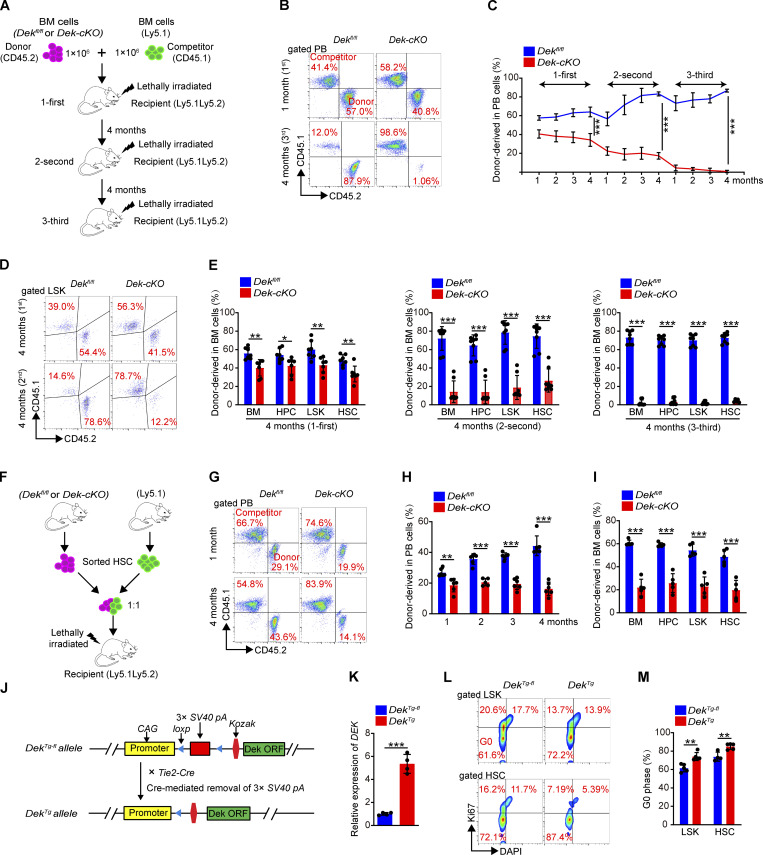
To further evaluate the self-renewal of DEK-deficient HSCs, we performed serial BM transplantation assays (Fig. 2 A). The DEK-deficient HSCs showed considerably impaired reconstitution capacity, as the recipient mice displayed a strikingly reduced number of BM cells, HPCs (common myeloid progenitors [CMPs], granulocyte-monocyte progenitors [GMPs], and megakaryocyte-erythroid progenitors [MEPs]), LSK cells, and HSCs at 4 mo after the second transplantation (Fig. 2, B–H). The survival capacity is critical for self-renewal of HSCs (Alenzi et al., 2009; Liu et al., 2019). Here, we found that DEK-deficient LSKs showed more apoptosis in recipient mice (Fig. 2 I), suggesting that loss of DEK might reduce the survival of LSK cells under stress. Moreover, the frequencies of myeloid, B, and T cells were comparable in donor-derived peripheral blood (PB) cells of recipient mice (Fig. 2 J), excluding the possibility that DEK deletion blocked the differentiation of HSCs in recipient mice.
Figure 2.
DEK deficiency impairs HSC self-renewal. (A) Experimental schematic for the serial transplantation assay. (B) BM cell count of recipient mice at 4 mo after first or second transplantation (n = 5–6). (C) FACS analysis of HPCs and LSK cells in BM cells of recipient mice at 4 mo after second transplantation. (D) Count of HPCs in BM cells of recipient mice at 4 mo after first or second transplantation (n = 5–6). (E) Count of CMPs, GMPs, and MEPs in BM cells of recipient mice at 4 mo after second transplantation (n = 5). (F–H) Count of LSK cells and HSCs (CD48−CD150+LSK or CD34−LSK) in BM cells of recipient mice at 4 mo after first or second transplantation (n = 4–6). (I) Analysis of apoptotic HPCs and LSK cells in BM cells of recipient mice at 4 mo after second transplantation. Annexin V+ cells represent early and later apoptotic cells (n = 3–4). (J) The ratio analysis of myeloid cells, B cells, and T cells in PB cells of recipient mice at 4 mo after first and second transplantation (n = 6). (K) Experimental schematic for the competitive transplantation assay. (L) FACS analysis of PB cells from recipient mice in serial competitive transplantation assay (donor-derived CD45.1−CD45.2+ cells, competitor-derived CD45.1+CD45.2− cells). (M) Percentage of donor-derived PB cells at the indicated time points in serial competitive transplantation assay (n = 6–7). (N) Percentage of donor-derived myeloid cells, B cells, and T cells in PB cells of recipient mice at 5 mo after first transplantation (n = 6), or at 4 mo after second transplantation (n = 6–7). (O) FACS analysis of LSK cells from recipient mice BM in serial competitive transplantation assay at 5 mo after first transplantation, or at 4 mo after second transplantation. (P) Percentage of donor-derived BM cells, HPCs, LSK cells, and HSCs at 5 mo after first transplantation (n = 6), or at 4 mo after second transplantation (n = 5–7). (Q) Experimental schematic for the homing assay. (R) FACS analysis of CFSE+ cells in BM Lin−Sca-1+ cells of recipient mice. The histogram indicates in vivo homing percentage of Dekfl/fl and Dek-cKO Lin− Sca-1+ cells in recipient mice at 6 h after transplantation (n = 5). Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test. Data are representative of three independent experiments.
We also assessed the long-term self-renewal capacity of DEK-deficient HSCs by performing a competitive serial transplantation assay using BM cells (Fig. S3 A). We monitored donor-derived (CD45.2+) PB cells from DEK-deficient HSCs and found that these cells showed continuously decreased chimeric percentages (Fig. S3, B and C). Moreover, DEK-deficient BM cells, HPCs, LSK cells, and HSCs exhibited a striking reduction in their long-term repopulation ability at 4 mo after the third transplantation (Fig. S3, D and E). To substantiate these data, we transplanted sorted Dekfl/fl or Dek-cKO HSCs (CD45.2+) into recipient mice with competitive HSCs (CD45.1+; Fig. S3 F). We found that donor-derived cells from DEK-deficient HSCs showed considerably reduced chimeric percentages in PB and BM cells (Fig. S3, G–I). These data suggest that DEK deletion results in the impaired self-renewal of HSCs. To explore whether DEK regulated the self-renewal of HSCs in a cell-intrinsic manner, Dekfl/fl or Dekfl/flMx1-Cre BM cells were first transplanted without inducing DEK deletion. The recipient mice were then injected with pIpC at 1 mo after transplantation, followed by PB chimerism analysis and second transplantation (Fig. 2 K). The results confirmed that DEK deletion impaired the self-renewal capacity of HSCs, as shown by the decreased PB chimeric percentages (Fig. 2, L and M). Moreover, the chimeric percentages of mature cells in PB showed a unanimous decrease (Fig. 2 N). The reduced repopulation ability of HPCs, LSK cells, and HSCs in BM cells of recipient mice demonstrated that DEK loss impaired the self-renewal of HSCs in a cell-intrinsic manner (Fig. 2, O and P). Moreover, we did not detect an obvious defect in the homing capacity of DEK-deficient HSPCs compared with control cells (Fig. 2, Q and R). Altogether, these data indicate that loss of DEK impairs the long-term self-renewal of HSCs under various stresses in a cell-intrinsic manner.
Figure S3.
DEK deletion impairs the reconstitution capacity of HSCs. (A) Experimental schematic for the competitive transplantation assay. BM cells of Dekfl/fl and Dek-cKO mice were transplanted into lethally irradiated recipient mice with competitor BM cells from WT Ly5.1 mice. The reconstituted BM cells of recipient mice were harvested (at 4 mo after transplantation) for analysis and the next transplantation. (B) FACS analysis of PB cells from recipient mice in serial competitive transplantation assay (first, third, for the first or third competitive transplantation; donor-derived CD45.1−CD45.2+ cells, competitor-derived CD45.1+CD45.2− cells). (C) Percentage of donor-derived PB cells at the indicated time points in serial competitive transplantation assay (n = 7). (D) FACS analysis of LSK cells from recipient mice BM in serial competitive transplantation assay at 4 mo after first or second transplantation. (E) Percentage of donor-derived BM cells, HPCs, LSK cells, and HSCs at the indicated time points in serial competitive transplantation assay (n = 7). (F) Experimental schematic for the competitive transplantation assay using sorted HSCs (5 × 102). (G) FACS analysis of PB cells from recipient mice in competitive transplantation assay. (H) Percentage of donor-derived PB cells at the indicated time points (n = 5–6). (I) Percentage of donor-derived BM cells, HPCs, LSK cells, and HSCs at the indicated time points (n = 5–6). Error bars represent means ± SD. (J) Experimental schematic for the generation of mice with overexpression of DEK in hematopoietic lineage cells. (K) Relative mRNA expression of DEK in freshly sorted DekTg-fl and DekTg HSCs (n = 4). (L) FACS analysis of Ki-67 staining in LSK cells and HSCs of DekTg-fl and DekTg mice. (M) Quiescence (G0 phase) analysis of LSK cells and HSCs (n = 4–5). *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test. Data are representative of three independent experiments.
DEK deficiency decreases quiescence in HSCs
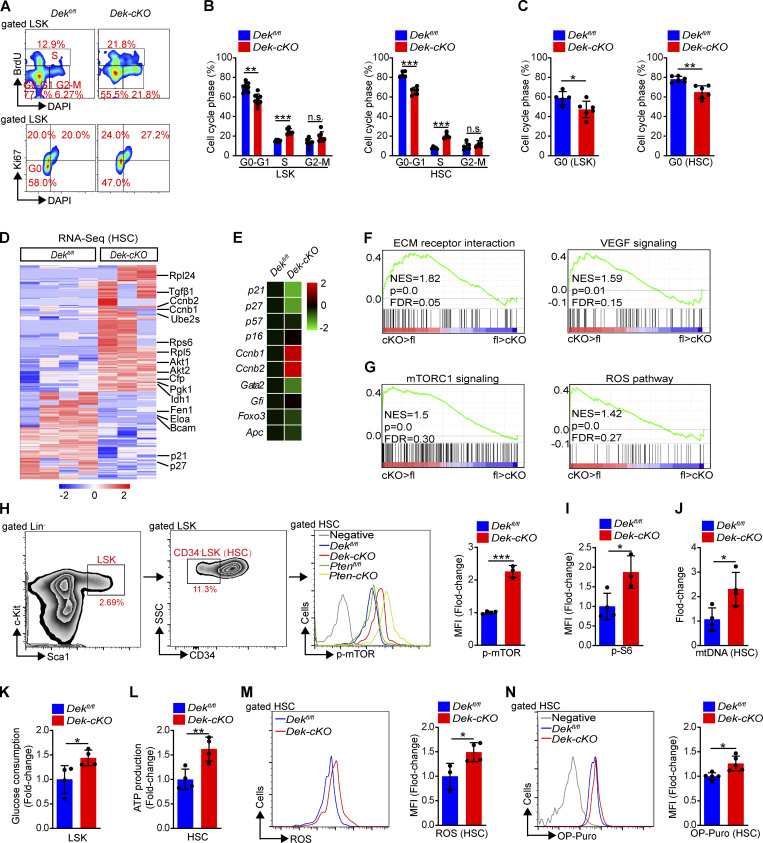
To acquire further insight into the depletion of HSC pool after DEK deletion, we assessed in vivo incorporation of the thymidine analogue BrdU. While 7.9% of Dekfl/fl HSCs incorporated BrdU, 20.6% of Dek-cKO HSCs were BrdU+, indicating that DEK deletion enhanced the proliferation of LSKs and HSCs (Fig. 3, A and B). Furthermore, staining with Ki-67 and DAPI showed that the proportion of quiescent LSKs and HSCs in Dek-cKO mice (HSC: 65.0%) was lower than that in control mice (HSC: 78.1%; Fig. 3, A and C), suggesting that DEK deficiency impairs quiescence maintenance of HSCs. To further confirm the regulatory role of DEKon HSC quiescence and proliferation, we generated a conditional overexpression transgenic line—defined as DekTg-fl mice—in which an open reading frame (ORF) of mouse DEK gene was inserted following a floxed STOP cassette (3 × SV40 pA) driven by the CAG promoter (Fig. S3 J). To achieve tissue-specific overexpression of DEK, DekTg-fl mice were crossed with Tie2-Cre mice to generate a DekTg-flTie2-Cre mouse model, defined as DekTg mice (Fig. S3, J and K). As expected, staining with Ki-67 and DAPI showed that the proportion of quiescent LSKs and HSCs in DekTg mice was considerably increased compared with control mice (Fig. S3, L and M), demonstrating the critical role of DEK in maintaining HSC quiescence.
Figure 3.
DEK loss decreases quiescence and activates mTOR signaling in HSCs. (A) FACS analysis of BrdU incorporation and Ki-67 staining in LSK cells of Dekfl/fl and Dek-cKO mice at 3 mo of age. (B) Cell-cycle analysis of LSK cells and HSCs in Dekfl/fl and Dek-cKO mice at 3 mo of age (n = 4–8). (C) Quiescence (G0 phase) analysis of LSK cells and HSCs in Dekfl/fl and Dek-cKO mice at 3 mo of age (n = 5–6). (D) Representative heatmap of upregulated genes or downregulated genes by 1.5-fold or more in Dek-cKO HSCs compared with Dekfl/fl HSCs (n = 3–4; P < 0.01). (E) qRT-PCR analysis of the indicated genes from freshly sorted HSCs (n = 4). (F and G) GSEA of the selected gene sets. (H and I) FACS analysis of p-mTOR and p-S6 in HSCs of the indicated mice. The histograms indicate the mean fluorescence intensity (MFI) analysis of p-mTOR and p-S6 (n = 3–4). (J) Quantitative PCR analysis of mitochondrial DNA (mtDNA) in freshly sorted HSCs (n = 4). (K) Glucose consumption analysis of LSK cells. Sorted LSK cells were in vitro for 24 h, and the change in glucose concentration in the culture medium was measured (n = 4). (L) ATP level in freshly sorted HSCs (n = 4). (M) FACS analysis of ROS level in HSCs. The histogram indicates the MFI analysis of ROS in HSCs (n = 3–4). (N) FACS analysis of OP-Puro incorporation in HSCs. The histogram indicates the MFI analysis of OP-Puro in HSCs (n = 5). *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test. Data in A–C, E, and H–N are representative of three independent experiments. NES, normalized enrichment score; FDR, false discovery rate; ECM, extracellular matrix; VEGF, vascular endothelial growth factor.
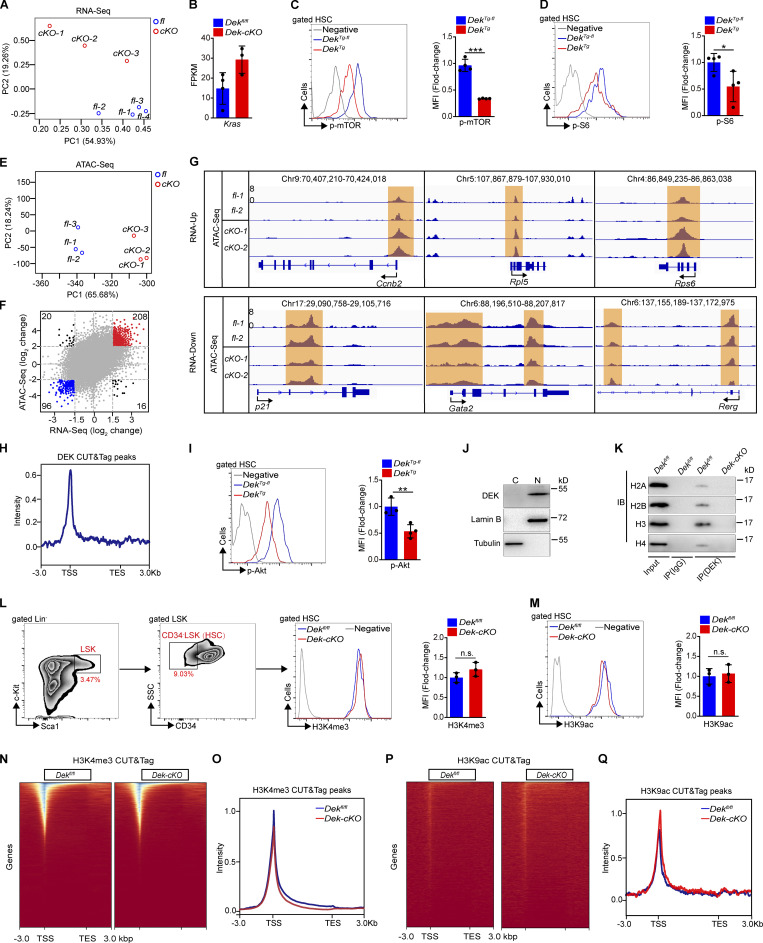
To explore the underlying molecular mechanisms of DEK-regulated HSC homeostasis and function, we performed RNA sequencing (RNA-seq) analysis on freshly sorted HSCs. The principal component assay showed that the samples have obvious discrimination (Fig. S4 A). We found that 785 genes in Dek-cKO HSCs were significantly upregulated compared with Dekfl/fl HSCs, while 542 genes were considerably downregulated (>1.5-fold change; P < 0.01; Fig. 3 D). To verify the changed gene expression, we performed quantitative RT-PCR (qRT-PCR) and discovered that DEK loss promoted the expression of cell cycling–associated genes (Ccnb1 and Ccnb2) but inhibited the expression of quiescence maintenance–associated genes (p21, p27) in HSCs (Fig. 3 E). Gene-set enrichment analysis (GSEA) of the RNA-seq data further revealed that the set of genes upregulated in HSCs showed enrichment for extracellular matrix receptor interaction and vascular endothelial growth factor signaling (Fig. 3 F). It has been reported that the oncogene Kras expands and exhausts HSCs (Sasine et al., 2018). Here, we observed that the expression of Kras in Dek-cKO HSC was upregulated (Fig. S4 B). Together, these results reveal that programs related to proliferation of HSCs are activated, while programs related to maintaining HSC quiescence are suppressed in DEK-deficient HSCs.
Figure S4.
DEK interacts with H3 and regulates chromatin accessibility. (A) Principal component analysis showing the relation between RNA-seq samples. (B) The FPKM value of Kras. Data are from RNA-seq (n = 3–4). (C and D) FACS analysis of p-mTOR and p-S6 in HSCs of the indicated mice. The histograms indicate the mean fluorescence intensity (MFI) analysis of p-mTOR and p-S6 (n = 4). (E) Principal component analysis showing the relation between ATAC-seq samples. (F) Intersection of ATAC-seq accessibility and nearby transcription of genes. Each point indicates a separate ATAC-seq peak. (G) Accessible chromatin located at gene loci, including mRNA-increased genes (Ccnb2, Rps6, and Rpl5) and mRNA-decreased genes (p21, Gata2, and Rerg). (H) Average diagram of genome-wide DEK CUT&Tag peaks in HSCs at TSS regions (±3,000 bp). (I) FACS analysis of p-Akt in HSCs of the indicated mice. The histograms indicate the MFI analysis (n = 3–4). (J) Western blot for DEK in lysates prepared from cytoplasmic (C) and nucleus (N) of freshly sorted Lin−c-Kit+ cells from WT mice. Lamin B and tubulin were used as a loading control. (K) Lin−c-Kit+ cells were freshly sorted from WT mice. DEK protein was immunoprecipitated (IP) from cell lysates, followed by immunoblotting (IB). (L and M) FACS analysis of H3K4me3 and H3K9ac level in HSCs. The histogram indicates the MFI analysis (n = 3). (N) Representative heatmap of genome-wide H3K4me3 CUT&Tag signal around genes. (O) Average diagram of genome-wide H3K4me3 CUT&Tag peaks at TSS regions (±3,000 bp). (P) Representative heatmap of genome-wide H3K9ac CUT&Tag signal around genes. (Q) Average diagram of genome-wide H3K9ac CUT&Tag peaks at TSS regions (±3,000 bp). Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t-test. Data in G–J are representative of three independent experiments. SSC, side scatter; TES, transcriptional end site.
DEK deficiency activates mTOR signaling and promotes metabolism in HSCs
The PI3K-Akt-mTOR axis is an important regulator of ribosome biogenesis, mitochondrial metabolism, and quiescence maintenance of HSCs (Qian et al., 2016; Martelli et al., 2010; Huang et al., 2019). Constitutively active Akt1 promotes cell proliferation and depletes the HSC pool (Kharas et al., 2010); however, Akt1/2 double-deficient LT-HSCs were found to persist in quiescence and have impaired long-term function (Juntilla et al., 2010). Loss of phosphatase and tensin homologue (PTEN), an upstream inhibitor of the PI3K-Akt-mTOR pathway, results in exhaustion of HSCs (Yilmaz et al., 2006). Such functional characteristics of HSCs with excessively activated PI3K-Akt-mTOR signaling are similar to those of DEK-deficient HSCs. Moreover, the set of genes for mTORC1 signaling and the downstream ROS pathway were significantly enriched in HSCs upon DEK deletion (Fig. 3 G). We used phospho-flow analysis and observed an elevated level of phospho-mTOR (p-mTOR) in DEK-deficient HSCs compared with Dekfl/fl HSCs, as well as for the p-S6 intensity in HSCs, a direct substrate of mTOR by phosphorylation (Fig. 3, H and I), indicating that DEK deficiency activates mTOR signaling. To further confirm the above results, we analyzed the activation of mTOR signaling in DEK-overexpressed HSCs and found a decreased level of p-mTOR and p-S6 in DekTg HSCs compared with DekTg-fl HSCs (Fig. S4, C and D), suggesting the inhibitory effect of DEK on mTOR activity in HSCs.
The mTOR activity governs HSC mitochondrial metabolism and ribosome biogenesis (Qian et al., 2016; Martelli et al., 2010; Huang et al., 2019). To acquire further insight into the metabolic activity of HSCs after DEK deletion, we analyzed the mitochondrial count in HSCs using mitochondrial DNA measurement. We observed a significant increase in mitochondrial mass in DEK-deficient HSCs compared with control HSCs (Fig. 3 J). We analyzed the metabolic profile and found that DEK-deficient LSK cells consumed more glucose than their Dekfl/fl counterparts when cultured in vitro (Fig. 3 K). Correspondingly, DEK-deficient HSCs produced more ATP than their fl/fl counterparts (Fig. 3 L), suggesting that DEK deficiency increases mitochondrial metabolism in HSCs. To substantiate these data, we analyzed the ROS level produced by mitochondrial metabolism, and flow cytometry results indicated that DEK deficiency promoted the ROS level in HSCs (Fig. 3 M). Moreover, we examined the rate of protein synthesis and found that DEK-deficient HSCs incorporated significantly more O-propargyl-puromycin (OP-Puro) than control cells (Fig. 3 N). Altogether, these data demonstrate that DEK is essential for quiescence maintenance of HSCs and that loss of DEK promotes activation of mTOR signaling and metabolism of HSCs.
DEK targets Akt1 and Akt2 by altering the chromatin accessibility in HSCs
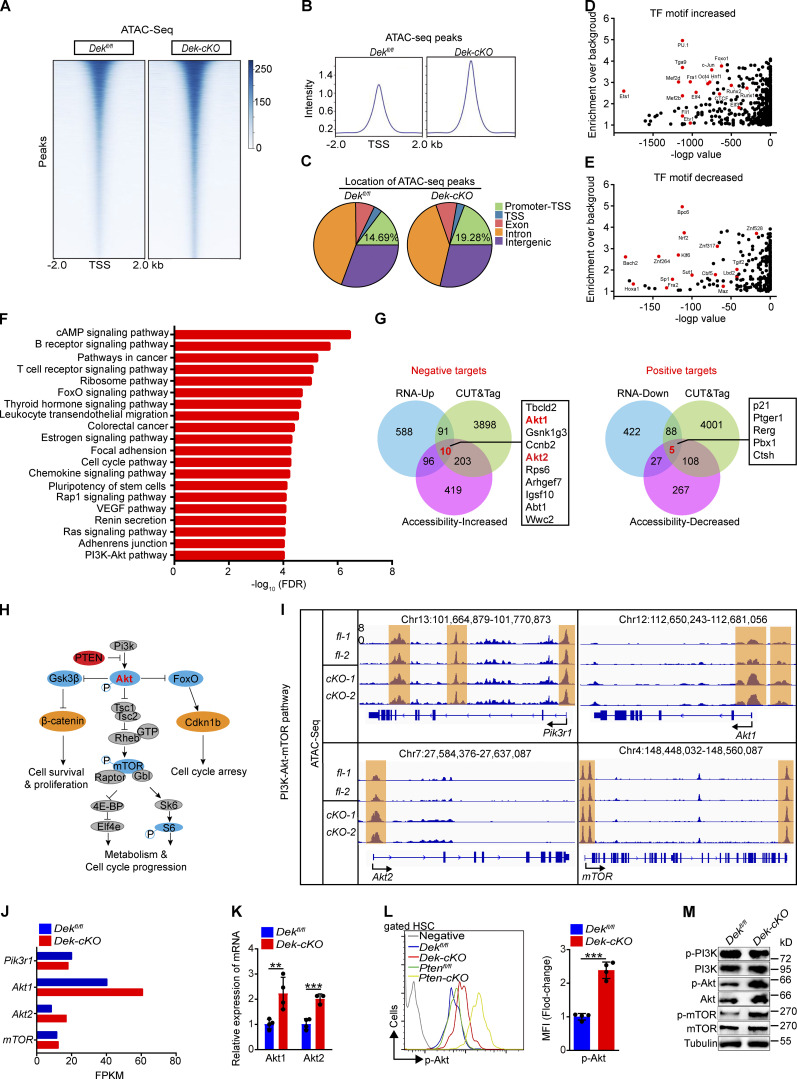
The large population of differentially expressed genes in Dek-cKO HSCs led us to hypothesize that DEK regulates gene expression by altering chromatin structure. To test this, we performed an assay for transposase-accessible chromatin (ATAC) sequencing (ATAC-seq) in freshly sorted HSCs. The principal component assay showed that the samples have obvious discrimination (Fig. S4 E). An increase of the average intensity of ATAC-seq signal was observed in Dek-cKO HSCs compared with Dekfl/fl HSCs (Fig. 4, A and B). We found that 1,231 ATAC peaks in Dek-cKO HSCs were significantly upregulated, while 483 ATAC peaks were significantly downregulated (>1.5-fold change; P < 0.01). Notably, more ATAC signals in Promoter-TSS (transcription start site) regions were identified in Dek-cKO HSCs (3,528 peaks) compared with Dekfl/fl HSCs (2,774 peaks; Fig. 4 C), suggesting enhanced transcriptional activity in DEK-deficient HSCs. Next, we sought to explore how DEK influences the activity of transcription factors (TFs) by performing known motif analysis in the differential accessibility regions. The analysis revealed that when deleting DEK in HSCs, enrichment of Pu.1, c-Jun, Elf4, and Runx1 binding motifs was increased (Fig. 4 D), whereas enrichment of Nrf2, Klf6, and Bpc6 binding motifs was decreased (Fig. 4 E). Together, these data demonstrate that DEK loss leads to increased chromatin accessibility of HSCs, affecting the landscape of TF binding.
Figure 4.
Akt1 and Akt2 are direct targets of DEK in HSCs by altering the chromatin accessibility landscape. (A) Representative heatmap of genome-wide ATAC-seq signal around genes in HSCs freshly sorted from Dekfl/fl and Dek-cKO mice (n = 3 biological independent samples per group). (B) Average diagram of genome-wide chromatin accessibility at TSS regions (±2,000 bp). (C) Location of all ATAC-seq peaks in Dekfl/fl and Dek-cKO HSCs. Promoter-TSS, the region between promoter and TSS (−1 kb to 100 bp). (D) Enrichment of the increased known TF binding motifs in different chromatin-accessible regions. (E) Enrichment of the decreased known TF binding motifs in different chromatin-accessible regions. (F) Pathway analysis of the genes with significant changed ATAC peaks (over twofold change; P < 0.01) in DEK-deficient HSCs. (G) Integrative analysis to identify transcriptome-wide potential targets of DEK in HSCs. Left: Potential negative targets of DEK. Right: Potential positive targets of DEK. RNA-Up and RNA-Down indicate genes with significantly increased and decreased expression, respectively, upon DEK deletion in HSCs as detected by RNA-seq (FPKM > 1, fold change > 1.5). CUT&Tag indicates genes with significant enrichment in DEK binding (reads per kilobase per million > 1). Accessibility-Increased and Accessibility-Decreased indicates genes with significantly increased and decreased accessibility, respectively, upon DEK deletion in HSCs as detected by ATAC-seq (reads per kilobase per million > 1, fold change > 2). (H) Summary of the PI3K-Akt-mTOR pathway. (I) Accessible chromatin located at gene loci, including Pi3kr1, Akt1, Akt2, and mTOR. (J) The FPKM value of indicated genes. Data are from RNA-seq. (K) Relative mRNA expression of Akt1 and Akt2 in freshly sorted Dekfl/fl and Dek-cKO HSCs (n = 4). (L) FACS analysis of p-Akt in HSCs of the indicated mice. The histograms indicate the mean fluorescence intensity analysis (n = 4). (M) Western blot of PI3K-Akt-mTOR pathway proteins in lysates prepared from freshly sorted Lin−c-Kit+ cells. **, P < 0.01; ***, P < 0.001; Student’s t test. Data in K–M are representative of three independent experiments. VEGF, vascular endothelial growth factor.
The major signaling pathways enriched with the genes whose loci were associated with significantly changed ATAC-seq peaks upon DEK deletion are shown in Fig. 4 F. Notably, many of the same pathways were also detected by RNA-seq, including cell-cycle, ribosome, and PI3K-Akt pathways, suggesting that they are the main pathways enriched with potential direct targets of DEK. We found that most of the differentially expressed genes exhibited altered accessibility (Fig. S4 F). For the up-expressed genes (Ccnb2, Rpl5, and Rps6), which promote HSC activation and ribosome biogenesis, we found elevated ATAC peaks (Fig. S4 G). We also observed that a fraction of down-expressed genes showed decreased ATAC peaks, including p21, Gata2, and Rerg (Fig. S4 G). To search the direct targets of DEK, we profiled the genome-wide DEK binding sites by using DEK cleavage under the targets and tagmentation (CUT&Tag) strategy with sorted HSCs. The data showed that DEK mainly bound TSS regions of the gene (Fig. S4 H), suggesting the transcriptionally regulatory roles of DEK in HSCs. Through integrative analysis of RNA-seq, ATAC-seq, and DEK CUT&Tag data, we identified genes that are significantly positively (RNA-Down and Accessibility-Decreased) and negatively (RNA-Up and Accessibility-Increased) regulated by DEK, respectively (Fig. 4 G). Importantly, we found that Akt1 and Akt2 were potential targets of DEK in HSCs (Fig. 4 G). PI3K-Akt is critical signaling to govern activity of mTOR, and the upregulated expression of Akt or activated Pi3k will result in an accumulation of p-Akt, leading to activation of Akt-mTOR signaling (Fig. 4 H). The accessibility of Pi3kr1 and mTOR gene loci did not change; however, the accessibility of Akt1 and Akt2 gene loci was increased in Dek-cKO HSCs compared with Dekfl/fl HSCs (Fig. 4 I). Moreover, RNA-seq data showed that DEK deletion increased the transcription of Akt1 and Akt2, rather than Pik3r1 and mTOR (Fig. 4 J), suggesting that DEK governs activity of the AKT-mTOR pathway by regulating the accessibility of Akt1/2 gene loci and subsequent transcription. To further strengthen the above results, we performed qRT-PCR and discovered that DEK loss promoted the expression of Akt1 and Akt2 (Fig. 4 K). We used phospho-flow analysis and observed an elevated level of p-Akt in DEK-deficient HSCs compared with Dekfl/fl HSCs (Fig. 4 L), but a decreased level of p-Akt in DEK-overexpressed HSCs compared with DekTg-fl HSCs (Fig. S4 I). Moreover, the Western blot results confirmed that DEK deletion upregulated the protein level of Akt and p-Akt (Fig. 4 M). Taken together, these data suggest that DEK regulates the chromatin accessibility landscape of HSCs, and that Akt1 and Akt2 are direct targets of DEK.
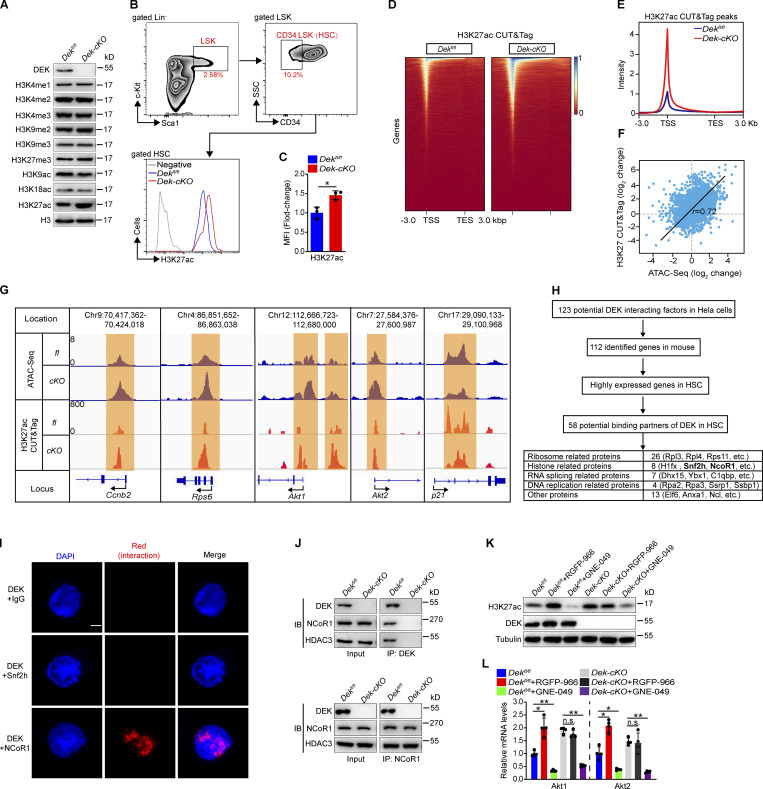
DEK regulates chromatin accessibility by recruiting the corepressor NCoR1 to induce deacetylation of H3K27
Chromatin accessibility is tightly associated with histone modification (ENCODE Project Consortium, 2012; Cabal-Hierro et al., 2020; Wu et al., 2018; Wu et al., 2016). We inferred that the increased chromatin accessibility of HSCs in DEK-deficient mice might represent an alteration in histone modification. Indeed, the DEK protein primarily located in the nucleus of Lin−c-kit+ cells (Fig. S4 J). The coimmunoprecipitation assay revealed that DEK primarily bound histone 3 in Lin−c-kit+ cells (Fig. S4 K). We found that the intensity of H3K27ac (acetylated H3K27) in Dek-cKO Lin−c-kit+ cells showed marked differences compared with control mice, but the intensity of other modifications had no obvious change (Fig. 5 A). To substantiate these data, we used flow cytometry to detect H3 modification in HSCs. The intensity of H3K4me3 (tri-methyl-histone H3 [Lys4]) and H3K9ac (acetyl-histone H3 [Lys9]) in HSCs in Dek-cKO mice was comparable to control mice (Fig. S4, L and M); however, the intensity of H3K27ac was considerably increased in DEK-deficient HSCs (Fig. 5, B and C), indicating a regulatory role for DEK in H3K27 acetylation in hematopoietic progenitor and stem cells.
Figure 5.
DEK induces deacetylation of H3K27 by recruiting the corepressor NCoR1. (A) Western blot for modified histone 3 in lysates prepared from freshly sorted Lin−c-Kit+ cells. (B and C) FACS analysis of H3K27ac level in HSCs (CD34−LSK). The histograms indicate the mean fluorescence intensity (MFI) analysis of H3K27ac in HSCs (n = 3). (D) Representative heatmap of genome-wide H3K27ac CUT&Tag signal around genes. (E) Average diagram of genome-wide H3K27ac CUT&Tag peaks at TSS regions (±3,000 bp). (F) Correlation of changes between ATAC peaks and H3K27ac CUT&Tag peaks. Correlation coefficient (r) and P values (r = 0.72, P < 1 × 10−50) were calculated by Pearson’s correlation analysis. (G) Distribution of ATAC peaks and H3K27ac CUT&Tag peaks across the indicated gene loci. (H) Schematic representation of the workflow for DEK partners’ discovery. Potential DEK interacting factors from published mass spectrometry data (Smith et al., 2018). The highly expressed genes in HSCs have an FPKM value of >30 (data from RNA-seq). (I) In situ ligation assay to detect DEK/Snf2h and DEK/NCoR1 interaction. As a negative control, proximity ligation was performed using a rabbit anti-DEK antibody and a mouse IgG. Nuclei were visualized using DAPI staining. Scale bar: 5 µm. (J) Lin−c-Kit+ cells were freshly isolated from mice. DEK or NCoR1 protein was immunoprecipitated from cell lysates, followed by immunoblotting (IB). (K) Western blot for H3K27ac, DEK, and tubulin in lysates prepared from Lin−c-Kit+ cells. Lin−c-Kit+ cells were sorted and cultured in vitro, with treatment of RGFP-966 (5 µM) or GNE-049 (500 nM) for 24 h. (L) qRT-PCR analysis of the indicated transcripts from HSCs (n = 4). HSCs were sorted and cultured in vitro, with treatment with RGFP-966 (5 µM) or GNE-049 (500 nM) for 24 h. Error bars represent means ± SD. *, P < 0.05, **, P < 0.01; Student’s t test or one-way ANOVA. Data in A–C and I–L are representative of three independent experiments. TES, transcriptional end site.
To further examine the genome-wide occupancy of histone modifications associated with chromatin accessibility, we performed H3K4me3, H3K9ac, and H3K27ac CUT&Tag in freshly sorted LSK cells or HSCs. We found that the average intensity of H3K4me3-marked and H3K9ac-marked signal was comparable in chromatin domains of Dek-cKO cells and Dekfl/fl cells (Fig. S4, N-Q); however, an increase in the average intensity of H3K27ac-marked signal was observed in chromatin domains of Dek-cKO cells compared with Dekfl/fl cells (Fig. 5, D and E). In line with previous reports that accessible sites are enriched with H3K27ac occupancy (Cabal-Hierro et al., 2020; Wu et al., 2016), we found that most of the peaks with accessibility increases exhibited increased H3K27ac CUT&Tag peaks (r = 0.72; P < 1 × 10−50; Fig. 5 F), indicating a similar pattern between ATAC-seq and CUT&Tag signals. Indeed, we found that the negative target genes of DEK—CCnb2, Rps6, Akt1, and Akt2—showed elevated ATAC peaks and increased H3K27ac CUT&Tag peaks at specific loci (Fig. 5 G), and the positive target gene (p21) showed both decreased ATAC peaks and H3K27ac CUT&Tag peaks at specific loci (Fig. 5 G), suggesting that DEK governs chromatin accessibility and subsequent gene expression, which largely depends on the regulation of H3K27 acetylation.
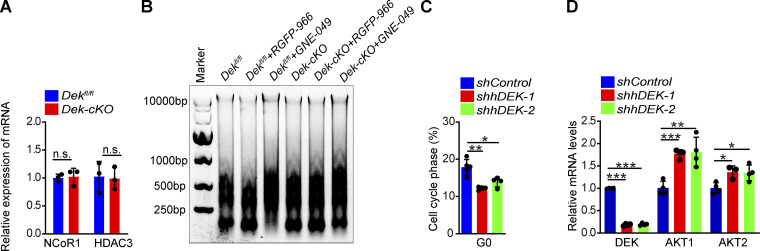
To test the mechanism underlying DEK-repressed acetylation of H3K27, we analyzed potential DEK interacting factors using previously published mass spectrometry data generated in human HeLa cells (Smith et al., 2018). We identified 58 potential binding partners of DEK in HSCs, including proteins related to ribosome, histone, and RNA splicing (Fig. 5 H). Novel epigenetic modulators were also identified (Fig. 5 H), including the ATP-dependent chromatin remodeling factor Snf2h and NCoR1—and NCoR1 modulates histone acetylation and gene transcription with its partner, HDAC3 (Perissi et al., 2010; Mottis et al., 2013). We performed the in situ proximity ligation assay (Söderberg et al., 2006) and found that DEK physically interacts with NCoR1, rather than Snf2h in HSC (Fig. 5 I). We further confirmed the interaction of DEK, NCoR1, and HDAC3 through an immunoprecipitation assay in Lin−c-kit+ cells (Fig. 5 J). The expression of NCoR1 and HDAC3 was comparable in DEK-deficient HSCs and control HSCs (Fig. S5 A), indicating that the increased H3K27ac in DEK-deficient HSCs was not attributed to the transcriptional change in NCoR1 and HDAC3.
Figure S5.
Knockdown of DEK in human primitive hematopoietic cells decreases quiescence. (A) Relative mRNA expression of NCoR1 and HDAC3 in freshly sorted HSCs of Dekfl/fl and Dek-cKO mice (n = 3). (B) Gel electrophoretic analysis of DNA recovered from MNase-digested nucleus of Lin−c-Kit+ cells, which were sorted and cultured in vitro, with treatment of RGFP-966 (5 µM) or GNE-049 (500 nM) for 24 h. (C) Analysis of quiescence of hHSPCs by Ki-67 + DAPI staining (n = 4–5). Human BM CD34+ cells (hHSPCs) were cultured in vitro and transduced to express control shRNA (shControl) or shRNA targeting DEK (either of two constructs: shDEK-1 and shDEK-2). (D) Relative mRNA expression for DEK, AKT1, and AKT2 in hHSPCs (n = 4). Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test or one-way ANOVA. Data are representative of three independent experiments.
To further investigate whether DEK modulates acetylation of H3K27ac by recruiting the NCoR1-HDAC3 complex, we treated Lin−c-kit+ cells with a specific inhibitor targeting HDAC3 (RGFP966) in vitro (Dai et al., 2019). We observed an efficient increase of H3K27ac level in Dekfl/fl cells, but the RGFP966 treatment had no obvious effect on H3K27ac level in Dek-cKO cells (Fig. 5 K), suggesting that DEK-repressed acetylation of H3K27 largely depends on the NCoR1-HDAC3 complex. To determine whether DEK governs chromatin accessibility by repressing acetylation of H3K27, we treated Lin−c-kit+ cells with a CBP/EP300-bromodomin inhibitor, GNE-049, which selectively blocks H3K27ac (Raisner et al., 2018). GNE-049 effectively decreased the H3K27ac level in Dekfl/fl and Dek-cKO cells (Fig. 5 K). Notably, the micrococcal nuclease (MNase) digestion assays revealed that GNE-049 treatment abolished the increased chromatin accessibility of Dek-cKO cells (Fig. S5 B). We also found that RGFP966 treatment led to increased chromatin accessibility observed in Dekfl/fl cells (Fig. S5 B). Moreover, treatment with GNE-049 abolished the effect of DEK deletion on gene expression in HSCs, including Akt1 and Akt2 (Fig. 5 L), and RGFP-966 treatment significantly promoted the expression of Ak1 and Akt2 in Dekfl/fl HSCs (Fig. 5 L). Together, these data demonstrate that DEK regulates chromatin accessibility and gene expression in a manner of repressing acetylation of H3K27 by recruiting the NCoR1-HDAC3 complex.
Targeting the Akt-mTOR pathway partially rescues defective Dek-cKO HSCs
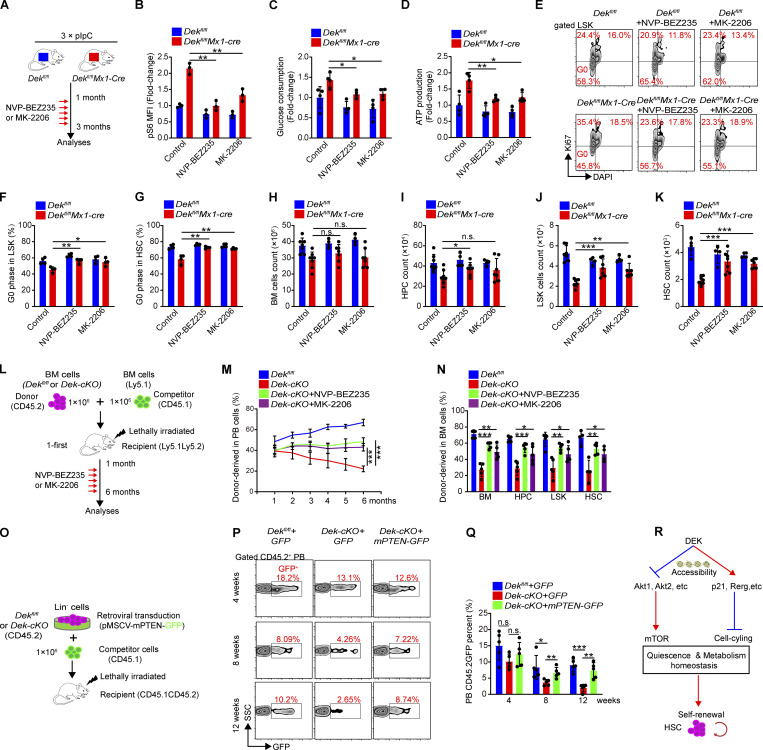
To determine whether the dysregulation of PI3K-Akt-mTOR activity in DEK-deficient HSCs contributes to the imbalance of quiescence and metabolism, we administered pIpC injection to induce DEK deletion and treated these mice with inhibitors of the PI3K-Akt-mTOR pathway: NVP-BEZ235 (NVP; dual PI3K and mTOR inhibitor) or MK-2206 (MK; pan-Akt inhibitor; Fig. 6 A). Both NVP and MK efficiently restricted the hyperactivation of mTOR in Dek-cKO HSCs (Fig. 6 B). Notably, the increased glucose consumption of LSK cells and ATP level of HSCs were partially abolished by NVP and MK treatment (Fig. 6, C and D). Moreover, the abnormal proliferation of DEK-deficient LSK cells and HSCs was partially inhibited by NVP or MK treatment (Fig. 6, E–G). Notably, the numbers of LSK cells and HSCs were efficiently increased in DEK-deficient mice compared with their counterparts (Fig. 6, H–K). To further confirm that the hyperactivated Akt-mTOR pathway leads to the impaired self-renewal of HSCs, we performed competitive transplantation assays (Fig. 6 L) and found that NVP or MK treatment partially restored the reconstitution capacity of Dek-cKO donor-derived HSCs in PB and BM cells compared with vehicle treatment (Fig. 6, M and N).
Figure 6.
Targeting the Akt-mTOR pathway partially rescues Dek-cKO defective HSCs. (A) Experimental schematic for administration of PI3K-mTOR pathway inhibitors. 1 mo after pIpC injection, Dekfl/fl and Dekfl/flMx1-Cre mice were orally gavaged with NVP-BEZ235 or MK-2206 for 2 mo. (B) FACS and mean fluorescence intensity (MFI) analysis of p-S6 in HSCs of Dekfl/flMx1-Cre mice at 2 mo after inhibitor treatment (n = 3). (C) Glucose consumption of LSK cells sorted from the indicated mice. Cells were cultured in vitro for 24 h and the change in glucose concentration in the culture medium was measured (n = 4). (D) ATP level in freshly sorted HSCs (n = 4). (E) FACS analysis of Ki-67 + DAPI staining in LSK cells. (F and G) G0 phase analysis of LSK cells and HSCs in mice (n = 4). (H–K) BM cell, HPC, LSK cell, and HSC count in mice BM (n = 4–8). (L) Experimental schematic for the competitive transplantation assay. BM cells of Dekfl/fl and Dek-cKO mice were transplanted into lethally irradiated recipient mice with competitor BM cells from WT Ly5.1 mice. Recipient mice were subjected to inhibitor treatment at 1 mo after transplantation. The reconstituted BM cells of recipient mice were harvested (at 6 mo after transplantation) for analysis. (M) Percentage of donor-derived PB cells at the indicated time points in competitive transplantation assay (n = 4–5). (N) Percentage of donor-derived BM cells, HPCs, LSK cells, and HSCs at 6 mo after first transplantation (n = 4–5). (O) Experimental schematic for competitive transplantation assay. Lin− cells were freshly isolated from mice and subjected to retroviral transduction to enforce the expression of PTEN with GFP fluorescence. (P) FACS analysis of GFP+ cells in donor-derived PB cells of recipient mice. (Q) Contribution of retrovirally transduced donor cells (CD45.2+GFP+) to recipient mouse PB cells after primary transplantation (n = 5). (R) Proposed model demonstrating the role and underlying mechanisms of DEK regulating quiescence and metabolic hemostasis, and self-renewal of HSCs. Error bars represent means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way ANOVA. Data are representative of three independent experiments.
PTEN is a phosphatase that dephosphorylates phosphatidylinositol 3,4,5-triphosphate produced by PI3K and negatively regulates the PI3K-Akt-mTOR pathway (Stiles et al., 2004; Cully et al., 2006). As direct deletion of Akt1 and Akt2 impairs self-renewal and long-term function of HSCs (Juntilla et al., 2010), we generated retrovirally expressed constructs to enforce PTEN-GFP expression in stem/progenitor cells to repress excessive activation of Akt-mTOR signaling in Dek-cKO HSCs (Fig. 6 O). After transplantation for 12 wk, the percentages of donor-derived GFP+ PB cells from PTEN-overexpressing HSCs showed a considerable increase compared with their counterparts (Fig. 6, P and Q), indicating that overexpression of PTEN efficiently rescues the repopulation capacity of HSCs.
To determine the function of DEK in the cell-cycle regulation of human HSPCs (hHSPCs), we isolated CD34+ hematopoietic cells from human BM and knocked down the expression of DEK using two independent shRNAs. We observed that DEK knockdown cells exhibited reduced quiescent percentages compared with their counterparts (Fig. S5 C). Notably, knockdown of DEK expression in human CD34+ hematopoietic cells led to significant upregulation of AKT1 and AKT2 (Fig. S5 D). These data confirm the effect of DEK on hHSPC cell cycle and the expression of specific genes. Altogether, our data reveal that DEK governs quiescence and metabolic hemostasis of HSCs by regulating the expression of cell-cycle and metabolism-associated genes in a manner of shaping chromatin accessibility (Fig. 6 R).
Discussion
Previous evidence implies that DEK has crucial functions in hematopoiesis and leukemogenesis. Here, we have identified a previously unrecognized role for nuclear DEK in the maintenance of HSCs and HPCs. Deletion of DEK in HSCs led to transient expansion but eventual depletion of the HSC pool, resulting in defective hematopoiesis. We discovered the intrinsic roles of DEK in maintaining quiescence of HSCs and preserving low metabolic activity of HSCs. Mechanistically, DEK functioned as a chromatin remodeler and directly regulated chromatin accessibility of quiescence-associated genes in HSCs by recruiting the corepressor NCoR1 to repress acetylation of H3K27. Akt1 and Akt2 were identified as the downstream effectors of DEK and were able to mediate the function of DEK in governing HSC homeostasis mainly via the Akt-mTOR axis.
In this study, we conditionally ablated the DEK alleles in hematopoietic cells using two different strategies: pIpC-induced deletion in adult Dekfl/flMx1-Cre mice and hematopoietic lineage-specific deletion in Dekfl/flTie2-Cre mice. All these strains demonstrated that DEK loss results in considerable depletion of the HSC pool. The discrepancy between our study and that of previous work (Serrano-Lopez et al., 2018) might be due to the following differences in experimental designs: (1) we used tissue-specific deletion in the hematopoietic lineage to exclude the possible influence from nonhematopoietic tissues; (2) the previous study used mice 8–10 wk of age, whereas we used Dekfl/flTie2-Cre mice with strictly the same age (6 or 12 wk of age); (3) we also used inducible deletion to precisely monitor the HSC pool in response to DEK deletion and found that DEK-deficient HSCs underwent transient expansion but eventual depletion, which may explain why Dek−/− mice showed unchanged hematopoiesis at 8–10 wk of age; and (4) we identified HSCs by three different phenotypic characterizations (CD48−CD150+LSK, CD34−CD135−LSK, or CD34−LSK) to test the effect of DEK deletion on HSC maintenance. Our results revealed that nuclear DEK is essential for quiescence maintenance of HSCs and preserves HSC potential. Importantly, recombinant DEK protein acts as a hematopoietic cytokine, which increases long-term HSC numbers via the CXC chemokine receptor CXCR2, independent of the nuclear function of DEK (Capitano et al., 2019). It seems that extranuclear DEK and nuclear DEK have different functions. Intracellular DEK promotes expansion of human and mouse HSCs, while nuclear DEK maintains HSC quiescence and potential. As intrinsic DEK protein locates in the nucleus and exogenous DEK can penetrate cells, eventually translocating to the nucleus (Saha et al., 2013), this suggests that it is necessary to design cytokine DEK with or without nuclear translocation signal. Preserving HSC quiescence and potential in vitro will benefit HSC clinical applications, including HSC transplantation. Nuclear DEK is an excellent candidate to protect HSCs against extra physiological oxygen shock and maintains HSC stemness. Additional studies are needed to systematically investigate the applications of DEK protein to improve hematopoietic regeneration in clinic.
Akt-mTOR signaling plays a critical role in cell metabolism, proliferation, and survival, especially for the balance of quiescence and activation in HSCs (Qian et al., 2016; Huang et al., 2019). Akt1/2 deletion results in inhibited proliferation of HSCs and impaired long-term function (Juntilla et al., 2010), while aberrant activation of Akt-mTOR signaling by PTEN deletion or overexpression of Akt1 leads to quiescence exit and eventual exhaustion of HSCs (Yilmaz et al., 2006; Kharas et al., 2010). Here, we found that DEK deficiency activates Akt-mTOR signaling in HSCs mainly by promoting transcription of Akt1/2. The increased mitochondrial metabolism, accelerated proliferation, and impaired self-renewal in DEK-deficient HSCs are similar to the phenomena observed in PTEN-deficient HSCs and Akt-overexpressed HSCs; however, DEK-deficient mice did not show some of phenomena observed in PTEN-deficient mice, including myeloid progenitor expansion and B cell depletion. It has been demonstrated that mTORC2, rather than mTORC1, is a major mediator of the effects of PTEN deletion on hematopoietic cells (Magee et al., 2012), and PTEN deletion or overexpression of Akt1 results in hyperactivation of Akt-mTOR signaling (Yilmaz et al., 2006; Kharas et al., 2010). We speculated that the difference in activation intensity of Akt-mTOR signaling between DEK-deficient HSCs and PTEN-deficient HSCs, as well as the diverse regulator roles of DEK and PTEN in Akt-mTOR signaling, result in the phenomena discrepancies.
The condensed structure of chromatin naturally represses gene expression, in part by preventing access of transcription machinery and other associated factors to DNA (Lorch and Kornberg, 2015; Venkatesh and Workman, 2015; Klemm et al., 2019). Histone chaperones patient SE translocation and PARP1 permit chromatin accessibility by removing DEK from chromatin (Gamble and Fisher, 2007), suggesting that DEK may affect chromatin accessibility. In Arabidopsis, DEK specifically binds H3 and H4, and MNase results reveal that DEK-overexpressed plants have more compact chromatin (Waidmann et al., 2014). To our knowledge, the role of DEK in regulating chromatin accessibility remains largely unknown. Here, we discovered specifically high expression of DEK in HSCs and demonstrated that DEK restricts the global chromatin accessibility of HSCs by ATAC-seq. Moreover, our results explain how DEK regulates chromatin accessibility, in which DEK recruits the NCoR1/HDAC3 complex to deacetylate H3K27. Deletion of NCoR1 results in the expansion of the HSC pool due to aberrant cell-cycle entry, and NCoR1 mainly interacts with its partner, HDAC3, to modulate histone acetylation and gene transcription by binding the promoter regions of myeloid-differentiation genes (Wan et al., 2019). Here, we discovered that DEK acts as a recruiter to guide NCoR1/HDAC3 to self-renewal–associated gene regions, such as Akt1/2; thus, our results uncover the upstream and downstream factors of NCoR1/HDAC3 in HSCs. Chromatin accessibility is precisely regulated by multiple factors and is tightly associated with histone modifications (Wu et al., 2018; Wu et al., 2016; Klemm et al., 2019). Clinical response to histone deacetylase inhibitors is strongly associated with a concurrent gain in chromatin accessibility in leukemic and host CD4+ T cells (Qu et al., 2017). Here, we found that an HDAC3 inhibitor—RGFP966—efficiently increased chromatin accessibility of hematopoietic cells in Dekfl/fl mice, and a CBP/P300 bromodomain inhibitor—GEN-049—selectively blocked H3K27ac, abolishing the increased chromatin accessibility of hematopoietic cells in Dek-cKO mice. Thus, our results support the tight correlations between histone modifications and chromatin accessibility and uncover the crucial roles of DEK in regulating histone acetylation and HSC homeostasis.
Materials and methods
Mice
All mice used had a C57BL/6J background. Ptenfl/+ (Ptenem1(flox)Smoc) mice were purchased from Shanghai Model Organisms. Dekfl/+and DekTg-fl mice were purchased from the Cyagen. Dekfl/+ mice carried a DEK loci in which exons 3–5 were flanked by two loxp sites. To achieve inducible or tissue-specific deletion of a gene, Floxed mice were crossed with Mx1-Cre or Tie2-Cre mice. DEK deletion in Dekfl/flMx1-Cre mice was induced by three i.p. injections of 6–10 µg pIpC (GE Healthcare) per gram of body weight every second for a total of three injections. DekTg-fl mice carried transgenic loci in which an ORF of mouse DEK gene was inserted following a floxed STOP cassette (3 × SV40 pA) driven by the CAG promoter. To achieve tissue-specific overexpression of DEK, DekTg-flmice were crossed with Tie2-Cre mice. All compound genetically engineered mice were a result of breeding the above mice followed by appropriate PCR-based genotyping with specific primers (Table S1). For drug treatments, mice were orally gavaged with NVP-BEZ235 (40 mg/kg; Selleck) or MK-2206 (10 mg/kg; Selleck) for indicated weeks. All mice experiments were approved by the Animal Committee of the Institute of Zoology, Third Military Medical University.
Blood cell counts
PB samples were collected from the tail vein and subjected to Hemavet 950FS (Drew Scientific) for blood cell counts.
Flow cytometry analysis and cell sorting
Total white blood cells were obtained after lysis of PB and BM with red cell lysis buffer (Solarbio). Single-cell suspensions from BM, PB, spleen, and thymus were stained with panels of fluorochrome-conjugated antibodies (Table S2). PB cells, spleen cells, thymus cells, and BM cells were stained with surface markers. Analyses were performed using BD FACSCanto II. For the intracellular staining, BM cells were stained with the surface marker and then subjected to Fixation/Permeabilization Kit (BD Biosciences) according to the manufacturer’s instructions, and then cells were stained with fluorochrome-conjugated antibodies (Table S2). For cell sorting, Dynabeads biotin binder (Invitrogen) was used to delete the mature cells of BM cells, and the remaining cells were stained with antibodies and sorted by a BD FACSAria II. All data were analyzed using FlowJo software (version 10).
Cell culture and purification of human CD34+ hematopoietic cells
Sorted mouse Lin−c-Kit+ cells, LSK cells, and HSCs were cultured in vitro in Stem Span medium (Stemcell Technologies). For drug treatment, 5 µM RGFP-966 or 500 nM GNE-049 was added into the cultured medium for indicated times.
Human BM CD34+ cells were isolated with the EasySep Human CD34 Positive Selection Kit II according to the manufacturer’s instructions (Stemcell Technologies). The purity of CD34+ cells was >90%, as determined by flow cytometry. Isolated CD34+ cells were cultured in vitro in Stem Span medium (Stemcell Technologies) with Stem Span CD34+ Expansion Supplement (Stemcell Technologies). The use of human cells was approved by the institutional review board of Third Military Medical University.
BrdU incorporation analysis
Mice were given an i.p. injection of 100 µl of 10 mg/ml BrdU (Sigma-Aldrich) 24 h before being killed for analysis. BM cells were stained with fluorochrome-conjugated antibodies (Table S2), followed by fixation and permeabilization with Cytofix/Cytoperm (BD Bioscience), treatment with Dnase I (Sigma-Aldrich), and staining with a BrdU-specific antibody according to the instruction manual of the BrdU flow kit (BD Pharmigen). Cells were analyzed by BD FACSCanto II.
Glucose consumption and ATP production
Sorted LSK cells were cultured in vitro for 24 h in Stem Span medium (Stemcell Technologies). Glucose concentration in the medium was analyzed using the Amplex Red Glucose/Glucose Oxidase Assay Kit (Invitrogen). The cellular ATP level in sorted HSCs was measured using Enhanced ATP Assay Kit (Beyotime) according to the manufacturer’s instructions.
Protein synthesis assay
To measure protein synthesis rates in vivo, mice were i.p. injected with OP-Puro (50 mg/kg). 1 h later, BM cells were collected and stained with surface markers, and we detected the incorporation of OP-Puro using the Click-iT Plus OPP Protein Synthesis Assay Kit (Molecular Probes).
Colony-forming assay
About 6 × 104 BM cells or 300 HSCs were isolated from mice and plated in 35-mm tissue culture dishes containing Mouse Methylcellulose Complete Media (R&D Systems). After 10–12 d of incubation, the number of granulocyte, erythroid, macrophage, and megakaryocyte colonies were counted. For second plating, cells in the first culture cycle were collected and replated in new culture dishes.
BM transplantation assay
For noncompetitive transplantation assay, BM cells (1 × 106) from mice were transplanted into lethally irradiated Ly5.1+ mice. For competitive transplantation assay, BM cells (1 × 106) or CD48−CD150+LSK cells (5 × 102) from Dek-cKO or Dekfl/flMx1-Cre mice or their corresponding control littermates (CD45.2+) were transplanted into lethally irradiated Ly5.1+Ly5.2+ (CD45.1+CD45.2+) recipient mice, together with competitor BM cells (1 × 106) or CD48−CD150+ LSK cells (5 × 102) from Ly5.1 (CD45.1+) mice. For inducible deletion of DEK in Dekfl/flMx1-Cre donor cells, recipient mice were subjected to three injections of pIpC at 1 mo after transplantation. For second transplantation, BM cells (1 × 106) were transplanted into lethally irradiated recipient mice.
Limiting-dilution competitive repopulation assay
The competitive repopulation cell (CRU) frequency was determined using 5 × 103, 1 × 104, 5 × 104, and 1 × 105 unfractionated BM cells from DEK-deficient mice or their corresponding control littermates (CD45.2+) that were transplanted into lethally irradiated Ly5.1+ (CD45.1+CD45.2+) recipient mice, together with competitor BM cells (5 × 105) from Ly5.1 (CD45.1+) mice. The cell dose was considered to contain at least one CRU if donor engraftment in the BM of recipient mice exceeded 1% of both lymphoid and myeloid lineages at 12 wk after transplantation. The CRU frequency was calculated and plotted using extreme limiting dilution analysis, as previously described (Hu and Smyth, 2009).
Homing assay
20 × 106 BM cells from Dekfl/fl or Dek-cKO mice were suspended in 1 ml PBS with added CFSE (2.5 µM) and incubated at 37°C for 15 min. Then an equal volume of prewarmed FBS was added to stop labeling. Cells were washed twice with PBS, followed by injection into the retro-orbital sinus of recipient mice that had been irradiated 24 h before transplantation. BM cells of recipient mice were harvested 6 h after transplantation and stained with antibodies against lineage markers and Sca-1. The frequency of CFSE+ cells in the Lin−Sca-1+ population was determined by FACS.
5-FU treatment
5-FU (Sigma-Aldrich) was administrated to mice intraperitoneally at a dose of 150 mg per kg body weight, once per week for 3 wk, and the survival of individual mice was monitored daily.
RNA extraction and qRT-PCR analysis
Total RNA was isolated using the Total RNA isolation Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. cDNA was reverse transcribed using PrimeScript RT reagent Kit (Takara) and subjected to real-time PCR with SYBR Green Supermix (Bio-Rad) in an iCycler iQ Real-Time PCR Detection System (Bio-Rad). All primers are listed in Table S1. All samples were run in triplicate. β-Actin (Actinb) or GAPDH was used as an internal control for mRNA.
Western blot and immunoprecipitation assay
For Western blot, BM cells or freshly sorted Lin−c-Kit+ cells were extracted in radio immunoprecipitation assay lysis buffer (1× PBS, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM EDTA). Protein extracts were subjected to electrophoresis on polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were first incubated in blocking buffer and then incubated with antibodies. The antibodies used in this study were against the following: DEK (1:1,000; 16448–1-AP; Proteintech), DEK (1:1,000; 66194–1-Ig; Proteintech), PI3K (1:1,000; AF1549; Beyotime), p-PI3K (1:1,000; 4228T; Cell Signaling Technology), Akt (1:1,000; 10176–2-AP; Proteintech), p-Akt (1:1,000; 66444–1-Ig; Proteintech), mTOR (1:1,000; 20657–1-AP; Proteintech), p-mTOR (1:1,000; 610301; BioLegend), Snf2h (1:1,000; 13066–1-AP; Proteintech), NCoR1 (1:1,000; sc-515934; Santa Cruz Biotechnology), HDAC3 (1:1,000; sc-376957; Santa Cruz Biotechnology), HDAC3 (1:1,000; 10255–1-AP; Proteintech), mono-methyl-histone H3 (Lys4; 1:1,000; 5326T; Cell Signaling Technology), di-methyl-histone H3 (Lys4; 1:1,000; 9725T; Cell Signaling Technology), H3K4me3 (1:1,000; 9751T; Cell Signaling Technology), di-methyl-histone H3 (Lys9; 1:1,000; AH438; Beyotime), tri-methyl-histone H3 (Lys9; 1:1,000; 13969T; Cell Signaling Technology), tri-methyl-histone H3 (Lys27; 1:1,000; 9733S; Cell Signaling Technology), H3K9ac (1:1,000; 9649T; Cell Signaling Technology), acetyl-histone H3 (Lys18; 1:1,000; 9675T; Cell Signaling Technology), H3K27ac (1:1,000; 8173T; Cell Signaling Technology), tubulin (1:1,000; AF0001; Beyotime), Lamin B1 (1:1,000; AF1408; Beyotime), H3 (1:1,000; AH433; Beyotime), H2A (1:1,000; AH419; Beyotime), H2B (1:1,000; AH426; Beyotime), and H4 (1:1,000; AH458; Beyotime). The HRP-conjugated secondary antibodies were goat anti-rabbit and goat anti-mouse (1:1,000; Beyotime). Immunoprecipitates were analyzed by Western blot.
Duolink proximity ligation assay (PLA)
Duolink PLA was performed according to the manufacturer’s instructions (Duolink In Situ Red Starter Kit; Sigma-Aldrich). Briefly, HSCs were sorted onto poly-lysine–coated coverslips and then fixed in 4% paraformaldehyde for 20 min before permeabilization in PBS containing 0.1% Triton X-100 for 5 min. Cells were then blocked in 5% BSA and incubated with primary antibodies (anti-DEK, anti-NCoR1, and anti- Snf2h). After washing the cells, PLA probes were added, followed by hybridization, ligation, and amplification for 90 min at 37°C. DNA and DEK-NCoR1 were visualized after incubation with the detection solution. Slides were analyzed by fluorescence microscopy.
MNase assay
About 0.5 million Lin−c-Kit+ cells were collected and resuspended in 1 ml of lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40) and incubated on ice for 10 min with three times vortex. The nucleus was isolated by centrifugation (500 g, 5 min). Finally, nuclei were resuspended in 50 µl of digestion buffer (15 mM Tris-HCl, pH7.4, 1 mM CaCl2) and digested with MNase (0.01 U/ml; Thermo Fisher Scientific) at 37°C for 5 min. The reactions were stopped by adding 10 µl of stop solution (2% SDS, 50 mM EDTA). DNA was purified using Hipure Tissue DNA Mini Kit (Magen) and subjected to agarose gel electrophoresis.
RNA-seq and data analysis
About 800 HSCs were sorted by FACS, and the mRNA library was constructed using the Smart-Seq V4 Ultra Low Input RNA Kit for sequencing (Takara) according to the manufacturer’s instruction. Libraries were sequenced by the Illumina HiSeq 2000 platform as 150-bp pair-ended reads. Reads were aligned using bowtie v0.12.9. Fragments per kilobase per million (FPKM) estimation was performed with Cufflinks v2.1.1, aligned reads were counted with HTSeq (a Python framework to work with high-throughput sequencing data), and differential expression analysis was performed with DESeq2. Differentially expressed genes were selected using a cutoff P value of <0.01 (false discovery rate adjusted for multiple testing). For functional profiling of changes in mRNA by RNA-seq, GSEA analysis was performed using the GSEA software.
ATAC-seq and data processing
ATAC-seq was performed following the protocol of Xu et al. (2019). Briefly, 50,000 HSCs were sorted and resuspended in 35 µl of lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and incubated on ice for 10 min with three times vortex. DNA fragmentation was conducted by adding 10 µl 5 × TruePrep Tagment Buffer L, 5 µl TruePrep Tagment Enzyme Mix and shaking at 37°C for 30 min. Fragmented DNA was purified using 1.8 × AMPure beads, barcoded with dual indexes, and PCR amplified. Size selection and purification of DNA fragments were done using AMPure beads. Size distribution and molarity of these sequencing libraries were determined by Agilent 2100 bioanalyzer and Qubit (Thermo Fisher Scientific). Sequencing was performed using Hiseq-PE150 (Illumina) by Novogene.
Low-quality reads and adaptor sequences were removed by Trim Galore v0.4.4 with parameters “-q 10 -length 30 -stringency 5′.” Paired-end reads were mapped to the mm10 reference genome using Bowtie2 version 2.2.9. Reads that aligned to the mitochondrial genome were filtered, and PCR duplicates were removed with picard MarkDuplicates. To identify peaks, the bam files containing unique, nonchrM reads were used to call peaks with MACS2 using parameters ‘‘-nomodel-keep-dup all -q 0.01-shift -100-extsize 200-call-summits -g mm.’’ For differential coverage, corresponding bam files were merged to call peaks to get a union peak set. For each peak in the peak set, raw ATAC-seq reads were counted by featureCounts. Differentially expressed peaks were identified by at least a 1.5-fold change and false discovery rate adjusted P value of 0.01. For downstream analysis, HOMER was used to find TF motifs. BedTools was used to find a common intersection between identified peaks (1 bp minimum overlap).
CUT&Tag and data processing
The CUT&Tag assay was performed as described previously (Kaya-Okur et al., 2019). In brief, 50,000 LSK cells or 20,000 HSCs were harvested and washed twice in 1 ml of wash buffer (20 mM Hepes, pH 7.5, 150 mM Nacl, 0.5 mM Spermidine, 1× protease inhibitor cocktail [PIC; Bimake]) and then resuspended in 300 µl of wash buffer. Concanavalin A–coated beads were activated by washing twice in binding buffer (20 mM Hepes, pH 7.5, 10 mM KCl, 1 mM MnCl2, 1 mM CaCl2). 10 µl activated beads were then added to the cell suspension. After 30 min of incubation at room temperature, the supernatant was removed and the beads were resuspended in 50 µl antibody buffer (20 mM Hepes, pH 7.5, 150 mM Nacl, 0.5 mM Spermidine, 0.0125% Digitonin, 2 mM EDTA, 0.1% BSA, 1× PIC). Then, 0.5 µl antibody (H3K27ac, H3K9ac, H3K4me3, or DEK) was added and incubated for 2 h at room temperature. After the supernatant was gently removed on DynaMag (Thermo Fisher Scientific), the beads were resuspended in 100 µl Dig wash buffer (20 mM Hepes, pH 7.5, 150 mM Nacl, 0.5 mM Spermidine, 0.0125% Digitonin, 1× PIC) containing 1 µl guinea pig anti-rabbit IgG antibody. After incubation for 1 h at room temperature, the beads were then washed three times in 800 µl Dig wash buffer, followed by resuspension in 100 µl Dig 300 wash buffer (20 mM Hepes, pH 7.5, 300 mM NaCl, 0.5 mM Spermidine, 0.0125% Digitonin, 1× PIC). Then, 0.04 µM pA-Tn5 adapter complex (Vazyme S603) was added and incubated for 1 h at room temperature. Beads were washed three times in 800 µl Dig 300 wash buffer and resuspended in 300 µl tagmentation buffer (20 mM Hepes, pH 7.5, 300 mM Nacl, 0.5 mM Spermidine, 0.0125% Digitonin, 10 mM MgCl2, 1× PIC) and incubated for 1 h at 37°C. Then, 10 µl of 0.5 M EDTA, 3 µl of 10% SDS, and 1 µl of 20 mg/ml Proteinase K were added and incubated at 55°C for 1 h to stop tagmentation. DNA was extracted by phenol chloroform iso-amyl alcohol and amplified with i5 and i7 primers (Vazyme, TD202) by 16–18 cycles’ PCR. The amplified products were purified by AMPure XP beads to remove >1 kb fragment and to enrich the 200–1,000 bp fragments for high-throughput sequencing (PE150 on Illumina platform).
The quality control of each sample was accomplished using FASTQC V0.11.5. Clean reads were aligned to mouse reference genome (mm10) using Bowtie 2 version 2.2.9. MarkDuplicates was used to remove PCR duplicate and unmapped reads. Only unique mapped reads were used in further analysis. Peak calling and annotation were performed using MACS2 and ChIPseeker package, respectively.
Lentivirus production and transduction
To generate the vectors for the expression of PTEN and specific shRNA targeting DEK, we designed the primers for PCR of mouse PTEN coding sequences and the oligos for shRNAs (Table S1). The PTEN coding sequences was cloned into the vector pMSCV-IRES-GFP, and the shRNAs oligos were cloned into the vector pLKO.1-puro.
For lentivirus production, 293T cells were transfected with the helper plasmid Ecopack. The medium was replaced with fresh medium at 10 h after transfection. The culture supernatants were collected at 48 h after transfection and filtered by 0.22-µm membrane. Virus was stored at −80°C until use. For lentiviral infection of cells in vitro, the hematopoietic progenitors were spin infected at 250 g at room temperature for 2 h in the presence of polybrene (5 µg/ml). For in vivo transplantation, cells were incubated for a further 1 h at 37°C.
Statistics
Comparisons between two groups were analyzed using two-tailed Student’s t test. Differences among more than two groups were analyzed using one-way ANOVA followed by Tukey-Kramer post-hoc tests. Values of P < 0.05 were considered statistically significant. All data are means ± SD.
Data availability
All sequencing data are deposited in the National Center for Biotechnology Information GEO database. RNA-seq, ATAC-seq, and CUT&Tag data are available under accession no. GSE166434.
Online supplemental material
Fig. S1 shows that DEK deletion impairs hematopoiesis in mice. Fig. S2 shows that DEK deletion decreases the HSPC pool in mice. Fig. S3 shows that DEK deletion impairs the reconstitution capacity of HSCs. Fig. S4 shows that DEK interacts with H3 and regulates chromatin accessibility. Fig. S5 shows that knockdown of DEK in human primitive hematopoietic cells decreases quiescence. Table S1 shows sequences of primers. Table S2 shows fluorochrome-conjugated antibodies.
Supplementary Material
lists primers used in this study.
lists antibodies used in this study.
Acknowledgments
We thank Dr. Wen-shu Wu for helping revise the manuscript.
This work was supported by grants from National Key Research and Development Program of China Stem Cell and Translational Research (2017YFA0106700), National Science Foundation of China (81970100), and Army Major Scientific Research Projects (AWS17J007).
Author contributions: Y. Hou, X. Wu, and J. Chen conceived the project, analyzed data, and wrote the paper. Z. Chen performed experiments, analyzed data, and wrote the paper. D. Huo, and L. Li performed some experiments and analyzed data. S. Xu, Z. Liu, Z. Li, C. Zhou, Y. Liu, W. Wu, Y. Huang, M. Kuang, F. Wu, H. Li, P. Qian, and G. Song contributed to the data analysis and paper revision.
References
- Alenzi, F.Q., Alenazi B.Q., Ahmad S.Y., Salem M.L., Al-Jabri A.A., and Wyse R.K.. 2009. The haemopoietic stem cell: between apoptosis and self renewal. Yale J. Biol. Med. 82:7–18. [PMC free article] [PubMed] [Google Scholar]
- Bigas, A., and Waskow C.. 2016. Blood stem cells: from beginning to end. Development. 143:3429–3433. 10.1242/dev.142828 [DOI] [PubMed] [Google Scholar]
- Böhm, F., Kappes F., Scholten I., Richter N., Matsuo H., Knippers R., and Waldmann T.. 2005. The SAF-box domain of chromatin protein DEK. Nucleic Acids Res. 33:1101–1110. 10.1093/nar/gki258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer, H.E., Kappes F., Mor-Vaknin N., Legendre M., Kinzfogl J., Cooper S., Hangoc G., and Markovitz D.M.. 2012. DEK regulates hematopoietic stem engraftment and progenitor cell proliferation. Stem Cells Dev. 21:1449–1454. 10.1089/scd.2011.0451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer, H.E., Mor-Vaknin N., Kappes F., Legendre M., Saha A.K., Ou X., O’Leary H., Capitano M., Cooper S., and Markovitz D.M.. 2013. Concise review: role of DEK in stem/progenitor cell biology. Stem Cells. 31:1447–1453. 10.1002/stem.1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal-Hierro, L., van Galen P., Prado M.A., Higby K.J., Togami K., Mowery C.T., Paulo J.A., Xie Y., Cejas P., Furusawa T., et al. 2020. Chromatin accessibility promotes hematopoietic and leukemia stem cell activity. Nat. Commun. 11:1406. 10.1038/s41467-020-15221-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi, L.M., and Link D.C.. 2015. The hematopoietic stem cell niche in homeostasis and disease. Blood. 126:2443–2451. 10.1182/blood-2015-07-533588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitano, M.L., Mor-Vaknin N., Saha A.K., Cooper S., Legendre M., Guo H., Contreras-Galindo R., Kappes F., Sartor M.A., Lee C.T., et al. 2019. Secreted nuclear protein DEK regulates hematopoiesis through CXCR2 signaling. J. Clin. Invest. 129:2555–2570. 10.1172/JCI127460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully, M., You H., Levine A.J., and Mak T.W.. 2006. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 6:184–192. 10.1038/nrc1819 [DOI] [PubMed] [Google Scholar]
- Dai, J., Huang Y.J., He X., Zhao M., Wang X., Liu Z.S., Xue W., Cai H., Zhan X.Y., Huang S.Y., et al. 2019. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 176:1447–1460.e14. 10.1016/j.cell.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak, E., and Bigas A.. 2018. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 22:639–651. 10.1016/j.stem.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Eaves, C.J. 2015. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 125:2605–2613. 10.1182/blood-2014-12-570200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium . 2012. An integrated encyclopedia of DNA elements in the human genome. Nature. 489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, G.K., Grosveld G., and Markovitz D.M.. 1997. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc. Natl. Acad. Sci. USA. 94:1811–1815. 10.1073/pnas.94.5.1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble, M.J., and Fisher R.P.. 2007. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat. Struct. Mol. Biol. 14:548–555. 10.1038/nsmb1248 [DOI] [PubMed] [Google Scholar]
- Ho, T.T., Warr M.R., Adelman E.R., Lansinger O.M., Flach J., Verovskaya E.V., Figueroa M.E., and Passegué E.. 2017. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 543:205–210. 10.1038/nature21388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., and Smyth G.K.. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 347:70–78. 10.1016/j.jim.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Huang, D., Chen C., Hao X., Gu H., Xie L., Yu Z., and Zheng J.. 2019. Metabolic Regulations in Hematopoietic Stem Cells. Adv. Exp. Med. Biol. 1143:59–74. 10.1007/978-981-13-7342-8_3 [DOI] [PubMed] [Google Scholar]
- Ito, K., and Ito K.. 2018. Hematopoietic stem cell fate through metabolic control. Exp. Hematol. 64:1–11. 10.1016/j.exphem.2018.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., Bonora M., and Ito K.. 2019. Metabolism as master of hematopoietic stem cell fate. Int. J. Hematol. 109:18–27. 10.1007/s12185-018-2534-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntilla, M.M., Patil V.D., Calamito M., Joshi R.P., Birnbaum M.J., and Koretzky G.A.. 2010. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 115:4030–4038. 10.1182/blood-2009-09-241000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes, F., Scholten I., Richter N., Gruss C., and Waldmann T.. 2004. Functional domains of the ubiquitous chromatin protein DEK. Mol. Cell. Biol. 24:6000–6010. 10.1128/MCB.24.13.6000-6010.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes, F., Waldmann T., Mathew V., Yu J., Zhang L., Khodadoust M.S., Chinnaiyan A.M., Luger K., Erhardt S., Schneider R., and Markovitz D.M.. 2011. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 25:673–678. 10.1101/gad.2036411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya-Okur, H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., and Henikoff S.. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10:1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharas, M.G., Okabe R., Ganis J.J., Gozo M., Khandan T., Paktinat M., Gilliland D.G., and Gritsman K.. 2010. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood. 115:1406–1415. 10.1182/blood-2009-06-229443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm, S.L., Shipony Z., and Greenleaf W.J.. 2019. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20:207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Liang, R., Arif T., Kalmykova S., Kasianov A., Lin M., Menon V., Qiu J., Bernitz J.M., Moore K., Lin F., et al. 2020. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell. 26:359–376.e7. 10.1016/j.stem.2020.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Zhao M., Jin X., Ney G., Yang K.B., Peng F., Cao J., Iwawaki T., Del Valle J., Chen X., and Li Q.. 2019. Adaptive endoplasmic reticulum stress signalling via IRE1α-XBP1 preserves self-renewal of haematopoietic and pre-leukaemic stem cells. Nat. Cell Biol. 21:328–337. 10.1038/s41556-019-0285-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch, Y., and Kornberg R.D.. 2015. Chromatin-remodeling and the initiation of transcription. Q. Rev. Biophys. 48:465–470. 10.1017/S0033583515000116 [DOI] [PubMed] [Google Scholar]
- Magee, J.A., Ikenoue T., Nakada D., Lee J.Y., Guan K.L., and Morrison S.J.. 2012. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 11:415–428. 10.1016/j.stem.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli, A.M., Chiarini F., Evangelisti C., Grimaldi C., Ognibene A., Manzoli L., Billi A.M., and McCubrey J.A.. 2010. The phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin signaling network and the control of normal myelopoiesis. Histol. Histopathol. 25:669–680. [DOI] [PubMed] [Google Scholar]
- Mor-Vaknin, N., Punturieri A., Sitwala K., Faulkner N., Legendre M., Khodadoust M.S., Kappes F., Ruth J.H., Koch A., Glass D., et al. 2006. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol. Cell. Biol. 26:9484–9496. 10.1128/MCB.01030-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Vaknin, N., Kappes F., Dick A.E., Legendre M., Damoc C., Teitz-Tennenbaum S., Kwok R., Ferrando-May E., Adams B.S., and Markovitz D.M.. 2011. DEK in the synovium of patients with juvenile idiopathic arthritis: characterization of DEK antibodies and posttranslational modification of the DEK autoantigen. Arthritis Rheum. 63:556–567. 10.1002/art.30138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottis, A., Mouchiroud L., and Auwerx J.. 2013. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 27:819–835. 10.1101/gad.214023.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Ishizu, A., Takizawa H., and Suda T.. 2014. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development. 141:4656–4666. 10.1242/dev.106575 [DOI] [PubMed] [Google Scholar]
- Oancea, C., Rüster B., Henschler R., Puccetti E., and Ruthardt M.. 2010. The t(6;9) associated DEK/CAN fusion protein targets a population of long-term repopulating hematopoietic stem cells for leukemogenic transformation. Leukemia. 24:1910–1919. 10.1038/leu.2010.180 [DOI] [PubMed] [Google Scholar]
- Perissi, V., Jepsen K., Glass C.K., and Rosenfeld M.G.. 2010. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 11:109–123. 10.1038/nrg2736 [DOI] [PubMed] [Google Scholar]
- Qian, P., He X.C., Paulson A., Li Z., Tao F., Perry J.M., Guo F., Zhao M., Zhi L., Venkatraman A., et al. 2016. The Dlk1-Gtl2 loci preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 18:214–228. 10.1016/j.stem.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, K., Zaba L.C., Satpathy A.T., Giresi P.G., Li R., Jin Y., Armstrong R., Jin C., Schmitt N., Rahbar Z., et al. 2017. Chromatin accessibility landscape of cutaneous T cell lymphoma and dynamic response to HDAC inhibitors. Cancer Cell. 32:27–41.e4. 10.1016/j.ccell.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner, R., Kharbanda S., Jin L., Jeng E., Chan E., Merchant M., Haverty P.M., Bainer R., Cheung T., Arnott D., et al. 2018. Enhancer activity requires CBP/P300 bromodomain-dependent Histone H3K27 acetylation. Cell Rep. 24:1722–1729. 10.1016/j.celrep.2018.07.041 [DOI] [PubMed] [Google Scholar]
- Riveiro-Falkenbach, E., and Soengas M.S.. 2010. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin. Cancer Res. 16:2932–2938. 10.1158/1078-0432.CCR-09-2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, A.K., Kappes F., Mundade A., Deutzmann A., Rosmarin D.M., Legendre M., Chatain N., Al-Obaidi Z., Adams B.S., Ploegh H.L., et al. 2013. Intercellular trafficking of the nuclear oncoprotein DEK. Proc. Natl. Acad. Sci. USA. 110:6847–6852. 10.1073/pnas.1220751110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandén, C., Järvstråt L., Lennartsson A., Brattås P.L., Nilsson B., and Gullberg U.. 2014. The DEK oncoprotein binds to highly and ubiquitously expressed genes with a dual role in their transcriptional regulation. Mol. Cancer. 13:215. 10.1186/1476-4598-13-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasine, J.P., Himburg H.A., Termini C.M., Roos M., Tran E., Zhao L., Kan J., Li M., Zhang Y., de Barros S.C., et al. 2018. Wild-type Kras expands and exhausts hematopoietic stem cells. JCI Insight. 3:e98197. 10.1172/jci.insight.98197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Lopez, J., Nattamai K., Pease N.A., Shephard M.S., Wellendorf A.M., Sertorio M., Smith E.A., Geiger H., Wells S.I., Cancelas J.A., and Privette Vinnedge L.M.. 2018. Loss of DEK induces radioresistance of murine restricted hematopoietic progenitors. Exp. Hematol. 59:40–50.e3. 10.1016/j.exphem.2017.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer, R.A., Magee J.A., Salic A., and Morrison S.J.. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 509:49–54. 10.1038/nature13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E.A., Krumpelbeck E.F., Jegga A.G., Prell M., Matrka M.M., Kappes F., Greis K.D., Ali A.M., Meetei A.R., and Wells S.I.. 2018. The nuclear DEK interactome supports multi-functionality. Proteins. 86:88–97. 10.1002/prot.25411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, L.M., Zanier K., Mackereth C., Sattler M., and Valcárcel J.. 2006. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 312:1961–1965. 10.1126/science.1128659 [DOI] [PubMed] [Google Scholar]
- Söderberg, O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Soekarman, D., von Lindern M., Daenen S., de Jong B., Fonatsch C., Heinze B., Bartram C., Hagemeijer A., and Grosveld G.. 1992. The translocation (6;9) (p23;q34) shows consistent rearrangement of two genes and defines a myeloproliferative disorder with specific clinical features. Blood. 79:2990–2997. 10.1182/blood.V79.11.2990.2990 [DOI] [PubMed] [Google Scholar]
- Stiles, B., Groszer M., Wang S., Jiao J., and Wu H.. 2004. PTENless means more. Dev. Biol. 273:175–184. 10.1016/j.ydbio.2004.06.008 [DOI] [PubMed] [Google Scholar]
- Venkatesh, S., and Workman J.L.. 2015. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16:178–189. 10.1038/nrm3941 [DOI] [PubMed] [Google Scholar]
- Waidmann, S., Kusenda B., Mayerhofer J., Mechtler K., and Jonak C.. 2014. A DEK domain-containing protein modulates chromatin structure and function in Arabidopsis. Plant Cell. 26:4328–4344. 10.1105/tpc.114.129254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, X., Liu L., Zhou P., Hui X., He Q., Yu F., Zhang W., Ding X., Yuan X., Zhang N., et al. 2019. The nuclear receptor corepressor NCoR1 regulates hematopoiesis and leukemogenesis in vivo. Blood Adv. 3:644–657. 10.1182/bloodadvances.2018022756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W., et al. 2016. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 534:652–657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Wu, J., Xu J., Liu B., Yao G., Wang P., Lin Z., Huang B., Wang X., Li T., Shi S., et al. 2018. Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature. 557:256–260. 10.1038/s41586-018-0080-8 [DOI] [PubMed] [Google Scholar]
- Xu, W., Zhao X., Wang X., Feng H., Gou M., Jin W., Wang X., Liu X., and Dong C.. 2019. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 51:826–839.e5. 10.1016/j.immuni.2019.10.006 [DOI] [PubMed] [Google Scholar]
- Yilmaz, O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., and Morrison S.J.. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 441:475–482. 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
- Yue, L., Wan R., Luan S., Zeng W., and Cheung T.H.. 2020. Dek Modulates Global Intron Retention during Muscle Stem Cells Quiescence Exit. Dev. Cell. 53:661–676.e6. 10.1016/j.devcel.2020.05.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primers used in this study.
lists antibodies used in this study.
Data Availability Statement
All sequencing data are deposited in the National Center for Biotechnology Information GEO database. RNA-seq, ATAC-seq, and CUT&Tag data are available under accession no. GSE166434.