Abstract
Background
Early-onset and severe atopic dermatitis (AD) in patients increase the probability of the development of allergic rhinitis or asthma. Treatment and prevention strategies in infants and young children with AD are targeted toward treating the symptoms, restoring skin barrier functions, and reducing the absorption of environmental allergens in an attempt to attenuate or block the onset of asthma and food allergy.
Objective
Given that the initiating events in AD remain poorly understood, identifying those at risk and implementing strategies to prevent AD is necessary.
Methods
Whole-exome sequencing (WES) was performed in a 43 control group and a disease group with 20 AD patients without atopic march (AM) and 20 with AM. Sanger sequencing was carried out to validate found variants in cohorts.
Results
DOCK8, IL17RA, and KLK12 single-nucleotide polymorphisms were identified by WES as missense mutations: c.1289C>A, p.P97T (rs529208); c.1685C>A, p.P562G (rs12484684); and c.457+27>C, rs3745540, respectively. A case-control study show that total immunoglobulin E (IgE) level was significantly increased in the AA genotype of DOCK8 compared to the CA genotype in allergic patients. The rs12484684 of IL17RA increased risk of adult-onset AD (odds ratio: 1.63) compared to the control for (A) allele frequency. AD and AM Patients with the IL17RA CA genotype also had elevated IgE levels. rs3745540 of KLK12 was associated with AD in dominant model (odds ratio: 2.86).
Conclusion
DOCK8 (rs529208), IL17RA (rs12484684), and KLK12 (rs3745540), were identified using a new WES filtering method. the result suggests that polymorphism of DOCK8 and IL17RA might be related to increase the total IgE level.
Keywords: Atopic dermatitis, DOCK8, Exome sequencing, IL17RA, KLK12, Sanger sequencing
INTRODUCTION
The pathophysiology of atopic dermatitis (AD) is complex and multifactorial, involving elements of alterations in cell-mediated immune responses, immunoglobulin E (IgE)-mediated hypersensitivity, environmental factors, and barrier dysfunction including increased transepidermal water loss (TEWL), pH alterations, dehydration, and filaggrin (FLG) gene mutations. Early-onset and severe AD in patients increase the probability of the development of allergic rhinitis or asthma, a phenomenon known as the ‘atopic march’ (AM)1,2. Treatment and prevention strategies in infants and young children with AD are targeted toward treating the symptoms, restoring skin barrier functions, and reducing the absorption of environmental allergens in an attempt to attenuate or block the onset of asthma and food allergy. Given that the initiating events in AD remain poorly understood, identifying those at risk and implementing strategies to prevent AD is impractical.
A genome-wide association study (GWAS) and epigenetic studies found chromosomal susceptibility loci and the epigenetic modulation associated with AD3. Profilaggrin degradation produces natural moisturizing factors (NMF) and flattens keratinocytes to maintain skin homeostasis. Loss-offunction FLG mutations have been reported as a major risk factor in skin barrier damage and are known to elevate AD severity4,5. allowing the prediction of early-onset AD from such genetic information or family history6. Hence, the genetic diversity of FLG mutations can be mapped in East Asian countries, and visualized using phylogenetic trees7, and the search for new genetic candidate markers can continue. Identification of genetic variants for early diagnosis will prevent AD development and being a major part of personalized therapy. Meanwhile, whole-exome sequencing (WES) for clinical diagnostics has increased steadily to identify casual variants in Mendelian and rare diseases, including cancer8. Eighty five percent disease-causing mutations are detected in coding regions (exome), which is comprised of about 180,000 exons9. WES is also a preferred analysis method with a better filtering system for identifying common and rare variants in a large cohort study than next-generation sequencing10,11.
In an attempt to eliminate certain variables and identify potential causative variants of AD, we divided three study groups as follows; (1) an adult-AD group that has not developed allergic rhinitis or allergic asthma, (2) an AM group, and (3) the Korean Personal Genome Project (KPGP) control group.
In evolutionary biology, it is known that there is a strong probability that conserved amino acid sites are likely to have a significant effect on protein function12. Thus, candidate risk variants, extracted from WES, were validated in a population-based case-control study, using Sanger sequencing. This study suggests that IL17RA, DOCK8, and KLK12 polymorphisms are novel candidate variants associated with allergic diseases.
MATERIALS AND METHODS
Patients
All patients with AD were given a diagnosis according to the revised criteria of Williams et al.13 The AM group was previously diagnosed with allergic rhinitis, asthma and allergic conjunctivitis along with AD. This study included a KPGP control group consisting of 43 patients and a disease group with 20 AD patients without AM and 20 with AM. Adult-onset (age≥18 years) of AD were recruited. KPGP is the Korean reference genome mapping project using the Illumina HiSeq platform14. Adult-AD means the development of AD in an adulthood, not a children, and the onset of childhood has a weakness to be dependent on memory. The development of AD in adulthood significantly reduces the incidence of asthma and allergic rhinitis compared to early-onset AD in infancy15. AM group were recruited both adults and children group who were progressed to allergic asthma and rhinitis. The clinical phenotype of the AM patients included 19 with AD plus allergic rhinitis and one with AD plus asthma (Table 1).
Table 1. Clinical information of the patients in whole-exome sequencing.
| Variable | Control (KPGP) | AD |
|---|---|---|
| Total | 43 | 40 |
| Age (yr) | - | 26.0±7.4 |
| Male/female | - | 25/15 |
| Total immunoglobulin E (KU/L) | 2,336±2,062 | |
| Eosinophils (mm3) | - | 438.7±369.7 |
| Clinical phenotype | - | |
| AD | 20 | |
| AD+AR | 19 | |
| AD+AS | 1 |
Values are presented as number only or mean±standard deviation. KPGP: Korean Personal Genome Project, AD: atopic dermatitis, AR: allergic rhinitis, AS: asthma, −: not available.
Eighty one healthy control, 87 adult AD and 72 AM patients were enrolled, and the characteristics of the study cohorts are presented in Table 2. Sanger sequencing was performed to validate the risk of AD for DOCK8, IL17RA, and KLK12 polymorphisms. Peripheral whole blood samples were obtained from all subjects.
Table 2. Clinical characteristic of subjects in Sanger sequencing.
| Variable | Control | AD | AM | p-value* |
|---|---|---|---|---|
| Total | 81 | 87 | 72 | |
| Age (yr) | 27.2±12.6 | 28.8±9.2 | 24.9±9.6 | |
| Male/female | 52/29 | 47/40 | 47/25 | |
| Total immunoglobulin E (KU/L) | - | 1,430±1,670 | 1,833±1,888 | 0.1620† |
| Eosinophils (mm3) | - | 459.4±377.8 | 449.4±376.1 | 0.8701† |
| Clinical phenotype | - | |||
| AD | 87 | |||
| AD+AR | 59 | |||
| AD+AS | 2 | |||
| AD+AR+AS | 6 | |||
| AD+AR+AC | 2 |
Values are presented as number only or mean±standard deviation. AD: atopic dermatitis, AM: atopic march, AR: allergic rhinitis, AS: asthma, AC: allergic conjunctivitis, −: not available. *By student's t-test between AD and AM, †DOCK8, IL17RA, and KLK12 polymorphisms in atopic dermatitis.
The present study protocol was reviewed and approved by the Chung-Ang University Hospital Institutional Review Board, IRB No. C2015258 (1716). Informed consent was obtained from all subjects when they were enrolled.
Whole-exome sequencing
Whole blood samples were collected in ethylenediaminetetraacetic acid tubes from all subjects and the QIAamp DNA Mini Kit (Qiagen Inc., Valencia, CA, USA) was used to isolate genomic DNA. A Qubit fluorometer (Life Technologies, Grand Island, NY, USA) and Nanodrop spectrometer (Nanodrop Technologies, Wilmington, DE, USA) were used to assess DNA concentration and purity. According to the manufacturer's standard protocol, WES was performed using SureSelect Human All Exon V4+UTR 71 Mb (Agilent, Santa Clara, CA, USA), and DNA samples were mechanically sheared using Covaris (Covaris, Woburn, MA, USA). A paired-end (PE) and deep sequencing DNA library was prepared through classic protocol steps (shearing, end-repair, A-tailing, peak detection, PE adaptor ligation, and amplification). After the library was hybridized with bait sequences for 24 hours, the captured library was purified and amplified with an index barcode tag. Then, library quality and quantity were measured. Sequencing of the exome library was carried out using the 100-bp PE mode of the HiSeq SBS kit.
WES processing and alignment
Millions of base sequence reads in FASTQ format were mapped to UCSC hg19 of the human assembly using the Burrows-Wheeler Aligner (BWA, v0.7.7)16 with “mem” and seed value parameters “−k 45” to create SAM files with correct mate-pair information for the genome. Picard (v1.92) was then used to convert the sequence alignment map (SAM) files to compressed binary alignment map (BAM) files, and then sort the BAM files by chromosome coordinates. The Genome Analysis Toolkit (v2.3.9Lite)17 was used for local realignment of the BAM files at intervals, corresponding to potential insertion and deletion alignment errors. Single-nucleotide polymorphisms (SNPs) and indels were annotated using snpEff (v3.6c; http://snpeff.sourceforge.net/index.html)18, which classified the variants.
Annotation
The dbNSFP is an integrated database of functional alteration predictions from multiple algorithms (scale invariant feature transform [SIFT], Polyphen2, PhyloP, PhastCons, 1000 Genome).
Filter 1: SnpEff annotates types of variants and predicts their effects on genomic regions (e.g., synonymous, nonsynonymous, nonsense, missense, stop-gain, frameshift, point mutations, and indels).
Filter 2: A simple assessment of putative variant impact was done using SnpEff. (High: Frame_Shift, Start_Lost, Splice_Site, Stop_Gained; Moderate: Non_Synonymous_coding, Codon_Insertion and _Deletion, Low: Start_gained, etc.).
Filter 3: SIFT and Polyphen2
The probability that an amino acid substitution will affect protein function (SIFT score) was predicted based on the degree of amino acid conservation derived from related sequences through PSI-BLAST. The SIFT score ranges from 0 to 1; a value lower than 0.05 or not significant is considered detrimental, otherwise, it is considered tolerable.
The Polyphen2 HDIV score was based on HumDiv, i.e., hdiv_prob ranged from 0 to 1, and categorized as “probably damaging, 0.957~1”, “possibly damaging, 0.453~0.956”, and “benign, 0~0.452.”
Filter 4: The PhyloP of dbNSFP reflects the score of evolutionary conservation sites under negative or positive selection over branches of the phylogenetic tree. A higher score indicates a more conserved site (http://varianttools.sourceforge.net/Annotation/DbNSFP). PhastCons scores contain multiple alignments of 99 vertebrate genomes to the human genome and predict conserved elements. A high score (>0.2) represents a functionally important genomic region.
Filter 5: The 1000 Genome Project was design to select variants with minor allele frequencies (MAFs, less than 0.01 or unknown) in the global population.
Filter 6: The in-house Korean variation database at the Theragen Etex Bio Institute was used to select variants (MAFs, less than 0.02 or unknown).
Sanger sequencing
Polymerase chain reaction (PCR) amplification conditions were as follows: 95℃ for 10 minutes, followed by 35 cycles at 95℃ for 30 seconds, 55℃~58℃ for 30 seconds, and 72℃ for 40 seconds, with a final extension at 72℃ for 1 minutes 30 seconds. The PCR mix (50 µl) contained 25 µl of 2× EF-Taq premix (SolGent, Seoul, Korea), 18 µl of distilled water, 2.5 µl of oligonucleotide primer (10 pmol/µl), and 2 µl of template containing 20 ng genomic DNA. The PCR products were purified, using a PCR purification kit (Favorgen, Pingtung, Taiwan), and sequenced on an Applied Biosystems 3500 DNA sequencer (Applied Biosystems, Foster City, CA, USA), according to the manufacturers' instructions.
Statistical analysis
Using logistic regression analysis, crude odds ratios (ORs), 95% confidence intervals (CIs), and associations between AD and the SNPs under investigation, were calculated. Associations between polymorphisms (rs529208, rs12484684, and rs3745540) and AD were assessed using Fisher's exact test. To compare total IgE and eosinophil count among three variants in AD+AM patients, a Mann–Whitney U-test was conducted. The significance level was set at p-value <0.05.
RESULTS
Whole-exome sequencing in 40 AD
WES was conducted to identify candidate variants that are processed to AM. The control KPGP group data were compared to AD at the Theragen Etex Bio Institute. KPGP is a reference group, and there is no clinical information for AD (Table 1). To overcome this limitation, we carried out a large-cohort study to validate extracted variants, using Sanger sequencing (Table 2).
After DNA shearing, variant type identification excluding meaningless variants proceeded through the six-stage filtering process (Supplementary Fig. 1). To identify susceptible variants for AD and AM, fast and convenient filters for dominant, recessive, and compound hetero models were used. Several variants were filtered out, and conserved and functional variants remained in filters 5 to 6. All variants were counted after each filtering step (Supplementary Table 1).
We compared raw exome sequencing data with the difference between 40 AD cases and 43 KPGP control group. Considerable numbers of missense SNPs that influence a codon via amino acid substitution were observed in filter 1 (Supplementary Table 1). We focused on common functional variants (MAF greater than 1%) in filter 4, rare variants (MAF lower than 1%) for global population in filter 5 and rare variants (MAF lower than 2%) for Koreans in filter 6. Among these filtered candidate genes, functions related to allergic skin disease were inferred through a literature search. DOCK8, IL17RA, and KLK12 SNPs were identified as missense mutations: c.1289C>A, p.P97T (rs529208); c.1685C>A, p.P562G (rs12484684); and c.457+27>C, rs3745540, respectively. DOCK8 (rs529208) was located in chromosome 9, and IL17RA (rs12484684) was identified in chromosome 22. KLK12 (rs3745540), a splice donor variant that is located in RNA splicing site in intron region, was located in chromosome 19. Low SIFT scores of less than DOCK8 (0.05) and IL17RA (0.03) variants predicted high protein damage via amino acid substitution. The polyphen2 scores close to 1 in the DOCK8 (0.996) and IL17RA (1.0) variants predicted harmful effects on protein structure due to substitution. The PhyloP positive scores of the DOCK8 (3.149) and IL17RA (4.583) variants indicated that two sites were evolutionarily conserved in multiple alignments of 99 vertebrate genomes to the human genome. The phastcons scores of the DOCK8 (0.975) and IL17RA (1) variants showed strong conservation in evolutionary events. KLK12 (rs3745540) is splice donor variant that is located in RNA splicing site and predicted to be fatal impact in filter 2. There are no SIFT and Polyphen2 scores because it is an intron lesion. About 53% to 75% (DOCK8), 11% to 33% (IL17RA), and 53% to 65% (KLK12) MAF were identified in 1,000 global, 286 East Asians, and 800 Koreans, respectively (Table 3).
Table 3. Effects of DOCK8, IL17RA, and KLK12 SNPs and protein function prediction scores.
| Gene | RS# | Chr | POS | AAC* | Type | SIFT† | Polyphen2‡ | PhyloP§ | Phast-Cons∥ | Global¶ | East Asian | Korean** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DOCK8 | rs529208 | chr9 | 286593 | P97T | cSNP | 0.05 | 0.996 | 3.149 | 0.975 | 0.529 | 0.721 | 0.746 |
| IL17RA | rs12484684 | chr22 | 17589794 | P562G | cSNP | 0.03 | 1.0 | 4.583 | 1 | 0.113 | 0.298 | 0.337 |
| KLK12 | rs3745540 | chr19 | 51535130 | - | Splice donor variant | - | 0.937 | 0.78 | 0.534 | 0.637 | 0.655 |
SNP: single-nucleotide polymorphisms, RS: reference SNP, Chr: chromosome, POS: position, AAC: amino acid changes, cSNP: SNP in coding regions, SIFT: scale invariant feature transform, −: not available. *Single-letter codes for amino acids. †Prediction scores for amino acid substitutions that affect protein function (damaging<0.05, tolerance>0.05; scores range from 0 to 1). ‡Prediction of the possible impact of amino acid substitutions (0.957<probably damaging<1, 0.453<possibly damaging<0.956, 0<benign<0.452; scores range from 0 to 1). §Prediction of conserved sites across species; a higher score indicates a more conserved site (values>0). ∥Predicts the possibility that a nucleotide belongs to a conserved element in the phylogenetic tree (value>0.2). ¶Global frequency, variants with minor allele frequencies (MAFs) as low as 1% or with an unknown frequency (value<0.01). **Korean frequency, variants with MAFs as low as 2% or in unknown genes (value>0.02).
Sanger sequencing in 81 control, 87 AD, and 72 AM
A case-control study was performed to determine whether DOCK8 (rs529208), IL17RA (rs12484684), and KLK12 (rs3745540) are candidate risk factors for only AD and AM. Eighty-seven patients aged greater 18 years with adult-onset AD, without an early-onset, and 81 control subjects, 72 atopic march patients were enrolled. A total IgE level of 1,430±1,670 KU/L, and a total eosinophil level of 459.4±377.8 mm3 for AD patients, and 1,833±1,888 KU/L, and 449.4±376.1 mm3 for AM patients were recruited in this study (Table 2).
Sanger sequencing was carried out, and the genotype and allele frequencies of Koreans were determined by direct variant counting using the BioEdit software. The A minor alleles of IL17RA (rs12484684) increased from 26% to 36% in AD compared to the control group. The genotype (GG+AG) of KLK12 (rs3745540) increased from 83% to 93% in AD compared to the control group. The minor allele frequency of DOCK8 (rs529208) showed no significant differences among AD, AM and control groups (Table 4).
Table 4. Allele and genotype frequency of DOCK8, IL17RA, and KLK12 polymorphisms*.
| Gene | SNP | Allele & genotype | Control | AD | AM |
|---|---|---|---|---|---|
| DOCK8 | rs529208 (c.1289C>A) | Allele | |||
| C | 44 (28) | 47 (28) | 38 (27) | ||
| A | 114 (72) | 121 (72) | 102 (73) | ||
| Genotype | |||||
| CC | 3 (4) | 6 (7) | 4 (6) | ||
| CA | 38 (48) | 35 (42) | 30 (43) | ||
| AA | 38 (48) | 43 (51) | 36 (51) | ||
| IL17RA | rs12484684 (c.1685C>A) | Allele | |||
| C | 119 (74) | 110 (64) | 99 (69) | ||
| A | 41 (26) | 62 (36) | 45 (31) | ||
| Genotype | |||||
| CC | 43 (54) | 34 (40) | 32 (44) | ||
| CA | 33 (41) | 42 (49) | 35 (49) | ||
| AA | 4 (5) | 10 (12) | 5 (7) | ||
| KLK12 | rs3745540 (g.51031874A>G) | Allele | |||
| A | 62 (38) | 52 (30) | 57 (40) | ||
| G | 100 (62) | 122 (70) | 87 (60) | ||
| Genotype | |||||
| AA | 14 (17) | 6 (7) | 12 (17) | ||
| AG | 34 (42) | 40 (46) | 33 (46) | ||
| GG | 33 (41) | 41 (47) | 27 (38) |
Values are presented as number (%). SNP: single-nucleotide polymorphisms, AD: atopic dermatitis, AM: atopic march. *81 healthy controls, 87 adult-AD, and 72 AM patients.
ORs with 95% CI were estimated for individual risk alleles. The rs529208 of DOCK8 showed no correlation with AD and AM in any of the allele and genotype (dominant and recessive) models (Table 5).
Table 5. Odds ratios (ORs) and 95% confidence intervals (CIs) for AD associated with DOCK8 polymorphisms in an allele model.
| DOCK8 rs529208 | Allele | Genotype (dominant) | Genotype (recessive) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor allele | Major allele | OR (95% CI) | p-value | Case | Control | OR (95% CI) | p-value | Case | Control | OR (95% CI) | p-value | |
| Con vs. AD | A | C | 0.99 (0.61~1.61) | 0.97 | AA+CA | CC | 0.51 (0.12~2.13) | 0.36 | AA | CA+CC | 1.13 (0.61~2.09) | 0.7532 |
| Con vs. AM | A | C | 1.03 (0.62~1.72) | 0.89 | AA+CA | CC | 0.65 (0.14~3.01) | 0.58 | AA | CA+CC | 1.14 (0.60~2.17) | 0.68 |
| Con vs. AD+AM | A | C | 1.01 (0.66~1.55) | 0.95 | AA+CA | CC | 0.57 (0.15~2.13) | 0.4 | AA | CA+CC | 1.13 (0.66~1.95) | 0.64 |
| AD vs. AM | A | C | 1.04 (0.63~1.72) | 0.87 | AA+CA | CC | 1.04 (0.63~1.72) | 0.87 | AA | CA+CC | 1.00 (0.53~1.90) | 0.98 |
Con: control, AD: atopic dermatitis, AM: atopic march.
The rs12484684 of IL17RA increased risk of adult-onset AD (OR, 1.63; 95% CI, 1.02~2.62; p=0.041) compared to the control for the A allele frequency. The AA+CA genotype was also associated with a trend towards an increased risk of AD with an OR of 1.77 (95% CI, 0.95~3.29; p=0.067; Table 6).
Table 6. Odds ratios (ORs) and 95% confidence intervals (CIs) for AD associated with IL17RA polymorphisms in an allele model.
| IL17RA rs12484684 | Allele | Genotype (dominant) | Genotype (recessive) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor allele | Major allele | OR (95% CI) | p-value | Case | Control | OR (95% CI) | p-value | Case | Control | OR (95% CI) | p-value | |
| Con vs. AD | A | C | 1.63 (1.02~2.62) | 0.041* | AA+CA | CC | 1.77 (0.95~3.29) | 0.067 | AA | CA+CC | 2.5 (0.75~8.31) | 0.135 |
| Con vs. AM | A | C | 1.31 (0.8~2.17) | 0.277 | AA+CA | CC | 1.45 (0.76~2.75) | 0.25 | AA | CA+CC | 1.41 (0.36~5.49) | 0.613 |
| Con vs. AD+AM | A | C | 1.48 (0.97~2.27) | 0.067 | AA+CA | CC | 1.62 (0.94~2.78) | 0.08 | AA | CA+CC | 1.99 (0.64~6.21) | 0.23 |
| AD vs. AM | A | C | 0.80 (0.50~1.29) | 0.369 | AA+CA | CC | 0.81 (0.43~1.54) | 0.32 | AA | CA+CC | 0.57 (0.18~1.74) | 0.32 |
Con: control, AD: atopic dermatitis, AM: atopic march. *Statistically significant (p<0.05).
rs3745540 of KLK12 was associated with AD in dominant model (Table 7). The GG+AG genotype showed a significantly increased risk of AD development (OR, 2.86; 95% CI, 1.04~7.83, p=0.041). However, there was no significant correlation when using a recessive model.
Table 7. Odds ratios (ORs) and 95% confidence intervals (CIs) for AD associated with KLK12 polymorphisms in an allele model.
| KLK12 (rs3745540) | Allele | Genotype (dominant) | Genotype (recessive) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor allele | Major allele | OR (95% CI) | p-value | Case | Control | OR (95% CI) | p-value | Case | Control | OR (95% CI) | p-value | |
| Con vs. AD | G | A | 1.45 (0.92~2.28) | 0.1 | GG+AG | AA | 2.86 (1.04~7.83) | 0.041* | GG | AG+AA | 1.32 (0.72~2.44) | 0.36 |
| Con vs. AM | G | A | 0.95 (0.6~1.5) | 0.81 | GG+AG | AA | 1.04 (0.45~2.43) | 0.92 | GG | AG+AA | 0.87 (0.46~1.67) | 0.682 |
| Con vs. AD+AM | G | A | 1.2 (0.81~1.77) | 0.36 | GG+AG | AA | 1.64 (0.77~3.51) | 0.195 | GG | AG+AA | 1.10 (0.64~1.90) | 0.723 |
| AD vs. AM | G | A | 0.64 (0.40~1.02) | 0.06 | GG+AG | AA | 0.36 (0.13~1.03) | 0.056 | GG | AG+AA | 0.66 (0.34~1.24) | 0.194 |
Con: control, AD: atopic dermatitis, AM: atopic march.
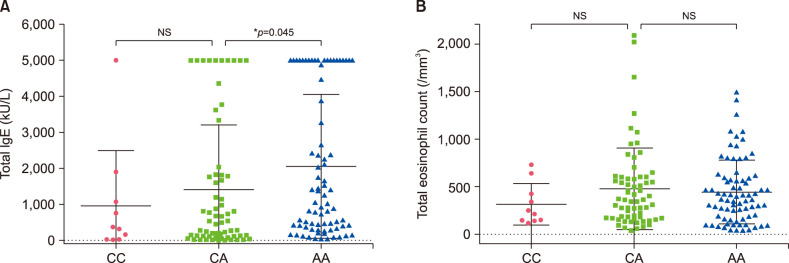
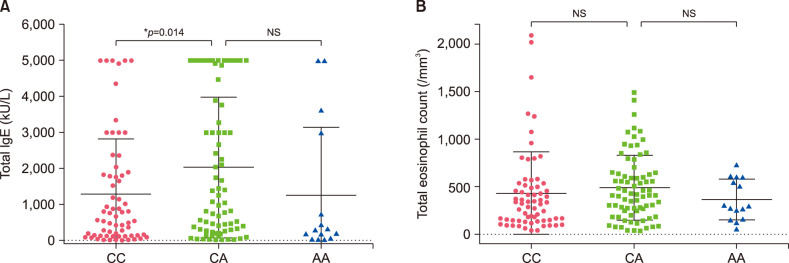
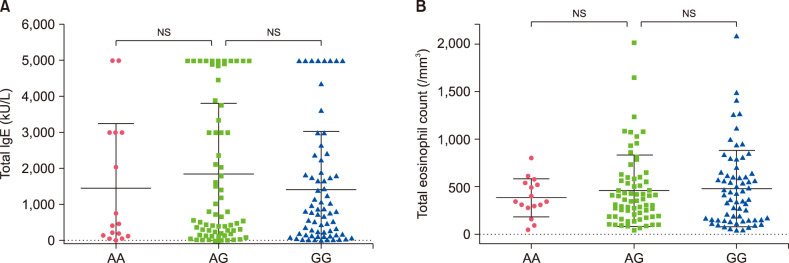
The relationship between the genotype of three SNPs (rs529208, rs12484684, rs3745540) and the clinical characteristics (total IgE level and eosinophil count) of AD+AM patients has been compared. Total IgE level was significantly increased in the AA genotype of DOCK8 compared to the CA genotype (p=0.045). There was no association between genotypes and eosinophil count (Fig. 1). AD and AM Patients with the IL17RA CA genotype also had elevated IgE levels (Fig. 2) and were not associated with the total eosinophil count. The KLK12 variant was not related to the clinical characteristics of AD (Fig. 3).
Fig. 1. The relationship of the DOCK8 polymorphism with total immunoglobulin E (IgE) and eosinophil counts in atopic dermatitis+atopic march patients. Dot-plot graphs showing total IgE levels (A) and eosinophil counts (B) for the CC, CA, and AA alleles. Values are presented as mean±standard deviation. *p<0.05 by Mann–Whitney U-test for statistical significance. NS: not significant.
Fig. 2. The relationship of the IL17RA polymorphism with total immunoglobulin E (IgE) and eosinophil counts in atopic dermatitis+atopic march patients. Dot-plot graphs showing total IgE levels (A) and eosinophil counts (B) for the CC, CA, and AA alleles. Values are presented as mean±standard deviation. *p<0.05 by Mann–Whitney U-test for statistical significance. NS: not significant.
Fig. 3. The relationship of the KLK12 polymorphism with total immunoglobulin E (IgE) and eosinophil counts in atopic dermatitis+atopic march patients. Dot-plot graphs showing total IgE levels (A) and eosinophil counts (B) for the AA, AG, and GG alleles. Values are presented as mean±standard deviation. *p<0.05 by Mann–Whitney U-test for statistical significance. NS: not significant.
DISCUSSION
FLG protein has an essential role in normal skin barrier function and epidermal homeostasis. Loss-of-function genetic mutation and copy-number variant in the FLG gene disrupts the integrity of the epidermal barrier that helps the development of AD. FLG mutation is a higher risk factor for AD. There are several FLG genetic studies in various ethnic groups7. Our target was to find other genetic mutations that can affect skin barrier dysfunction and the immune system.
Several susceptibility loci have been identified in GWAS19, so finding a novel genetic variant for AD using exome sequencing is essential. AD is a common and complex disease involving environmental factors, thus, finding a rare variant that directly relates to disease is difficult. The common disease-common variant (CD-CV) hypothesis in the field of genetics was the first dominant theory. We focused on the common variants and genes related to skin immunity through a literature search.
To detect candidate variants associated with AD and AM, allele frequency among groups and protein prediction scores were used through six whole-exome sequencing filtering steps before the population-based study. Functional sequences can be predicted by confirming evolutionary conserved elements across species20. Our WES data suggest that rs529208 and rs12484684 are strongly conserved protein coding regions among 100 vertebrate species.
The DOCK8 gene is a dedicator of cytokinesis 8 located in 9p24.3. The protein is an activator of small G protein involved in intracellular signaling networks. It was reported that DOCK8 deficiency associated with hyper-IgE syndrome impairs T cell activation21,22. Increased Th2 and low Th1 signals, along with increased IgE levels, are found as early as in the cord blood of newborns with AD23,24. Recent studies suggest an association between atopic phenotypes and serum IgE levels. Children with severe AD and high IgE levels are at risk for sensitization to food allergens and aeroallergens25. Our study shows that a high IgE level was induced at the AA homo genotype of rs529208 compared to the CA hetero genotype in AD+AM patients. This represents the possibility of causing hyper-IgE in patients with the rs529208 genotype (AA) of DOCK8.
The IL17RA gene is an interleukin-17 receptor A located in 22q11.1 and a key regulator of type 2 inflammatory responses, and it protects the skin barrier against allergic inflammation. IL17RA-deficiency mice have a defective skin barrier with altered FLG and increased TSLP expression aberrant skin inflammation. IL17RA-deficiency mice crossed with FLG mutation mice induce severe skin inflammation26. Our data show that total IgE level was further increased in the CA hetero genotype than in the CC homo genotype IL17RA (rs12484684). Thus, this variant may affect exacerbation of AD. A more functional genetic study is necessary to validate its influence.
KLK12 is a kallikrein-related peptidase 12 protein located in 19q13.4 and a subgroup of serine proteases. Co-expression of various KLKs and SPINK5 messenger RNA is suspected to affect serine protease activities in the stratum corneum27. KLK12 messenger RNAs in both normal human epidermal keratinocyte cells and skin tissue were not amplified by RT-PCR. The functional role of the KLK12 gene is limited in AD. However, it might be worth using variation as a tool for early-onset diagnosis of AD. Eosinophil counts of all three variants were not associated with AD and AM. Our data was limited as there was no clinical data for the 43 KPGP control group individuals. To overcome this, we performed Sanger sequencing in a large cohort to validate our variants, which were filtered by WES.
To overcome small sample size, the frequency of DOCK8, IL17RA, and KLK12 polymorphisms in Sanger sequencing was also compared with the frequency of 1,000 global, 286 East Asian, and 800 Korean subjects without clinical information. The 72% to 74% MAF of DOCK8 (rs529208) in the 286 East Asian and the 800 Koreans surveyed almost identical that of the 82 health controls (72% MAF) in Sanger sequencing. Other variants (IL17RA and KLK12) also nearly matched as well (Supplementary Table 2). These data demonstrate the credibility of the data despite the small study size.
Taken together, three novel AD candidate variants, DOCK8 (rs529208), IL17RA (rs12484684), and KLK12 (rs3745540), were identified using a new WES filtering method. The high PolyPhen and evolutionary conservation PhyloP scores of these two SNPs predict deleterious protein function. Although the relationship with AM was not discovered in this study, our case-control study suggests that polymorphism of DOCK8 and IL17RA might be related to increase the total IgE level. The A minor allele of IL17RA allele and the (GG+AA) KLK12 genotype seem to enhance the risk of AD. In conclusion, three novel AD candidate variants, DOCK8 (rs529208), IL17RA (rs12484684), and KLK12 (rs3745540), were identified using a new WES filtering method. Although the relationship with AM was not discovered in this study, the result suggests that polymorphism of DOCK8 and IL17RA might be related to increase the total IgE level.
ACKNOWLEDGMENT
This study was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2019R1A2C1090226). This research was supported by the Chung-Ang University Research Scholarship Grants in 2018.
This study was supported by a grant from the Korean Healthcare Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI14C2687).
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
SUPPLEMENTARY MATERIALS
Supplementary data can be found via http://anndermatol.org/src/sm/ad-32-197-s001.pdf.
Flow chart of the process for filtering variants.
The filtered WES variants for 40 AD cases vs. 43 KPGP control individuals
Minor allele frequency of DOCK8, IL17RA, and KLK12 polymorphisms in 1000 genome project and Sanger sequencing data
References
- 1.Bantz SK, Zhu Z, Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014;5:202. doi: 10.4172/2155-9899.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomsen SF. Atopic dermatitis: natural history, diagnosis, and treatment. ISRN Allergy. 2014;2014:354250. doi: 10.1155/2014/354250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng W, Novak N. Recent developments in atopic dermatitis. Curr Opin Allergy Clin Immunol. 2014;14:417–422. doi: 10.1097/ACI.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 4.Pyun BY. Natural history and risk factors of atopic dermatitis in children. Allergy Asthma Immunol Res. 2015;7:101–105. doi: 10.4168/aair.2015.7.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaniboni MC, Samorano LP, Orfali RL, Aoki V. Skin barrier in atopic dermatitis: beyond filaggrin. An Bras Dermatol. 2016;91:472–478. doi: 10.1590/abd1806-4841.20164412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uehara M, Kimura C. Descendant family history of atopic dermatitis. Acta Derm Venereol. 1993;73:62–63. doi: 10.2340/00015555736263. [DOI] [PubMed] [Google Scholar]
- 7.Li K, Oh WJ, Park KY, Kim KH, Seo SJ. FLG mutations in the East Asian atopic dermatitis patients: genetic and clinical implication. Exp Dermatol. 2016;25:816–818. doi: 10.1111/exd.13063. [DOI] [PubMed] [Google Scholar]
- 8.Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 9.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59:5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- 10.Carson AR, Smith EN, Matsui H, Brækkan SK, Jepsen K, Hansen JB, et al. Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinformatics. 2014;15:125. doi: 10.1186/1471-2105-15-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heo WI, Park KY, Lee MK, Kim JH, Moon NJ, Seo SJ. Association of CDKAL1 polymorphisms with early-onset atopic dermatitis in Koreans. Ann Dermatol. 2018;30:276–283. doi: 10.5021/ad.2018.30.3.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas PD, Kejariwal A. Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci U S A. 2004;101:15398–15403. doi: 10.1073/pnas.0404380101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams HC, Burney PG, Hay RJ, Archer CB, Shipley MJ, Hunter JJ, et al. The U.K. working party's diagnostic criteria for atopic dermatitis. I. Derivation of a minimum set of discriminators for atopic dermatitis. Br J Dermatol. 1994;131:383–396. doi: 10.1111/j.1365-2133.1994.tb08530.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Meehan J, Su Z, Ng HW, Shu M, Luo H, et al. Whole genome sequencing of 35 individuals provides insights into the genetic architecture of Korean population. BMC Bioinformatics. 2014;15 Suppl 11:S6. doi: 10.1186/1471-2105-15-S11-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garmhausen D, Hagemann T, Bieber T, Dimitriou I, Fimmers R, Diepgen T, et al. Characterization of different courses of atopic dermatitis in adolescent and adult patients. Allergy. 2013;68:498–506. doi: 10.1111/all.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marenholz I, Esparza-Gordillo J, Rüschendorf F, Bauerfeind A, Strachan DP, Spycher BD, et al. Meta-analysis identifies seven susceptibility loci involved in the atopic march. Nat Commun. 2015;6:8804. doi: 10.1038/ncomms9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boos AC, Hagl B, Schlesinger A, Halm BE, Ballenberger N, Pinarci M, et al. Atopic dermatitis, STAT3- and DOCK8-hyper-IgE syndromes differ in IgE-based sensitization pattern. Allergy. 2014;69:943–953. doi: 10.1111/all.12416. [DOI] [PubMed] [Google Scholar]
- 22.Engelhardt KR, McGhee S, Winkler S, Sassi A, Woellner C, Lopez-Herrera G, et al. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124:1289–1302.e4. doi: 10.1016/j.jaci.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scirica CV, Gold DR, Ryan L, Abulkerim H, Celedón JC, Platts-Mills TA, et al. Predictors of cord blood IgE levels in children at risk for asthma and atopy. J Allergy Clin Immunol. 2007;119:81–88. doi: 10.1016/j.jaci.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Herberth G, Heinrich J, Röder S, Figl A, Weiss M, Diez U, et al. Reduced IFN-gamma- and enhanced IL-4-producing CD4+ cord blood T cells are associated with a higher risk for atopic dermatitis during the first 2 yr of life. Pediatr Allergy Immunol. 2010;21(1 Pt 1):5–13. doi: 10.1111/j.1399-3038.2009.00890.x. [DOI] [PubMed] [Google Scholar]
- 25.Laske N, Niggemann B. Does the severity of atopic dermatitis correlate with serum IgE levels? Pediatr Allergy Immunol. 2004;15:86–88. doi: 10.1046/j.0905-6157.2003.00106.x. [DOI] [PubMed] [Google Scholar]
- 26.Floudas A, Saunders SP, Moran T, Schwartz C, Hams E, Fitzgerald DC, et al. IL-17 receptor A maintains and protects the skin barrier to prevent allergic skin inflammation. J Immunol. 2017;199:707–717. doi: 10.4049/jimmunol.1602185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu N, Takata M, Otsuki N, Toyama T, Ohka R, Takehara K, et al. Expression and localization of tissue kallikrein mRNAs in human epidermis and appendages. J Invest Dermatol. 2003;121:542–549. doi: 10.1046/j.1523-1747.2003.12363.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow chart of the process for filtering variants.
The filtered WES variants for 40 AD cases vs. 43 KPGP control individuals
Minor allele frequency of DOCK8, IL17RA, and KLK12 polymorphisms in 1000 genome project and Sanger sequencing data