eTOC Blurb:
Reese et al. profile the gut microbiota of wild chimpanzees over the life course. High microbial diversity among infants <2 years old contrasts sharply with the low diversity observed in human infants. This divergence could relate to interspecific differences in lactation, diet and immunity, and may contribute to lifelong differences in physiology.
SUMMARY
Survival in primates is facilitated by commensal gut microbes that ferment otherwise indigestible plant matter, resist colonization by pathogens, and train the developing immune system [1, 2]. However, humans are unique among primates in that we consume highly digestible foods, wean early, mature slowly, and exhibit high lifelong investments in maintenance [3–6]. These adaptations suggest that lifetime trajectories of human-microbial relationships could differ from those of our closest living relatives. Here, we profile the gut microbiota of 166 wild chimpanzees aged 8 months to 67 years in the Kibale National Park, Uganda, and compare the patterns of gut microbial maturation to those previously observed in humans. We found that chimpanzee gut microbial alpha-diversity, composition, interindividual variation, and change over time varied significantly with age. Notably, gut microbial signatures in infants <2 years old were distinct across all four metrics. Infant chimpanzee guts were enriched in some of the same taxa prevalent in infant humans (e.g., Bifidobacterium, Streptococcus, Bacteroides), and, like humans, chimpanzee gut microbial communities exhibited higher interindividual variation in infancy versus later in life. However, in direct contrast to human infants, chimpanzee infants harbored surprisingly high-diversity rather than low-diversity gut bacterial communities compared with older conspecifics. These data indicate differential trajectories of gut microbiota development in humans and chimpanzees that are consistent with interspecific differences in lactation, diet, and immune function. Probing the phenotypic consequences of differential early life gut microbial diversity in chimpanzees and other primates will illuminate the life history impacts of the hominid-microbiome partnership.
RESULTS
We collected 618 fresh fecal samples over three years from 166 known individuals in two groups of wild chimpanzees (Kanyawara and Ngogo) in the Kibale National Park, Uganda. We assessed changes in gut microbial community composition over the chimpanzee life course, focusing on patterns of alpha-diversity (i.e. the community of bacterial taxa within a given sample) and beta-diversity (i.e. differences in gut microbial community membership between individuals or between timepoints for a given individual). We grouped chimpanzees into 6 age classes (young infant: <2 years old [yo], late infant: 2–5 yo, juvenile: >5–10 yo, adolescent: >10–15 yo, adult: >15–35 yo, post-prime adult: >35 yo) that roughly correspond to behavioral and reproductive milestones [7]. Our dataset compares advantageously to previous studies [8–12] in its larger sample, longitudinal tracking of known individuals over multiple years, and sampling over the full range of the chimpanzee lifespan, including nine individuals under 2 years old at Kanyawara. By using three years of field collections, we could assess variation among age groups and variability within individuals over time. Because gut microbial maturation in early life has lifelong consequences for aspects of ecology and life history that differ between humans and chimpanzees, we paid special attention to early life transitions.
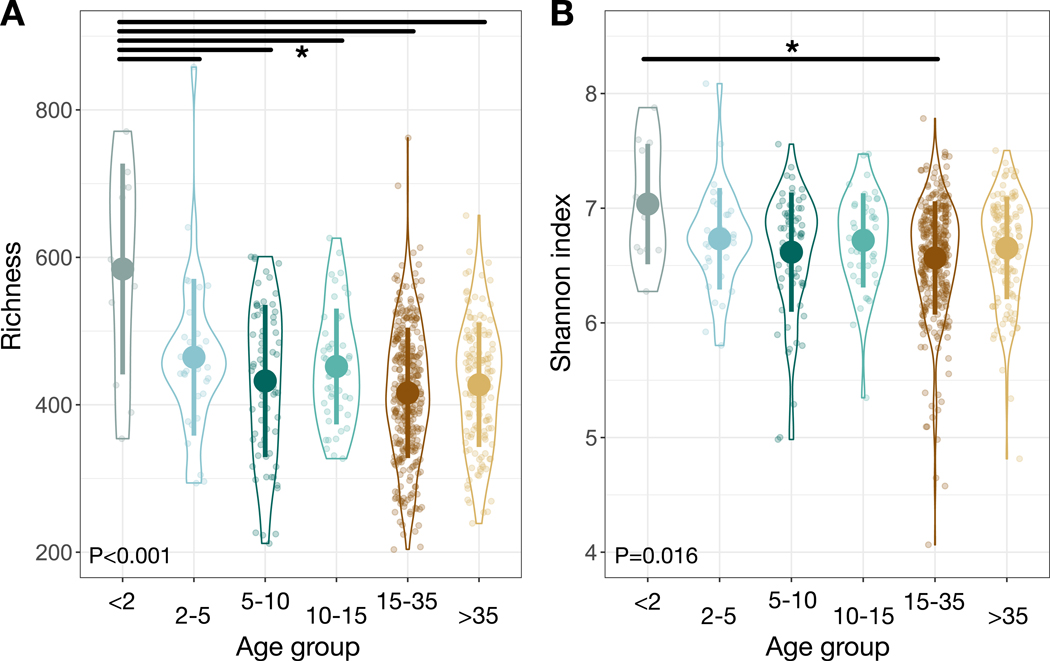
Gut microbial communities of young infants (<2 yo) were distinct compared to those of other age groups. Within-sample alpha-diversity of microbial amplicon sequence variants (ASVs) varied among age groups, whether measured by observed operational taxonomic unit richness or Shannon index (P<0.001 and P=0.016 respectively, likelihood tests of linear mixed effects models; Figure 1), with the highest average values (776.0±44.5; 4.7±0.2) observed in young infants. Post-hoc tests confirmed that young infants had significantly higher observed operational taxonomic unit richness and Shannon diversity values than other age groups (P<0.05, contrast tests for estimated marginal means of linear mixed effects model; Data S1A).
Figure 1: Gut microbial community alpha-diversity varies among chimpanzee age groups.
(A) Observed OTU richness of individual chimpanzees as a factor of age group. (B) Shannon index of individual chimpanzees as a factor of age group. Large circles are means; bars show standard deviations. P values reported for linear mixed effects model likelihood tests. * indicates P<0.05 contrast for estimated marginal means of linear mixed effects model. See also Data S1 and Table S2.
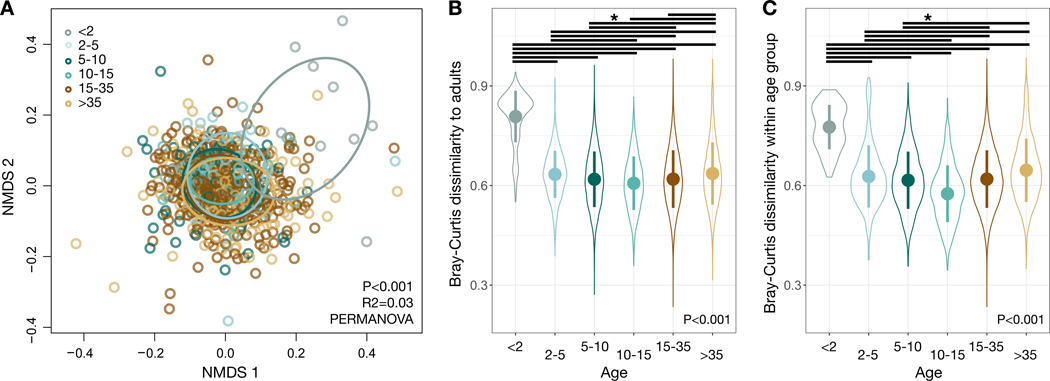
Age group had the largest explanatory power of the non-temporal factors considered as possible drivers of divergence in gut microbial communities (P<0.001, R2=0.030, PERMANOVA). Considering only age group effects, we found that there were significant differences between all but two pair-wise age group comparisons (P<0.05, pairwise PERMANOVA test; Data S1B). Significant variation could also be ascribed to group membership (P<0.001, R2=0.021) and sex (P<0.001, R2=0.016), as well as month or year of sampling (P<0.001, R2=0.054 and R2=0.053 respectively; Figure S1), as previously observed in chimpanzees at Gombe National Park, Tanzania [11]. A NMDS ordination highlights the unique nature of young infant microbiota, in that the youngest chimpanzees were all consistent outliers (Figure 2A). Bray-Curtis distances between the gut microbial communities of adults and other individuals varied by age group (P<0.001, bootstrapped Kruskal-Wallis test; Figure 2B, Data S1C), with young infants being most dissimilar to adults (P<0.001, contrast tests for estimated marginal means of linear mixed effects models; Data S1D). Gut microbiota distances between individuals of the same age group also varied across age groups (P<0.001, bootstrapped Kruskal-Wallis test; Figure 2C), with consistently higher interindividual variation among young infants (P<0.001, contrast tests for estimated marginal means of linear mixed effects model; Data S1E).
Figure 2: Gut microbial community composition varies among chimpanzee age groups.
(A) Nonmetric multidimensional scaling (NMDS) ordination plot illustrates differences in gut microbial community composition, based on Bray-Curtis dissimilarities as a factor of age group. Ellipses illustrate standard deviation for age groups. (B) Bray-Curtis dissimilarities between individual chimpanzees and each adult chimpanzee as a factor of age group. (C) Bray-Curtis dissimilarities between individual chimpanzees and other members of their age group, as a factor of age group. Large circles are means; bars show standard deviations. P values reported for (A) PERMANOVA and (B,C) bootstrapped Kruskal-Wallis. * indicates P<0.05 contrast for estimated marginal means of linear mixed effects model. See also Figures S1-S3, Data S1 and Table S2.
Surprisingly, given evidence of human mother to infant vertical transmission of microbes [13, 14] and expected dilutions of this signature over time [15], the gut microbial communities of young infants were not more similar to the microbiotas of their mothers than were the gut microbial communities of offspring in older age classes (Figure S2A). In fact, young infants were significantly more dissimilar from their mothers than were offspring in other age groups (P=0.016, likelihood tests of linear mixed effects models), a result that reinforces the distinctiveness of the infant chimpanzee gut microbiota. Nevertheless, we did find that the gut microbiota of offspring were more similar to those of their mothers than to those of randomly selected females of reproductive age (P=0.016, likelihood tests of linear mixed effects models; Figure S2B). Despite higher gut microbial similarity in mother-offspring pairs, relatedness, where it could be assigned by pedigree, was not significantly associated with microbial community dissimilarity generally when compared among pairs of chimpanzees with known levels of genetic relatedness (P=0.153, bootstrapped Kruskal-Wallis test; Figure S3A, Data S1C). In contrast, chimpanzee group membership and sex did predict levels of microbial dissimilarity (P<0.001, bootstrapped Kruskal-Wallis tests; Figure S3B, C, Data S1C).
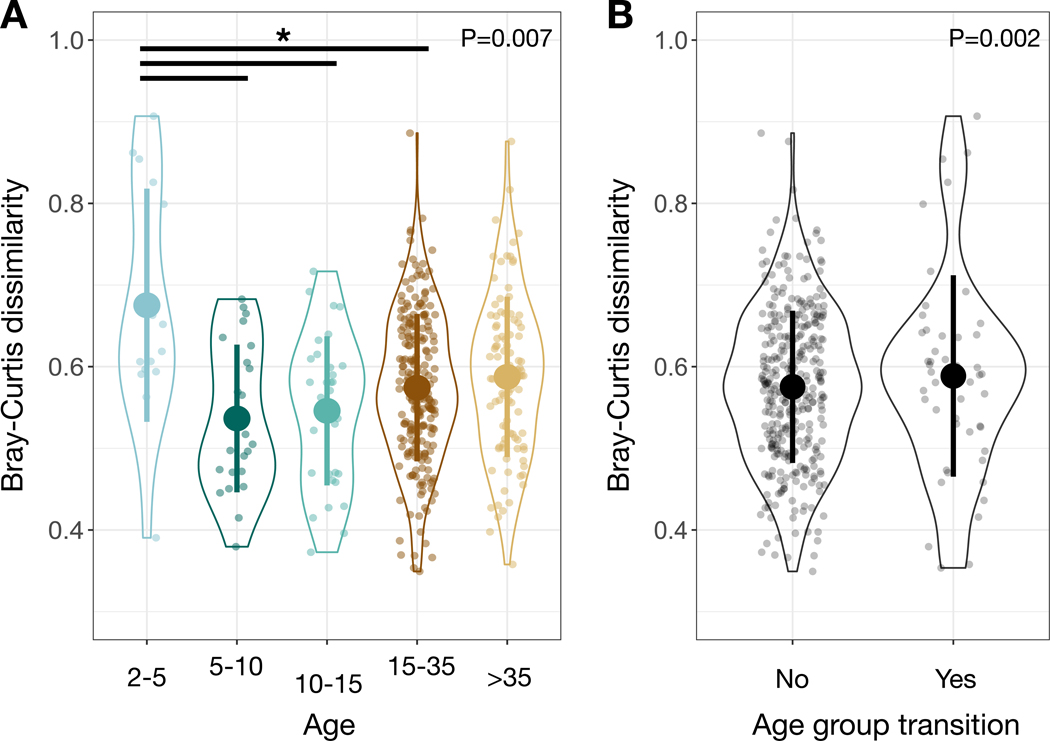
To understand further how aging impacted the gut microbiota, we analyzed a subsample of the full dataset that included only repeat measurements of individuals with greater than one year between sampling dates (N=106 samples from 87 individuals sampled during 2015–2018). Overall, we did not find a significant correlation between the time elapsed between samplings and the level of compositional dissimilarity (P=0.788, Spearman correlations). However, we did find significant variation among age groups in the extent to which individual microbiotas changed over time, with infants showing the greatest change in microbiota with age (P=0.006, likelihood tests of linear mixed effects models; Figure 3A). Post-hoc tests demonstrate that infants changed more with age than did all other age groups except post-prime adults (P<0.05, contrast tests for estimated marginal means of linear mixed effects model; Data S1F. Moreover, individuals who underwent an age transition (e.g., went from the young infant to the late infant group or from adult to post-prime adult) exhibited significantly more change per unit time than individuals who stayed within the same age group (P=0.002, likelihood test of linear mixed effects models; Figure 3B), suggesting increased stability in the gut microbiota within developmental stages versus between them.
Figure 3: Change over time in an individual’s gut microbial community composition depends on age and transition between age groups.
(A) Bray-Curtis dissimilarities for an individual chimpanzee compared to itself over time (with greater than one year between samples) as a factor of age group. (B) Bray-Curtis dissimilarities for an individual chimpanzee compared to itself over time (with greater than one year between samples) as a factor of whether the elapsed time led to them transitioning between two age groups. Large circles are means; bars show standard deviations. P values reported for linear mixed effects model likelihood tests. * indicates P<0.05 contrast for estimated marginal means of linear mixed effects model. See also Data S1 and Table S2.
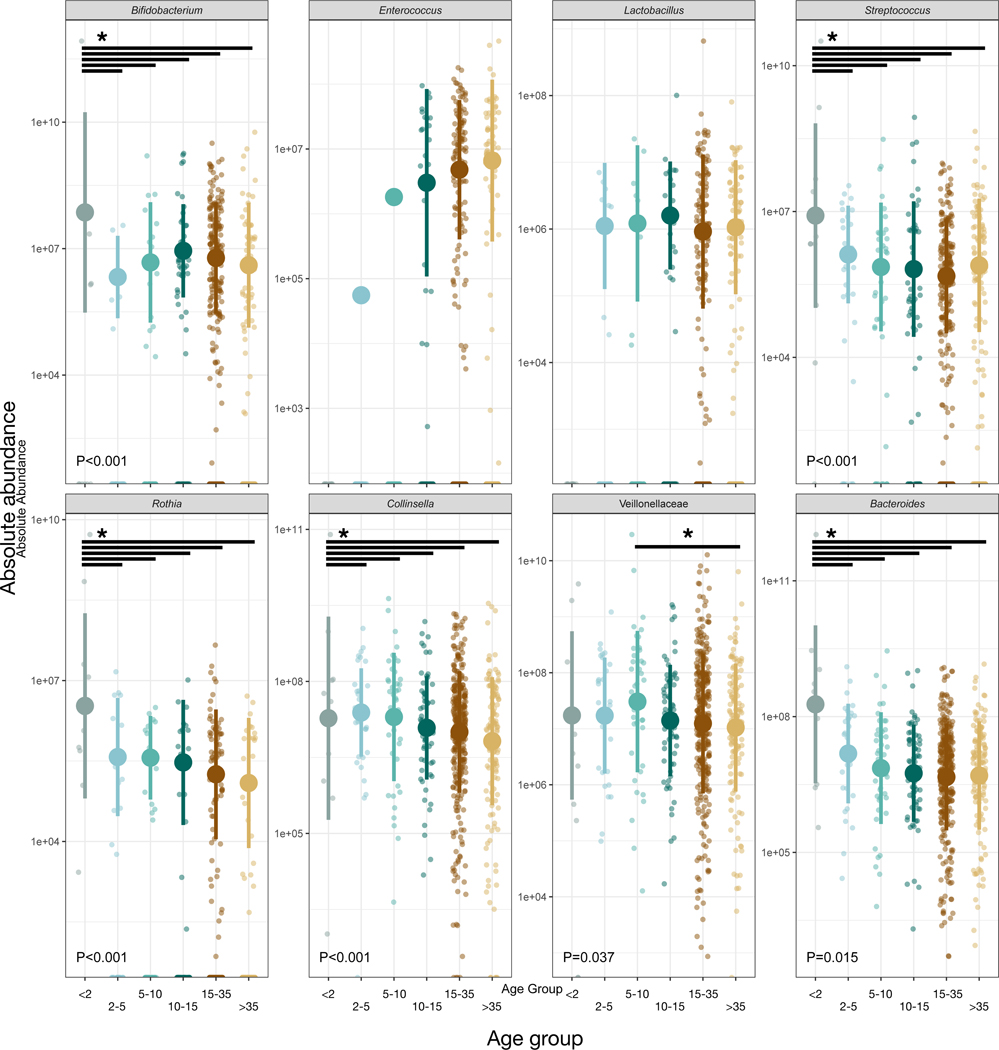
We used all the samples in an indicator species analysis to identify microbial ASVs that distinguished among age groups through significant enrichment. Less than 6% of taxa were uniquely associated with a single age group, but of the 899 taxa that were, 659 (73%) were most abundant in young infants, while another 161 (18%) were most abundant in infants ages 2–5 (Table S1). Thus, the vast majority (91%) of all microbial taxa assessed to be useful for distinguishing among chimpanzee age groups were harbored by the youngest individuals. We also tested whether the absolute abundances of bacterial taxa associated with human infants [16] were similarly elevated in young chimpanzees. We found that the abundances of Bifidobacterium, Streptococcus, Rothia, Collinsella, Veillonellaceae, and Bacteroides varied with age group (P<0.05, likelihood test linear mixed effects models; Figure 4). Of these taxa, all except Veillonellaceae reached their highest abundances in young infants (P<0.05, contrast tests for estimated marginal means of linear mixed effects models; Data S1G). Young infants also had higher overall microbial density as estimated by universal qPCR targeting the 16S rRNA gene (P<0.001, contrast tests for estimated marginal means of linear mixed effects model; Data S1H.
Figure 4: Several bacterial taxa associated with human infants (as per Ref. 16) were also enriched in chimpanzee infants.
Relative abundance of bacterial taxa by chimpanzee age group. Large circles are means; bars show standard deviations. P values reported for linear mixed effects model likelihood tests. * indicates P<0.05 contrast for estimated marginal means of linear mixed effects model. See also Data S1 and Tables S1, S2.
DISCUSSION
Human infants harbor gut microbial communities distinct from those of adults, notably because of their high interindividual variation, low alpha-diversity, and dominance by a few stereotypical taxa [16]. With the introduction of solid foods and cessation of breastfeeding, these microbial communities transition to a more diverse and speciose state, generally with reduced interindividual variation [16–20]. The gut microbiota retains this pattern until late in life, when interindividual variation rises, alpha-diversity drops, and opportunistic pathogens gain stronger footholds. These age-related changes have been attributed to declining immune function and have been documented in diverse human populations [20, 21], although patterns may be blunted in populations with especially high rates of microbial dispersal between individuals [22].
Whether this pattern of change over the life course is unique to humans has remained unclear. Although previous research comparing human and non-human primate gut microbiotas has revealed differences that parallel phylogeny [8] and ecology [10, 23], the studies did not sample the youngest individuals, precluding a test of how these differences originate and develop. For instance, a previous study of wild chimpanzees (Pan troglodytes schweinfurthii) in Tanzania found higher gut microbial alpha-diversity in young than in older individuals [11], but the “younger” sample included only three individuals less than 7 years old, and none less than 3 years old, the age at which the gut microbiota in humans assumes its adult-like patterning [20]. Another study that included infants under 3 years old did not directly examine age effects on diversity [12].
To address this gap, we investigated patterns of gut microbial maturation in wild chimpanzees and compared them to patterns previously observed in humans. Analyzing more than three years of samples from two groups of chimpanzees ranging in age from 8 months to 67 years, we found that chimpanzee gut microbiota composition and diversity varied with age in biologically meaningful ways, with microbial shifts occurring during age group transitions, and the largest shifts seen in young individuals as they underwent weaning.
The infant gut microbiota of Kibale chimpanzees shared certain features with that of the human infant gut microbiota. The high interindividual variation we document among infant chimpanzee gut microbial communities relative to older conspecifics is comparable to that among infant humans [20]. In addition, chimpanzee and human infant gut microbial communities are enriched in many of the same bacterial genera, including those known to degrade milk oligosaccharides (e.g., Bifidobacterium and Bacteroides) [24]. However, in direct contrast to the pattern that has typically been documented in humans [20, 21], the gut microbial communities of infant chimpanzees in our sample exhibited higher rather than lower alpha-diversity compared to those of older conspecifics. Indeed, gut microbial alpha-diversity in the youngest chimpanzees (<2 yo) exceeded that of adults, with alpha-diversity rapidly declining as young chimpanzees aged.
Higher gut microbial diversity in infant chimpanzees compared to older conspecifics could arise from age-specific variation in exogenous exposures, but likely also reflects incomplete immune development. Among infants still consuming milk, oligosaccharides can directly protect against pathogen colonization [25, 26] and microbial metabolites derived from these sugars train the infant immune system [27]. Such effects appear to have been sufficiently important to have allowed for the evolution of investment in oligosaccharide production during lactation, the most energetically demanding period of female life [28]. Nevertheless, infants generally have less mature immune systems than do adults [29–31] and thus may be more permissive of microbial colonization.
However, the high diversity observed in infant chimpanzees compared with older conspecifics raises questions about why humans exhibit comparatively low microbial diversity in early life. One proposed constraint on gut microbial diversity in human infants is the low efficiency of vertical transmission of the gut microbiota from mother to offspring [32–34] (although there is certainly evidence of vertical transmission occurring [e.g., 13, 14]). In theory, relatively high gut microbiota diversity in young chimpanzees compared with older conspecifics could arise from more efficient vertical transmission, but this appears not to be the case. Contrary to the expected direction of effects, previous studies have not yielded detectable signatures of vertical transmission in chimpanzees [11, 12] and we report here that mother-young infant pairs did not harbor especially similar gut microbial communities. Moreover, gut microbiota diversity was higher in young infant chimpanzees than in adults, suggesting that vertical transmission alone cannot directly account for early-life diversity. Alternatively, vertical transmission might indirectly impose constraints on gut microbiota diversity if mothers are preferentially transferring putatively beneficial taxa that are able to dominate available niche space in the infant gut. However, this hypothesis has not yet been tested in either host species.
Our findings do lend support to the idea that changes in bacterial diversity may be linked to lactation. We observed that the largest age-related shifts in gut microbial alpha- and beta-diversity correspond to typical ages at weaning [35, 36] and that some of the indicator taxa in the microbiota of young infants are oligosaccharide specialists. However, the mechanistic link between lactation and diversity remains unclear.
One possibility is that there is a direct effect of milk composition on diversity. The low diversity of the human infant gut microbiota is often attributed to the highly selective prebiotic effects of human milk oligosaccharides [37–39]. Milk oligosaccharides cannot be digested by infant enzymes, and instead pass into the colon, where they are metabolized by a select number of bacterial taxa with important roles in immune development [38–40]. Great ape milk is thought to resemble human milk in terms of macronutrient content [41], but human milk contains unique oligosaccharide structures that may exert stronger overall constraints on the developing gut microbiome [40]. The reducing end of milk oligosaccharides carries a lactose molecule that can be elongated by type I units of lacto-N-biose (Galβ1–3GlcNAc) or type II units of N-acetyllactosamine (Galβ1–4GlcNAc). Human milk predominantly contains oligosaccharides with the type I unit, whereas the milks from chimpanzees, bonobos, gorillas, orangutans – and indeed all other non-human mammals tested – predominantly or exclusively bear the type II unit [40, 42]. Type I oligosaccharides appear to be more efficiently metabolized by the human gut microbiota, with a lower fraction of type I versus type II oligosaccharides excreted in feces [42, 43]. In addition, type I but not type II oligosaccharides efficiently promote the growth of Bifidobacterium [42, 44] a dominant genus in the guts of both human infants [16] and our chimpanzee infants. These observations suggest that human milk could wield more selective power than does chimpanzee milk over gut microbiota composition, although this hypothesis remains to be tested.
An alternative, but not mutually exclusive, possibility is that diversity is regulated not by milk but instead by the diets onto which infant humans and chimpanzees are weaned. Weaning is not an event, but rather a process in which increasing quantities of supplemental foods are introduced over time. Humans and chimpanzees both begin to incorporate supplemental foods into the diet before 6 months of age [7, 35], but these foods differ profoundly in content and form. Mirroring differences in the adult diets of humans and chimpanzees [45], supplemental foods given to human infants are comparatively lower in fiber, richer in fat, and highly processed by cooking and/or non-thermal techniques that soften foods and reduce particle size [46]. Together, these features suggest two mechanisms that may contribute to constraining early life gut microbial diversity in humans in a manner not applicable in chimpanzees.
First, low fiber content, high fat content, and extensive processing/cooking render human supplemental foods easily digested in the small intestine [46]. Increasing the fraction of nutrients digested in the small intestine reduces the fraction entering the large intestine, where the majority of the gut microbiota resides [47]. Indeed, by altering the nutritional milieu of the large intestine, food selection [48, 49] and cooking [50] have both been shown to exert effects on gut microbial community structure and function. Importantly, higher small intestinal digestibility suggests that human supplemental foods may wield less power than chimpanzee supplemental foods to shape the distal gut microbiota. Thus, especially under conditions of high oligosaccharide-mediated constraint, gut microbial diversity in human infants may not be substantially enriched by the incorporation of supplemental foods. This hypothesis is consistent with data showing that the human infant gut microbiome retains an infant-like signature until the cessation of breastfeeding, as opposed to the first incorporation of supplemental foods [16], although parallel data are not available for chimpanzees. Second, the widespread human practice of cooking supplemental foods is expected to minimize colonization by foodborne microbes, including pathogens [51, 52], thus limiting exogenous contributions to early life microbiota diversity.
Ultimately, humans and chimpanzees differ anatomically, physiologically, ecologically, and behaviorally in ways that can be expected to affect endogenous and exogenous regulation of the gut microbiota [53]. Addressing these factors and conducting comparative studies across different populations of each species will be required to understand the roots of human-chimpanzee divergence in early life gut microbial diversity. We expect studies that bridge the transition to supplemental foods and capture variation in weaning practices and environmental pathogen loads to be especially instructive. The question also remains as to which pattern is derived. Resolving this question will require demographic studies of gut microbial development in closely related primates. If bonobos (Pan paniscus) and outgroup species such as gorillas (Gorilla spp.) or orangutans (Pongo spp.) exhibit high infant gut microbial diversity akin to that seen in chimpanzees, parsimony would suggest the human pattern is derived. Comparisons to baboons (Papio spp.) could also be informative as, amongst primates analyzed to date, baboons have been shown to harbor gut microbiota most similar to humans, likely due to their ecological similarities [9, 10].
Taken together, our data suggest that differences between the human gut microbiota and the chimpanzee gut microbiota [8, 54] are present early in life and show differential trajectories during maturation. Microbial influences in early life, in particular, have lifelong consequences for aspects of host biology as fundamental as metabolism [55], immunity [56] and behavior [57]. Therefore, future efforts to understand early life differences among humans and closely related species like chimpanzees hold substantial promise for advancing our understanding of comparative human biology and health, as well as the evolutionary underpinnings of the host-microbial partnership.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Additional information and requests for resources should be directed to the Lead Contact, Rachel Carmody (carmody@fas.harvard.edu).
Materials Availability
This study did not generate new reagents.
Data and Code Availability
Sequence data are available through the European Nucleotide Archive under accession PRJEB39807 at https://www.ebi.ac.uk/ena/browser/view/PRJEB39807.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Wild chimpanzee subjects
Fecal samples were collected from habituated chimpanzees in two wild groups in Kibale National Park, Uganda between July 2015 and December 2018. Studied continuously since 1987, the Kanyawara group was comprised of 49 individuals in 2015 and 55 in 2018. Studied continuously since 1995, the Ngogo group was comprised of 192 individuals in 2015 and 200 in 2018. All chimpanzees were individually recognized, and none were directly provisioned with human foods, although those at Kanyawara occasionally forage from nearby farms. Chimpanzees at both sites are monitored daily by researchers and Ugandan research assistants. While these groups are only separated by 10–15 km, they exhibit important differences: the Ngogo group is considerably larger, with a higher population density, and has a higher concentration of fruiting trees [58], leading to improved foraging efficiency [59] and higher energy balance [60]. A summary of the metadata for the samples collected can be found in Table S2.
Wild chimpanzee metadata
Because the chimpanzees had been monitored on a daily basis for at least 20 years prior to data collection, all immature chimpanzees and young adult males have ages known to within one year. Females disperse within a narrow age range, so immigrant females are assigned to the average age of dispersal (13 with an assumed error of ±2 years). Some females do not immigrate and, for those, ages to within one year are known. The ages of individuals that were adult at the onset of observations were estimated by physical features [for details, see 61, 62].
Age classes were specified for these study groups based on prior work [7]. The age classes align with average age of behavioral milestones related to feeding (e.g., weaning, independent foraging) and reproductive maturity. Early infants were classified as individuals under 2 years old, when infants rely on breastmilk most heavily [35, 63, 64]; late infants were classified as individuals between 2 and 5 years old; juveniles as individuals between 5.01 and 10 years old; adolescents as individuals between 10.01 and 15 years old; adults as individuals between 15.01 and 35 years old; and post-prime adults as individuals older than 35 years of age. In total, we analyzed 11 samples from 9 individuals under 2; 33 samples from 14 late infants; 43 samples from 20 juveniles; 73 samples from 33 adolescents; 322 samples from 86 adults; and 136 samples from 33 post-prime adults.
Relatedness values for an individual to a parent or full sibling (0.5); grandparent, aunt/uncle, or half siblings (0.25); and half-aunt/uncle (0.125) were drawn from known pedigrees based on microsatellite-based parentage analysis for individuals in the Kanyawara population. Individuals for which pedigree data were not available or for which the relationship was more distant than those described above were not included in analyses of relatedness.
METHOD DETAILS
Sample collection
Fresh fecal samples were collected within minutes of deposit. Sterile wooden spatulas were used to scoop feces immediately into 2 ml of RNALater (Ambion, Thermo Fisher) to reach a volume of approximately 4 ml. Samples were shaken, sealed with parafilm, and then stored in a −20 °C solar freezer at the end of the observation day and until shipment to the U.S.
DNA isolation
Fecal samples stored at −20 °C were thawed on ice and homogenized by vortexing prior to use. Genomic DNA was extracted using a DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. 150 mg of starting material was used, samples were lysed by vortexing at maximum speed for 10 min and DNA was eluted in 100 μl of Buffer C6. DNA concentration was measured using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
16S rRNA gene sequencing
We performed 16S rRNA gene amplicon sequencing on fecal samples to determine gut microbial community structure. We used custom barcoded primers (515F to 806R) [65] targeting the V4 region of the 16S rRNA gene following published protocols [65–67]. Three separate libraries were prepared and sequenced on an Illumina HiSeq with single-end 150 bp reads in the Bauer Core Facility at Harvard University. Data were processed in QIIME2 (version 2020.2) [68] using the demux command to demultiplex raw reads and the DADA2 pipeline to generate ASV feature tables for each sequencing run separately. The three ASV tables were then merged, resulting in 15,263 unique features with 160,568 ± 86,906 assigned features per sample (median = 138,926). Taxonomy was mapped against the GreenGenes database (v. 13.8) [69].
Universal 16S quantitative PCR
To estimate total bacterial density and the absolute abundance of certain bacterial taxa, we performed universal 16S quantitative PCR (qPCR) on fecal DNA employing non-barcoded primers that targeted the same region used for sequencing (515F to 806R) [65]. qPCR assays were run using PerfeCTa SYBR Green SuperMix Reaction Mix (QuantaBio) on a BioRad CFX384 Touch (Applied Biosystems, Foster City, CA) in the Bauer Core Facility at Harvard University. Cycle-threshold values were standardized against a dilution curve of known concentration and then adjusted for the weight of fecal matter extracted. All reactions were run in triplicate and the results were averaged across these technical replicates.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were carried out in R (R core team, version 3.6) [70]. Statistical details can be found in the results section and in figure legends.
Analysis of alpha-diversity
Alpha-diversity metrics (observed operational taxonomic unit richness, Shannon index) were calculated in QIIME2. To assess what predictors affect gut microbial diversity, we used the lmer and anova functions in the package lme4 [71] to perform likelihood tests comparing a linear mixed effects model that included the variable of interest (e.g. age group) to a model that included only time variables. In both models, chimpanzee identity and sequencing run were included as random effects. For variables with significant effects on diversity, we used the emmeans package [72] to compute the pairwise comparison TukeyHSD contrasts for the estimated marginal means of the linear mixed effects models.
Analysis of beta-diversity
To assess beta-diversity among samples, we computed a Bray-Curtis dissimilarity table from the ASV table. Permutational MANOVA (PERMANOVA) on this table was carried out with the adonis2 function in the vegan package [73] to test for associations among gut microbial beta-diversity and variables of interest (e.g., age group, community, sex). We performed pairwise PERMANOVA calculations for the age group variable with the calc_pairwise_permanova function in the package mctoolsr [74]. We used a bootstrapping approach to test how beta-diversity varied within or between groups while correcting for the non-independence of dissimilarity measurements that include the same individuals in multiple comparisons. In short, we permuted the Kruskal-Wallis test statistic and p value, resampling with stratification specified by chimpanzee identity, using the boot package. We carried out these tests for beta-diversity measurements of individuals within their age group (70,000 permutations) and individuals compared to adults (100,000 permutations). We used the same approach to test whether Bray-Curtis dissimilarities among individuals with known relatedness (e.g., sibling, parent/offspring, grandparent, etc.) from the Kanyawara group differed based on the degree of relatedness or whether dissimilarity varied based on community membership or sex (25,000 permutations for each). For relatedness, community membership and sex-based analyses, we only compared individuals sampled during the same year and quarter to control for temporal variation.
To analyze mother-offspring relationships, we first performed likelihood tests to compare a linear mixed effects model including age group as a variable to a model that only included time variables. In both models, mother and offspring identities were included as random effects. We computed the pairwise comparison TukeyHSD contrasts for the estimated marginal means of the linear mixed effects model including the age group variable. We also randomly selected one other community-, age-class-, and sampling-quarter- matched female to stand in as the non-mother for each offspring with a known and sampled mother in the dataset. We tested for overall differences in Bray-Curtis dissimilarities between the offspring-mother pair and offspring-random pair with a likelihood test comparing a model including the mother/random term and the age group term to a model including only the age group term. For both types of analyses, we only included offspring-mother or offspring-random pairs where the individuals had been sampled during the same year and quarter to control for temporal variability.
Identification of indicator taxa
Positively associated indicator taxa for different age groups were identified with the signassoc function in the indicspecies package [75]. We stratified by chimpanzee identity and ran 1,000 permutations to correct for multiple measurements of the same individuals. All p values for the indicator taxa analyses were adjusted with the Sidak correction to address multiple hypothesis testing.
Analysis of bacterial absolute abundance
To test whether the absolute abundances of bacterial genera previously associated with human infancy varied significantly among chimpanzee age groups, we used permuted Kruskal-Wallis tests, as described above. We estimated absolute abundance by multiplying relative abundance data derived from 16S rRNA gene amplicon sequencing by bacterial load as indexed by universal 16S qPCR for the following taxa identified in [16]: Bacteroides, Bifidobacterium, Collinsella, Enterococcus, Lactobacillus, Streptococcus, Rothia, and Veillonellaceae. We used the abundance of the family Veillonellaceae instead of the genus Veillonella because we did not specifically identify the genus in the chimpanzee samples.
Supplementary Material
Statistical detail. A. Contrast tests for alpha-diversity metrics, related to Figure 1 and Table S2. B. Pairwise PERMANOVA tests for Bray-Curtis dissimilarity as a function of age group, related to Figure 2 and Table S2. C. Permuted Kruskal-Wallis statistics for Bray-Curtis dissimilarities, related to Figure 2 and Table S2. D. Contrast tests for Bray-Curtis dissimilarity to adults, related to Figure 2 and Table S2. E. Contrast tests for Bray-Curtis dissimilarity within age group, related to Figure 2 and Table S2. F. Contrast tests for change over 1+ year, related to Figure 3 and Table S2. G. Contrast tests for bacterial taxa associated with human infants, related to Figure 4 and Table S2. H. Contrast tests for qPCR-based bacterial absolute abundance, related to Figure 4 and Table S2.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Chimpanzee fecal samples | Kanyawara Chimpanzee Project | https://kibalechimpanzees.wordpress.com/ |
| Chimpanzee fecal samples | Ngogo Chimpanzee Project | http://ngogochimpanzeeproject.org/ |
| Critical Commercial Assays | ||
| DNeasy Powersoil Kit | Qiagen | Cat. #12888–100 |
| Deposited Data | ||
| Chimpanzee fecal sample sequence data | This Study | ENA Accession # PRJEB39807 https://www.ebi.ac.uk/ena/browser/view/PRJEB39807 |
| Greengenes database 13.8 | [69] | https://data.qiime2.org/2020.2/common/gg-13–8-99–515-806-nb-classifier.qza |
| Oligonucleotides | ||
| 16S V4 amplicon primers | [66] | 515F: GTGYCAGCMGCCGCGGTAA 806R: GGACTACNVGGGTWTCTAAT |
| Software and Algorithms | ||
| qiime2 | [68] | https://docs.qiime2.org/2020.8/ |
| R | [70] | https://www.r-project.org/ |
| lme4 | [71] | https://github.com/lme4/lme4/ |
| emmeans | [72] | https://github.com/rvlenth/emmeans |
| vegan | [73] | https://github.com/vegandevs/vegan |
| mctoolsr | [74] | http://leffj.github.io/mctoolsr/ |
| indicspecies | [75] | https://www.rdocumentation.org/packages/indicspecies/versions/1.7.9 |
Highlights:
We profiled changes in the wild chimpanzee gut microbiota over the life course
600+ samples collected over 3 years from 166 individuals aged 8 months to 67 years
Unlike humans, gut microbial diversity in chimpanzees is highest in infancy (<2 yo)
Divergent patterns may reflect species differences in lactation, diet and immunity
ACKNOWLEDGMENTS
For assistance in collecting and processing wild chimpanzee samples we thank Emily Otali, the staff of Kibale Chimpanzee Project, and the staff of Ngogo Chimpanzee Project. For helpful discussions of oligosaccharide biology, we thank Cary Allen-Blevins. For productive comments on the manuscript we thank our three peer-reviewers and members of the Carmody lab.
Funding: This work was supported by the National Institute on Aging and NIH Office for Research on Women’s Health (R01AG049395).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Marques TM, Wall R, Ross RP, Fitzgerald GF, Ryan CA, and Stanton C. (2010). Programming infant gut microbiota: influence of dietary and environmental factors. Curr. Opin. Biotechnol 21, 149–156. [DOI] [PubMed] [Google Scholar]
- 2.Sommer F, and Bäckhed F. (2013). The gut microbiota: masters of host development and physiology. Nat. Rev. Microbiol 11, 227–238. [DOI] [PubMed] [Google Scholar]
- 3.Aiello LC, and Wheeler P. (1995). The brain and the digestive system in human and primate evolution. Curr. Anthropol 36, 199–221. [Google Scholar]
- 4.Carmody RN, and Wrangham RW (2009). The energetic significance of cooking. J. Hum. Evol 57, 379–391. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan H, Hill K, Lancaster J, and Hurtado AM (2000). A theory of human life history evolution: Diet, intelligence, and longevity. Evol. Anthropol 9, 156–185. [Google Scholar]
- 6.Kramer KL, and Ellison PT (2010). Pooled energy budgets: Resituating human energy-allocation trade‐offs. Evol. Anthropol 19, 136–147. [Google Scholar]
- 7.Bray J, Emery Thompson M, Muller MN, Wrangham RW, and Machanda ZP (2018). The development of feeding behavior in wild chimpanzees (Pan troglodytes schweinfurthii). Am. J. Phys. Anthropol 165, 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moeller AH, Li Y, Mpoudi Ngole E, Ahuka-Mundeke S, Lonsdorf EV, Pusey AE, Peeters M, Hahn BH, and Ochman H. (2014). Rapid changes in the gut microbiome during human evolution. Proc. Natl. Acad. Sci 111, 16431–16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez A, Sharma AK, Mallott EK, Petrzelkova KJ, Robinson CAJ, Yeoman CJ, Carbonero F, Pafco B, Rothman JM, Ulanov A, et al. (2019). Plasticity in the human gut microbiome defies evolutionary constraints. mSphere 4, e00271–00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amato KR, Mallott EK, McDonald D, Dominy NJ, Goldberg T, Lambert JE, Swedell L, Metcalf JL, Gomez A, Britton GA, et al. (2019). Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 20, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degnan PH, Pusey AE, Lonsdorf EV, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, and Ochman H. (2012). Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc. Natl. Acad. Sci 109, 13034–13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moeller AH, Foerster S, Wilson ML, Pusey AE, Hahn BH, and Ochman H. (2016). Social behavior shapes the chimpanzee pan-microbiome. Sci. Adv 2, e1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, et al. (2018). Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, Ferretti P, Gorfer V, Pedrotti A, Tett A, et al. (2017). Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2, e00164–00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar A, Harty S, Johnson KVA, Moeller AH, Archie EA, Schell LD, Carmody RN, Clutton-Brock TH, Dunbar RIM, and Burnet PWJ (2020). Microbial transmission in animal social networks and the social microbiome. Nat. Ecol. Evol 4, 1020–1035. [DOI] [PubMed] [Google Scholar]
- 16.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. [DOI] [PubMed] [Google Scholar]
- 17.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, and Ley RE (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci 108, 4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eggesbø M, Moen B, Peddada S, Baird D, Rugtveit J, Midtvedt T, Bushel PR, Sekelja M, and Rudi K. (2011). Development of gut microbiota in infants not exposed to medical interventions. Apmis 119, 17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer C, Bik EM, DiGiulio DB, Relman DA, and Brown PO (2007). Development of the Human Infant Intestinal Microbiota. PLoS Biol. 5, e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, et al. (2014). Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayeni FA, Biagi E, Rampelli S, Fiori J, Soverini M, Audu HJ, Cristino S, Caporali L, Schnorr SL, Carelli V, et al. (2018). Infant and adult gut microbiome and metabolome in rural Bassa and urban settlers from Nigeria. Cell Rep. 23, 3056–3067. [DOI] [PubMed] [Google Scholar]
- 23.Gomez A, Sharma AK, Mallott EK, Petrzelkova KJ, Robinson CAJ, Yeoman CJ, Carbonero F, Pafco B, Rothman JM, Ulanov A, et al. (2019). Plasticity in the human gut microbiome defies evolutionary constraints. mSphere 4, e00271–00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Palacio SD, Montes SA, Mancabelli L, et al. (2017). The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev 81, e00036–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bode L. (2015). The functional biology of human milk oligosaccharides. Early Hum. Dev 91, 619–622. [DOI] [PubMed] [Google Scholar]
- 26.Newburg DS, Ruiz-Palacios GM, Altaye M, Chaturvedi P, Meinzen-Derr J, Guerrero MDL, and Morrow AL (2004). Innate protection conferred by fucosylated oligosaccharides of human milk against diarrhea in breastfed infants. Glycobiology 14, 253–263. [DOI] [PubMed] [Google Scholar]
- 27.Thum C, Cookson AL, Otter DE, McNabb WC, Hodgkinson AJ, Dyer J, and Roy NC (2012). Can nutritional modulation of maternal intestinal microbiota influence the development of the infant gastrointestinal tract? J. Nutr 142, 1921–1928. [DOI] [PubMed] [Google Scholar]
- 28.Allen-Blevins CR, Sela DA, and Hinde K. (2015). Milk bioactives may manipulate microbes to mediate parent–offspring conflict. Evol. Med. Public Health 2015, 106–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machado CSM, Rodrigues MAM, and Maffei HVL (1994). Gut intraepithelial lymphocyte counts in neonates, infants and children. Acta Paediatr. 83, 1264–1267. [DOI] [PubMed] [Google Scholar]
- 30.Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, and Farber DL (2014). Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thome JJ, Bickham KL, Ohmura Y, Kubota M, Matsuoka N, Gordon C, Granot T, Griesemer A, Lerner H, Kato T, et al. (2016). Early-life compartmentalization of human T cell differentiation and regulatory function in mucosal and lymphoid tissues. Nat. Med 22, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, Ferretti P, Gorfer V, Ferrario C, Milani C, et al. (2017). Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, and Knight R. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci 107, 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, et al. (2015). Exploring vertical transmission of bifidobacteria from mother to child. Appl. Environ. Microbiol 81, 7078–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bădescu I, Katzenberg MA, Watts DP, and Sellen DW (2017). A novel fecal stable isotope approach to determine the timing of age‐related feeding transitions in wild infant chimpanzees. Am. J. Phys. Anthropol 162, 285–299. [DOI] [PubMed] [Google Scholar]
- 36.Lonsdorf EV, Stanton MA, Pusey AE, and Murray CM (2020). Sources of variation in weaned age among wild chimpanzees in Gombe National Park, Tanzania. Am. J. Phys. Anthropol 171, 419–429. [DOI] [PubMed] [Google Scholar]
- 37.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, and Mills DA (2010). Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem 58, 5334–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zivkovic AM, German JB, Lebrilla CB, and Mills DA (2011). Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc. Natl. Acad. Sci 108, 4653–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sela DA, and Mills DA (2010). Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urashima T, Fukuda K, and Messer M. (2012). Evolution of milk oligosaccharides and lactose: a hypothesis. Animal 6, 369. [DOI] [PubMed] [Google Scholar]
- 41.Garcia M, Power ML, and Moyes KM (2017). Immunoglobulin A and nutrients in milk from great apes throughout lactation. Am. J. Primatol 79, e22614. [DOI] [PubMed] [Google Scholar]
- 42.Urashima T, Odaka G, Asakuma S, Uemura Y, Goto K, Senda A, Saito T, Fukuda K, Messer M, and Oftedal OT (2009). Chemical characterization of oligosaccharides in chimpanzee, bonobo, gorilla, orangutan, and siamang milk or colostrum. Glycobiology 19, 499–508. [DOI] [PubMed] [Google Scholar]
- 43.Albrecht S, Schols HA, van den Heuvel EG, Voragen AG, and Gruppen H. (2010). CE‐LIF‐MSn profiling of oligosaccharides in human milk and feces of breast‐fed babies. Electrophoresis 31, 1264–1273. [DOI] [PubMed] [Google Scholar]
- 44.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, and Yamamoto K. (2008). Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl. Environ. Microbiol 74, 3996–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmody RN (2017). Evolution of the human dietary niche – quest for high-quality. In Chimpanzees & Human Evolution, Muller MN, Wrangham RW and Pilbeam DR, eds. (Cambridge, MA: Harvard University Press; ), pp. 311–338. [Google Scholar]
- 46.Wrangham R, and Carmody R. (2010). Human adaptation to the control of fire. Evol. Anthropol 19, 187–189. [Google Scholar]
- 47.Sender R, Fuchs S, and Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, and Sonnenburg JL (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carmody RN, Bisanz JE, Bowen BP, Maurice CF, Lyalina S, Louie KB, Treen D, Chadaideh KS, Rekdal VM, Bess EN, et al. (2019). Cooking shapes the structure and function of the gut microbiome. Nat. Microbiol 4, 2052–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith AR, Carmody RN, Dutton RJ, and Wrangham RW (2015). The significance of cooking for early hominin scavenging. J. Hum. Evol 84, 62–70. [DOI] [PubMed] [Google Scholar]
- 52.Ragir S, Rosenberg M, and Tierno P. (2000). Gut morphology and the avoidance of carrion among chimpanzees, baboons, and early hominids. J. Anthropol. Res 56, 477–512. [Google Scholar]
- 53.Muller MN, Wrangham RW, and Pilbeam DR (2017). Chimpanzees and Human Evolution, (Cambridge, MA: Harvard University Press; ). [Google Scholar]
- 54.Moeller AH, Caro-Quintero A, Mjungu D, Georgiev AV, Lonsdorf EV, Muller MN, Pusey AE, Peeters M, Hahn BH, and Ochman H. (2016). Cospeciation of gut microbiota with hominids. Science 353, 380–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cox LM, and Blaser MJ (2015). Antibiotics in early life and obesity. Nat. Rev. Endocrinol 11, 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gensollen T, Iyer SS, Kasper DL, and Blumberg RS (2016). How colonization by microbiota in early life shapes the immune system. Science 352, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sampson TR, and Mazmanian SK (2015). Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 17, 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potts KB, Watts DP, and Wrangham RW (2011). Comparative feeding ecology of two communities of chimpanzees (Pan troglodytes) in Kibale National Park, Uganda. Int. J. Primatol 32, 669–690. [Google Scholar]
- 59.Potts KB, Baken E, Ortmann S, Watts DP, and Wrangham RW (2015). Variability in population density is paralleled by large differences in foraging efficiency in chimpanzees (Pan troglodytes). Int. J. Primatol 36, 1101–1119. [Google Scholar]
- 60.Thompson ME, Muller MN, Wrangham RW, Lwanga JS, and Potts KB (2009). Urinary C-peptide tracks seasonal and individual variation in energy balance in wild chimpanzees. Horm. Behav 55, 299–305. [DOI] [PubMed] [Google Scholar]
- 61.Wood BM, Watts DP, Mitani JC, and Langergraber KE (2017). Favorable ecological circumstances promote life expectancy in chimpanzees similar to that of human hunter-gatherers. J. Hum. Evol 105, 41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller MN, and Wrangham RW (2014). Mortality rates among Kanyawara chimpanzees. J. Hum. Evol 66, 107–114. [DOI] [PubMed] [Google Scholar]
- 63.Thompson ME, Muller MN, and Wrangham RW (2012). The energetics of lactation and the return to fecundity in wild chimpanzees. Behav. Ecol 23, 1234–1241. [Google Scholar]
- 64.Clark CB (1977). A preliminary report on weaning among chimpanzees of the Gombe National Park, Tanzania. In Primate Bio-Social Development: Biological, Social and Ecological Determinants, Chevalier-Skolnikoff S and Poirier F, eds. (New York: Garland Press; ), pp. 235–260. [Google Scholar]
- 65.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci 108, 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maurice CF, Haiser HJ, and Turnbaugh PJ (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol 37, 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, and Hugenholtz P. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Team, R.C. (2017). R: a language and environment for statistical computing. (Vienna, Austria: R Foundation for Statistical Computing; ). [Google Scholar]
- 71.Bates D, Maechler M, Bolker B, and Walker S. (2015). Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw 67, 1–48. [Google Scholar]
- 72.Lenth R. (2020). emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.4.8. (https://CRAN.R-project.org/package=emmeans). [Google Scholar]
- 73.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, Dan McGlinn, Minchin PR, O’Hara RB, Simpson GL, Solymos P, et al. (2017). vegan: Community Ecology Package. R package version 2.4–2. (https://github.com/vegandevs/vegan) [Google Scholar]
- 74.Leff JW (2017). mctoolsr: Microbial Community Data Analysis Tools. R package version 0.1.1.2. (https://github.com/leffj/mctoolsr). [Google Scholar]
- 75.De Caceres M, and Legendre P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical detail. A. Contrast tests for alpha-diversity metrics, related to Figure 1 and Table S2. B. Pairwise PERMANOVA tests for Bray-Curtis dissimilarity as a function of age group, related to Figure 2 and Table S2. C. Permuted Kruskal-Wallis statistics for Bray-Curtis dissimilarities, related to Figure 2 and Table S2. D. Contrast tests for Bray-Curtis dissimilarity to adults, related to Figure 2 and Table S2. E. Contrast tests for Bray-Curtis dissimilarity within age group, related to Figure 2 and Table S2. F. Contrast tests for change over 1+ year, related to Figure 3 and Table S2. G. Contrast tests for bacterial taxa associated with human infants, related to Figure 4 and Table S2. H. Contrast tests for qPCR-based bacterial absolute abundance, related to Figure 4 and Table S2.
Data Availability Statement
Sequence data are available through the European Nucleotide Archive under accession PRJEB39807 at https://www.ebi.ac.uk/ena/browser/view/PRJEB39807.