ABSTRACT
Cancer is a heterogeneous disease and its treatment remains unsatisfactory with inconstant therapeutic responses. This variability could be related, at least in part, to different and highly personalized gut microbiota compositions. Different studies have shown an impact of microbiota on antitumor therapy. It has been demonstrated that some gut bacteria influences the development and differentiation of immune cells, suggesting that different microbiota compositions could affect the efficacy of the antitumor vaccine. Emerging data suggest that recognition of neoantigens for the generation of neoantigen cancer vaccines (NCVs) could have a key role in the activity of clinical immunotherapies. However, it is still unknown whether there is a crosstalk between microbiota and NCV. This study aimed to understand the possible mechanisms of interaction between gut microbiota and NCV delivered by DNA-electroporation (DNA-EP). We found that decreased microbiota diversity induced by prolonged antibiotic (ATB) treatment is associated with higher intratumor specific immune responses and consequently to a better antitumor effect induced by NCV delivered by DNA-EP.
KEYWORDS: Microbiota, neoantigens, vaccine, T cells
INTRODUCTION
Immunotherapy is revolutionizing the treatment of cancer. The clinical success of immunotherapy has been demonstrated in a growing number of cancer types, including melanoma, lung cancer, bladder cancer, and hematological malignancies. However, response rates to immunotherapy are generally low and vary among cancer types and patients with the same type of malignancy. Recent evidence suggests that this variability may depend upon the composition of intestinal microbiota.1 The host-microbiota is highly personalized, depending on the host’s genetic background, physiology, diet, and lifestyle. Evidence regarding the impact of gut microbiota composition on the efficacy of immune checkpoint inhibitors (ICI) has been reported with monoclonal antibodies against programmed cell death protein 1 (PD1) (nivolumab, pembrolizumab), programmed death-ligand 1 (PD-L1)2 (atezolizumab), and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4)3 (ipilimumab). Several studies showed that some bacterial strains can modulate cancer progression and treatment efficacy.1 Gut microbiota may also play a role in adoptive T cell therapy (ACT). Paulos et al. revealed that total body irradiation leads to gut bacteria translocation into mesenteric lymph nodes thus increasing antitumor CD8+T cell adoptive transfer efficacy in a melanoma model.4 Recently, this was confirmed in the cervix and lung tumor mice models by showing that ACT efficacy depends on native gut bacteria composition and ATB treatment.5 In particular, vancomycin treatment increased ACT efficacy as a result of a high level of systemic CD8α+ dendritic cells (DCs). Different hypotheses have been formulated to explain the antitumor effects of gut microbiota on tumors growing at distant sites. Bacterial peptides or bacteria themselves can activate DCs cells, which migrate to the draining lymph nodes to prime T cells. T cells diffuse systemically and induce immune response at distant sites6,7 against the same organism or other organisms expressing cross-reacting antigens.8,9 Besides, dysbiosis induced by chemotherapy can breakdown mucosal barriers, allowing translocation of gut bacteria in lymph nodes modulating cytokine production.10 Viaud et al. demonstrated that the effect of cyclophosphamide treatment can be due to translocation of specific Gram− bacteria, which facilitates Th17 response and induced memory Th1 immune response.11 Overall these data provide a solid basis for the existence of a crosstalk between microbiota and the antitumor immunotherapy and suggest that the modulation of gut microbiota with ATB, probiotic administration or fecal microbiota transplantation can be utilized to improve immunotherapy.12
The role of microbiota in the immune response induced by prophylactic vaccines has been characterized mainly in infectious diseases.13 A high level of microbial diversity correlated with a more stable microbial community following vaccination and challenge with Shigella disenteriae in macaques.14 In human volunteers, vaccination with attenuated Salmonella Typhi was assessed by measuring IFN-γ+ in CD8 T cells. Most of the individuals showing an increased T cell response harbored a greater community richness and diversity of microbiota.14 Recently, microbiota composition was correlated with memory T cell response. The transfer of activated CD8+ T cells in germ-free mice resulted in impaired transition into long-lived memory cells, which was associated with a lack of microbiota-derived short-chain fatty acid (SFCA) metabolism.15 Broad-spectrum ATB treatment in healthy adults with low preexisting antibody titers strongly reduced the humoral response induced by the influenza vaccine.16
In the last decade, thanks to next-generation sequencing, it became feasible to identify cancer-specific mutations, which can be recognized by the T cells. This new class of cancer antigens is known as neoantigens. Somatic mutations are, by definition, not expressed in healthy tissues, and their cognate T cells are not shaped by central tolerance. Thus, neoantigens provide a unique opportunity for inducing potent tumor-specific T cells. Preclinical studies have shown the efficacy of the neoantigen cancer vaccine (NCV) using a variety of vaccine technologies in melanoma,17 colon carcinoma,18–21 sarcoma,22 and glioma.23 The promising results of preclinical studies have encouraged the development of clinical trials with neoantigen vaccines.24–26 Different vaccination platforms were utilized including dendritic cells, long peptide vaccine, or RNA vaccine delivered alone or in combination with ICIs. Although neoantigen-specific immune responses were observed in most patients clinical benefits were not reported,27,28supporting the need for further characterization of NCV mediated anti-tumor effects. It is reasonable to suppose that external factors, such as the microbiota may affect this type of immunotherapy.
In contrast, limited information is available on the T cell response to cancer vaccines, especially to personalized ones. Previous evidence using the ovalbumin antigen system ectopically expressed in the MC38 tumor model revealed an adjuvant effect of microbiota translocation, which was induced by chemotherapy treatment, on vaccine-specific T cell response.29 Here, we interrogated the potential interaction of the microbiota with a DNA vaccine targeting cancer-specific neoantigens naturally expressed by the MC38 cells.19,30 We discovered that changes in the microbiota composition may favorably affect the efficacy of the DNA vaccine delivered by electroporation.
RESULTS
Changes in microbiota correlate with improvement of the antitumor effect of a NCV
To evaluate the impact of microbiota on the NCV delivered by DNA-EP, we used the MC38 tumor model and, as a DNA vaccine, the M2 vector, which encodes for 10 neoantigens specifically expressed in this cell line.19 As expected, mice treated with broad-spectrum ATB showed a dramatic reduction of gut bacterial colonies11 at two weeks (Suppl. Figure 1) . After two weeks of ATB administration, mice were treated with the M2 vaccine weekly for four weeks (Figure 1). Nine days after the last immunization, mice were challenged subcutaneously with MC38 cells and tumor growth followed overtime. Surprisingly, tumor growth was significantly reduced in the DNA-EP + ATB group (p < 0.001) as compared to the DNA-EP group or the control (i.e. untreated) group and survival was increased by more than 50% (Figure 1a, 1b).
Figure 1.
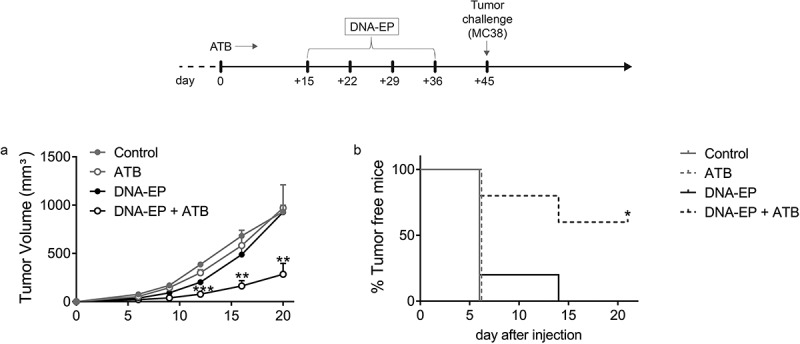
ATB treatment enables NCV to delay tumor growth. C57Bl/6 mice were treated or not with ATB starting from day zero for the whole experiment, vaccinated with M2 DNA-EP (see M&M) once a week for four weeks, and injected with MC38 cells on day 45. a, Tumor volumes were measured twice a week by caliper. Tumor growth was significantly reduced in vaccinated and ATB treated mice as compared to the control group (* p < 0.05) and vaccinated mice (** p < 0.01). Unpaired two-tailed Student’s t-tests were conducted. b, Percentage of tumor-free mice. Sixteen mice per group were utilized in three independent experiments. The results of one representative experiment out of three are shown. The tumor-bearing mice curve of DNA-EP and ATB treated mice is significantly different (p < 0.0298) compared to the DNA-EP group. Log-rank (Mantel-Cox) test was conducted
Reduction of tumor growth was observed neither in ATB-only treated mice nor in vaccinated mice suggesting that NCV was effective only in combination with ATB treatment. Recent evidence in mice15 and humans16 showed the role of the microbiota in modulating the memory T cell response induced by different vaccines. To verify the role of the memory immune response, we performed a tumor challenge 28 days after the last vaccination. In line with previous evidence that antibiotic treatment diminishes the effect of immunotherapy. the memory T cell response against NCV was significantly reduced in the DNA-EP + ATB treated mice but it did not correlate with tumor growth (Suppl Figure 2). These results raised the question of how the microbiota composition interacts with the NCV delivered by DNA-EP.
Figure 2.
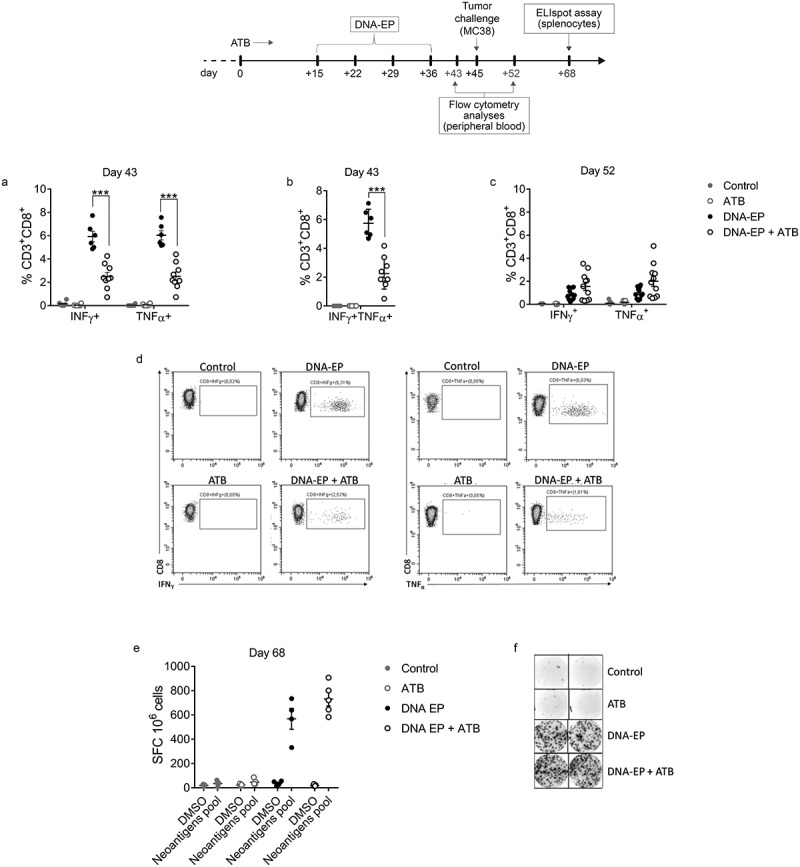
ATB treatment affects neoantigen-specific immune responses induced by DNA-EP. a, b, Immune response was evaluated one week after the last vaccination on day 43 (a, b) and one week after tumor challenge on day 52 (c) by flow cytometry in the PBMCs. To analyze neoantigen-specific cytokine production by intracellular staining live cells were gated on CD3+CD8+ for IFNγ and TNFα production. Unpaired two-tailed Student’s t-tests were conducted (*** p < 0.0001). c, At the end of the experiment on day 65 mice were sacrificed and IFNγ producing cells were evaluated by IFNγ ELIspot assay with splenocytes restimulated with neoantigens pool (e, f). Unpaired two-tailed Student’s t-tests were conducted. Data are from two independent experiments
To understand how the combination of the DNA-EP vaccine plus ATB elicited a delay in tumor growth, immune responses were evaluated by flow cytometry (FC) one week after the last vaccination and one week after tumor challenge. The neoantigen specific-immune response was analyzed in PBMC samples after stimulation with the pool of neoantigen peptides. One week after the last vaccination, we observed a significant reduction of neoantigen-specific CD3+CD8+IFNγ+ T cells, CD3+CD8+TNFα+ T cells, and also of the polyfunctional double-positive T cell population CD3+CD8+IFNγ+TNFα+ in DNA-EP + ATB group as compared to DNA-EP (Figure 2a, b). The analysis performed in splenocytes showed similar results (Suppl. Figure 3). It seems that upon tumor challenge, neoantigen-specific CD8 response remains at the steady-state level in DNA-EP and ATB treated mice while a decrease in circulating immune response was observed in DNA-EP treated mice. The result suggests a different mobilization of tumor-specific responses when microbiota composition is altered. To verify the impact of tumor growth on the NCV-specific immune responses PBMCs were analyzed again one week after the tumor challenge. Differently from the previous time point, this analysis showed a slight, albeit not statistically significant, increase in cytokine production by CD8 T cells in the DNA-EP + ATB group as compared to DNA-EP (Figure 2c). To further measure the neoantigen-specific immune responses in tumor-bearing mice, IFNγ producing cells were evaluated by IFNγ ELIspot assay in splenocytes at the time of sacrifice. This analysis suggests a slight increase in IFNγ production in DNA-EP + ATB as compared to DNA-EP (Figure 2e, f).
NCV increases the diversity of ATB reduced microbiota
To check the impact of ATB on bacterial composition, we analyzed the bacterial load the day before the tumor challenge on day 44 (Supp. Figure 1). The strong reduction of bacterial colonies observed at day 15 was not confirmed at day 44 suggesting that a prolonged ATB treatment selected a different bacterial population.31 To correlate the reduced tumor growth with gut bacteria composition, DNA from samples of stools on day 44 was extracted and analyzed by NGS on 16S rRNA (Suppl. Fig. 4a). As expected, ATB administration produced a reduction in the number of operational taxonomic units (Figure 3a). Principal component analysis (PCA) did not separate DNA-EP treated mice from untreated mice, while DNA-EP+ATB treated mice were separated from the other groups (Figure 3b). Furthermore, this analysis made it possible to identify some families of bacteria not shared by the two groups (Suppl. Tab. 1). A reduction in α- and β- diversity, measured by Shannon index and distance, respectively, was observed in ATB treated mice. However, in DNA-EP+ATB treated mice, a greater diversity of bacteria species in the gut as compared to ATB alone was revealed (Figure 3c, 3d).
Figure 3.
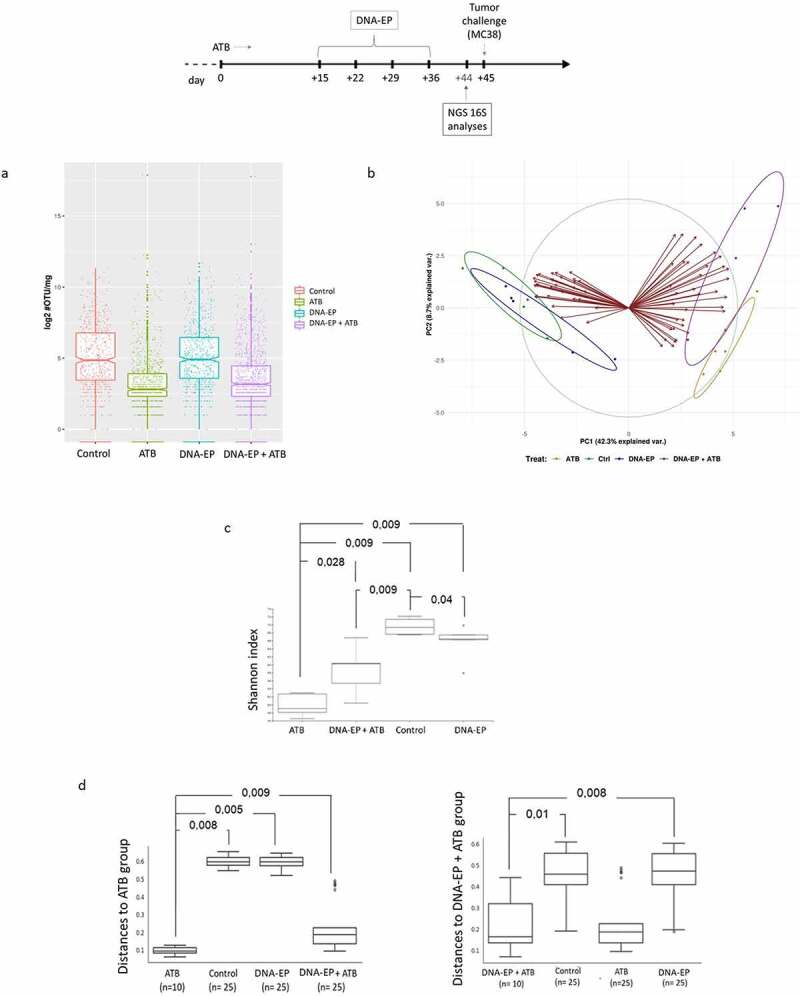
Vaccination increases the diversity reduced by ATB administration. Stool samples were analyzed before the tumor challenge on day 44. a, Bacteria load was expressed by operational taxonomic unit (OTU)/mg of stools and measured by NGS 16 S. b, Principal component (PC) analysis between groups. The PC1 explains 42,3% of the total variance, PC2 explains 8,7%. c, Alpha diversity, measured by Shannon’s index, represents species richness. d, Beta diversity represents the different microbiota composition between two groups. The value can be from 0 to 1, where 0 represents groups with similar species composition and 1 represents groups with no species in common. In the beta diversity, a weighted average was considered. Data are from two independent experiments
Treatment with ATB promoted the survival of some bacteria strains that were poorly or not normally existing in mice untreated with ATB. Gut microbiota in untreated mice was constituted prominently of Firmicutes and Bacteroides (Suppl. Tab. 2). ATB administration reduced both, in particular Bacteroides, in favor of Proteobacteria. DNA-EP vaccine, in ATB treated mice, restored Firmicutes phyla and increased Bacteroides and Proteobacteria compared to ATB treated mice. Some of the families identified by PCA, such as Bosea, (Beijerinckiaceae), Thermoactinomyces (Thermoactinomycetaceae), Bacillus (Bacillaceae), and Romboutsia (Peptostreptococcaceae) were also the most abundant in DNA-EP + ATB group (Suppl. Figure 4b, Suppl. Tab. 3). DNA-EP+ATB group presented some bacteria strains that were not present in ATB-treated mice, such as Muribaculaceae and Bacillaceae (Suppl. Tab. 3). In addition, differences in the relative frequency of bacteria composition between the DNA-EP group and DNA-EP+ATB group were observed, such as Bosea, Thermoactinomyces, Bacillus, Muribaculaceae, Lactobacillus, Lachnospiraceae, (Suppl. Tab. 3). Hence, the DNA-EP vaccine modifies the microbiota composition of ATB treated mice and this effect might contribute to the observed antitumor effect.
Changes in short-chain fatty acids composition does not correlate with tumor growth
We next asked whether the modified microbiota composition is acting on NCV-specific immune responses through the release of soluble factors. Different studies have shown that the influence of microbiota on the immune response is mediated by metabolites produced by microbiota, among which the most studied are SCFAs.32 Therefore, one week after the tumor challenge, we evaluated whether there were differences in propionate, butyrate, and acetate in the mice serum. The results showed a significant difference in the concentration of acetate between the group of mice treated with ATB and mice treated only with DNA-EP (Figure 4). Although it was shown that acetate concentration correlates with CD3+IFNγ+ T immune responses,33 in our experimental condition this observation did not associate with different tumor growth.
Figure 4.
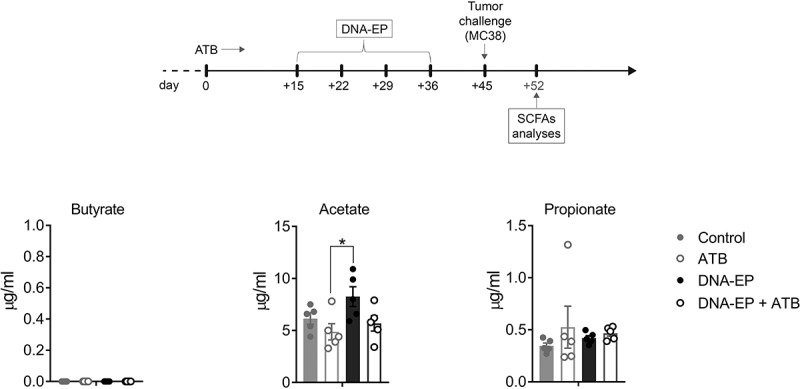
SCFAs metabolites do not correlate with tumor growth. Level of SCFAs analyzed in the serum of mice one week after tumor challenge on day 52 determined using LC-SIM-MS (n = 5 mice for group). Data are from one out of two experiments. Unpaired two-tailed Student’s t-tests were conducted (*p < 0.05)
Increased TILs functionality in DNA-EP and ATB treated mice
To further characterize the correlation between microbiota, NCV, and tumor growth, analysis of gene expression by RNAseq was carried out on tumor samples 12 days after tumor challenge (tumor volume ≅ 100 mm3). This analysis showed that changes in microbiota composition, in association with the vaccine, modified the expression levels of different genes. Of particular interest were the changes in Arg1, Gzmc and Gzmb genes (Figure 5a). The expression of Arg1, encoding for the enzyme arginase involved in the urea cycle, was reduced in vaccinated mice compared to control and further reduced in DNA-EP + ATB group (Figure 5b). Moreover, increased expression of T cell functionality markers (Gzmc) was observed in DNA-EP+ATB treated mice (Figure 5b). These data were further supported by RT-PCR, which showed similar trends (Figure 5c), and a strong correlation with RNAseq results (Suppl. Figure 5). Moreover, to characterize the immune responses, tumor-infiltrating lymphocytes (TILs) and splenocytes were analyzed 12 days after tumor challenge (tumor volume ≅ 100 mm3) by FC. The frequency of infiltrating lymphocytes was not affected by NCV or ATB treatments (Suppl. Figure 6), however, the frequency of CD8+IFNγ+, CD8+TNFα+, CD4+IFNγ+, CD4+TNFα+ cells was significantly increased in TILs of DNA-EP+ATB treated mice (Figure 5d). Moreover, we observed a significant increase of innate immune populations, such as NK+IFNγ+ and TNFα-producing monocytes (Figure 5e), in DNA-EP + ATB group compared to the DNA-EP group. Although not statistically significant, neoantigen-specific cytokine production by CD8 was increased in the DNA-EP+ATB group in the TILs population (Figure 5f) and slightly reduced in splenocytes (Figure 5g), in comparison to the DNA-EP group.
Figure 5.
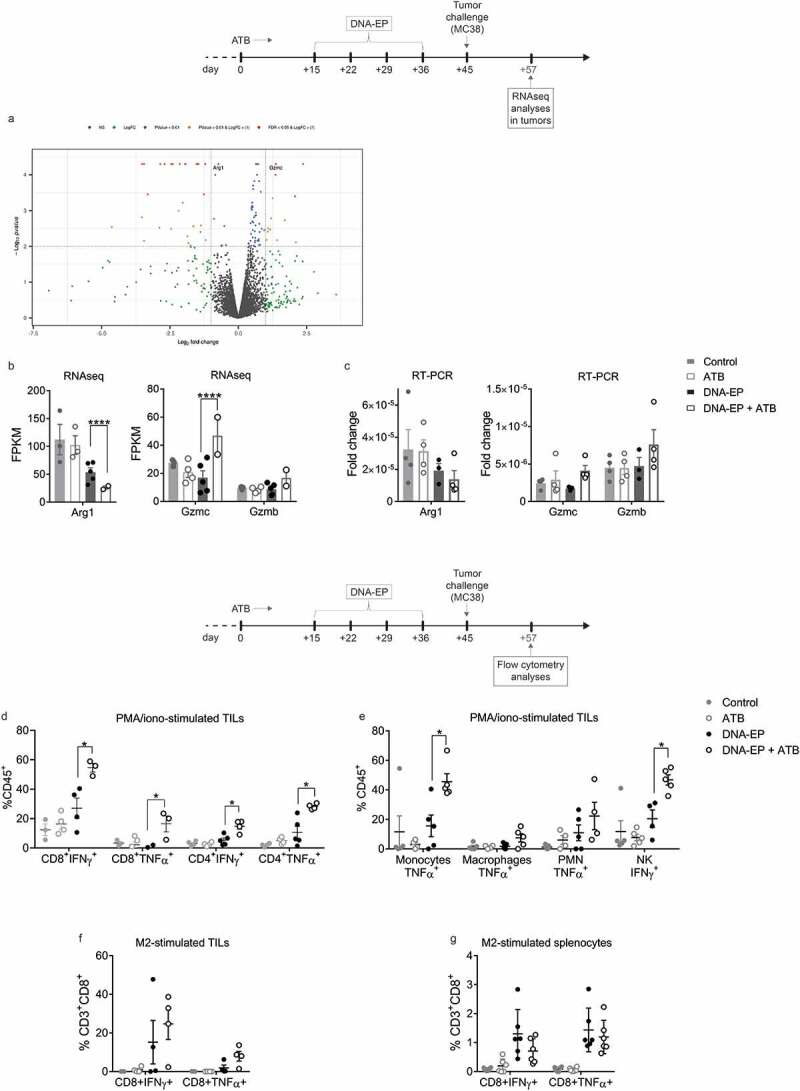
Increased TIL effector functions in DNA-EP and ATB treated mice a, Volcano plot demonstrating the significantly differentially expressed genes, highlighting Arg1 and Gzmc. b, Level of RNA expression (FPMK, fragments per kilobase of exon model per million reads mapped) of Gzmc, Gzmb, and Arg1 measured by RNAseq in tumors of mice sacrificed one week after tumor challenge. c, Tumor expression of Gzmc, Gzmb, and Arg1 genes assessed by RT-PCR. d, Flow cytometry (FC) analysis of IFNγ and TNFα expression in TILs (CD4 and CD8) gated on CD45+CD3+ and stimulated with PMA/iono. e, FC analysis of cytokines expression of innate immune cells gated on CD45+CD3−CD11b+CD11c−; monocytes (Ly6ChighLy6G−), macrophages (Ly6ClowLy6G−), and polymorphonucleated (PMN, Ly6ChighLy6G+). Natural killer (NK, NK1.1+) cells were gated on CD45+CD3−. f, FC analysis of IFNγ and TNFα expression in TILs (CD3+CD8+) gated on CD45+ and stimulated with M2-specific peptides. g, FC analysis of IFNγ and TNFα expression in splenocytes (CD3+CD8+) gated on CD45+ and stimulated with M2-specific peptides. Unpaired two-tailed Student’s t-tests were conducted. (*p < 0.05). Data are from two independent experiments
DISCUSSION
A growing body of evidence indicates that the host microbiota may affect cancer progression and response to therapy, including cancer immunotherapy, although data on the impact of the microbiota on cancer vaccines are generally limited.29 Therefore, we wondered if NCV-induced immune response and antitumoral effect could be affected by the modulation of microbiota composition. To this end, we used the M2 vaccine that we previously demonstrated to be effective in preventing tumor growth when delivered one month before tumor challenge.19 Here, we show that early point, i.e. one week after the last vaccination, the antitumor effect mediated by the M2 vaccine is observed only in DNA-EP+ATB treated mice (Figure 1a) while there was no impact at four weeks (Suppl. Figure 2). The observation that a reduced T cell response does not impact on tumor growth at four weeks post-vaccination may suggest that quality and not the quantity of T cells was relevant.34 An alternative explanation may be the so-called depot effect. A sort of vicious loop effect induced by a strong expression of the antigen at the vaccination site that diverts the anti-tumor immune response.35 This effect is likely to wane over time given that the transgene expression is less prominent at four weeks. However, the observation that a reduced peripheral T cell response is associated with a significant increase of effector function in the TIL population suggests that external factors such as tumor microenvironment may play a role in the tumor delay.
The antitumor effects correlate with microbiota alterations induced by DNA-EP treatment in association with ATB (Figure 3d). Indeed, we observed that ATB administration in vaccinated mice causes a reduction in the relative distribution of bacteria strains such as Lachnospiracee (Suppl. Table 3), involved in Treg development, and an increase of Bacillales (including Thermoactinomyces and Bacillus) and Alphaproteobacteria (including Bosea) (Figure 4a), of which very little is known in the literature. FC analysis on peripheral blood showed that these microbiota modulations are associated with different NCV-specific immune responses among the groups. Indeed, in ATB treated mice, NCV-specific cytokine production by CD8 T cells is significantly reduced one week after the last vaccination (Figure 2a), and this immunological observation is in line with previous evidence that supports a role of the healthy microbiota as adjuvant of humoral13 and T cell response.11 However, after the tumor challenge, we observed a reduced percentage of cytokine-producing CD8 T cells in the DNA-EP group, while it remained the same in DNA-EP + ATB mice (Figure 2c). Therefore, these results suggest a role of ATB-induced dysbiosis in protecting vaccine-induced immune response. Nevertheless, further studies are necessary to clarify why tumor cell injection in DNA-EP vaccinated mice reduces NCV-specific CD8+ T response. The interaction between gut microbiota and host immune response is complex. Several studies have suggested that this relationship is mediated by metabolites produced by microbiota, among which the most studied are SCFAs.36 However, in our study, we did not observe any difference in serum levels of SCFAs between DNA-EP mice and DNA-EP + ATB mice. On the other hand, bacteria strains identified only in ATB treated mice and enriched in DNA-EP+ATB treated mice, such as Bacillales (gen. Bacillus) and Alphaproteobacteria (gen. Bosea), have been correlated with amino acid metabolism.6,7
In line with the immune responses and tumor growth, the analysis of gene expression on tumor samples revealed a reduced Arg1 gene expression level in DNA-EP+ATB mice compared to the DNA-EP group. Arg1 gene is involved in L-arginine metabolism and it has been shown that it can have a role in immune response regulation. A recent study showed that L-Arginine is important for T cell metabolism, enhancing their survival and anti-tumor activity,37 and it is expressed in some immune populations, such as myeloid-derived suppressor cells, resulting in the inhibition of T-cell receptor expression and consequent antigen-specific T cell response, thus promoting tumor evasion. Moreover, granulocytes-associated arginase, through suppression of T cell proliferation and cytokine production, inhibits immune reactivity in different tumor models.38 In our model, the combination of DNA-EP+ATB resulted in an increased expression of Gzmc and Gzmb genes, which are important for cytotoxic lymphocyte functions.39 Therefore, our data suggest that in DNA-EP + ATB group, microbiota alterations could inhibit Arg1, resulting in a higher amount of arginine and intratumoral functional T cells able to destroy tumor cells. Besides, FC analyses in TILs confirmed that CD8+IFNγ+ and CD8+TNFα+ T cells were significantly increased in DNA-EP + ATB group compared to the DNA-EP group in the tumors (Figure 5d). This could suggest an increased homing of activated and functional CD8 + T cells in the tumor of DNA-EP + ATB mice. Indeed, the percentage of NCV-specific CD8 T response (CD8+IFNγ+ and CD8+TNFα+) was slight, though not significantly, increased In the TILs of the DNA-EP+ATB group (Figure 5f) but not in the spleen (Figure 5g). Moreover, although solid tumor-infiltrating NK cells have been shown to become dysfunctional in a mouse model and human cancers,40 we observed a significant increase of NK+IFNγ+ cells in the tumors of the DNA-EP+ATB group compared to DNA-EP group (Figure 5e). A significant increase of monocytes expressing TNF was also observed, further experiments are necessary in order to understand the role of these cells in the tumor. Our data are in line with previous evidence showing the interplay of gut microbiota and antitumor effects mediated by natural killer T cells.41 Further studies are required to rule out the antitumor role of NK cells and the interplay with ATB induced dysbiosis.
Our data suggest a crosstalk between microbiota, NCV-specific immune response, and tumor growth with an altered microbiota composition enabling a more effective anti-cancer T cell response (Figure 6). The simple ATB treatment resulted in an improved NCV efficacy suggesting the possibility of improving NCV specific immune response by modulating the microbiota composition. The mechanisms underlying these complex interactions remain to be determined and deserve further investigation.
Figure 6.
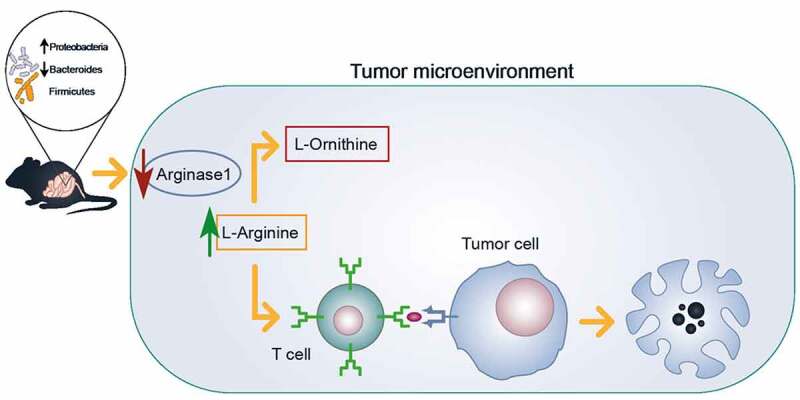
Microbiota alteration elicits T cell activation in DNA-EP treated mice. A schematic model of possible crosstalk between microbiota and tumor is represented. ATB treatment, in combination with a vaccine, modifies microbiota composition reducing some bacteria species in favor of others that are not normally present in mice. This microbiota alteration induces a reduction of Arginase1 in tumor lysate, possibly related to reported TIL activation in the DNA EP+ATB group
MATERIAL & METHODS
Cell lines and mice
The MC38 colon carcinoma cell line was purchased from Kerafast (ENH204-FP). Master and working cell banks were generated upon receipt and used at the third and fourth passage for all tumor challenge experiments. Cells were mycoplasma free as per internal regular controls. Six week old C57BL/6 female mice (Envigo, USA) were housed in the Plaisant animal house according to national legislation and kept in standard conditions according to Takis ethical committee approval. Animal studies were authorized by the Italian Minister of Health (586/2019-PR).
Antibiotics treatments and immunization schedule
Mice were treated with or without an ATB cocktail (1 mg/ml ampicillin, 1 mg/ml colistin, 5 mg/ml streptomycin) in their drinking water and the solution was changed twice a week.
The M2 DNA vaccine vector was generated as previously described.19 50 μg of plasmid DNA was injected in a 50 μL volume into the tibialis muscle followed by electroporation, as previously described.42 Tumor challenge was performed by injecting 3 × 105 MC38 cells s.c. in the right flank of the mice.
Immune responses
The neoantigen-specific T-cell response was determined using intracellular cytokine staining (ICS) performed by FC detection and IFNγ ELIspot as previously described19 For the FC analysis, the following antibodies were utilized according to different panels: anti-CD3-Alexafluor488 (cat. 53–0031-82, eBioscience), anti-CD3-PEefluor610 (cat. 61–0031-82, eBioscience), anti-CD4-PerCP-Cy5.5 (cat. 45–0042-82, eBioscience), anti-CD8-APCeFluor780 (cat 47–0081-82, eBioscience), anti-CD45-efluor450 (cat. 48–0451-82, eBioscience), anti-CD45-APC (cat. 559,864, BD), NK1.1-BV786 (cat. 740,853, Bio Legend), anti-CD11b-FITC (cat. 53,310, BD), anti-CD11c-SB645 (cat. 64–0114-82, Ebioscience), anti-LY6C-PE-Cy7 (cat. 560,593, BD), anti-LY6G-Alexafluor700 (cat. 561,236, BD), anti-IFN-γ-PE (cat. 12–7311-82, eBioscience), anti-TNF-α-PE-Cy7 (cat. 25–7321-82, eBioscience), anti-TNF-α-eFluor450 (cat. 48–7321-82, eBioscience FC was carried out with a Citoflex flow cytometer (Beckman Coulter) and data analyzed with Cytexpert software (Beckman Coulter). For the IFNγ ELIspot cells were plated at 4 × 105 and 2 × 105 cells/well in duplicate and spots were counted using an automated ELISPOT reader (Aelvis ELIspot reader, A.EL.VIS Gmbh, Germany).
Metabolomic analysis of short-chain fatty acids (SCFA)
Metabolomic analysis was performed by Tuscano Life Science. Each serum sample (25 ul) was processed, adding deuterated internal standard and acetonitrile: methanol mixture (50%:50%, v/v) and centrifuged at 1300 rpm for 10 minutes (4°C). The supernatant was analyzed at LC-SIM-MS. For the analyses, Acquity UPLC HSST T3 1,8 μm 2.1 × 50 mm (Waters) column in UHPLC Ultimate 3000 was used. Used eluent phases were 0.1% formic acid in water (A-phase) and 0.1% formic acid in acetonitrile (B phase). Mass spectrometer Q-Exactive Plus (ThermoFisher) was used. It was possible to analyze all six species, monitoring ions mass: m/z 59.01385 and 62.03210 for acetic acid and D4-acetic acid, respectively; m/z 73.02950 e 78.06030 for propionic acid and D5-propionic acid; m/z 87.04515 e 94.08850 for butirric acid and D8-butirric acid. The area generated by each pair of signals was used for the external calibration curve and the quantitative determination of the analytes in the murine serum samples.
Next-generation sequencing (NGS) analysis
DNA extraction from fecal samples was performed by QIAamp PowerFecal DNA Kit (QIAGEN) according to the manufacturer’s instructions. NGS analyses were performed by Biofab research. DNA was amplified to enrich the bacterial 16S V3-V4 rRNA region by PCR with a fusion primer containing the sequence to incorporate Illumina adapters and indexing barcodes. The gene‐specific sequences used have been selected from the Klindworth et al. publication.43 The Illumina adapter overhang nucleotide sequences are added to the gene‐specific sequences. The full-length primer sequences are:
16S Amplicon PCR Forward Primer = V3
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG
16S Amplicon PCR Reverse Primer = V4
GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC.
Analysis was performed according to published literature43
RNAseq and RT-PCR
RNA was extracted by 30 mg of frozen tumor tissues according to the manufacturer’s instructions (RNeasy kit, QIAGEN). To smash and homogenize a TissueLyser LT (QIAGEN) was used and operated at 50 Hz for 2 minutes. RNA extracted was used for RNAseq. Moreover, RNA extracted was retro-transcribed to cDNA using high capacity cDNA Reverse Transcription Kit (applied biosystem) according to the manufacturer’s instructions. Real-time (RT)-PCR was performed using TaqMan™ Gene Expression Master Mix (applied biosystem) and commercial Taq-Man probes for Arg1, Gzmc, and Gzmb genes (applied biosystem). Gene expression was normalized to 18S and expressed using the 2-Δct method.
Statistical analysis
Statistical analysis was performed with the Prism 7.04 Graphpad software. The nonparametric t-test (Mann–Whitney test) was applied to compare the results between groups of treatment and tumor growth curves at different time points. Log-rank (Mantel-Cox) test was conducted for the tumor-free curve. A two-tailed p-value of 0.05 was considered statistically significant.
Supplementary Material
Disclosure statement
Authors do not have a conflict of interest to be declared.
Supplementary Material
Supplementary data for this article can be accessed here.
References
- 1.Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G.. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382–11. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 2.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (80-). 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CPM. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science (80-). 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.T CD, Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, Palmer DC, Boni A, Muranski P. Microbial translocation augments the function of adoptively transferred self/tumor-specific. J Clin Invest. 2007;117(8):2197–2204. doi: 10.1172/JCI32205.mens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uribe-Herranz M, Bittinger K, Rafail S, Guedan S, Pierini S, Tanes C, Ganetsky A, Morgan MA, Gill S, Tanyi JL. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight. 2018;3(4):1–18. doi: 10.1172/jci.insight.94952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leyn SA, Kazanov MD, Sernova NV, Ermakova EO, Novichkov PS, Rodionova DA. Genomic reconstruction of the transcriptional regulatory network in bacillus subtilis. J Bacteriol. 2013;195(11):2463–2473. doi: 10.1128/JB.00140-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai Z, Wu Z, Hang S, Zhu W, Wu G. Amino acid metabolism in intestinal bacteria and its potential implications for mammalian reproduction. Mol Hum Reprod. 2014;21(5):389–409. doi: 10.1093/molehr/gav003. [DOI] [PubMed] [Google Scholar]
- 8.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Littman DR. If so, is there potential fohttps://mail.google.com/mail/u/0?ui=2&ik=b0e31beded&attid=0.8&permmsgid=msg-f:1672298554166362643&th=173532acbf7c4213&view=att&disp=inliner efficacious microbiota-based vaccines?. CSH Perspect. 2017;(Do the Microbiota Influence Vaccines and Protective Immunity to Pathogens ?):1–9. doi: 10.1101/cshperspect.a028860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17(4):219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 11.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science (80-). 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, Narushima S, Vlamakis H, Motoo I, Sugita K. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565(7741):600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann P, Curtis N. The influence of the intestinal microbiome on vaccine responses. Vaccine. 2018;36(30):4433–4439. doi: 10.1016/j.vaccine.2018.04.066. [DOI] [PubMed] [Google Scholar]
- 14.Seekatz AM, Panda A, Rasko DA, Toapanta FR, Eloe-Fadrosh EA, Khan AQ, Liu Z, Shipley ST, DeTolla LJ, Sztein MB. Differential response of the cynomolgus macaque gut microbiota to shigella infection. PLoS One. 2013;8(6). doi: 10.1371/journal.pone.0064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachem A, Makhlouf C, Binger KJ, De Souza DP, Tull D, Hochheiser K, Whitney PG, Fernandez-Ruiz D, Dähling S, Kastenmüller W. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity. 2019;51(2):285–297.e5. doi: 10.1016/j.immuni.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, Das J, Wang H, Guthmiller J, Zheng NY. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178(6):1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreiter S, Vormehr M, Van De Roemer N, Diken M, Löwer M, Diekmann J, Boegel S, Schrörs B, Vascotto F, Castle JC. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuai R, Ochyl LJ, Bahjat KS, Schwendeman A, Moon JJ. Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat Mater. 2017 Apr;16(4):489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aurisicchio L, Salvatori E, Lione L, Bandini S, Pallocca M, Maggio R, Fanciulli M, De NF, Goeman F, Ciliberto G. Poly-specific neoantigen-targeted cancer vaccines delay patient derived tumor growth. J Exp Clin Cancer Res. 2019;4:1–13. doi: 10.1186/s13046-019-1084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duperret EK, Perales-Puchalt A, Stoltz R, G H H, Mandloi N, Barlow J, Chaudhuri A, Sardesai NY, Weiner DB. A synthetic DNA, multi-neoantigen vaccine drives predominately MHC class I CD8(+) T-cell responses, impacting tumor challenge. Cancer Immunol Res. 2019;7(2):174–182. doi: 10.1158/2326-6066.CIR-18-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tondini E, Arakelian T, Oosterhuis K, Camps M, Van Duikeren S, Han W, Arens R, Zondag G, Van Bergen J, Ossendorp F. A poly-neoantigen DNA vaccine synergizes with PD-1 blockade to induce T cell-mediated tumor control. Oncoimmunology. 2019;8(11). doi: 10.1080/2162402X.2019.1652539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber W-J. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, Quandt J, Menn O, Osswald M, Oezen I, Ott M. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 24.Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie W-R, Hildebrand WH, Mardis ER. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science (80-). 2015;348(6236):803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z, Ott PA, Wu CJ. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat Publ Gr. 2017. doi: 10.1038/nri.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, Bukur V, Tadmor AD, Luxemburger U, Schrörs B. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 27.Fang Y, Mo F, Shou J, Wang H, Luo K, Zhang S, Han N, Li H, Ye S, Zhou Z. A pan-cancer clinical study of personalized neoantigen vaccine monotherapy in treating patients with various types of advanced solid tumors. Clin Cancer Res. 2020. doi: 10.1158/1078-0432.ccr-19-2881. clincanres.2881.2019. [DOI] [PubMed] [Google Scholar]
- 28.Ott PA, Hu-Lieskovan S, Chmielowski B, Govindan R, Naing A, Bhardwaj N, Margolin K, Awad MM, Hellmann MD, Lin JJ. A phase Ib trial of personalized neoantigen therapy plus anti-PD-1 in patients with advanced melanoma, non-small cell lung cancer, or bladder cancer. Cell. 2020;183(2):347–362.e24. doi: 10.1016/j.cell.2020.08.053. [DOI] [PubMed] [Google Scholar]
- 29.Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, Duong CPM, Flament C, Lepage P, Roberti MP. Enterococcus hirae and barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic Immunomodulatory effects. Immunity. 2016;45(4):931–943. doi: 10.1016/j.immuni.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J, Weinschenk T. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 31.Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, Young GP, Marshall HS, Wesselingh SL, Rogers GB. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. 2018;23(5):653–660.e5. doi: 10.1016/j.chom.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Skelly AN, Sato Y, Kearney S, Honda K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol. 2019;19(5):305–323. doi: 10.1038/s41577-019-0144-5. [DOI] [PubMed] [Google Scholar]
- 33.Luu M, Weigand K, Wedi F, Breidenbend C, Leister H, Pautz S, Adhikary T, Visekruna A. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci Rep. 2018;8(1):1–10. doi: 10.1038/s41598-018-32860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baharom F, Ramirez-Valdez RA, Tobin KKS, Yamane H, Dutertre CA, Khalilnezhad A, Reynoso GV, Coble VL, Lynn GM, Mulè MP. Intravenous nanoparticle vaccination generates stem-like TCF1+ neoantigen-specific CD8+ T cells. Nat Immunol. 2020;(1). doi: 10.1038/s41590-020-00810-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hailemichael Y, Woods A, Fu T, He Q, Nielsen MC, Hasan F, Roszik J, Xiao Z, Vianden C, Khong H. Cancer vaccine formulation dictates synergy with CTLA-4 and PD-L1 checkpoint blockade therapy. J Clin Invest. 2018;128(4):1338–1354. doi: 10.1172/JCI93303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5(4):1–8. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meer F, Mann M. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167(3):829–842.e13. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro H, Thaiss CA, Levy M, Elinav E. The cross talk between microbiota and the immune system: metabolites take center stage. Curr Opin Immunol. 2014;30(1):54–62. doi: 10.1016/j.coi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Revell PA, Grossman WJ, Thomas DA, Cao X, Behl R, Ratner JA, Lu ZH, Ley TJ. Granzyme B and the downstream granzymes C and/or F are important for cytotoxic lymphocyte functions. J Immunol. 2005;174(4):2124–2131. doi: 10.4049/jimmunol.174.4.2124. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt L, Eskiocak B, Kohn R, Dang C, Joshi NS, DuPage M, Lee DY, Jacks T. Enhanced adaptive immune responses in lung adenocarcinoma through natural killer cell stimulation. Proc Natl Acad Sci U S A. 2019;116(35):17460–17469. doi: 10.1073/pnas.1904253116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science (80-). 2018;360(6391). doi: 10.1126/science.aan5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elia L, Mennuni C, Storto M, Podda S, Calvaruso F, Salucci V, Aurisicchio L, Scarito A, Ciliberto G, La Monica N. Genetic vaccines against Ep-CAM break tolerance to self in a limited subset of subjects: initial identification of predictive biomarkers. Eur J Immunol. 2006;36(5):1337–1349. doi: 10.1002/eji.200535514. [DOI] [PubMed] [Google Scholar]
- 43.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41(1):1–11. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


