Abstract
Strict anaerobes are undeniably important residents of the cystic fibrosis (CF) lung but are still unknowns. The main objectives of this study were to describe anaerobic bacteria diversity in CF airway microbiota and to evaluate the association with lung function. An observational study was conducted during eight months. A hundred and one patients were enrolled in the study, and 150 sputum samples were collected using a sterile sample kit designed to preserve anaerobic conditions. An extended-culture approach on 112 sputa and a molecular approach (quantitative PCR targeting three of the main anaerobic genera in CF lung: Prevotella, Veillonella, and Fusobacterium) on 141 sputa were developed. On culture, 91.1% of sputa were positive for at least one anaerobic bacterial species, with an average of six anaerobic species detected per sputum. Thirty-one anaerobic genera and 69 species were found, which is the largest anaerobe diversity ever reported in CF lungs. Better lung function (defined as Forced Expiratory Volume in one second > 70%) was significantly associated with higher quantification of Veillonella. These results raise the question of the potential impact of anaerobes on lung function.
Subject terms: Cystic fibrosis, Clinical microbiology
Introduction
Cystic fibrosis (CF) is a genetic disease due to CF transmembrane conductance regulator (cftr) gene mutations (mostly p.F508del mutation). From another point of view, CF can also be considered as an infectious disease, as lung colonisation and infection aggravate the clinical condition and are the main causes of morbidity-mortality in people with CF (PWCF). Key bacterial pathogens (Staphylococcus aureus, Haemophilus influenzae, Pseudomonas aeruginosa and Burkholderia cepacia complex) are known to chronically colonise CF lungs and cause pulmonary exacerbation1. However, CF pulmonary microbiota harbours a wider range of bacterial communities that include certain species thought to play an important role in respiratory disease2. An accurate description of the lung bacterial species, many of which have yet to be identified3,4, could lead to a better understanding of the lung disease2. Strict anaerobes play an important role among these as yet unknown species, and are undeniably important residents of the CF lung5,6, that have been detected in respiratory samples at all ages4. Anaerobes’ growth is stimulated by anoxic conditions, provided partially by mucus hyperviscosity and oxygen consumption by neutrophil afflux or bacterial proliferation5,7. The most frequently detected genera in CF lungs are Prevotella, Veillonella, Fusobacterium, Atopobium, Peptostreptococcus and Porphyromonas4,5,8. However, in spite of their wide diversity and abundance, the role of airway anaerobic bacteria is still controversial. On the one hand, anaerobes exhibit their own virulence factors such as proteases9,10, can enhance others pathogens virulence11–14 or can promote antibiotic resistance15,16. On the other hand, they can be associated with less lung inflammation6,17 and better lung function4,6,18–20. Hence, it remains under debate whether the presence of anaerobes is beneficial and whether targeted antibiotic therapy against anaerobic bacteria is needed or should rather be avoided in case of CF pulmonary exacerbation4,21. In the light of these contradictory observations, we conducted an observational study to go deeper in deciphering anaerobe diversity at species level within the CF lung microbiota. We implemented a dual approach, combining extended-culture and targeted molecular techniques for optimally exhaustive and accurate identification and quantification of anaerobic communities.
Results
Subjects and samples
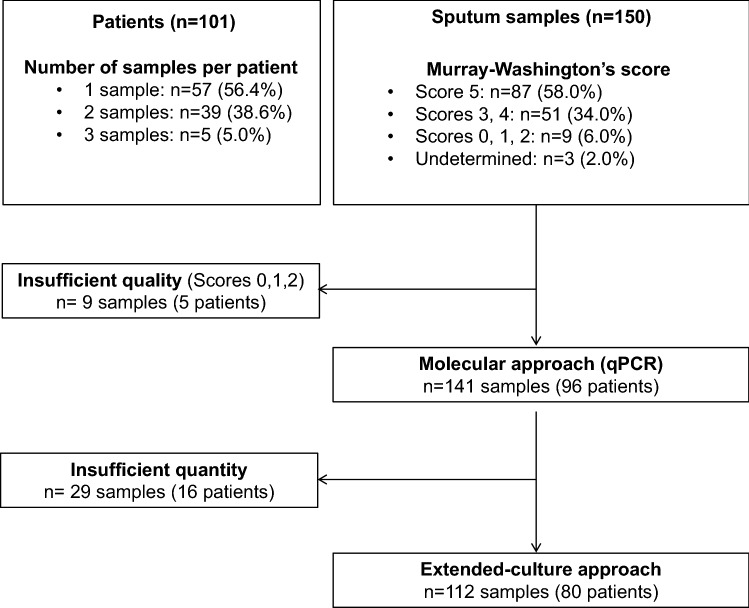
From March to October 2018, 150 sputa from 101 PWCF were collected in the study: one sample for 57 patients, and two or three samples for 54 patients. Based on Murray-Washington criteria22,23 (Table S1), salivary contamination was low in 58.0% of sputa (87/150), intermediate in 34.0% (51/150), and high in 6.0% (9/150); 2.0% of sputa (3/150) were undetermined because of poor quantity (Fig. 1). The nine samples with high salivary contamination were excluded from further analysis (five patients) and 29 sputa underwent molecular analyses but not extended-culture due to insufficient volume (16 patients) (Fig. 1). After exclusion, a total of 96 PWCF were enrolled in the study; subject characteristics are shown in Table 1 (and in Table S2 for the 80 PWCF whose samples were analysed only by extended-culture approach). According to the Leeds definition for P. aeruginosa colonisation24, the majority of the cohort were categorised as chronic or intermittent (72.9%; 70/96). Concerning lung function, 80.3% of patients (77/96) presented Forced Expiratory Volume in one second (FEV1) inferior or equal to 70%.
Figure 1.
Flow chart of the patients and samples included in the study.
Table 1.
Study cohort (96 people with cystic fibrosis): demographic and clinical data.
| Patient characteristics | n (%) |
|---|---|
| Age group (years) | |
| < 13 | 7 (7.3) |
| 13–< 18 | 2 (2.1) |
| 18–< 25 | 30 (31.3) |
| 25–< 30 | 14 (14.6) |
| ≥ 30 | 43 (44.7) |
| Gender | |
| Female | 47 (49.0) |
| Male | 49 (51.0) |
| cftr mutation | |
| p.F508del homozygote | 53 (55.2) |
| p.F508del heterozygote | 33 (34.4) |
| Other mutation | 10 (10.4) |
| Pancreas status | |
| Pancreatic sufficiency | 8 (8.3) |
| Pancreatic insufficiency | 88 (91.7) |
| Body-mass index▲ (kg/m2) | |
| Underweight (< 18.5) | 22 (22.9) |
| Reference value (18.5–24.9) | 63 (65.6) |
| Overweight/Obesity (> 24.9) | 11 (11.5) |
| Lung function (Forced Expiratory Volume in one second, %) | |
| < 40 | 23 (24.0) |
| 40–70 | 54 (56.3) |
| > 70 | 19 (19.7) |
| Chronic antibiotic therapy (inhaled and/or oral) | |
| Azithromycin | 53 (55.2) |
| Tobramycin | 19 (19.8) |
| Colistin | 49 (51.0) |
| Aztreonam | 8 (8.3) |
| Oral antibiotic therapy (one month before) | |
| Yes | 39 (40.6) |
| No | 57 (59.4) |
| Corticosteroids (inhaled and/or oral) | |
| Yes | 73 (76.0) |
| No | 23 (24.0) |
| Diabetes | |
| Yes | 38 (39.6) |
| No | 58 (60.4) |
| CFTR modulators (ivacaftor, lumacaftor) | |
| Yes | 29 (30.2) |
| No | 62 (64.6) |
| Change during study | 5 (5.2) |
| Leeds status (Pseudomonas aeruginosa colonisation) | |
| Never | 5 (5.2) |
| Free | 21 (21.9) |
| Intermittent | 11 (11.5) |
| Chronic | 59 (61.4) |
Extended-culture approach for anaerobic bacteria isolation and identification at species level
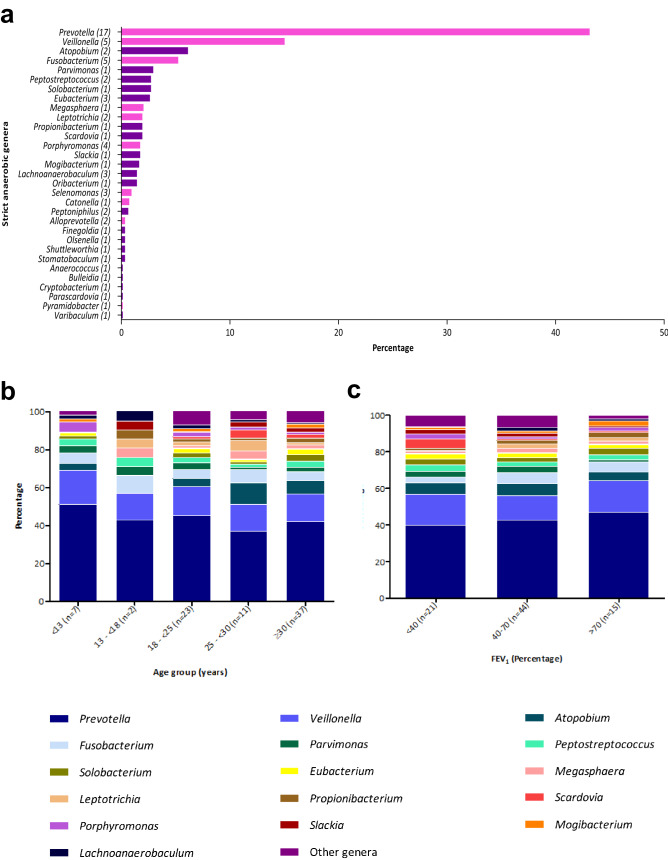
On the extended-culture approach, 692 strict anaerobic strains were isolated from the 112 sputa analysed: 84.8% were identified by MALDI-TOF MS (587/692), and 15.2% by 16S rRNA gene sequencing (105/692). A total of 91.1% of sputum samples (102/112) were positive for at least one anaerobic species. After the exclusion of duplicate species per patient (Table S3), an average of six anaerobic bacteria were isolated per patient (range 0–16). Thirty-one strict anaerobic genera were cultured from the 112 sputum samples, comprising 69 species (Fig. 2a; Table S4). The 10 most frequent genera were: Prevotella (43.1%; 298/692), Veillonella (15.0%;104/692), Atopobium (6.1%; 42/692), Fusobacterium (5.2%; 36/692), Parvimonas (2.9%; 20/692), Peptostreptococcus (2.7%; 19/692), Solobacterium (2.7%; 19/692), Eubacterium (2.6%; 18/692), Megasphaera (2.0%; 14/692), and Leptotrichia (1.9%; 13/692) (Fig. 2a). Anaerobic genera composition within the different age- and lung function- groups is detailed in Fig. 2b/2c. Two anaerobic genera, poorly represented but both isolated from high-quality sputum (Murray-Washington score at 5), were first detected in the CF lung: Pyramidobacter (0.1%; 1/692; species Pyramidobacter piscolens), and Varibaculum (0.1%; 1/692; species Varibaculum anthropi). At a finer taxonomic level, a wide diversity was found: 17 Prevotella species (P. baroniae, P. buccae, P. denticola, P. histicola, P. intermedia, P. loescheii, P. maculosa, P. marshii, P. melaninogenica, P. nanceiensis, P. nigrescens, P. oralis, P. oulorum, P. pallens, P. salivae, P. timonensis, and P. veroralis) five Veillonella species (V. atypica, V. denticariosi, V. dispar, V. parvula, and Veillonella sp.), and five Fusobacterium species (F. canifelinum, F. naviforme, F. nucleatum, F. periodonticum, and Fusobacterium sp.) (Table S4).
Figure 2.
Extended-culture approach results. (a) Description of strict anaerobic genera identified in 112 sputum samples: Gram-positive genera are shown in purple, Gram-negative genera in pink, and number of species in brackets. (b) Prevalence of strict anaerobic genera identified according to patient age group (80 patients). (c) Prevalence of strict anaerobic genera identified according to patient lung function evaluated by Forced Expiratory Volume in one second (FEV1) (80 patients).
Molecular approach for detection and quantification of Prevotella, Veillonella and Fusobacterium genera
qPCR targeting these three anaerobic genera in the lung confirmed that Prevotella was the most abundant genus in sputa (median of quantification: 1.34 × 106 CFU/mL), compared to Veillonella (median of quantification: 2.71 × 104 CFU/mL) and Fusobacterium (median of quantification: 2.55 × 102 CFU/mL). The molecular approach was significantly more positive than culture for the detection of the less abundant genera, Veillonella and Fusobacterium (p < 0.01 respectively, McNemar test) (Table S5).
Relationship between lung function and Prevotella, Veillonella and Fusobacterium quantification
Lung function was evaluated by FEV1, with a 70% clinical threshold25. Statistical analysis (univariate analysis) of results for the 96 patients (141 samples) showed that a one-log increase of Prevotella, Veillonella and Fusobacterium quantification (CFU/mL) was significantly associated with a FEV1 superior to 70% (OR = 1.41, p = 0.045; OR = 1.92, p < 0.001; OR = 1.45, p = 0.032 respectively). Multivariate analysis confirmed that a one-log increase of the quantity of the genus Veillonella was associated with a better lung function (FEV1 > 70%) (p = 0.02, logistic regression) (Table 2). Except for patient age, no other significant associations were found between lung function and the clinical parameters (gender, body-mass index, oral antibiotic therapy, CFTR modulator treatment, or P. aeruginosa positive sputum culture) (Table 2). Moreover, all 13 samples in which Veillonella was not detected by qPCR came from patients with FEV1 < 70% (10 patients, median age: 36 years). Focusing on the three most identified Veillonella species by culture approach (V. atypica, n = 34; V. dispar, n = 21; V. parvula, n = 47), V. dispar was the most associated with a better lung function (FEV1 > 70%) but this association was not significant (OR = 3.50, p = 0.08).
Table 2.
Multivariate analysis: logistic regression modelling the probability of Forced Expiratory Volume in one second (FEV1) > 70%.
| OR▲ (95% confidence interval) | p value | |
|---|---|---|
| Quantification of Veillonellaa | 2.14 (1.30–3.53) | < 0.01 |
| Ageb | 0.86 (0.80–0.92) | < 0.01 |
| Genderc | 1.48 (0.42–5.19) | 0.54 |
| Body-mass indexd | 0.34 (0.11–1.03) | 0.06 |
| CFTR modulatorse | 1.07 (0.29–3.94) | 0.92 |
| Oral antibiotic treatmente | 0.92 (0.23–3.62) | 0.90 |
| Pseudomonas aeruginosa positive culturee | 0.56 (0.17–1.79) | 0.33 |
aOdds-ratio relative to a one-log increase.
bOdds-ratio relative to a one-year increase.
cWoman versus man.
d < 18.5 kg/m2 versus > 18.5 kg/m2.
eYes versus no.
Discussion
CF lungs harbour diverse communities of bacteria, the abundance and variety of which fluctuate depending on multiple factors: clinical status, treatment and environment3,26–28. Within this rich and diversified bacterial microbiota, anaerobes are able to grow in anoxic areas in CF lungs and have been detected in higher quantities in PWCF than in healthy individuals5. These oxygen-sensitive bacteria come from diverse reservoirs (mainly oral and digestive)29 and can be detected in equal or higher proportions than some CF pathogens in lung4. Moreover, numerous anaerobic genera have been identified as part of the CF core lung microbiota: Prevotella, Veillonella, Porphyromonas, Fusobacterium, Catonella and Peptostreptococcus20,28,30. The recent literature emphasised the importance of anaerobic bacteria in lung, but results and hypotheses concerning their role in the pathophysiology and progression of CF disease are contradictory4,6,9–21,31,32. Anaerobes’ impact is mainly evaluated according to genus rather than species, which may explain these conflicting results, as some species within a given genus may generate contrasting effects33. Thus, anaerobic microbiota description should be exhaustive, as accurate as possible and frequently implemented.
To this end, culture approaches seem to be appropriate to provide descriptive information of anaerobe diversity at species level34. However, due to the fastidiousness of implementing a strictly anaerobic atmosphere, lower sensitivity than molecular approaches, the effect of previous antibiotic therapy, and growth limited to “culturable” bacteria, most descriptive studies of the lung “anaerobiome” are currently provided by 16S-targeted metagenomics. However, the targeting of 16S variable regions with most of the sequencing platforms used did not allow the optimal taxonomic resolution to be achieved35. We decided to conduct an observational study, based on a dual approach combining extended-culture and targeted molecular techniques to go deeper in deciphering anaerobe diversity within the CF lung microbiota. To evaluate sputum quality, we used the Murray-Washington cytological score, which showed that 94.0% of the sputum samples (141/150) were properly sampled and informative. Our findings supplement previous culture data on anaerobe diversity4–6,8. Thanks to the innovative collecting device and the culture-extensive protocol (21 days of incubation) coupled with both MALDI-TOF MS and 16S rRNA gene sequencing, 31 strict anaerobic genera and 69 species were described in 112 CF sputa. Considering the seven studies that described strict anaerobes in the CF lung microbiota based on an extensive-culture approach (Table 3), our study provided the greatest diversity ever described so far. Mirkovic et al.13 found 52.3% positive respiratory samples with 8 anaerobic genera detected from 109 samples, Muhlebach et al.4 found 67.0% positive sputa with 18 anaerobic genera detected from 200 samples; O’Neill et al.6 found 12 genera from 41 sputum, Paganin et al.36 found five genera from 78 sputum, Sherrard et al.37 found 23 anaerobic genera from 199 sputa; Sibley et al.38 found 15 genera from 246 sputum, and Tunney et al.5 found 64.0% positive sputa with seven genera from 66 samples. Focusing on the most recent study which reported 18 anaerobic genera on culture in CF lung4, all of them were also detected in the present study, supplemented by description of 13 additional genera. To our knowledge, two anaerobic genera (Pyramidobacter and Varibaculum) were described here in CF lung for the first time in sputa with low salivary contamination. These culture results were contributive as they demonstrate the viability of anaerobes in CF lung, which might not be clear using molecular techniques, and they shed light on anaerobic communities in CF lung according to species. On the molecular side of the study, qPCR improved detection compared with culture for two anaerobic genera (Veillonella and Fusobacterium) which were represented at lower rates in sputum samples. These results showed that the combination of both approaches is essential for optimally exhaustive anaerobic microbiota description.
Table 3.
Comparison of studies based on an extended-culture approach for the CF “anaerobiome” description.
| References | Number of patients/samples (type of samples) | Clinical state | Taxonomic affiliation detailed to the genus/species level | Number of strict anaerobic genera/species/ isolatesb | Delay between the sampling and anaerobiosis | Incubation time (days) | Identification methods applied on colonies |
|---|---|---|---|---|---|---|---|
| This study | 80/112 (sputum) | All patients | Yes/yes | 31/69/692 | No delay | 21 | MALDI-TOF MSc, targeted qPCR and 16S rRNA gene sequencing |
| Sherrard et al.37 | 80/199 (sputum) | All patients | Yes/no | 23/NR/NM | No delay | 2–5 | 16S rRNA gene sequencing |
| Muhlebach et al.4 | 255/255 (sputum, BAL) | All patients | Yes/yesa | 18/49/NM | No delay | 2–7 | 16S rRNA gene sequencing |
| Mirković et al.13 | 109/109 (sputum, BAL) | Only stable patients | Yes/yesa | 8/22/NM | No delay | 5–7 | 16S rRNA gene sequencing |
| O’Neill et al.6 | 41/41 (sputum) | Only stable patients | Yes/no | 12/NM/NM | No delay | 5–7 | 16S rRNA gene sequencing |
| Paganin et al.36 | 78/78 (sputum) | All patients | Yes/yes | 5/18/NM | 15 min | 5–7 | MALDI-TOF MS and 16S rRNA/recA-gene sequencing |
| Sibley et al.38 | 117/246 (sputum) | All patients | Yes/yes | 15/28/157 | 2 min | 7 | 16S rRNA gene sequencing |
| Tunney et al.5 | 60/76 (sputum, BAL) | Only stable patients | Yes/yesa | 7/12/NM | 15 min | 5–7 | 16S rRNA gene sequencing and RapID Ana II identification system |
NR not realized, NM not mentioned, BAL bronchoalveolar lavage fluid, MALDI-TOF MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry.
aNot available for all isolates.
bOnly strict anaerobes, according to the Bergey’s Manual of Systematic Bacteriology definition46, were considered (if data available).
bIdentification according to Li et al.47
As well as being descriptive, this study demonstrated a significant positive association between Veillonella quantification and lung function (FEV1 > 70%). This was already seen in previous studies, for the genus Veillonella6 and also for the genus Prevotella18. Moreover, although the observation was based on a small number of samples (n = 13), the genus Veillonella was absent only in sputa from PWCF with poor respiratory function (FEV1 < 70%). Consequently, the genus Veillonella and associated species detected on culture (V. parvula, V. atypica, V. dispar) could in future be used as biomarkers to predict lung function, as previously shown for the anaerobic genus Porphyromonas which could be used to predict earlier P. aeruginosa infection39. In our study, there is a tendency of association of the species V. dispar with a better lung function (non-significant association). The lung role of this species in PWCF is as yet unknown but a recent study in healthy people has shown that lung bacteria are the same as oral or nasal bacteria, but that some, like V. dispar, are enriched in the healthy lung40. This finding suggests that V. dispar may play an important role in lung health40, which needs further investigation. These results also pave the way for discussion of the protective role of anaerobic bacteria. However, little is known about the pathophysiology and mechanisms involved in this potential positive impact, although significant positive correlations were demonstrated between anaerobes and better lung function or/and less airway inflammation, especially for the genera Prevotella, Veillonella and Porphyromonas1,4,6,18,20.
The present study and the methods used here are subject to some limitations. Firstly, the cohort was composed of relatively old PWCF (median age, 27 years; only 12 patients under 18 years of age), which might constitute a recruitment bias for the description of anaerobic bacteria in CF lung, as diversity may be impacted by patient age17,41. Secondly, as the design was single-centre, geographical background was not taken into account, although it may have an impact on the richness and diversity of the anaerobic microbiota. Thirdly, molecular studies were performed only on three anaerobic genera (Prevotella, Veillonella and Fusobacterium). The anaerobe panel needs to be extended, and should target species rather than genera to accurately evaluate the impact of anaerobes on lung function. Fourthly, the Murray-Washington criteria were used to determine sputum quality and to exclude samples suspected of high salivary contamination. However this score does not make it possible to assert the only pulmonary location of the anaerobic bacteria detected. Fifthly, FEV1 may not be the most relevant lung parameter to evaluate lung function. Lung clearance index (LCI) was reported to show greater sensitivity than FEV1 and better correlation with clinical outcome. However, LCI is now indicated mainly for trials in young PWCF or with early or mild lung disease42, which were not the main conditions in our cohort. All in all, new longitudinal studies are required to better understand anaerobic microbiota dynamics and impact on respiratory function, depending on the patient’s clinical status, age (focusing on paediatric patients) and antimicrobial treatments, with inclusion in several CF centres.
In conclusion, this cohort study revealed the largest anaerobe diversity ever reported in CF lungs, thanks to preservation of anaerobic atmosphere in sputa and the powerful performance of the extended-culture approach. Moreover, the study highlighted that a greater quantity of the genus Veillonella was significantly positively associated with better lung function. Consequently, anaerobes should not be underestimated within the lung microbiota. Subsequent longitudinal studies are needed to determine and understand anaerobe impact and pathophysiology according to species and to evaluate anaerobe antibiotic sensitivity under antibiotic pressure.
Methods
People with cystic fibrosis (PWCF) and sample processing
This observational study was conducted between March and October 2018. Samples were collected in the Western Brittany CF centre (Roscoff, France) during follow-up consultations. As samples consisted of sputa, only PWCF able to expectorate were enrolled. Pulmonary transplant patients were excluded. The following sociodemographic and clinical parameters were recorded: age, gender, cftr gene mutation, pancreas status, body-mass index, lung function, diabetes, and treatments (oral antibiotic therapy one month before sampling, chronic antibiotic therapy, corticosteroids, CFTR modulators). A collecting device was specifically designed and patented for the study to ensure preservation of an anaerobic atmosphere for the sputa (EP 20305133.9) (Fig. S1). Between collection and transfer to the anaerobic chamber, samples were conserved at + 4 °C.
Extended-culture approach for bacterial isolation and identification
Sputa quality and salivary contamination were evaluated on Murray-Washington score22,23 (Table S1). Each sputum sample was mixed with an equal volume of dithiothreitol (Digest-EUR, Eurobio, Courtaboeuf, France), and incubated at 37 °C for 30 min to one hour in accordance with the manufacturer’s instructions. One-thousand-fold dilution was prepared in physiological saline supplemented with 0.05% (wt/vol) L-cystein (Sigma-Aldrich, Dorset, UK). One hundred microliters of liquefied diluted sputum was plated onto three different media: anaerobe basal agar with sheep blood (ABA-SB) (ThermoFisher, Waltham, USA), kanamycin-vancomycin ABA-SB, and colistin-nalidixic acid ABA-SB. Plates were incubated in an anaerobic chamber (90% N2, 5% H2, 5% CO2) (Bactron, Sheldon Manufacturing, Cornelius, USA) for 21 days at 37 °C and readings were made every 2–5 days. Isolates of each distinct colony type were subcultured onto ABA-SB, and identified by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF MS Biotyper MBT) (Bruker, Billerica, USA) (Supplementary Information). All strict anaerobes isolates were stored at − 80 °C (Microbank, Pro-Lab Diagnostics, Ontario, Canada). In case of mass spectrometry failure (database or technical problems), a 16S rRNA gene (rrs) sequencing method was used for isolate identification. The following primers were used for amplification of the rrs gene (996 bp): sense 5′-CCAGCAGCCGCCGTAATACG-3′; antisense 5′-TACGGYTACCTTGTTACGACT-3′. The PCR reaction was conducted using the Eppendorf Mastercycler Gradient instrument (Eppendorf, Hamburg, Germany). PCR sequencing used the Big Dye Terminator Cycle Sequencing Kit Ready Reaction version 3.1 (Applied Biosystems, Courtaboeuf, France), according to the manufacturer’s recommendations, on an Applied Biosytems 3130xl instrument (ThermoFisher, Waltham, USA). Sequences were then aligned and corrected using the software BioEdit version 7.0.5.3 (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Comparison with the NCBI BLAST database (Basic Local Alignment Search Tool, http://blast.ncbi.nlm.nih.gov/Blast.cgi) enabled bacterial identification, with ≥ 98% similarity determining homology of identity. In parallel to the anaerobic extended-culture approach, all samples were cultured in aerobic conditions to detect usual pathogens such as P. aeruginosa.
Detection and quantification of Prevotella, Veillonella and Fusobacterium genera by quantitative PCR (qPCR)
qPCR was performed for detection and quantification of three anaerobic genera (Prevotella, Veillonella, Fusobacterium) in each sputum sample. 200 µL aliquots of liquefied sputum were stored at − 80 °C. Samples were treated by five minutes-sonication using a bath sonicator (Elmasonic S10, Elma Schmidbauer GmbH, Singen, Germany). After 10 min-centrifugation (5000 g) and prior lysis by proteinase K, total DNA was extracted using the QIAamp DNA Minikit (Qiagen, Courtaboeuf, France) according to the manufacturer’s instructions, with an elution volume of 100 µL. The qPCR reaction was conducted using an ABI Prism 7500 Fast Real-Time PCR System instrument (ThermoFisher, Waltham, USA). Primers (and probe) used for qPCR are detailed in Table S643–45. Specificity and efficiency were evaluated for each primer and probe pairs (Supplementary Information).
Statistical analysis
McNemar’s test was used to compare the percentage of positive samples according to culture and molecular approaches. A logistic regression was used for binary outcome (FEV1 > 70%) to estimate odds-ratios relative to a one-log increase of the quantification of the anaerobic genera and to the absence/presence of Veillonella species (V. atypica, V. dispar, V. parvula) in univariate analysis. Only the most associated genus (Veillonella) and species (V. dispar) were considered in the multivariate model, adjusting for potential confounders (age, gender, body-mass index, CFTR modulators, oral antibiotic treatment and P. aeruginosa positive culture). Intra-patient correlation due to repeated samples was handled thanks to an exchangeable correlation structure, using the SAS software GENMOD procedure (SAS Institute, Cary, USA) with binomial distribution and logit link function.
Ethics
The study was approved by the French Ethical Research Committee in March 2018 (2018-A00624-51). All experiments were performed in accordance with relevant guidelines and regulations. Informed consent was obtained from all participants and/or their legal guardians.
Conference presentation
The study was presented in part at the 39th “Réunion Interdisciplinaire de Chimiothérapie Anti-Infectieuse-RICAI”, 16–17 December 2019 in Paris, France.
Supplementary Information
Acknowledgements
The authors gratefully acknowledge Morgane Floch and Jocelyne Pengam for their precious help with patient inclusion. The authors thank Iain McGill for revision of the English manuscript.
Author contributions
C.L., C.A.G. and G.H.A. contributed in every aspect of this research work including conception, study design, data analysis and interpretation. C.L. and S.G. performed the laboratory experiments. C.L. and G.H.A. wrote the manuscript. C.A.G., C.B., S.G., R.L., S.V., R.L.B. and E.N. participated in interpretation of the data and review of the manuscript. E.N. carried out all the statistical analysis. S.R., A.D., J.L.B., K.R. and T.R. participated in the coordination of patients’ inclusion. All authors read and approved the final manuscript.
Funding
This study was supported by a grant from the French Cystic Fibrosis Association ‘Vaincre la Mucoviscidose’ (Contract No. RC20180502218).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-85592-w.
References
- 1.Carmody LA, et al. Fluctuations in airway bacterial communities associated with clinical states and disease stages in cystic fibrosis. PLoS ONE. 2018;13:e0194060. doi: 10.1371/journal.pone.0194060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Toole GA. Cystic fibrosis airway microbiome: overturning the old, opening the way for the new. J. Bacteriol. 2017;200:e00561–e617. doi: 10.1128/JB.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang YJ, LiPuma JJ. The microbiome in cystic fibrosis. Clin. Chest Med. 2016;37:59–67. doi: 10.1016/j.ccm.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muhlebach MS, et al. Anaerobic bacteria cultured from cystic fibrosis airways correlate to milder disease: a multisite study. Eur. Respir. J. 2018;52:1800242. doi: 10.1183/13993003.00242-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tunney MM, et al. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2008;177:995–1001. doi: 10.1164/rccm.200708-1151OC. [DOI] [PubMed] [Google Scholar]
- 6.O’Neill K, et al. Reduced bacterial colony count of anaerobic bacteria is associated with a worsening in lung clearance index and inflammation in cystic fibrosis. PLoS ONE. 2015;10:e0126980. doi: 10.1371/journal.pone.0126980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley ES, Kopf SH, LaRiviere A, Ziebis W, Newman DK. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. MBio. 2015;6:e00767. doi: 10.1128/mBio.00767-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worlitzsch D, et al. Antibiotic-resistant obligate anaerobes during exacerbations of cystic fibrosis patients. Clin. Microbiol. Infect. 2009;15:454–460. doi: 10.1111/j.1469-0691.2008.02659.x. [DOI] [PubMed] [Google Scholar]
- 9.Jansen H-J, Grenier D, Hoeven JS. Characterization of immunoglobulin G-degrading proteases of Prevotella intermedia and Prevotella nigrescens. Oral Microbiol. Immunol. 1995;10:138–145. doi: 10.1111/j.1399-302X.1995.tb00134.x. [DOI] [PubMed] [Google Scholar]
- 10.Ulrich M, et al. Relative contribution of Prevotella intermedia and Pseudomonas aeruginosa to lung pathology in airways of patients with cystic fibrosis. Thorax. 2010;65:978–984. doi: 10.1136/thx.2010.137745. [DOI] [PubMed] [Google Scholar]
- 11.Duan K, Dammel C, Stein J, Rabin H, Surette MG. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 2003;50:1477–1491. doi: 10.1046/j.1365-2958.2003.03803.x. [DOI] [PubMed] [Google Scholar]
- 12.Field TR, Sibley CD, Parkins MD, Rabin HR, Surette MG. The genus Prevotella in cystic fibrosis airways. Anaerobe. 2010;16:337–344. doi: 10.1016/j.anaerobe.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Mirković B, et al. The role of short-chain fatty acids, produced by anaerobic bacteria, in the cystic fibrosis airway. Am. J. Respir. Crit. Care Med. 2015;192:1314–1324. doi: 10.1164/rccm.201505-0943OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn R, et al. Metabolomics of pulmonary exacerbations reveals the personalized nature of cystic fibrosis disease. PeerJ. 2016;4:e2174. doi: 10.7717/peerj.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherrard LJ, et al. Antibiotic resistance in Prevotella species isolated from patients with cystic fibrosis. J. Antimicrob. Chemother. 2013;68:2369–2374. doi: 10.1093/jac/dkt191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherrard LJ, et al. Production of extended-spectrum β -lactamases and the potential indirect pathogenic role of Prevotella isolates from the cystic fibrosis respiratory microbiota. Int. J. Antimicrob. Agents. 2016;47:140–145. doi: 10.1016/j.ijantimicag.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zemanick ET, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur. Respir. J. 2017;50:1700832. doi: 10.1183/13993003.00832-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zemanick ET, et al. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS ONE. 2013;8:e62917. doi: 10.1371/journal.pone.0062917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zemanick ET, et al. Assessment of airway microbiota and inflammation in cystic fibrosis using multiple sampling methods. Ann. Am. Thorac. Soc. 2015;12:221–229. doi: 10.1513/AnnalsATS.201407-310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernarde C, et al. Impact of the CFTR-potentiator Ivacaftor on airway microbiota in cystic fibrosis patients carrying a G551D mutation. PLoS ONE. 2015;10:e0124124. doi: 10.1371/journal.pone.0124124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chmiel JF, et al. Antibiotic management of lung infections in cystic fibrosis. II. Nontuberculous mycobacteria, anaerobic bacteria, and fungi. Ann. Am. Thorac. Soc. 2014;11:1298–1306. doi: 10.1513/AnnalsATS.201405-203AS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong LK, Barry AL, Horgan SM. Comparison of six different criteria for judging the acceptability of sputum specimens. J. Clin. Microbiol. 1982;16:627–631. doi: 10.1128/JCM.16.4.627-631.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray PR, Washington JA. Microscopic and bacteriologic analysis of expectorated sputum. Mayo Clin. Proc. 1975;50:339–344. [PubMed] [Google Scholar]
- 24.Lee TWR, Brownlee KG, Conway SP, Denton M, Littlewood JM. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J. Cyst. Fibros. 2003;2:29–34. doi: 10.1016/S1569-1993(02)00141-8. [DOI] [PubMed] [Google Scholar]
- 25.Konstan MW, Wagener JS, VanDevanter DR. Characterizing aggressiveness and predicting future progression of CF lung disease. J. Cyst. Fibros. 2009;8:15–19. doi: 10.1016/S1569-1993(09)60006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coburn B, et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 2015;5:10241–10252. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stressmann FA, et al. Analysis of the bacterial communities present in lungs of patients with cystic fibrosis from American and British centers. J. Clin. Microbiol. 2011;49:281–291. doi: 10.1128/JCM.01650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Einarsson GG, et al. Community analysis and co-occurrence patterns in airway microbial communities during health and disease. ERJ Open Res. 2019;5:00128–02017. doi: 10.1183/23120541.00128-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guilloux C-A, Lamoureux C, Héry-Arnaud G. Anaerobic bacteria, the unknown members of the lung microbiota. Med. Sci. 2018;34:253–260. doi: 10.1051/medsci/20183403014. [DOI] [PubMed] [Google Scholar]
- 30.van der Gast CJ, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011;5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamoureux C, Guilloux C-A, Beauruelle C, Jolivet-Gougeon A, Héry-Arnaud G. Anaerobes in cystic fibrosis patients’ airways. Crit. Rev. Microbiol. 2019;45:103–117. doi: 10.1080/1040841X.2018.1549019. [DOI] [PubMed] [Google Scholar]
- 32.Layeghifard M, et al. Microbiome networks and change-point analysis reveal key community changes associated with cystic fibrosis pulmonary exacerbations. NPJ Biofilms Microbiomes. 2019;5:4. doi: 10.1038/s41522-018-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones AM. Anaerobic bacteria in cystic fibrosis: pathogens or harmless commensals? Thorax. 2011;66:558–559. doi: 10.1136/thx.2010.157875. [DOI] [PubMed] [Google Scholar]
- 34.Françoise A, Héry-Arnaud G. The microbiome in cystic fibrosis pulmonary disease. Genes. 2020;11:536. doi: 10.3390/genes11050536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JS, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paganin P, et al. Changes in cystic fibrosis airway microbial community associated with a severe decline in lung function. PLoS ONE. 2015;10:e0124348. doi: 10.1371/journal.pone.0124348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherrard LJ, et al. Assessment of stability and fluctuations of cultured lower airway bacterial communities in people with cystic fibrosis. J. Cyst. Fibros. 2019;18:808–816. doi: 10.1016/j.jcf.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sibley CD, et al. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS ONE. 2011;6:e22702. doi: 10.1371/journal.pone.0022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keravec M, et al. Porphyromonas, a potential predictive biomarker of Pseudomonas aeruginosa pulmonary infection in cystic fibrosis. BMJ Open Respir. Res. 2019;6:e000374. doi: 10.1136/bmjresp-2018-000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ibironke O, et al. Species-level evaluation of the human respiratory microbiome. GigaScience. 2020;9:giaa038. doi: 10.1093/gigascience/giaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox MJ, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS ONE. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kent L, et al. Lung clearance index: Evidence for use in clinical trials in cystic fibrosis. J. Cyst. Fibros. 2014;13:123–138. doi: 10.1016/j.jcf.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Chalmers NI, Palmer RJ, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 2008;190:8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gall A, et al. Bacterial composition of the human upper gastrointestinal tract microbiome is dynamic and associated with genomic instability in a Barrett’s esophagus cohort. PLoS ONE. 2015;10:e0129055. doi: 10.1371/journal.pone.0129055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki N, Yoshida A, Saito T, Kawada M, Nakano Y. Quantitative microbiological study of subgingival plaque by real-time PCR shows correlation between levels of Tannerella forsythensis and Fusobacterium spp. J. Clin. Microbiol. 2004;42:2255–2257. doi: 10.1128/JCM.42.5.2255-2257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitman WB, Rainey F, Kämpfer P, et al. Bergey’s Manual of Systematics of Archaea and Bacteria. London: Wiley; 2015. [Google Scholar]
- 47.Li Y, et al. Application of MALDI-TOF MS to rapid identification of anaerobic bacteria. BMC Infect. Dis. 2019;19:941. doi: 10.1186/s12879-019-4584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.