Abstract
During the COVID‐19 pandemic, dermatologists reported an array of different cutaneous manifestations of the disease. It is challenging to discriminate COVID‐19‐related cutaneous manifestations from other closely resembling skin lesions. The aim of this study was to generate and evaluate a novel CNN (Convolutional Neural Network) ensemble architecture for detection of COVID‐19‐associated skin lesions from clinical images. An ensemble model of three different CNN‐based algorithms was trained with clinical images of skin lesions from confirmed COVID‐19 positive patients, healthy controls as well as 18 other common skin conditions, which included close mimics of COVID‐19 skin lesions such as urticaria, varicella, pityriasis rosea, herpes zoster, bullous pemphigoid and psoriasis. The multi‐class model demonstrated an overall top‐1 accuracy of 86.7% for all 20 diseases. The sensitivity and specificity of COVID‐19‐rash detection were found to be 84.2 ± 5.1% and 99.5 ± 0.2%, respectively. The positive predictive value, NPV and area under curve values for COVID‐19‐rash were 88.0 ± 5.6%, 99.4 ± 0.2% and 0.97 ± 0.25, respectively. The binary classifier had a mean sensitivity, specificity and accuracy of 76.81 ± 6.25%, 99.77 ± 0.14% and 98.91 ± 0.17%, respectively for COVID‐19 rash. The model was robust in detection of all skin lesions on both white and skin of color, although only a few images of COVID‐19‐associated skin lesions from skin of color were available. To our best knowledge, this is the first machine learning‐based study for automated detection of COVID‐19 based on skin images and may provide a useful decision support tool for physicians to optimize contact‐free COVID‐19 triage, differential diagnosis of skin lesions and patient care.
Keywords: artificial intelligence, COVID‐19 cutaneous manifestations, COVID‐19 skin lesions, deep learning
1. INTRODUCTION
The clinical spectrum of COVID‐19 is very heterogeneous ranging from mild symptoms to severe symptoms like respiratory failure or multi‐organ dysfunction. Dermatologists in Europe, particularly in Italy, Spain and France, who were on the frontlines of managing the deluge of hospitalized COVID‐19‐positive patients, reported various skin rashes that appeared to be correlated with the disease. 1 , 2 , 3 Literature reports show, however, a great deal of variation in the skin manifestations, their latency periods and the percentage of patients who develop these. For instance, in the first study to draw attention to skin lesions, nearly 20.4% (18 of 88) of patients, with a confirmed diagnosis of COVID‐19, developed cutaneous manifestations. 4 This number contrasts with 0.2%, 7.25%, and 12.7% of cases that developed skin abnormalities in a study from China and two studies from India, respectively. 5 , 6 , 7 Subsequently, various reports of skin manifestations in both adults (with severe forms of COVID‐19) and younger paucisymptomatic patients have been published. 8 , 9 Marzanoet al. divided the reported skin lesions into six main clinical patterns: (a) urticarial rash, (b) erythematous/maculopapular/morbilliform rash, (c) papulovesicular exanthem, (d) chilblain‐like acral pattern, (e) livedo reticularis/racemosa‐like pattern, (f) purpuric “vasculitic” pattern. 8 The scientific understanding of COVID‐19 and associated dermatological symptoms is currently evolving and the diagnostic and/or prognostic value of these lesions is a subject of exploration, but researchers agree that observation of cutaneous symptoms, without another explanation, should prompt confirmatory testing. 1 , 9 , 10 To collate cases of dermatoses in COVID‐19‐positive and COVID‐19‐suspected cases from a global network and inform doctors on the frontlines, the American Academy of Dermatology (AAD), in collaboration with the International League of Dermatologic Societies (ILDS), has launched an online COVID‐19 dermatology registry. 9 Diagnosing skin manifestations in patients with COVID‐19 remains a challenge even for dermatologists because it is unclear whether the skin lesions are related to the virus. 10 A decision support tool that assists physicians in differentiating skin lesions known to be associated with COVID‐19 from incidental skin findings may be helpful in disease management.
In this study, we present a novel machine‐learning model for the identification of COVID‐19‐associated skin manifestations based on a single clinical image of skin lesions on both Caucasian (white) and Indian skin (skin of color). The model can discriminate COVID‐19‐associated skin lesions from normal skin and 18 common skin conditions including COVID‐19 mimics. Although the utility of this algorithm in COVID‐19 screening is limited by the observation that only a fraction of COVID‐19 patients exhibit cutaneous manifestations and lesions often appear late in the infection course, this proof‐of‐concept study shows the potential for the model to be used as a high‐level diagnostic aid in the differential diagnosis of skin lesions in COVID‐19‐suspected patients.
2. MATERIALS AND METHODS
2.1. Model architectures
We implemented a neural architecture that was an ensemble of three models‐ Densenet‐161, 11 SeresNext‐101 12 and EfficientNet‐B4, 13 trained on the same training set with predictions combined at the corresponding final output layers. These models were selected based on their best individual performances across their architecture families, limited by the available GPU machine (2 gtx‐1080ti cards in multi node training mode). The batch sizes were 32 for Densenet‐161, 24 for EfficientNet‐B4 and 32 for SeresNext‐101. The number of layers were 809 for Densenet‐161, 470 for EfficientNet‐B4 and 2724 for SeresNext‐101.The number of epoch were 70. We used pre trained weights and fine‐tuned them on our dataset. All training and testing images were segmented, so a bounding box surrounded the lesion of interest. These images were split into five equal parts (folds) and five iterations of training and validation were performed so that a different fold of the data was held‐out for validation, while the remaining four‐folds were used for learning. A five‐fold cross‐validation test ensured that each image had a chance of being validated.
We initially experimented with just Densenet‐161 for fine‐tuning the model parameters such as loss functions and augmentation, and inference schemes. After we achieved the best possible configuration, we scaled these to SeresNext‐101 and EfficientNet‐B4. A schematic of the inference and training pipelines are shown in Figure 1A,B. In the training stage, each of the three models was modified at the final output layer and trained separately with a uniquely optimized augmentation and preprocessing pipeline (the training schematic refers to Densenet‐161, but is representative of all three models). Image data augmentation is a technique commonly employed to artificially expand the size of a training dataset by creating modified versions of images in the dataset. Another benefit of augmentation is to generate a model that is generalizable to different kinds of images. In the inference stage, we used six replicas of the source image to be inferred, with six unique, pre‐selected augmentation schemes applied on each of them and passed them through six copies of each of the three models to get six unique prediction scores from each of those three modes. These pre‐selected augmentation schemes were derived through rigorous experiments and were attuned to clinical skin images. We combined the predictions of each of these unique replicas to get a single prediction per model, which were again combined using max‐pooling across all three models to arrive at the final output score.
FIGURE 1.
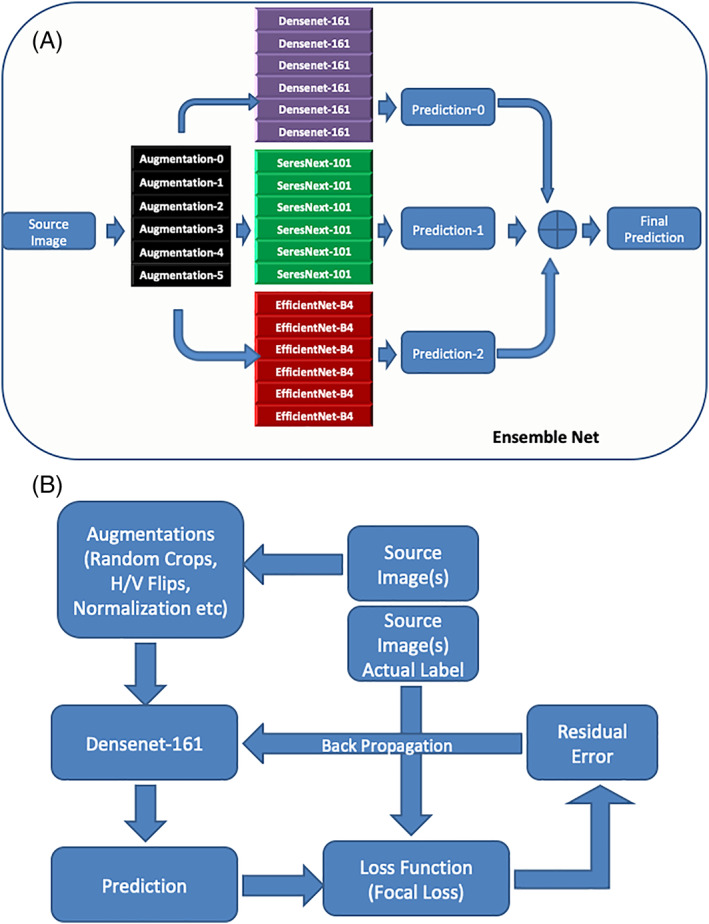
A, depicts a schematic of the inference pipeline. B, depicts the training pipelines in the CNN ensemble architecture. Note, the training schematic refers to Densenet‐161, but is representative of all three models
2.2. Disease classes
In this study, a convolutional neural network (CNN)‐based algorithm was trained with clinical images of skin lesions from confirmed COVID‐19‐positive patients, healthy controls as well as 18 other common skin conditions, that included COVID‐19‐rash disease differentials described in the literature, such as urticaria, 10 , 14 chickenpox, 4 , 15 herpes zoster, 1 pityriasis rosea, 1 bullous pemphigoid, psoriasis, 1 and fixed drug eruption. 1 The remaining diseases were acne, lichen planus, normal skin, pemphigus, pityriasis versicolor, rosacea, secondary syphilis, tinea corporis, cruris or faciei, tinea manuum, tinea pedis, and paronychia. The COVID‐19 associated skin lesions, used for training and testing, were further divided into 12 categories based on their morphology (Supplementary Table 1).
2.3. Dataset
Our complete clinical image dataset was comprised of 7053 total, pre‐augmentation images, of which 2904 were from Indian patients and 4141 were from Caucasian patients (Table 1). Our COVID‐19 dataset was acquired from publicly available literature and private data and consisted of 259 clinical images from 129 males and 130 females with RT‐PCR confirmed COVID‐19 disease. The sources for public databases are shown in Supplementary Table 2. The median age of patients was 39 years and the average age was 42 years. We also collected images from suspected COVID‐19 patients (who were described in the literature as having either close contact with positive patients or with COVID‐19‐like symptoms). These 177 images were from 99 males and 101 females with a median age of 20. Our training set only included images from RT‐PCR‐positive COVID‐19 patients. But we generated two separate test sets. The first test set only included images from COVID‐19‐confirmed patients, while a second set was comprised of only images from COVID‐19‐suspected patients. Informed consent was received from patients whose images were acquired by direct communication with dermatologists for our private dataset. All private patients underwent confirmation of COVID‐19 diagnosis by RT‐PCR testing.
TABLE 1.
Number of images (pre‐augmentation) per skin disease class from public databases (Caucasian skin) and private database (Indian skin)
| Disease class | Total | Indian | Caucasian |
|---|---|---|---|
| Acne | 576 | 337 | 239 |
| Bullous pemphigoid | 155 | 54 | 101 |
| Chicken pox | 191 | 77 | 114 |
| COVID‐19 confirmed | 259 | 12 | 247 |
| COVID‐19 suspected | 177 | 0 | 177 |
| Fixed drug eruption | 165 | 65 | 100 |
| Herpes zoster | 259 | 98 | 153 |
| Impetigo and Pyodermas | 475 | 198 | 277 |
| Lichen planus | 427 | 205 | 222 |
| Normal skin | 339 | 80 | 259 |
| Paronychia | 104 | 60 | 44 |
| Pemphigus | 247 | 99 | 148 |
| Pityriasis rosea | 257 | 95 | 162 |
| Pityriasis versicolor | 179 | 67 | 112 |
| Psoriasis | 897 | 452 | 445 |
| Rosacea | 442 | 184 | 258 |
| Secondary syphilis | 199 | 100 | 99 |
| Tinea cruris, corporis or faciei | 1093 | 603 | 490 |
| Tinea manuum | 131 | 1 | 130 |
| Tinea pedis | 262 | 0 | 262 |
| Urticaria | 219 | 117 | 102 |
| Grand total | 7053 | 2904 | 4141 |
2.4. Determination of model performance
We performed both a binary analysis of COVID‐19 rash and non‐COVID‐19 skin lesions as well as a multi‐class analysis of COVID‐19 rash, normal skin and 18 other skin diseases. The model was evaluated using performance metrics of sensitivity, specificity, accuracy. positive predictive values (PPV), negative predictive values (NPV) and area under the curve (AUC). Mean values with standard deviation were shown for disease‐specific sensitivities, specificities, PPV, NPV, and overall top‐1 and top‐3 sensitivity/accuracy of the model were calculated. The output of the model in binary analysis was either COVID‐19 rash or Non‐COVID skin. In the multi‐class analysis, the outputs were three predictions ordered by their descending probabilities. If the first prediction was correct, it was included in top‐1 sensitivity calculations. For top‐3 sensitivity, the correct class only needed to be in the top three predicted classes to be counted. Receiver operating characteristics (ROC) curves were plotted using probability scores for each of the disease classes by varying the cutoff threshold as described by Pedregosa et al. 16
3. RESULTS
The performance of the CNN ensemble model for binary classification of skin lesions showed a sensitivity of 76.81±6.25 %, specificity of 99.77±0.14, accuracy of 98.91±0.17%, PPV of 94.58±5.00% and NPV of 97.57±3.41% for all COVID‐19 confirmed skin lesions (Table 2). The diagnosis of 20 skin diseases showed a top‐1 overall sensitivity of 87.65 ± 0.01% and top‐3 sensitivity of 96.72±0.19% (Table 3). High specificities and NPV were observed for all the 20 disease classes (Table 3). Laboratory confirmed COVID‐19‐rash was diagnosed at a mean top‐1 sensitivity of 84.15±5.12%, top‐3 sensitivity 95.74±4.89% and top‐1 specificity of 99.53±0.24%, PPV of 88.01±5.59%, NPV of 99.38±0.20, and AUC of 0.97. Notably, the top‐1 and top‐3 sensitivity of COVID‐19‐suspected lesion was significantly lower at 63.26±1.26% and 88.24±1.69%, respectively. The ROC plots for COVID‐19‐confirmed rash and other skin conditions are shown in Supplementary Figure 1.
TABLE 2.
The sensitivity, PPV and NPV of binary classification of skin lesions into COVID‐19 and non‐COVID‐19 categories from 5‐fold validation are depicted below
| COVID‐19‐confirmed skin lesions | |||||
|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Accuracy (%) | PPV (%) | NPV (%) | |
| Fold 0 | 78.18 | 99.71 | 98.87 | 91.49 | 91.49 |
| Fold 1 | 72.55 | 99.77 | 98.76 | 92.50 | 98.94 |
| Fold 2 | 68.63 | 100.00 | 98.83 | 100.00 | 98.80 |
| Fold 3 | 80.39 | 99.62 | 98.90 | 89.13 | 99.24 |
| Fold 4 | 84.31 | 99.77 | 99.19 | 99.77 | 99.39 |
| Mean ± SD | 76.81 ± 6.25 | 99.77 ± 0.14 | 98.91 ± 0.17 | 94.58 ± 5.00 | 97.58 ± 3.41 |
TABLE 3.
The disease‐specific top‐1 sensitivity, top‐3 sensitivity, specificity, PPV, NPV and AUC from 5‐fold validation are depicted below. The overall top‐1 accuracy and top‐3 accuracy for COVID‐19‐suspected lesions is also shown. NA: not available, ND: not determined
| Disease class | Top1 Sensitivity (mean ± SD) | Top3 Sensitivity (mean ± SD) | Specificity (mean ± SD) | PPV (mean ± SD) | NPV (mean ± SD) | AUC (mean) |
|---|---|---|---|---|---|---|
| Acne | 92.35 ± 2.65 | 97.91 ± 1.91 | 99.16 ± 0.23 | 91.02 ± 2.14 | 99.30 ± 0.24 | 0.99 |
| Bullous pemphigoid | 80.65 ± 7.90 | 93.55 ± 4.56 | 99.70 ± 0.14 | 86.58 ± 4.62 | 99.56 ± 0.18 | 0.96 |
| COVID‐19 confirmed | 84.15 ± 5.12 | 95.74 ± 4.89 | 99.53 ± 0.24 | 88.01 ± 5.59 | 99.38 ± 0.20 | 0.97 |
| Chicken pox | 79.60 ± 4.97 | 90.07 ± 4.26 | 99.52 ± 0.26 | 83.33 ± 6.73 | 99.42 ± 0.14 | 0.94 |
| Fixed drug eruption | 86.67 ± 10.19 | 92.73 ± 7.30 | 99.67 ± 0.24 | 87.00 ± 9.06 | 99.67 ± 0.25 | 0.96 |
| Herpes zoster | 86.47 ± 3.70 | 95.35 ± 2.26 | 99.41 ± 0.18 | 85.28 ± 3.73 | 99.47 ± 0.14 | 0.97 |
| Impetigo & Pyodermas | 88.63 ± 1.73 | 95.37 ± 3.12 | 99.23 ± 0.27 | 89.67 ± 3.22 | 99.16 ± 0.12 | 0.97 |
| Lichen planus | 81.26 ± 3.23 | 96.26 ± 1.90 | 99.02 ± 0.24 | 84.77 ± 3.15 | 98.76 ± 0.21 | 0.97 |
| Normal skin | 98.24 ± 1.21 | 98.81 ± 1.25 | 99.72 ± 0.07 | 94.88 ± 1.22 | 99.91 ± 0.06 | 0.99 |
| Paronychia | 94.33 ± 1.49 | 99.00 ± 2.24 | 99.85 ± 0.07 | 90.97 ± 3.96 | 99.91 ± 0.03 | 0.99 |
| Pemphigus | 77.73 ± 6.62 | 94.32 ± 2.69 | 99.31 ± 0.14 | 80.72 ± 3.37 | 99.17 ± 0.24 | 0.96 |
| Pityriasis rosea | 84.08 ± 4.45 | 96.50 ± 1.63 | 99.18 ± 0.27 | 80.32 ± 4.83 | 99.38 ± 0.17 | 0.98 |
| Pityriasis versicolor | 83.66 ± 4.61 | 95.49 ± 2.61 | 99.66 ± 0.19 | 86.87 ± 7.20 | 99.57 ± 0.11 | 0.97 |
| Psoriasis | 85.39 ± 2.23 | 96.99 ± 1.09 | 97.91 ± 0.28 | 86.01 ± 1.45 | 97.81 ± 0.33 | 0.97 |
| Rosacea | 95.72 ± 3.10 | 99.10 ± 0.93 | 99.57 ± 0.17 | 93.83 ± 2.42 | 99.71 ± 0.21 | 0.99 |
| Secondary syphilis | 88.40 ± 8.67 | 95.95 ± 3.91 | 99.58 ± 0.22 | 86.49 ± 6.51 | 99.66 ± 0.25 | 0.98 |
| Tinea cruris, corporis or faciei | 90.03 ± 1.35 | 98.35 ± 0.53 | 97.94 ± 0.38 | 89.23 ± 1.80 | 98.11 ± 0.26 | 0.98 |
| Tineamanuum | 86.24 ± 5.88 | 96.98 ± 4.14 | 99.70 ± 0.14 | 85.16 ± 6.42 | 99.73 ± 0.11 | 0.98 |
| Tinea pedis | 91.60 ± 4.44 | 98.48 ± 0.85 | 99.46 ± 0.11 | 86.97 ± 2.50 | 99.67 ± 0.18 | 0.99 |
| Urticaria | 79.47 ± 5.05 | 94.96 ± 4.17 | 99.50 ± 0.12 | 84.04 ± 3.82 | 99.33 ± 0.17 | 0.97 |
| Overall accuracy | 87.65 ± 0.00 | 96.72 ± 0.19 | NA | NA | NA | NA |
| COVID‐19 suspected | 63.26 ± 1.26 | 88.24 ± 1.69 | ND | ND | ND | ND |
A representative confusion matrix from fold 0 is shown in Figure 2A. Examination of the confusion matrices from all five‐fold experiments showed that the number of false‐positives and false‐negatives for COVID‐19‐rash are relatively few at 39 instances (0.59% of all COVID‐19‐negative images) and 30 instances (10.7% of all COVID‐19‐positive lesions), respectively (Figure 2B). The false‐positives are derived mainly from urticaria, described as a close disease differential of COVID‐19‐rash by dermatologists. 1 , 10 , 14 Interestingly, there were also a few false‐positive cases from chickenpox, psoriasis, pityriasis rosea, all of which have been described previously in the literature as resembling COVID‐19‐rash. 1 , 8 , 15 , 17 Similarly, the false‐negative cases stem largely from tinea corporis, cruris or faciei, acne and urticaria (Figure 2B). These lesions can be discriminated further by analyses of patients' history.
FIGURE 2.
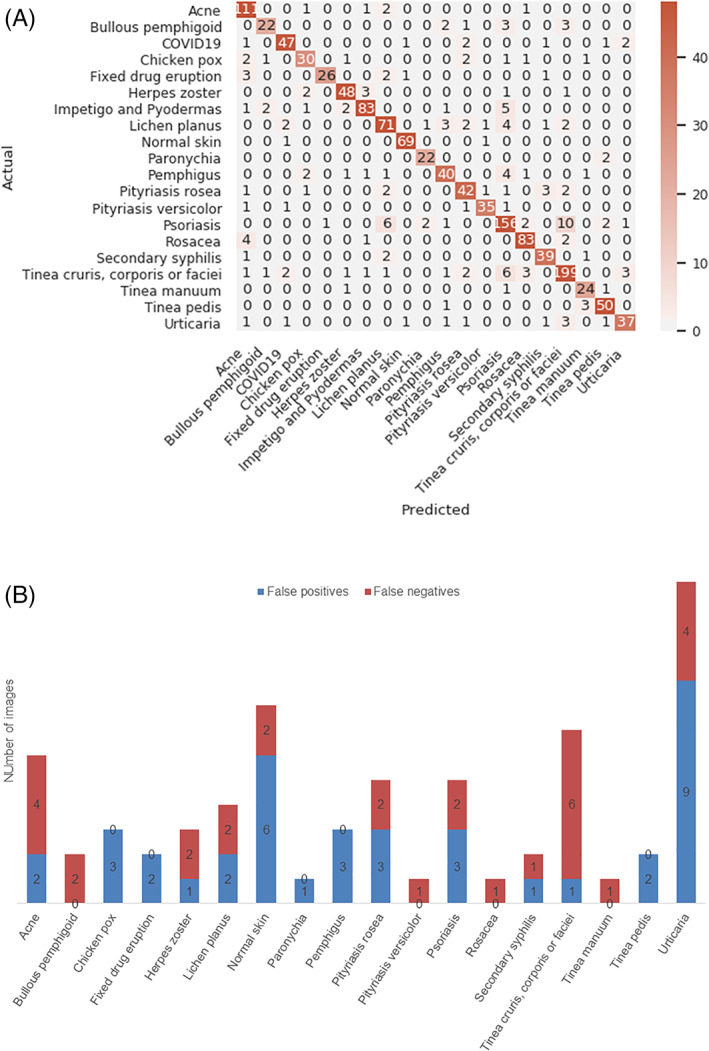
A, shows a representative top‐1 confusion matrix for all 20 skin conditions. The numbers indicate image numbers. The y‐axis represents the actual disease class and x‐axis represents the predicted disease class for fold 0. The color gradient depicts the degree of correct predictions. B, shows the disease categories that represent the false‐positive and false‐negative cases in all 5‐folds for COVID‐19‐rash
4. DISCUSSION
In this study, we included images from COVID‐19‐confirmed and suspected patients that fall into the six main clinical categories of urticarial rash, maculopapular rash, vesicular eruptions, chilblain‐like rash, livedo and purpura‐like rash and found high sensitivity, specificity, NPV and AUC values for COVID‐19‐rash in an in‐silico validation study across 20 skin conditions (Supplementary Table 1 and Table 3). The utility of this algorithm is highlighted by the observation that cutaneous COVID‐19 is differentiated from non‐COVID skin lesions with a sensitivity of ~77% and accuracy of ~99%. Moreover, differential diagnosis of skin lesions is feasible as COVID‐19 rash appears as a correct prediction in approximately 96% of cases if the first three disease differentials are shown as an output to users (Table 3). Our algorithms have the potential to improve with more images and can be extended to include other disease differentials. It is also important to consider that our dataset is composed of lesions on both white and dark skin. As of yet, very few case studies have reported skin lesions in COVID‐19 patients from the Indian subcontinent. 6 , 7 This work was constrained by the small number of images of COVID‐19 lesions from skin‐of‐color populations, used for training as well as for validation and the model is likely biased towards white skin (Table 1). Another limitation was that the validation of the algorithm could not be done in actual clinical settings. In the future, this model could be enhanced by combining image analysis with automated patient history analysis to provide a more accurate, integrated inference. 18
In patients where COVID‐19 skin lesions manifest before systemic symptoms, machine‐learning (ML) algorithms may help in diagnosing these lesions and quarantine of patients with suspected SARS‐CoV‐2 infection. In other cases, where COVID‐19 skin lesions occur alongside systemic symptoms but before testing results, ML‐based detection of skin lesions may serve as an additional diagnostic clue and aid in early intervention and quarantine. In addition, certain skin rashes may also be useful for disease prognosis, if some correlation between the lesion and disease severity can be established via large‐scale studies.
A particularly interesting finding from a large‐scale prospective study, conducted on 375 patients in Spain, was that while vesicular eruptions appeared early in the course of the disease, a pseudo‐chilblain pattern frequently appeared late in the evolution of the COVID‐19 disease, and the rest seemed to coincide with other classic symptoms of COVID‐19. 1 The authors hypothesized that pseudo‐chilblains (also known as COVID‐19‐associated perniosis) and vesicular lesions may be useful as indicators of disease. Similar to the timing of skin lesions, the association of skin manifestations with illness severity is also difficult to ascertain. 19 Casas et al. noted that the severity of COVID‐19 showed a gradient from less severe in pseudo‐chilblain lesions to more severe disease in livedoid and necrotic groups. 2 A related finding was that pseudo‐chilblains affected a greater proportion of younger patients, which has been corroborated by other studies in COVID‐19‐confirmed individuals and COVID‐19‐suspected young patients. 2 , 3 , 20
Another compelling suggestion was that since these chilblains were not associated with severe disease, they could be useful for prognosis. 1 , 3 Infact, the PCR‐negative status of many reported patients has been tied to the late appearance of these lesions. 3 , 20 , 21 Other groups have refuted the association of pseudo‐chilblains with COVID‐19 disease. 22 , 23 Nonetheless, it is likely that cutaneous lesions have been underestimated as they are not as pressing as other symptoms, are either self‐limiting or with relatively short duration and not part of the restrictive screening policies of many countries. 1
While the clinical value of these patterns for diagnosis needs to be confirmed, many investigators have raised the important possibility that these cutaneous manifestations may be helpful for remote and contact‐free diagnosis in areas where testing is scarce. 1 , 10 , 14 , 24 , 25 In addition, certain manifestations that predict a more severe disease course may guide early, aggressive intervention. 24 To answer the question if skin lesions can inform treatment regimens, Young et al. proposed documenting cutaneous abnormalities present at diagnosis and during the course of COVID‐19 using photographs captured by mobile phones. 24 We envision that algorithms like ours could be integrated into smartphone apps to provide a decision support tool for dermatologists to recognize COVID‐19‐related skin lesions, guide patient care and research efforts. 18
CONFLICT OF INTEREST
Rashi Pangti and Somesh Gupta have no financial interests to declare. Jyoti Mathur, Sharad Kumar and Vikas Chouhan own stock in Nurithm Labs Private Limited. The COVID‐19 skin project is a non‐commercial endeavor, purely for research collaboration between Nurithm Labs and AIIMS hospital.
AUTHOR CONTRIBUTIONS
Jyoti Mathur, Vikas Chouhan, Somesh Gupta and Sharad Kumar conceived the study and designed the experiments; Jyoti Mathur, Rashi Pangti and Somesh Gupta curated patient data; Vikas Chouhan performed experiments; Jyoti Mathur and Vikas Chouhan performed data analysis and Jyoti Mathur, Vikas Chouhan, Rashi Pangti and Somesh Gupta wrote the manuscript.
Supporting information
Supplementary Table 1 This table shows the numbers of images from COVID‐19 confirmed patients and their respective subcategories, based on lesion morphology.
Supplementary Table 2 Supporting information
Supplementary Figure 1 Representative ROC plots for all 20 skin conditions showing the AUC values from fold 0
Mathur J, Chouhan V, Pangti R, Kumar S, Gupta S. A convolutional neural network architecture for the recognition of cutaneous manifestations of COVID‐19. Dermatologic Therapy. 2021;34:e14902. 10.1111/dth.14902
Jyoti Mathur and Vikas Chouhan are equal contributors
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are provided in the supplementary files which are submitted with the manuscript. Rest of the data is available from the corresponding author upon reasonable request.
REFERENCES
- 1. Galván Casas C, Català A, Carretero Hernández G, et al. Classification of the cutaneous manifestations of COVID‐19: a rapid prospective nationwide consensus study in Spain with 375 cases. Br J Dermatol. 2020;183(1):71‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Masson A, Bouaziz JD, Sulimovic L, et al. Chilblains is a common cutaneous finding during the COVID‐19 pandemic: a retrospective nationwide study from France. J Am Acad Dermatol. 2020;83(2):667‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Freeman EE, McMahon DE, Lipoff JB, et al. Pernio‐like skin lesions associated with COVID‐19: a case series of 318 patients from 8 countries. J Am Acad Dermatol. 2020;83(2):486‐492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Recalcati S. Cutaneous manifestations in COVID‐19: a first perspective. J Eur Acad Dermatol Venereol. 2020;34(5):e212‐e213. [DOI] [PubMed] [Google Scholar]
- 5. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pangti R, Gupta S, Nischal N, Trikha A. Recognizable vascular skin manifestations of SARS‐CoV‐2 (COVID‐19) infection are uncommon in patients with darker skin phototypes. Clin Exp Dermatol. 2021;46(1):180‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dalal A, Jakhar D, Agarwal V, Beniwal R. Dermatological findings in SARS‐CoV‐2 positive patients: an observational study from North India. Dermatol Ther. 2020;33(6):e13849. 10.1111/dth.13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marzano AV, Cassano N, Genovese G, Moltrasio C, Vena GA. Cutaneous manifestations in patients with COVID‐19: a preliminary review of an emerging issue. Br J Dermatol. 2020;183:431‐442. 10.1111/bjd.19264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Freeman EE, McMahon DE, Fitzgerald ME, et al. The American Academy of Dermatology COVID‐19 registry: crowdsourcing dermatology in the age of COVID‐19. J Am Acad Dermatol. 2020;83(2):509‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Damme C, Berlingin E, Saussez S, Accaputo O. Acute urticaria with pyrexia as the first manifestations of a COVID‐19 infection. J Eur Acad Dermatol Venereol. 2020;34(7):e300‐e301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang G, Liu Z, Weinberger KQ. Densely connected convolutional networks. IEEE Conf Comput Vision Pattern Recog (CVPR). 2017;2261‐2269. 10.1109/CVPR.2017.243. [DOI] [Google Scholar]
- 12. Hu J, Shen L, Albanie S, et al. Squeeze‐and‐excitation networks. IEEE Conf Comput Vision Pattern Recog. 2018:7132–7141. 10.1109/CVPR.2018.00745. [DOI] [PubMed] [Google Scholar]
- 13. Tan M, Le QV. Efficientnet: rethinking model scaling for convolutional neural networks. 2019; arXiv:1905.11946.
- 14. Henry D, Ackerman M, Sancelme E, et al. Urticarial eruption in COVID‐19 infection. J Eur Acad Dermatol Venereol. 2020;34(6):e244‐e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Genovese G, Colonna C, Marzano AV. Varicella‐like exanthem associated with COVID‐19 in an 8‐year‐old girl: a diagnostic clue? Pediatr Dermatol. 2020;37(3):435‐436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit‐learn: machine learning in python. J Machine Learn Res. 2011;12:2825‐2830. [Google Scholar]
- 17. Sachdeva M, Gianotti R, Shah M, et al. Cutaneous manifestations of COVID‐19: report of three cases and a review of literature. J Dermatol Sci. 2020;98(2):75‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pangti R, Mathur J, Chouhan V, et al. A machine learning‐based, decision support, mobile phone application for diagnosis of common dermatological diseases. J Eur Acad Dermatol Venereol. 2021;35(2):536‐545. [DOI] [PubMed] [Google Scholar]
- 19. Suchonwanit P, Leerunyakul K, Kositkuljorn C. Cutaneous manifestations in COVID‐19: lessons learned from current evidence. J Am Acad Dermatol. 2020;83(1):e57‐e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernandez‐Nieto D, Jimenez‐Cauhe J, Suarez‐Valle A, et al. Characterization of acute acral skin lesions in nonhospitalized patients: a case series of 132 patients during the COVID‐19 outbreak. J Am Acad Dermatol. 2020;83(1):e61‐e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hubiche T, Le Duff F, Chiaverini C, et al. Negative SARS‐CoV‐2 PCR in patients with chilblain‐like lesions. 2020. Lancet Infect Dis. 2020;20(20):30518‐30511. 10.1016/S1473-3099(20)30518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Torres‐Navarro I, Abril‐Pérez C, Roca‐Ginés J, Sánchez‐Arráez J, Botella‐Estrada R, Évole‐Buselli M. Comment on 'Two cases of COVID‐19 presenting with a clinical picture resembling chilblains: first report from the Middle East': pernio unrelated to COVID‐19. Clin Exp Dermatol. 2020;45(6):752‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herman A, Peeters C, Verroken A, et al. Evaluation of chilblains as a manifestation of the COVID‐19 pandemic. JAMA Dermatol. 2020;9:e202368. 10.1001/jamadermatol.2020.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young S, Fernandez AP. Skin manifestations of COVID‐19. Cleve Clin J Med. 2020. 10.3949/ccjm.87a.ccc031. [DOI] [PubMed] [Google Scholar]
- 25. Amatore F, Macagno N, Mailhe M, et al. SARS‐CoV‐2 infection presenting as a febrile rash. J Eur Acad Dermatol Venereol. 2020;34(7):e304‐e306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 This table shows the numbers of images from COVID‐19 confirmed patients and their respective subcategories, based on lesion morphology.
Supplementary Table 2 Supporting information
Supplementary Figure 1 Representative ROC plots for all 20 skin conditions showing the AUC values from fold 0
Data Availability Statement
The data that supports the findings of this study are provided in the supplementary files which are submitted with the manuscript. Rest of the data is available from the corresponding author upon reasonable request.


