Abstract
Background
The emergence and spread of multidrug‐resistant organisms (MDRO) present a threat to human and animal health.
Objectives
To assess acquisition, prevalence of and risk factors for MDRO carriage in dogs and cats presented to veterinary clinics or practices in Switzerland.
Animals
Privately owned dogs (n = 183) and cats (n = 88) presented to 4 veterinary hospitals and 1 practice.
Methods
Prospective, longitudinal, observational study. Oronasal and rectal swabs were collected at presentation and 69% of animals were sampled again at discharge. Methicillin‐resistant (MR) staphylococci and macrococci, cephalosporinase‐, and carbapenemase‐producing (CP) Enterobacterales were isolated. Genetic relatedness of isolates was assessed by repetitive sequence‐based polymerase chain reaction and multilocus sequence typing. Risk factors for MDRO acquisition and carriage were analyzed based on questionnaire‐derived and hospitalization data.
Results
Admission prevalence of MDRO carriage in pets was 15.5% (95% confidence interval [CI], 11.4‐20.4). The discharge prevalence and acquisition rates were 32.1% (95% CI, 25.5‐39.3) and 28.3% (95% CI, 22‐35.4), respectively. Predominant hospital‐acquired isolates were extended spectrum β‐lactamase‐producing Escherichia coli (ESBL‐E coli; 17.3%) and β‐lactamase‐producing Klebsiella pneumoniae (13.7%). At 1 institution, a cluster of 24 highly genetically related CP (bla oxa181 and bla oxa48) was identified. Multivariate analysis identified hospitalization at clinic 1 (odds ratio [OR], 5.1; 95% CI, 1.6‐16.8) and days of hospitalization (OR 3‐5 days, 4.4; 95% CI, 1.8‐10.9; OR > 5 days, 6.2; 95% CI, 1.3‐28.8) as risk factors for MDRO acquisition in dogs.
Conclusions
Veterinary hospitals play an important role in the selection and transmission of MDRO among veterinary patients.
Keywords: carbapenemase‐producing enterobacterales, extended‐spectrum β‐lactamase, risk factors, transmission
Abbreviations
- 3GCR
third‐generation cephalosporin‐resistant
- 3GCR‐E
third‐generation cephalosporin‐resistant enterobacterales
- CI
confidence interval
- CLSI
clinical and laboratory standards institute
- CP
carbapenemase‐producing
- CRE
carbapenem‐resistant enterobacterales
- ERIC‐PCR
enterobacterial repetitive intergenic consensus polymerase chain reaction
- ESBL
extended spectrum β‐lactamase
- ESBL‐E coli
ESBL‐producing Escherichia coli
- ESBL‐KP
ESBL‐producing Klebsiella pneumoniae
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- ICU
intensive care unit
- IPC
infection prevention and control
- IQR
interquartile range
- KP
Klebsiella pneumoniae
- MALDI‐TOF MS
matrix‐assisted laser desorption/ionization time of flight mass spectrometry
- MDR
multidrug‐resistant
- MDRO
multidrug‐resistant organisms
- MICs
minimal inhibitory concentrations
- MLST
multilocus sequence typing
- MR
methicillin‐resistant
- MRCoNS
methicillin‐resistant coagulase‐negative staphylococci
- MRSA
methicillin‐resistant Staphylococcus aureus
- MRSP
methicillin‐resistant Staphylococcus pseudintermedius
- OR
odds ratio
- pAmpC
plasmid‐encoded AmpC
- REP‐PCR
repetitive element palindromic polymerase chain reaction
- WGS
whole‐genome sequencing
1. INTRODUCTION
Emergence of multidrug‐resistant organisms (MDRO) poses an important threat to human and animal health. 1 Like human hospitals, veterinary hospital environments favor the selection and transmission of MDRO because of a high density of susceptible patients and the use of broad‐spectrum antimicrobials. 2 During their hospitalization, small animals therefore may become asymptomatic carriers of MDRO or may develop life‐threatening nosocomial infections. These infections may be difficult to treat because of resistance of MDRO to several, if not all, commonly used classes of antimicrobials. 3
Several MDRO are considered to be of particular relevance to both human and animal health. Methicillin‐resistant (MR) Staphylococcus aureus (S aureus; MRSA) generally is isolated at low frequency from dogs and cats, 4 , 5 and pet owners may be the primary source. 6 In contrast, MR Staphylococcus pseudintermedius (S pseudintermedius; MRSP) clones are widespread in veterinary settings worldwide 5 , 7 and show resistance to many antibiotics licensed in veterinary medicine. 5 , 8 Methicillin‐resistant S pseudintermedius cause a wide range of infections in dogs and cats including skin and postoperative infections. 7 , 8 , 9 It is also increasingly reported as cause of severe infections in humans. 9 Macrococcus spp. (M canis; M caseolyticus) have been isolated from the skin of healthy dogs and infection sites, and have the potential to acquire methicillin resistance genes (mecB; mecD). 10 , 11 , 12 Their relevance in veterinary settings so far has not been evaluated in detail.
Multidrug‐resistant Enterobacterales including Escherichia coli (E coli), Klebsiella spp. and Enterobacter spp. presently are considered highly problematic in both veterinary 13 , 14 and human 13 , 15 hospital settings. Enterobacterales with third‐generation cephalosporin resistance (3GCR‐E) are widespread 16 and often display coresistance to other classes of antibiotics including tetracyclines, sulfonamides, phenicols, aminoglycosides, and fluoroquinolones. 17 , 18 The use of carbapenems to treat such infections has led to the selection of carbapenem‐resistant Enterobacterales (CRE) in human medicine, 19 whereas up until very recently, reports of carbapenem resistance have been very rare in veterinary medicine. 20 , 21 However, as molecular mechanisms of resistance continue to evolve, the epidemiology of MDRO colonization and infection and relevant risk factors are changing. 22
Our aims were to assess risk factors for prevalence and acquisition of MDRO carriage in dogs and cats presented to veterinary clinics or practices in Switzerland and to describe relevant MDRO in veterinary care settings.
2. MATERIALS AND METHODS
2.1. ETHICS STATEMENT
Ethical approval for collection of samples and clinical data from cats and dogs was granted by the Veterinary Office (Ref. BE 16/18). Written owner consent for participation in the study was obtained before enrollment.
2.2. Study Design and Setting
This prospective multicenter longitudinal observational study was part of a large project to assess the role of small animal clinics in the dissemination of MDRO (Ref FSVO 1.8.10). Dogs and cats presented to 3 large referral hospitals (clinics 1‐3), 1 smaller clinic (clinic 4), and a small practice (practice 1) across Switzerland between May and September 2018 were enrolled sequentially regardless of their clinical focus. Clinics 1 to 3 are large referral hospitals with an annual caseload of approximately 6000, 12 000, and 60 000 cases, respectively. Clinic 4 is a moderately sized small animal clinic providing a large range of services including orthopedic surgery and practice 1 a small practice for companion animals providing predominantly outpatient services and routine surgery. Animals were included if they were expected to be hospitalized for at least 48 hours, except for practice 1, where hospitalizations rarely occurred and samples therefore were collected from dogs and cats presented for outpatient care.
2.3. Data collection
2.3.1. Questionnaires
At presentation, owners were asked to fill out a questionnaire that contained questions regarding the animal's origin, lifestyle, living environment, preventative health care, diet, current or previous medical treatments, travel history, and contact with other animals, including farm animals. The questionnaire also contained owner‐centered questions (results not included here). The questionnaire was available in paper format and via an online link (Supplemental data Questionnaire). Study data were collected and managed using the Research Electronic Data Capture system (REDCap) hosted at the University of Bern. 23 , 24
2.3.2. Animals
Demographic data including breed, age, weight, sex, neuter status, hospitalizations, and treatments within the past 12 months were recorded for all enrolled animals. Clinical diagnoses and hospitalization data including admission ward, clinical problems or diagnoses, duration of hospitalization, days in intensive care unit (ICU), medical interventions, and treatments including antimicrobial treatments were extracted from the medical records of dogs and cats hospitalized at the 2 large university clinics only (clinics 1 and 2), where the most cases were enrolled.
2.3.3. Sampling
Rectal and oronasal swabs were collected from dogs and cats within the first 6 hours of admission and again on the day of discharge using dry sterile swabs with Amies transport medium (Sarstedt AG & Co. KG, Nümbrecht, Germany). Rectal swabs were inserted 1 to 2 cm into the rectum and rotated gently until fecal material adhered to the swab. In dogs, nasal swabs were gently inserted in 1 naris and rotated; the same swab was then introduced in to the mouth lateral to the tongue. In cats, small dogs, and uncooperative animals, the swab was only inserted orally. Swabs were stored for a maximum of 5 days at 4°C or 2 days at ambient temperature and then sent by batch to the laboratory.
2.3.4. Isolation and identification of bacteria
Rectal swabs and fecal samples were tested for the presence of Gram‐negative MDRO as previously described. 25 , 26 Swabs were placed into 5 mL of nonselective Luria‐Bertani (LB) enrichment broth at 37°C for 24 hours. A loopful of the culture was then streaked on ChromID extended spectrum β‐lactamase (ESBL) plates to select for 3GCR‐E, or ChromID OXA‐48 and ChromID CARBA plates (BioMérieux SA, Marcy‐l'Étoile, France), to select for CRE. Plates were incubated at 37°C for 24 hours under aerobic conditions. Colonies were subcultivated onto tryptone soy agar plates containing 5% sheep blood (TSA‐S; Becton & Dickinson Company, Franklin Lakes, New Jersey). Isolates were identified to the species level by using the matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI‐TOF MS; Bruker Daltonics GmbH, Bremen, Germany). Carbapenemase production was detected using the Blue‐Carba test (BCT). 27 Oronasal swabs were tested for the presence of MR staphylococci and macrococci using a 2‐step selective enrichment 28 followed by selection on selective agar (BBL CHROMagar MRSA II, Becton, Dickinson and Company, New Jersey) at 37°C for 24 hours.
2.3.5. Antimicrobial susceptibility testing
Minimal inhibitory concentrations (MICs) of antimicrobials were determined by broth microdilution using Sensititre EUST, EUVSEC, EUVSEC2, and GNX2F plates (Thermo Fisher Scientific, Waltham, Massachusetts). For Gram‐negative bacteria, 14 antibiotics were tested (Thermo Fisher Scientific) following the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST). 29 Results were interpreted using the EUCAST criteria, except for nalidixic acid, sulfamethoxazole, and tetracycline, for which the criteria from the Clinical and Laboratory Standards Institute (CLSI) were used. 30 Extended‐spectrum β‐lactamase and carbapenemase genes were identified using the CT103XL microarray (Check‐Points, Wageningen, The Netherlands). 31 Before whole genome sequencing (WGS), carbapenemase‐producing (CP) isolates were tested for the presence of bla OXA‐48‐like genes by polymerase chain reaction (PCR). 32 The MR genes mecA, mecB, and mecD were identified by PCR as previously described. 33 , 34 , 35
2.3.6. REP‐PCR and whole‐genome sequencing
Genetic relationships and clonality among isolates of the same species were determined by repetitive element palindromic polymerase chain reaction (REP‐PCR) or enterobacterial repetitive intergenic consensus‐polymerase chain reaction (ERIC‐PCR) 36 , 37 and by multilocus sequence typing (MLST). For MRSA, MRSP, methicillin‐resistant coagulase‐negative staphylococci (MRCoNS), and MR Macrococcus spp., the corresponding schemes published in the pubMLST database (https://pubmlst.org/databases/) were used and for colistin‐resistant (COL‐R), ESBL‐producing, and CP Enterobacterales, the Center for Genomic Epidemiology database was used (http://www.genomicepidemiology.org/).
Whole genome sequencing was used to confirm the identity of selected isolates and the presence of specific resistance genes. Parts of these analyses have been reported previously. 26
2.3.7. Data analysis and statistical methods
Statistical analysis was performed using the NCSS program (NCSS11 Statistical Software. 2016. NCSS, LLC, Kaysville, Utah. ncss.com/software/ncss). Post hoc sample size calculations were performed using the online calculator Epitools (https://epitools.ausvet.com.au).
The overall prevalence of MDRO carriage in dogs and cats at admission and discharge and the rate of MDRO acquisition during hospitalization were estimated with a 95% confidence interval (CI). Acquisition was defined as the presence of a new or a genetically unrelated MDRO in discharge samples. Persistence was defined as isolation of MDRO with matching molecular profiles (REP‐PCR, WGS) using repeated samples from the same individual.
Associations between MDRO carriage at admission and questionnaire‐derived variables, as well as associations between MDRO acquisition during hospitalization and hospitalization data, were examined using univariate and multivariate logistic regression analyses. First, univariate analysis was performed for all independent variables to assess suitability of inclusion into the multivariate regression model. Variables with a P‐value of <.1 were included in the full models. The final model of each multivariate analysis was achieved by backward stepwise elimination, including assessment of interaction and confounding. Variables were kept in the final model if the P‐value was <.05 or if removing the variable changed the effect of another risk factor by >20%. Odds ratios (OR) and 95% CIs were reported; values were considered significant if the 95% CI of the OR did not include 1.
3. RESULTS
3.1. Study population
A total of 271 animals, including 183 (67.5%) dogs and 88 (32.5%) cats, were sampled at presentation and 187 (69%) animals, including 124 (66.3%) dogs and 63 (33.7%) cats, were sampled again at discharge. Most animals (221/271; 81.5%) were enrolled at clinics 1 and 2 (Supplemental Table 1).
The median age of dogs was 7 years (interquartile range [IQR], 3‐9 years) and the median weight was 14.4 kg (IQR, 7.1‐28.4 kg). There were 86 females (28 intact/58 neutered) and 97 males (36 intact/61 neutered). The majority of dogs (155/183, 85.7%) were purebred of 68 different breeds, the most frequent being Labrador Retriever (6.6%), Jack Russell Terrier (5.5%), Yorkshire Terrier (4.4%), French Bulldog (3.9%), and Chihuahua (3.9%).
Cats had a median age of 6 years (IQR, 2‐11 years) and a median weight of 4.3 kg (IQR, 3.6‐5.1). There were 39 neutered females and 49 males (5 intact/44 neutered). The most common breeds were European Shorthair (68.2%), Maine Coon (11.4%), and Ragdoll (4.6%).
Questionnaire‐derived lifestyle data and hospitalization and treatment details of cats and dogs included in the study are shown in Supplemental Tables 2 to 5. For organizational reasons, most dogs and cats were recruited from the emergency services of the participating hospitals. In dogs, the most common disease groups were neurologic, gastrointestinal, orthopedic, and neoplastic, whereas in cats gastrointestinal, orthopedic, and urinary tract disease groups were most common. Although 154 (82.4%) animals had been hospitalized ≥48 hours according to inclusion criteria, 33 (17.6%) were in the hospital between 7 and 47 hours only, resulting in a median duration of hospitalization of 2 days (IQR, 2; range, 0‐14 days). This diversion from initial inclusion criteria resulted from unexpected early discharges or death and a loosening of inclusion criteria to increase enrollment numbers. It led to the observation that some animals seemingly acquired MDRO as early as 24 hours after admission (2 cases). The majority of dogs (78/152, 51.3%) and cats (42/77, 54.6%) were treated with antimicrobials during their hospitalization, but none of them received carbapenems.
3.2. Prevalence of MDRO carriage at presentation
The overall estimated admission prevalence of MDRO carriage was 15.5% (95% CI, 11.4‐20.4) with 18% (33/183; 95% CI, 12.8‐24.4) of sampled dogs and 10.2% (9/88; 95% CI, 4.8‐18.5) of sampled cats carrying MDRO. The prevalence varied among the 5 veterinary care facilities and was estimated at 14% (95% CI, 8.9‐20.6) in clinic 1, 9.9% (95% CI, 4.1‐19.3) in clinic 2, 7.7% (95% CI, 0.2‐36) in clinic 3, 0% (95% CI, 0‐41) in clinic 4, and 43.3% (95% Cl, 25.5‐62.6) in the outpatient population at practice 1. The numbers of patients enrolled in each clinic and the proportions of MDRO‐positive cases are shown in Figure 1.
FIGURE 1.
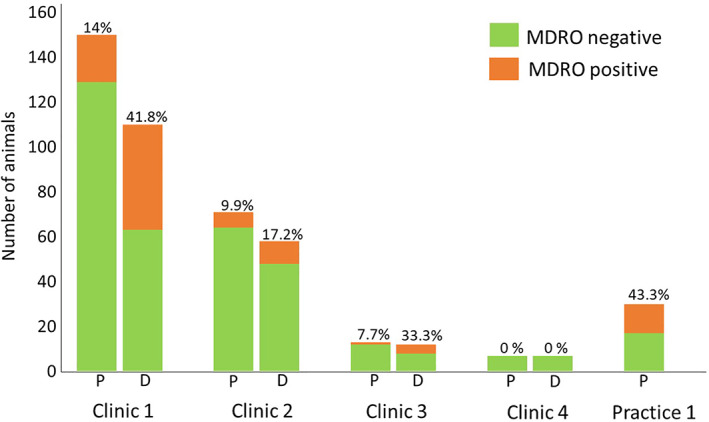
Total number of animals tested at presentation to (P) and discharge from (D) participating clinics/practice. Proportions of MDRO‐positive animals are indicated above the columns. MDRO, multidrug‐resistant organisms
Of the 271 animals, 1 variety of MDRO strain was isolated in 42 animals and 1 cat and 1 dog carried an ESBL‐producing E coli (ESBL‐E coli) in addition to a MR Staphylococcus warneri (S warneri) or a MR Staphylococcus haemolyticus (S haemolyticus).
The numbers of MDRO isolated in each clinic or practice at admission are shown in Supplemental Table 1. Third‐generation cephalosporin‐resistant E coli predominated, accounting for 33.3% of all isolates. Methicillin‐resistant Staphylococcus aureus only was isolated in 1 cat, and MRSP in none of the animals. Methicillin‐resistant coagulase‐negative staphylococci were isolated from 9.6% of the animals and were predominately found in 1 practice, where MR S warneri was isolated from nearly one‐third (9/30) of the sampled animals. At presentation, MR macrococci were isolated only in 1.1% of cases. The numbers of MDRO isolated at presentation are shown in Figure 2.
FIGURE 2.
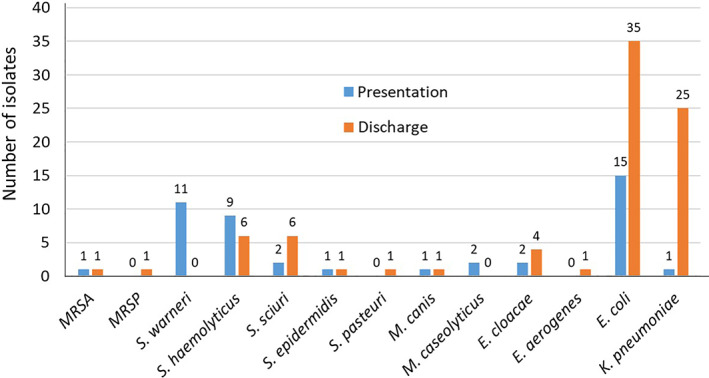
Number of multidrug‐resistant organisms isolated from dogs and cats at presentation to and discharge from 3 veterinary clinics and 1 practice. MRSA, methicillin‐resistant Staphylococcus aureus; MRSP, methicillin‐resistant Staphylococcus pseudintermedius; S, Staphylococcus; M, Macrococcus; E, Enterococcus; E coli, Escherichia coli; K pneumoniae, Klebsiella pneumoniae
3.3. Prevalence MDRO carriage at discharge and acquisition rate
The overall numbers of MDRO isolated at discharge are shown in Figure 2. The numbers of MDRO isolated in each clinic are shown in Supplemental Table 1. Sixty animals (26.7% cats; 73.3% dogs) carried MDRO at discharge, corresponding to an overall discharge prevalence of 32.1% (95% CI, 25.5‐39.3). The MDRO discharge prevalence for cats was 25.4% (95% CI, 15.3‐38) and for dogs 35.5% (95% CI, 27.1‐44.6).
Of the 42 animals that carried MDRO at admission, 11.9% (5/42; 95% CI, 4‐25.5) carried a genetically related strain and 16.7% (7/42; 95% CI, 7‐31.4) a genetically unrelated strain at discharge. In 19% of animals (8/42; 95% CI, 8.6‐34.1), no MDRO was isolated at discharge, and 52.4% (22/42; 95% CI, 36.4‐68) either were outpatients (practice 1; n = 15) or could not be resampled at discharge because of unexpected early discharge, death, or withdrawal from the study.
The overall rate of MDRO acquisition was 28.3% (95% CI, 22‐35.4): 20.6% (95% CI, 11.5‐33) for cats and 32.3% (95% CI, 24.1‐41.2) for dogs. Discharge prevalence and acquisition rates varied considerably among care facilities with clinic 1 showing a particularly high rate of MDRO acquisition (39.1%; 95% CI, 29.9‐48.9) compared with clinic 2 (12.1%; 95% CI, 5‐23.3), clinic 3 (25%; 95% CI, 5.48‐57.2), and clinic 4 (0%; 95% CI, 0‐41).
The most common hospital‐acquired isolates were 3GCR‐E coli and 3GCR‐K pneumoniae (KP) isolated from 15% (29/187) and 12.3% (23/187) of animals. One dog (0.5%) acquired MRSP and 8% of the animals (15/187) MRCoNS. In clinic 1, in‐hospital acquisition of 3GCR‐E coli, 3GCR‐KP, and MRCoNS was particularly frequent.
3.4. Antimicrobial resistance profiles
Resistance profiles of all isolates are shown in Supplemental Tables 6 to 9. Third‐generation cephalosporin‐resistant‐E coli producing either ESBL (CTX‐M‐1/−3/−15, CTX‐M‐9/−14, CTX‐M‐1) or plasmid‐encoded AmpC (pAmpC; CMY‐42; CMY‐2‐like) predominated, making up 84% of all E coli isolates (Supplemental Table 6).
Twenty‐two E coli isolates from clinic 1 displayed similar antimicrobial resistance patterns and a common bla OXA‐181 carbapenemase gene. One further E. coli isolate (101.2) from clinic 1 carried a bla OXA‐48 carbapenemase gene. This isolate had MICs below the resistance breakpoint set by EUCAST for meropenem, but was resistant to ertapenem. Two E. coli isolates producing NDM‐5 carbapenemase were isolated at clinic 2. These isolates were broadly resistant including resistance to meropenem. All isolates were sensitive to colistin. The numbers and proportions of E coli isolates resistant to the tested antimicrobials are shown in Table 1.
TABLE 1.
Resistance of third‐generation cephalosporin‐resistant E coli, K pneumoniae, and Enterobacter spp. isolated from admission and discharge samples of 271 dogs and cats
| Antimicrobial type | E coli, n = 50 | K pneumoniae, n = 26 | Enterobacter spp, n=8 |
|---|---|---|---|
| Ampicillin | 50 (100) | 26 (100) | 8 (100) |
| Cefotaxime | 49 (98) | 25 (96.2) | 7 (87.5) |
| Ceftazidime | 41 (82) | 23 (88.5) | 8 (100) |
| Meropenem | 2 (4) | 0 (0) | 0 (0) |
| Sulfamethoxazole | 24 (48) | 24 (92) | 7 (87.5) |
| Trimetoprim | 24 (48) | 22 (84.6) | 5 (62.5) |
| Ciprofloxacin | 32 (64) | 24 (92) | 4 (50) |
| Tetracyclin | 22 (44) | 16 (61.5) | 5 (62.5) |
| Azitromycin | 3 (6) | 6 (23.1) | NI |
| Nalidixic acid | NI | 26 (100) | NI |
| Chloramphenicol | 11 (22) | 7 (26.9) | 6 (75) |
| Gentamycin | 13 (26) | 18 (69.2) | 5 (62.5) |
| Colistin | 0 (0) | 0 (0) | 0 (0) |
| Tigecycline | 0 (0) | 0 (0) | 0 (0) |
Note: Minimal inhibitory concentrations (MIC) were interpreted using the criteria of the European Committee on Antimicrobial Susceptibility Testing 27 except for nalidixic acid, sulfamethoxazole, and tetracycline, for which the criteria from the Clinical and Laboratory Standards Institute were used. 28 No MIC was available for azithromycin; therefore, an MIC of >64 mg/L was tentatively used. Data are reported as No. (%) of isolates. NI indicates the minimum inhibitory concentrations of the antimicrobials that were not interpreted. Numbers are given in bold when ≥50% of isolates showed resistance.
Abbreviations: E coli, Escherichia coli; K pneumoniae, Klebsiella pneumoniae.
The majority of KP were of the CTX‐M‐1/‐3/‐15 ESBL and DHA‐1 pAmpC subtypes and 1 isolate from clinic 1 (142.2) possessed the bla OXA‐48 carbapenemase gene (Supplemental Table 7). Enterobacter cloacae (E cloacae) isolates were resistant to several classes of antimicrobials (Table 1; Supplemental Table 8).
The resistance profiles of MR Gram‐positive isolates are shown in Supplemental Table 9. The 2 MRSA isolates (carried at admission and discharge by the same animal) carried the mecA gene and showed resistance to penicillin, cefotaxime, and ciprofloxacin. One M caseolyticus isolate (31.2) contained the mecD gene and showed resistance to 5 different classes of antimicrobials.
3.5. Molecular relatedness of MDRO
The clonality of all isolates is listed in Supplemental Tables 6 to 9. Overall, 3GCR‐E coli isolates were diverse, belonging to 24 different phylogenetic groups, with the exception of a cluster of 24 ST410 CMY‐42 isolates from clinic 1, which displayed similar antimicrobial resistance patterns and a common bla OXA‐181 carbapenemase. The detailed molecular characterization of this cluster of isolates has been reported recently. 26
Two carbapenem‐resistant ST167 E coli isolates were obtained from clinic 2. The two isolates shared the bla NDM‐5 gene and were highly genetically related. The detailed molecular characterization of these strains has been reported. 26
Of the 26 3GCR‐KP isolates, 20 expressed CTX‐M‐1/‐3/‐15 and 7 DHA‐1 ß‐lactamases. All isolates showed close genetic relatedness belonging to REP‐PCR group A. Among animals hospitalized in clinic 1, a cluster of 10 isolates (CTX‐M‐1/‐3/‐15) with similar antimicrobial resistance pattern was identified. Three K pneumoniae isolates from clinic 2 resembled each other in terms of ESBL resistance type and antimicrobial resistance pattern. One single KP isolated at discharge from a dog from clinic 1 carried a carbapenemase‐encoding gene (blaOXA48).
The MDR E. cloacae isolates belonged to REP‐PCR group C, with the exception of 1 isolate from clinic 2, that was typed as REP‐PCR group E. Two isolates from clinic 1 (REP‐group C) shared a similar antibiotic resistance profile and were found in 2 dogs, which had been admitted to clinic 1 by the emergency service on 2 consecutive days.
3.6. Risk factors for MDRO carriage in dogs and cats
The questionnaire‐derived variables included in the risk factor analysis for MDRO carriage at admission for dogs and cats are shown in Supplemental Tables 2 and 3. Univariate analysis identified antimicrobial treatment before admission as a significant risk factor for MDRO carriage in dogs (OR, 2.7; 95% CI, 1.2‐5.9; P = .01), but not in cats. Of the 7 cats fed a so‐called Bones and Raw Food (BARF) diet, 3 were carrying MDRO at presentation and this association was significant in the univariate analysis (OR, 7.3; 95% CI, 1.2‐45; P = .04).
The variables included in the risk factor analysis for in‐hospital MDRO acquisition in dogs and cats are shown in Supplemental Tables 4 and 5. Univariate analysis showed the following significant associations for MDRO acquisition during hospitalization: hospitalization at clinic 1 (dogs OR, 4.4; 95% CI, 1.4‐13.8; cats OR, 5.1; 95% CI, 1.2‐21.7), esophageal feeding tube (esophagostomy or nasoesophageal) placement in cats (OR, 8; 95% CI, 1.3‐50.8), days of hospitalization in dogs (OR 3‐5 days, 3.7; 95% CI, 1.6‐8.6; OR > 5 days, 5.3; 95% CI, 1.2‐22.5), and administration of buprenorphine in dogs (OR, 4.2; 95% CI, 1.8‐10.1). The administration of antimicrobials during hospitalization did not increase the odds of becoming MDRO carriers in the population of cats but increased it in the population of dogs (OR, 2.5; 95% CI, 1.1‐5.7).
Multivariate analysis confirmed hospitalization at clinic 1 (OR, 5.1; 95% CI, 1.6‐16.8) and days of hospitalization (OR 3‐5 days, 4.4; 95% CI, 1.8‐10.9; OR > 5 days, 6.2; 95% CI, 1.3‐28.8) as risk factors for MDRO acquisition in dogs.
4. DISCUSSION
Our results identified acquisition of MDRO in approximately one‐third of cats and dogs treated at veterinary referral hospitals. Although 1 clinic showed a particularly high acquisition rate, MDRO acquisition was documented in all but 1 of the participating clinics. Of 180 animals hospitalized at referral clinics, 49 (27.2%) were discharged carrying at least 1 MDR Enterobacterales, nearly half of which expressed carbapenemases. This finding represents a very worrisome development, because ongoing colonization of these animals likely contributes to the dissemination of MDRO in the community.
The overall admission prevalence of 15.5% was calculated including all dogs and cats that carried Gram‐positive MR, Gram‐negative 3GCR isolates, or both across all 5 participating clinics and practices. In most studies, prevalence in healthy dogs and cats or veterinary‐visiting animals is limited to MDR E coli or MDR Enterobacterales with admission prevalence of 10.7% to 24.8% for dogs, 13 , 38 , 39 , 40 1.4% to 15% for cats, 38 , 40 and 7% to 8.6% for both. 41 , 42 Studies that examined colonization with MR Gram‐positives 5 , 41 often exclude MRCoNS because of their questionable clinical relevance as pathogens. 43 Considering the methodical differences across different studies, the admission prevalence found in our cohort was similar to what has been reported previously.
Third‐generation cephalosporin‐resistant E coli were the most common isolates found at presentation (34% of all MDRO isolates), suggesting that these bacteria may already be part of the gut flora of a large proportion of the small animal referral population in Switzerland. The 3GCR‐E coli isolated at admission were diverse, including pAmpC (CMY‐42, CMY‐2‐like) and ESBL (CTX‐M‐1/‐3/‐15, CTX‐M‐9/‐14, and CTX‐M‐1) genotypes. These genotypes are found in up to 10% of the Swiss human population 44 and also have been isolated from urban birds (CTX‐M‐1, CTX‐M‐15), 45 chicken meat (CMY‐2, CTX‐M‐1), 46 and pet food (CTX‐M‐1, CTX‐M‐3, and CTX‐M‐15). 47 A large percentage of 3GCR‐E coli isolates displayed additional resistance to other classes of antimicrobials; notably, two‐thirds of the isolates were resistant to trimethoprim/sulfamethoxazole (TMP‐S), 53.3% to tetracyclines and 26.7% to fluoroquinolones, aminoglycosides, or both. Carbapenem resistance at admission only was detected in 1 E. coli strain isolated from a dog. This dog had not been treated with antimicrobials before presentation and had never been presented at the clinic before the admission date. 26
At admission, carriage of MRSA, MRSP, or macrococci was very rare in this cohort of cats and dogs, which is in agreement with some 41 , 48 but not all recent studies. 7 , 49 In contrast, there was a high rate of isolation of MRCoNS (S warneri; S haemolyticus; ERIC‐PCR group D) in the outpatient population of practice 1. As previously reported, sampling of the practice environment and staff in this practice identified contamination of 3/37 sampling sites with S haemolyticus (ST49; mecA). 25 Based on these results, it cannot be excluded that MRCoNS were acquired in the practice. However, a high prevalence of carriage of MRCoNS also has been documented in a population of healthy nonveterinary‐visiting Labrador retrievers in the United Kingdom, which had no contact with veterinary practice environments and had not received antimicrobial treatments, 50 thus suggesting sources of acquisition outside of veterinary care. Beyond their role as opportunistic pathogens, MRCoNS can act as reservoirs for resistance genes and a source of infection or colonization of in‐contact humans. 43
Dogs having received systemic antimicrobial treatments before presentation had increased odds of carrying MDRO upon admission. This finding is in agreement with other studies in pets, 51 farm animals, 52 and humans, 53 , 54 suggesting that systemic antimicrobial treatments select for MDR bacteria within the host microbiome. The fact most animals presented at referral institutions already have been treated with antimicrobials puts these institutions at increased risk of continuously having MDRO introduced into their hospital environments. 2 Screening of patients with risk factors for MDRO carriage in large referral institutions therefore should be considered part of routine animal admission procedures.
An additional risk factor for MDRO carriage at presentation in cats was a BARF diet. Extended spectrum β‐lactamase‐producing E coli and other MDRO have been isolated from farm animals at slaughter, 55 as well as from fresh meat 56 and milk, 57 and the association between BARF and MDRO colonization, in particular by MDR Enterobacterales, previously has been described in both cats 58 , 59 and dogs. 38 , 59 The feeding of a BARF diet to dogs and cats therefore should be discouraged.
Acquisition of MDRO was common in referral hospitals in our study and still amounted to 12.1% in the institution with the lowest acquisition rate among all 3 institutions. In a previous study, the hospital acquisition rate for MDR E coli was reported at 6.8%. 41 In comparison, the acquisition rate of MDR E coli in this set of hospitals ranged between 6.9% and 20.9% and thus was markedly higher in some of the hospitals than previously reported.
In clinic 1, the MDRO acquisition rate was strikingly high (39.1%), predominantly because of the acquisition of 3GCR‐E coli and 3GCR‐KP. More than half of E coli isolates (26/50) and 1/26 K pneumoniae isolates carried carbapenemase‐encoding genes (bla oxa‐181, bla oxa‐48, bla NDM5). The closely related bla oxa‐181, bla oxa‐48 genes have been associated with low‐level resistance to carbapenems and may go undetected during routine diagnostic testing if the EUCAST screening breakpoints for meropenem or imipenem are not used. 60 As carbapenemase producers regrettably may become more widespread in veterinary medicine, it is important that veterinary bacteriology laboratories extend their routine diagnostic testing to the identification of these isolates, in particular in view of their public health relevance. 32
The molecular relatedness and mobile genetic elements of a cluster of 21 E. coli isolates (ST410; bla OXA‐181) from clinic 1 subsequently were analyzed further using WGS and shown to be clonal, displaying shared plasmidic resistance genes. 26 The same E coli strain could be linked to the transitory colonization of 1 staff member, 61 and a closely related E coli strain (ST410, bla CMY‐42, bla OXA‐181) was found in 1 environmental sample from clinic 1. 25 Overall, these findings are indicative of nosocomial spread.
Interestingly, in this clinic, the environment was also shown to be extensively contaminated with K pneumoniae (ST11; bla OXA‐48, bla DHA‐1) and MRSP (ST551; mecA). 25 A closely related KP strain (ST11; bla OXA‐48) was isolated only from 1 cat. The MRSP was only acquired by 1 dog during hospitalization and was unrelated (ST1337; mecA).
As a consequence of the high acquisition rate, hospitalization at clinic 1 represented a substantial risk factor for MDRO acquisition. In this institution, 30% of environmental sampling sites were positive for MDRO, the infrastructure for hand hygiene was outdated, cleaning and disinfection protocols had not been updated for some time, and the wearing of gloves had largely replaced standard hand disinfection. 25 These factors likely contributed to the spread of MDRO in this institution and have since been addressed.
In clinic 2, acquisition of CP E coli (ST167; bla NDM‐5) also occurred, and closely genetically related strains were carried by staff members, 61 but the MDRO acquisition rate (12.1%) was markedly lower than in clinic 1. In clinic 2, stringent infection prevention and control (IPC) bla NDM‐5 standards, reflected by a high IPC score, were in place and only 8% of environmental sampling sites identified MDRO colonization. 25
Additional risk factors for MDRO acquisition from the univariate analysis were days of hospitalization, administration of buprenorphine, and use of antimicrobials during hospitalization in dogs. Of these, only days of hospitalization remained significant in the multivariate model. This finding concurs with what has been reported previously in pets 41 , 62 and humans. 62 , 63
The high rate of isolation of CP organisms was unexpected, because carbapenems are not used in clinics 1 and 2. 64 , 65 However, the restricted use of a specific class of antimicrobials itself is not sufficient to avoid emergence of relative resistance, because the selection pressure exerted by the use of any broad‐spectrum antimicrobial may co‐select for carbapenem resistance. 66 Antimicrobial prescriptions for selected conditions in dogs and cats in 2016 recently have been reported, and these studies included prescription data from clinics 1 and 2. 64 , 65 Comparison of the data subsets from these 2 clinics did not identify any substantial differences in the frequency or appropriateness of antimicrobial prescriptions (data not shown). Therefore, differences in IPC standards are more likely the driving force behind the differences in environmental colonization 25 and MDRO acquisition between the 2 clinics. Nonetheless, besides active surveillance of resistance and implementation of stringent environmental and hand hygiene protocols, antimicrobial stewardship plays an important role in limiting the emergence and spread of MDRO in referral hospitals.
Limitations of our study include the limited number of animals enrolled, which might have led to a failure to detect significant risk factors for MDRO carriage because of type II error. Furthermore, the statistical power of the comparison of acquisition rates among clinics was limited because of unequal sample sizes.
Differences in sample storage conditions (4°C vs room temperature) and duration (1‐5 days) may have influenced the likelihood of MDRO recovery. Although some investigators have found that samples could be stored at room temperature or at 4°C for up to 14 days without a decrease in recovery, 67 others have described decreased recovery of bacteria after storage at 4°C for 1 or 4 weeks using direct plating without prior enrichment. 68 We are confident however that the short storage times and use of enrichment culture in our study would have minimized any potential effect of storage at 4°C.
In conclusion, screening of dogs and cats presented to veterinary clinics in Switzerland identified an unexpectedly high rate of acquisition of 3GCR‐Enterobacterales, including strains with carbapenemase resistance. These findings emphasize that small animal veterinary clinics may contribute to the selection and spread of MDRO. Active surveillance of resistance, stringent IPC, and antimicrobial stewardship are key elements to ensure patient safety and decrease the public health risk.
5. CONFLICT OF INTEREST DECLARATION
Authors declare no conflict of interest.
OFF‐LABEL ANTIMICROBIAL DECLARATION
Authors declare no off‐label use of antimicrobials.
INSTITUTIONAL ANIMAL CARE AND USE COMMITTEE (IACUC) OR OTHER APPROVAL DECLARATION
Ethical approval for collection of samples and data from cats and dogs was obtained from the Federal Food Safety and Veterinary Office (Ref. BE 16/18).
HUMAN ETHICS APPROVAL DECLARATION
Ethical approval for collection of samples and data from humans was obtained from the regional Swiss Association of Research Ethics Committees on research involving humans (Ref. 2018‐00866). Data was pseudonymized and managed via a secure platform (REDcap) hosted by the University of Bern.
Supporting information
Data S1: Supporting information
ACKNOWLEDGMENT
Funding provided by the Swiss Federal Food Safety and Veterinary Office [FSVO Grant No. 1.18.10; due to SS, BW, AE, and VP] and by the NRP‐72, “National Research Programme, Antimicrobial Resistance” of Swiss National Science Foundation [SNSF Grant No. 177378; due to AE and VP]. We thank Mathieu Clément, Nina Gutzwiller and Alexandra Collaud for technical assistance. We are grateful to all pet owners for their participation in the study.
Dazio V, Nigg A, Schmidt JS, et al. Acquisition and carriage of multidrug‐resistant organisms in dogs and cats presented to small animal practices and clinics in Switzerland. J Vet Intern Med. 2021;35:970–979. 10.1111/jvim.16038
Funding information National Research Programme, Antimicrobial Resistance of Swiss National Science Foundation, Grant/Award Number: 177378; Swiss Federal Food Safety and Veterinary Office (FSVO), Grant/Award Number: 1.18.10
REFERENCES
- 1. WHO Library cataloguing‐in‐publication data global action plan on antimicrobial resistance [internet]. 2015. https://www.who.int/antimicrobial‐resistance/publications/global‐action‐plan/en/. Accessed January 27, 2021.
- 2. Walther B, Tedin K, Lübke‐Becker A. Multidrug‐resistant opportunistic pathogens challenging veterinary infection control. Vet Microbiol. 2017;200:71‐78. [DOI] [PubMed] [Google Scholar]
- 3. Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42‐51. [DOI] [PubMed] [Google Scholar]
- 4. Rahman MM, Amin KB, Rahman SMM, et al. Investigation of methicillin‐resistant Staphylococcus aureus among clinical isolates from humans and animals by culture methods and multiplex PCR 11 medical and health sciences 1103 clinical sciences 11 medical and health sciences 1108 medical Microbiolog. BMC Vet Res. 2018;14(1):1‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Worthing KA, Brown J, Gerber L, Trott DJ, Abraham S, Norris JM. Methicillin‐resistant staphylococci amongst veterinary personnel, personnel‐owned pets, patients and the hospital environment of two small animal veterinary hospitals. Vet Microbiol. 2018;223:79‐85. [DOI] [PubMed] [Google Scholar]
- 6. Pfeiffer DU, Loeffler A, Mccarthy A, Lloyd DH, Musilov E, Lindsay JA. Whole‐genome comparison of meticillin‐resistant Staphylococcus aureus CC22 SCC mec IV from people and their in‐contact pets. Vet Dermatol. 2013;24:538–e‐128. 10.1111/vde.12062. [DOI] [PubMed] [Google Scholar]
- 7. Duim B, Verstappen KMHW, Kalupahana RS, Ranathunga L, Fluit AC, Wagenaar JA. Methicillin‐resistant Staphylococcus pseudintermedius among dogs in the description of novel SCCmec variants. Vet Microbiol. 2018;213(April 2017):136‐141. [DOI] [PubMed] [Google Scholar]
- 8. Perreten V, Kadlec K, Schwarz S, et al. Clonal spread of methicillin‐resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother. 2010;65(6):1145‐1154. [DOI] [PubMed] [Google Scholar]
- 9. Somayaji R, Priyantha MAR, Rubin JE, Church D. Human infections due to staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis. 2016;85(4):471‐476. [DOI] [PubMed] [Google Scholar]
- 10. Gobeli Brawand S, Cotting K, Gómez‐Sanz E, et al. Macrococcus canis sp. nov., a skin bacterium associated with infections in dogs. Int J Syst Evol Microbiol. 2017. Mar;67(3):621‐626. [DOI] [PubMed] [Google Scholar]
- 11. Cotting K, Strauss C, Rodriguez‐Campos S, et al. Macrococcus canis and M. caseolyticus in dogs: occurrence, genetic diversity and antibiotic resistance. Vet Dermatol. 2017;28(6):559‐e133. [DOI] [PubMed] [Google Scholar]
- 12. Chanchaithong P, Perreten V, Schwendener S. Macrococcus canis contains recombinogenic methicillin resistance elements and the mecB plasmid found in Staphylococcus aureus . J Antimicrob Chemother. 2019;74(9):2531‐2536. [DOI] [PubMed] [Google Scholar]
- 13. Wedley AL, Dawson S, Maddox TW, et al. Carriage of antimicrobial resistant Escherichia coli in dogs: Prevalence, associated risk factors and molecular characteristics. Vet Microbiol. 2017;199:23‐30. [DOI] [PubMed] [Google Scholar]
- 14. Adams RJ, Kim SS, Mollenkopf DF, et al. Antimicrobial‐resistant Enterobacteriaceae recovered from companion animal and livestock environments. Zoonoses Public Health. 2018;65(5):519‐527. [DOI] [PubMed] [Google Scholar]
- 15. Rubin JE, Pitout JDD. Extended‐spectrum β‐lactamase, carbapenemase and AmpC producing Enterobacteriaceae in companion animals. Vet Microbiol. 2014;170(1–2):10‐18. [DOI] [PubMed] [Google Scholar]
- 16. Wohlwend N, Endimiani A, Francey T, Perreten V. Isolates from humans and companion animals in Switzerland: spread of a DHA‐producing sequence type 11 clone in a veterinary setting. Antimicrob Agents Chemother. 2015;59(5):2949‐2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu X, Thungrat K, Boothe DM. Occurrence of OXA‐48 carbapenemase and other β‐lactamase genes in ESBL‐producing multidrug resistant Escherichia coli from dogs and cats in the United States, 2009–2013. Front Microbiol. 2016;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akiba M, Sekizuka T, Yamashita A, et al. Distribution and relationships of antimicrobial resistance determinants among extended‐spectrum‐cephalosporin‐resistant or carbapenem‐resistant Escherichia coli isolates from rivers and sewage treatment plants in India. Antimicrob Agents Chemother. 2016;60(5):2972‐2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4(MAR):1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grönthal T, Österblad M, Eklund M, et al. Sharing more than friendship – transmission of NDM‐5 ST167 and CTX‐M‐9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill. 2018;23(27):1700497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith A, Wayne AS, Fellman CL, Rosenbaum MH. Usage patterns of carbapenem antimicrobials in dogs and cats at a veterinary tertiary care hospital. J Vet Intern Med. 2019;33(4):1677‐1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Logan LK, Weinstein RA. The epidemiology of carbapenem‐resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28‐S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JGNIH. Research Electronic Data Capture. J Biomed Inform. 2009;42(2):377‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmidt JS, Kuster SP, Nigg A, et al. Poor infection prevention and control standards are associated with environmental contamination with carbapenemase‐producing Enterobacterales and other multidrug‐resistant bacteria in Swiss companion animal clinics. Antimicrob Resist Infect Control. 2020;9(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nigg A, Brilhante M, Dazio V, et al. Shedding of OXA‐181 carbapenemase‐producing Escherichia coli from companion animals after hospitalisation in Switzerland: an outbreak in 2018. Euro Surveill. 2019;24(39):1‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pires J, Novais A, Peixe L. Blue‐Carba, an easy biochemical test for detection of diverse Carbapenemase producers directly from bacterial cultures. J Clin Microbiol. 2013;51(12):4281‐4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Overesch G, Büttner S, Rossano A, Perreten V. The increase of methicillin‐resistant Staphylococcus aureus (MRSA) and the presence of an unusual sequence type ST49 in slaughter pigs in Switzerland. BMC Vet Res. 2011;7(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) . Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0, 2019. Växjö: EUCAST; 2019. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdforg. Accessed October, 2019.
- 30. CLSI , Dolinsky AL, Ohiro RK, et al. Performance standard for antimicrobial susceptibility testing. Document M100–S10. J Int Med Res. 2017;46:18. [Google Scholar]
- 31. Bernasconi OJ, Principe L, Tinguely R, et al. Evaluation of a new commercial microarray platform for the simultaneous detection of β‐lactamase and mcr‐1 and mcr‐2 genes in Enterobacteriaceae. J Clin Microbiol. 2017;55(10):3138‐3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poirel L, Potron A, Nordmann P. OXA‐48‐like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67(7):1597‐1606. [DOI] [PubMed] [Google Scholar]
- 33. Louie L, Goodfellow J, Mathieu P, Glatt A, Louie M, Simor AE. Rapid detection of methicillin‐resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J Clin Microbiol. 2002;40(8):2786‐2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gómez‐Sanz E, Schwendener S, Thomann A, Brawand SG, Perreten V. First staphylococcal cassette chromosome mec containing a mecB‐carrying gene complex independent of transposon Tn6045 in a macrococcus caseolyticus isolate from a canine infection. Antimicrob Agents Chemother. 2015;59(8):4577‐4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwendener S, Cotting K, Perreten V. Novel methicillin resistance gene mecD in clinical macrococcus caseolyticus strains from bovine and canine sources. Sci Rep. 2017;7(2016):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Endimiani A, Hujer KM, Hujer AM, et al. Acinetobacter baumannii isolates from pets and horses in Switzerland: molecular characterization and clinical data. J Antimicrob Chemother. 2011;66(10):2248‐2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Büdel T, Kuenzli E, Clément M, et al. Polyclonal gut colonization with extended‐spectrum cephalosporin‐ and/or colistin‐resistant Enterobacteriaceae: a normal status for hotel employees on the Island of Zanzibar, Tanzania. J Antimicrob Chemother. 2019;74(10):2880‐2890. [DOI] [PubMed] [Google Scholar]
- 38. Van Den Bunt G, Fluit AC, Spaninks MP, et al. Faecal carriage, risk factors, acquisition and persistence of ESBL‐producing Enterobacteriaceae in dogs and cats and co‐carriage with humans belonging to the same household. J Antimicrob Chemother. 2020;75(2):342‐350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Melo LC, Oresco C, Leigue L, et al. Prevalence and molecular features of ESBL/pAmpC‐producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet Microbiol. 2018;221(May):59‐66. [DOI] [PubMed] [Google Scholar]
- 40. Murphy C, Reid‐Smith RJ, Prescott JF, et al. Occurrence of antimicrobial resistant bacteria in healthy dogs and cats presented to private veterinary hospitals in southern Ontario: a preliminary study. Can Vet J. 2009;50(10):1047‐1053. [PMC free article] [PubMed] [Google Scholar]
- 41. Hamilton E, Kruger JM, Schall W, Beal M, Manning SD, Kaneene JB. Acquisition and persistence of antimicrobial‐resistant bacteria isolated from dogs and cats admitted to a veterinary teaching hospital. J Am Vet Med Assoc. 2013;243(7):990‐1000. [DOI] [PubMed] [Google Scholar]
- 42. Pires J, Bernasconi OJ, Kasraian S, Hilty M, Perreten V, Endimiani A. Intestinal colonisation with extended‐spectrum cephalosporin‐resistant Escherichia coli in Swiss pets: molecular features, risk factors and transmission with owners. Int J Antimicrob Agents. 2016;48(6):759‐760. [DOI] [PubMed] [Google Scholar]
- 43. Gandolfi‐Decristophoris P, Regula G, Petrini O, Zinsstag J, Schelling E. Prevalence and risk factors for carriage of multi‐drug resistant staphylococci in healthy cats and dogs. J Vet Sci. 2013;14(4):449‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pires J, Kuenzli E, Hauser C, et al. Intestinal colonisation with extended‐spectrum cephalosporin‐resistant Enterobacteriaceae in different populations in Switzerland: prevalence, risk factors and molecular features. J Glob Antimicrob Resist. 2018;12:17‐19. [DOI] [PubMed] [Google Scholar]
- 45. Ngaiganam EP, Pagnier I, Chaalal W, et al. Investigation of urban birds as source of β‐lactamase‐producing Gram‐negative bacteria in Marseille city, France. Acta Vet Scand. 2019;61(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vogt D, Overesch G, Endimiani A, Collaud A, Thomann A, Perreten V. Occurrence and genetic characteristics of third‐generation cephalosporin‐resistant Escherichia coli in Swiss retail meat. Microb Drug Resist. 2014;20(5):485‐494. [DOI] [PubMed] [Google Scholar]
- 47. Seiffert SN, Carattoli A, Tinguely R, Lupo A, Perreten V, Endimiani A. High prevalence of extended‐spectrum β‐lactamase, plasmid‐mediated AmpC, and carbapenemase genes in pet food. Antimicrob Agents Chemother. 2014;58(10):6320‐6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davis JA, Jackson CR, Fedorka‐Cray PJ, et al. Carriage of methicillin‐resistant Staphylococci by healthy companion animals in the US. Lett Appl Microbiol. 2014;59(1):1‐8. [DOI] [PubMed] [Google Scholar]
- 49. Ma GC, Worthing KA, Gottlieb T, Ward MP, Norris JM. Molecular characterization of community‐associated methicillin‐resistant Staphylococcus aureus from pet dogs. Zoonoses Public Health. 2019;67:1‐9. [DOI] [PubMed] [Google Scholar]
- 50. Schmidt VM, Williams NJ, Pinchbeck G, et al. Antimicrobial resistance and characterisation of staphylococci isolated from healthy Labrador retrievers in the United Kingdom. BMC Vet Res. 2014;10(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vincze S, Brandenburg AG, Espelage W, et al. Risk factors for MRSA infection in companion animals: results from a case–control study within Germany. Int J Med Microbiol. 2014. Oct;304(7):787‐793. [DOI] [PubMed] [Google Scholar]
- 52. Burow E, Käsbohrer A. Risk factors for antimicrobial resistance in Escherichia coli in pigs receiving oral antimicrobial treatment: a systematic review. Microb Drug Resist. 2017;23(2):194‐205. [DOI] [PubMed] [Google Scholar]
- 53. Predic M, Delano JP, Tremblay E, Iovine N, Brown S, Prins C. Evaluation of patient risk factors for infection with carbapenem‐resistant Enterobacteriaceae. Am J Infect Control. 2020;48(9):1028‐1031. [DOI] [PubMed] [Google Scholar]
- 54. Gandolfi‐Decristophoris P, De Benedetti A, Petignat C, et al. Evaluation of pet contact as a risk factor for carriage of multidrug‐resistant staphylococci in nursing home residents. Am J Infect Control. 2012;40(2):128‐133. [DOI] [PubMed] [Google Scholar]
- 55. Normanno G, Dambrosio A, Lorusso V, Samoilis G, Di Taranto P, Parisi A. Methicillin‐resistant Staphylococcus aureus (MRSA) in slaughtered pigs and abattoir workers in Italy. Food Microbiol. 2015;51:51‐56. [DOI] [PubMed] [Google Scholar]
- 56. Barilli E, Vismarra A, Villa Z, Bonilauri P, Bacci CESLE. Coli isolated in pig's chain: genetic analysis associated to the phenotype and biofilm synthesis evaluation. Int J Food Microbiol. 2019;289(2018):162‐167. [DOI] [PubMed] [Google Scholar]
- 57. Yu ZN, Wang J, Ho H, Wang YT, Huang SN, Han RW. Prevalence and antimicrobial‐resistance phenotypes and genotypes of Escherichia coli isolated from raw milk samples from mastitis cases in four regions of China. J Glob Antimicrob Resist. 2019;22:94‐101. [DOI] [PubMed] [Google Scholar]
- 58. Baede VO, Broens EM, Spaninks MP, et al. Raw pet food as a risk factor for shedding of extended‐spectrum beta‐lactamase‐producing Enterobacteriaceae in household cats. PLoS One. 2017;12(11):1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nüesch‐Inderbinen M, Treier A, Zurfluh K, Stephan R. Raw meat‐based diets for companion animals: a potential source of transmission of pathogenic and antimicrobial‐resistant Enterobacteriaceae. R Soc Open Sci. 2020;6(10):191170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V. Identification and screening of carbapenemase‐producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18(5):432‐438. [DOI] [PubMed] [Google Scholar]
- 61. Endimiani A, Brilhante M, Bernasconi OJ, et al. Employees of Swiss veterinary clinics colonized with epidemic clones of carbapenemase‐producing Escherichia coli . J Antimicrob Chemother. 2020;75(3):766‐768. [DOI] [PubMed] [Google Scholar]
- 62. Pomba C, Rantala M, Greko C, et al. Public health risk of antimicrobial resistance transfer from companion animals. J Antimicrob Chemother. 2017;72(4):957‐968. [DOI] [PubMed] [Google Scholar]
- 63. De Waele JJ, Boelens J, Leroux‐Roels I. Multidrug‐resistant bacteria in ICU: fact or myth. Curr Opin Anaesthesiol. 2020;33(2):156‐161. [DOI] [PubMed] [Google Scholar]
- 64. Lutz B, Lehner C, Schmitt K, et al. Antimicrobial prescriptions and adherence to prudent use guidelines for selected canine diseases in Switzerland in 2016. Vet Rec Open. 2020;7(1):e000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schmitt K, Lehner C, Schuller S, et al. Antimicrobial use for selected diseases in cats in Switzerland. BMC Vet Res. 2019;15(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gentilini F, Turba ME, Pasquali F, et al. Hospitalized pets as a source of carbapenem‐resistance. Front Microbiol. 2018;9:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Robinson GL, Harris AD, Morgan DJ, et al. Survival of methicillin‐resistant Staphylococcus aureus and vancomycin‐resistant Enterococcus spp. for an extended period of transport. J Clin Microbiol. 2012. Jul;50(7):2466‐2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mende K, Beckius ML, Hospenthal DR. Recovery of multidrug‐resistant bacteria from swabs stored for durations of 1 and 4 weeks under conditions mimicking long‐distance‐shipping conditions. J Clin Microbiol. 2014;52(5):1798‐1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Supporting information


