Significance
The misfolding and aggregation into amyloid fibrils of the prion protein (PrP) have been strongly linked with a group of neurodegenerative disorders that include the mad cow disease. Currently, the molecular origins of the prion diseases are unknown, including the underlying mechanisms of PrP misfolding and the regions promoting its aggregation. Here, we identified the structural basis by which the folded domain of the human PrP converts into amyloids. We showed that this process is promoted by intermediate species forming as a result of the pathological mutation T183A, and that POM antibodies are able to suppress completely the aggregation process by blocking the misfolding mechanism. This study thereby suggests possible molecular strategies to inhibit PrP aggregation into amyloids.
Keywords: neurodegenerative diseases, prion protein, protein misfolding, amyloid, transmissible spongiform encephalopathies (TSEs)
Abstract
The misfolding and aggregation of the human prion protein (PrP) is associated with transmissible spongiform encephalopathies (TSEs). Intermediate conformations forming during the conversion of the cellular form of PrP into its pathological scrapie conformation are key drivers of the misfolding process. Here, we analyzed the properties of the C-terminal domain of the human PrP (huPrP) and its T183A variant, which is associated with familial forms of TSEs. We show that the mutation significantly enhances the aggregation propensity of huPrP, such as to uniquely induce amyloid formation under physiological conditions by the sole C-terminal domain of the protein. Using NMR spectroscopy, biophysics, and metadynamics simulations, we identified the structural characteristics of the misfolded intermediate promoting the aggregation of T183A huPrP and the nature of the interactions that prevent this species to be populated in the wild-type protein. In support of these conclusions, POM antibodies targeting the regions that promote PrP misfolding were shown to potently suppress the aggregation of this amyloidogenic mutant.
The misfolding and aggregation of the human prion protein (PrP) is associated with a number of fatal neurodegenerative disorders designated as transmissible spongiform encephalopathies (TSEs), including Kuru, Creutzfeldt–Jakob disease, fatal familial insomnia, and mad cow disease (1). In its physiological form, the cellular PrP (PrPC) is a 23-kDa monomeric glycosylated protein that is linked to the outer surface of the neuronal plasma membrane via a glycophosphatidylinositol (GPI) anchor (2). Under conditions associated with TSEs, PrPC misfolds into a nonnative conformation (scrapie PrPSc) that is prone to aggregation into insoluble amyloid fibrils (3). The conversion into PrPSc has been proposed to be able to propagate from one cell to another (4). Genetic traits also exist linking point mutations of PrP and familial forms of TSEs. These pathological mutations are mostly located in the C-terminal globular domain of the PrPC and have been shown to generally enhance the aggregation propensity of PrP (5, 6).
While the pathological relevance of PrP is now established, its function remains highly debated. PrPC has been shown to be involved in the maintenance of myelin on peripheral nerves (7), and evidence exists for a number of other putative physiological roles, including calcium modulation, copper sensing, long-term potentiation, and long-term memory (8). A well-characterized property of the physiological PrPC is its native structure, which is composed of a disordered N-terminal flexible tail (residues 23–124) and a structured C-terminal region (125–230) composed of three α-helices and a short antiparallel β-sheet (9–11). Recently, structures of amyloid states of PrP generated in vitro have been resolved, providing insights about the possible in vivo form of PrPSc (12, 13). Despite the progress made in the structural characterization of PrPSc, there are major gaps in our understanding of the mechanisms that trigger the misfolding and aggregation of PrPC under conditions associated with the insurgence and development of TSEs. A major challenge in this context is the study of metastable misfolding intermediates, which are elusive to conventional experimental techniques for structure determination because of their transient and heterogeneous nature (14). It is now generally acknowledged that the initial step for the misfolding of the C-terminal domain of PrPC is the disruption of the native packing of the region formed by the two β-strands S1 and S2 and the α-helix H1, against the region composed of the helices H2–H3 (15). The detachment of these two subdomains, denoted as S1–H1–S2 and H2–H3, respectively, is prevented under native conditions by four main intramolecular “gatekeeper” interactions (D178–R164, T183–Y162, H187–R156, D202–R156). Each of these interactions is altered by specific pathological mutations associated with inherited forms of TSEs (D178N, T183A, H187R, D202N), suggesting a destabilization of the native interface between the two subdomains (15). Among these PrP variants, a mutation associated with very early-onset dementia and spongiform encephalopathy in patients, namely T183A (16), abolishes a crucial hydrogen bond between Y162 from the β-strand S2 and T183 from the α-helix H3. Under physiological conditions, this H-bond stabilizes the packing at the interface between the subdomains S1–H1–S2 and H2–H3 (SI Appendix, Fig. S1) (17), and its depletion by T183A induces the strongest destabilization of the PrPC structure among the TSE-associated PrP variants (18).
We addressed in the present study the fundamental early molecular mechanism of human PrPC (huPrPC) misfolding induced by T183A using NMR experiments in combination with biophysical investigations and enhanced molecular metadynamics simulations. The study compared the properties of wild-type (WT) and T183A huPrPC and identified a misfolded intermediate species that acts as a precursor to the formation of amyloid aggregates by the C-terminal domain of the mutant (residues 125–230) despite the fact that this construct lacks the amyloidogenic region 106–126 (19). We provide conclusive evidence of this aggregation mechanism using POM antibodies (Abs) targeting the specific epitope that was here found to initiate the misfolding of T183A huPrPC. Taken together, these results generate a new detailed understanding of the structural transitions in huPrPC that trigger its amyloid formation and provide proof of principle for a structure-based identification of targeted molecular strategies to prevent huPrPC misfolding and aggregation.
Results
T183A Promotes a Misfolded Intermediate in huPrPC125–230.
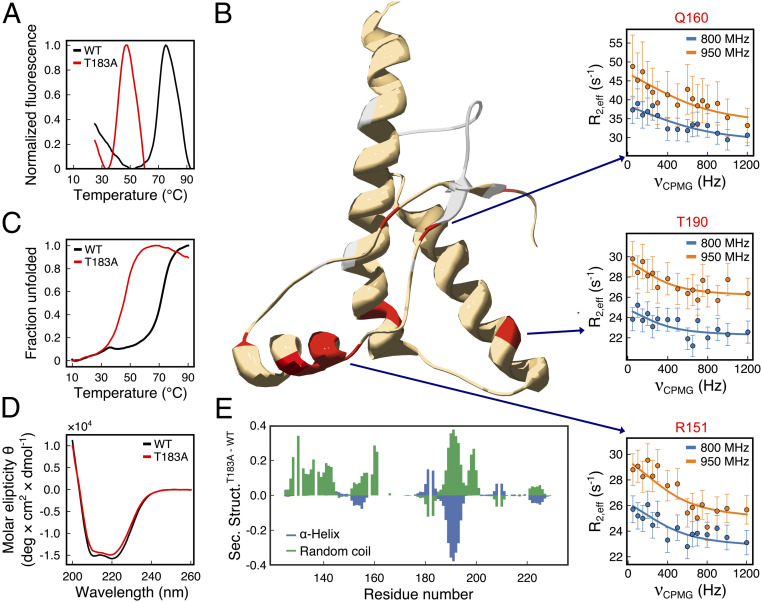
We purified the recombinant C-terminal globular domain (residues 125–230) of WT huPrPC125–230 and compared its structural and thermodynamic properties to those of the T183A variant. Differential scanning fluorimetry (DSF) revealed that T183A strongly destabilizes this huPrPC domain, resulting in a reduction of 28 °C in the melting temperature (Tm, Fig. 1A and SI Appendix, Table S1). Similar experimental observations were obtained using circular dichroism (CD) (Fig. 1C and SI Appendix, Fig. S2), indicating a reduction of 26 °C in the Tm of the mutant. The CD curves were fitted with the Gibbs–Helmholtz equation (20), providing values for ΔΔHT183A-WT (+2.1 kcal/mol) and ΔΔGT183A-WT (+2.1 kcal/mol) at 37 °C.
Fig. 1.
Properties of WT and T183A huPrPC125–230. (A) Melting curves of WT (black) and T183A (red) huPrPC125–230 probed using differential scanning fluorimetry (DSF) in combination with the SYPRO orange dye. Fluorescence at 570 nm is shown as a function of the temperature. The resulting melting temperatures (Tm) are 69 and 41 °C for WT and T183A, respectively. (B) Conformational exchange in T183A huPrPC125–230 as probed by NMR CPMG experiments. Three hot-spot regions were identified featuring residues in conformational exchange (red). Representative relaxation dispersion curves of residues from each hot spot (R151, Q160, and T190; one for each hot spot) are shown. Data measured at 800 and 950 MHz are plotted in blue and orange, respectively. Residues whose resonances are broadened beyond detection are colored in gray. (C) Melting curves probed using CD at 222 nm as a function of the temperature. Tm are 71 and 45 °C for WT (black) and T183A (red), respectively. (D) CD spectra of WT (black) and T183A (red) variants. (E) Residue-specific secondary structure populations from the δ2D (21) analysis of NMR chemical shifts. α-Helix (blue) and coil (green) populations in T183A minus WT are reported along the sequence.
The significant perturbation in the folding stability of T183A huPrPC125–230 does not correlate with changes in the secondary structure elements of the protein, which were found to be mostly preserved in the mutant. In particular, CD spectra measured at 16 °C for WT and T183A huPrPC125–230 overlap considerably (Fig. 1D), indicating only a minor reduction in the α-helical content in the mutant. To obtain higher resolution data on the protein secondary structure, we measured the NMR resonances of the backbone atoms and analyzed these observables using the δ2D method (21) (SI Appendix, Fig. S3). The results revealed, at a residue-specific resolution, the secondary-structure populations in the two PrP variants, indicating that the T183A mutation induces considerable loss of α-helix content in the C-terminal segment of the α-helix H2 (residues 184–194, Fig. 1E). This threonine-rich region of PrP was shown to have ambivalent structural propensity in vitro (22) and to be a hot spot for local misfolding (15). In addition, the α-helix H1 and the short antiparallel β-sheet region showed a partial loss of secondary structure population in T183A huPrPC125–230. Overall, the NMR analyses revealed, in agreement with CD measurements, that T183A induces a reduction of 7.5% in the secondary structure content of huPrPC125–230.
The local perturbation in the secondary structure elements of T183A huPrPC125–230 does not appear to affect the overall structural fold of the protein, as revealed by similar distributions of peaks in the 1H–15N HSQC spectra of the two variants (SI Appendix, Fig. S4). Some chemical shift differences (Δδ, Methods), however, were observed (SI Appendix, Fig. S5), specifically for amino acids that are spatially close to residue 183 (V180, I184, and V210; SI Appendix, Fig. S5C). In addition, the NMR spectra of T183A huPrPC125–230 were generally associated with larger line widths (Δν) than the spectra of the WT (SI Appendix, Fig. S6), with resonances of nine residues found to be broadened beyond detection. These residues are located in the β-strands S2 (V161, Y162, Y163, and M166) and S1 (M129 and G131) and in regions of the α-helixes H2 and H3 that are close the site of the mutation in the huPrP structure (I182, V209, and M213). The broadening of these NMR signals indicates faster transverse relaxations in the T183A variant, which are generally associated with enhanced conformational dynamics in protein molecules. We verified, however, whether the observed line broadening is associated with a monomer–oligomer equilibrium (23) by measuring two 1H–15N HSQC spectra with a 10-fold difference in the protein concentration. The data showed no additional peaks as a result of the protein dilution, with the two spectra featuring very similar line widths (SI Appendix, Fig. S7). This finding indicates that the line broadening observed in the NMR spectra of T183A huPrPC125–230 is not a consequence of monomer–oligomer equilibrium but can be attributed to enhanced structural fluctuations of the protein.
To further characterize the structural dynamics of T183A huPrPC125–230, we performed Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion experiments (24, 25). CPMG is an accurate probe of the equilibrium between two or more distinct macromolecular conformations exchanging in the submillisecond timescale. These experiments identified 11 residues of T183A huPrPC125–230 undergoing conformational exchange and clustering in three specific “hot spot” regions of the protein (Fig. 1B and SI Appendix, Fig. S8). A large hot spot includes residues from the α-helix H1 (S143, Y149, R151, E152, and N153) and its interface with the α-helix H3 (V203 and K204). A second hot spot involves the antiparallel β-sheet, with three residues (Y128, M134, and Q160) showing conformational exchange in CPMG. These hot spot are flanked by some of the residues whose resonances are broadened beyond NMR detection (M129, G131, V161, Y162, Y163, and M166), collectively indicating strong conformational exchange in the β-sheet region of the mutant. A third hot spot was found in the C-terminal region of the α-helix H2, where conformational exchange in CPMG was observed for residue T190. In conjunction with the observed loss of α-helical content in the region 184–194, this finding suggests that in T183A huPrPC125–230 the C terminus of the α-helix H2 is highly dynamical and structurally unstable. Using a two-state model (26), the fitting of the relaxation dispersion curves revealed that T183A huPrPC125–230 exists in equilibrium between two conformations with uneven populations (98.6% and 1.4%) and exchanging with a of 2,686 ± 509 s−1. The 15N chemical shift differences (Δω) between the resonances of the two conformations are substantial (in average, 1.84 ± 0.67 ppm), which is indicative of considerable rearrangements in the transition between the two structural states (SI Appendix, Table S2).
The enhanced structural dynamics observed in T183A huPrPC125–230 is not mirrored by the WT construct, as shown in CPMG data indicating essentially no conformational exchange by any residue in the protein (SI Appendix, Fig. S8). Taken together, these experimental observations suggest that the T183A mutation causes minor disruption of specific secondary structure regions, destabilizes the huPrPC125–230 fold, and induces conformational exchange between a major (native) and a minor conformation.
Mechanism of Misfolding of huPrPC125–230 into an Amyloidogenic State.
Having identified a low population species that accumulates in the T183A huPrPC125–230 ensemble, we investigated its dynamical properties using metadynamics simulations (27) to identify rare structural transitions of the protein by sampling its conformational free energy landscape. Metadynamics simulations were carried out using five different collective variables (Methods) across five replicas (SI Appendix, Fig. S9), each simulated for 800 ns until proven convergence (SI Appendix, Fig. S10 and Methods), under the well-tempered bias-exchange approach (Methods). As a starting configuration for both variants we utilized the atomic structure of huPrPC (28) (Protein Data Bank ID code 1HJM) and, in the case of the mutant, substituted in silico the threonine 183 of this structure into an alanine residue.
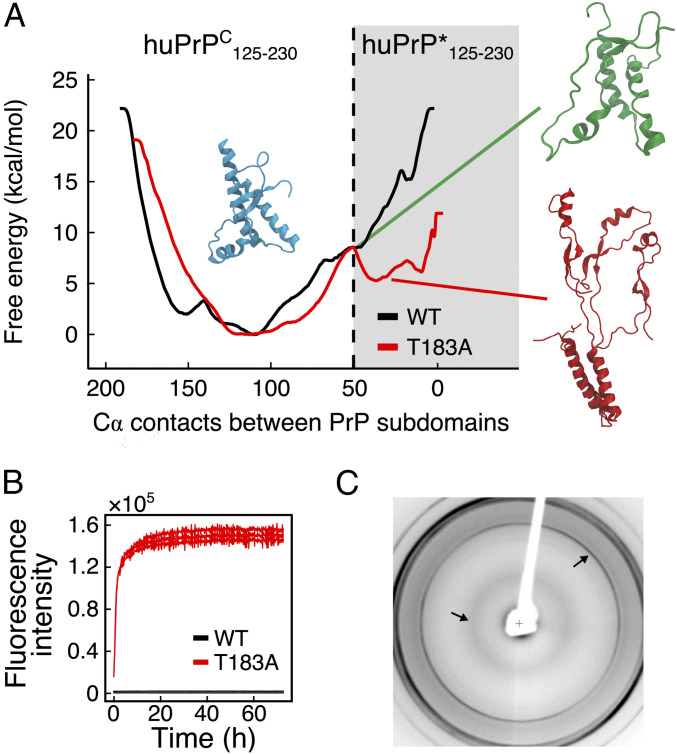
The resulting free energy landscapes of WT and T183A huPrPC125–230 revealed that the native structure is the most stable conformation in both variants. In addition to this major conformation, a high energy state designated as huPrP*125–230 was observed in the simulations of the mutant. This species showed distinctive conformational properties, such as the detachment of the subdomains S1–H1–S2 and H2–H3 and a partial loss of α-helical structure, specifically in the C-terminal region of the α-helix H2 and in the α-helix H1, which becomes solvent exposed in huPrP*125–230. The equilibrium between huPrPC125–230 and huPrP*125–230 in the simulations is substantiated by several NMR data, including the conformational exchange of hot-spot regions identified in CPMG experiments and the destabilization of specific elements of secondary structure as highlighted by chemical shift analyses (Fig. 1). In order to further characterize the nature of T183A huPrP*125–230, we calculated its chemical shifts using SPARTA+ (29) and compared these values with those calculated for the native structure. The average chemical shift difference (ΔωMD) between these two metadynamics states (1.46 ± 1.16 ppm) was found to be in close match, and within the SPARTA+ SE (30), with the average ΔωNMR (1.84 ± 0.67 ppm) obtained by CPMG experiments. To further compare the simulations and the NMR experiments, we estimated the rate constants expected from the metadynamics free energy surfaces. In particular, the surfaces provided the free energy barriers intervening between the native T183A huPrPC125–230 and huPrP*125–230 conformations, being 8.5 ± 0.3 and 3.2 ± 0.3 kcal/mol for the forward and reverse reactions, respectively. These energy values were used in the Eyiring theory (Methods) to estimate the rate of exchange between native and intermediate metadynamics basins, resulting in a value of 2,642 s−1, in excellent agreement with the experimental exchange rate (2,686 ± 509 s−1).
In contrast to the mutant, the WT protein was found to have an energy barrier of 14.7 ± 0.5 kcal/mol, which effectively prevents the protein from accessing the conformations of huPrP*125–230 (Fig. 2A) and allows large-scale fluctuations resulting only in a minor and reversible detachment of the α-helix H1 from the native interface (SI Appendix, Fig. S11). We further tested whether huPrP*125–230 can be accessed under physiological conditions by the WT protein. In particular, we run two unbiased molecular dynamics simulations, respectively sampling T183A and WT huPrP*125–230 (SI Appendix, Fig. S12). The results showed that WT huPrP*125–230 is unstable and readily evolves into the native conformation by repacking the β-sheet onto the α-helix H3 and forming the native contacts between the subdomains S1–H1–S2 and H2–H3. By contrast, T183A huPrP*125–230 resulted to be metastable in the simulated time, due to the inability to lock the β-strand S2 in the native interface. These simulations suggested therefore that, unlike the WT construct, the mutant has a tendency to accumulate misfolded huPrP*125–230 species under physiological conditions.
Fig. 2.
Misfolding and aggregation of T183A huPrPC125–230. (A) Metadynamics free energy landscape of WT (black) and T183A (red) huPrPC125–230 reconstructed on the number of residue contacts between subdomains S1–H1–H2 and H2–H3. A representative native huPrPC125–230 structure is shown in light blue (number of contacts > 100), whereas huPrP*125–230 is shown with a red structure (number of contacts < 50). The transition between these two conformations requires energy barriers of 8.5 ± 0.3 kcal/mol for the transition from T183A huPrPC125–230 to T183A huPrP*125–230 and 3.2 ± 0.3 kcal/mol for the reverse conversion. The conformation in the saddle point (green structure; number of contacts = 50) features a partial detachment of α-helix H1 from the native interface and the disruption of the C-terminal region of α-helix H2 (residues 184–194). (B) ThT fluorescence assay of WT (black) and T183A (red) huPrPC125–230 incubated at 37 °C in PBS, pH 7.4, under orbital shaking at 200 rpm and at a concentration of 10 μM. (C) X-ray diffraction pattern of T183A huPrP125–230 fibers formed by incubation at 50 μM in 100 mM Na2HPO4 overnight at 37 °C and 200 rpm shaking. The arrows indicate the characteristic amyloid peaks at 4.8 Å and at 10 Å.
Amyloid Formation by T183A huPrPC125–230 Is Suppressed by Anti-PrPC Antibodies.
The amyloid formation by huPrPC has been shown to depend on the presence of the aggregation-prone region 106–126 (19). The shortest PrPC construct currently known to convert into amyloids from the native structure indeed spans residues 90–231 (D178N variant) (31, 32). Since the T183A mutation was here found to promote the accumulation of a misfolded intermediate in huPrP125–230 under native conditions, we employed an established Thioflavin T (ThT) fluorescence assay (33) to probe the amyloidogenic propensity of the mutant. By incubating 10 μM WT huPrPC125–230 at 37 °C in PBS buffer and under shaking at 200 rpm, no aggregation into amyloid fibrils was observed for 5 d (Fig. 2B); however, under the same experimental conditions, the mutant induced a rapid increase in ThT fluorescence, indicating amyloid formation with no apparent lag phase (Fig. 2B). The aggregates formed by T183A huPrPC125–230 showed the characteristic signatures of amyloids as probed by X-ray diffraction experiments, with a 4.8-Å peak of the β-sheet periodicity and a 10-Å peak arising from the packing of facing β-sheets in the amyloid core (34) (Fig. 2C). This is an observation of the conversion into amyloids by the sole globular domain of PrPC (residues 125–230), which was not observed for other variants of PrP including the WT.
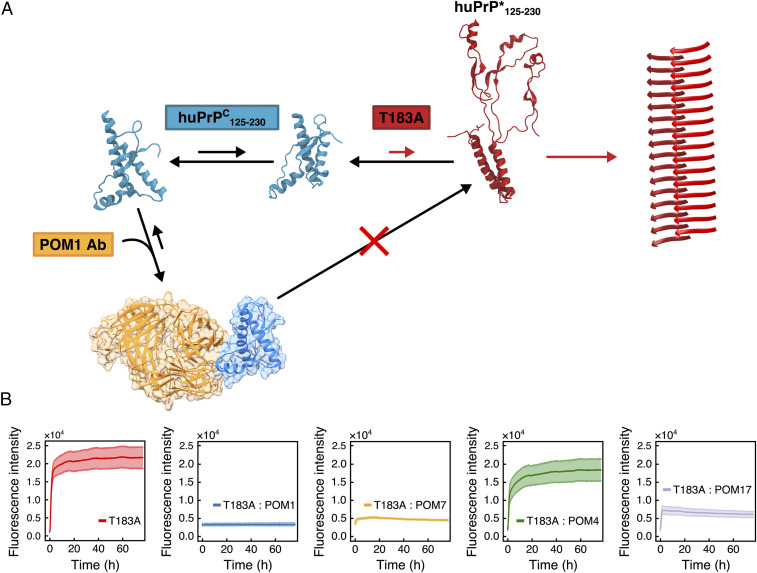
To investigate the relevance of the misfolding intermediate huPrP*125–230 in the aggregation of T183A huPrPC125–230, we used monoclonal POM Abs (35) to lock the protein in its native structure by specifically preventing the detachment of the α-helix H1 from the native packing. These Abs were developed against a variety of epitopes of PrPC, including the interface between the subdomains S1–H1–S2 and H2–H3 that is recognized by POM1 via the simultaneous binding of the α-helixes H1 and H3 (36). First, the affinity of POM1 scFv for WT and T183A huPrPC125–230 was probed by surface plasmon resonance. Sensograms were analyzed with a simple 1:1 Langmuir interaction model. As this model did not fit the data in a perfectly accurate manner, we additionally analyzed the sensograms with a “heterogeneous ligand” model that better fits heterogenous binding surfaces. Both models revealed comparable dissociation constants (KD) for the two variants (SI Appendix, Fig. S13 and Table S3). Subsequently, the aggregation of T183A huPrPC125–230 was monitored in the presence of POM1 at a 1:1 molar ratio PrP:POM1. The presence of POM1 completely suppressed the amyloid formation by T183A huPrPC125–230 (Fig. 3B). Consistent results were obtained using the POM1 single-chain variable fragment (scFv), a construct of 26 kDa that combines the variable regions of the light and heavy chains (35) (SI Appendix, Fig. S14).
Fig. 3.
POM Abs interfere with the misfolding process of T183A huPrPC125–230. (A) Model of the predicted suppression of the T183A huPrPC125–230 amyloid formation by POM1. The binding of POM1 at the interface between the α-helixes H1 and H3 prevents the detachment of subdomains S1–H1–S2 and H2–H3 and suppresses the accumulation of the misfolding intermediate huPrP* that leads to the amyloid formation. (B) ThT fluorescence traces of T183A huPrPC125–230 (red) and T183A huPrPC125–230 incubated with different POM Abs at a 1:1 molar ratio. The mean fluorescence trace from triplicate experiments is represented, with error bars corresponding to the mean ± SD. POM1 (blue) binds to α-helixes H1 and H3, POM4 (green) recognizes mouse PrPC only, POM7 (orange) recognizes α-helixes H1 and H2, and POM17 (purple) binds to α-helix H1.
Additional aggregation experiments of T183A huPrPC125–230 were carried out in the presence of POM4, POM7, and POM17 (35) (Fig. 3B). The results showed that the control POM4, which recognizes only the mouse PrPC and has no affinity for huPrPC, does not inhibit the aggregation of T183A huPrPC125–230. Conversely, POM7, which as POM1 is able to prevent the detachment of the two PrPC subdomains by interacting simultaneously with both helices H1 and H2, induces strong inhibition of the aggregation of T183A huPrPC125–230. Finally, an intermediate suppression of T183A huPrPC125–230 aggregation is induced by POM17, which binds only the α-helix H1 of huPrPC. The intermediate efficiency in preventing the aggregation of T183A huPrPC125–230 by POM17 is attributed to the stabilization of the α-helix H1, which was here shown to become unstable in huPrP*125–230. Taken together, these findings indicate that POM1, POM1 scFv, and POM7 are strong suppressors of the aggregation of T183A huPrPC125–230 as a result of the locking of subdomains S1–H1–S2 and H2–H3 into the native packing. The results thereby provide evidence for a link between the structural dynamics promoting the accumulation of huPrP*125–230 and the amyloid formation (Fig. 3A).
Discussion
Prion diseases are among the most relevant neurodegenerative disorder involving the misfolding and aggregation of otherwise-functional proteins. It is now established that the conversion of PrPC into PrPSc is strongly associated with the onset and development of TSEs (1). Currently, a limiting factor in the characterization of the underlying molecular mechanism of PrP misfolding is associated with the heterogeneity of the intermediate species forming in this process (37–39). We here generated considerable advancement in this challenge by providing evidence that a misfolding intermediate of huPrP, denoted huPrP*, is responsible for the aggregation into amyloids of the TSE-associated PrP mutant T183A. We characterized the structural properties of T183A huPrP*125–230 and identified the molecular basis of its enhanced propensity to promote amyloid formation. More specifically, the amyloidogenic intermediate species was shown to adopt a complete detachment of the subdomains S1–H1–H2 and H2–H3. The equilibrium between native species and huPrP*125–230 was observed to occur in the submillisecond timescale in both CPMG NMR experiments and metadynamics simulations, resulting in enhanced structural dynamics in T183A huPrPC125–230 that generate line broadening in the NMR spectra. The intermediate conformation also displays a reduced secondary structure, primarily in correspondence to the α-helix H1 and in the highly conserved TVTTTT motif at the C-terminal end of the α-helix H2. These observations are of key relevance for PrP misfolding in view of the role of the destabilization of the α-helix H1 (40–43) as well as of the local unfolding of helical segment TVTTTT (44) under acidic conditions (45, 46).
A relevant finding of the present study was that the C-terminal region of huPrPC can form amyloid aggregates under native conditions despite lacking the amyloidogenic segment 106–126, which is an essential element in the kinetics of aggregation of PrPC (19, 47). Amyloid conversion of the C-terminal domain under native conditions was not observed before and is likely to be unique to the T183A mutant. We found that by lowering of the energetic barrier to access the conformation of huPrP*125–230, the T183A mutation of huPrP promotes fast aggregation of the C-terminal domain without an apparent lag phase. Most of the biophysical studies (31, 48, 49) and cryo-EM structures (12) have indicated that the core of the PrP amyloids primarily includes residues ranging from 160 to 220. In contrast, a recent cryo-EM structure of PrP amyloids showed a core composed of residues 106–145 (13). Both alternative morphologies in PrPSc, however, require the separation of the S1–H1–S2 and H2–H3 subdomains from the native structure, indicating that the identified huPrP* conformation may be of general relevance for the conversion into pathological PrP aggregates.
We also provided evidence of how the characterization of the structural mechanisms of misfolding is fundamental to identify new approaches to suppress the amyloidogenic aggregation of PrP, as here illustrated using a set of POM (35) anti-PrPC monoclonal Abs that specifically suppress the structural fluctuations that generate huPrP* from huPrPC. The suppression of the aggregation of T183A huPrPC125–230 is total when using POMs binding epitopes of the native globular C-terminal structure that are at the interface between the subdomains S1–H1–H2 and H2–H3 (36), thus preventing the detachment of these regions from the native packing. This inhibition is in line with previous studies using other Abs to stop the propagation of PrPSc in full-length PrP strains by binding to the hydrophobic region of PrP (50, 51).
In conclusion, this study provides the details of a specific underlying mechanism of misfolding and aggregation of T183A huPrPC under physiological conditions, and indicates that the C-terminal structured domain can form amyloids even in the absence of the amyloidogenic segment 106–126. In addition to their value for understanding the origin of PrP misfolding, the experiments with POM Abs indicate that the suppression of the abnormal structural fluctuations of T183A huPrPC125–230 is a promising strategy to abolish the accumulation of huPrP* and to prevent PrP misfolding and aggregation.
Materials and Methods
huPrPC125–230 Sample Preparation.
WT and T183A huPrPC125–230 were expressed recombinantly in Escherichia coli, refolded, and purified up to a concentration of 100 μM in a buffer containing 100 mM Na2HPO4, pH 7.0. Details on the expression and purification are provided in SI Appendix, Methods.
CD Spectroscopy.
CD spectra were measured at a temperature of 16 °C and a huPrPC125–230 concentration of 20 μM in a 20 mM Na2HPO4, pH 7.0, buffer. Experiments were carried out using a Chirascan instrument (Applied Photophysics), on a quartz cuvette with path length of 1 mm, and a sample volume of 300 μL. Spectra were acquired between 200 and 260 nm with 0.5-nm intervals, a bandwidth of 1 nm, and scanning speed of 30 nm/s. CD melting curves were measured under the same buffer conditions and protein concentration, and were obtained by acquiring the CD signal intensity at 222 nm, starting at 10 °C and raising to 90 °C in 1 °C steps. Analysis of the melting profiles was performed by fitting the data to a two-state Gibbs–Helmholtz model (20).
DSF.
huPrPC125–230 samples for DSF experiments were diluted to a concentration of 10 μM into a 100 mM Na2HPO4, pH 7.0, buffer and combined with SYPRO Orange fluorophore at a 2× dye concentration. Melting curves were measured on a sealed 96-well plate, using a real-time PCR instrument, by following the fluorescence emission intensity in the 560- to 580-nm range after excitation at 450–490 nm. The curves were acquired in the temperature range 25 to 94 °C, at a 1 °C interval.
NMR.
All NMR spectra and experiments described were recorded at 16 °C and pH 7.0, using a huPrPC125–230 concentration of 100 μM in 100 mM Na2HPO4, 10% D2O. Spectral assignment of the backbone resonances was achieved using a combination of 1H–15N HSQC, HNCA, HN(CO)CA, CBCANH, CBCA(CO)NH, HNCO, and HN(CA)CO using an 800-MHz Advance III HD Bruker spectrometer. CPMG relaxation dispersion experiments (24, 25) were recorded also using a 950-MHz Advance III HD Bruker spectrometer. Details on the NMR measurements and data analysis are provided in SI Appendix, Methods.
ThT Assay.
ThT assays were performed at 37 °C in PBS, pH 7.4, under orbital shaking at 200 rpm and a huPrPC125–230 concentration of 10 μM. ThT was added at a concentration of 10 μM. Experiments were performed using an Omega FLUOstar microplate reader, with sealed 96-well plate with bottom optic, using a 440-nm excitation and 480-nm emission filter. Measurements were run for a minimum of 72 h, with a cycle time of 4 min. POM Abs and POM1-scFv employed in the assays were produced as described previously (35, 36). All assays with POM Abs were performed at a 1:1 molar ratio.
X-Ray Diffraction.
T183A huPrPC125–230 were incubated at 50 μM in 100 mM Na2HPO4 overnight at 37 °C and 200 rpm shaking in order to obtain a solution containing amyloid fibers. The solution was mixed, and a droplet of 10 μL was placed between two wax-tipped capillary tubes, and left to dry overnight (52). X-ray diffraction images were collected using a Rigaku Micromax 007HF-M high-flux generator, on a Rigaku Saturn 944+ CCD detector.
Surface Plasmon Resonance.
Measurements were performed with a Biacore T100 at 20 °C. POM1 scFv was immobilized on a CMD200M chip (Xantec) using standard NHS/EDC (N-hydroxy-succinimide/1-ethyl-3,3-dimethylaminopropyl-carbodiimide) chemistry with a density of 705 resonance units. Various concentrations of WT and T183A huPrPC125–230 (1.9, 3.8, 7.6, 15, and 30.5 nM) were diluted in HBS buffer (Teknova). The same buffer was also used as running buffer at a flow rate of 30 µL⋅min−1. For regeneration, the chip was washed with 10 mM glycerin buffer, pH 2.0. Data were evaluated with the 1:1 Langmuir and heterogeneous ligand model (BIAevaluation Software 3.1; Biacore) to determine the kinetic data. The heterogeneous ligand model describes the interaction of one analyte with two independent ligands or ligand sites on the same molecule, which show different binding behaviors based on their different immobilization. The observed binding is the sum of the interaction with the two ligands.
Metadynamics Simulations.
Metadynamics simulations were performed using five parallel replicas under the bias-exchange scheme. Details of the sampling, convergence, and collective variables are reported in SI Appendix, Methods. Simulations of the WT and T183A huPrPC125–230 were extended for 800 ns per replica, resulting in a total sampling time of 4 μs per construct. Energy barriers between native and intermediate basins of the metadynamics free energy landscape were used to estimate exchange rates () of exchange between T183A huPrPC125–230 and huPrP*125–230. The energies were employed in the Eyiring equation using a preexponential factor of 5 × 105 s−1, a value that has been established through the analysis of different downhill protein folders (53–55).
Extended materials and methods are in SI Appendix.
Supplementary Material
Acknowledgments
We thank Rita Moos and Marc Morgan for technical assistance and Stefano Gianni for discussions. A.D.S. acknowledges European Research Council (Consolidator Grant “BioDisOrder” 819644) and Alzheimer’s Research UK (Grant ARUK-PG2018B-013) for supporting this research. A.A. is the recipient of an Advanced Grant of the European Research Council (PRION2020) and grants from the Swiss National Research Foundation, the Nomis Foundation, and a donation from the estate of Dr. Hans Salvisberg. M.S.-H. is recipient of the Engineering and Physical Sciences Research Council Doctorate Prize Fellowship (EP1579441), which supported his research in this study. We acknowledge support by the Francis Crick Institute through provision of access to the Medical Research Council Biomedical NMR Centre (core funding Grant FC001029 by Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019631118/-/DCSupplemental.
Data Availability
NMR chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) (entries 50527 and 50528).
References
- 1.Prusiner S. B., Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A., Baumann F., Bremer J., The prion’s elusive reason for being. Annu. Rev. Neurosci. 31, 439–477 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Chiti F., Dobson C. M., Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Telling G. C., et al., Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 274, 2079–2082 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Vanik D. L., Surewicz W. K., Disease-associated F198S mutation increases the propensity of the recombinant prion protein for conformational conversion to scrapie-like form. J. Biol. Chem. 277, 49065–49070 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Apetri A. C., Surewicz K., Surewicz W. K., The effect of disease-associated mutations on the folding pathway of human prion protein. J. Biol. Chem. 279, 18008–18014 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Küffer A., et al., The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 536, 464–468 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wulf M.-A., Senatore A., Aguzzi A., The biological function of the cellular prion protein: An update. BMC Biol. 15, 34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riek R., et al., NMR structure of the mouse prion protein domain PrP(121–231). Nature 382, 180–182 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Zahn R., et al., NMR solution structure of the human prion protein. Proc. Natl. Acad. Sci. U.S.A. 97, 145–150 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knaus K. J., et al., Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 8, 770–774 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Wang L.-Q., et al., Cryo-EM structure of an amyloid fibril formed by full-length human prion protein. Nat. Struct. Mol. Biol. 27, 598–602 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Glynn C., et al., Cryo-EM structure of a human prion fibril with a hydrophobic, protease-resistant core. Nat. Struct. Mol. Biol. 27, 417–423 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobson C. M., Protein folding and misfolding. Nature 426, 884–890 (2003). [DOI] [PubMed] [Google Scholar]
- 15.De Simone A., Zagari A., Derreumaux P., Structural and hydration properties of the partially unfolded states of the prion protein. Biophys. J. 93, 1284–1292 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nitrini R., et al., Familial spongiform encephalopathy associated with a novel prion protein gene mutation. Ann. Neurol. 42, 138–146 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Lee S., et al., Conformational diversity in prion protein variants influences intermolecular β-sheet formation. EMBO J. 29, 251–262 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liemann S., Glockshuber R., Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry 38, 3258–3267 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Muramoto T., Scott M., Cohen F. E., Prusiner S. B., Recombinant scrapie-like prion protein of 106 amino acids is soluble. Proc. Natl. Acad. Sci. U.S.A. 93, 15457–15462 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenfield N. J., Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 1, 2527–2535 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camilloni C., De Simone A., Vranken W. F., Vendruscolo M., Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry 51, 2224–2231 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Tizzano B., et al., The human prion protein α2 helix: A thermodynamic study of its conformational preferences. Proteins 59, 72–79 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Rennella E., Morgan G. J., Yan N., Kelly J. W., Kay L. E., The role of protein thermodynamics and primary structure in fibrillogenesis of variable domains from immunoglobulin light chains. J. Am. Chem. Soc. 141, 13562–13571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carr H. Y., Purcell E. M., Effects of diffusion on free precession in nuclear magnetic resonance experiments. Phys. Rev. 94, 630–638 (1954). [Google Scholar]
- 25.Meiboom S., Gill D., Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 29, 688–691 (1958). [Google Scholar]
- 26.Tollinger M., Skrynnikov N. R., Mulder F. A., Forman-Kay J. D., Kay L. E., Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 123, 11341–11352 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Piana S., Laio A., A bias-exchange approach to protein folding. J. Phys. Chem. B 111, 4553–4559 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Calzolai L., Zahn R., Influence of pH on NMR structure and stability of the human prion protein globular domain. J. Biol. Chem. 278, 35592–35596 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Shen Y., Bax A., SPARTA+: A modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR 48, 13–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanz-Hernández M., De Simone A., The PROSECCO server for chemical shift predictions in ordered and disordered proteins. J. Biomol. NMR 69, 147–156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cobb N. J., Sönnichsen F. D., McHaourab H., Surewicz W. K., Molecular architecture of human prion protein amyloid: A parallel, in-register beta-structure. Proc. Natl. Acad. Sci. U.S.A. 104, 18946–18951 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cobb N. J., Apetri A. C., Surewicz W. K., Prion protein amyloid formation under native-like conditions involves refolding of the C-terminal α-helical domain. J. Biol. Chem. 283, 34704–34711 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biancalana M., Koide S., Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta 1804, 1405–1412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson R., et al., Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polymenidou M., et al., The POM monoclonals: A comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One 3, e3872 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonati T., et al., The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 501, 102–106 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Apetri A. C., Surewicz W. K., Kinetic intermediate in the folding of human prion protein. J. Biol. Chem. 277, 44589–44592 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Honda R. P., Xu M., Yamaguchi K. I., Roder H., Kuwata K., A native-like intermediate serves as a branching point between the folding and aggregation pathways of the mouse prion protein. Structure 23, 1735–1742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornemann S., Glockshuber R., A scrapie-like unfolding intermediate of the prion protein domain PrP(121–231) induced by acidic pH. Proc. Natl. Acad. Sci. U.S.A. 95, 6010–6014 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viles J. H., et al., Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry 40, 2743–2753 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Hosszu L. L., et al., Structural mobility of the human prion protein probed by backbone hydrogen exchange. Nat. Struct. Biol. 6, 740–743 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Honda R. P., Yamaguchi K., Kuwata K., Acid-induced molten globule state of a prion protein: Crucial role of strand 1-helix 1-strand 2 segment. J. Biol. Chem. 289, 30355–30363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camilloni C., Schaal D., Schweimer K., Schwarzinger S., De Simone A., Energy landscape of the prion protein helix 1 probed by metadynamics and NMR. Biophys. J. 102, 158–167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi K., Matsumoto T., Kuwata K., Critical region for amyloid fibril formation of mouse prion protein: Unusual amyloidogenic properties of the helix 2 peptide. Biochemistry 47, 13242–13251 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Hosszu L. L. P., et al., Definable equilibrium states in the folding of human prion protein. Biochemistry 44, 16649–16657 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Bjorndahl T. C., et al., Detailed biophysical characterization of the acid-induced PrP(c) to PrP(β) conversion process. Biochemistry 50, 1162–1173 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi K. I., Kuwata K., Formation and properties of amyloid fibrils of prion protein. Biophys. Rev. 10, 517–525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X., Wintrode P. L., Surewicz W. K., Beta-sheet core of human prion protein amyloid fibrils as determined by hydrogen/deuterium exchange. Proc. Natl. Acad. Sci. U.S.A. 104, 1510–1515 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tycko R., Savtchenko R., Ostapchenko V. G., Makarava N., Baskakov I. V., The α-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel β-sheet structure in PrP fibrils: Evidence from solid state nuclear magnetic resonance. Biochemistry 49, 9488–9497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abskharon R. N. N., et al., Probing the N-terminal β-sheet conversion in the crystal structure of the human prion protein bound to a nanobody. J. Am. Chem. Soc. 136, 937–944 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Abskharon R., et al., Structural evidence for the critical role of the prion protein hydrophobic region in forming an infectious prion. PLoS Pathog. 15, e1008139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris K. L., Serpell L. C., X-ray fibre diffraction studies of amyloid fibrils. Methods Mol. Biol. 849, 121–135 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Chan C.-K., et al., Submillisecond protein folding kinetics studied by ultrarapid mixing. Proc. Natl. Acad. Sci. U.S.A. 94, 1779–1784 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muñoz V., Eaton W. A., A simple model for calculating the kinetics of protein folding from three-dimensional structures. Proc. Natl. Acad. Sci. U.S.A. 96, 11311–11316 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eaton W. A., Searching for “downhill scenarios” in protein folding. Proc. Natl. Acad. Sci. U.S.A. 96, 5897–5899 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) (entries 50527 and 50528).