Significance
The aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor. It was originally characterized as a sensor, responding to environmental pollutants and inducing enzymes able to eliminate toxins. AhR is now known to be activated by endogenous ligands as well, such as the tryptophan metabolite, kynurenine. Its activation regulates numerous cell functions, including immunity and proliferation, that impact multiple diseases, such as cancer and infection. Here we show that AhR is activated following infection with human cytomegalovirus (HCMV). Its activation impacts both viral and cellular gene expression. Of note, AhR induces a cell cycle block, providing a mechanistic explanation for a long-known consequence of infection that is believed to reserve metabolic resources for the virus by preventing cellular proliferation.
Keywords: kynurenine, aryl hydrocarbon receptor, human cytomegalovirus, cell cycle
Abstract
The tryptophan metabolite, kynurenine, is known to be produced at elevated levels within human cytomegalovirus (HCMV)-infected fibroblasts. Kynurenine is an endogenous aryl hydrocarbon receptor (AhR) ligand. Here we show that the AhR is activated following HCMV infection, and pharmacological inhibition of AhR or knockdown of AhR RNA reduced the accumulation of viral RNAs and infectious progeny. RNA-seq analysis of infected cells following AhR knockdown showed that the receptor alters the levels of numerous RNAs, including RNAs related to cell cycle progression. AhR knockdown alleviated the G1/S cell cycle block that is normally instituted in HCMV-infected fibroblasts, consistent with its known ability to regulate cell cycle progression and cell proliferation. In sum, AhR is activated by kynurenine and perhaps other ligands produced during HCMV infection, it profoundly alters the infected-cell transcriptome, and one outcome of its activity is a block to cell cycle progression, providing mechanistic insight to a long-known element of the virus–host cell interaction.
Human cytomegalovirus (HCMV), a β-herpesvirus, is a major cause of birth defects, a life-threatening opportunistic infection in immunodeficient individuals, and a potential oncomodulatory agent (1–3). The virus manipulates numerous cellular processes, including cell metabolism (4, 5), to facilitate its replication and spread. One metabolite that is markedly elevated following infection of fibroblasts with both laboratory and clinical HCMV isolates, e.g., ∼25-fold increase for the TB40/E clinical strain at 24 hours post infection (hpi), is kynurenine (Kyn) (6), a tryptophan catabolite produced by indoleamine-2,3-dioxygenase (IDO) 1 and 2 and tryptophan-2,3-dioxygenase (TDO) (7). Kyn and its metabolite, quinolinic acid, are also elevated in the plasma of immunosuppressed kidney transplant patients with active HCMV infections, and their levels correlate with the severity of viral disease (8).
Kyn is an endogenous aryl hydrocarbon receptor (AhR) ligand (9). AhR was first described as a sensory component of a chemical surveillance system that moderates the adverse effects of xenobiotic toxins via the induction of cellular functions that transform and eliminate them (10, 11). Subsequent work has shown that AhR responds to both xenobiotic and endogenous ligands and functions in numerous biological processes, including development, homeostasis, immunity, cell proliferation, and apoptosis, impacting many disease states, including viral infections (12–17).
AhR is the only ligand-activated member of the basic helix–loop–helix/PER-ARNT-SIM (bHLH-PAS) superfamily of transcription factors (18). Inactive AhR resides in the cytoplasm, and upon ligand binding it translocates to the nucleus (19), where it partners with different proteins to regulate gene expression. It can form a heterodimeric complex with the AhR nuclear translocator (ARNT; also known as HIF1-β) through its N-terminal bHLH and PAS-A domains, which then binds DNA at so-called xenobiotic response elements (XREs; also known as dioxin response elements, DREs) in the vicinity of responsive genes (20, 21). AhR can also interact with other transcriptional regulatory proteins, such as the retinoblastoma protein and Kruppel-like factor 6 (22, 23). AhR is activated by a diverse range of ligands, including xenobiotics and endogenous molecules, and even closely related ligands may differentially affect the nuclear interactome of the transcription factor (24, 25). Once the liganded receptor leaves the nucleus, it undergoes proteasome-mediated degradation (26, 27).
In addition to its role as a transcription factor, AhR acts as a ligand-dependent adapter independently of ARNT that directs the CUL4B E3 ubiquitin ligase to targets (28). Interestingly, IDO expression is transcriptionally induced by activated AhR, and it has been proposed that it might be subject to AhR-directed degradation via CUL4B (29).
The Kyn-AhR axis has been most intensively studied in the context of cancer. Tumors often express high levels of IDO1 and TDO, which activate Kyn-AhR signaling and enhance the malignant phenotype, affecting tumor cell behaviors such as proliferation and motility (30–32). Further, extracellular Kyn in the tumor microenvironment can suppress T cell activity (9, 33, 34) and recruit immunosuppressive macrophages (35).
Since aspects of tumor phenotypes arising from Kyn-AhR signaling might also contribute to HCMV pathogenesis, we explored the cell-autonomous consequences of Kyn production in HCMV-infected fibroblasts. AhR is activated following infection, and pharmacological inhibition or knockdown of AhR reduced the accumulation of viral RNAs and virus yield and altered the levels of numerous cellular RNAs. Of note, knockdown of the receptor impacted cell cycle progression in infected fibroblasts. HCMV normally institutes a cell cycle block near the G1/S boundary in fibroblasts, where viral gene expression and DNA replication are facilitated, while cellular DNA replication is suppressed (36–42). AhR knockdown antagonized the ability of HCMV to induce the cell cycle block, consistent with its known ability to modulate cell growth in a cell-specific context (12) and arguing that AhR normally contributes to viral subversion of cell cycle progression.
Results
HCMV Infection Modulates AhR Activity.
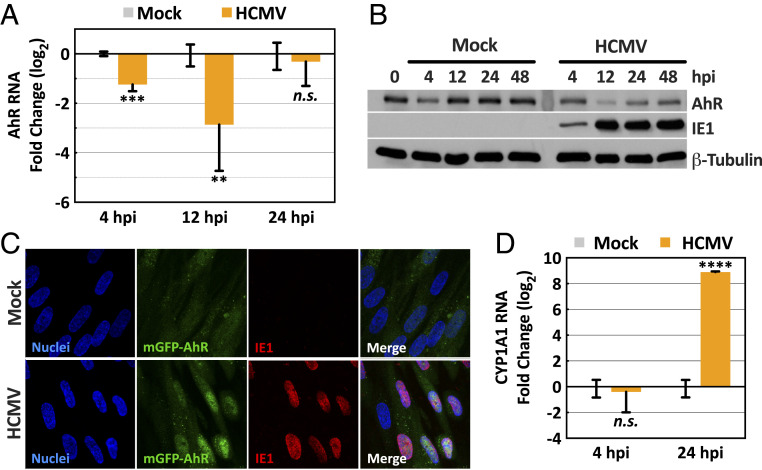
Since Kyn is induced by HCMV infection (6, 8) and serves as a ligand for AhR (9), we tested the impact of infection on AhR levels and activity. To avoid possible effects of AhR ligands in serum (43), confluent, serum-starved fibroblasts were mock infected or infected with the AD169 strain of HCMV prepared in serum-free medium, and AhR RNA and protein levels were monitored after various time intervals. AhR RNA was assayed by RT-qPCR analysis (Fig. 1A). Although AhR transcript levels were reduced in infected versus mock-infected cells at 4 and 12 hpi by as much as eightfold, there was no significant difference at 24 hpi. AhR protein was measured by immunoblot assay (Fig. 1B). With the exception of a modest reduction at 4 h, the level of receptor was stable at 12, 24, and 48 h after mock infection. In contrast, AhR protein was reduced in infected cells, especially at 12 hpi.
Fig. 1.
HCMV infection of fibroblasts modulates AhR expression and activity. Confluent HFFs were starved in serum-free medium for 48 h, and then mock infected or infected at a multiplicity of 3 IU/cell. (A) Modulation of AhR RNA levels by HCMV. Cultures were harvested at the indicated times and AhR RNA was quantified relative to actin RNA by RT-qPCR assay with infected normalized to mock-infected samples (n = 2, assayed in triplicate). (B) Modulation of AhR protein levels by HCMV. Cultures were harvested at the indicated times and AhR protein was quantified by immunoblot assay. IE1 was monitored as a marker of infection and β-tubulin served as a loading control. (C) Accumulation of AhR in the nuclei of HCMV-infected cells. Cells were fixed at 16 hpi and mGFP-tagged AhR fluorescence was monitored. Infected cells were identified by indirect immunofluorescence assay of the viral IE1 protein, and nuclei were counterstained using Hoechst 33342 dye (representative of three independent experiments). (D) Induction of AhR-responsive CYP1A1 RNA by HCMV. Cultures were harvested at the indicated times and CYP1A1 RNA was quantified relative to actin RNA by RT-qPCR assay with infected normalized to mock-infected samples (n = 2, assayed in triplicate). All RT-qPCR data, normalized to mock-infected samples, are shown as mean ± SD; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired Student’s t test).
It’s not clear why AhR RNA is transiently reduced following infection, but the change is not likely responsible for the fairly rapid reduction in AhR protein, since AhR has been estimated to have a half-life of ∼28 h in mouse hepatoma cells (26, 27). Rather, the loss of AhR protein might result from its proteasomal degradation, which occurs rapidly following its interaction with an activating ligand, reducing its half-life to about 3 h (26, 27). Thus, the reduced level of AhR protein following infection raises the possibility that the receptor has been activated, possibly by its endogenous ligand, Kyn, which accumulates following HCMV infection (6). To test for receptor activation, fibroblasts were prepared expressing AhR fused to monomeric GFP, and its cellular localization was monitored during the early phase of infection, at 16 hpi (Fig. 1C). Activated AhR translocates from cytoplasm to nucleus, and while it was predominantly cytoplasmic in mock-infected cells, it localized to the nucleus following infection. To further assess AhR activation, expression of cytochrome P450 1A1 (CYP1A1) RNA, an AhR target (44), was monitored at 24 hpi (Fig. 1D). It was induced by a factor of nearly 500 in infected compared to mock-infected cells, again arguing that AhR is activated by HCMV.
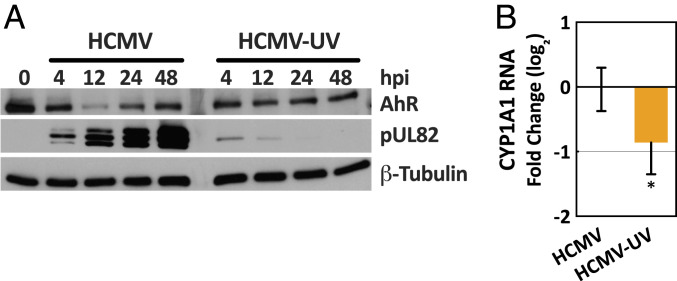
The rapid effect of infection on AhR levels suggested that a virion constituent and/or immediate-early/early viral gene expression might be responsible. To ascertain whether viral gene expression is required for AhR activation, ultraviolet (UV)-inactivated virus was compared to active HCMV. The viral pUL82 protein was monitored to confirm successful UV treatment (Fig. 2A). Whereas the protein accumulated to increasingly higher levels with time after infection by active virus, it did not accumulate following infection with the UV-inactivated virus. Rather, a small amount of the protein, delivered to cells in virions, was initially evident and then lost. Thus, the UV inactivation allowed delivery of virion components to cells, but damaged virion DNA to prevent viral gene expression. In contrast to active virus, the inactivated virus failed to modulate AhR protein levels (Fig. 2A) and modulated CYP1A1 RNA expression to a reduced extent (Fig. 2B).
Fig. 2.
HCMV gene expression is required to activate AhR. Confluent HFFs were starved in serum-free medium for 48 h, and then mock infected or infected at a multiplicity of 3 IU/cell. (A) Modulation of AhR protein levels by HCMV but not UV-inactivated HCMV (HCMV-UV). Cultures were harvested at the indicated times and AhR protein was quantified by immunoblot assay. pUL82 was monitored as a marker of infection and β-tubulin served as a loading control. (B) Induction of AhR-responsive CYP1A1 RNA by HCMV versus HCMV-UV. Cultures were harvested at 24 hpi and CYP1A1 RNA was quantified relative to actin RNA by RT-qPCR assay with HCMV-UV-infected normalized to HCMV-infected samples. Data are shown as mean ± SD (n = 3, assayed in triplicate); *P < 0.05 (unpaired Student’s t test).
We conclude that HCMV infection activates AhR, and full activation requires viral gene expression following cell entry.
AhR Supports the Efficient Production of HCMV Progeny.
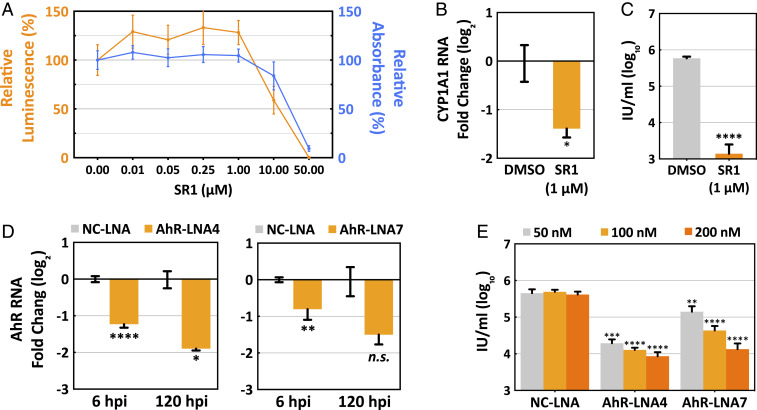
To determine whether AhR activity influences viral replication, we initially tested the effect of an AhR antagonist, StemRegenin 1 (SR1) (45). SR1 (1 μM) was not toxic to serum-starved human fibroblasts following treatment for 96 h (Fig. 3A), it markedly reduced the induction of AhR-responsive CYP1A1 RNA following HCMV infection (Fig. 3B), and it reduced virus yield by a factor of about 400 (Fig. 3C). To confirm the drug effect, AhR RNA was knocked down in serum-starved fibroblasts by using two different locked nucleic acid (LNA) RNAs (46). With the knockdown treatment beginning 48 h before infection, LNA4 (100 nM) or LNA7 (200 nM) reduced AhR RNA by ∼75% and 65%, respectively, at 120 hpi (Fig. 3D). The same LNA4 or LNA7 knockdown treatment reduced the yield of HCMV by about 35- and 34-fold, respectively, at 120 hpi (Fig. 3E). Inhibition of AhR by two different methods—drug treatment and knockdown—substantially reduced HCMV yield, demonstrating that the receptor is required for efficient virus growth. To avoid possible off-target effects of the drug, further experiments used knockdown by the two different LNAs to target AhR activity.
Fig. 3.
AhR supports the efficient production of HCMV progeny. Confluent HFFs were starved in serum-free medium for 48 h before the initiation of experiments. (A) Toxicity of the AhR inhibitor, SR1. Serum-starved HFFs were treated with DMSO (1% wt/wt) and increasing concentrations of SR1 in DMSO. After 96 h, cell viability (orange) and cell proliferation (blue) were monitored (n = 4). (B) SR1 (1 μM) inhibits CYP1A1 RNA accumulation. Serum-starved HFFs were infected at a multiplicity of 3 IU/cell, drug or solvent was added 2 h post viral absorption, and 24 h later cells were harvested and CYP1A1 RNA was quantified relative to actin RNA by RT-qPCR assay with drug-treated normalized to DMSO-treated samples (n = 3, assayed in triplicate). (C) SR1 (1 μM) reduces HCMV yield at 96 hpi. Serum-starved HFFs were infected at a multiplicity of 1 IU/cell, drug or solvent was added at 2 h post viral absorption, and cell-free virus was assayed (n = 3, assayed in triplicate). (D) LNAs reduce AhR RNA levels. Serum-starved HFFs were treated with nonspecific control NC-LNA (200 nM), AhR-LNA4 (100 nM, Left) or AhR-LNA7 (200 nM, Right) for 48 h, then infected and harvested for analysis of AhR RNA by RT-qPCR at the times indicated. (E) AhR-specific LNAs reduce HCMV yield at 120 hpi. Serum-starved HFFs were treated with LNAs at indicated concentrations for 48 h, then infected at a multiplicity of 1 IU/cell, and cell-free virus was assayed (n = 3, assayed in triplicate). Data are shown as mean ± SD; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant (unpaired Student’s t test).
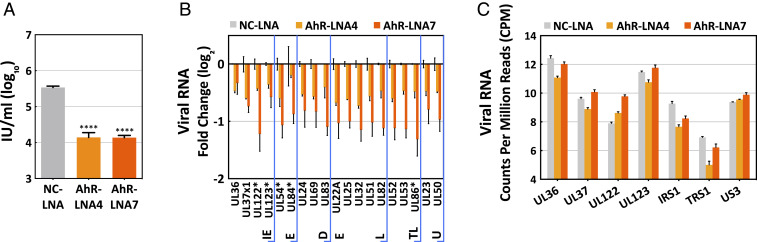
The effect of AhR knockdown on viral RNA levels was quantified by RT-qPCR at 120 hpi by testing expression levels for representatives of all classes of viral transcripts. The two AhR-specific LNAs again reduced the yield of extracellular virus, confirming that they were active (Fig. 4A); and the levels of all 19 viral RNAs tested were reduced by 25 to 60% at 120 hpi (Fig. 4B). Fourteen of the viral transcription units tested contained potential XRE/DRE AhR-binding motifs, while five did not contain an identifiable motif (marked with asterisks in Fig. 4B). There was no difference in their response to AhR knockdown. This result could be explained by indirect effects of reduced AhR function at viral promoters lacking the motif. It could also result from a direct or indirect effect on a viral immediate early (IE) transcriptional activator that in turn impacts downstream viral promoters that depend on the activator.
Fig. 4.
AhR supports efficient accumulation of HCMV RNAs during the late but not immediate-early phase of infection. (A) Treatment with AhR-specific (LNA4 and LNA7), but not nonspecific control (NC) LNAs, reduce HCMV yield at 120 hpi. Serum-starved HFFs were treated with NC-LNA and LNA7 at 200 nM and LNA4 at 100 nM for 48 h, then infected at a multiplicity of 3 IU/cell, and cell-free virus was assayed (n = 3, assayed in triplicate). (B) Treatment with AhR-specific LNAs reduce the accumulation of HCMV RNAs at 120 hpi. Serum-starved HFFs were infected at a multiplicity of 3 IU/cell, and viral RNAs were quantified relative to peptidylprolyl isomerase A (PPIA) RNA by RT-qPCR assay with AhR-specific LNA normalized to nonspecific LNA samples (n = 2, assayed in triplicate). Kinetic classes of viral RNAs are designated: IE, immediate early; E, early; DE, delayed early; L, late; TL, true late; U, unclassified. RNAs marked with an asterisk are coded by genes that do not contain a potential XRE/DRE motif in their known regulatory regions. (C) Treatment with AhR-specific LNAs does not perturb the accumulation of HCMV immediate-early RNAs at 6 hpi. Serum-starved HFFs were infected at a multiplicity of 3 IU/cell, and viral RNAs were quantified by RNA-seq analysis. Data are shown as mean ± SD; ****P < 0.0001 (unpaired Student’s t test).
In sum, AhR activity is required for maximal virus yield, and loss of activity has a modest effect on the accumulation of the subset of virus-coded RNAs assayed.
AhR Broadly Impacts the Infected Cell Transcriptome.
To gain insight into the mechanisms underlying the contribution of AhR to the production of viral progeny, its effect on the infected-cell transcriptome was evaluated. It appeared likely that AhR acts relatively early during the replication cycle, because AhR protein levels were reduced by 12 hpi (Fig. 1B), which is a marker for its activation (26, 27). Accordingly, we assayed the effect of AhR knockdown on the cellular transcriptome at 6 hpi, reasoning that by assaying during the immediate-early phase, changes would more likely be a direct consequence of the knockdown than changes occurring later in the replication cycle. Following a 48-h pretreatment with negative control LNA (NC-LNA) (200 nM), AhR-specific LNA4 (100 nM) or AhR-specific LNA7 (200 nM), serum-starved fibroblasts were infected with HCMV, cultures were harvested at 6 hpi, and poly(A)+ RNA was isolated for RNA-seq analysis. RNA expression data were compared for the control and AhR knockdowns. In contrast to the modest reductions observed at 120 hpi (Fig. 4B), HCMV immediate-early transcripts were not significantly affected by the knockdown at 6 hpi (Fig. 4C).
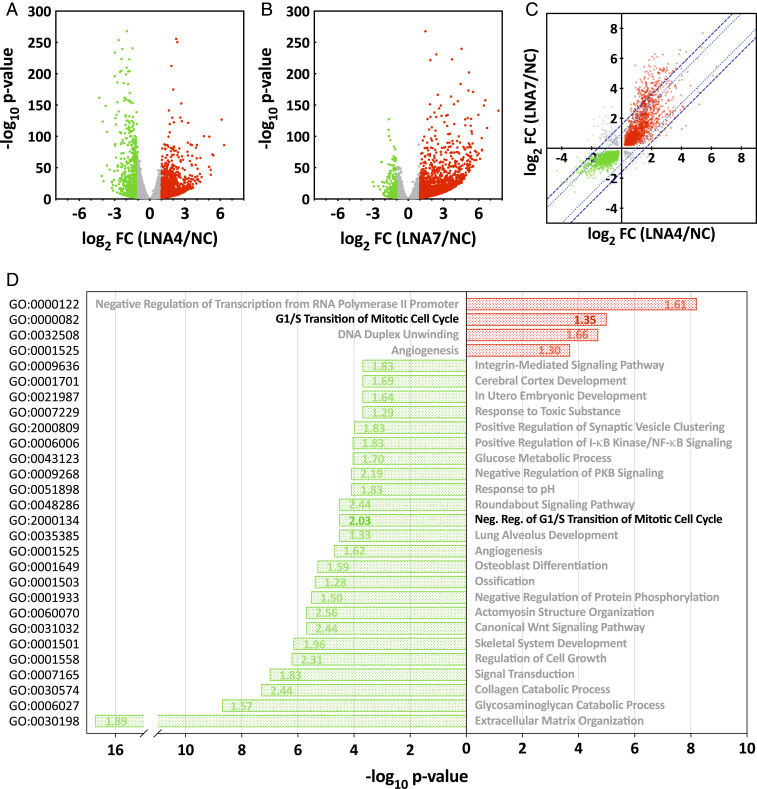
Despite nearly normal levels of HCMV immediate-early RNAs monitored, nearly 15% of the ∼25,000 infected-cell RNAs detected were significantly modulated by AhR knockdown at 6 hpi (Fig. 5 A and B and Dataset S1), with similar numbers of transcripts increased (∼48.5%) and decreased (∼51.5%) in abundance. Further, the transcriptomic effects observed in cells receiving the two different AhR-targeting LNAs were significantly correlated, with Pearson’s r = 0.6217, arguing against off-target effects (Fig. 5C). The broad effect of AhR knockdown on RNA levels is consistent with the large number of AhR/ARNT XRE-binding motifs in the human genome, the interaction of AhR with other DNA-binding transcription factors, and downstream effects of AhR targets (13, 20). Since the majority of changes in the HCMV-infected-cell transcriptome are reflected in the proteome (47), the RNA-expression data were used to identify functions that were likely modulated by AhR at this immediate-early time after infection. Annotation of AhR-LNA-altered RNAs was followed by biological process gene ontology (GO) and AhR-target gene enrichment analyses to identify the range of biological processes impacted by AhR during infection. Representative processes that illustrate the broad range of infected-cell functions affected by AhR are presented in Fig. 5D.
Fig. 5.
AhR modulates the HCMV infected-cell transcriptome. (A and B) Volcano plots showing the effect of AhR KD with two different AhR-specific LNAs. Fibroblasts were transfected with an AhR-specific LNA (A, LNA4 or B, LNA7) or a nonspecific control LNA (NC), and maintained in serum-free medium for 48 h, after which cultures were infected with HCMV at a multiplicity of 3 IU/cell. At 6 hpi, poly(A)+ RNA was prepared from infected cells and subjected to RNA-seq analysis (n = 2). Fold-change ratios (AhR-LNA/NC-LNA) and P values were determined. RNAs with a significant (P < 0.01) change of ≥ 2-fold in their expression levels are depicted by red (elevated) and green (lowered) dots; RNAs that did not meet these criteria are gray. (C) Fold change of cell-coded transcripts with significantly altered expression levels (q < 0.01) were plotted for LNA4- versus LNA7-treated cultures. RNAs with a significant (q < 0.01) change in their expression levels are depicted by red (elevated in response to both LNAs), green (lowered in response to both LNAs) and gray dots (LNAs generated opposing changes). The dotted (light blue) and dashed (dark blue) lines show the thresholds for two- and threefold variance between the AhR-KD LNA treatments, respectively. (D) Bar graph showing enriched biological process gene ontology terms of differentially expressed genes following treatment with AhR-specific LNAs. Green bars show GOs enriched within the group of transcripts negatively affected by AhR knockdown and red bars show GOs enriched within the group of transcripts up-regulated in AhR knockdown cells. The numbers within bars report the AhR target gene fold-enrichment values. Data are derived from the RNA-seq results presented in A–C.
Although it is not possible to discriminate between transcripts directly or indirectly affected by AhR knockdown, the results nevertheless illustrate the extensive impact of AhR activity across numerous processes during the very early stage of HCMV infection.
AhR Facilitates the HCMV-Induced G1/S Block to Cell Cycle Progression.
Two GO categories directly related to cell cycle progression were altered by AhR knockdown in the context of infected cells (Fig. 5D, bold): GO:2000134, negative regulation of G1/S transition of mitotic cell cycle, was enriched within the group of transcripts negatively affected by AhR knockdown; and GO:0000082, G1/S transition of mitotic cell cycle, was enriched within the group of transcripts up-regulated in AhR knockdown cells. These changes in cellular gene expression are consistent with the known modulatory effects of AhR on cell cycle regulation and cell proliferation (12, 48). Since HCMV manipulates the fibroblast cell cycle, instituting a block near the G1/S boundary and preventing cellular DNA replication (36–42), we tested for a role of AhR in the process.
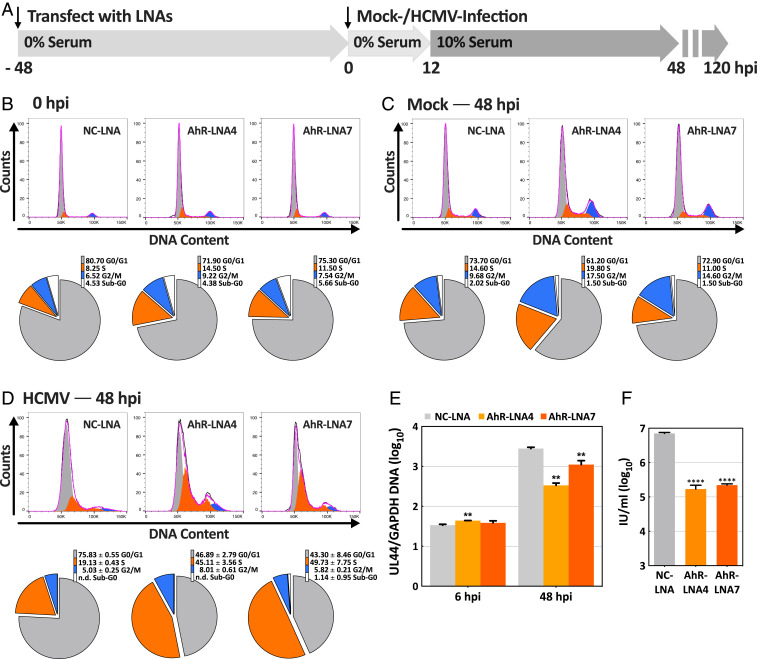
The protocol used to assess the role of AhR in the HCMV cell cycle block is diagrammed in Fig. 6A. In a control experiment, confluent, serum-starved fibroblasts were treated with NC-LNA or AhR-specific LNAs for 48 h and then stained with propidium iodide for the analysis of DNA content by flow cytometry (Fig. 6B). The cell cycle distribution was very similar for cultures subjected to knockdown with nonspecific versus AhR-specific LNAs with the majority of cells in the G0/G1 compartment and relatively few in the S phase. We can conclude that AhR knockdown doesn’t influence the cell cycle status of confluent, serum-starved fibroblasts. Next, the knockdowns were repeated and, after 48 h, cells were mock infected for an additional 48 h before assaying their DNA content (Fig. 6C). To facilitate potential cell cycle progression following AhR knockdown and mock or HCMV infection, cells received medium with 10% serum for the last 36 h before analysis (Fig. 6A). Mock-infected cells subjected to the knockdown treatments were substantially diploid (2n) in DNA content, indicative of the G0/G1 compartment and, again, many fewer cells were in the S phase. Although the primary fibroblast cultures were stimulated with serum, they were confluent, so they did not progress into the S phase in response to serum. Infected cells (Fig. 6D) that received the nonspecific knockdown treatment also remained substantially diploid (NC-LNA = 75.8 ± 0.6%), consistent with the ability of the virus to institute a block in late G1. In contrast, a much smaller portion of infected cell populations subjected to AhR-specific knockdown treatments exhibited diploid DNA content, indicative of the G0/G1 compartment (LNA4 = 46.9 ± 2.8%; LNA7 = 43.3 ± 8.5%). Rather, AhR knockdown generated markedly increased numbers of cells in the S phase. Whereas 19.1 ± 0.4% of infected cells receiving the control LNA contained 2n+ DNA, 45.1 ± 3.6% of LNA4-treated and 49.7 ± 7.8% of LNA7-treated cultures contained 2n+ DNA, characteristic of the S phase.
Fig. 6.
AhR facilitates the HCMV-induced cell cycle block in fibroblasts. (A) Experimental plan. After treatment with AhR-specific LNA (LNA4 or LNA7) or a nonspecific control LNA (NC) for 48 h in serum-free medium, HFFs were mock-infected or HCMV-infected at a multiplicity of 3 IU/cell and fed with serum-free medium. At 12 hpi, cultures were refed with medium containing 10% serum. Cell cycle distributions were determined at 48 h after mock- or HCMV-infection by staining DNA with propidium iodide and flow cytometry. (B) Cell cycle analysis of uninfected fibroblasts following treatment with LNAs for 48 h in serum-free medium (n = 1). Flow cytometry data (Upper) and pie charts showing the percentages of cells in different cell cycle compartments (Lower) are displayed. (C) Cell cycle analysis of LNA-treated fibroblasts that were mock infected in serum-free medium and maintained in medium with 10% serum from 12 to 48 h after mock infection (n = 1). (D) Cell cycle analysis of LNA-treated fibroblasts that were infected with HCMV in serum-free medium and maintained in medium with 10% serum from 12 to 48 hpi (n = 3). The pie charts report the averages of three determinations; n.d., not detected. (E) Control experiment monitoring the relative amounts of cell versus viral DNA in infected cells following knockdown of AhR. Cells harvested at 48 hpi were infected and fed with serum-free medium, refed with medium containing 10% serum at 12 hpi. Viral UL44 and host GAPDH DNA copy numbers were quantified by qPCR (n = 3). (F) Control experiment monitoring HCMV yield following knockdown of AhR. Cells were infected and fed with serum-free medium, refed with medium containing 10% serum at 12 hpi and cell-free virus was assayed at 120 hpi (n = 3). **P < 0.01; ****P < 0.0001. Unpaired Welch and Student’s t test were used in E and F, respectively.
The increase in cells with 2n+ DNA content is not due to increased viral DNA accumulation, because knockdown of AhR reduced viral DNA accumulation by a factor of about 4 (LNA7) to 10 (LNA4) in cells receiving medium with 10% serum for the last 36 h before analysis (Fig. 6E). As a further control for their activity, the AhR-specific LNAs were confirmed to substantially reduce the production of extracellular infectious progeny (Fig. 6F).
Knockdown of AhR with two independent LNAs compromised the G1/S block that is normally instituted by HCMV. We conclude that AhR function is required to efficiently prevent progression of infected fibroblasts into the S compartment.
Discussion
In 1996, rat CMV was reported to grow to higher titers than normal in the salivary glands of animals that were fed herrings contaminated with the AhR activator, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD; dioxin), providing the first indication that AhR might influence the growth of a cytomegalovirus (49). Subsequent work showed that dioxin increased HCMV DNA replication and yield following infection of human MRC5 fibroblasts (50); whereas certain flavonoids, known to act as AhR ligands (51), inhibited HCMV growth (52, 53). Although any of these AhR modulators could have off-target effects that influence viral growth, the opposite effects of different ligands on HCMV yield might be explained by known ligand-specific effects, including persistent versus transient AhR responses, as well as cell-type- and context-dependent AhR activity (24, 54–56).
AhR is activated following HCMV infection, as evidenced by its nuclear localization (Fig. 1C) and the accumulation of CYP1A1 RNA (Fig. 1D), a known target of AhR transcriptional activation (44). Viral gene expression is required for full AhR activation, because UV-irradiated virus particles activate CYP1A1 RNA expression to a significantly reduced extent (Fig. 2B). There is, however, a partial activation by UV-irradiated virus, raising the possibility that multiple viral gene products contribute to the induction of AhR activity. The drop in AhR protein levels that becomes most pronounced at 12 hpi (Figs. 1B and 2A) is consistent with its activation, since AhR is known to be rapidly degraded following persistent activation (24, 26, 27).
Kyn is an endogenous AhR ligand (9), it is produced following HCMV infection (6, 8) and is likely responsible for AhR activity in infected cells. IDO1, IDO2, and TDO oxidize tryptophan to generate N-formylkynurenine, which is hydrolyzed to produce Kyn (7). Ectopically expressed HCMV IE1 protein induces IDO1 RNA accumulation in fibroblasts (57) and decreases kynureninase RNA accumulation in fibroblasts (58). Kynureninase converts Kyn to anthranilic acid. Thus, IE1 could drive elevated Kyn levels by increasing its synthesis and/or reducing its consumption. However, HCMV infection has been reported to block accumulation of IDO1 RNA and protein in response to interferon-γ treatment in fibroblasts (59). This would suggest that another viral or induced cellular protein supersedes the IE1 IDO-1-inducing function, and perhaps IE1-reduced kynureninase levels are sufficient to elevate Kyn. It is also possible that IDO2 or TDO are induced and activated by infection to increase Kyn levels. Likewise, the l-amino acid oxidase, interleukin-4-induced-1, could be induced and generate the AhR ligands kynurenic acid and indole-3-aldehyde (60); and additional Trp metabolites that serve as AhR ligands, such as 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (61), might also contribute.
AhR inhibition or knockdown revealed that the receptor supports viral replication in fibroblasts, significantly increasing the yield of infectious progeny (Figs. 3 C and E and 6F), consistent with an earlier report that dioxin increases HCMV DNA yield (50). Although it is not clear which of the myriad AhR activities influence viral growth, it is very likely that transcriptional activation plays a major role. AhR knockdown reduced the level of RNAs encoded by all temporal classes of viral genes, but to a limited extent (Fig. 4B). The 19 viral RNAs tested were each reduced by 25 to 60% at 120 hpi. As noted earlier, the viral genome contains multiple XRE/DRE AhR-binding motifs. So, it is possible that AhR directly activates transcription of some viral genes while indirectly influencing others.
RNA-seq analysis demonstrated that AhR broadly impacts the HCMV-infected-cell transcriptome during the immediate-early phase of the replication cycle (Fig. 5 A and B and Dataset S1). Knockdown of AhR with two different LNAs generated very similar changes to the transcriptome (Fig. 5C), arguing against extensive contributions from off-target effects. Although the consequences of compromised AhR activity are likely modulated by viral products within infected cells, knockdown of AhR impacted many functions known to be influenced by the receptor (Fig. 5D), including cell growth and cell cycle regulation, NF-κB-driven immune functions, and cell-extracellular matrix interactions (12, 13). Given the role of AhR in cell cycle progression, the G1/S block in HCMV-infected fibroblasts and the observation that cell cycle-related RNAs are modulated following AhR knockdown in infected cells, we tested the cell cycle status of HCMV-infected, AhR-deficient fibroblasts. AhR knockdown markedly compromised the virus-induced cell cycle block (Fig. 6).
HCMV infection has long been known to block cell cycle progression in fibroblasts, within G1, near the S boundary (36–42). G1, S, and M phase cell products are expressed in blocked cells, so the cell cycle position has been referred to as pseudo-G1 (37). Knockdown of AhR relieves the G1/S block in infected cells allowing progression into the S phase of the cycle (Fig. 6D). However, the mechanism by which the receptor contributes to the block remains uncertain and may be multifactorial.
Different HCMV proteins have been shown to stimulate or antagonize cell cycle progression. This makes sense, because the virus generally infects quiescent, G0 cells, first inducing progression from G0 and through G1 and then instituting the G1/S block. pUL82 (pp71) targets hypophosphorylated retinoblastoma protein (pRB) for degradation (62) and pUL97 stimulates phosphorylation of pRB (63), supporting cell cycle progression. In contrast, pUL69 (64, 65) and pUL122 (IE2) (66, 67) each induces a block at G1/S. Knockdown of AhR had modest effects on the levels of all viral RNAs tested, so it is possible that AhR influences cell cycle progression in part via effects on viral gene expression.
Since it modulates cell cycle progression in uninfected cells, it is very likely that AhR also supports the infection-induced G1/S block through additional mechanisms. There are seemingly contradictory reports describing both proliferative and antiproliferative AhR effects, which likely reflect ligand status and cell-type-specific effects. For example, in the absence of ligand, AhR has been reported to interact with cyclin D1/cyclin-dependent kinase 4 (CDK4) to enhance phosphorylation of the retinoblastoma protein (pRB) and favor cell cycle progression, whereas ligand-bound AhR did not interact with the complex (68). Sustained versus transient AhR activation might influence the effect on cell cycle progression (54), and multiple antiproliferative mechanisms have been detailed for the activated receptor (69). The AhR signaling pathway mediates the transcriptional induction of the cyclin-dependent kinase inhibitors, p21Cip1 and p27Kip1, reducing pRB phosphorylation and blocking cell cycle progression at the G1/S boundary (70–72). AhR also interacts directly with pRB (73), preventing its hyperphosphorylation (74), again enforcing a block at the G1/S boundary.
The failure to institute a G1/S block likely contributes to the reduction in viral RNA accumulation in AhR-deficient cells during the late phase of infection (Fig. 4B), because HCMV RNA and protein accumulation are reduced when cells are infected in the S phase (39, 75, 76). This provides a second mechanism, beyond action at viral promoters through XRE/DRE motifs, by which AhR deficiency potentially influences HCMV gene expression, i.e., via a failure to block progression into the S phase.
In addition to cell cycle progression, AhR regulates numerous aspects of immune responses, integrating the immune response with the levels of its endogenous ligands as well as xenobiotic toxins (13, 77–79). For example, Kyn- or dioxin-activated AhR can inhibit the activity of effector T cells and increase numbers of regulatory T (Treg) cells directly by modifying the transcriptional program and differentiation of T cells and indirectly by modulating the activity of dendritic cells. This can suppress antitumor and antiviral cell immune responses. AhR also profoundly impacts innate immune mechanisms. As a case in point, AhR modulates Toll-like receptor signaling (80). AhR has also been shown to constrain the type I interferon response to both RNA and DNA viruses by inducing expression of the TCDD-inducible poly (ADP ribose) polymerase, which then blocks the pathway favoring the production of IFN-β (17). As predicted by this earlier work, AhR knockdown elevated IFN-β RNA expression in HCMV-infected fibroblasts (Dataset S1); and, assuming that IFN-β protein expression correlates with the levels of its RNA, suppression of this interferon response is likely one mechanism by which AhR supports the production of viral progeny (Figs. 3 C and E and 6F). Of note, AhR is induced in some cell types by type I interferons (81), so the residual levels of IFN-β normally expressed in HCMV-infected cells might serve to sustain AhR levels, further enforcing an AhR/IFN-β inhibitory loop.
Active AhR has been shown to increase IDO1 expression via an autocrine feedback loop in tumor cells (82). AhR induces production of IL-6 that activates STAT3, which then induces expression of IDO1, which in turn generates Kyn that activates AhR. IL-6 is one of the most abundant components of the HCMV secretome (83), and, not surprisingly, STAT3 is activated in HCMV-infected cells (84). Thus, it is likely that an AhR-sponsored autocrine feedback loop is activated by infection to increase IDO expression following infection. However, AhR knockdown increased the level of IDO1 RNA in HCMV-infected cells (Dataset S1), suggesting that a negative feedback loop predominates following infection. It has been proposed that activated AhR might also negatively regulate IDO1 at the level of protein stability through its interaction with the CUL4B ubiquitin ligase (28); but, to our knowledge, this possibility has not been proven.
In conclusion, our study demonstrates a profound effect of AhR activity on the HCMV-infected-cell transcriptome, impacting multiple functions fundamental to the virus–host interaction, including cell cycle regulation. This work sets the stage for further dissection of the roles of AhR in the HCMV replication cycle and pathogenesis.
Materials and Methods
Cells, Virus, and Reagents.
Primary human foreskin fibroblasts (HFFs) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum. HFFs stably expressing monomeric GFP (mGFP) (85)-tagged AhR were generated by transduction with the pHR-MCS lentivirus vector expressing AhR (86), using full-length AhR cDNA (Dharmacon Clone ID 30342582). The fusion protein was generated by using overlap PCR positioning mGFP at the N terminus with the following primers: mGFP forward 5′-GCGCAGATCTATGGTGAGCAAGGGCGAGGAG, mGFP reverse 5′-TGCCGCAGCACTCCCTGAGCCGGATCCGCTCTTGTACAGCTCGTCCATGCCG; AhR forward, 5′-AGCGGATCCGGCTCAGGGAGTGCTGCGGCAATGAACAGCAGCAGCGCC, AhR reverse 5′-GAACGCGGCCGCTTACAGGAATCCACTGGATGTCAAATC.
The AD169 laboratory strain of HCMV was derived from a BAC clone, pAD/Cre (87). Virus stocks were prepared from the supernatant of infected HFFs. Virus was partially purified and concentrated by centrifugation through a sorbitol cushion (20% sorbitol, 1 mM MgCl2, 50 mM Tris⋅HCl, pH 7.2), resuspended in serum-free DMEM containing 3% bovine serum albumin (BSA), stored at −80 °C and used as inoculum in experiments without the addition of serum. Virus titers were determined by end-point dilution assay (TCID50) on HFFs. UV inactivation was performed by exposing virus in DMEM with 3% BSA to three 25-s exposures at 120 mJ/cm2 of 254 nM light using auto cross link settings on a Stratalinker 2400 (Strategene).
For knockdown of AhR, antisense LNAs with a phosphorothioate backbone were obtained from Qiagen, and their sequences are as follows: NC-LNA, 5′-AACACGTCTATACGC; AhR-LNA-4, 5′-ACGGATGATGAAGTGG; AhR-LNA-7, 5′-GCTGTGGACAATTGAA. LNAs were used to transfect HFFs following the manufacturer’s protocols in the Lipofectamine RNAiMax Transfection Reagent (Thermo Fisher Scientific). The AhR antagonist, StemRegenin 1 (SR1; Sigma-Aldrich) (45), was dissolved in DMSO to produce a stock solution that was stored at −20 °C.
RNA and Protein Analysis.
For RT-qPCR analysis, cells were lysed in TRIzol, and total RNA was extracted using either a Direct-zol RNA MiniPrep kit (Zymo Research) or miRNeasy Mini kit (Qiagen). Genomic DNA was eliminated using TURBO DNase (Invitrogen) according to the manufacturer’s protocol. cDNA was prepared by reverse transcription, using either TaqMan Reverse Transcription Reagents or SuperScript III Reverse Transcriptase (Thermo Fisher Scientfic). SYBR Green PCR Mastermix (Applied Biosystems) and appropriate primers (Dataset S2) were used for assaying RNA levels, and results were normalized to beta-actin (ACTB) or peptidylprolyl isomerase A (PPIA).
For RNA-seq analysis, total RNA was isolated from TRIzol-lysed HFFs using the above-mentioned miRNeasy Mini Kit. DNA was removed from samples using TURBO DNase and RNA quality was analyzed using the Bioanalyzer 2100 (Agilent Technologies). The cDNA libraries were prepared using the NEXTflex Illumina Rapid Directional RNA-Seq Library Prep Kit (BioO Scientific) according to the manufacturer’s instructions. Briefly, polyadenylated RNA was purified from 200 ng of total RNA using oligo (dT) beads. The extracted mRNA fraction was subjected to fragmentation, reverse transcription, end repair, 3′-end adenylation, and adaptor ligation, followed by PCR amplification and SPRI bead purification (Beckman Coulter). The unique index sequences were incorporated in the adaptors for multiplexed high-throughput sequencing. The final product was assessed for its size distribution and concentration using BioAnalyzer High Sensitivity DNA Kit (Agilent Technologies). cDNA sequencing libraries were prepared at the Penn State College of Medicine Genome Sciences Facility and subjected to multiplexed sequencing (RNA-seq) using a Rapid HiSeq2500 sequencer (Illumina) for 100 cycles in single-read (1 × 100 bp), rapid mode. To calculate the annotated ratios for the sequenced libraries, data analysis was performed substantially as described previously (88). Human and HCMV fasta and annotation (.gtf) files were created for mapping and feature counting by combining sequences and annotations from gencode human genome (GRCh38.5) and AD169 (FJ5275630). Quality-filtered reads were mapped to that concatenated human-virus genome using HISAT2 (89), requiring a minimum mapping quality score of 10. Fragment counts were generated at the gene level using featureCounts (90). AhR-LNA-4/NC-LNA and AhR-LNA-7/NC-LNA ratio determinations and significance calling was determined using DESeq2 (91). Fold changes in gene expression were considered significant when q-value for multiple testing with the Benjamini–Hochberg procedure was <0.01. Data were then analyzed using DAVID (92) and REVIGO (93) software. Statistical significance analysis for the enrichment of AhR-target genes was performed using the hypergeometric test.
To assay proteins by immunoblotting, cells were harvested in radio-immunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors [150 mM NaCl; 1% Nonidet P-40; 0.5% sodium deoxycholate; 0.1% sodium dodecyl sulfate (SDS); 50 mM Tris, pH 8.0; 5 μg/mL aprotinin; 10 μg/mL leupeptin; 50 mM Hepes, pH 7.4], freeze-thawed once or twice, sonicated, and clarified by centrifugation. Protein concentrations were assessed by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific), and an equal amount of total protein, i.e., 10 to 20 μg per sample, was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to nitrocellulose membranes (120 mA, overnight) and blocked with 5% BSA in PBST (0.1% Tween-20, Dulbecco’s phosphate buffered saline, pH 7.4) overnight. Primary antibodies were as follows: anti-HCMV pUL123 (IE1) (Clone 1B12) (94), anti-HCMV pUL82 (Clone 10G11) (62), anti-AhR (sc-5579, Santa Cruz Biotechnology), and anti-β-tubulin (E7, Developmental Studies Hybridoma Bank). Secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse and goat anti-rabbit antibodies were from Jackson ImmunoResearch.
Protein localization was evaluated by direct and indirect immunofluorescence assay for mGFP-AhR and IE1, respectively. For IE1, the 1B12 primary antibody was used, followed by a secondary goat anti-mouse antibody conjugated with Alexa Fluor-568 (A-11031, Invitrogen). Nuclei were counterstained with Hoechst 33342. Images were acquired using a TCS SP5 confocal laser scanning microscope (Leica Microsystems).
Cell Cycle Analysis.
At 0 or 48 hpi, cells were collected by trypsinization, washed twice with cold PBS (4 °C), fixed with 70% ethanol (−20 °C), and stored at 4 °C overnight. Cellular DNA was stained by treatment with propidium iodide (Sigma-Aldrich) and RNase A (200 μg/mL, FEREN0531, Thermo Fisher Scientific) 1 to 2 h before analysis. At least 20,000 cells per sample were assayed on a Bio-Rad S3 Cell Sorter and classified into four categories based on DNA content: G0/G1, S, G2/M, and sub-G0 (95).
Statistical Analysis.
Quantitative results are shown as mean ± SD of at least two independent experiments assayed in triplicate, unless otherwise noted. Statistical significance was evaluated using GraphPad Prism 8 (GraphPad Software), except for RNA-seq analysis with previously described methods (88).
Supplementary Material
Acknowledgments
We acknowledge J. Hwang (Yale University) for guiding PN-N in the early stages of this project, and we thank A. Oberstein (University of Illinois) for guidance in the RNA-seq analysis. The work reported here was supported by NIH Grant AI142520.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026336118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo, under accession number GSE159375 (96). All study data are included in the article and/or supporting information.
References
- 1.Britt W. J., Maternal immunity and the natural history of congenital human cytomegalovirus infection. Viruses 10, 405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffiths P., Baraniak I., Reeves M., The pathogenesis of human cytomegalovirus. J. Pathol. 235, 288–297 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Cobbs C., Cytomegalovirus is a tumor-associated virus: Armed and dangerous. Curr. Opin. Virol. 39, 49–59 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Shenk T., Alwine J. C., Human cytomegalovirus: Coordinating cellular stress, signaling, and metabolic pathways. Annu. Rev. Virol. 1, 355–374 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Rodríguez-Sánchez I., Munger J., Meal for two: Human cytomegalovirus-induced activation of cellular metabolism. Viruses 11, 273 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vastag L., Koyuncu E., Grady S. L., Shenk T. E., Rabinowitz J. D., Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 7, e1002124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platten M., Nollen E. A. A., Röhrig U. F., Fallarino F., Opitz C. A., Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 18, 379–401 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Sadeghi M., et al., Strong association of phenylalanine and tryptophan metabolites with activated cytomegalovirus infection in kidney transplant recipients. Hum. Immunol. 73, 186–192 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Mezrich J. D., et al., An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 185, 3190–3198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hankinson O., The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35, 307–340 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Rowlands J. C., Gustafsson J.-Å., Aryl hydrocarbon receptor-mediated signal transduction. Crit. Rev. Toxicol. 27, 109–134 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Larigot L., Juricek L., Dairou J., Coumoul X., AhR signaling pathways and regulatory functions. Biochim. Open 7, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothhammer V., Quintana F. J., The aryl hydrocarbon receptor: An environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 19, 184–197 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Mehraj V., Routy J.-P., Tryptophan catabolism in chronic viral infections: Handling uninvited guests. Int. J. Tryptophan Res. 8, 41–48 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser H., Parker E., Hamrick M. W., Kynurenine signaling through the aryl hydrocarbon receptor: Implications for aging and healthspan. Exp. Gerontol. 130, 110797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badawy A. A. B., Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int. J. Tryptophan Res. 10, 1178646917691938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada T., et al., Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 17, 687–694 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Gu Y.-Z., Hogenesch J. B., Bradfield C. A., The PAS superfamily: Sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 40, 519–561 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Wright E. J., De Castro K. P., Joshi A. D., Elferink C. J., Canonical and non-canonical aryl hydrocarbon receptor signaling pathways. Curr. Opin. Toxicol. 2, 87–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo R., Matthews J., High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol. Sci. 130, 349–361 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Swanson H. I., Chan W. K., Bradfield C. A., DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins. J. Biol. Chem. 270, 26292–26302 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Elferink C. J., Ge N. L., Levine A., Maximal aryl hydrocarbon receptor activity depends on an interaction with the retinoblastoma protein. Mol. Pharmacol. 59, 664–673 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Wilson S. R., Joshi A. D., Elferink C. J., The tumor suppressor Kruppel-like factor 6 is a novel aryl hydrocarbon receptor DNA binding partner. J. Pharmacol. Exp. Ther. 345, 419–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denison M. S., Faber S. C., And now for something completely different: Diversity in ligand-dependent activation of Ah receptor responses. Curr. Opin. Toxicol. 2, 124–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuti R., et al., Ligand binding and functional selectivity of L-tryptophan metabolites at the mouse aryl hydrocarbon receptor (mAhR). J. Chem. Inf. Model. 54, 3373–3383 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Ma Q., Baldwin K. T., 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced degradation of aryl hydrocarbon receptor (AhR) by the ubiquitin-proteasome pathway. Role of the transcription activaton and DNA binding of AhR. J. Biol. Chem. 275, 8432–8438 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Pollenz R. S., Buggy C., Ligand-dependent and -independent degradation of the human aryl hydrocarbon receptor (hAHR) in cell culture models. Chem. Biol. Interact. 164, 49–59 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Ohtake F., et al., Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446, 562–566 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Pallotta M. T., Fallarino F., Matino D., Macchiarulo A., Orabona C., AhR-mediated, non-genomic modulation of Ido1 function. Front. Immunol. 5, 497 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Amato N. C., et al., A TDO2-AhR signaling axis facilitates anoikis resistance and metastasis in triple-negative breast cancer. Cancer Res. 75, 4651–4664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novikov O., et al., An aryl hydrocarbon receptor-mediated amplification loop that enforces cell migration in ER-/PR-/Her2- human breast cancer cells. Mol. Pharmacol. 90, 674–688 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opitz C. A., et al., An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Greene L. I., et al., A role for tryptophan-2,3-dioxygenase in CD8 T-cell suppression and evidence of tryptophan catabolism in breast cancer patient plasma. Mol. Cancer Res. 17, 131–139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., et al., Tumor-repopulating cells induce PD-1 expression in CD8+ T cells by transferring kynurenine and AhR activation. Cancer Cell 33, 480–494.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Takenaka M. C., et al., Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci. 22, 729–740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marschall M., Feichtinger S., Milbradt J., Regulatory roles of protein kinases in cytomegalovirus eplication. Adv. Virus Res. 80, 69–101 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Spector D. H., Human cytomegalovirus riding the cell cycle. Med. Microbiol. Immunol. (Berl.) 204, 409–419 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Jault F. M., et al., Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69, 6697–6704 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salvant B. S., Fortunato E. A., Spector D. H., Cell cycle dysregulation by human cytomegalovirus: Influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72, 3729–3741 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bresnahan W. A., Boldogh I., Thompson E. A., Albrecht T., Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224, 150–160 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Dittmer D., Mocarski E. S., Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71, 1629–1634 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu M., Shenk T., Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70, 8850–8857 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hestermann E. V., Stegeman J. J., Hahn M. E., Serum withdrawal leads to reduced aryl hydrocarbon receptor expression and loss of cytochrome P4501A inducibility in PLHC-1 cells. Biochem. Pharmacol. 63, 1405–1414 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Ma Q., Induction of CYP1A1. The AhR/DRE paradigm: Transcription, receptor regulation, and expanding biological roles. Curr. Drug Metab. 2, 149–164 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Boitano A. E., et al., Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 329, 1345–1348 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petersen M., Wengel J., LNA: A versatile tool for therapeutics and genomics. Trends Biotechnol. 21, 74–81 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Tirosh O., et al., The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog. 11, e1005288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bock K. W., Aryl hydrocarbon receptor (AHR): From selected human target genes and crosstalk with transcription factors to multiple AHR functions. Biochem. Pharmacol. 168, 65–70 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Ross P. S., et al., Host resistance to rat cytomegalovirus (RCMV) and immune function in adult PVG rats fed herring from the contaminated Baltic Sea. Arch. Toxicol. 70, 661–671 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Murayama T., Inoue M., Nomura T., Mori S., Eizuru Y., 2,3,7,8-Tetrachlorodibenzo-p-dioxin is a possible activator of human cytomegalovirus replication in a human fibroblast cell line. Biochem. Biophys. Res. Commun. 296, 651–656 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Kajta M., Domin H., Grynkiewicz G., Lason W., Genistein inhibits glutamate-induced apoptotic processes in primary neuronal cell cultures: An involvement of aryl hydrocarbon receptor and estrogen receptor/glycogen synthase kinase-3β intracellular signaling pathway. Neuroscience 145, 592–604 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Evers D. L., et al., Human cytomegalovirus-inhibitory flavonoids: Studies on antiviral activity and mechanism of action. Antiviral Res. 68, 124–134 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cotin S., et al., Eight flavonoids and their potential as inhibitors of human cytomegalovirus replication. Antiviral Res. 96, 181–186 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Mitchell K. A., Elferink C. J., Timing is everything: Consequences of transient and sustained AhR activity. Biochem. Pharmacol. 77, 947–956 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swedenborg E., et al., The aryl hydrocarbon receptor ligands 2,3,7,8-tetrachlorodibenzo-p-dioxin and 3-methylcholanthrene regulate distinct genetic networks. Mol. Cell. Endocrinol. 362, 39–47 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Bock K. W., Human and rodent aryl hydrocarbon receptor (AHR): From mediator of dioxin toxicity to physiologic AHR functions and therapeutic options. Biol. Chem. 398, 455–464 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Knoblach T., Grandel B., Seiler J., Nevels M., Paulus C., Human cytomegalovirus IE1 protein elicits a type II interferon-like host cell response that depends on activated STAT1 but not interferon-γ. PLoS Pathog. 7, e1002016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harwardt T., et al., Human cytomegalovirus immediate-early 1 protein rewires upstream STAT3 to downstream STAT1 signaling switching an IL6-type to an IFNγ-Like response. PLoS Pathog. 12, e1005748 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmermann A., et al., Checks and balances between human cytomegalovirus replication and indoleamine-2,3-dioxygenase. J. Gen. Virol. 95, 659–670 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Sadik A., et al., IL4I1 is a metabolic immune checkpoint that activates the AHR and promotes tumor progression. Cell 182, 1252–1270.e34 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Song J., et al., A ligand for the aryl hydrocarbon receptor isolated from lung. Proc. Natl. Acad. Sci. U.S.A. 99, 14694–14699 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalejta R. F., Bechtel J. T., Shenk T., Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol. Cell. Biol. 23, 1885–1895 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hume A. J., et al., Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 320, 797–799 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Lu M., Shenk T., Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73, 676–683 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayashi M. L., Blankenship C., Shenk T., Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. U.S.A. 97, 2692–2696 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murphy E. A., Streblow D. N., Nelson J. A., Stinski M. F., The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74, 7108–7118 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noris E., et al., Cell cycle arrest by human cytomegalovirus 86-kDa IE2 protein resembles premature senescence. J. Virol. 76, 12135–12148 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barhoover M. A., Hall J. M., Greenlee W. F., Thomas R. S., Aryl hydrocarbon receptor regulates cell cycle progression in human breast cancer cells via a functional interaction with cyclin-dependent kinase 4. Mol. Pharmacol. 77, 195–201 (2010). [DOI] [PubMed] [Google Scholar]
- 69.Huang G., Elferink C. J., Multiple mechanisms are involved in Ah receptor-mediated cell cycle arrest. Mol. Pharmacol. 67, 88–96 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Jackson D. P., Li H., Mitchell K. A., Joshi A. D., Elferink C. J., Ah receptor-mediated suppression of liver regeneration through NC-XRE-driven p21Cip1 expression. Mol. Pharmacol. 85, 533–541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pang P. H., et al., Molecular mechanisms of p21 and p27 induction by 3-methylcholanthrene, an aryl-hydrocarbon receptor agonist, involved in antiproliferation of human umbilical vascular endothelial cells. J. Cell. Physiol. 215, 161–171 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Dever D. P., Opanashuk L. A., The aryl hydrocarbon receptor contributes to the proliferation of human medulloblastoma cells. Mol. Pharmacol. 81, 669–678 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puga A., et al., Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J. Biol. Chem. 275, 2943–2950 (2000). [DOI] [PubMed] [Google Scholar]
- 74.Barnes-Ellerbe S., Knudsen K. E., Puga A., 2,3,7,8-Tetrachlorodibenzo-p-dioxin blocks androgen-dependent cell proliferation of LNCaP cells through modulation of pRB phosphorylation. Mol. Pharmacol. 66, 502–511 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Zydek M., et al., General blockade of human cytomegalovirus immediate-early mRNA expression in the S/G2 phase by a nuclear, Daxx- and PML-independent mechanism. J. Gen. Virol. 92, 2757–2769 (2011). [DOI] [PubMed] [Google Scholar]
- 76.Fortunato E. A., Sanchez V., Yen J. Y., Spector D. H., Infection of cells with human cytomegalovirus during S phase results in a blockade to immediate-early gene expression that can be overcome by inhibition of the proteasome. J. Virol. 76, 5369–5379 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheong J. E., Sun L., Targeting the IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy–Challenges and opportunities. Trends Pharmacol. Sci. 39, 307–325 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Shinde R., McGaha T. L., The aryl hydrocarbon receptor: Connecting immunity to the microenvironment. Trends Immunol. 39, 1005–1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gutiérrez-Vázquez C., Quintana F. J., Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 48, 19–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kado S., et al., Aryl hydrocarbon receptor signaling modifies Toll-like receptor-regulated responses in human dendritic cells. Arch. Toxicol. 91, 2209–2221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rothhammer V., et al., Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Litzenburger U. M., et al., Constitutive IDO expression in human cancer is sustained by an autocrine signaling loop involving IL-6, STAT3 and the AHR. Oncotarget 5, 1038–1051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dumortier J., et al., Human cytomegalovirus secretome contains factors that induce angiogenesis and wound healing. J. Virol. 82, 6524–6535 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Slinger E., et al., HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci. Signal. 3, ra58 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Zacharias D. A., Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 (2002). [DOI] [PubMed] [Google Scholar]
- 86.Diner B. A., et al., The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Mol. Syst. Biol. 11, 787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu D., Smith G. A., Enquist L. W., Shenk T., Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76, 2316–2328 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oberstein A., Shenk T., Cellular responses to human cytomegalovirus infection: Induction of a mesenchymal-to-epithelial transition (MET) phenotype. Proc. Natl. Acad. Sci. U.S.A. 114, E8244–E8253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 91.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Supek F., Bošnjak M., Škunca N., Šmuc T., REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, e21800 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu H., Shen Y., Shenk T., Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69, 7960–7970 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krishan A., Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 66, 188–193 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shenk T., Naseri-Nosar P., Differential genome expression in AhR-KD and control HCMV-infected fibroblasts. National Center for Biotechnology Information Gene Expression Omnibus. https://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159375. Deposited 12 October 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo, under accession number GSE159375 (96). All study data are included in the article and/or supporting information.