Abstract
The fruit of Lycium barbarum L. (goji berry) is used as traditional Chinese medicine, and has the functions of immune regulation, anti-tumor, neuroprotection, anti-diabetes, and anti-fatigue. One of the main bioactive components is L. barbarum polysaccharide (LBP). Nowadays, LBP is widely used in the health market, and it is extracted from the fruit of L. barbarum. The planting of L. barbarum needs large amounts of fields, and it takes one year to harvest the goji berry. The efficiency of natural LBP production is low, and the LBP quality is not the same at different places. Goji berry-derived LBP cannot satisfy the growing market demands. Engineered Saccharomyces cerevisiae has been used for the biosynthesis of some plant natural products. Recovery of LBP biosynthetic pathway in L. barbarum and expression of them in engineered S. cerevisiae might lead to the yeast LBP production. However, information on LBP biosynthetic pathways and the related key enzymes of L. barbarum is still limited. In this review, we summarized current studies about LBP biosynthetic pathway and proposed the strategies to recover key enzymes for LBP biosynthesis. Moreover, the potential application of synthetic biology strategies to produce LBP using engineered S. cerevisiae was discussed.
Keywords: lycium barbarum polysaccharide, Saccharomyces cerevisiae, synthetic biology, Goji Berry
1. Introduction
There are more than 80 Lycium species in the world, and they are widely distributed in Asia, Europe, North America, and other regions [1]. Among them, Lycium barbarum L. is a perennial deciduous shrub, and its fruits, goji berries, have been used as one of the traditional Chinese drugs [2,3]. Currently, goji berry is recorded in the Pharmacopoeia of the People’s Republic of China [4,5]. Goji berry has nourishing effects on the kidneys and lungs [6]. It is regarded as a nourishing Chinese medicine, and can be used as medicine and food [7]. Goji berry has diverse biological activities, including anti-inflammatory [8], anti-tumor [9], anti-oxidation [7], anti-aging [10], hypoglycemic [11], and hypolipidemic [12].
Goji berry is rich in nutrition and abundant in natural products, such as L. barbarum polysaccharide (LBP), betaine, flavone, and vitamin [3]. Among its bioactive components, LBP composes 5%–8% of the dried goji berry [13]. LBP is a water-soluble complex with carbohydrate chains and proteins; the carbohydrate chains are mainly composed of six saccharides, including arabinose, galactose, glucose, rhamnose, mannose, and xylose, which accounts for about 70% of all the saccharides [14,15]. LBP can modulate gut microbiota to improve nutrient utilization and health [16,17]. Nowadays, LBP is mainly extracted from goji berry. It takes one year to obtain goji berry, and climate, diseases as well as insect pests might affect the yield of goji berry [18]. Moreover, planting areas for high-quality goji berry with high-levels of LBP are limited. Besides, fresh goji berry is highly perishable, which limits the acquisition of high-quality LBP [19,20]. Extraction of LBP from goji berry is unable to meet the rapidly increasing commercial LBP demands. Therefore, it is of great interest to find other sustainable and stable LBP supplies [7].
With the rapid development of yeast synthetic biology, Saccharomyces cerevisiae and other yeasts have been used as cell factories for the production of plant natural products [21]. One advantage of producing polysaccharides using yeast cell factories is that the production is not affected by seasons, regions, and pests [22]. S. cerevisiae is generally recognized as a safe microorganism (GRAS) and has been widely used in food and drug production. Some plant natural products have been produced in engineered S. cerevisiae, such as cocoa butter and ginsenosides [23,24,25,26,27,28,29,30,31]. Therefore, S. cerevisiae is an ideal microbial host to produce LBP. Introducing the LBP biosynthetic pathway and rewiring the metabolic pathway of S. cerevisiae might provide a green and feasible way for LBP production [32,33].
2. LBP Biosynthetic Pathway in L. barbarum
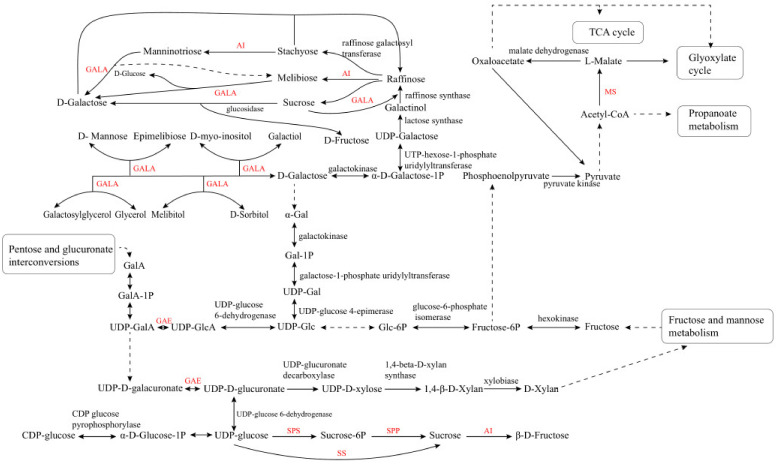
LBP is constituted of six saccharides of α-(1→4)-GalA, α-(1→6)-Glc, β-(1→3)-Galp, β-(1→6)-Galp, α-(1→5) -Ara, and β-(1→4)–Galp. Enzymatic selectivity is essential to combine the diverse saccharides into LBP. After the glycosidic bond is formed, the stereochemistry of LBP can be retained or reversed by glycosyltransferases [34,35,36]. In fact, multiple metabolic pathways might participate in LBP biosynthesis in goji berry, including the galactose metabolism pathway, tricarboxylic acid cycle, glyoxylic acid cycle, propionic acid metabolism, amino sugars metabolism, starch metabolism, and sucrose metabolism. These pathways provide precursors for the anabolism of various carbohydrates [37,38]. Among these pathways, sucrose synthetic pathway is mainly responsible for plant cell wall formation and biomass accumulation. Sucrose phosphate synthase (SPS) and sucrose phosphate phosphatase (SPP) are the two essential enzymes in sucrose biosynthesis [39]. In addition, the acid invertase (AI), sucrose synthase (SS), and SPS involved in sucrose metabolism are the key enzymes of the plant carbon metabolic pathway [40]. Sucrose synthesis is closely associated with the metabolism of amino sugar, nucleotide sugar, and galactose [41]. The fruits of goji berry mainly synthesize glucose and fructose, and the LBP content is mainly determined by fructose content in goji fruits.
The α-galactosidase (GALA) participates in several steps of galactose metabolism. These carbohydrate anabolic pathways are the prerequisite for LBP synthesis, and they can regulate carbohydrate accumulation. UDP-glucose pyrophosphorylase (UGP) is an essential enzyme involved in carbohydrate metabolism, which can affect normal cell development, polysaccharide synthesis, and stress response in S. cerevisiae and other fungi [42,43]. The key enzymes in the LBP synthetic pathway, UDP-glucuronate 4-epimerase (GAE), malate synthase (MS), and α-galactosidase (GALA), have not been characterized yet [37]. These key genes play important roles in the synthesis of LBP, and the information might be obtained by analyzing the transcriptome of goji berry (Figure 1).
Figure 1.
Metabolic pathways of saccharides in goji berry. AI, acid invertase; GAE, UDP-glucuronate 4-epimerase; GALA, α-galactosidase; MS, malate synthase; SPS, sucrose phosphate synthase; SPP, sucrose phosphate phosphatase; SS, sucrose synthase.
3. Mining of Key Enzymes for LBP Biosynthesis
The transcriptome of goji berry can help recover genes in the LBP biosynthetic pathway. Several studies about transcriptome of goji berry have been applied. Wang et al. prepared the leaf transcriptome of goji berry and elucidated the mechanisms of carotenoid biosynthesis in goji berry [44]. Chen et al. obtained 139,333 predicted genes of goji berry, and they identified genes in the flavonoid and taurine biosynthetic pathways [45]. Ma et al. analyzed the transcriptome of goji berry and identified the candidate genes involved in sugar metabolism under elevated CO2, which helped recover the LBP biosynthetic pathway [37]. However, current omics data in public databases have not recovered the whole LBP biosynthetic pathway in goji berry.
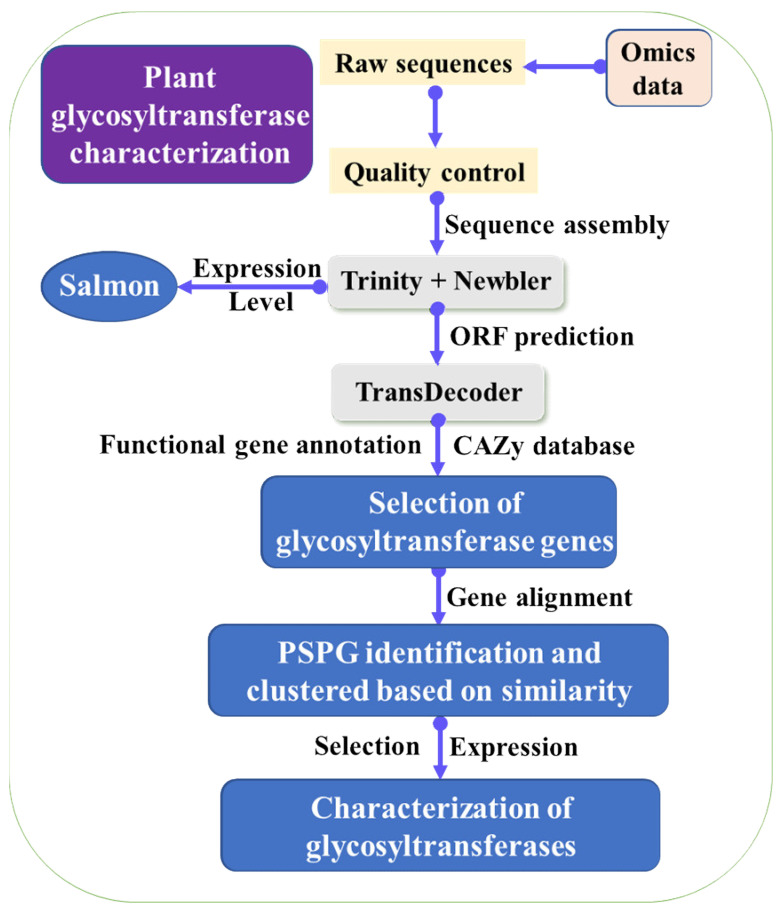
To explore the key enzymes in the LBP biosynthetic pathway, collecting fresh tissues of goji berry at different growth phases and different planting areas are necessary. Sequencing of these fresh tissues would give an array of transcriptome data, and de novo assembly of these data would obtain full gene profiles of goji berry. Further transcriptome comparison analysis would give insights into the details of the LBP biosynthetic pathway (Figure 2). To predict the key enzymes in LBP synthetic pathways, phylogenetic, gene similarity network analysis, and other bioinformatic methods can be used to identify the candidate key genes [46]. Diverse glycosyltransferase genes were identified from public omics data by using phylogenetic analyses and plant secondary product glycosyltransferase (PSPG) homolog [28,29,30]. Thus, finding homologous in goji genomes is possible to predict potential glycosyltransferases for LBP synthesis. As the sugar metabolic pathway is complex, characterizing candidate LBP synthetic genes identified with bioinformatic strategies will help identify efficient enzymes for LBP biosynthesis in engineered yeasts.
Figure 2.
Strategy used for identification and characterization of glycosyltransferase genes from transcriptomic data.
The glycosyltransferases can determine saccharide compositions and arrangements of the LBP. The bioinformatic strategy was used for the recovery of glycosyltransferases functioned in ginsenoside biosynthesis. The omics data in the public database were downloaded and assembled. The open reading frames (ORFs) were predicted and annotated with the help of CAZy and other databases. The candidate glycosyltransferase genes were selected for sequence alignment. Only predicted glycosyltransferase genes with more than 1320 bp and PSPG motif were used for further analysis. The glycosyltransferase genes were clustered based on the similarity, in order to reduce the redundancy. Besides, gene expression level can be determined at different growth stages, which can help identify real glycosyltransferase genes. The identified potential glycosyltransferase genes for ginsenosides were selected for expression. The enzymatic characterization of the glycosyltransferases was identified (Figure 2), and the characterized glycosyltransferases were used for adding activated sugars to the substrates [28,29,30,31]. Recently, another two cellulose synthases were identified to function as glycosyltransferases in saponin biosynthesis [47,48], showing identification of glycosyltransferases for LBP synthesis is complex. Nowadays, the use of gene similarity and network analysis have been widely used for gene identification [46].
4. S. cerevisiae is an Efficient Cell Factory for the Production of Plant Natural Products
Polysaccharides are nontoxic and natural biodegradable biopolymers. Normally, the main components of plant cell wall and other biomass are polysaccharides. Besides, the extracellular polysaccharides play essential roles in biofilm formation. It is possible to use microorganisms to produce polysaccharides. S. cerevisiae is a popular organism for industrial-scale production of plant natural products, such as artemisinic acid (the precusor of artemisinin), ginsenosides, and cocoa butter [49,50,51]. Genetic engineering and synthetic biology strategies, including homologous recombination and CRISPR-Cas9 system, are easy to implement in S. cerevisiae [52,53].
As an ideal microbial cell factory for the production of plant natural products, S. cerevisiae is often used to characterize key genes of plant natural products. The cell wall of S. cerevisiae is mainly composed of glucan and mannose [54,55]. Furthermore, arabinose, rhamnose, xylose, and galactose can be synthesized in S. cerevisiae directly [56,57,58,59]. S. cerevisiae can use cheap substrates and expressing key LBP synthetic genes in S. cerevisiae are easy, therefore, converting cheap sugar substrates into LBP are possible.
Levan is a bacterial extracellular polysaccharide, and it can be used as prebiotic fibers in the production of functional foods and pharmaceutical formulation. Franken et al. engineered S. cerevisiae for the biosynthesis of levans. In order to synthesize levan, an invertase of S. cerevisiae was deleted to build an invertase null S. cerevisiae mutant. The levansucrase expressed in invertase null S. cerevisiae mutants or strains with sucrose accumulation strains led to the intracellular levan accumulation [60]. Co-expression of the levansucrase and the spinach sucrose transporter achieved hyper-production of extracellular levans [61]. Overexpression of the levansucrase gene of Rahnella aquatilis in S. cerevisiae and enhancing its secretion by expression with a secretion signal resulted in the final levan titer of 76 g/L in a 50-L fermenter [62]. The substrate sucrose conversion efficiency is 80% and the production rate is 3.17 g/L/h [62]. The β-Glucan is one of the main components of yeast. Overexpression of PGM2 (encoding phosphoglucomutase) and RHO1 (encoding a GTPase for activating glucan synthesis) enhanced β-Glucan production in S. cerevisiae. Optimization of the culture condition further led to a >53.1% increase [55]. Moreover, S. cerevisiae was discussed as one potential producer of chitosan, which is a β-1,4-linked glucosamine polymer [63].
5. Engineering Strategies for High-Level LBP Production in S. cerevisiae
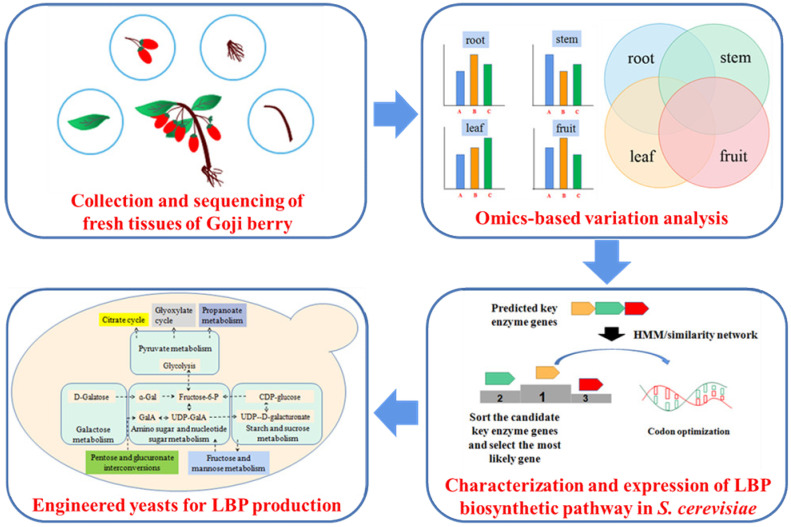
With the rapid development of the next-generation sequencing technology, plant transcriptome and genomic data were used to determine key genes in the LBP pathway, such as the integration of transcriptomic and metabonomic strategy for gene identification, and the variation analysis of different omics data [64]. AI and SPS are the key enzymes involved in sucrose metabolism. AI breaks down sucrose into glucose and fructose, and SPS catalyzes the synthesis of sucrose [48,65]. Balance expression of these genes in S. cerevisiae may help to synthesize LBP. Using bioinformatics and systems biology, we may characterize unknown key enzymes of GAE, MS, and GALA functioned in complex natural product biosynthesis (Figure 3) [66,67]. The identification of genes in the LBP biosynthetic pathway is the first step for LBP biosynthesis (Figure 3). Though LBP biosynthetic genes of goji berry might be the most efficient ones for LBP biosynthesis, the genes of goji berry might not be well expressed and functioned in S. cerevisiae. Codon-optimized and engineering of the key enzymes can possibly help obtain efficient enzymes for LBP biosynthesis in S. cerevisiae.
Figure 3.
Construction of the Lycium barbarum polysaccharide (LBP) metabolic pathways in Saccharomyces cerevisiae through extraction of total RNA from different tissues of goji berry, analyzing the different expression strength in different parts. Using HMM and gene similarity network to analyze the predicted key enzymes, optimize the codons of the key enzyme would lead to the LBP production in engineered S. cerevisiae using synthetic biology strategies.
When the engineered S. cerevisiae strain can synthesize LBP, selecting suitable substrates, enhancing the supply of cofactors and precursors, and the heterologous expression of key genes will direct the metabolic flux to LBP synthesis and improve the production of LBP. For industrial-scale production, optimization of yeast cultivation medium and conditions might further increase LBP production in S. cerevisiae. Some plant natural products produced with engineered yeasts, such as artemisinin, have been approved for medicinal use [43,44], but using products derived from engineered microorganisms as foods is rarely reported. Further demonstrating the safety of products derived from engineered microorganisms and discussing the possible revision of the regulatory rules are essential before providing engineered yeast-based products to consumers. As S. cerevisiae is GRAS, the S. cerevisiae biomass might be directly used as prebiotics for poultry or other animals without further bioprocess. In the future, the synthetic biology will help introduce the redesigned LBP biosynthetic pathway in yeast and lead to the high-level production of LBP in the future [68].
6. Conclusions and Future Perspective
As the main bioactive component, LBP has a variety of pharmacological effects, but its supply depends on the extraction from goji berry. Production of LBP by engineered S. cerevisiae provides a possible green strategy for LBP supply. The completed synthetic pathways and key enzymes are unclear now, which limits yeast production of LBP. Finding the key enzymes and designing the intact pathways using omics data are possible now. In the near future, construction of an optimized redesigned LBP synthetic pathway in S. cerevisiae will lead to large-scale production of LBP, which would provide a sustainable way for LBP supply.
Acknowledgments
In this section, you can acknowledge any support given which is not covered by the author contribution or funding sections. This may include administrative and technical support, or donations in kind (e.g., materials used for experiments).
Author Contributions
Y.W. conceived the project. J.P., L.W., M.W., and R.D. contributed to figures and manuscript draft preparations. Y.W., C.-Y.J., and S.Q. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the National Natural Science Foundation of China (NO. 31800079 for Y. W), Clinical Laboratories, Shenyou Bio, Henan Muyi Animal Pharmaceutical Co., Ltd, Kaifeng Muyi Huamiao Biological Technology Co., Ltd, and Institute of Henan modern biotechnology Co. Ltd (No. HMB0066).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yao R., Heinrich M., Weckerle C.S. The genus Lycium as food and medicine: A botanical, ethnobotanical and historical review. J. Ethnopharmacol. 2018;212:50–66. doi: 10.1016/j.jep.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Amagase H., Farnsworth N.R. A review of botanical characteristics, phytochemistry, clinical relevance in efficacy and safety of Lycium barbarum fruit (Goji) Food Res. Int. 2011;44:1702–1717. doi: 10.1016/j.foodres.2011.03.027. [DOI] [Google Scholar]
- 3.Gao Y., Wei Y., Wang Y., Gao F., Chen Z. Lycium Barbarum: A Traditional Chinese Herb and A Promising Anti-Aging Agent. Aging Dis. 2017;8:778–791. doi: 10.14336/AD.2017.0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang B., Jin M., Liu H. Water-soluble polysaccharide from dried Lycium barbarum fruits: Isolation, structural features and antioxidant activity. Carbohydr. Polym. 2011;83:1947–1951. doi: 10.1016/j.carbpol.2010.10.066. [DOI] [Google Scholar]
- 5.Wetters S., Horn T., Nick P. Goji Who? Morphological and DNA Based Authentication of a "Superfood". Front Plant Sci. 2018;9:1859. doi: 10.3389/fpls.2018.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwok S.S., Bu Y., Lo A.C., Chan T.C., So K.F., Lai J.S., Shih K.C. A Systematic Review of Potential Therapeutic Use of Lycium Barbarum Polysaccharides in Disease. Biomed Res. Int. 2019;2019:4615745. doi: 10.1155/2019/4615745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Z.F., Zhang H., Teh S.S., Wang C.W., Zhang Y., Hayford F., Wang L., Ma T., Dong Z., Zhang Y., et al. Goji Berries as a Potential Natural Antioxidant Medicine: An Insight into Their Molecular Mechanisms of Action. Oxidative Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/2437397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avila C.N., Uecker J.N., Ribeiro Trindade F.M., Alvarado Rincon J.A., Penteado J.O., de Barros C.C., Janke F., Andreazza R., Schneider J.P., Pieniz S. Anti-inflammatory Effect of a Goji Berry Extract (Lycium barbarum) in Rats Subjected to Inflammation by Lipopolysaccharides (LPS) Braz. Arch. Biol. Technol. 2020;63 doi: 10.1590/1678-4324-2020180612. [DOI] [Google Scholar]
- 9.Tang W.M., Chan E., Kwok C.Y., Lee Y.K., Wu J.H., Wan C.W., Chan R.Y., Yu P.H., Chan S.W. A review of the anticancer and immunomodulatory effects of Lycium barbarum fruit. Inflammopharmacology. 2012;20:307–314. doi: 10.1007/s10787-011-0107-3. [DOI] [PubMed] [Google Scholar]
- 10.Xia G., Xin N., Liu W., Yao H., Hou Y., Qi J. Inhibitory effect of Lycium barbarum polysaccharides on cell apoptosis and senescence is potentially mediated by the p53 signaling pathway. Mol. Med. Rep. 2014;9:1237–1241. doi: 10.3892/mmr.2014.1964. [DOI] [PubMed] [Google Scholar]
- 11.Cai H., Liu F., Zuo P., Huang G., Song Z., Wang T., Lu H., Guo F., Han C., Sun G. Practical Application of Antidiabetic Efficacy of Lycium barbarum Polysaccharide in Patients with Type 2 Diabetes. Med. Chem. 2015;11:383–390. doi: 10.2174/1573406410666141110153858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Q., Cai Y., Yan J., Sun M., Corke H. Hypoglycemic and hypolipidemic effects and antioxidant activity of fruit extracts from Lycium barbarum. Life Sci. 2004;76:137–149. doi: 10.1016/j.lfs.2004.04.056. [DOI] [PubMed] [Google Scholar]
- 13.Skenderidis P., Lampakis D., Giavasis I., Leontopoulos S., Petrotos K., Hadjichristodoulou C., Tsakalof A. Chemical Properties, Fatty-Acid Composition, and Antioxidant Activity of Goji Berry (Lycium barbarum L. and Lycium chinense Mill.) Fruits. Antioxidants. 2019;8:60. doi: 10.3390/antiox8030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng G.-Q., Zheng Z.-Y., Xu X., Hu Z.-H. Variation in fruit sugar composition of Lycium barbarum L. and Lycium chinense Mill. of different regions and varieties. Biochem. Syst. Ecol. 2010;38:275–284. doi: 10.1016/j.bse.2010.01.008. [DOI] [Google Scholar]
- 15.Masci A., Carradori S., Casadei M.A., Paolicelli P., Petralito S., Ragno R., Cesa S. Lycium barbarum polysaccharides: Extraction, purification, structural characterisation and evidence about hypoglycaemic and hypolipidaemic effects. A review. Food Chem. 2018;254:377–389. doi: 10.1016/j.foodchem.2018.01.176. [DOI] [PubMed] [Google Scholar]
- 16.Tian X., Liang T., Liu Y., Ding G., Zhang F., Ma Z. Extraction, Structural Characterization, and Biological Functions of Lycium Barbarum Polysaccharides: A Review. Biomol. 2019;9:389. doi: 10.3390/biom9090389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Lv X., Wu T., Ma Q., Teng A., Zhang Y., Zhang M. Composition of Lycium barbarum polysaccharides and their apoptosis-inducing effect on human hepatoma SMMC-7721 cells. Food Nutr. Res. 2015;59:28696. doi: 10.3402/fnr.v59.28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W.-F., Yang J.-L., Shi Y.-P. Quality evaluation of six bioactive constituents in goji berry based on capillary electrophoresis field amplified sample stacking. Electrophoresis. 2018;39:2117–2124. doi: 10.1002/elps.201800102. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y., Lai Y., Chen Z., Qu H., Ma S., Wang Y., Jiang Y. Evolution of physiological characteristics and nutritional quality in fresh goji berry (Lycium barbarum) stored under different temperatures. J. Food Process. Preserv. 2020;44:e14835. doi: 10.1111/jfpp.14835. [DOI] [Google Scholar]
- 20.Song H.H., Bi J.F., Chen Q.Q., Zhou M., Wu X.Y., Song J.X. Structural and health functionality of dried goji berries as affected by coupled dewaxing pre-treatment and hybrid drying methods. Int. J. Food Prop. 2018;21:2527–2538. doi: 10.1080/10942912.2018.1536148. [DOI] [Google Scholar]
- 21.Parapouli M., Vasileiadis A., Afendra A.S., Hatziloukas E. Saccharomyces cerevisiae and its industrial applications. Aims Microbiol. 2020;6:1–31. doi: 10.3934/microbiol.2020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid J., Sieber V. Enzymatic transformations involved in the biosynthesis of microbial exo-polysaccharides based on the assembly of repeat units. Chembiochem. 2015;16:1141–1147. doi: 10.1002/cbic.201500035. [DOI] [PubMed] [Google Scholar]
- 23.Wei Y., Gossing M., Bergenholm D., Siewers V., Nielsen J. Increasing cocoa butter-like lipid production of Saccharomyces cerevisiae by expression of selected cocoa genes. Amb Express. 2017;7:34. doi: 10.1186/s13568-017-0333-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei Y., Siewers V., Nielsen J. Cocoa butter-like lipid production ability of non-oleaginous and oleaginous yeasts under nitrogen-limited culture conditions. Appl. Microbiol. Biotechnol. 2017;101:3577–3585. doi: 10.1007/s00253-017-8126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergenholm D., Gossing M., Wei Y., Siewers V., Nielsen J. Modulation of saturation and chain length of fatty acids in Saccharomyces cerevisiae for production of cocoa butter-like lipids. Biotechnol. Bioeng. 2018;115:932–942. doi: 10.1002/bit.26518. [DOI] [PubMed] [Google Scholar]
- 26.Wei Y., Bergenholm D., Gossing M., Siewers V., Nielsen J. Expression of cocoa genes in Saccharomyces cerevisiae improves cocoa butter production. Microb. Cell Factories. 2018;17:11. doi: 10.1186/s12934-018-0866-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Y., Ji B., Siewers V., Xu D., Halkier B.A., Nielsen J. Identification of genes involved in shea butter biosynthesis from Vitellaria paradoxa fruits through transcriptomics and functional heterologous expression. Appl. Microbiol. Biotechnol. 2019;103:3727–3736. doi: 10.1007/s00253-019-09720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., Zhang L., Zhao G., Yue J., Zhou Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P., Wei Y., Fan Y., Liu Q., Wei W., Yang C., Zhang L., Zhao G., Yue J., Yan X., et al. Production of bioactive ginsenosides Rh2 and Rg3 by metabolically engineered yeasts. Metab. Eng. 2015;29:97–105. doi: 10.1016/j.ymben.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Wei W., Wang P., Wei Y., Liu Q., Yang C., Zhao G., Yue J., Yan X., Zhou Z. Characterization of Panax ginseng UDP-glycosyltransferases catalyzing protopanaxatriol and biosyntheses of bioactive ginsenosides F1 and Rh1 in metabolically engineered yeasts. Mol. Plant. 2015;8:1412–1424. doi: 10.1016/j.molp.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Yang C., Li C., Wei W., Wei Y., Liu Q., Zhao G., Yue J., Yan X., Wang P., Zhou Z. The unprecedented diversity of UGT94-family UDP-glycosyltransferases in Panax plants and their contribution to ginsenoside biosynthesis. Sci. Rep. 2020;10:15394. doi: 10.1038/s41598-020-72278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlean P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics. 2012;192:775–818. doi: 10.1534/genetics.112.144485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bzducha-Wróbel A., Błażejak S., Kieliszek M., Pobiega K., Falana K., Janowicz M. Modification of the cell wall structure of Saccharomyces cerevisiae strains during cultivation on waste potato juice water and glycerol towards biosynthesis of functional polysaccharides. J. Biotechnol. 2018;281:1–10. doi: 10.1016/j.jbiotec.2018.06.305. [DOI] [PubMed] [Google Scholar]
- 34.Yuan Y., Wang Y.B., Jiang Y., Prasad K.N., Yang J., Qu H., Wang Y., Jia Y., Mo H., Yang B. Structure identification of a polysaccharide purified from Lycium barbarium fruit. Int. J. Biol. Macromol. 2016;82:696–701. doi: 10.1016/j.ijbiomac.2015.10.069. [DOI] [PubMed] [Google Scholar]
- 35.Liu W., Liu Y., Zhu R., Yu J., Lu W., Pan C., Yao W., Gao X. Structure characterization, chemical and enzymatic degradation, and chain conformation of an acidic polysaccharide from Lycium barbarum L. Carbohydr. Polym. 2016;147:114–124. doi: 10.1016/j.carbpol.2016.03.087. [DOI] [PubMed] [Google Scholar]
- 36.Mestrom L., Przypis M., Kowalczykiewicz D., Pollender A., Kumpf A., Marsden S.R., Bento I., Jarzębski A.B., Szymańska K., Chruściel A., et al. Leloir Glycosyltransferases in Applied Biocatalysis: A Multidisciplinary Approach. Int. J. Mol. Sci. 2019;20:5263. doi: 10.3390/ijms20215263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma Y., Reddy V.R., Devi M.J., Song L., Cao B. De novo characterization of the Goji berry (Lycium barbarium L.) fruit transcriptome and analysis of candidate genes involved in sugar metabolism under different CO2 concentrations. Tree Physiol. 2019;39:1032–1045. doi: 10.1093/treephys/tpz014. [DOI] [PubMed] [Google Scholar]
- 38.Chen J.H., Zhang D.Z., Zhang C., Xu M.L., Yin W.L. Physiological characterization, transcriptomic profiling, and microsatellite marker mining of Lycium ruthenicum. J. Zhejiang Univ. Sci. B. 2017;18:1002–1021. doi: 10.1631/jzus.B1700135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J.W., Zhang S.L., Zhang L.C. Sugar transport, metabolism, accumulation and their regulation in fruits. J. Plant Physiol. Mol. Biol. 2004;30:1–10. [PubMed] [Google Scholar]
- 40.Koch K. Sucrose metabolism: Regulatory mechanisms and pivotal roles in sugar sensing and plant development. Curr. Opin. Plant Biol. 2004;7:235–246. doi: 10.1016/j.pbi.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Stein O., Granot D. An Overview of Sucrose Synthases in Plants. Front Plant Sci. 2019;10:95. doi: 10.3389/fpls.2019.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng S., Yao C., Zhang X., Jia Z., Shan C., Luo X., Lin L. Involvement of UDP-glucose pyrophosphorylase from Verticillium dahliae in cell morphogenesis, stress responses, and host infection. Fungal Biol. 2020;124:648–660. doi: 10.1016/j.funbio.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Zan X.Y., Wu X.H., Cui F.J., Zhu H.A., Sun W.J., Jiang L.H., Tao T.L., Zhao X. UDP-glucose pyrophosphorylase gene affects mycelia growth and polysaccharide synthesis of Grifola frondosa. Int. J. Biol. Macromol. 2020;161:1161–1170. doi: 10.1016/j.ijbiomac.2020.06.139. [DOI] [PubMed] [Google Scholar]
- 44.Wang G., Du X., Ji J., Guan C., Li Z., Josine T.L. De novo characterization of the Lycium chinense Mill. leaf transcriptome and analysis of candidate genes involved in carotenoid biosynthesis. Gene. 2015;555:458–463. doi: 10.1016/j.gene.2014.10.058. [DOI] [PubMed] [Google Scholar]
- 45.Chen C., Xu M., Wang C., Qiao G., Wang W., Tan Z., Wu T., Zhang Z. Characterization of the Lycium barbarum fruit transcriptome and development of EST-SSR markers. PLoS ONE. 2017;12:e0187738. doi: 10.1371/journal.pone.0187738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M., Wei Y., Ji B., Nielsen J. Advances in Metabolic Engineering of Saccharomyces cerevisiae for Cocoa Butter Equivalent Production. Front. Bioeng. Biotechnol. 2020;8:1194. doi: 10.3389/fbioe.2020.594081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung S.Y., Seki H., Fujisawa Y., Shimoda Y., Hiraga S., Nomura Y., Saito K., Ishimoto M., Muranaka T. A cellulose synthase-derived enzyme catalyses 3-O-glucuronosylation in saponin biosynthesis. Nat. Commun. 2020;11:5664. doi: 10.1038/s41467-020-19399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jozwiak A., Sonawane P.D., Panda S., Garagounis C., Papadopoulou K.K., Abebie B., Massalha H., Almekias-Siegl E., Scherf T., Aharoni A. Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat. Chem. Biol. 2020;16:740–748. doi: 10.1038/s41589-020-0541-x. [DOI] [PubMed] [Google Scholar]
- 49.Nimpiboon P., Tumhom S., Nakapong S., Pongsawasdi P. Amylomaltase from Thermus filiformis: Expression in Saccharomyces cerevisiae and its use in starch modification. J. Appl. Microbiol. 2020;129:1287–1296. doi: 10.1111/jam.14675. [DOI] [PubMed] [Google Scholar]
- 50.Gao Q., Wang L., Zhang M., Wei Y., Lin W. Recent Advances on Feasible Strategies for Monoterpenoid Production in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 2020;8:1372. doi: 10.3389/fbioe.2020.609800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guan R., Wang M., Guan Z., Jin C.-Y., Lin W., Ji X., Wei Y. Metabolic engineering for glycyrrhetinic acid production in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 2020;8:1318. doi: 10.3389/fbioe.2020.588255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai Z., Huang M., Chen Y., Siewers V., Nielsen J. Global rewiring of cellular metabolism renders Saccharomyces cerevisiae Crabtree negative. Nat. Commun. 2018;9:3059. doi: 10.1038/s41467-018-05409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mans R., van Rossum H.M., Wijsman M., Backx A., Kuijpers N.G., van den Broek M., Daran-Lapujade P., Pronk J.T., van Maris A.J., Daran J.M. CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. Fems Yeast Res. 2015:15. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jigami Y., Odani T. Mannosylphosphate transfer to yeast mannan. Biochim. Biophys. Acta. 1999;1426:335–345. doi: 10.1016/S0304-4165(98)00134-2. [DOI] [PubMed] [Google Scholar]
- 55.Zhou X., He J., Wang L., Wang Y., Du G., Kang Z. Metabolic Engineering of Saccharomyces cerevisiae to Improve Glucan Biosynthesis. J. Microbiol. Biotechnol. 2019;29:758–764. doi: 10.4014/jmb.1812.12049. [DOI] [PubMed] [Google Scholar]
- 56.Ye S., Kim J.W., Kim S.R. Metabolic Engineering for Improved Fermentation of L-Arabinose. J. Microbiol. Biotechnol. 2019;29:339–346. doi: 10.4014/jmb.1812.12015. [DOI] [PubMed] [Google Scholar]
- 57.Oka T., Jigami Y. Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. Febs J. 2006;273:2645–2657. doi: 10.1111/j.1742-4658.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 58.Bahia F.M., de Almeida G.C., de Andrade L.P., Campos C.G., Queiroz L.R., da Silva R.L.V., Abdelnur P.V., Corrêa J.R., Bettiga M., Parachin N.S. Rhamnolipids production from sucrose by engineered Saccharomyces cerevisiae. Sci. Rep. 2018;8:2905. doi: 10.1038/s41598-018-21230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van den Brink J., Akeroyd M., van der Hoeven R., Pronk J.T., de Winde J.H., Daran-Lapujade P. Energetic limits to metabolic flexibility: Responses of Saccharomyces cerevisiae to glucose-galactose transitions. Microbiology. 2009;155 Pt 4:1340–1350. doi: 10.1099/mic.0.025775-0. [DOI] [PubMed] [Google Scholar]
- 60.Lafraya Á., Sanz-Aparicio J., Polaina J., Marín-Navarro J. Fructo-oligosaccharide synthesis by mutant versions of Saccharomyces cerevisiae invertase. Appl. Env. Microbiol. 2011;77:6148. doi: 10.1128/AEM.05032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Franken J., Brandt B.A., Tai S.L., Bauer F.F. Biosynthesis of Levan, a Bacterial Extracellular Polysaccharide, in the Yeast Saccharomyces cerevisiae. PLoS ONE. 2013;8:e77499. doi: 10.1371/journal.pone.0077499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ko H., Bae J.H., Sung B.H., Kim M.J., Kim C.H., Oh B.R., Sohn J.H. Efficient production of levan using a recombinant yeast Saccharomyces cerevisiae hypersecreting a bacterial levansucrase. J. Ind. Microbiol. Biotechnol. 2019;46:1611–1620. doi: 10.1007/s10295-019-02206-1. [DOI] [PubMed] [Google Scholar]
- 63.Ghormade V., Pathan E.K., Deshpande M.V. Can fungi compete with marine sources for chitosan production? Int. J. Biol. Macromol. 2017;104 Pt B:1415–1421. doi: 10.1016/j.ijbiomac.2017.01.112. [DOI] [PubMed] [Google Scholar]
- 64.Ambardar S., Gupta R., Trakroo D., Lal R., Vakhlu J. High Throughput Sequencing: An Overview of Sequencing Chemistry. Indian J. Microbiol. 2016;56:394–404. doi: 10.1007/s12088-016-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li C., Liu Y., Tian J., Zhu Y., Fan J. Changes in sucrose metabolism in maize varieties with different cadmium sensitivities under cadmium stress. PLoS ONE. 2020;15:e0243835. doi: 10.1371/journal.pone.0243835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scossa F., Benina M., Alseekh S., Zhang Y., Fernie A.R. The Integration of Metabolomics and Next-Generation Sequencing Data to Elucidate the Pathways of Natural Product Metabolism in Medicinal Plants. Planta Med. 2018;84:855–873. doi: 10.1055/a-0630-1899. [DOI] [PubMed] [Google Scholar]
- 67.Han R., Rai A., Nakamura M., Suzuki H., Takahashi H., Yamazaki M., Saito K. De Novo Deep Transcriptome Analysis of Medicinal Plants for Gene Discovery in Biosynthesis of Plant Natural Products. Methods Enzym. 2016;576:19–45. doi: 10.1016/bs.mie.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 68.Voigt C.A. Synthetic biology 2020–2030: Six commercially-available products that are changing our world. Nat. Commun. 2020;11:6379. doi: 10.1038/s41467-020-20122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.