Significance
The lack of viable tumor-specific targets continues to thwart efforts to implement selective anticancer drugs in the clinic. Clonal loss of heterozygosity (LOH) occurs in the great majority of human tumors and represents an irreversible genetic alteration present in cancer cells that unequivocally distinguishes them from normal cells. Here, we report the development of NASCAR (Neoplasm-targeting Allele-Sensing CAR), a platform comprising pairwise chimeric receptors for detecting and targeting LOH events in cancer. As proof-of-concept, we demonstrate specific NASCAR T cell responses in models of HLA LOH in vitro and in vivo. This work lays the foundation for future exploration and exploitation of LOH-mediated vulnerabilities for precision cellular immunotherapy.
Keywords: loss of heterozygosity, human leukocyte antigen, cell engineering, cancer immunotherapy, chimeric antigen receptor
Abstract
Developing therapeutic agents with potent antitumor activity that spare normal tissues remains a significant challenge. Clonal loss of heterozygosity (LOH) is a widespread and irreversible genetic alteration that is exquisitely specific to cancer cells. We hypothesized that LOH events can be therapeutically targeted by “inverting” the loss of an allele in cancer cells into an activating signal. Here we describe a proof-of-concept approach utilizing engineered T cells approximating NOT-gate Boolean logic to target counterexpressed antigens resulting from LOH events in cancer. The NOT gate comprises a chimeric antigen receptor (CAR) targeting the allele of human leukocyte antigen (HLA) that is retained in the cancer cells and an inhibitory CAR (iCAR) targeting the HLA allele that is lost in the cancer cells. We demonstrate that engineered T cells incorporating such NOT-gate logic can be activated in a genetically predictable manner in vitro and in mice to kill relevant cancer cells. This therapeutic approach, termed NASCAR (Neoplasm-targeting Allele-Sensing CAR), could, in theory, be extended to LOH of other polymorphic genes that result in altered cell surface antigens in cancers.
A major challenge in both cancer diagnosis and therapy is specificity. The paucity of reagents in the clinic that target properties exquisitely specific for cancer cells presents a substantial obstacle for the development of potent therapeutic modalities. Given the success of early oncogene-targeted therapies, it was hoped that cancer genome sequencing initiatives would yield a plethora of novel actionable targets (1–5). Unfortunately, such sequencing efforts identified few unique oncogene targets and instead revealed that the cancer genome landscape was dominated by mutations of tumor suppressor genes, with most cancers having a limited number of mutated oncogenes (6). Compounding these issues, many of the most frequently mutated oncogenes have proved resilient to therapeutic intervention (7–10).
One of the most pervasive and specific genetic characteristics in cancers involves chromosomal gains and losses, which are nearly always associated with aneuploidy (11–14). Aneuploidy is found in all major human tumor types and was the first genetic abnormality identified in cancers (15–18). A molecular manifestation of many chromosomal aberrations is loss of heterozygosity (LOH). LOH occurs when a heterozygous locus loses one of its two parental alleles, typically as a result of losses of large chromosomal regions, often involving an entire chromosome arm (19–23). Such allelic deletions are observed in over 90% of human cancers and are commonly associated with tumor suppressor gene loss (6, 24–31). Importantly, loss of genetic material through LOH is irreversible. LOH events thus represent frequent, predictable, and irreversible genetic events that can distinguish cancer cells from normal cells in an unequivocal fashion.
While a variety of approaches to detect genetic abnormalities such as LOH in cancers have been developed (32, 33), therapeutically targeting these recurrent tumor-specific alterations has proven to be challenging. A range of modalities have been explored to target LOH at the DNA (CRISPR), RNA (antisense oligonucleotide, short hairpin RNA, small interfering RNA), and protein (small-molecule inhibitors) levels (34–40). Although promising in principle, these approaches have distinct practical barriers that have precluded their clinical realization, ranging from issues surrounding reagent delivery to proteins that lack targetable binding pockets for small molecules.
Advances in immunotherapeutic approaches have ushered in a new era of cancer treatment and, with it, the exciting prospect for the specific targeting of cancer cells. Much of the success with immunotherapy has resulted from unleashing preexisting antitumor immune responses toward somatic mutation-associated neoantigens by immune checkpoint inhibitors (41–56). Attempts to redirect the immune system toward cancer cells de novo have also been successful but have largely relied on targeting tumor-associated antigens (TAAs) (57–59). Although such targets are typically overexpressed on cancer cells—sometimes as a result of genetic amplification—their expression in various normal tissues can result in major “on-target, off-tumor” toxicities (60–63).
Chimeric antigen receptor (CAR) T cells are an emerging class of immunotherapeutic agents that permit immune cell redirection and have generated remarkable clinical responses in patients with B cell malignancies (64–70). Despite such progress, CAR T cells have yet to be widely applied to patients with solid tumors (71–73). Among the many obstacles that remain in translating the impressive success of CAR therapy to solid tumors is the dearth of tumor-specific antigens (74). Similar to other targeted therapeutic modalities, CAR T cell therapies have largely been developed against TAAs, such as CD19 (58). Unlike CD19 CAR therapy whereby simultaneous ablation of cancerous and healthy B cells alike results in manageable adverse effects (61), toxicity profiles for other TAA targets are often unacceptable for clinical implementation and, in rare cases, fatal (75–78). In the absence of suitable targets similar to CD19 for non-B cell malignancies, CAR T cells capable of combinatorial antigen recognition have been developed to increase on-tumor specificity (79–85). However, the lack of truly tumor-specific antigens will continue to present a challenge for bringing CAR T cells to fruition for most cancer patients (74).
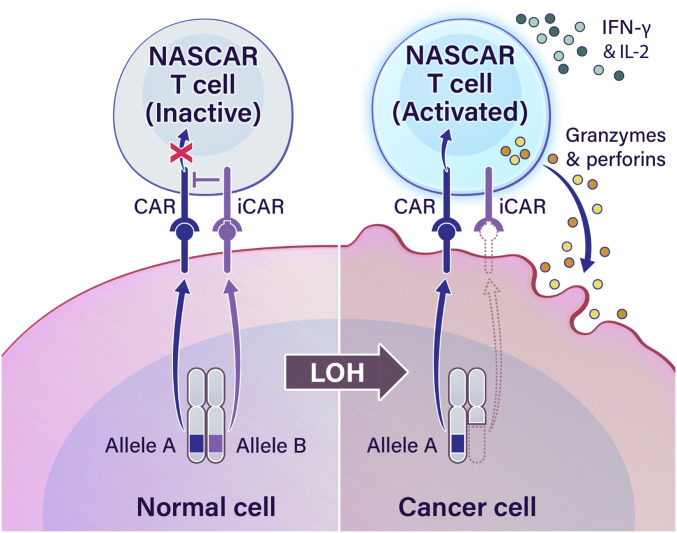
On the premise that LOH provides an opportunity for tumor-specific targeting, we sought to develop a method to target LOH of heterozygous alleles. We hypothesized that implementation of a NOT gate could permit target discrimination between a normal cell expressing products from both alleles and a cancer cell expressing products from the one retained allele following LOH. One innovative strategy approximating NOT-gate logic was described by Fedorov et al. (86), where a CAR and inhibitory CAR (iCAR) targeting two different antigens were simultaneously introduced into a T cell. While CARs contain activating T cell receptor (TCR)-derived domains, iCARs contain inhibitory domains derived from immune inhibitory receptors such as programmed cell death protein-1 (PD-1) and cytotoxic T lymphocyte-associated protein-4 (CTLA-4). Accordingly, CAR- and iCAR-expressing T cells will kill cells expressing only the CAR antigen while sparing cells harboring both the CAR and iCAR antigens. In this work, we investigated whether such a logic-gated system could be applied to LOH in cancer by targeting the iCAR to the allele lost through LOH while targeting the CAR to the retained allele (Fig. 1). As proof-of-concept, we focused on targeting human leukocyte antigen (HLA) genes owing to the seminal work of George D. Snell in identifying the major histocompatibility complex (MHC) gene complex as a potent mediator of tumor rejection (87, 88) and because HLA genes are highly polymorphic and have been observed to commonly undergo LOH in cancer (89–93).
Fig. 1.
An immunotherapeutic approach for targeting LOH. A schema describing the proposed cellular engineering strategy to target LOH events in cancer. Arrows labeled “Allele A” and “Allele B” depict the production of a polymorphic protein as a result of transcription and translation of a polymorphic gene subject to LOH in cancer. The NASCAR platform comprises pairwise CAR and iCAR receptors in T cells. Concurrent engagement of both receptors will result in iCAR-mediated quenching of proximal CAR signaling and divert T cell activation away from normal cells expressing both alleles (Left). However, cancer cells that have undergone LOH will trigger the CAR but not the iCAR, resulting in NASCAR T cell activation (Right).
Here we report the development of NASCAR (Neoplasm-targeting Allele-Sensing CAR), a platform comprising pairwise chimeric receptors that, together, are capable of allele sensing and detecting LOH events in cancer. Utilizing CARs and iCARs specific to two common alleles (HLA-A*02:01 or HLA-A*03:01) of the same gene (HLA-A), we demonstrate LOH-specific NASCAR T cell responses in models of HLA LOH in vitro and in vivo.
Results
Identification of HLA-A Allele-Specific Detection Moieties.
LOH of the short arm of chromosome 6 (6p)—where the HLA gene complex resides—has been reported in as many as 24%, 40%, 41%, 41%, and 50% of breast, colon, lung, brain (glioblastoma), and pancreatic cancers, respectively (SI Appendix, Table S1) (94–98). As HLA-A*02:01 and HLA-A*03:01 (henceforth referred to as “A2” and “A3”) are among the most common HLA-A alleles represented in the human population (99, 100), we focused our efforts on developing reagents targeting these two alleles to maximize the patient population that could conceivably benefit from an LOH-directed therapy.
A cell-based therapy platform was chosen due to the innate ability of a cell to integrate a multitude of signals and inputs to drive a coordinated cellular response program. Accordingly, we developed chimeric receptors in T cells, targeting either A2 or A3, that could permit allelic discrimination (i.e., NASCAR T cells). A messenger RNA (mRNA) electroporation-based expression system was initially employed to allow for facile and rapid iteration of candidate constructs. An optimized protocol for primary human T cells was developed, which resulted in virtually all cells expressing the desired proteins following electroporation (SI Appendix, Figs. S1 and S2).
HLA allele-specific antibodies have previously been developed for A2 and A3, clones BB7.2 and GAP.A3, respectively (101, 102). In contrast to the BB7.2 single-chain variable fragment (scFv), we were unable to functionally graft the GAP.A3 scFv onto a second-generation CAR comprising a cluster of differentiation 28 (CD28) and CD3ζ cytoplasmic domains (Materials and Methods). We therefore turned to identifying an alternative A3-specific scFv. An scFv phage display library, with an estimated complexity of 3.6 × 1010, was screened for binders that could selectively target A3 but not other HLA-A alleles (103). Positive selection was conducted with a different A3 peptide-HLA (pHLA) monomer during each round of panning so as to enforce specificity to the HLA molecule itself and not the associated peptide, while negative selection was performed with a mixture of non-A3 pHLA monomers. Enriched candidate phage clones were amplified and assessed for their ability to bind to cells with either A2 alone (T2 cells) or A2 and A3 (T2A3 cells) via flow cytometry. Clone 13 was chosen for its strong ability to bind to T2A3 cells but not to T2 cells relative to the other phage clones tested (SI Appendix, Fig. S3B). We next sought to confirm whether this in-house selected A3-specific scFv could be functionally grafted onto a CAR molecule while maintaining specificity. As expected, clone 13- and BB7.2-engineered CAR T cells were only activated when exposed to COS-7 target cells transfected with A3 or A2, respectively, as assessed by interferon (IFN)-γ release (SI Appendix, Fig. S3C).
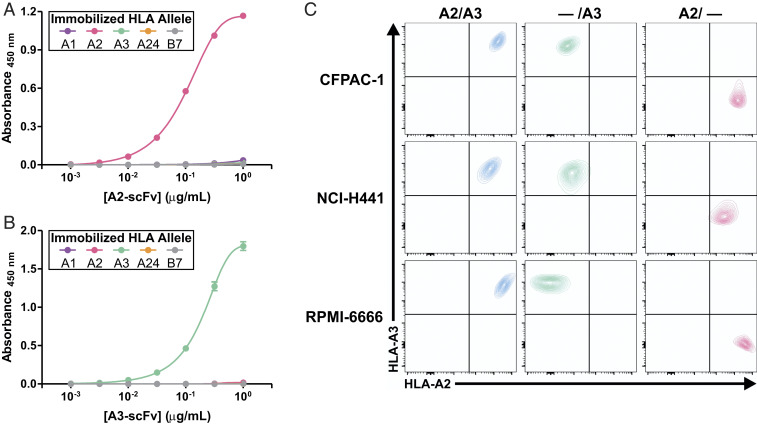
The specificities of the targeting moieties were further evaluated by titration enzyme-linked immunosorbent assay (ELISA) using recombinantly expressed scFvs. Bacterial expression vectors for the A2- and A3-specific scFvs were generated based on the BB7.2 and clone 13 sequences in the pAP-III6 backbone, respectively. The pHLA monomers from each of the four HLA-A superfamilies were tested, as was a common HLA-B allele (HLA-B*07:02). As expected, the A2 and A3 scFvs specifically bound to their cognate HLA allele but did not bind to any of the other alleles tested (Fig. 2 A and B). The pHLA monomer complexes were confirmed to be comparably folded, as verified by ELISA via detection with W6/32, a pan-HLA class I antibody (SI Appendix, Fig. S4).
Fig. 2.
Generation of HLA-A allele-targeting scFvs and isogenic cell line models. (A) A2 scFv and (B) A3 scFv binding to various immobilized HLA alleles was assessed by ELISA. Data represent means ± SD of three technical replicates. (C) Flow cytometric evaluation with α-A2 (BB7.2-PE) and α-A3 (GAP.A3-APC) antibodies of HLA KO isogenic cancer cell lines following CRISPR-mediated HLA-A locus disruption.
Generation of HLA LOH Isogenic Cell Line Models.
Next, we employed CRISPR technology to generate isogenic knockout (KO) clones from cancer cell lines expressing endogenous A2 and A3 alleles. We selected three cell lines of differing cancer types with varied HLA expression levels—CFPAC-1 (pancreatic), NCI-H441 (lung), and RPMI-6666 (Hodgkin lymphoma)—and obtained HLA single-allele KO clones for all three cell lines (Fig. 2C). The clones showed 100% identity match with their originating parental cell line as assessed by short tandem repeat (STR) profiling (SI Appendix, Table S2).
Development and Optimization of LOH Detection.
To enable LOH detection, we grafted the HLA-A allele-specific scFvs to chimeric receptors with either activating (CAR) or inhibitory (iCAR) signaling domains (86). Previous demonstrations of iCAR functionality relied on model antigen combinations with skewed expression levels. However, as we employed cell lines with endogenous levels of A2 and A3 expression to model HLA LOH allele loss, further engineering efforts to tune the system were necessary. To maximize specificity, a systematic optimization of various parameters for each component of the NASCAR targeting platform (i.e., iCAR format, CAR hinge, stoichiometry between CAR and iCAR) was performed.
A second-generation CAR was employed as the activating construct, comprising CD28 hinge, transmembrane, and cytoplasmic domains fused to the CD3ζ cytoplasmic domain (SI Appendix, Fig. S5). Four iCAR constructs incorporating CTLA-4 or PD-1 inhibitory domains with various hinge and transmembrane combinations were screened. For each inhibitory domain, two combinations were tested. Either the CTLA-4 or PD-1 cytoplasmic domain was fused to CD8α hinge and transmembrane domains, or the CTLA-4 or PD-1 cytoplasmic domain was used with each inhibitory receptor’s cognate hinge and transmembrane domains (SI Appendix, Fig. S5). Control constructs wherein the targeting scFv moiety for each iCAR construct was deleted were also created. To evaluate iCAR performance, T cells coexpressing a CAR in combination with each individual iCAR were coincubated with A2 or A2+A3-transfected COS-7 cells. Only the PD-1 iCAR constructs resulted in substantial allele-specific inhibition as evidenced by a reduction in IFN-γ secretion toward A2+A3-transfected COS-7 cells relative to A2-transfected cells (SI Appendix, Fig. S6). The iCAR construct comprising the PD-1 cytoplasmic domain with CD8α hinge and transmembrane domain exhibited the most potent allele specificity and was therefore selected as the NASCAR inhibitory module for the experiments described below.
We postulated that adjusting the CAR hinge domain from CD28 to CD8α would allow for greater iCAR-mediated quenching of proximal CAR signaling by permitting CD8α hinge heterodimerization between CAR and iCAR (SI Appendix, Fig. S5). Indeed, iCAR-mediated inhibition of CAR activity was strengthened with a CD8α-hinged CAR when engaged with CFPAC-1 target cells expressing both A2 and A3, as assessed by IFN-γ release (SI Appendix, Fig. S7). As the CD8α-hinged CAR conferred an increased window of allele specificity, it was selected as the NASCAR activating module for the experiments described below.
We next considered what additional parameters could be modulated to impart greater specificity and divert immune responses away from normal, HLA-heterozygous cells. We observed that the mRNA-based expression system for introducing chimeric receptors into primary human T cells provided a linear correlation between the amount of mRNA electroporated and the corresponding CAR expression level (SI Appendix, Fig. S8). Thus, we explored whether adjusting the stoichiometry between CAR and iCAR could allow for greater specificity to the system. While a 1:1 ratio of CAR:iCAR mRNA displayed considerable allele specificity, there was still significant off-target activation as evidenced by detectable IFN-γ secretion with CFPAC-1 A2/A3 target cells. However, decreasing the CAR:iCAR ratio to 1:3 resulted in pronounced suppression of T cell activation toward off-target A2/A3 cells (SI Appendix, Fig. S9). The 1:3 ratio of CAR:iCAR was therefore selected for the NASCAR experiments described below.
Determination of NASCAR Specificity In Vitro.
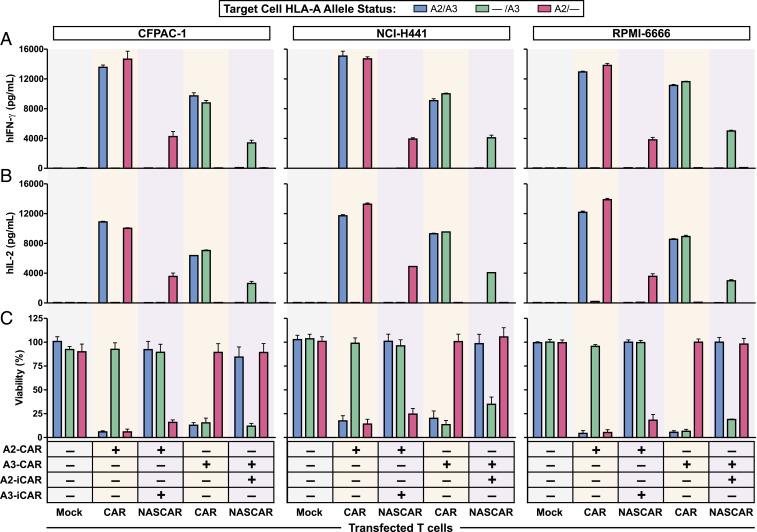
To thoroughly demonstrate the modularity, specificity, and symmetry of the NASCAR approach, both combinations of CAR and iCAR were tested (i.e., A2-CAR+A3-iCAR, A3-CAR+A2-iCAR) against all three sets of isogenic cell lines. T cell activation, as assessed by IFN-γ and interleukin-2 (IL-2) cytokine release, was remarkably similar across all three cell line backgrounds and revealed the expected allele-specific targeting profiles (Fig. 3 A and B). Consistently, the degree of cytotoxicity mirrored that of cytokine release (Fig. 3C).
Fig. 3.
Determination of NASCAR specificity in vitro. Three isogenic cancer cell line models of LOH were employed to determine NASCAR specificity. Cancer cells with the indicated HLA-A allele status were coincubated with CAR or NASCAR T cells configured with the indicated allele-targeting CAR and iCAR receptor combinations at an E:T ratio of 2:1. T cell activation was assessed by ELISA for (A) human IFN-γ, hIFN-γ and (B) human IL-2, hIL-2 release. Data represent means ± SD of three technical replicates. (C) Cytotoxicity mediated by CAR or NASCAR T cells was measured by CellTiter-Glo (CFPAC-1, NCI-H441) or Steady-Glo (RPMI-6666). Data represent means ± SD of three technical replicates.
The assays described above employed primary human T cells from A2/A3-negative donors as effector cells. Otherwise, in the control conditions with the CAR but without the iCAR, the effector T cells would commit fratricide due to self-expression of the target antigen. In practice, however, effector T cells would likely be derived from autologous sources that express both A2 and A3, and it was therefore important to confirm that fratricide would not occur. To that end, we assayed for autoreactivity in model “autologous” antigen-positive (A2/A3) versus model “allogeneic” antigen-negative (A1/A24) effector donor T cells. While A1/A24 donor T cells elicited no IFN-γ signal as they lacked the activating antigen, introduction of the CAR alone into A2/A3 donor T cells resulted in the expected autoreactivity and fratricide. By contrast, NASCAR expression of the inhibitory module together with the activating module in A2/A3 donor T cells did not result in fratricide (SI Appendix, Fig. S10A). Furthermore, NASCAR-engineered A2/A3 donor T cells were as effective in targeting cancer cells with LOH as A1/A24 donor T cells, as assessed by IFN-γ release (SI Appendix, Fig. S10B).
Evaluation of NASCAR Antitumor Activity In Vivo.
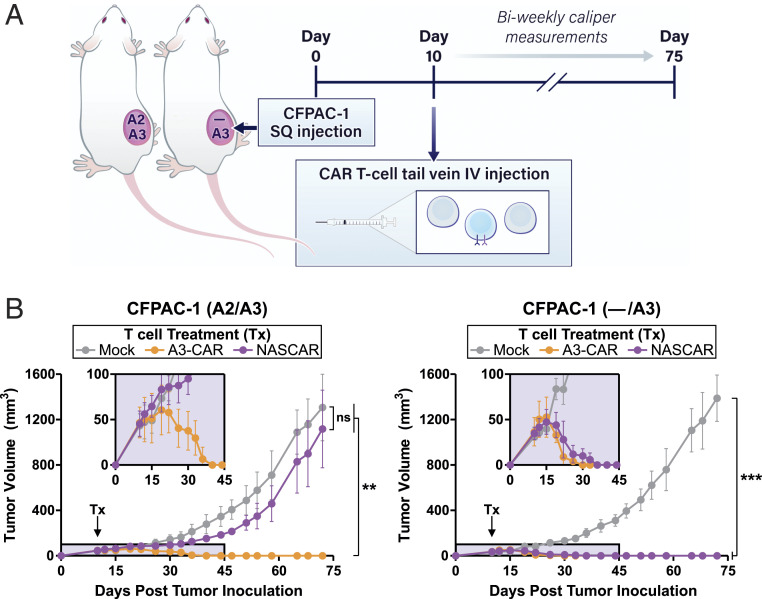
To determine whether the NASCAR approach could be extended to an in vivo setting, we converted our constructs from the transient mRNA-based expression system to a stable CRISPR-based knock-in expression system. A knock-in strategy was chosen due to its eported ability to produce tight expression distribution patterns for introduced CAR constructs in primary human T cells, a property we surmised was important as a result of experiences from our in vitro mRNA optimization efforts (104). Homology-directed repair (HDR) templates comprising an A3-CAR, or a bicistronic NASCAR construct containing an A3-CAR and A2-iCAR with an intervening 2A self-cleaving peptide sequence, were generated. The iCAR was placed 5′ to the CAR to skew the ratio in favor of the antecedent receptor (105). The HDR template (HDRT) was targeted to the B2M locus to allow for high levels of expression, and a guide RNA targeting the TCR α constant (TRAC) locus was simultaneously included in the electroporation mixture to inactivate TRAC and consequently reduce alloreactivity toward the human cancer cells used to establish tumors in the mice. In vitro characterization of this stable NASCAR expression system revealed the near-complete ablation of TCR expression, an ∼30% editing efficiency of the introduced transgenes, and the expected allelic recognition pattern when coincubated with CFPAC-1 HLA KO isogenic target cell lines (SI Appendix, Fig. S11 A–C). In vivo characterization employed a subcutaneous xenograft model of NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice with CFPAC-1 A2/A3 or — /A3 tumors. For treatment, CRISPR-engineered CAR or NASCAR T cells were administered via tail vein 10 d following tumor inoculation once tumors were established and palpable (Fig. 4A). While treatments with CAR T cells resulted in regression of both tumors, NASCAR T cells eliminated the — /A3 tumor and spared the A2/A3 heterozygous tumor representing normal tissues (Fig. 4B). The treatments were well tolerated as evidenced by the absence of significant deviations from normal body weight gains (SI Appendix, Fig. S12).
Fig. 4.
Evaluation of NASCAR antitumor activity in vivo. (A) A single-flank, subcutaneous (SQ) xenograft model of NSG mice was employed, and CRISPR-engineered A3-CAR T cells or NASCAR T cells targeting A2 loss were introduced via tail vein intravenous (IV) injection 10 d following tumor inoculation. Tumors were measured biweekly for 75 d following tumor inoculation. (B) Tumor growth curves were serially monitored by external caliper measurements. (Insets) Magnified window of the first 45 d of treatment; n = 6 mice per group. Data represent means ± SD; ** and *** denote P ≤ 0.01 and P ≤ 0.001, respectively, as determined by one-way ANOVA with Tukey’s multiple comparison test; ns, not significant.
Discussion
The sparsity of tumor-specific antigens presents a major obstacle for the wider implementation of powerful immunotherapeutic agents in the clinic. Clonal genetic alterations in cancer confer unparalleled specificity given their presence in tumor but not normal cells. Here, we describe an immunotherapeutic approach to exploit and specifically target LOH, one of the most ubiquitous somatic alterations in human cancers.
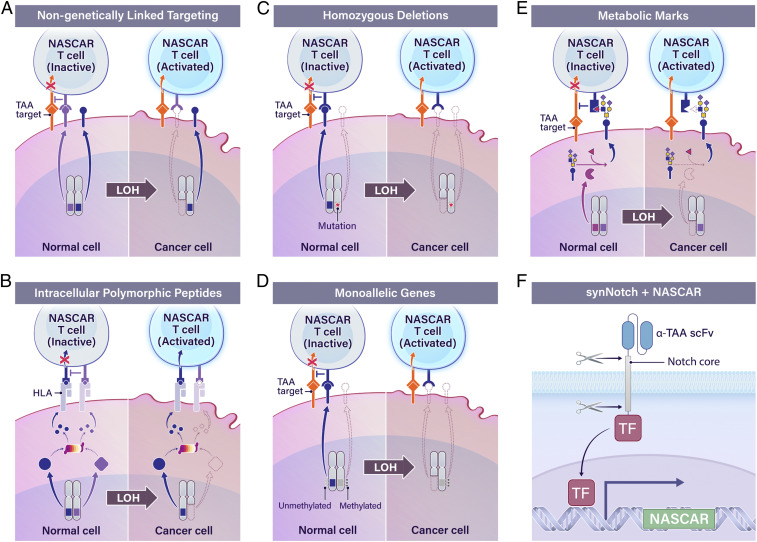
There are two broad approaches where we envision the NASCAR LOH-targeting system being applied. The first, as demonstrated here, is where both the activating and inhibitory modules target polymorphic forms of the same molecule (Fig. 1). This approach, while potentially more challenging, has the highest probability of maximizing specificity by ensuring that the activating and inhibitory molecules are always coexpressed, thereby circumventing issues related to “on-target, off-tumor” toxicity. The second approach is to target the activating and inhibitory modules to different, non-genetically linked molecules (Fig. 5A). Here, the activating antigen need not be genetically tied to the lost allele; for example, the CAR antigen could instead be a TAA while the iCAR could still be directed to a polymorphic allele that is lost in the cancer cells but present on all normal cells. For both approaches (Figs. 1 and 5A), the polymorphic molecules sensed by NASCAR do not have to be restricted to those residing on the cell surface. Either approach could be extended to target intracellular polymorphic peptides that are presented on the cell surface in the context of HLA molecules (Fig. 5B). For example, antibodies and TCRs that selectively react to HLA-presented peptides differing by only a single amino acid residue have been described (103, 106–109). Furthermore, NASCAR inhibitory antigens could also be expanded beyond the direct targeting of polymorphic forms lost through LOH to include other antigen loss mechanisms exposed by LOH (110–112). For example, the NASCAR approach could be adapted to target the complete loss of a marker revealed by LOH due to a genetic deletion in the retained allele (e.g., homozygous deletions; Fig. 5C), loss of normal monoallelic expression (e.g., epigenetic imprinting or allelic expression; Fig. 5D), or loss of metabolic marks (Fig. 5E).
Fig. 5.
Next-generation LOH targeting. (A) NASCAR targeting as applied to genetically unlinked molecules. (B) NASCAR targeting as applied to intracellular polymorphic peptides that are presented on the cell surface in the context of HLA molecules. (C–E) A model of tumor-specific inhibitory markers revealed by LOH that are amenable to NASCAR-based targeting. (F) TAA-specific synNotch receptor-driven conditional expression of a NASCAR expression cassette. TF, transcription factor.
Our study also carries implications beyond targeting tumors with LOH as a primary therapy. Recently, LOH has been implicated as both a resistance mechanism and a negative clinical correlate for immunotherapies (113, 114). For example, HLA LOH has been clinically correlated with poor prognosis in several cancer types, is associated with reduced patient survival following immune checkpoint blockade, and provides a pathway for resistance to tumor-infiltrating lymphocyte therapies targeting KRASG12D and TP53R175H driver mutation neoantigens (97, 115–121). In such instances, HLA allelic loss presumably reflects genetic loss of the HLA restriction element responsible for presenting the dominant cancer-targeting antigens required to mount an effective antitumor immune response. Thus, the NASCAR approach described herein could be applied to refractory tumors that develop in the context of an LOH event encompassing the targeted antigen.
Several limitations of our study should be acknowledged. First, there are inherent risks associated with any immunotherapeutic approach. In the example presented here, these challenges are compounded by the near-ubiquitous expression of HLA, thus requiring exquisitely specific and potent performance of the allele-sensing component of NASCAR. Second, clinical administration of NASCAR T cells targeting antigens expressed on all normal cells could conceivably result in tonic signaling of both receptors, as well as possible reduction in T cell trafficking due to interactions with the endothelium and other tissues. One solution to address both concerns would be to conditionally express the NASCAR signaling molecules (Fig. 5F). An elegant way to implement such regulation would be to incorporate LOH targeting into a synthetic Notch (synNotch) system, for example, by conditionally driving a NASCAR expression cassette with a constitutively expressed TAA-specific synNotch receptor (80). This would theoretically allow for spatially and temporally restricted NASCAR expression in T cells at the local tumor environment and also introduce an additional layer of AND-gated specificity with the TAA-activated synNotch receptor. In addition, future iterations of the NASCAR approach could incorporate suicide or elimination gene systems to impart greater safety to the system, particularly when targeting ubiquitously expressed antigens (61, 73, 74).
In summary, we have shown that it is possible to generate pairwise chimeric receptors that are capable of discriminating and targeting cancers cells harboring LOH events. An independent study targeting CD19 with exogenously expressed A2 as the inhibitory antigen (non-genetically linked targeting; Fig. 5A) was published during final preparation of this manuscript and reached similar conclusions (122). These studies build on the clinical success of CAR T cells by targeting a unique class of tumor-specific antigens and lay the conceptual and practical foundation for harnessing the immune system to react against one of the most frequent types of genetic alterations in human cancers. While subsequent studies will clarify whether NASCAR can successfully treat cancer patients, the present work described herein sets the stage for such future exploration.
Materials and Methods
pHLA Monomer Complexes.
HLA molecules were recombinantly expressed and refolded with peptide and β2-microglobulin, followed by size-exclusion purification and biotinylation [Fred Hutchinson Cancer Research Center (FHCRC) Immune Monitoring Lab; Baylor MHC Tetramer Core]. The pHLA monomer complexes were folded as confirmed by ELISA via detection with α-HLA class I (W6/32) antibody (BioLegend) prior to use (123). The pHLA tetramers used for flow cytometry were generated by combining folded pHLA monomer complexes with fluorescently labeled streptavidin (FHCRC Immune Monitoring Lab).
Conversion of Commercially Available HLA Allele-Specific Antibodies to CAR Format.
HLA allele-specific antibodies have previously been developed [e.g., clone BB7.2 for A2 (101); clone GAP.A3 for A3 (102)]. Hybridomas for both clones were commercially acquired (American Type Culture Collection [ATCC]), and hybridoma sequences were obtained by next-generation sequencing (GenScript). The variable chain regions were converted to scFvs, which were then grafted onto a second-generation CAR comprising a cluster of differentiation 28 (CD28) and CD3ζ cytoplasmic domains. Allele-selective CAR T cell activation was assessed by IFN-γ release after coincubation with target cells with either A2 alone (T2 cells) or A2 and A3 (T2A3 cells) on their cell surfaces. As expected, the BB7.2 CAR T cells were activated by T2 as well as T2A3 cells, both expressing A2. However, the GAP.A3 CAR failed to recognize T2A3 cells (SI Appendix, Fig. S3A). We confirmed, by flow cytometry, that the GAP.A3 CAR was indeed expressed, suspected that the lack of activity was due to grafting failure, and thus turned to identifying an alternative scFv specific for HLA-A3 with phage display technology.
Phage Display.
Experimental procedures to isolate phage clones reactive to HLA-A3 followed methods that were previously described (103, 106), with the following exceptions. For positive selection, a different HLA-A3 pHLA monomer complex was employed during each round of panning, to enrich for allele-specific phage; conversely, for negative selection, a mixture of non−HLA-A3 pHLA monomer complexes was used to deplete non−allele-specific phage. For each round of panning, the composition of positive and negative selection mixtures was modified such that each round of panning exposed the phage to a new set of pHLA monomer complexes.
scFv Expression and Purification.
The scFv sequences were cloned into the pAP-III6 vector (124) containing C-terminal FLAG- and HIS-tags, recombinantly expressed in Escherichia coli, and purified via nickel chromatography (AxioMx, Abcam).
ELISA.
For scFv pHLA monomer-based ELISAs, biotinylated monomers were first coated onto EvenCoat Streptavidin Coated Plates (R&D Systems) and allowed to bind overnight. Recombinant scFvs were diluted to the indicated concentrations, allowed to bind to the monomer-coated plate, and detected by α-FLAG-HRP (M2) antibody (Abcam). ELISAs were developed by addition of TMB Substrate (BioLegend) and stopped with an equal volume of 1N Sulfuric Acid Solution (Thermo Fisher Scientific).
Flow Cytometry.
Flow cytometry was performed either on an LSRII cytometer (BD Biosciences) or an IntelliCyt iQue Screener PLUS (Sartorius). α-HLA-A2 (BB7.2), α-CD3 (SK7), and α-Rabbit IgG were obtained from BioLegend. α-HLA-A3 (GAP.A3) and biotinylated Protein L were obtained from Thermo Fisher Scientific. α-fd/M13 bacteriophage was obtained from Novus Biologicals, and α-Biotin (Bio3-18E7) was obtained from Miltenyi Biotec (Bergisch Gladbach). Flow cytometry analysis was performed either with FACSDiva or FlowJo software (BD).
mRNA Generation.
Human codon-optimized constructs were synthesized (GeneArt, Thermo Fisher Scientific) and cloned into the mammalian expression vector pCI (Promega). Cap 1 mRNA was synthesized with the T7 mScript Standard mRNA Production System Kit (CELLSCRIPT) and purified by the MEGAclear Transcription Clean-Up Kit (Thermo Fisher Scientific) per manufacturers’ instructions. The mRNA purity, integrity, and transcript size were confirmed by RNA TapeStation analysis (Agilent) prior to electroporation.
T Cell Activation, Culture, and Electroporation.
Engineered T cells via mRNA electroporation were generated as follows. Peripheral blood mononuclear cells (PBMCs) were obtained by Ficoll-Paque PLUS (GE Healthcare) gradient centrifugation of whole blood from healthy volunteer donors. CD3 cells were isolated from PBMCs by negative selection (STEMCELL Technologies; Astarte Biologics). CD3 purity following selection was confirmed to be >97% by flow cytometry. T cells were activated and expanded with Dynabeads Human T-Activator CD3/CD28 (Thermo Fisher Scientific). Unless otherwise noted, primary human CD3 T cells were maintained in RPMI-1640 (ATCC) supplemented with 10% fetal bovine serum (FBS) (HyClone Defined, GE Healthcare), 1% Penicillin-Streptomycin (Thermo Fisher Scientific), and 100 IU/mL recombinant human IL-2 (Proleukin, Prometheus Laboratories). Electroporation into primary human CD3 T cells was performed in a 0.2-cm cuvette (Bio-Rad) with the BTX ECM 2001 Electro Cell Manipulator (Harvard Apparatus) at least 10 d post bead activation. In brief, an electroporation volume totaling 125 μL containing the indicated amount(s) of mRNA at a concentration of 2 × 107 cells per mL, diluted in Opti-MEM (Thermo Fisher Scientific), was electroporated with a square wave pulse of 200 V and 16 ms.
Engineered T cells via CRISPR HDR electroporation were generated as follows. Stably integrated CARs and iCARs were generated via electroporation of DNA HDRTs targeted to the B2M locus and Cas9 ribonucleoproteins (RNPs) targeting the B2M and TRAC genes (125). HDRTs were prepared by PCR amplification from a plasmid template using the Q5 Hot Start High-Fidelity 2X Master Mix (New England BioLabs) with primers containing truncated Cas9 target sequences (IDT) (125). Amplicons were purified with 1× AMPure beads (Beckman Coulter Life Sciences), eluted in water, and quantified with a NanoDrop Spectrophotometer (Thermo Fisher Scientific). The purified PCR products were also electrophoresed (1% agarose, 1× TAE) to assess amplicon size and purity. Prior to electroporation, purified CD3 T cells were activated with Dynabeads Human T-Activator CD3/CD28 (Thermo Fisher Scientific) at a 1:1 bead-to-cell ratio in the presence of 100 IU/mL recombinant human IL-2 (Proleukin, Prometheus Laboratories) and 5 ng/mL recombinant human IL-7 (BioLegend). After 48 h to 56 h, the CD3/CD28 beads were removed with a magnet. To prepare electroporation master mixes, individual RNPs targeting B2M (CGTGAGTAAACCTGAATCTT) and TRAC (AGAGTCTCTCAGCTGGTACA) were first assembled individually by mixing each single-guide RNA (IDT) with Cas9 nuclease (IDT) and single-stranded DNA Electroporation Enhancer (IDT) at a molar ratio of 2:1:1.3 and allowed to incubate at room temperature for 15 min. Then, equal volumes of B2M and TRAC RNPs (totaling 50 pmol of Cas9 nuclease) were mixed with 0.5 μg of the specified HDRT and incubated for at least 5 min at room temperature. Finally, 20 μL of activated T cells resuspended at 5 × 107 cells per mL in P3 buffer (Lonza) were added to the electroporation mixture. Electroporations were performed with a 4D-Nucleofector X Unit (Lonza) in 16-well cuvettes using pulse code EH115. After electroporation, T cells were recovered by immediately adding 80 μL of warm, cytokine-free T cell growth media to the cuvettes and incubating at 37 °C for 20 min. Then, T cells were diluted in T cell growth media containing 100 IU/mL recombinant human IL-2 (Proleukin, Prometheus Laboratories) and 5 ng/mL recombinant human IL-7 (BioLegend) and plated at 1 × 106 cells per mL.
Cell Lines.
All cell lines (CFPAC-1, NCI-H441, RPMI-6666, COS-7, T2), except for T2A3 cells (a kind gift from Eric Lutz and Elizabeth Jaffee, Johns Hopkins University, Baltimore, MD) that were procured from ATCC. CFPAC-1 and NCI-H441 cell lines were maintained in McCoy’s 5A (Modified) media (Thermo Fisher Scientific) supplemented with 10% FBS (HyClone Defined, GE Healthcare) and 1% Penicillin-Streptomycin (Thermo Fisher Scientific). The COS-7 cell line was maintained in Dulbecco’s modified Eagle’s medium (high glucose, pyruvate) media (Thermo Fisher Scientific) supplemented with 10% FBS (HyClone Defined, GE Healthcare) and 1% Penicillin-Streptomycin (Thermo Fisher Scientific). T2 and T2A3 cells were maintained in RPMI-1640 (ATCC) supplemented with 10% FBS (HyClone Defined, GE Healthcare) and 1% Penicillin-Streptomycin (Thermo Fisher Scientific). The RPMI-6666 cell line was maintained in RPMI-1640 (ATCC) supplemented with 20% FBS (HyClone Defined, GE Healthcare) and 1% Penicillin-Streptomycin (Thermo Fisher Scientific). All cells, including primary human T cells, were maintained at 37 °C under 5% CO2.
Target Cell Line Generation.
For exogenous target cell line generation, COS-7 cells were seeded in a 96-well plate format and transfected with various combinations of pcDNA3.1 (Thermo Fisher Scientific) plasmids encoding HLA-A2, HLA-A3, HLA-B7, and vector only with Lipofectamine 3000 (Thermo Fisher Scientific). DNA−lipid complexes were incubated with the cells for 24 h, then washed away prior to use as target cells for T cell coculture assays.
For endogenous target cell line generation, CRISPR KOs of the endogenous HLA-A alleles were performed using the Alt-R CRISPR system (IDT). An HLA-A Alt-R Cas9 CRISPR RNA (crRNA) GCTGCGACGTGGGGTCGGAC (IDT) and the Alt-R CRISPR-Cas9 trans-activating crRNA (tracrRNA) (IDT) were each resuspended at 100 μM with Nuclease-Free Duplex Buffer (IDT). The tracrRNA and crRNA were mixed at a 1:1 molar ratio, heated to 95 °C for 5 min, and then allowed to cool to room temperature. Then, 50 pmols of duplexed tracrRNA/crRNA were mixed with 40 pmols of Cas9 Nuclease (IDT) and allowed to complex at room temperature for 15 min. The Cas9 RNP solution was mixed with 2 × 105 NCI-H441 cells, 2 × 105 CFPAC-1 cells, or 1 × 106 RPMI-6666 cells in 20 μL of Opti-MEM (Thermo Fisher Scientific) in a 0.1-cm cuvette (Bio-Rad). The cuvettes were electroporated at 150 V for 10 ms (NCI-H441 and CFPAC-1) or 100 V for 10 ms (RPMI-6666) using the BTX ECM 2001 Electro Cell Manipulator (Harvard Apparatus). Cells were recovered in prewarmed complete growth media. NCI-H441 and CFPAC-1 polyclonal pools were single-cell plated at a density of 0.5 to 2 cells per well and grown for 3 wk. Individual clones were screened for expression of HLA-A2 and HLA-A3 alleles by staining with allele-specific antibodies described above. Single cells of the RPMI-6666 polyclonal pool were sorted with a FACSAria Fusion (BD Biosciences) into 96-well plates for clonal expansion after staining with HLA-A2 and HLA-A3 allele-specific antibodies.
To generate luciferase-expressing RPMI-6666 HLA isogenic cell lines, cells were transduced with RediFect Red-Fluc-GFP lentiviral particles (PerkinElmer). Transduced cells were sorted for green fluorescent protein expression with a FACSAria Fusion (BD Biosciences) to obtain a purely transduced population.
All engineered endogenous target cell lines were authenticated and confirmed to be of parental origin (100% exact match of eight core STR loci) by ATCC Cell Line Authentication Service.
In Vitro T Cell Assays.
Engineered T cells, both mRNA modified and CRISPR modified, were allowed to expand until at least 10 d post bead activation prior to use in in vitro T cell assays. The mRNA-modified T cells were used for T cell assays after being allowed to recover overnight in cytokine-free media following electroporation; similarly, CRISPR-modified T cells were incubated in cytokine-free media the day prior to use for T cell assays. Engineered T cells were combined with target cell lines at the indicated effector-to-target (E:T) ratios in flat-bottom 96-well plates and allowed to incubate for 4 h or overnight. Cytokine levels from clarified cell culture supernatant were quantified by Quantikine ELISA kits (R&D Systems) per manufacturer’s instructions. Target cell line viability was measured either by CellTiter-Glo Luminescent Cell Viability Assay (for CFPAC-1 and NCI-H441 cells) following six washes to the plate, or by Steady-Glo Luciferase Assay System (for luciferase-expressing RPMI-6666 cells), per manufacturer’s instructions (Promega).
Murine Xenograft Model.
The 6- to 8-wk-old female NSG mice were obtained from The Jackson Laboratory; 5 × 105 CFPAC-1 cells with the indicated HLA-A allele status were inoculated subcutaneously into mice, and 5 × 106 CRISPR-modified T cells were infused intravenously via lateral tail vein 10 d following tumor inoculation. Gross tumor volume measurements were obtained biweekly with an external caliper using the modified ellipsoidal formula of V = 0.5(A × B2), where A represents the greatest longitudinal diameter and B represents the greatest transverse diameter. Mice were cared for in accordance with a Johns Hopkins University Animal Care and Use Committee approved research protocol.
Statistical Analysis.
Where applicable, data are presented as means ± SD. Statistical analyses were carried out using specific tests indicated in the figure legends. A P value of < 0.05 was used to denote statistical significance. All analyses were performed using Prism Version 5.01 (GraphPad).
Supplementary Material
Acknowledgments
We thank Evangeline Watson for expert technical assistance with animal experiments. We thank Richard L. Blosser, Ada J. Tam, Maria Popoli, Joshua D. Cohen, Surojit Sur, Nicolas Wyhs, Ashley Cook Morgan, Abby Sukman, and José Rodríguez Molina for insightful discussions and assistance with this study. This work was supported by The Virginia and D. K. Ludwig Fund for Cancer Research, Lustgarten Foundation for Pancreatic Cancer Research, The Commonwealth Fund, The Burroughs Wellcome Career Award For Medical Scientists, The Bloomberg∼Kimmel Institute for Cancer Immunotherapy, NIH Cancer Center Support Grant P30 CA006973, and National Cancer Institute Grant R37 CA230400. B.J.M., J.D., A.H.P., and S.R.D. were supported by NIH T32 Grant GM73009. S.P. was supported by NIH T32 Grant 5T32CA009071-38, and the Society for Immunotherapy of Cancer−Amgen Cancer Immunotherapy in Hematologic Malignancies Fellowship. M.F.K. was supported by NIH T32 Grant AR048522.
Footnotes
Competing interest statement: B.V., K.W.K., and N.P. are founders of Thrive Earlier Detection. K.W.K. and N.P. are consultants to and were on the Board of Directors of Thrive Earlier Detection. B.V., K.W.K., N.P., and S.Z. own equity in Exact Sciences. B.V., K.W.K., N.P., S.Z., and D.M.P. are founders of, hold or may hold equity in, and serve or may serve as consultants to ManaT Bio. B.V., K.W.K., N.P., and S.Z. are founders of, hold equity in, and serve as consultants to Personal Genome Diagnostics. S.Z. has a research agreement with BioMed Valley Discoveries. K.W.K. and B.V. are consultants to Sysmex, Eisai, and CAGE Pharma and hold equity in CAGE Pharma. B.V. is also a consultant to Catalio. K.W.K., B.V., S.Z., and N.P. are consultants to and hold equity in NeoPhore. N.P. is an advisor to and holds equity in CAGE Pharma. C.B. is a consultant to Depuy-Synthes and Bionaut Pharmaceuticals. S.B.G. is a founder and holds equity in AMS. The terms of all these arrangements are being managed by Johns Hopkins University in accordance with its conflict of interest policies. M.F.K. received personal fees from Bristol Myers Squibb and Celltrion. D.M.P. reports grant and patent royalties through institution from Bristol Myers Squibb, a grant from Compugen, stock from Trieza Therapeutics and Dracen Pharmaceuticals, and founder equity from Potenza; being a consultant for Aduro Biotech, Amgen, AstraZeneca (MedImmune/Amplimmune), Bayer, DNAtrix, Dynavax Technologies Corporation, Ervaxx, FLX Bio, Rock Springs Capital, Janssen, Merck, Tizona, and Immunomic Therapeutics; being on the scientific advisory board of Five Prime Therapeutics, Camden Nexus II, and WindMIL; and being on the board of directors for Dracen Pharmaceuticals.
See QnAs, e2102936118, in vol. 118, issue 12.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022410118/-/DCSupplemental.
Data Availability
All study data are included in the article and SI Appendix.
References
- 1.Capdeville R., Buchdunger E., Zimmermann J., Matter A., Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 1, 493–502 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Nahta R., Esteva F. J., Trastuzumab: Triumphs and tribulations. Oncogene 26, 3637–3643 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Weisberg E., Manley P. W., Cowan-Jacob S. W., Hochhaus A., Griffin J. D., Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat. Rev. Cancer 7, 345–356 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Camidge D. R., Pao W., Sequist L. V., Acquired resistance to TKIs in solid tumours: Learning from lung cancer. Nat. Rev. Clin. Oncol. 11, 473–481 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Oh D.-Y., Bang Y.-J., HER2-targeted therapies—A role beyond breast cancer. Nat. Rev. Clin. Oncol. 17, 33–48 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Vogelstein B., et al., Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostrem J. M. L., Shokat K. M., Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Ryan M. B., Corcoran R. B., Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 15, 709–720 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Moore A. R., Rosenberg S. C., McCormick F., Malek S., RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 19, 533–552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beroukhim R., et al., The landscape of somatic copy-number alteration across human cancers. Nature 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garraway L. A., Lander E. S., Lessons from the cancer genome. Cell 153, 17–37 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Zack T. I., et al., Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knouse K. A., Davoli T., Elledge S. J., Amon A., Aneuploidy in cancer: Seq-ing answers to old questions. Annu. Rev. Cancer Biol. 1, 335–354 (2017). [Google Scholar]
- 15.Nowell P. C., The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). [DOI] [PubMed] [Google Scholar]
- 16.Mitelman F., Klinger H. P., Catalogue of Chromosome Aberrations in Cancer (Karger Medical and Scientific Publishers, 1983). [Google Scholar]
- 17.Mitelman F., Mertens F., Johansson B., A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat. Genet. 15, 417–474 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Boveri T., Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 121, 1–84 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Vogelstein B., et al., Allelotype of colorectal carcinomas. Science 244, 207–211 (1989). [DOI] [PubMed] [Google Scholar]
- 20.Seymour A. B., et al., Allelotype of pancreatic adenocarcinoma. Cancer Res. 54, 2761–2764 (1994). [PubMed] [Google Scholar]
- 21.Radford D. M., et al., Allelotyping of ductal carcinoma in situ of the breast: Deletion of loci on 8p, 13q, 16q, 17p and 17q. Cancer Res. 55, 3399–3405 (1995). [PubMed] [Google Scholar]
- 22.Boige V., et al., Concerted nonsyntenic allelic losses in hyperploid hepatocellular carcinoma as determined by a high-resolution allelotype. Cancer Res. 57, 1986–1990 (1997). [PubMed] [Google Scholar]
- 23.Lengauer C., Kinzler K. W., Vogelstein B., Genetic instabilities in human cancers. Nature 396, 643–649 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Haber D. A., et al., An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms’ tumor. Cell 61, 1257–1269 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Stoler D. L., et al., The onset and extent of genomic instability in sporadic colorectal tumor progression. Proc. Natl. Acad. Sci. U.S.A. 96, 15121–15126 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shih I.-M., et al., Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res. 61, 818–822 (2001). [PubMed] [Google Scholar]
- 27.Thiagalingam S., et al., Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl. Acad. Sci. U.S.A. 98, 2698–2702 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo L., Li B., Pretlow T. P., DNA alterations in human aberrant crypt foci and colon cancers by random primed polymerase chain reaction. Cancer Res. 63, 6166–6169 (2003). [PubMed] [Google Scholar]
- 29.Donahue S. L., Lin Q., Cao S., Ruley H. E., Carcinogens induce genome-wide loss of heterozygosity in normal stem cells without persistent chromosomal instability. Proc. Natl. Acad. Sci. U.S.A. 103, 11642–11646 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solimini N. L., et al., Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science 337, 104–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue W., et al., A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl. Acad. Sci. U.S.A. 109, 8212–8217 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douville C., et al., Detection of aneuploidy in patients with cancer through amplification of long interspersed nucleotide elements (LINEs). Proc. Natl. Acad. Sci. U.S.A. 115, 1871–1876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douville C., et al., Assessing aneuploidy with repetitive element sequencing. Proc. Natl. Acad. Sci. U.S.A. 117, 4858–4863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basilion J. P., et al., Selective killing of cancer cells based on loss of heterozygosity and normal variation in the human genome: A new paradigm for anticancer drug therapy. Mol. Pharmacol. 56, 359–369 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Fluiter K., et al., Tumor genotype-specific growth inhibition in vivo by antisense oligonucleotides against a polymorphic site of the large subunit of human RNA polymerase II. Cancer Res. 62, 2024–2028 (2002). [PubMed] [Google Scholar]
- 36.Fluiter K., et al., In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 31, 953–962 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fluiter K., Housman D., Ten Asbroek A. L. M. A., Baas F., Killing cancer by targeting genes that cancer cells have lost: Allele-specific inhibition, a novel approach to the treatment of genetic disorders. Cell. Mol. Life Sci. 60, 834–843 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mook O. R. F., Baas F., de Wissel M. B., Fluiter K., Allele-specific cancer cell killing in vitro and in vivo targeting a single-nucleotide polymorphism in POLR2A. Cancer Gene Ther. 16, 532–538 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Nichols C. A., et al., Loss of heterozygosity of essential genes represents a widespread class of potential cancer vulnerabilities. Nat. Commun. 11, 2517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rendo V., et al., Exploiting loss of heterozygosity for allele-selective colorectal cancer chemotherapy. Nat. Commun. 11, 1308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellman I., Coukos G., Dranoff G., Cancer immunotherapy comes of age. Nature 480, 480–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pardoll D. M., The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D. S., Mellman I., Oncology meets immunology: The cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Schumacher T. N., Schreiber R. D., Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Topalian S. L., Drake C. G., Pardoll D. M., Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khalil D. N., Smith E. L., Brentjens R. J., Wolchok J. D., The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 13, 273–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gotwals P., et al., Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat. Rev. Cancer 17, 286–301 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Yarchoan M., Johnson B. A. III, Lutz E. R., Laheru D. A., Jaffee E. M., Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 17, 209–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran E., Robbins P. F., Rosenberg S. A., ‘Final common pathway’ of human cancer immunotherapy: Targeting random somatic mutations. Nat. Immunol. 18, 255–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ribas A., Wolchok J. D., Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schumacher T. N., Scheper W., Kvistborg P., Cancer neoantigens. Annu. Rev. Immunol. 37, 173–200 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Wei S. C., Duffy C. R., Allison J. P., Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 8, 1069–1086 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Wolchok J., Putting the immunologic brakes on cancer. Cell 175, 1452–1454 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Zappasodi R., Merghoub T., Wolchok J. D., Emerging concepts for immune checkpoint blockade-based combination therapies. Cancer Cell 33, 581–598 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haen S. P., Löffler M. W., Rammensee H.-G., Brossart P., Towards new horizons: Characterization, classification and implications of the tumour antigenic repertoire. Nat. Rev. Clin. Oncol. 17, 595–610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waldman A. D., Fritz J. M., Lenardo M. J., A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goebeler M.-E., Bargou R. C., T cell-engaging therapies—BiTEs and beyond. Nat. Rev. Clin. Oncol. 17, 418–434 (2020). [DOI] [PubMed] [Google Scholar]
- 58.MacKay M., et al., The therapeutic landscape for cells engineered with chimeric antigen receptors. Nat. Biotechnol. 38, 233–244 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Yu J. X., Upadhaya S., Tatake R., Barkalow F., Hubbard-Lucey V. M., Cancer cell therapies: The clinical trial landscape. Nat. Rev. Drug Discov. 19, 583–584 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Batlevi C. L., Matsuki E., Brentjens R. J., Younes A., Novel immunotherapies in lymphoid malignancies. Nat. Rev. Clin. Oncol. 13, 25–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neelapu S. S., et al., Chimeric antigen receptor T-cell therapy—Assessment and management of toxicities. Nat. Rev. Clin. Oncol. 15, 47–62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teachey D. T., Hunger S. P., Acute lymphoblastic leukaemia in 2017: Immunotherapy for ALL takes the world by storm. Nat. Rev. Clin. Oncol. 15, 69–70 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Parker K. R., et al., Single-cell analyses identify brain mural cells expressing CD19 as potential off-tumor targets for CAR-T immunotherapies. Cell 183, 126–142.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neelapu S. S., et al., Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sadelain M., Rivière I., Riddell S., Therapeutic T cell engineering. Nature 545, 423–431 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.June C. H., Sadelain M., Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maude S. L., et al., Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schuster S. J.et al.; JULIET Investigators , Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 380, 45–56 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Mikkilineni L., Kochenderfer J. N., CAR T cell therapies for patients with multiple myeloma. Nat. Rev. Clin. Oncol., 1–14 (2020). [DOI] [PubMed] [Google Scholar]
- 70.Wang M., et al., KTE-X19 CAR T-cell therapy in relapsed or refractory mantle-cell lymphoma. N. Engl. J. Med. 382, 1331–1342 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown C. E., Mackall C. L., CAR T cell therapy: Inroads to response and resistance. Nat. Rev. Immunol. 19, 73–74 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Majzner R. G., Mackall C. L., Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med. 25, 1341–1355 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Rafiq S., Hackett C. S., Brentjens R. J., Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 17, 147–167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watanabe K., Kuramitsu S., Posey A. D. Jr, June C. H., Expanding the therapeutic window for CAR T cell therapy in solid tumors: The knowns and unknowns of CAR T cell biology. Front. Immunol. 9, 2486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamers C. H. J., et al., Treatment of metastatic renal cell carcinoma with autologous T-lymphocytes genetically retargeted against carbonic anhydrase IX: First clinical experience. J. Clin. Oncol. 24, e20–e22 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Morgan R. A., et al., Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol. Ther. 18, 843–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parkhurst M. R., et al., T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 19, 620–626 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamers C. H., et al., Treatment of metastatic renal cell carcinoma with CAIX CAR-engineered T cells: Clinical evaluation and management of on-target toxicity. Mol. Ther. 21, 904–912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kloss C. C., Condomines M., Cartellieri M., Bachmann M., Sadelain M., Combinatorial antigen recognition with balanced signaling promotes selective tumor eradication by engineered T cells. Nat. Biotechnol. 31, 71–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roybal K. T., et al., Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 164, 770–779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim W. A., June C. H., The principles of engineering immune cells to treat cancer. Cell 168, 724–740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Perna F., et al., Integrating proteomics and transcriptomics for systematic combinatorial chimeric antigen receptor therapy of AML. Cancer Cell 32, 506–519.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cho J. H., Collins J. J., Wong W. W., Universal chimeric antigen receptors for multiplexed and logical control of T cell responses. Cell 173, 1426–1438.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fry T. J., et al., CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nat. Med. 24, 20–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Srivastava S., et al., Logic-gated ROR1 chimeric antigen receptor expression rescues T cell-mediated toxicity to normal tissues and enables selective tumor targeting. Cancer Cell 35, 489–503.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fedorov V. D., Themeli M., Sadelain M., PD-1– and CTLA-4–based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Trans. Med. 5, 215ra172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Snell G. D., Jackson R. B., Histocompatibility genes of the mouse. II. Production and analysis of isogenic resistant lines. J. Natl. Cancer Inst. 21, 843–877 (1958). [PubMed] [Google Scholar]
- 88.Snell G. D., Studies in Histocompatibility. Scand. J. Immunol. 36, 514–525 (1992). [DOI] [PubMed] [Google Scholar]
- 89.Jiménez P., et al., Chromosome loss is the most frequent mechanism contributing to HLA haplotype loss in human tumors. Int. J. Cancer 83, 91–97 (1999). [DOI] [PubMed] [Google Scholar]
- 90.Little A. M., Parham P., Polymorphism and evolution of HLA class I and II genes and molecules. Rev. Immunogenet. 1, 105–123 (1999). [PubMed] [Google Scholar]
- 91.Mazurenko N. N., et al., Genetic alterations at chromosome 6 associated with cervical cancer progression. Mol. Biol. 37, 404–411 (2003). [PubMed] [Google Scholar]
- 92.Robinson J., et al., The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 43, D423–D431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McGranahan N.et al.; TRACERx Consortium , Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hahn S. A., Seymour A. B., Allelotype of pancreatic adenocarcinoma using xenograft enrichment. Cancer Res. 55, 4670–4675. [PubMed] [Google Scholar]
- 95.Girard L., Zöchbauer-Müller S., Virmani A. K., Gazdar A. F., Minna J. D., Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 60, 4894–4906 (2000). [PubMed] [Google Scholar]
- 96.Maleno I., et al., Frequent loss of heterozygosity in the β2-microglobulin region of chromosome 15 in primary human tumors. Immunogenetics 63, 65–71 (2011). [DOI] [PubMed] [Google Scholar]
- 97.Yeung J. T., et al., LOH in the HLA class I region at 6p21 is associated with shorter survival in newly diagnosed adult glioblastoma. Clin. Cancer Res. 19, 1816–1826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garrido M. A., et al., HLA class I alterations in breast carcinoma are associated with a high frequency of the loss of heterozygosity at chromosomes 6 and 15. Immunogenetics 70, 647–659 (2018). [DOI] [PubMed] [Google Scholar]
- 99.Maiers M., Gragert L., Klitz W., High-resolution HLA alleles and haplotypes in the United States population. Hum. Immunol. 68, 779–788 (2007). [DOI] [PubMed] [Google Scholar]
- 100.Gonzalez-Galarza F. F., et al., Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 48, D783–D788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Parham P., Brodsky F. M., Partial purification and some properties of BB7.2. A cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum. Immunol. 3, 277–299 (1981). [DOI] [PubMed] [Google Scholar]
- 102.Berger A. E., Davis J. E., Cresswell P., Monoclonal antibody to HLA-A3. Hybridoma 1, 87–90 (1982). [DOI] [PubMed] [Google Scholar]
- 103.Miller M. S., et al., An engineered antibody fragment targeting mutant β-catenin via major histocompatibility complex I neoantigen presentation. J. Biol. Chem. 294, 19322–19334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eyquem J., et al., Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Z., et al., Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 7, 2193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Skora A. D., et al., Generation of MANAbodies specific to HLA-restricted epitopes encoded by somatically mutated genes. Proc. Natl. Acad. Sci. U.S.A. 112, 9967–9972 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tran E., et al., Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leko V., Rosenberg S. A., Identifying and targeting human tumor antigens for T cell-based immunotherapy of solid tumors. Cancer Cell 38, 454–472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu D., Gallagher D. T., Gowthaman R., Pierce B. G., Mariuzza R. A., Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen. Nat. Commun. 11, 2908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fountain J. W., et al., Homozygous deletions within human chromosome band 9p2l in melanoma. Med. Sci. 89, 10557–10561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Narimatsu H., et al., Lewis and secretor gene dosages affect CA19-9 and DU-PAN-2 serum levels in normal individuals and colorectal cancer patients. Cancer Res. 58, 512–518 (1998). [PubMed] [Google Scholar]
- 112.Yan H., Yuan W., Velculescu V. E., Vogelstein B., Kinzler K. W., Allelic variation in human gene expression. Science 297, 1143 (2002). [DOI] [PubMed] [Google Scholar]
- 113.Sade-Feldman M., et al., Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 8, 1136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Orlando E. J., et al., Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat. Med. 24, 1504–1506 (2018). [DOI] [PubMed] [Google Scholar]
- 115.Tran E., et al., T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chowell D., et al., Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Montes P., et al., Genomic loss of HLA alleles may affect the clinical outcome in low-risk myelodysplastic syndrome patients. Oncotarget 9, 36929–36944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen H., et al., Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat. Commun. 10, 5472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lo W., et al., Immunologic recognition of a shared p53 mutated neoantigen in a patient with metastatic colorectal cancer. Cancer Immunol. Res. 7, 534–543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tamaki K., et al., Somatic mutations and loss of heterozygosity of HLA genes are frequently occurred and tightly associated with poor prognosis in adult T cell leukemia-lymphoma. Blood 134, 2785 (2019). [Google Scholar]
- 121.Shim J. H., et al., HLA-corrected tumor mutation burden and homologous recombination deficiency for the prediction of response to PD-(L)1 blockade in advanced non-small-cell lung cancer patients. Ann. Oncol. 31, 902–911 (2020). [DOI] [PubMed] [Google Scholar]
- 122.Hamburger A. E., et al., Engineered T cells directed at tumors with defined allelic loss. Mol. Immunol. 128, 298–310 (2020). [DOI] [PubMed] [Google Scholar]
- 123.Martayan A., et al., Class I HLA folding and antigen presentation in β 2-microglobulin-defective Daudi cells. J. Immunol. 182, 3609–3617 (2009). [DOI] [PubMed] [Google Scholar]
- 124.Haidaris C. G., et al., Recombinant human antibody single chain variable fragments reactive with Candida albicans surface antigens. J. Immunol. Methods 257, 185–202 (2001). [DOI] [PubMed] [Google Scholar]
- 125.Nguyen D. N., et al., Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 38, 44–49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.