Significance
Many photosynthetic cyanobacteria are capable of migrating in response to light in a process known as phototaxis. In all of the cyanobacteria where this process has been characterized at the genetic and molecular level, motility has been shown to be powered by type IV pili and influenced by chemotaxis-like systems with methyl-accepting chemotaxis proteins containing light-sensing GAF domains. However, the means by which the light-sensing systems modulate T4P activity has not been defined. In this study we provide evidence that cyanobacteria possess a second, distinct system for sensing light, the Hmp chemotaxis-like system, which lacks a GAF domain and modulates direct interaction between the cyanobacterial taxis protein HmpF and the T4P to regulate motility in response to light.
Keywords: cyanobacteria, gliding motility, hormogonia, type IV pili, phototaxis
Abstract
Motility is ubiquitous in prokaryotic organisms including the photosynthetic cyanobacteria where surface motility powered by type 4 pili (T4P) is common and facilitates phototaxis to seek out favorable light environments. In cyanobacteria, chemotaxis-like systems are known to regulate motility and phototaxis. The characterized phototaxis systems rely on methyl-accepting chemotaxis proteins containing bilin-binding GAF domains capable of directly sensing light, and the mechanism by which they regulate the T4P is largely undefined. In this study we demonstrate that cyanobacteria possess a second, GAF-independent, means of sensing light to regulate motility and provide insight into how a chemotaxis-like system regulates the T4P motors. A combination of genetic, cytological, and protein–protein interaction analyses, along with experiments using the proton ionophore carbonyl cyanide m-chlorophenyl hydrazine, indicate that the Hmp chemotaxis-like system of the model filamentous cyanobacterium Nostoc punctiforme is capable of sensing light indirectly, possibly via alterations in proton motive force, and modulates direct interaction between the cyanobacterial taxis protein HmpF, and Hfq, PilT1, and PilT2 to regulate the T4P motors. Given that the Hmp system is widely conserved in cyanobacteria, and the finding from this study that orthologs of HmpF and T4P proteins from the distantly related model unicellular cyanobacterium Synechocystis sp. strain PCC6803 interact in a similar manner to their N. punctiforme counterparts, it is likely that this represents a ubiquitous means of regulating motility in response to light in cyanobacteria.
Motility is ubiquitous in prokaryotic organisms, including both swimming motility in aqueous environments and twitching or gliding motility on solid surfaces, and enables these organisms to optimize their position in response to various environmental factors. Among the photosynthetic cyanobacteria, surface motility is widespread and facilitates phototaxis to seek out favorable light environments (1, 2), and, for multicellular filamentous cyanobacteria, plays a key role in dispersal as well as the establishment of nitrogen-fixing symbioses with eukaryotes (3) and the formation of supracellular structures (3–5).
Current understanding of cyanobacterial surface motility at the molecular level has been informed primarily by studies of two model organisms, the unicellular strain Synechocystis sp. strain PCC6803 (herein Synechocystis) and the filamentous strain Nostoc punctiforme ATCC29133/PCC73102, where motility is exhibited only by differentiated filaments termed “hormogonia.” Motility in both organisms is powered by a type IV pilus (T4P) system where the ATPases PilB and PilT drive the extension and subsequent retraction, respectively, of pili which adhere to the substrate and pull the cells forward (for review, see ref. 6). In Synechocystis, the T4P motors are distributed throughout the entire cell, allowing a 360 ° range of motion (7), whereas in N. punctiforme they are confined to rings at the cell poles (8), resulting in movement only along the long axis of the filament. Comparative genomics implies that this mechanism of motility is widely conserved among cyanobacteria (9).
Both Synechocystis and N. punctiforme employ chemotaxis-like systems to regulate motility. One of these systems, the Hmp chemotaxis-like system of N. punctiforme (3, 10), and its orthologous counterpart, the Pil chemotaxis-like system of Synechocystis (11), includes homologs to the canonical Escherichia coli chemotaxis complex (for review, see ref. 12), including the histidine kinase CheA, the adaptor protein CheW, the response regulator CheY, and the methyl-accepting chemotaxis protein MCP. These systems are essential for motility in their respective organisms and appear to regulate the T4P motors, although there are distinct differences in the phenotypes for inactivation of the components from each. In Synechocystis, null mutations either enhance or reduce the level of surface piliation (11), whereas in N. punctiforme they disrupt the coordinated polarity, but not the overall level of piliation, and affect various other aspects of hormogonium development (3, 10). In N. punctiforme, the subcellular localization of this system has been determined and has been found arrayed in static, bipolar rings similar to the T4P motors (3). However, the signals that are perceived by the MCPs and the precise mechanism by which these systems modulate T4P activity is currently undefined.
Recently, an additional component of the Hmp system, HmpF, was characterized (9). HmpF is a predicted coiled-coil protein and is ubiquitous to, but confined within, the cyanobacterial lineage (9). It is essential for accumulation of surface pili and exhibits dynamic, unipolar localization to the leading poles of most cells in hormogonium filaments (9). Based on these findings, a model has been proposed where the localization of HmpF is regulated by the other components of the Hmp system, and in turn, the unipolar accumulation of HmpF leads to the activation of the T4P motors on one side of the cell to facilitate directional movement.
A second chemotaxis-like system in each organism, the Ptx system of N. punctiforme (13) and the Pix system of Synechocystis (14, 15), is essential for positive phototaxis. These systems contain MCPs with cyanobacteriochrome sensory domains capable of perceiving light (for review, see ref. 16). Disruption of the Pix system results in negative phototaxis under light conditions that normally produce a positive phototactic response (14). Several other proteins containing cyanobacteriochromes, and one containing a BLUF domain, also modulate phototaxis in Synechocystis (for review, see ref. 6). In N. punctiforme, disruption of the Ptx system abolishes the phototactic response completely, resulting in uniform movement in all directions regardless of the light conditions (13), and there are currently no other proteins reported to modulate phototaxis. More recently, a motile, wild isolate of the model unicellular cyanobacterium Synechococcus elongatus sp. PCC7942 was shown to possess a chemotaxis-like system that modulates phototaxis in a manner similar to that of the N. punctiforme Ptx system (17). How these systems influence T4P activity to facilitate phototaxis is also currently unknown.
There is also a substantial body of literature on motility and phototaxis in cyanobacteria, primarily based on observational studies of various filamentous strains, that predates the development of genetically tractable model organisms (for review, see ref. 18). These reports suggested that the photosystems may serve a sensory role in modulating phototaxis and that proton motive force (PMF) powers motility (19, 20), a finding that is inconsistent with the theory that cyanobacteria possess a common T4P-based gliding motor driven by ATP hydrolysis. In this study, we help reconcile this historical data with more recent molecular studies by providing evidence that the Hmp chemotaxis-like system senses light, possibly indirectly through alterations in PMF, and in turn modulates the interaction of HmpF with the T4P base to activate the motors.
Results
HmpF Interacts with the T4P Motors via Hfq, PilT1, and PilT2.
As part of an ongoing transposon mutagenic screen (21) a nonmotile isolate was identified containing a transposon insertion in hfq (Npun_F5230). Previous studies in Synechocystis have shown that Hfq is essential for motility and directly interacts with the pilus extension ATPase PilB (22, 23). To confirm that Hfq plays a similar role in N. punctiforme, an hfq in-frame deletion strain was constructed and its ability to differentiate motile hormogonium was assessed. Deletion of hfq completely abolished motility, as assessed by the loss of colony spreading in plate motility assays (SI Appendix, Fig. S1A) and by the absence of motility for individual filaments in time-lapse microscopy (Movie S1), but did not prevent the development of morphologically distinct hormogonia (SI Appendix, Fig. S1C) or the accumulation of the hormogonium-specific proteins HmpD or PilA (SI Appendix, Fig. S1D). Reintroduction of hfq and its putative promoter region on a replicative shuttle vector restored motility (SI Appendix, Fig. S1A), although at levels much lower than observed for the wild-type strain, consistent with other nonmotile mutants complemented in a similar manner (9, 21, 24). Introduction of an hfq-gfp allele at the native chromosomal locus restored motility to levels more comparable to that of the wild-type strain (SI Appendix, Fig. S1A), and immunoblot analysis detected a protein of the predicted size for Hfq-GFP (SI Appendix, Fig. S1B), indicating that a full-length, functional Hfq-GFP protein is produced in this strain. Although the Δhfq strain produced cellular PilA at levels comparable to the wild type, immunofluorescence (SI Appendix, Fig. S1D) and lectin staining (SI Appendix, Fig. S1E) indicated that the accumulation of extracellular PilA or hormogonium polysaccharide (HPS) was abolished. Collectively, the phenotype of the Δhfq strain is indistinguishable from that previously reported for deletion of pilB (8) or hmpF (9).
In N. punctiforme, both the T4P proteins and HmpF localize to rings at the cell poles with T4P proteins static at both poles and HmpF exhibiting dynamic localization to the leading poles in motile hormogonia (8, 9). A recent study has also found sigC-dependent up-regulation of hfq in developing hormogonia (25). To determine the subcellular localization and confirm enhanced expression of Hfq in hormogonia, the hfq-gfp strain was visualized with fluorescence microscopy (SI Appendix, Fig. S1F). In vegetative filaments, Hfq-GFP was visible as bright foci with no obvious bias for the cell poles, whereas in hormogonia, there was a substantial increase in fluorescence with diffuse accumulation in the cytoplasm as well as clear bipolar localization.
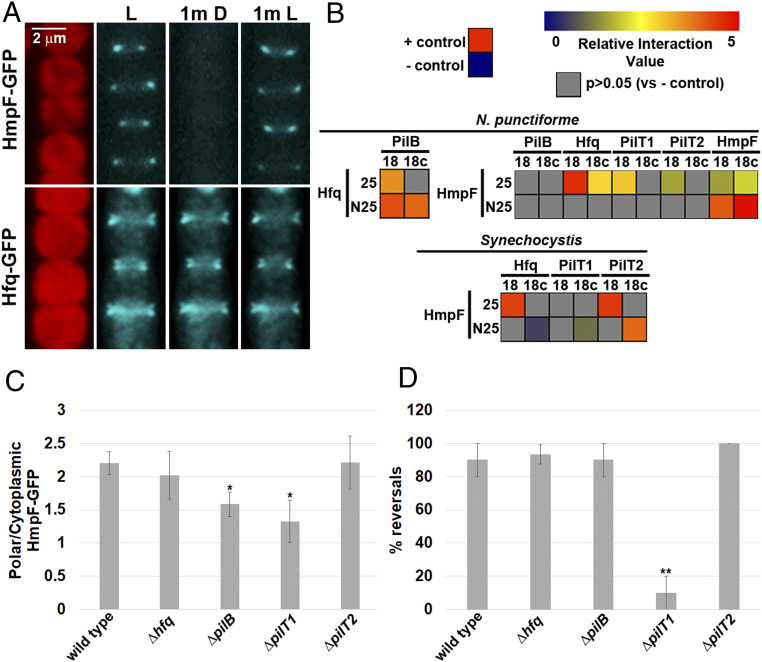
To determine if the polar localization of Hfq-GFP was static or dynamic, its localization was compared to that of HmpF-GFP under light conditions that trigger dynamic relocalization of HmpF-GFP. Exposure of motile hormogonia to illumination with a wide spectrum of lasers has been shown to trigger reversals and relocalization of HmpF-GFP to the new leading poles (9), but the movement of the filaments during imaging makes precisely assigning localization of HmpF-GFP within the cell challenging. Therefore, a light regimen was established that triggered polar relocalization of HmpF-GFP in immobilized hormogonium filaments (SI Appendix, Fig. S2 A and B). A 1-min exposure of immobilized filaments to 405 nm light used for imaging GFP caused HmpF-GFP to completely dissociate from the poles and accumulate in the cytoplasm. HmpF-GFP remained in the cytoplasm following an additional 1 min of incubation in the dark, but a subsequent 1 min of exposure to white light resulted in relocalization of HmpF-GFP at the opposite pole in most filaments. Therefore, to assess the dynamic localization of proteins in subsequent experiments, filaments were incubated in white light for 1 min followed by darkness for 1 min and subsequently re-exposed to white light for 1 min, with exposure to 405 nm light for GFP imaging at the end of each 1-min incubation period. HmpF-GFP exhibited dynamic localization under these conditions, dissociating from the poles upon exposure to 405 nm light followed by incubation in darkness and then reaccumulating at the opposite pole when white light was reintroduced (Fig. 1A). In contrast, localization of Hfq-GFP was unaffected by this light regimen, indicating static localization at both poles (Fig. 1A).
Fig. 1.
Interaction of HmpF with T4P components. (A) Localization of HmpF-GFP and Hfq-GFP in response to light. Depicted are fluorescence micrographs of cellular autofluorescence (red) and GFP fluorescence (cyan) at each interval of a light regiment with exposure to white light for 1 min (L), followed by darkness for 1 min (1m D), and subsequently white light for 1 min (1m L). (B) Heat maps depicting the results of BACTH analysis between various proteins of N. punctiforme and Synechocystis sp. strain PCC6803 fused to the C (25) and N (N25) terminus of the T25 fragment or C (18c) and N (18) terminus of the T18 fragment of B. pertussis adenylate cyclase. Data are derived from SI Appendix, Fig. S3. Relative interaction value = Log2(Experimental/– control). (C and D) Quantification of the fraction of HmpF-GFP localized to the poles (C) and HmpF-GFP polar reversals (D) in various T4P-gene deletion strains. Error bars = ±1 SD; *P value < 0.05 as determined by two-tailed Student’s t-test between the wild type and each deletion strain, **P value < 0.01, n = 3. C is derived from data depicted in SI Appendix, Fig. S6.
These data, along with the identical phenotypes for deletion of hmpF, pilB, and hfq, is consistent with a model where direct interaction between HmpF and one or more components of the T4P base activates cycles of pilus extension and retraction at the leading poles of motile hormogonia. The bacterial adenylate cyclase two-hybrid assay (BACTH) (26) was employed to test this hypothesis (Fig. 1B and SI Appendix, Fig. S3). As previously reported in Synechocystis (23), Hfq was found to interact with PilB. HmpF did not interact with PilB but interacted with Hfq, PilT1, and PilT2, as well as itself, indicating that HmpF forms homo-multimers. Similar interactions were detected among the orthologs of these proteins from Synechocystis (Fig. 1B), implying that these interactions are broadly conserved among distantly related cyanobacteria. Collectively, these results are consistent with a model where HmpF associates with Hfq, PilT1, and PilT2 at the T4P base, triggering pilus activation at the leading pole of motile filaments.
To further test this model, localization of HmpF-GFP was determined in strains containing deletions in genes encoding the interacting proteins. Each deletion strain produced the expected motility phenotype (8) as assessed by colony-spreading assays (SI Appendix, Fig. S4A). Deletion of these genes individually was insufficient to completely disrupt the dynamic, polar localization of HmpF-GFP (Fig. 1 C and D and SI Appendix, Fig. S5). However, deletion of pilB and pilT1 reduced the fraction of HmpF-GFP localized to the poles (Fig. 1C). This change in localization does not appear to be the result of a reduction in total levels of HmpF-GFP because cytoplasmic fluorescence in both strains was elevated compared to a wild-type genetic background and well above that of natural autofluorescence in a control strain lacking gfp (SI Appendix, Fig. S6). Furthermore, deletion of pilT1 reduced the frequency of HmpF-GFP polar reversals (Fig. 1D). These data are consistent with the finding that HmpF interacts with several components of the T4P system and implies that the status of the T4P motor influences polar reversals of HmpF.
The Hmp Chemotaxis-like Systems Regulates the Coordinated, Unipolar Localization of HmpF.
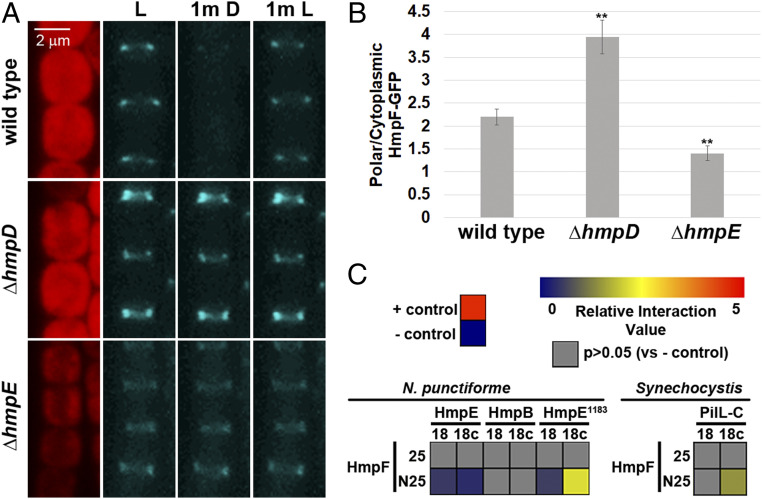
The current working model for the Hmp chemotaxis-like system posits that the homologs to canonical chemotaxis system components regulate the polar localization of HmpF-GFP to coordinate the activation of the T4P motors at the leading poles of cells in motile hormogonia. To test this hypothesis, the dynamic localization of HmpF-GFP was assessed in strains harboring deletions in hmpD and hmpE, which encode a methyl-accepting chemotaxis protein (MCP) and CheA homolog, respectively. Deletion of hmpD or hmpE in the hmpF-gfp strain produced the expected nonmotile phenotype (3) as assessed by colony-spreading assays (SI Appendix, Fig. S4A). Deletion of hmpD enhanced polar accumulation of HmpF-GFP with a concomitant decrease in cytoplasmic localization compared to the wild-type genetic background, whereas deletion of hmpE showed the opposite phenotype with diminished polar HmpF-GFP and enhanced cytoplasmic accumulation (Fig. 2 A and B and SI Appendix, Fig. S6). In both the ΔhmpD and ΔhmpE strains, HmpF-GFP exhibited uniform, bipolar fluorescence in hormogonium filaments and did not exhibit any light-responsive dynamic localization (Fig. 2A), although, for the ΔhmpE strain, the bipolar accumulation of HmpF-GFP was apparent only in filaments displaying the brightest levels of polar fluorescence, such as the example depicted in Fig. 2A.
Fig. 2.
Regulation of HmpF by the Hmp system. (A) Localization of HmpF-GFP in response to light in strains harboring deletions in genes encoding Hmp system components. Depicted are fluorescence micrographs of cellular autofluorescence (red) and GFP fluorescence (cyan) at each interval of a light regiment with exposure to white light for 1 min (L), followed by darkness for 1 min (1m D), and subsequently by white light for 1 min (1m L). (B) Quantification of the fraction of HmpF-GFP localized to the poles in various Hmp-gene deletion strains. Error bars = ±1 SD; **P value < 0.01 as determined by two-tailed Student’s t-test between the wild type and each deletion strain; n = 3. B is derived from data depicted in SI Appendix, Fig. S6. (C) Heat maps depicting the results of BACTH analysis between various proteins of N. punctiforme and Synechocystis sp. strain PCC6803 fused to the C (25) and N (N25) terminus of the T25 fragment or C (18c) and N (18) terminus of the T18 fragment of B. pertussis adenylate cyclase. Data are derived from SI Appendix, Fig. S7. Relative interaction value = Log2(Experimental/– control).
A direct interaction between HmpF and HmpE, or its cognate response regulator HmpB (3), could account for the reduction in polar HmpF-GFP in the ΔhmpE strain. To test this hypothesis, BACTH was employed to investigate interactions among these proteins (Fig. 2C and SI Appendix, Fig. S7A). No interaction was detected between HmpF and HmpB, but a very weak positive signal was detected between HmpF and HmpE. HmpE is a large protein containing an N-terminal Hpt domain followed by a number of repeats of unknown function, then the remaining CheA-like domains, and finally a REC domain at the C terminus. The large size of HmpE could limit expression or lead to problems with stability that interfere with the BACTH assay. Therefore, a C-terminal fragment of HmpE, containing amino acids 1183 to 1865 (HmpE1183), that encodes all of the domains found in a typical CheA protein, with the exception of the Hpt domain, and includes the C-terminal REC domain was subsequently tested for interaction with HmpF. A much stronger positive signal was detected between HmpF and HmpE1183, indicating that HmpF directly interacts with this portion of HmpE. The Synechocystis ortholog of HmpE is split into two separate proteins, PilL-N, which contains only the Hpt domain, and PilL-C, which contains the same domains as those of HmpE1183, plus a second C-terminal REC domain (11). The genome of Synechocystis does not encode an ortholog of the repeat region from HmpE. Therefore, HmpF and PilL-C from Synechocystis were probed for protein–protein interaction using BACTH as well (Fig. 2C and SI Appendix, Fig. S7A). HmpF and PilL-C from Synechocystis also interacted with one another, with the interaction exhibited by the same combination of T18 and T25 fusions as found in N. punctiforme, implying that this interaction is also broadly conserved in cyanobacteria. Immunoblot analysis with an antibody against the T18 fragment of adenylate cyclase indicated that HmpE is either highly unstable or subject to premature termination of transcription or translation in E. coli, given the detection of prominent bands well below the expected molecular weight for HmpE (SI Appendix, Fig. S7B). In contrast, HmpE1183, PilL-C, and HmpF were expressed primarily as full-length proteins of the expected molecular weight (SI Appendix, Fig. S7B). As suggested above, this could account for the weak interaction between HmpF and HmpE compared to HmpE1183. It should also be noted that, although no interaction was detected between HmpF and the response regulator HmpB, we cannot discount the possibility that these proteins interact only when HmpB is phosphorylated, assuming that HmpB is not efficiently phosphorylated in E. coli. Previous experiments have demonstrated that HmpB is a direct target for phosphorylation by HmpE (3).
The Hmp Chemotaxis-like System Regulates Localization of HmpF in Response to Light, Possibly via Light-Driven Changes in Proton Motive Force.
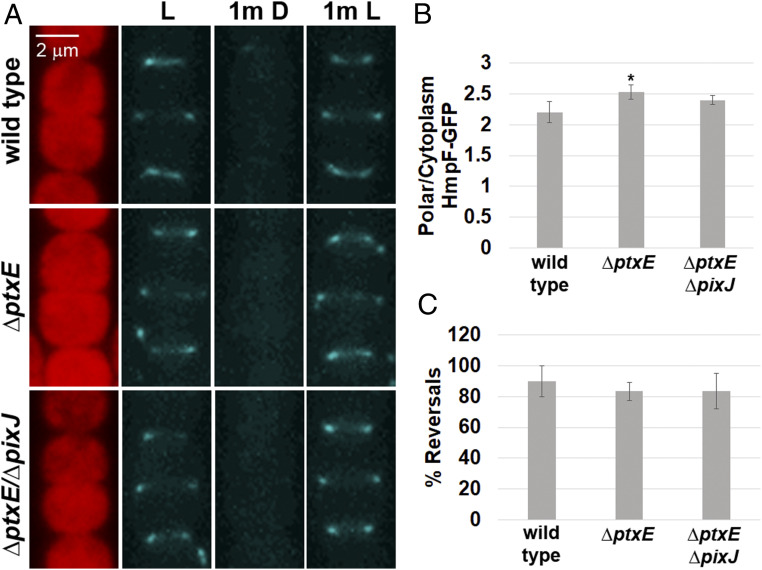
The Ptx chemotaxis-like system is essential for positive phototaxis in N. punctiforme and could exert directional control of the T4P motors by modulating localization of HmpF in response to light. To test this hypothesis, ptxE, which encodes a CheA homolog, was deleted in the hmpF-gfp strain. Deletion of ptxE in the hmpF-gfp strain abolished positive phototaxis in macroscopic colony-spreading assays under directional light (SI Appendix, Fig. S4B), as previously reported (12). Surprisingly, deletion of ptxE had no effect on the dynamic localization of HmpF-GFP in response to light, indicating that light-driven changes in HmpF localization do not require a functional Ptx system (Fig. 3 A–C). The only notable difference between the wild-type and ΔptxE genetic backgrounds is a very moderate increase in polar HmpF-GFP in the latter (Fig. 3B). The genome of N. punctiforme also harbors the pix locus, which encodes paralogs to that of the ptx locus (12) and may be responsible for modulating localization of HmpF in response to light instead or serve a redundant function to that of the Ptx system. Therefore, a strain harboring deletions in both ptxE and pixJ, which encodes the putative light-sensing MCP for the Pix system, was constructed from the hmpF-gfp genetic background. The phototactic response of this strain in macroscopic colony-spreading assays was comparable to the ΔptxE single-deletion mutant (SI Appendix, Fig. S4B). As with deletion of ptxE, deletion of both ptxE and pixJ in conjunction did not disrupt the light-driven dynamic localization of HmpF-GFP (Fig. 3 A–C). These results imply that another system, distinct from the prototypical chemotaxis-like systems encoding light-sensing MCPs with GAF domains, is responsible for influencing HmpF in response to changes in light.
Fig. 3.
Regulation of HmpF by the Ptx and Pix systems. (A) Localization of HmpF-GFP in response to light in strains harboring deletions in genes encoding Ptx and Pix system components. Depicted are fluorescence micrographs of cellular autofluorescence (red) and GFP fluorescence (cyan) at each interval of a light regiment with exposure to white light for 1 min (L), followed by darkness for 1 min (1m D), and subsequently by white light for 1 min (1m L). (B and C) Quantification of the fraction of HmpF-GFP localized to the poles (B) and HmpF-GFP polar reversals (C) in various Ptx and Pix deletion strains. Error bars = ±1 SD; *P value < 0.05 as determined by two-tailed Student’s t-test between the wild type and each deletion strain; n = 3. B is derived from data depicted in SI Appendix, Fig. S6.
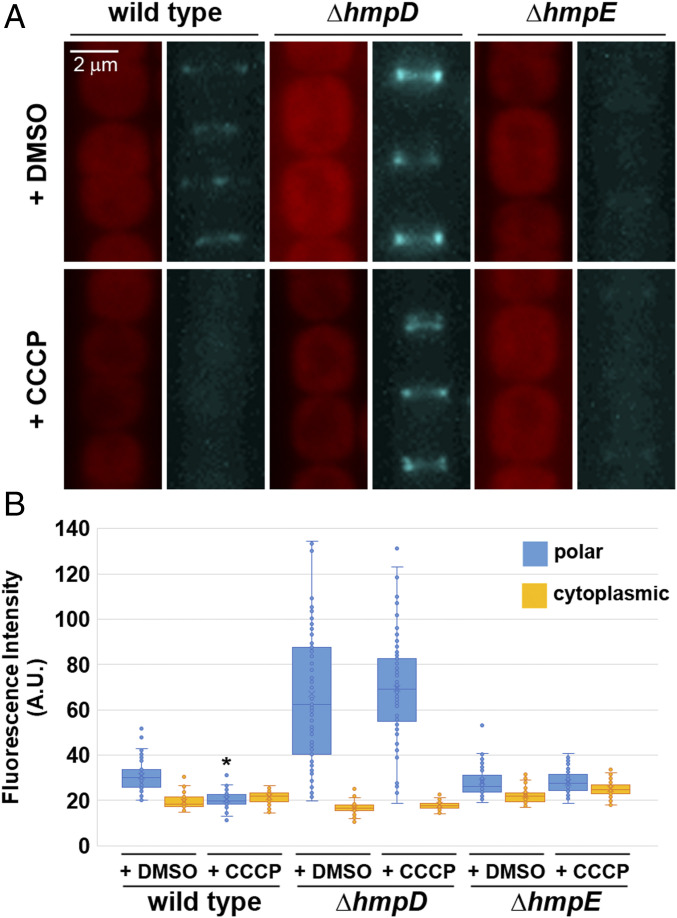
The proton ionophore carbonyl cyanide m-chlorophenyl hydrazine (CCCP) has been shown to abolish motility in hormogonia much faster than ATP synthesis inhibitors, leading to speculation that PMF, rather than ATP hydrolysis, powers motility in these organisms (20). However, this would appear to be incompatible with the finding that cyanobacteria possess a conserved T4P system that drives motility and is presumed to be powered by ATP hydrolysis. These two seemingly contradictory findings could be reconciled if disruption of PMF, and perhaps more generally, membrane polarity, plays a regulatory, rather than kinetic role in motility. To test this hypothesis, the effect of CCCP treatment on HmpF-GFP localization was determined (Fig. 4 A and B). Treatment with CCCP caused HmpF-GFP to dissociate from the poles and also lead to the cessation of motility (Movie S2). In contrast, when strains harboring deletions in either hmpD or hmpE were subjected to CCCP treatment, there was no apparent effect on the localization of HmpF-GFP (Fig. 4 A and B). These findings indicate that the Hmp chemotaxis-like system may be capable of indirectly sensing light via changes in PMF and subsequently influencing the localization of HmpF to modulate motility.
Fig. 4.
Effect of CCCP treatment on HmpF-GFP localization. (A) Localization of HmpF-GFP in response to treatment with CCCP or DMSO alone (– control) in a wild-type genetic background or strains harboring deletions in genes encoding Hmp system components. Depicted are fluorescence micrographs of cellular autofluorescence (red) and GFP fluorescence (cyan). (B) Quantification of the fraction of HmpF-GFP localized to the poles and cytoplasm. *P value < 0.05 as determined by two-tailed Student’s t-test between the CCCP and DMSO treatment for each strain and position measured; n = 3.
Discussion
Based on the findings of this study, we propose the following working model for the Hmp chemotaxis-like system (Fig. 5). Light absorbed by the photosystems results in changes to PMF across the cytoplasmic membrane, primarily due to the pumping of protons from the cytoplasm into the thylakoid space. These changes are perceived by the Hmp system, which in turn modulates the interaction between HmpF and Hfq/PilT1/PilT2 to regulate the T4P motors. Given the widespread conservation of the Hmp and T4P systems in cyanobacteria (9), and the evidence presented here that orthologs of HmpF, HmpE, Hfq, PilT1, and PilT2 from Synechocystis interact in a similar manner to their N. punctiforme counterparts, it is likely that this model applies to most cyanobacteria capable of surface motility. This model also reconciles older findings on the role of PMF and the photosystems in controlling cyanobacterial motility with more recent findings from genetic and molecular studies.
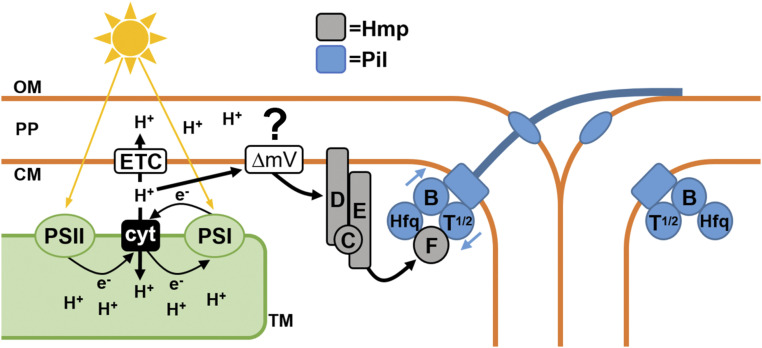
Fig. 5.
A working model of how HmpF regulates the T4P in response to light. Light absorbed by the photosystems (PSI + PSII) drives linear and cyclic electron flow, resulting in proton pumping via the cytochrome complex (cyt). The efflux of protons from the cytoplasm into the thylakoid space could result in a change to the membrane potential (ΔmV) across the cytoplasmic membrane and might serve as a signal sensed by HmpD, which in turn stimulates the Hmp system to regulate HmpF interaction with components at the base of the T4P. OM: outermembrane; PP: periplasm; CM: cytoplasmic membrane; TM: thylakoid membrane; ETC: respiratory electron transport chain.
Although the experiments with CCCP support the idea that PMF, and perhaps more generally, membrane potential is the signal most directly sensed by the Hmp system, this part of the model is highly speculative at this point, and it is possible that indirect effects on electron transport or ATP synthesis may account for the localization of HmpF-GFP in response to CCCP treatment. Assuming that membrane polarization does serve as the signal, the fact that both exposure to high-intensity illumination, which would be expected to hyperpolarize the membrane, and CCCP, which would be expected to depolarize the membrane, lead to delocalization of HmpF from the poles implies that a specific range of membrane polarity is required to facilitate the association of HmpF with the T4P motors. It should also be noted that it is conceivable that light alters PMF independently of the photosystems. A recent report has shown that all five E. coli MCPs are capable of regulating taxis in response to blue light, and based on indirect evidence, the authors speculate that this may be due to light-induced changes in PMF, possibly due to photoreduction of electron carriers (27). More work will be needed to determine precisely how the photosystems and respiration might contribute to light-induced changes in PMF and whether this is the signal directly sensed by the Hmp system.
Several additional lines of evidence from published reports support this model. HmpF has been shown to be essential for motility in Synechocystis (28), and an HmpF pull-down experiment in this organism recovered proteins from the T4P and Pil chemotaxis-like systems (29). Another report indicated that the T4P of Synechocystis possesses two distinct responses to light, one which is rapid and nondirectional, and a second that is slower but directional (30). It is possible that the rapid response is controlled by the Pil chemotaxis-like systems, whereas the slower response is regulated by the Pix system and other light-sensing proteins. Finally, direct measurements have also demonstrated that light alters membrane polarity in cyanobacteria (31), and exposure to electrical fields has been shown to trigger filament reversals (32), a finding consistent with the idea that membrane potential may serve a regulatory role in motility. The putative structure of the MCP HmpD is also consistent with a role in sensing membrane polarity given that HmpD is predicted to be anchored to the membrane by a pair of closely spaced transmembrane a-helices, but does not contain any obvious periplasmic domain.
The results from this study clearly indicate that N. punctiforme has two distinct systems with different light-sensing capabilities. The Hmp system senses light indirectly, possibly via alterations in PMF, and is probably responsive only to drastic increases, and perhaps decreases, in light intensity, whereas the Ptx system senses light more directly through its GAF domains and is responsive to more subtle changes in light intensity. This is consistent with the role of the Ptx system in macroscopic colony-spreading assays where filaments are exposed to a gradation in light intensity and also with the finding that the MCP of the Ptx system is a broad-spectrum power sensor (33). It is unclear at this point if the Ptx system also modulates T4P activity via HmpF or through another independent means of regulation. A recent study in Synechocystis has shown that at least one response regulator is capable of interacting directly with PilB to modulate T4P activity in response to light (34), indicating that there may be mechanisms for regulating the T4P motors independent of HmpF.
Furthermore, while the proposed model partially explains how HmpF localization is controlled by light, it fails to account for how coordinated unipolar localization of HmpF along the filament is initially established, nor why uniform alterations in light across the filament lead to polar reversals, rather than to reaccumulation at the same pole. Given that disruption of hmpD and hmpE results in bipolar, rather than unipolar localization, it is likely that the Hmp chemotaxis-like system is somehow involved in establishing coordinated polarity. The fact that reversals of HmpF were substantially reduced in the pilT1 mutant strain could also indicate that the status of the T4P motors on each side of the cell may influence polar switching. Additional experiments will be needed to address these outstanding questions.
Materials and Methods
Strains and Culture Conditions.
For a detailed description of the strains used in this study, refer to SI Appendix, Table S1. N. punctiforme ATCC 29133 and its derivatives were cultured in Allan and Arnon medium diluted four-fold (AA/4), without supplementation of fixed nitrogen, as previously described (35), with the exception that 4 and 10 mM of sucralose was added to liquid and solid medium, respectively, to inhibit hormogonium formation (36). For hormogonium induction, the equivalent of 30⋅μg ml−1 chlorophyll a (Chl a) of cell material from cultures at a Chl a concentration of 10 to 20 μg⋅ml−1 was harvested at 2,000 x g for 3 min, washed two times with AA/4, and resuspended in 2 mL of fresh AA/4 without sucralose. For selective growth, the medium was supplemented with 50 μg⋅ml−1 neomycin. E. coli cultures were grown in lysogeny broth (LB) for liquid cultures or LB supplemented with 1.5% (wt/vol) agar for plates. Selective growth medium was supplemented with 50 μg⋅ml−1 kanamycin, 50 μg⋅ml−1 ampicillin, and 15 μg⋅ml−1 chloramphenicol.
Plasmid and Strain Construction.
For a detailed description of the plasmids, strains, and oligonucleotides used in this study, refer to SI Appendix, Tables S1 and S2. All constructs were sequenced to insure fidelity.
To construct plasmids for in-frame deletion of target genes, ∼900 bp of flanking DNA on either side of the gene and several codons at the beginning and end of each gene were amplified via overlap extension PCR (see SI Appendix, Tables S1 and S2 for details) and cloned into pRL278 (37) as BamHI-SacI fragments using restriction sites introduced on the primers. It should be noted that hfq (Npun_F5230) is likely to be mis-annotated in the JGI IMG Integrated Microbial Genomes and Microbiomes database (https://img.jgi.doe.gov/) (38), which was used to retrieve the genetic sequence of the gene and flanking DNA. In the database it is annotated as 315 bp, with the start codon located within the coding sequence of the divergently transcribed upstream gene. However, read coverage from previous RNA sequencing data (25) indicates that hfq transcripts do not include this start codon. Instead, a second ATG start codon within this coding region is most likely utilized for translation of hfq, resulting in a 219-bp gene that produces a protein product more similar in length and sequence identity to Hfq from Synechocystis sp. strain PCC 6803. Therefore, all experiments involving hfq utilized this 219-bp coding region.
To construct plasmid pEZ102, a mobilizable shuttle vector containing hfq and its putative promoter region, the coding region and 241 bp upstream of the start codon were amplified via PCR (see SI Appendix, Tables S1 and S2, for details) and subsequently cloned into pAM504 (39) as a BamHI‐SacI fragment using restriction sites introduced in the primers.
To construct plasmid pEZ101 for replacement of the chromosomal allele of hfq with a C-terminal gfpuv-tagged variant, ∼900 bp of DNA downstream of the stop codon were amplified via PCR and cloned into pSCR569 (40) as SpeI-SacI fragments using restriction sites introduced on the primers. The coding region of hfq and ∼900 bp of DNA upstream of the start codon were then amplified via PCR and cloned into this plasmid as a BamHI-SmaI fragment using restriction sites introduced on the primers.
To construct plasmids encoding proteins of interest fused to either the T18 or T25 fragment of Bordetella pertussis adenylate cyclase for BACTH analysis (26, 41), the coding region of each gene was amplified via PCR and cloned into either pUT18/pUT18c or pKT25/pKNT25 using restriction sites introduced on the primers (see SI Appendix, Table S2, for details on restriction sites used for each gene).
Generation of transposon mutants and identification of transposon insertion sites was performed as previously described (21) using plasmid pRL1063a (42). Gene deletions and allelic replacements were performed as previously described (10) with N. punctiforme cultures supplemented with 4 mM sucralose to inhibit hormogonium development and enhance conjugation efficiency (21, 36). To construct UOP160, plasmid pDDR465 was introduced into UCD598 (13). To construct UOP161-164, UOP191, and UOP200, plasmids pDDR368, pSCR583, pDDR354, pDDR293, pDDR136, and pDDR476, respectively, were introduced into UOP157 (9). To create UOP173, plasmid pTVH102 was introduced into UOP160. To create UOP185, plasmid pDDR476 was introduced into the wild-type strain. To create UOP193, plasmid pEZ101 was introduced into UOP185.
Motility Assays.
Both plate and time-lapse motility assays were performed as previously described (8).
Immunoblot Analysis.
Preparation of N. punctiforme cell material, protein extraction, and detection of PilA, RbcL, HmpD, and GFPuv by immunoblot analysis were performed as previously described (9). For immunoblot analysis of T18 fusion proteins, pUT18c derivative plasmids were transformed into E. coli BL21(DE3). These experiments were performed in BL21(DE3) rather than in the BACTH reporter strain BTH101 because transcription from the lac operon in BTH101 is influenced by the strength of protein–protein interactions due to the absence of an endogenous cyaA gene. Transformed strains were cultured in LB containing 100 μg⋅ml−1 ampicillin and 0.5 mM isopropyl β-d-1-thiogalactopyranoside, 1.5 mL of culture was harvested by centrifugation, and the cell pellet was resuspended in 0.5 mL of phosphate-buffered saline and lysed by sonication. The lysate was cleared by centrifugation, and the soluble fraction was separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis on a 4 to 20% gel, followed by transfer to a nitrocellulose membrane and detection of the T18 fragment of B. pertussis adenylate cyclase with a 1:1,000 dilution of primary antibody sc-13582 (Santa Cruz Biotechnology) and 1:2,000 dilution of secondary antibody NXA931 (GE Healthcare Life Sciences).
Immunofluorescence and Fluorescent Lectin Staining.
Detection of PilA and HPS by immunofluorescence and fluorescent lectin staining was performed as previously described (9).
Bacterial Adenylate Cyclase Two-Hybrid Assays.
BACTH (26, 41) was employed to probe protein–protein interaction between various proteins. BTH101 (adenylate cyclase-deficient) E. coli strains transformed with appropriate plasmids were streaked onto LB agar plates containing 100 µg/mL ampicillin and 50 µg/mL kanamycin and incubated at 30 °C for 24 h. Qualitative assays on MacConkey agar and quantitative β-galactosidase assays were performed as previously described (43) with several modifications as described (24). OD600 and OD420 measurements were collected using the Synergy H1 microplate reader (BioTek). The average Miller Units were calculated for each strain as a standardized measure of β-galactosidase activity from two technical replicates for each of three biological replicates.
Microscopy.
Light microscopy of filament morphology was performed using a Leica DME light microscope with a 40× objective lens and equipped with a Leica DFC290 digital camera controlled by micromanager imaging software (44).
Fluorescence microscopy was performed with an EVOS FL fluorescence microscope (Life Technologies) equipped with a 10× or 63× objective lens. Excitation and emission were as follows: EVOS light cube, GFP (AMEP4651: excitation 470 ± 22 nm; emission 525 ± 50 nm) for UEA-fluorescein-labeled HPS; EVOS Light Cube, DAPI (AMEP4650: excitation 357 ± 44 nm; emission 447 ± 60 nm) for immunofluorescence-labeled PilA; EVOS light cube, Nrw 405 (AMEP4857: excitation 390 ± 18 nm; emission 525 ± 50 nm) for GFPuv; and EVOS Light Cube, RFP (AMEP4652: excitation 531 ± 40 nm; emission 593 ± 40 nm) for cellular autofluorescence. To image immobilized filaments expressing HmpF-GFP, 5 μL of culture was placed on a dehydrated 1% agarose pad on a glass slide and overlaid with a coverslip. For treatment with CCCP, 1 μL of 10 mM CCCP in dimethylsulfoxide (DMSO) or DMSO alone was added to 1 mL of culture and incubated for 15 min. Subsequently, 5 μL of culture was placed on a hydrated 1% agarose pad containing 10 μM CCCP or DMSO alone and overlaid with a coverslip.
Quantification of polar and cytoplasmic fluorescence derived from HmpF-GFP was performed using imageJ (NIH). A line was drawn perpendicular to the long axis of the filament across the width of the cell junction or the middle of cell for five contiguous cells for each of five filaments from three biological replicates, and the average pixel intensity was measured for these regions. To quantify polar reversals, 10 filaments with three contiguous cells displaying the same HmpF-GFP polarity were scored based on whether HmpF-GFP polarity reversed following exposure to the light regimen triggering HmpF-GFP reversals described in Results. This was repeated for three biological replicates.
Supplementary Material
Acknowledgments
We thank Carrie Kozina and the Microbiology (BIOL 145) laboratory course teaching assistants and students for purification of chromosomal DNA from nonmotile transposon mutants of N. punctiforme. This work was supported by NSF award #1753690 (to D.D.R.) and by a University of the Pacific Summer Undergraduate Research Fellowship (to T.V.H.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023988118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Castenholz R. W., “Motility and Taxes” in The Biology of Cyanobacteria, Carr N. G., Whitton B. A., Eds. (Blackwell Scientific, 1982), pp. 413–419. [Google Scholar]
- 2.Rippka R., Castenholz R. W., Herdman M., “Cyanobacteria” in Bergey’s Manual of Systematic Bacteriology, Boone D. R., Castenholz R. W., Garrity G. M., Eds. (Springer, 2001), pp. 562–589. [Google Scholar]
- 3.Risser D. D., Chew W. G., Meeks J. C., Genetic characterization of the hmp locus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme. Mol. Microbiol. 92, 222–233 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Shepard R. N., Sumner D. Y., Undirected motility of filamentous cyanobacteria produces reticulate mats. Geobiology 8, 179–190 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Tzubari Y., Magnezi L., Be’er A., Berman-Frank I., Iron and phosphorus deprivation induce sociality in the marine bloom-forming cyanobacterium Trichodesmium. ISME J. 12, 1682–1693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuergers N., Mullineaux C. W., Wilde A., Cyanobacteria in motion. Curr. Opin. Plant Biol. 37, 109–115 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Bhaya D., Bianco N. R., Bryant D., Grossman A., Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37, 941–951 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Khayatan B., Meeks J. C., Risser D. D., Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria. Mol. Microbiol. 98, 1021–1036 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Cho Y. W., et al., Dynamic localization of HmpF regulates type IV pilus activity and directional motility in the filamentous cyanobacterium Nostoc punctiforme. Mol. Microbiol. 106, 252–265 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Risser D. D., Meeks J. C., Comparative transcriptomics with a motility-deficient mutant leads to identification of a novel polysaccharide secretion system in Nostoc punctiforme. Mol. Microbiol. 87, 884–893 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Yoshihara S., Geng X., Ikeuchi M., pilG gene cluster and split pilL genes involved in pilus biogenesis, motility and genetic transformation in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 43, 513–521 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Sourjik V., Wingreen N. S., Responding to chemical gradients: Bacterial chemotaxis. Curr. Opin. Cell Biol. 24, 262–268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell E. L., et al., Genetic analysis reveals the identity of the photoreceptor for phototaxis in hormogonium filaments of Nostoc punctiforme. J. Bacteriol. 197, 782–791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhaya D., Takahashi A., Grossman A. R., Light regulation of type IV pilus-dependent motility by chemosensor-like elements in Synechocystis PCC6803. Proc. Natl. Acad. Sci. U.S.A. 98, 7540–7545 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshihara S., Suzuki F., Fujita H., Geng X. X., Ikeuchi M., Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 41, 1299–1304 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Wiltbank L. B., Kehoe D. M., Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 17, 37–50 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Yang Y., et al., Phototaxis in a wild isolate of the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. U.S.A. 115, E12378–E12387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nultsch W., Häder D. P., Photomovement in motile microorganisms-–II. Photochem. Photobiol. 47, 837–869 (1988). [DOI] [PubMed] [Google Scholar]
- 19.Glagoleva T. N., Glagolev A. N., Gusev M. V., Nikitina K. A., Proton motive force supports gliding in cyanobacteria. FEBS Lett. 117, 49–53 (1980). [Google Scholar]
- 20.Hirose M., Energy supply system for the gliding movement of hormogonia of the cyanobacterium Nostoc cycadae. Plant Cell Physiol. 28, 587–597 (1987). [Google Scholar]
- 21.Khayatan B., et al. , A putative O-linked beta-N-acetylglucosamine transferase is essential for hormogonium development and motility in the filamentous cyanobacterium Nostoc punctiforme. J. Bacteriol. 199, e00075-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dienst D., et al., The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology (Reading) 154, 3134–3143 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Schuergers N., et al., Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol. Microbiol. 92, 840–852 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Riley K. W., Gonzalez A., Risser D. D., A partner-switching regulatory system controls hormogonium development in the filamentous cyanobacterium Nostoc punctiforme. Mol. Microbiol. 109, 555–569 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez A., Riley K. W., Harwood T. V., Zuniga E. G., Risser D. D., A tripartite, hierarchical sigma factor cascade promotes hormogonium development in the filamentous cyanobacterium Nostoc punctiforme. mSphere 4, e00231-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karimova G., Pidoux J., Ullmann A., Ladant D., A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perlova T., Gruebele M., Chemla Y. R., Blue light is a universal signal for Escherichia coli chemoreceptors. J. Bacteriol. 201, e00762-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhaya D., Takahashi A., Shahi P., Grossman A. R., Novel motility mutants of synechocystis strain PCC 6803 generated by in vitro transposon mutagenesis. J. Bacteriol. 183, 6140–6143 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Springstein B. L., et al., Identification and characterization of novel filament-forming proteins in cyanobacteria. Sci. Rep. 10, 1894–1899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakane D., Nishizaka T., Asymmetric distribution of type IV pili triggered by directional light in unicellular cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 114, 6593–6598 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Häder D., Extracellular and intracellular determination of light-induced potential changes during photophobic reactions in blue-green algae. Arch. Microbiol. 119, 75–79 (1978). [Google Scholar]
- 32.Häder D., Electrical and proton gradients in the sensory transduction of photophobic responses in the blue-green alga, Phormidium uncinatum. Arch. Microbiol. 130, 83–86 (1981). [Google Scholar]
- 33.Kirpich J. S., et al., Noncanonical photodynamics of the orange/green cyanobacteriochrome power sensor NpF2164g7 from the PtxD phototaxis regulator of Nostoc punctiforme. Biochemistry 57, 2636–2648 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Jakob A., et al., The (PATAN)-CheY-like response regulator PixE interacts with the motor ATPase PilB1 to control negative phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 61, 296–307 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Campbell E. L., Summers M. L., Christman H., Martin M. E., Meeks J. C., Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 189, 5247–5256 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Splitt S., Risser D., The non-metabolizable sucrose analog sucralose is a potent inhibitor of hormogonium differentiation in the filamentous cyanobacterium Nostoc punctiforme. Arch. Microbiol. 198, 137–147 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Cai Y. P., Wolk C. P., Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172, 3138–3145 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz V. M., et al., IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei T. F., Ramasubramanian T. S., Golden J. W., Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J. Bacteriol. 176, 4473–4482 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Risser D. D., Wong F. C., Meeks J. C., Biased inheritance of the protein PatN frees vegetative cells to initiate patterned heterocyst differentiation. Proc. Natl. Acad. Sci. U.S.A. 109, 15342–15347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battesti A., Bouveret E., The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Wolk C. P., Cai Y., Panoff J. M., Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. U.S.A. 88, 5355–5359 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Bremer H., Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 270, 11181–11189 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Edelstein A. D., et al., Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.