Significance
Meiotic recombination promotes genetic diversity by shuffling parental chromosomes. As observed by the very first geneticists, crossovers inhibit the formation of another crossover nearby, an elusive phenomenon called crossover interference. Another intriguing observation is heterochiasmy, the marked difference in male and female crossover rates observed in many species. Here, we show that the synaptonemal complex, a structure that zips homologous chromosomes together during meiosis, is essential for crossover interference in Arabidopsis. This suggests that a signal that inhibits crossover formation nearby a first crossover propagates along this specific structure. Furthermore, in the absence of the synaptonemal complex, crossover frequencies become identical in both sexes, suggesting that heterochiasmy is due to variation of crossover interference imposed by the synaptonemal complex.
Keywords: crossover, interference, heterochiasmy, synaptonemal complex, meiosis
Abstract
Meiotic crossovers (COs) have intriguing patterning properties, including CO interference, the tendency of COs to be well-spaced along chromosomes, and heterochiasmy, the marked difference in male and female CO rates. During meiosis, transverse filaments transiently associate the axes of homologous chromosomes, a process called synapsis that is essential for CO formation in many eukaryotes. Here, we describe the spatial organization of the transverse filaments in Arabidopsis (ZYP1) and show it to be evolutionary conserved. We show that in the absence of ZYP1 (zyp1a zyp1b null mutants), chromosomes associate in pairs but do not synapse. Unexpectedly, in absence of ZYP1, CO formation is not prevented but increased. Furthermore, genome-wide analysis of recombination revealed that CO interference is abolished, with the frequent observation of close COs. In addition, heterochiasmy was erased, with identical CO rates in males and females. This shows that the tripartite synaptonemal complex is dispensable for CO formation and has a key role in regulating their number and distribution, imposing CO interference and heterochiasmy.
Two particularly prominent phenomena occur in parallel during meiotic prophase: meiotic recombination and chromosome pairing. On the one hand, meiotic recombination generates crossovers (COs) between homologous chromosomes. This is initiated by the formation of numerous DNA double-strand breaks, which are repaired using the homologous chromosome as a template. A small proportion of the breaks mature into COs (1). Two pathways for CO formation have been described. The class I pathway, which accounts for the majority of COs in most examined species, is promoted by a group of proteins called ZMMs (Saccharomyces cerevisiae Zip1 through 4, Msh4 through 5, and Mer3) and the MutL-γ complex (MLH1/3). The presence of class I COs inhibits the formation of additional class I COs nearby along the same chromosome pair, a phenomenon known as CO interference, whose mechanism has been elusive since its first description >100 y ago (2, 3). The second pathway, which accounts for a minority of COs in most species, including Arabidopsis thaliana, does not exhibit CO interference (1). The ZMM pathway is remarkably conserved from budding yeast to mice and A. thaliana with homologs of Zip2, Zip3, Zip4, Msh4, Msh5, and Mer3 being all required for class I CO formation (4).
The second prominent phenomenon of meiotic prophase is association of chromosomes in pairs, which culminates in their synapsis, an intimate juxtaposition in a structure called the synaptonemal complex (SC). The SC, which was described more than 60 y ago, closely opposes homologous chromosomes along their length (100 to 200 nm) (5). This highly conserved tripartite structure is assembled from two axial elements (called lateral elements after synapsis), each anchoring the two sister chromatids of a homolog and a central region (6, 7). Numerous transverse filaments (TFs) span the central region and attach the lateral elements together in a zipper-like structure. TF proteins have been functionally characterized in the fungi S. cerevisiae (Zip1) (8) and Sordaria macrospora (Sme4) (9), the animals mouse (Sycp1) (10, 11), Drosophila (C(3)G) (12), and Caenorhabditis elegans (SYPs) (13–17), and the plants A. thaliana (ZYP1) (18), rice (ZEP1) (19, 20), and barley (ZYP1) (21). Immunolocalization coupled with electron microscopy or super-resolution light microscopy elucidated the conserved organization of the TF in yeast, mammals, C. elegans, and Drosophila (22–29). The TF proteins form parallel dimers through their central coiled-coil region and then align between the axial elements of the chromosomes. The globular carboxyl termini of the dimer associate with the lateral axes of the homologs, whereas the N termini overlap in the central region of the SC, keeping the chromosomes synapsed to each other. TF proteins are associated in many organisms with central element proteins that are located along the center of the SC (Ecm11 and Gmc2 in S. cerevisiae, SYCE1-3 and TEX12 in mammals, and SYP-4 in C. elegans (5, 30–35). In mutants of the TF, synapsis does not occur as defined by close zipping of the chromosome axes, but homologous chromosomes can align at a distance that is less regular and twice or more than in wild-type synapsed chromosomes (8–10, 20).
The role of the TFs in regulating CO formation is still under investigation and appears to be dual, with pro- and anti-CO functions. The S. cerevisiae TF protein Zip1 is one of the originally defined ZMM proteins required for class I CO formation. TF proteins have also been shown to be required for most or all CO/chiasmata formation in S. macrospora, mice, Drosophila, C. elegans, and barley (9, 10, 12, 14–16, 21, 36), suggesting that TFs are universally essential for class I CO formation. Interestingly, a separation-of-function allele of Zip1 (zip1-N1) in S. cerevisiae is defective in tripartite SC assembly but makes class I COs (37), suggesting that the TF itself, but not the tripartite SC, is a prerequisite for class I CO formation. Actually, this S. cerevisiae zip1-N1 mutation, as well as a moderate decrease of SYP-1 levels in C. elegans, has the opposite effect compared to the null mutants, with an increase of COs and attenuated interference (37, 38). In rice, four zep1 mutants generated by tos17 insertions were described (20). One of them, which is clearly hypomorphic as it shows residual loading of ZEP1 on chromosomes, has increased CO numbers and reduced CO interference (19). The three other zep1 alleles have increased COs, but it is unclear if these alleles are complete loss of function or not (20). These data suggest that TFs participate in CO interference and limit CO numbers.
In A. thaliana, the TF is encoded by a tandemly duplicated gene, making its functional analysis difficult with classical genetics tools. ZYP1 RNA interference lines showed a minor decrease in chiasma number (18), raising the intriguing possibility that TFs are not essential for CO formation in this species. Here, we used genome editing to reexplore the function of TFs in A. thaliana. We generated a series of null zyp1a zyp1b double mutants and demonstrated that ZYP1 and the tripartite SC are dispensable for CO formation. We observed the opposite behavior, with zyp1 mutants showing an increased CO number and abolished CO interference. Furthermore, the marked difference between male and female CO rates observed in the wild type is erased in the zyp1 mutant. Altogether, this supports the conclusion that the SC limits COs and mediates both CO interference and heterochiasmy.
Results
Organization of ZYP1 within the SC.
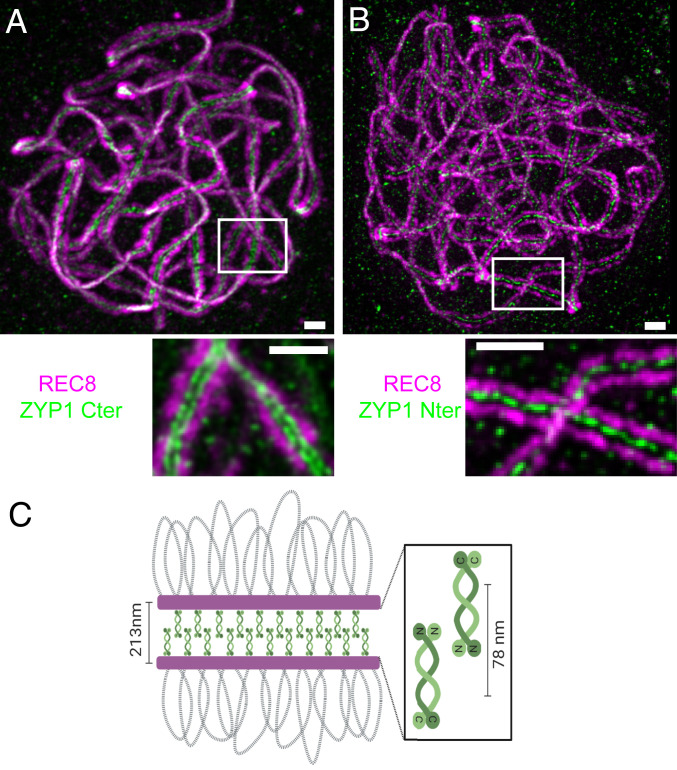
The TF proteins were shown to have a similar organization in budding yeast and animals, with their globular carboxyl termini associated with the lateral axes of the homologs and the N termini overlapping in the central region of the SC (22–28). In A. thaliana and closely related species, the TF element is encoded by a duplicated gene, ZYP1A and ZYP1B (87% identity of the encoded proteins; 871 and 856 aa, respectively). We used stimulated emission depletion (STED) microscopy and antibodies raised against the REC8 cohesin, which is a component of the lateral element, and two antibodies raised against two different parts of ZYP1A/B to explore their arrangement within the SC in A. thaliana. At pachytene, REC8 formed two parallel lines separated by a distance of 213 ± 17 nm (average ± SD, measured between the signal peaks, Fig. 1). Staining with an antibody raised against the amino acids 422 to 845 (total length = 871 aa) of ZYP1A and that recognize both ZYP1A and ZYP1B (18) revealed two parallel lines separated by a distance of 78 ± 8 nm running between the REC8 lines (Fig. 1A). In contrast, localizing ZYP1 with an antibody raised against the N terminus (amino acids 34 to 45) of ZYP1A/B revealed a single line running in the middle of the two REC8 signals (Fig. 1B). This shows that the general organization of the TF is conserved in plants, with the carboxyl terminus orientated toward the lateral elements and the N terminus lying in the central region (Fig. 1C).
Fig. 1.
Immunolocalization of the C and N termini of ZYP1 on male meiocytes. A double immunolocalization was performed against REC8 (purple) and ZYP1 (green). The ZYP1 antibody was raised against either the carboxyl (A) or N terminus (B) of the protein. The images were acquired with STED microscopy. The maximum intensity projection is shown. The complete series of plans is shown in Movies S1 and S2. (Scale bar, 0.5 µm.) (C) A schematic representation of the SC organization.
Generation of a Series of zyp1 Mutants Using Cas9.
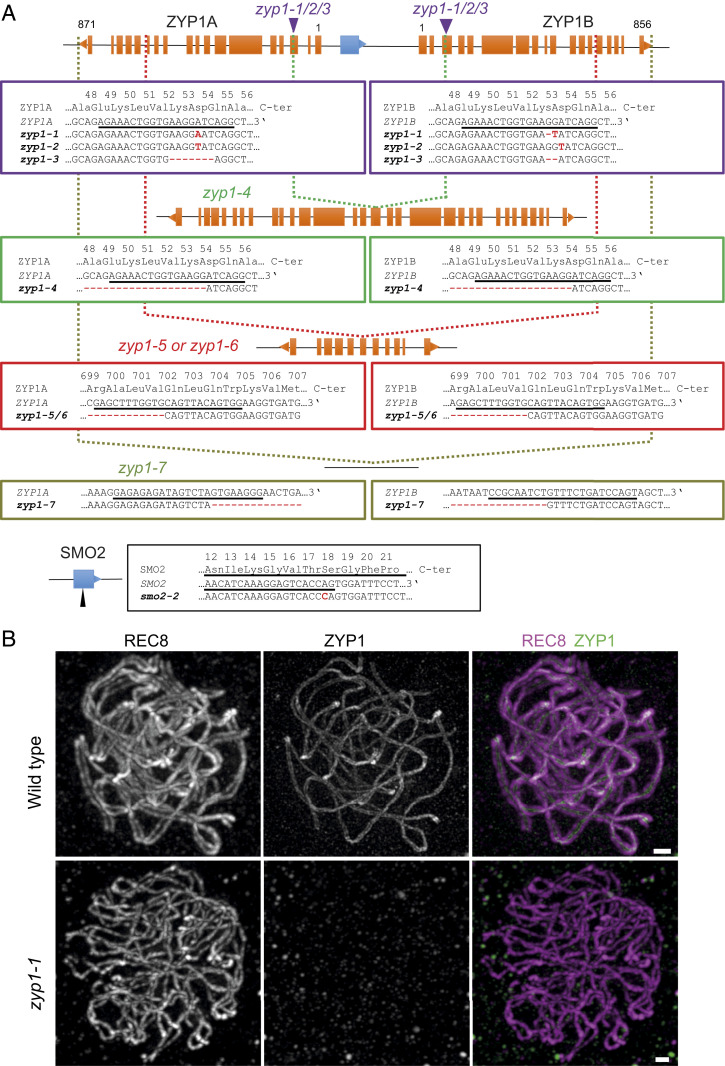
The ZYP1A and ZYP1B genes (AT1G22260 and AT1G22275) are arranged in a divergent tandem duplication (Fig. 2A) (18). The two start codons are separated by 2,102 base pairs (bp) that contain the two ZYP1 promoter regions in diverging directions and the SMALL ORGAN 2 gene ([SMO2], AT1G22270), which is required for normal growth and fertility (39). We generated a smo2 mutant, with a 1-bp insertion provoking a frameshift at codon 17 (smo2-2, Fig. 2A), and confirmed its reduced growth and strongly reduced fertility (Fig. 3). Examination of meiotic chromosome spreads did not reveal any meiotic defects in smo2-2 (SI Appendix, Fig. S1), suggesting that smo2 does not affect meiosis and that the origin of the quasisterility is postmeiotic.
Fig. 2.
Identification of a series of zyp1a zyp1b double mutants. (A) A diagram of the ZYP1 locus, showing the exon organization of the ZYP1A, ZYP1B, and SMO2 genes. The orientations of the genes are indicated by a triangle at the end. The two purple arrows show the positions of the mutations in zyp1-1, zyp1-2, and zyp1-3. The green, red, and khaki vertical dotted lines show the position of the deletions in zyp1-4, zyp1-5/6, and zyp1-7, respectively. (B) ZYP1 is not detected in zyp1-1 mutant. A double immunolocalization was performed against REC8 (purple) and ZYP1 (green). The images at pachytene were acquired with STED microscopy. Maximum intensity projection: The individual plans are shown in Movies S3 and S4. (Scale bar, 0.5 µm.)
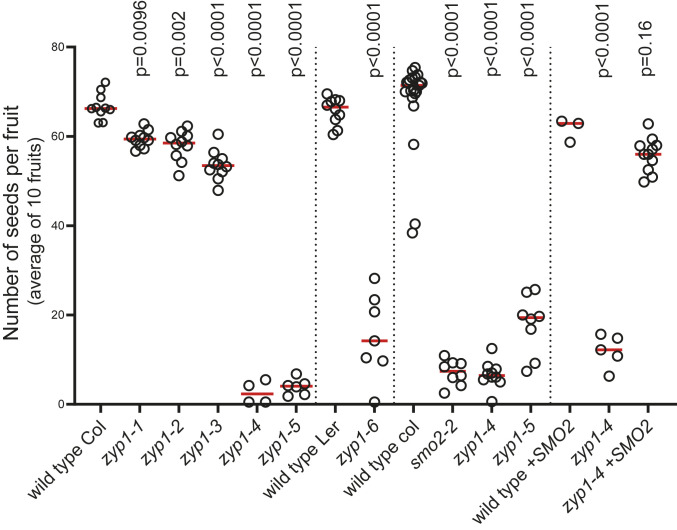
Fig. 3.
Analysis of fertility of zyp1 mutants. Each dot represents the fertility of an individual plant, measured as the number of seeds per fruit averaged on 10 fruits. The red bar shows the mean for a given genotype. The vertical lines separate independent experiments. In each experiment, all plants have been grown in parallel, and the wild-type controls are siblings of the mutants. The tests are one-way ANOVA followed by Fisher’s least significant difference (LSD) test compared with the respective wild-type control.
The single zyp1a and zyp1b mutants have normal synapsis and both proteins localize in the SC, showing that they are redundant (18). The close proximity of the two genes prohibited the generation of double mutants with classical genetics. Here, we produced a series of zyp1a zyp1b double mutants (hereafter called zyp1 for simplicity) with CRISPR-Cas9. Using a guide RNA targeting an identical site in ZYP1A and ZYP1B (underlined sequences in Fig. 2A), three independent double mutations were identified with various small insertions/deletions provoking frameshifts at codon Asp54 or Lys53 (zyp1-1, zyp1-2, and zyp1-3), as well as a line with a complete deletion between the two loci (zyp1-4), deleting the beginning of the ZYP1 genes (the first intact codon being Gln55 in both genes), the promoter regions of both genes, and SMO2. Using a guide further downstream in the coding region, a larger deletion was obtained (zyp1-5), deleting SMO2 and the ZYP1 promoter regions and most of the ZYP1 coding regions up to Val702 for both genes. Finally, using two guides in the 3′ untranslated region of the ZYP1 genes, we generated a complete deletion of the locus, where both ZYP1 genes and SMO2 are entirely absent (zyp1-7). While all of these alleles were obtained in the Col-0 strain, we produced an additional allele (zyp1-6) in the Ler strain with the exact same deletion as zyp1-5. Immunolocalization of ZYP1 (Cter) failed to detect any signal beyond background on all tested mutants (zyp1-1, zyp1-2, zyp1-3, zyp1-5, and zyp1-7), suggesting that all of the alleles are null (Fig. 2B).
ZYP1 Is Not Essential for CO Formation and Meiosis Completion.
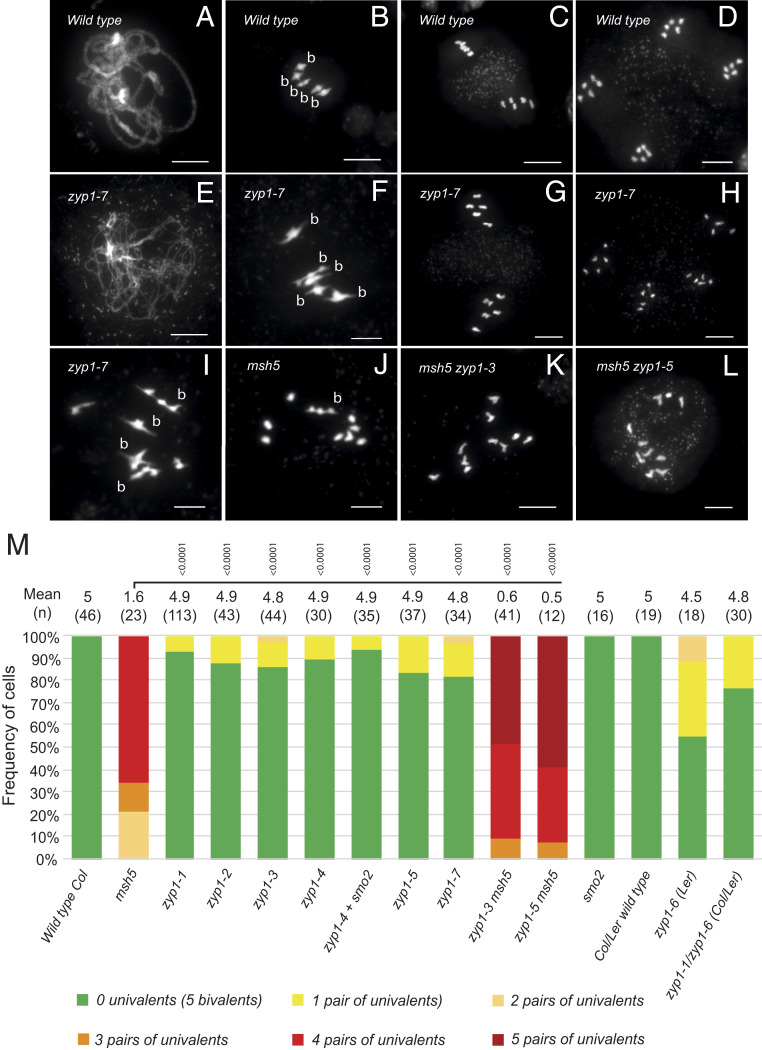
We then explored the meiotic behavior of this zyp1 allelic series. Chromosome spreads of male meiocytes stained with DAPI failed to identify classical pachytenes among prophase stages in all mutants (e.g., compare Fig. 4 A–E), suggesting that synapsis does not occur (see also below). However, ∼90% of the cells at metaphase I and diakinesis had five bivalents (Fig. 4 F and M), sharply contrasting with the strong defect in bivalent formation observed in msh5 (Fig. 4J) and all the other zmm mutants (msh4, shoc1/zip2, hei10, zip4, and mer3), in which class I COs are abolished (40–45). One or two pairs of univalents were observed in ∼10% of the cells at metaphase I (Fig. 4 I and M), showing that not all chromosomes receive the obligate CO in zyp1 mutants. Consistently, chromosome segregation was slightly affected in zyp1 with 17% (zyp1-1 2/24; zyp1-7 9/52) of metaphase II showing 6:4 chromosome distribution (compare Fig. 4 C–D to Fig. 4 G–H). This shows that CO and functional chiasmata, which are required to connect homologous chromosomes and their balanced segregation at meiosis I, are produced at high levels in the zyp1 mutants. In the msh5 zyp1 double mutant, bivalent formation was almost entirely abolished (Fig. 4 K–M), showing that the vast majority of COs that are produced in zyp1 mutants are class I/ZMM-dependent COs. This demonstrates that ZYP1, contrary to the ZMMs, is not required for class I CO formation but required for full implementation of the obligate CO. The number of bivalents in msh5 zyp1 is lower than in msh5 (Mann–Whitney U test, P < 10−4), suggesting that ZYP1 and/or the SC facilitates the formation of class II COs.
Fig. 4.
Chromosome spreads of male meiocytes. (A–D) Wild type. (A) Pachytene, (B) Metaphase I with 5 bivalents, (C) Metaphase II, and (D) telophase II. (E–I) zyp1-7. (E) Prophase I, (F) Metaphase I with five bivalents (B), (G) Metaphase II, (H) Telophase II, and (I) Metaphase I with four bivalents and one pair of univalents. (J) Metaphase I in msh5, (K) Metaphase I in msh5 zyp1-3, and (L) Metaphase I in msh5 zyp1-5. (Scale bar, 10 µm.) (M) Quantification of bivalents at metaphase I. Cells were categorized according to the number of pairs of univalents/bivalents. The average number of bivalents per cell and the number of analyzed cells are indicated above the bar. The tests are one-way ANOVA followed by Fisher’s LSD test compared with msh5.
The Absence of ZYP1 Only Marginally Reduces Fertility.
The zyp1-1, zyp1-2, and zyp1-3 mutants that contain frameshifts early in the ZYP1 genes showed fertility close to wild type with a modest (∼10%) but significant reduction in seed number (Fig. 3). The zyp1-4 to zyp1-7 mutants, which contain large deletions and lack the SMO2 gene, showed a reduced fertility similar to the single smo2 mutants. Furthermore, introducing a SMO2 genomic fragment in zyp1-4 restored fertility to the zyp1-1/zyp1-2/zyp1-3 levels. This shows that the absence of the TF element in A. thaliana only marginally reduces fertility. The low frequency of univalents and the associated frequency of nondisjunction is presumably at the origin of the mild reduction of fertility provoked by the zyp1 mutations.
In zyp1, Alignment Occurs, but Synapsis and ASY1 Remodeling Do Not.
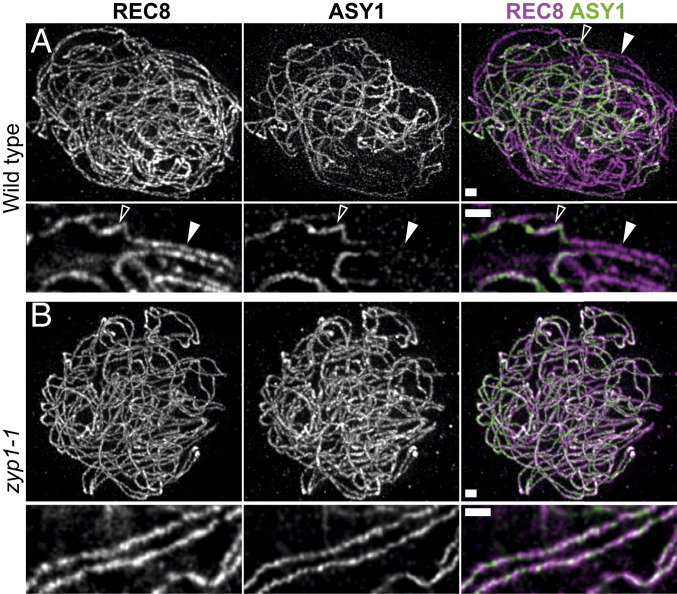
In the wild type, ZYP1 polymerizes between chromosome pairs, juxtaposing the two axes at ∼200 nm (see above, Fig. 1). We performed immunolocalization of REC8 and the HORMA domain meiotic protein ASY1 on male meiocytes (Fig. 5). In the zyp1 mutants, juxtaposition of the two REC8 axes at ∼200 nm, which is a hallmark of synapsis, was not observed (n = 90). However, we observed alignment of chromosome pairs all along their length at an increased and more variable distance of 415 ± 109 nm (average ± SD; Fig. 5B and Movie S1), showing that paring occurs. On wild-type chromosomes, as previously shown (46, 47), the ASY1 signal vanishes concomitantly with synapsis (Fig. 5A). In contrast, in the zyp1-1 mutants, ASY1 and REC8 both decorated chromosome axes on all prophase cells (n > 50) (Fig. 5B). Altogether, this shows that pairing occurs in the absence of ZYP1 but that the close axes juxtaposition that defines synapsis and the associated ASY1 remodeling are lost.
Fig. 5.
REC8 and ASY1 localization in wild type and zyp1. A double immunolocalization was performed against REC8 (purple) and ASY1 (green). The images were acquired with STED microscopy. In wild-type zygotene (A), the ASY1 signal (green) is associated with unsynapsed axes. When the two REC8 axes (purple) are synapsed (aligned at ∼200 nm), the ASY1 signal is barely detectable. The open and closed arrows show unsynapsed and synapsed axes, respectively. (Scale bar, 0.5 µM.) In the zyp1 mutant (B), synapsis is not observed, but axes are paired with a loose alignment at ∼400 nM. See Movie S6 to visualize the pairing more clearly. The ASY1 signal is still present on aligned chromosomes. Maximum intensity projection: The individual plans are shown in Movies S5 and S6. (Scale bar, 0.5 µm.)
Cytological Markers of Class I COs Are Increased in zyp1.
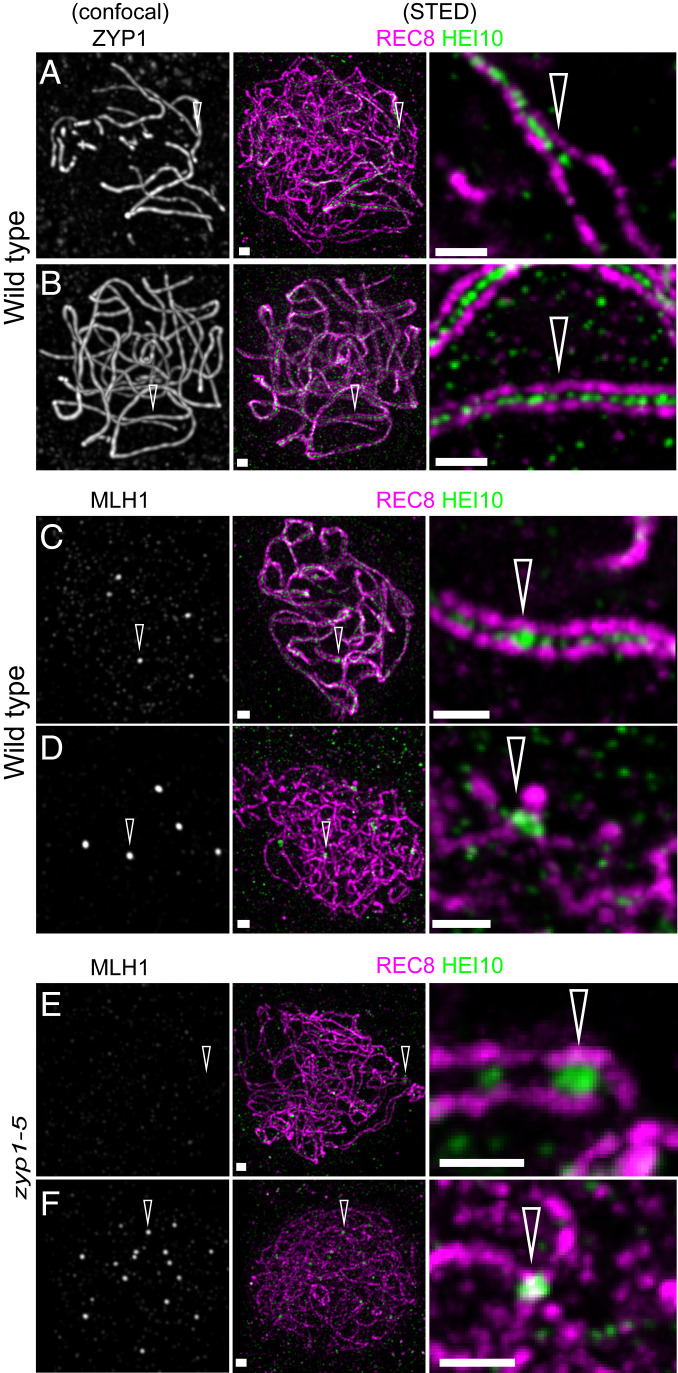
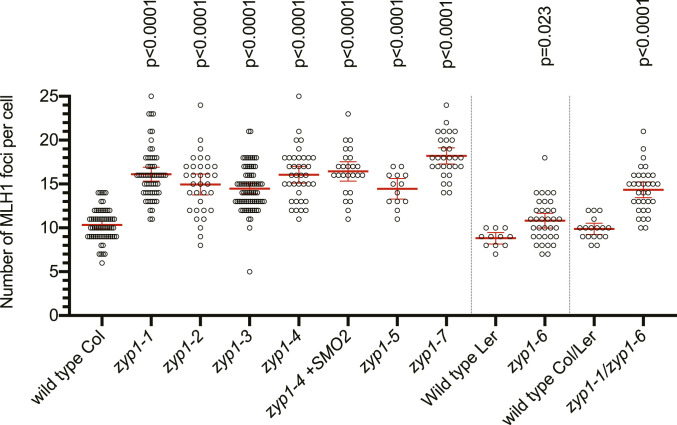
In the wild type, HEI10 is a classical marker of the progression of class I CO formation (45, 47). Indeed, HEI10 forms a hundred foci associated with the central element of the SC at zygotene and early pachytene (Fig. 6 A and B) and progressively concentrates into about 10 larger foci that colocalize with MLH1 (Fig. 6 C and D), specifically marking class I CO at late pachytene, diplotene, and diakinesis (45, 47). In contrast, the numerous HEI10 foci observed in the center of the SC in wild type were not detected in zyp1. However, large HEI10 foci were observed to form between the two axes (Fig. 6E). In cells presumably at a later stage, the large HEI10 foci become brighter and colocalize with MLH1 foci (Fig. 6F), suggesting that recombination progresses until maturation of COs in absence of the TF and synapsis and the associated numerous HEI10 foci. Furthermore, quantification of MLH1–HEI10 cofoci on male meiocytes (Fig. 7) revealed an increase of ∼50% of their number in all zyp1 alleles in the Col strain, +23% in the Ler zyp1-6 allele, and +45% in the zyp1-1/zyp1-6 hybrid, compared to their respective wild-type controls. This suggests that ZYP1 and/or the tripartite SC limit class I CO formation.
Fig. 6.
HEI10 localization in wild type and zyp1. A triple immunolocalization was performed against REC8 (purple), HEi10 (green), and either ZYP1 (A and B) or MLH1 (C–F) on male meiocytes of wild type (A–D) and zyp1-5 (E and F). REC8 and HEi10 were imaged with STED, while ZYP1 and MLH1 were imaged with confocal microscopy. (A) Wild-type zygotene nucleus: The arrow points at the synapsis forks. (B) Wild-type pachytene. (C) Wild-type late pachytene nucleus: The arrow points at MLH1-HEI10 foci. (D) Diplotene: The arrow points at MLH1–HEI10 foci. (E) Prophase stage with forming HEI10 foci, without corresponding MLH1 foci (arrow). (F) Prophase stage with HEI10–MLH1 foci (arrow). (Scale bar, 0.5 µm.)
Fig. 7.
MLH1–HEI10 foci are increased in zyp1 mutants. MLH1–HEI10 foci were quantified following a triple immunolocalization REC8–MLH1–HEI10 performed on male meiocytes and imaged with an epifluorescence microscope. Each dot is an individual cell, and the red bar is the mean. Tests are one-way ANOVA followed by Fisher’s LSD test, compared with the respective wild-type control. Fitting of the MLH1 number distribution to a Poisson distribution is shown in SI Appendix, Fig. S2.
COs Are Increased in Male and Female zyp1 Meiosis.
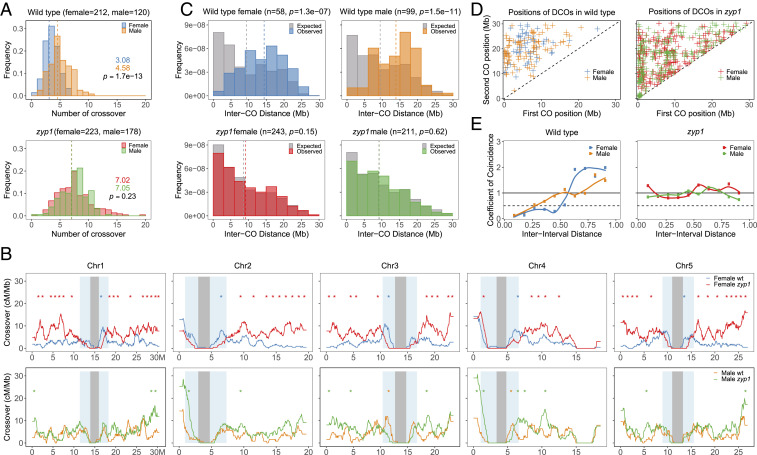
The increased number of HEI10/MLH1 foci in male meiosis suggested an increase in COs. To measure COs genetically, we produced F1 hybrids carrying two nonfunctional ZYP1 alleles, zyp1-1 (Columbia Col strain) and zyp1-6 (Landsberg Ler strain). These F1s and sibling wild-type controls were crossed as male or female to wild-type Col, and the progeny obtained was sequenced for CO analysis (wild-type female n = 212, wild-type male n = 120, zyp1 female n = 223, and zyp1 male n = 178). Note that because a given CO involves only two of the four chromatids of a bivalent and that a gamete inherits a single chromatid, the number of COs observed per gamete is on average one-half of the number of COs per meiocyte. In males, the average number of COs per gamete was increased from 4.58 ± 1.9 (mean ± SD) in wild type to 7.05 ± 2.2 in zyp1 (Fig. 8A), a 54% increase in observed COs, which is very close to the 45% increase of HEI10–MLH1 foci in male meiocytes in this background. In female wild type, the average number of COs was 3.08 ± 1.4 per gamete, which is markedly lower than in male wild type as previously reported (48), a phenomenon called heterochiasmy. Remarkably, the number of COs in female zyp1 was more than double compared to wild type (7.05 ± 3.1) and reached the same level as seen in male zyp1. Thus, zyp1 mutation increases COs in both sexes but disproportionally affects female recombination, erasing the heterochiasmy observed in wild type, suggesting that heterochiasmy is regulated by the TF.
Fig. 8.
Analysis of CO distribution in male and female zyp1. COs were detected following whole-genome sequencing of male and female backcrosses. (A) Distribution of CO number per gamete. The number of analyzed samples is indicated in brackets. The mean CO number per gamete is given and indicated by a dashed line. The difference was assessed by a two-sided Mann–Whitney test. The CO distribution per chromosome and fitting to a Poisson distribution is shown in SI Appendix, Fig. S3. (B) The distribution of COs along the five chromosomes. The centromere and pericentromeric regions are indicated by gray and blue shading, respectively. Analysis is done with 1-Mb windows and 50-kb sliding steps. Intervals with significant difference between wild type and mutants are indicated by stars (P < 0.05, not overlapping 1-Mb windows, Chi2 test, without correction for multiple testing). (C) Distribution of inter-CO distances for chromosomes having exactly two COs. The gray bars represent the expected distribution of COs in absence of interference, as calculated by permuting the CO positions between gametes. The number of analyzed events and the Mann–Whitney U test comparing observed and expected distributions are indicated in brackets. (D) The positions of first and second COs for double-CO pairs, according to their physical distance. By construction, close CO pairs appear next to the diagonal, and distant COs appear in the top left corner. (E) The CoC Curves. Chromosomes were divided into 11 intervals, and the mean CoC was calculated for pairs of intervals separated by a certain distance (proportion of chromosome length).
In addition to an increased number, CO distribution is also modified in zyp1 with COs occurring more distally than in wild type (Fig. 8B): In female zyp1 meiosis, CO numbers are significantly higher than wild type along chromosome arms and distal regions (Fig. 8C) (1-Mb intervals where recombination is significantly higher in zyp1 are indicated by red stars). In contrast, in intervals directly adjacent to centromeres, recombination is lower in female zyp1 than female wild type (intervals with significantly lower recombination in zyp1 are indicated with blue stars). In male meiosis, where the global CO increase induced by zyp1 is less marked than female, fewer individual intervals are significantly different, but the trend is similar, with recombination being significantly increased in arms and distal regions (green stars) and decreased in proximal intervals (yellow stars). This suggests that ZYP1, while globally limiting COs, favors the formation of COs in proximal regions. This contrast with the zip1 mutant in yeast where COs are increased in pericentromeric regions (49, 50).
CO Interference Is Abolished in zyp1 Meiosis.
To analyze interference in zyp1, we first looked at the distribution of the distance between two COs that occurred on the same chromosome. In wild type, the distributions are shifted to large distances compared to the expected distribution of independent COs, which is a manifestation of CO interference (Fig. 8C). In contrast, in male and female zyp1, the observed distributions of inter-CO distances are not different from the expected distribution in absence of CO interference, with notably numerous pairs of COs at a distance of less than 5 Mb that are rare in wild type. This can also be illustrated by plotting the position of the two COs as the X and Y positions on a two-dimensional plot (Fig. 8D). The lack of events close to the diagonal, as observed in the wild type, reflects the absence of close double COs. In contrast, close double COs (close to the diagonal) are numerous in both male and female zyp1, suggesting a diminution of CO interference. Interference can be accurately analyzed by a coefficient of coincidence (CoC) analysis, which divides the observed frequency of COs occurring concomitantly in two intervals by the expected frequency according to the frequency of COs in each interval (51, 52). A CoC of 1 means that COs are distributed independent of one another. A CoC close to 0 reveals an absence of double COs and thus of CO interference. The CoC is then plotted versus the distance between the two considered intervals (Fig. 8E). In wild-type A. thaliana, CoC curves are below 1 for short distances in both male and female, confirming the presence of CO interference, and then goes at 1 or above 1 for larger distances (∼50% of the chromosome length) where interference effects decline, as previously shown (Fig. 8E) (53, 54). In contrast, the CoC curves are flat, close to 1 for adjacent and distant pairs of intervals, for both male and female zyp1, suggesting that CO interference is abolished in absence of ZYP1.
Discussion
The synaptonemal complex is a prominent structure at meiosis, but its function is still under debate more than 60 y after its discovery (55). The TF of the SC has been shown to be required for class I CO formation in species as diverse as budding yeast, mice, S. Macrospora, Drosophila, C. elegans, and barley (8, 10, 11, 14–16, 21). It should be noted that the SC is not essential for all classes of COs, as, for example, Schizosaccharomyces pombe completely lacks both the SC and the class I pathway but produces COs by an alternative pathway (class II COs) (56). However, the SC seemed intimately associated with the class I pathway, which is dependent on the ZMMs that include the TF protein Zip1 (57). The role of the ZMMs homologs is remarkably conserved, and all, apart from ZYP1, were shown to be required for class I CO formation in A. thaliana (40–45). Here, we showed that, in the absence of ZYP1 in A. thaliana, synapsis does not occur, but class I COs (MSH5 dependent and marked by MLH1) are produced at high levels. This shows that neither ZYP1 nor synapsis is essential for class I CO maturation and suggests that the transverse element of the SC should not be considered as a universal pro-CO factor like the other ZMMs. The rice TF could be also dispensable for class I CO formation, as four zep1 alleles show high levels of COs. However, in contrast to our A. thaliana mutant series that includes a complete deletion, it is unclear if any of the rice mutants are null, as the four were generated by tos17 insertions and at least two show residual ZEP1 expression (20, 21).
One possible function of the tripartite SC is to provide a chromosome spatial arrangement that would be required for class I CO maturation and chiasmata formation (5), but our results argue against such a function. Indeed, the TF is dispensable for class I CO formation in A. thaliana and potentially in rice. This suggests that the major function of ZYP1 homologs in promoting CO formation could be independent of the SC. In favor of this hypothesis, budding yeast Zip1 contributes to CO formation independently of synapsis (36, 37, 58). This synapsis-independent pro-CO activity of the TF could be dispensable or ensured by alternative mechanisms in A. thaliana. This might also be related to the observation that ZMM proteins are not required for synapsis in A. thaliana (40–45).
The mechanism of CO interference remains elusive despite a century-old initial description (2). A current model postulates that a signal propagates from an initial designated CO along the chromosomes, preventing the designation of other COs nearby (59). The nature of the hypothesized signal is still mysterious and could be of physical (e.g., release of tension) (38, 60) or chemical nature (e.g., a wave of protein modification) (61). An important question is the scaffolding along which that signal propagates. While it seems established that the chromosome axes propagate CO interference (1, 3, 5, 62, 63), the requirement of a tripartite SC (synapsis) is a matter of debate. In S. cerevisiae, the decision of whether or not to make a CO, and thus interference, is made very early, before synapsis (36, 64–66). Cytological markers of COs still display interference in the zip1 mutant (67), and some mutants defective in SC extension still show wild-type interference (68). Altogether, this strongly suggests that CO interference is established before and independently from synapsis in S. cerevisiae. However, in the zip1-N1 and central element mutants, where synapsis is abolished, CO interference is still present but weakened, suggesting that synapsis may contribute to CO interference in S. cerevisiae (37). In C. elegans and rice, a partial depletion of the TF protein level led to an excess of CO formation and diminished CO interference (20, 38), strongly supporting a significant role of the tripartite SC in CO interference in these two species. Our conclusion that class I COs occur in the absence of ZYP1 and synapsis in A. thaliana offers an unprecedented opportunity to explore the role of ZYP1/synapsis in CO interference. We showed that, in zyp1 mutants, class I CO number is increased, and CO interference is no longer detected. This differs from other described mutants, in which the alternative noninterfering class II COs are enhanced, leading to an overall decrease of the observed interference (54, 69–72). We cannot exclude that some low level of CO interference is present in the A. thaliana zyp1 mutant, but it is dramatically reduced compared to the wild type, if not abolished. This suggests that the tripartite SC has a central role in CO interference in A. thaliana. It is therefore possible that the CO-discouraging signal, which propagates from a CO site and prevents other COs nearby, propagates along the axial element in some organisms while requiring a fully assembled SC in others, such as A. thaliana. Alternatively, the perception of the CO-discouraging signal and its translation into the formation of non-COs could be dependent on synapsis in some organisms but not in others.
While interference appears abolished in zyp1, the number of COs is still limited to ∼14 per meiosis on average, suggesting that CO interference is not the sole mechanism that limits class I CO. The existence of a limiting pro-CO factor would also provide a ground for the loss of the obligate CO in zyp1. Under this hypothesis, the loss of CO interference would allow COs to form in excess on some chromosomes, making the pro-CO factor unavailable for other chromosomes. Alternatively, homologous chromosomes might not be aligned enough to produce recombination products when there is a lack of a stable physical interaction provided by the SC. This would be independent of the transmission of CO interference and account for the lack of the obligatory CO.
The variation in recombination rate between sexes—heterochiasmy—is observed in many eukaryotes, with either male or female having higher levels and the ratio being a fast-evolving trait (73). Interestingly, it is also observed in hermaphrodite species, such as A. thaliana, in which recombination levels are markedly higher in male meiocytes than female meiocytes (48, 74), despite having an identical genetic makeup as cells of a single organism. The mechanism of heterochiasmy is elusive, but a striking covariation has been observed between SC length and recombination rates (75), notably when comparing male/female in species as diverse as humans (76, 77), mice (78), zebrafish (79), planarian (80), and A. thaliana (81). One proposed mechanism suggests that interference propagates with the same properties along the axis/SC in males and females, but because the SC is shorter in females, the interference propagates on a larger part of the genome and reduces CO number (75, 81). This is supported by the observation that class II COs, which are insensitive to CO interference, are not subjected to heterochiasmy in A. thaliana (54). One key prediction of this model is that the abolition of CO interference would erase heterochiasmy. Our observation that both CO interference and heterochiasmy are abolished in the zyp1 mutant supports the conclusion that a difference of the effect of CO interference is responsible for heterochiasmy.
The possibility of manipulating CO frequency is of interest for plant breeding (82). Mutation of the anti-CO genes RECQ4, FANCM, and FIGL1 increases class II CO, while over expression of HEI10 increases class I CO (54, 83). To date, the manipulation of RECQ4 is the most effective strategy to increase CO in both model and crop species (84). The identification of ZYP1 as an anti-CO factor in both A. thaliana and rice adds a tool to the box, and it would be of interest to test the effect of the manipulation on ZYP1—alone and in combination—in more crop species.
Materials and Methods
Plant Materials and Growth Conditions.
A. thaliana plants were grown in greenhouses or growth chambers (16-h day/8-h night, 20 °C). Wild-type Col-0 and Ler-1 are 186AV1B4 and 213AV1B1 from the Versailles A. thaliana stock center (http://publiclines.versailles.inra.fr/). The msh5-2 (N526553) mutant was previously described (44). Genotyping was performed by PCR (Dataset S1).
Generation of Mutants.
Guide RNAs targeting ZYP1 or SMO2 genes were designed with the TEFOR website (http://crispor.tefor.net). The guide RNAs were synthetized with U6 promotor (Dataset S1) and Gateway recombination sites and inserted in the pDE–Cas9–DSred vector (85, 86). Transformations were performed with floral dipping (87). Plant transformants (T1) were selected by seed fluorescence, and young plantlets were treated with heat cycles to increase mutagenesis efficiency (88). T2 seeds without fluorescence were selected and screened for mutations by PCR and Sanger sequencing of the targeted locus.
Cytology.
Chromosome spreads were performed as previously described (89). Immunolocalization was performed on cells with preserved three-dimensional structures as described in ref. 47 with modifications: Sepals and petals were removed from 0.35- to 0.45-mm flower buds and collected in buffer A (80 mM KCl, 20 mM NaCl, 15 mM Pipes–NaOH, 0.5 mM ethylene glycol tetraacetic acid (EGTA), 2 mM ethylenediaminetetraacetic acid (EDTA), 80 mM sorbitol, 1 mM dithiothreitol (DTT), 0.15 mM spermine, and 0.5 mM spermidine) and fixed by incubation in bufferA +2% formaldehyde for 25 min under vacuum. Buds were then washed in buffer A for 10 min and digested at 37 °C for 40 min (0.3% cellulase, 0.3% pectolyase Y23, 0.3% driselase, and 0.1% sodium azide in citrate buffer). After a wash in buffer A, digested buds were kept in buffer A on ice. To make the embedding, five to eight buds were placed in 12 µl of buffer A on an 18 mm × 18 mm high-precision coverslip, and anthers were dissected and squashed to extrude meiocytes.
A 6-µl drop of activated polyacrylamide solution (25 µl 15% polyacrylamide [SIGMA A3574] in buffer A + 1.25 µl of 20% sodium sulfite + 1.25 µl of 20% ammonium persulfate) is added to the meiocytes, and a second coverslip is placed on the top with gentle pressure. The polyacrylamide gels were left to polymerize for 1 h, and then the two coverslips were separated. The coverslips covered by a gel pad were incubated in 1× phosphate-buffered saline (PBS), 1% Triton X-100, and 1 mM EDTA for 1 h with agitation, followed by 2 h in blocking buffer (3% bovine serum albumin in 1× PBS + 0.1% Tween 20) at room temperature. Coverslips were then incubated with 200 µl of primary antibody in blocking buffer at 4 °C in a humid chamber for 48 h. Coverslips were washed four times for 30 min with 1× PBS, 0.1% Triton X-100. A total of 100 µl of the appropriate fluorophore-conjugated secondary antibodies in blocking buffer were applied (1:250) and incubated at room temperature for 2 h in the dark. Gels were washed four times for 20 min with 1× PBS, 0.1% Triton X-100. A total of 15 µl of Vectashield + 10 µM DAPI (for the epifluorescence microscopy) or SlowFade Gold (for super-resolution microscopy) was used for mounting the coverslip with a slide that was sealed with nail polish.
The primary antibodies used were as follows for both epifluorescence and super-resolution microscopy: anti-REC8 raised in rat (90) (laboratory code PAK036, dilution 1:250), anti-ASY1 raised in rabbit (91) (PAK006, 1:1,000), anti-MLH1 raised in rabbit (92) (PAK017, 1:200), anti-HEI10 raised in chicken (PAK046, 1:10,000) (45), and anti-ZYP1A carboxyl terminus raised in rabbit (18) (PAK042, 1:500). The anti-ZYP1N was raised against the peptide DSVSSGSFSNLKTA in guinea pig and affinity purified (Eurogentec speedy program) (PAK053, 1:500). Secondary antibodies were conjugated with Alexa 488, Alexa 568, and Alexa 647 for epifluorescence and confocal microscopy and Abberior StarRed and STAROrange for STED microscopy. Images for MLH1–HEI10 cofoci analysis were taken with a Zeiss Axio Imager Z2 epifluorescence microscope and deconvolved and analyzed with Zen 3.2 Lite. Super-resolution images were acquired with the Abberior instrument facility line (https://abberior-instruments.com/) using 561- and 640-nm excitation lasers (for STAR Orange and STAR Red, respectively) and a 775-nm STED depletion laser. Confocal images were taken with the same instrument with a 485-nm excitation laser (for Alexa 488). Images were deconvolved with Huygens Essential version 20.04 (Scientific Volume Imaging, https://svi.nl/) using the classic maximum likelihood estimation algorithm with lateral drift stabilization; signal-to-noise ratio: 7 for STED images and 20 for confocal images, 40 iterations, and quality threshold of 0.5. Maximum intensity projections and contrast adjustments were done with Huygens Essential.
CO Analysis.
Plants heterozygous for the zyp1-1 mutation (Col) were crossed as female with plants heterozygous for the zyp1-6 mutation (Ler). Wild type and zyp1-1/zyp-6 plants were selected among the F1s and crossed as male or as female with wild-type Col. Leaf samples from the four obtained back-cross populations were used for DNA purification and library preparation (93) for Illumina sequencing(HiSeq 3000 2 × 150 base pairs [bp]), performed by the Max Planck-Genome-center (https://mpgc.mpipz.mpg.de/home/). CO analysis suggests that a genome rearrangement is present on chromosome 4 (∼15 to 17 Mb, see Fig. 8B) that was not seen in previous analysis using the same strains. This rearrangement may have been produced during the genome editing procedure or be previously present in either our Col or Ler wild-type strain.
The whole-genome resequencing datasets of A. thaliana Col (SRX202246) and Ler (SRX202247) were downloaded from the National Center for Biotechnology Information Sequence Read Archive database (94). The raw reads were evaluated for quality by using FastQC version 0.11.9 (95), and then potential adapter sequences were trimmed and low-quality bases were filtered using Trimmomatic version 0.38 (96). Then, paired-end reads were aligned to the A. thaliana Col TAIR10 reference genome (97) using Burrows–Wheeler Aligner version 0.7.15-r1140 (98) with default parameters, and those with mapping quality larger than 20 were considered as uniquely mapped and were used in subsequent analysis. inGAP (99) and inGAP-sv (100) were used to identify single-nucleotide polymorphisms (SNPs) and structural variations from resequencing datasets. In addition, the whole-genome assembly of A. thaliana Ler was downloaded from 1,001 A. thaliana Genomes database (https://1001genomes.org/data/MPIPZ/MPIPZJiao2020/releases/current/) (101). SyRI version 1.2 (102) was used to detect SNPs in the syntenic region from the genome assembly of Ler. To obtain high-confidence SNP markers between Col and Ler, we collected both of SNPs identified by these two strategies and filtered using the methods described by Qi et al. (103). Finally, we obtained a set of 620, 115 high-confident SNP markers for subsequent analysis.
The raw reads of sequenced samples (with a mean depth of 1.6×) were processed by the same methods of quality control and alignment. The read count and genotype profile for SNPs were generated by inGAP (99) and used for CO identification. We employed a sliding-window–based method to identify COs across samples, with a window size of 70 kb and a sliding size of 35 kb. The genotype of each sliding window was defined as the genotype with the highest probability, calculated using a binomial model with an error rate of 0.001 substitutions per nucleotide. A CO breakpoint was validated if supported by five adjacent windows; two for ends of chromosomes and three for centromeric regions. Final CO breakpoint resolution was refined by examining the genotype information of individual SNPs nearby. Individuals with low coverage (<0.1× depth), with high percentage (>5%) of window with first-allele frequency in a range from 0.8 to 0.9 and, with extreme number of breakpoints were removed from further analysis. CO interference was evaluated using MADpattern (51, 52).
Supplementary Material
Acknowledgments
We thank the Max Planck-Genome-centre (https://mpgc.mpipz.mpg.de/home/) for performing library preparations and DNA sequencing in this study. We thank Chloé Girard, Wayne Crismani, Andrew Loyd, Christine Mézard, and Rajeev Kumar for critical reading of the manuscript. We are grateful to Arnaud de Muyt, Nicolas Christophorou, Bruno Huettel, Manish Goel, and Korbinian Schneeberger for fruitful discussions and technical advice. We thank Nathalie Vrielynck for her support in the production of antibodies. This work was supported by a core grant from the Max Planck Society to R.M. and has benefited from the support of Institut Jean-Pierre Bourgin’s (IJPB) Plant Observatory technological platforms. The IJPB benefits from the support of Saclay Plant Sciences (ANR-17-EUR-0007).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023613118/-/DCSupplemental.
Data Availability
The list of CO positions can be found in Dataset S2. Raw read data of Fig. 8 can be found in the ArrayExpress database at The European Bioinformatics Institute under accession number E-MTAB-9593. Sequencing data have been deposited in ArrayExpress (E-MTAB-9593) (104).
References
- 1.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, 1–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller H. J., The mechanism of crossing over II. Am. Nat. 50, 284–305 (1916). [Google Scholar]
- 3.Zickler D., Kleckner N., A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. Semin. Cell Dev. Biol. 54, 135–148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M., The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Zickler D., Kleckner N. E., Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Page S. L., Hawley R. S., The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525–558 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Cahoon C. K., Hawley R. S., Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 23, 369–377 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Sym M., Engebrecht J. A., Roeder G. S., ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365–378 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Espagne E., et al., Sme4 coiled-coil protein mediates synaptonemal complex assembly, recombinosome relocalization, and spindle pole body morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 10614–10619 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries F. A. T., et al., Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao H., et al., Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 8, e1002790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page S. L., Hawley R. S., c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 15, 3130–3143 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., et al., Multivalent weak interactions between assembly units drive synaptonemal complex formation. J. Cell Biol. 219, e201910086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacQueen A. J., Colaiácovo M. P., McDonald K., Villeneuve A. M., Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428–2442 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colaiácovo M. P., et al., Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463–474 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Smolikov S., et al., Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 176, 2027–2033 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurlock M. E., et al., Identification of novel synaptonemal complex components in C. elegans. J. Cell Biol. 219, e201910043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins J. D., Sanchez-Moran E., Armstrong S. J., Jones G. H., Franklin F. C., The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K., Wang C., Liu Q., Liu W., Fu Y., Increasing the genetic recombination frequency by partial loss of function of the synaptonemal complex in rice. Mol. Plant 8, 1295–1298 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Wang M., et al., The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22, 417–430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barakate A., et al., The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell 26, 729–740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmekel K., et al., Organization of SCP1 protein molecules within synaptonemal complexes of the rat. Exp. Cell Res. 226, 20–30 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Liu J.-G., et al., Localization of the N-terminus of SCP1 to the central element of the synaptonemal complex and evidence for direct interactions between the N-termini of SCP1 molecules organized head-to-head. Exp. Cell Res. 226, 11–19 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Dong H., Roeder G. S., Organization of the yeast Zip1 protein within the central region of the synaptonemal complex. J. Cell Biol. 148, 417–426 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernández-Hernández A., et al., The central element of the synaptonemal complex in mice is organized as a bilayered junction structure. J. Cell Sci. 129, 2239–2249 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Anderson L. K., et al., Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. U.S.A. 102, 4482–4487 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schücker K., Holm T., Franke C., Sauer M., Benavente R., Elucidation of synaptonemal complex organization by super-resolution imaging with isotropic resolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2029–2033 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobson M. J., et al. , Synaptonemal complex proteins: Occurrence, epitope mapping and chromosome disjunction. J. Cell Sci. 107, 2749–2760 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Schild-Prüfert K., et al., Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics 189, 411–421 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphryes N., et al., The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet. 9, e1003194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa Y., et al., Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J. Cell Sci. 118, 2755–2762 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Bolcun-Filas E., et al., SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 176, 741–747 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schramm S., et al., A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 7, e1002088 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamer G., et al., Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J. Cell Sci. 119, 4025–4032 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Smolikov S., Schild-Prüfert K., Colaiácovo M. P., A yeast two-hybrid screen for SYP-3 interactors identifies SYP-4, a component required for synaptonemal complex assembly and chiasma formation in Caenorhabditis elegans meiosis. PLoS Genet. 5, e1000669 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Börner G. V., Kleckner N., Hunter N., Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Voelkel-Meiman K., Cheng S.-Y., Morehouse S. J., MacQueen A. J., Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics 203, 1091–1103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libuda D. E., Uzawa S., Meyer B. J., Villeneuve A. M., Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502, 703–706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Z., et al., The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J. 61, 600–610 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Higgins J. D., Armstrong S. J., Franklin F. C. H., Jones G. H., The arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mercier R., et al., Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3,whereas the other one is not. Curr. Biol. 15, 692–701 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Chelysheva L., et al., Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 3, e83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macaisne N., et al., SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 18, 1432–1437 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Higgins J. D., et al., AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 55, 28–39 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Chelysheva L., et al., The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambing C., et al., Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genet. 11, e1005372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurel A., et al., A cytological approach to studying meiotic recombination and chromosome dynamics in Arabidopsis thaliana male meiocytes in three dimensions. Plant J. 95, 385–396 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Giraut L., et al., Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7, e1002354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S. Y., et al., Global analysis of the meiotic crossover landscape. Dev. Cell 15, 401–415 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincenten N., et al., The kinetochore prevents centromere-proximal crossover recombination during meiosis. eLife 4, 1–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L., Liang Z., Hutchinson J., Kleckner N., Crossover patterning by the beam-film model: Analysis and implications. PLoS Genet. 10, e1004042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White M. A., Wang S., Zhang L., Kleckner N., Quantitative modeling and automated analysis of meiotic recombination. Methods Mol. Biol. 1471, 305–323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lloyd A., Jenczewski E., Modelling sex-specific crossover patterning in arabidopsis. Genetics 211, 847–859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandes J. B., Séguéla-Arnaud M., Larchevêque C., Lloyd A. H., Mercier R., Unleashing meiotic crossovers in hybrid plants. Proc. Natl. Acad. Sci. U.S.A. 115, 2431–2436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moses M. J., The relation between the axial complex of meiotic prophase chromosomes and chromosome pairing in a salamander (Plethodon cinereus). J. Biophys. Biochem. Cytol. 4, 633–638 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cromie G. A., Smith G. R., Branching out: Meiotic recombination and its regulation. Trends Cell Biol. 17, 448–455 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Lynn A., Soucek R., Börner G. V., ZMM proteins during meiosis: Crossover artists at work. Chromosome Res. 15, 591–605 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Storlazzi A., Xu L., Schwacha A., Kleckner N., Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc. Natl. Acad. Sci. U.S.A. 93, 9043–9048 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hillers K. J., Crossover interference. Curr. Biol. 14, R1036–R1037 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Kleckner N., et al., A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. U.S.A. 101, 12592–12597 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.King J. S., Mortimer R. K., A polymerization model of chiasma interference and corresponding computer simulation. Genetics 126, 1127–1138 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L., et al., Topoisomerase II mediates meiotic crossover interference. Nature 511, 551–556 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lambing C., Kuo P. C., Tock A. J., Topp S. D., Henderson I. R., ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 117, 13647–13658 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allers T., Lichten M., Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Bishop D. K., Zickler D., Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15 (2004). [DOI] [PubMed] [Google Scholar]
- 66.Hunter N., Kleckner N., The single-end invasion: An asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Fung J. C., Rockmill B., Odell M., Roeder G. S., Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116, 795–802 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Shinohara M., Oh S. D., Hunter N., Shinohara A., Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat. Genet. 40, 299–309 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Girard C., et al., AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11, e1005369 (2015). Corrected in: PLoS Genet.11, e1005448 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crismani W., et al., FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Séguéla-Arnaud M., et al., Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. U.S.A. 112, 4713–4718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokoo R., et al., COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lenormand T., Dutheil J., Recombination difference between sexes: A role for haploid selection. PLoS Biol. 3, e63 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Toyota M., Matsuda K., Kakutani T., Terao Morita M., Tasaka M., Developmental changes in crossover frequency in Arabidopsis. Plant J. 65, 589–599 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Kleckner N., Storlazzi A., Zickler D., Coordinate variation in meiotic pachytene SC length and total crossover/chiasma frequency under conditions of constant DNA length. Trends Genet. 19, 623–628 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Wallace B. M., Hultén M. A., Meiotic chromosome pairing in the normal human female. Ann. Hum. Genet. 49, 215–226 (1985). [DOI] [PubMed] [Google Scholar]
- 77.Tease C., Hultén M. A., Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet. Genome Res. 107, 208–215 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Lynn A., et al. , Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296, 2222–2225 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Wallace B. M. N., Wallace H., Synaptonemal complex karyotype of zebrafish. Heredity 90, 136–140 (2003). [DOI] [PubMed] [Google Scholar]
- 80.Jones G. H., Croft J. A., Chromosome pairing and chiasma formation in spermatocytes and oocytes of Dendrocoelum lactem (Turbellaria, Tricladida); a cytogenetical and ultrastructural study. Heredity 63, 97–106 (1989). [Google Scholar]
- 81.Drouaud J., et al., Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 3, e106 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lambing C., Franklin F. C. H., Wang C. R., Understanding and manipulating meiotic recombination in plants. Plant Physiol. 173, 1530–1542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serra H., et al., Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc. Natl. Acad. Sci. U.S.A. 115, 2437–2442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mieulet D., et al., Unleashing meiotic crossovers in crops. Nat. Plants 4, 1010–1016 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Fauser F., Schiml S., Puchta H., Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Morineau C., et al., Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15, 729–739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clough S. J., Bent A. F., Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 88.LeBlanc C., et al., Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 93, 377–386 (2018). [DOI] [PubMed] [Google Scholar]
- 89.Cromer L., et al., Patronus is the elusive plant securin, preventing chromosome separation by antagonizing separase. Proc. Natl. Acad. Sci. U.S.A. 116, 16018–16027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cromer L., et al., Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr. Biol. 23, 2090–2099 (2013). [DOI] [PubMed] [Google Scholar]
- 91.Armstrong S. J., Caryl A. P. P. A., Jones G. H., Franklin F. C. H., Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115, 3645–3655 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Chelysheva L., et al., An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. 129, 143–153 (2010). [DOI] [PubMed] [Google Scholar]
- 93.Rowan B. A., Patel V., Weigel D., Schneeberger K., Rapid and inexpensive whole-genome genotyping-by-sequencing for crossover localization and fine-scale genetic mapping. G3 (Bethesda) 5, 385–398 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang S., et al., Great majority of recombination events in Arabidopsis are gene conversion events. Proc. Natl. Acad. Sci. U.S.A. 109, 20992–20997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Andrews S., Fast QC: A quality control tool for high throughput sequence data (2010). http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 8 March 2021.
- 96.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lamesch P., et al., The arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qi J., Zhao F., Buboltz A., Schuster S. C., inGAP: An integrated next-generation genome analysis pipeline. Bioinformatics 26, 127–129 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qi J., Zhao F., inGAP-sv: A novel scheme to identify and visualize structural variation from paired end mapping data. Nucleic Acids Res. 39, W567–W575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiao W. B., Schneeberger K., Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat. Commun. 11, 989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goel M., Sun H., Jiao W. B., Schneeberger K., SyRI: Finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20, 277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qi J., Chen Y., Copenhaver G. P., Ma H., Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic mapping. Proc. Natl. Acad. Sci. U.S.A. 111, 10007–10012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Capilla-Perez L., Durand S., Lian Q., Mercier R., Whole-genome resequencing of BC1 populations from Arabidopsis (Col x Ler) x Col crosses (wild type and zyp1). ArrayExpress. https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9593/. Deposited 24 September 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The list of CO positions can be found in Dataset S2. Raw read data of Fig. 8 can be found in the ArrayExpress database at The European Bioinformatics Institute under accession number E-MTAB-9593. Sequencing data have been deposited in ArrayExpress (E-MTAB-9593) (104).