Significance
ADAR1, an A-to-I RNA editing enzyme, is an essential gene and also an attractive target for cancer therapy. Mutations in ADAR1 cause devastating autoinflammatory diseases, and inhibition of ADAR1 remarkably reduces tumor growth. ADAR1 binds and edits RNA through two isoforms: p150 (150 kDa) and p110 (110 kDa), but the function and RNA substrates of each isoform are incompletely understood. Here we report that the p150 mRNA is capable of coexpressing p150 and p110 isoforms. This finding raises the importance of investigating the biological significance behind coupled expression of p150 and p110. To this end, we developed a genetic strategy to decouple p150 and p110 expression that allows for determination of isoform-selective RNA editing events.
Keywords: ADAR1, p110, p150, RNA editing
Abstract
Human adenosine deaminase acting on RNA 1 (ADAR1) catalyzes adenosine-to-inosine deamination reactions on double-stranded RNA molecules to regulate cellular responses to endogenous and exogenous RNA. Defective ADAR1 editing leads to disorders such as Aicardi-Goutières syndrome, an autoinflammatory disease that manifests in the brain and skin, and dyschromatosis symmetrica hereditaria, a skin pigmentation disorder. Two ADAR1 protein isoforms, p150 (150 kDa) and p110 (110 kDa), are expressed and can edit RNA, but the contribution of each isoform to the editing landscape remains unclear, largely because of the challenges in expressing p150 without p110. In this study, we demonstrate that p110 is coexpressed with p150 from the canonical p150-encoding mRNA due to leaky ribosome scanning downstream of the p150 start codon. The presence of a strong Kozak consensus context surrounding the p110 start codon suggests the p150 mRNA is optimized to leak p110 alongside expression of p150. To reduce leaky scanning and translation initiation at the p110 start codon, we introduced synonymous mutations in the coding region between the p150 and p110 start codons. Cells expressing p150 constructs with these mutations produced significantly reduced levels of p110. Editing analysis of total RNA from ADAR1 knockout cells reconstituted separately with modified p150 and p110 revealed that more than half of the A-to-I edit sites are selectively edited by p150, and the other half are edited by either p150 or p110. This method of isoform-selective editing analysis, making use of the modified p150, has the potential to be adapted for other cellular contexts.
Adenosine deaminases acting on RNA (ADARs) are enzymes that catalyze adenosine-to-inosine (A-to-I) editing of RNA in animal cells (1–4). ADAR1 binds double-stranded RNA (dsRNA) and is expressed as p150 (150 kDa) and p110 (110 kDa) protein isoforms (5). ADAR1 is an active A-to-I editor of RNA in humans, and the majority of edit sites are found in a class of short interspersed nuclear elements (SINEs) called Alu elements, many of which are located in introns and 3′ untranslated regions (UTRs) (6, 7). Although the functional consequences of A-to-I editing are varied, a total lack of functional ADAR1 is not tolerated. For example, removing the catalytic activity of ADAR1 leads to MDA5-mediated embryonic lethality in mice (8). In humans, mutations in ADAR1 lead to an autoinflammatory disease called Aicardi-Goutières syndrome (9). These mutations are thought to cause a partial loss of ADAR1 function. Complete deaminase null mutations have yet to be found in humans, likely because such mutations are embryonic lethal. Indeed, complete lack of ADAR1 in human cells causes up-regulation of interferon and cell death (10–12). On the other hand, overactive A-to-I editing has been associated with increased proliferation of certain cancer cells (13–16). With regard to the two ADAR1 isoforms, p150 and p110, both were found to have roles in the regulation of organ development (17). The p110 isoform, during cellular stress, was found to suppress apoptosis by binding to antiapoptotic genes and preventing their decay (18). Given the importance of ADAR1 in maintaining homeostasis in various contexts, there is a need to understand how the p150 and p110 isoforms are regulated and how they individually contribute to the A-to-I RNA editome.
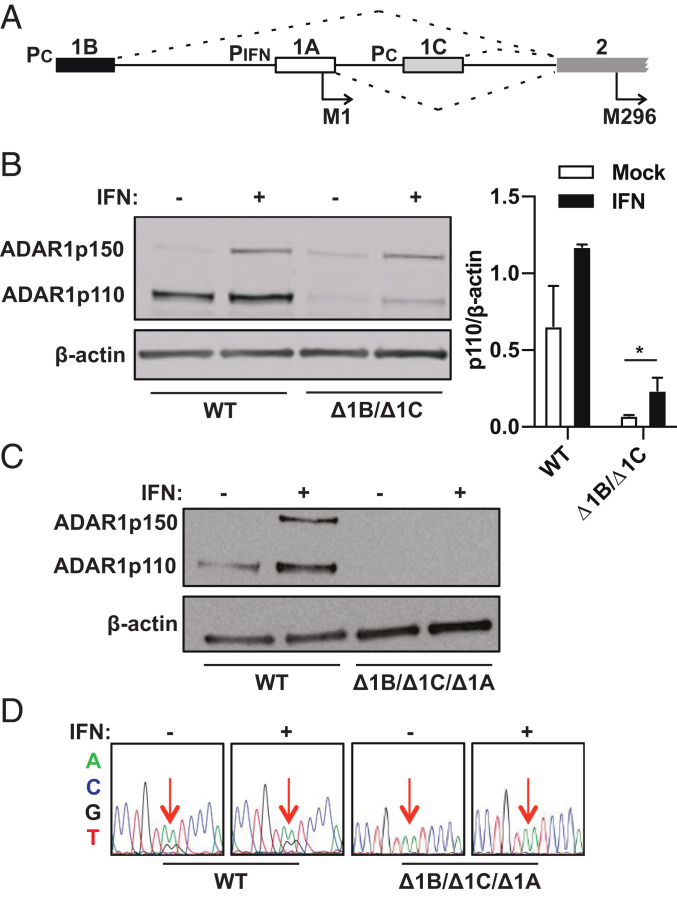
The ADAR1 gene maps to chromosome 1q21.3, and expression is controlled by three promoters—two are constitutively active, upstream of exon 1B and exon 1C, and one is an interferon-stimulated response element (ISRE), upstream of exon 1A (19–22) (Fig. 1A). Following transcription, splicing results in linkages between three splice donors, exons 1B, 1C, and 1A, and the common splice acceptor site in exon 2 (20, 21, 23). Exon 1A contains a methionine codon (M1), and when translation initiates within exon 1A, the entirety of exon 2 functions as a coding sequence, resulting in production of the interferon-inducible p150 isoform, which has the unique Z-alpha binding domain at its N-terminal end (24). Exons 1B and 1C lack methionine codons in the correct reading frame, so mRNA variants that begin with exons 1B or 1C have an extended 5′ UTR that extends into the first half of exon 2. Within exon 2, a methionine codon (M296) in the correct reading frame begins at nucleotide position 886. Hence, mRNA variants that start with exons 1B and 1C give rise to the constitutively expressed p110, which lacks the Z-alpha binding domain at its N terminus.
Fig. 1.
ADAR1 p110 persists following deletion of exons 1B and 1C. (A) The human ADAR1 locus on chromosome 1 consists of three alternative exon 1 structures that have individual promoters, including PIFN, which is inducible by type I interferon, and PC, which has constitutive activity. Exon 2 is the common splice acceptor for the three exon 1 structures. Exon 1A has a start codon for the p150 isoform (M1). Exon 2 has a start codon for the p110 isoform (M296). (B) Immunoblot shows ADAR1 isoforms probed with a C-terminal specific antibody (amino acids 1051 to 1226) in WT and exon 1B and exon 1C double-deletion (Δ1B/Δ1C) cell clones. Relative band intensities of p110 in WT cells and Δ1B/Δ1C cells were quantified relative to beta-actin levels using ImageJ for both mock- and interferonβ-treated conditions. Data points are shown as mean ± SEM and are gathered from three separate experiments. An unpaired t test comparing p110 levels in mock or IFNβ treated Δ1B/Δ1C cells gives a P value of 0.024. (C) Immunoblot shows ADAR1 isoforms in wild-type (WT) and exon 1B, exon 1C, and exon 1A triple-deletion (Δ1B/Δ1C/Δ1A) cell clones. (D) Sanger sequencing of cDNA from the ATM gene 3′ UTR region shows previously identified A-to-I ADAR1 edit sites (red arrows) in WT and Δ1B/Δ1C/Δ1A cell clones.
Localization of ADAR1 varies—p150 shuttles between the nucleus and cytoplasm, owing to the presence of a nuclear export signal at its amino terminus, and p110 is largely retained in the nucleus (5, 25–27). Homodimer formation has been reported for both p150 and p110, and in the case of p150, this was determined to be essential for efficient catalytic activity (28–30). The differential localization of p150 and p110 may influence the RNA targets that are selectively edited by either isoform. However, information regarding isoform-selective edit sites has been limited by the inability to express p150 without p110. We found that the exon 1A splice variant (the canonical p150-encoding mRNA isoform) contributes not only p150, but also p110, because of leaky ribosome scanning. We then created a modified p150 open-reading frame that significantly suppresses leaky p110 expression, and we individually expressed p110 or modified p150 in cells for in vivo A-to-I editing analysis. Our analysis revealed that p150 has a remarkably expansive editome compared to p110, and that a significant proportion of ADAR1-edit sites are also shared between both isoforms.
Results
ADAR1 p110 Persists following Deletion of Exons 1B and 1C.
CRISPR-Cas9 was used to delete exons 1B and 1C along with their promoters, and PCR was used to confirm deletion of the exons and respective promoters at the genomic DNA level (SI Appendix, Fig. S1). Immunoblotting for ADAR1 with a C-terminal specific antibody (amino acids 1051 to 1226) showed that while p110 levels were reduced following deletion of exons 1B and 1C, the protein was not entirely ablated. Quantification of p110 band intensities relative to beta-actin levels reveals a ninefold reduction in the p110 protein levels with deletion of exons 1B and 1C. Of note, p110 is up-regulated following treatment of exon 1B/1C knockout (Δ1B/Δ1C) cells with type I IFN (interferonβ), suggesting an interferon-inducible mRNA variant is contributing to p110 expression (Fig. 1B). Next, when exon 1A was deleted in the Δ1B/Δ1C background, p110 was no longer detectable by immunoblot, suggesting that the exon 1A mRNA variant contributes to p110 expression in Δ1B/Δ1C cells (Fig. 1C). As expected, p150 was also undetectable in Δ1B/Δ1C/Δ1A cells, and Sanger sequencing of reverse transcription (RT) PCR products from these cells confirmed a lack of A-to-I editing at known ADAR1-edit sites in the ATM gene 3′ UTR (Fig. 1D).
Downstream Translation Initiation on the p150 mRNA Creates p110.
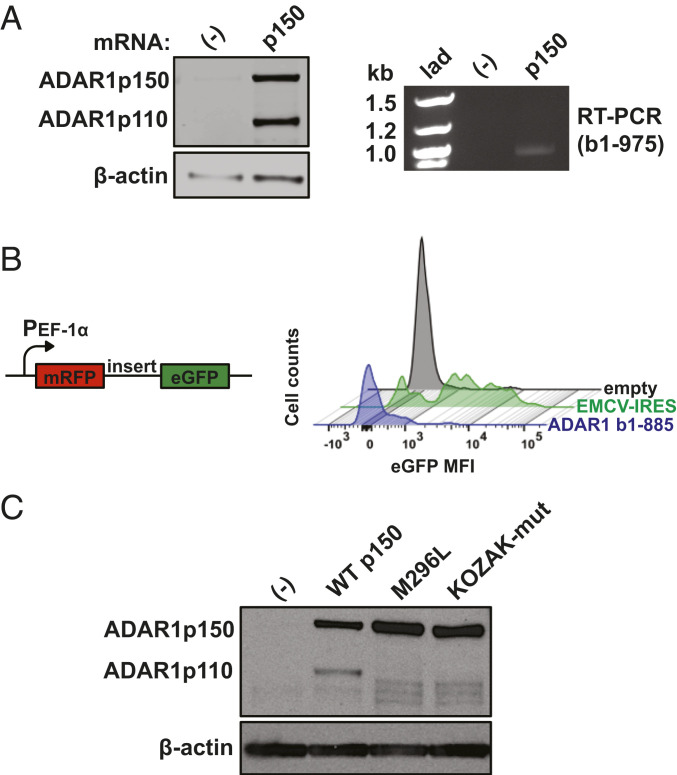
With the observation that p110 persists following deletion of exons 1B and 1C, we next aimed to understand how p110 is produced from the canonical p150-encoding exon 1A mRNA. First, to examine if the p150 transcript can express p110, in vitro transcribed p150 mRNA was generated and transfected into exon Δ1B/Δ1C/Δ1A cells (ADAR1 KO cells). Following transfection, immunoblot revealed presence of p110 in addition to p150, and RT-PCR using primers to amplify the coding region between the p150 and p110 start codons revealed a single band, suggesting that both isoforms can be produced from a single mRNA species (Fig. 2A).
Fig. 2.
Downstream translation initiation on the p150 mRNA creates p110. (A) Immunoblot shows 293T Δ1B/Δ1C/Δ1A cells, referred to as ADAR1 knockout (KO) cells, that are untransfected (-) or transfected for 24 h with in vitro transcribed mRNA that encodes p150 (Left). The RT PCR product corresponding to bases 1 to 975 of the p150 open-reading frame is also shown (Right). Of note, the p110 start codon on the p150 open-reading frame begins at base position 886. lad, ladder. (B) WT 293T cells were transfected for 24 h with a bicistronic reporter plasmid with a human elongation factor-1 alpha (PEF-1α) constitutive promoter that encodes for monomeric red fluorescent protein (mRFP) and a sequence of interest upstream of enhanced green fluorescent protein (eGFP). The histogram shows counts of eGFP mean fluorescent intensity (MFI) values for a 46-base random sequence (empty), the 575-base encephalomyocarditis virus internal ribosomal entry site sequence (EMCV-IRES), and the 885-base sequence upstream of the p110-AUG (ADAR1 b1-885). (C) Immunoblot shows ADAR1 isoforms in ADAR1 KO cells with stable expression of integrated cDNA constructs: untransduced (-), WT p150 sequence, p150 with p110-AUG mutated to CUC (M296L), and p150 with mutations (GACAUGGC→UUUAUGCU) in the Kozak consensus sequence surrounding the p110-AUG (KOZAK-mut).
Several mechanisms could contribute to expression of p110 from the p150 mRNA, including ribosomal skipping of the p150 start codon (leaky scanning) and/or direct binding of ribosomes to the p110 start codon. To investigate whether direct binding contributes significantly to p110 translation, a bicistronic reporter plasmid was used in which translation of enhanced green fluorescent protein (eGFP) is facilitated if the upstream cloned sequence contains an internal ribosomal entry site (IRES) (31). The cDNA sequence between the p150 and p110 start codons (bases 1 to 885) was introduced into the reporter plasmid, which was then transfected into 293T cells. The eGFP signal associated with this cDNA sequence (ADAR1 b1-885) is significantly lower compared to that (88% of mRFP+ cells) seen with the encephalomyocarditis virus (EMCV) IRES (Fig. 2B). We conclude that internal ribosome initiation between the p150 and p110 start codons does not contribute significantly to p110 expression.
Next, we devised strategies to test the hypothesis that leaky ribosome scanning on the p150 mRNA contributes to p110 expression. Changing the p110-AUG to CUC (M296L) significantly decreased p110 protein production and also resulted in production of smaller ADAR1 isoforms, which could result from leaky translation initiation at start codons downstream of M296 (Fig. 2C). Of note, translation initiation efficiency is known to be related to sequence motifs surrounding start codons (32). As such, we noticed that the p110 start codon is surrounded by an optimal Kozak consensus sequence: G at positions −3 and +4, and C at position −1. In contrast, the Kozak context surrounding the p150 start codon in endogenously transcribed mRNAs is GCAAUGA, which is suboptimal compared to the Kozak context surrounding the p110 start codon. Thus, these two start codon positions are organized in such a way to potentially give an advantage for the p110 initiation site. Changing the −3, +4, and −1 positions surrounding the p110 start codon (KOZAK-mut) to create a weakened translation initiation context resulted in a protein expression phenotype similar to that of M296L—significantly reduced p110 expression and production of ADAR1 isoforms smaller than p110 (Fig. 2C).
Collectively, these results suggest that leaky ribosome scanning downstream of the p150 start codon results in initiation of translation at M296, giving rise to p110 from the p150 mRNA. Furthermore, there are no in-frame start codons between the p150 and p110 start codons, suggesting that p150 mRNA is optimized to ensure that no isoforms other p110 are produced. The smaller proteins in the mutant constructs (M296L, KOZAK-mut) are likely produced by scanning ribosomes that translate from start codons downstream of L296 and M296, respectively, in the absence of efficient p110 translation.
Mutations Upstream of the p110 Start Codon Suppress Leaky p110 Expression.
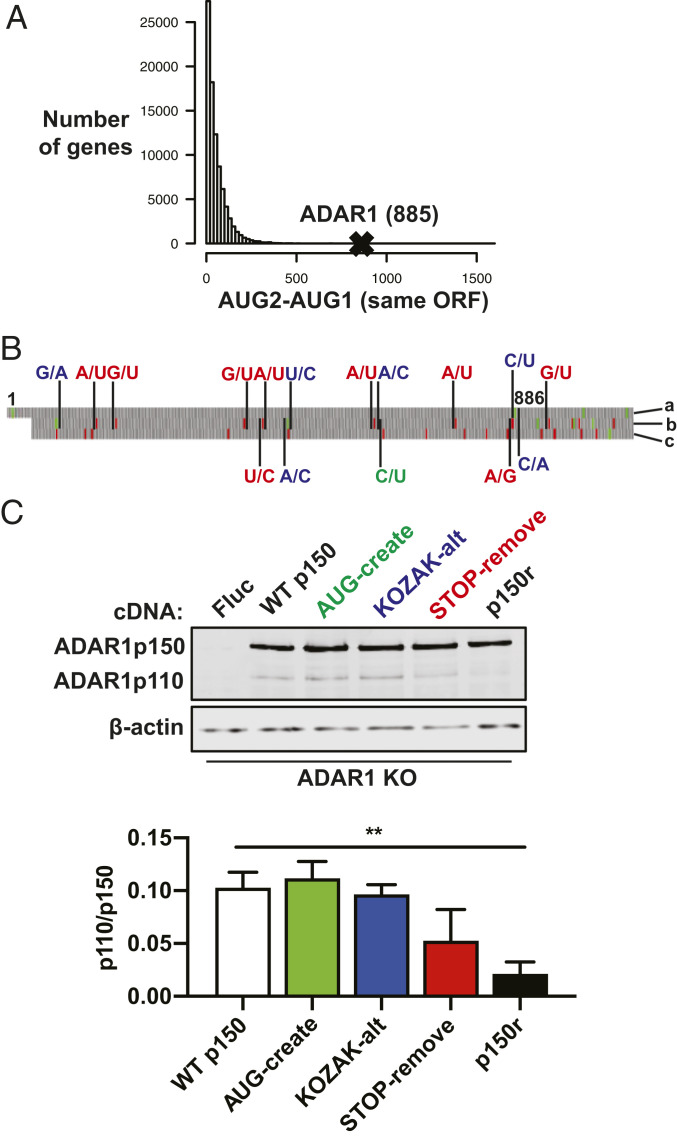
Of note, the first in-frame AUG after the p150-AUG is the p110-AUG, and the distance between these two codons: 885 bases, stands out among coding sequences—most have a smaller distance between the first two AUGs (Fig. 3A). We looked within this region for unique features that may relate to leaky expression of p110, and noted the presence of two start codons in an alternate reading frame (Fig. 3B). During the process of ribosome scanning, the translation initiation complex can assemble at downstream start codons, which could be in the same or a different reading frame as the annotated translational start site. We reasoned that increasing ribosome activity in an alternate reading frame would decrease the number of scanning ribosomes that initiate at the p110 start codon. To this end, synonymous mutations were introduced, color coded by the mutation category (Fig. 3B). The changes are as follows: 1) create a start codon in an alternate reading frame in optimal initiation context (AUG-create); 2) change the bases surrounding the two start codons in the alternate reading frame to increase likelihood of translation initiation at those start codons, and alter the bases surrounding the p110-AUG to decrease likelihood of translation initiation at that start codon (KOZAK-alt); and 3) remove stop codons that result in early termination of alternate reading frame translation (STOP-remove). Removing stop codons in the alternate reading frame may reduce the likelihood of scanning reinitiation at the p110 start codon following translation of small peptides (33, 34). Immunoblot revealed a significant decrease in p110 protein levels when the three groups of mutations are combined, called p150-reduced-p110 (p150r) (Fig. 3C). Importantly, this p150r construct now allows the ability to express p150 with suppressed leaky expression of p110.
Fig. 3.
Synonymous mutations upstream of the p110 start codon reduce p110 expression from the p150 mRNA. (A) Human cDNA sequences compiled from GRCh38 were analyzed in RStudio. The histogram displays counts of genes with different nucleotide distances between the first two AUG codons (AUG2–AUG1) in the same open-reading frame (ORF). The AUG2–AUG1 value for the ADAR1 p150 ORF is 885. (B) The start of the ADAR1 p150 sequence is shown and nucleotides 1 (p150-AUG) and 886 (p110-AUG) are labeled. On the Right side, the letter labels refer to the reading frames, with “a” referring to the ADAR1 ORF and letters “b” and “c” referring to the two alternate ORFs. Synonymous mutations introduced into the p150 ORF are color coded as follows: green creates an additional start codon in the “b” ORF; blue weakens the Kozak consensus sequence surrounding the p110-AUG in the “a” ORF and strengthens the Kozak consensus sequence surrounding the AUG codons in the “b” ORF; red removes stop codons upstream of the p110-AUG and also removes one stop codon downstream of the p110-AUG, both for the “b” ORF. Green and red blocks correspond to AUG and stop codons, respectively. (C) Immunoblot shows ADAR1 isoforms in ADAR1 KO cells that were transduced with lentivirus to create cell clones that have stable expression of various constructs: firefly luciferase (Fluc), WT p150, green mutations from B (AUG-create), blue mutations from B (KOZAK-alt), red mutations from B (STOP-remove), and combined green/blue/red mutations (p150r). Relative band intensities quantified in ImageJ show levels of p110 relative to p150. Data points are shown as mean ± SEM and are gathered from three separate experiments. An unpaired t test comparing the WT p150 and p150r groups gives a P value of 0.0016.
The A-to-I Editome Includes p150-Selective and p150/p110-Shared Edit Sites.
With the ability to express p150 with significantly reduced levels of p110, we next aimed to understand the individual contributions of p150 and p110 to the A-to-I RNA editome. Using a constitutive promoter-driven expression system, ADAR1 constructs (WT p150, p150r, and p110) and firefly luciferase (Fluc) control constructs were stably integrated into ADAR1 KO cells with lentiviral transduction (SI Appendix, Fig. S2A). The amount of ADAR1 p110 lentivirus was titrated so that p110 levels are similar to that of p110 leaking from the WT p150 expression construct, and p150r lentivirus was titrated so that p150 levels are also similar to that of the WT p150 expression construct. Compared to p150 and p110 protein levels produced from the activities of endogenous promoters in WT cells with interferon treatment, stably integrated p150 and p110 protein expression levels were twofold higher and twofold lower, respectively. Without interferon treatment, levels of p110 produced from the activities of endogenous promoters in WT cells were twofold higher than ectopic p110 expression levels (SI Appendix, Fig. S2B). Three different groups were analyzed: WT p150, to examine the editome when both isoforms are present; p150r, to examine the editome when p150 is the predominant editing isoform; and p110, to examine the editome when p110 is the predominant editing isoform. In each group, single-cell clones were selected in triplicate, with the selection criterion being that ADAR1 expression levels between the clones are similar, to minimize clone-to-clone variability.
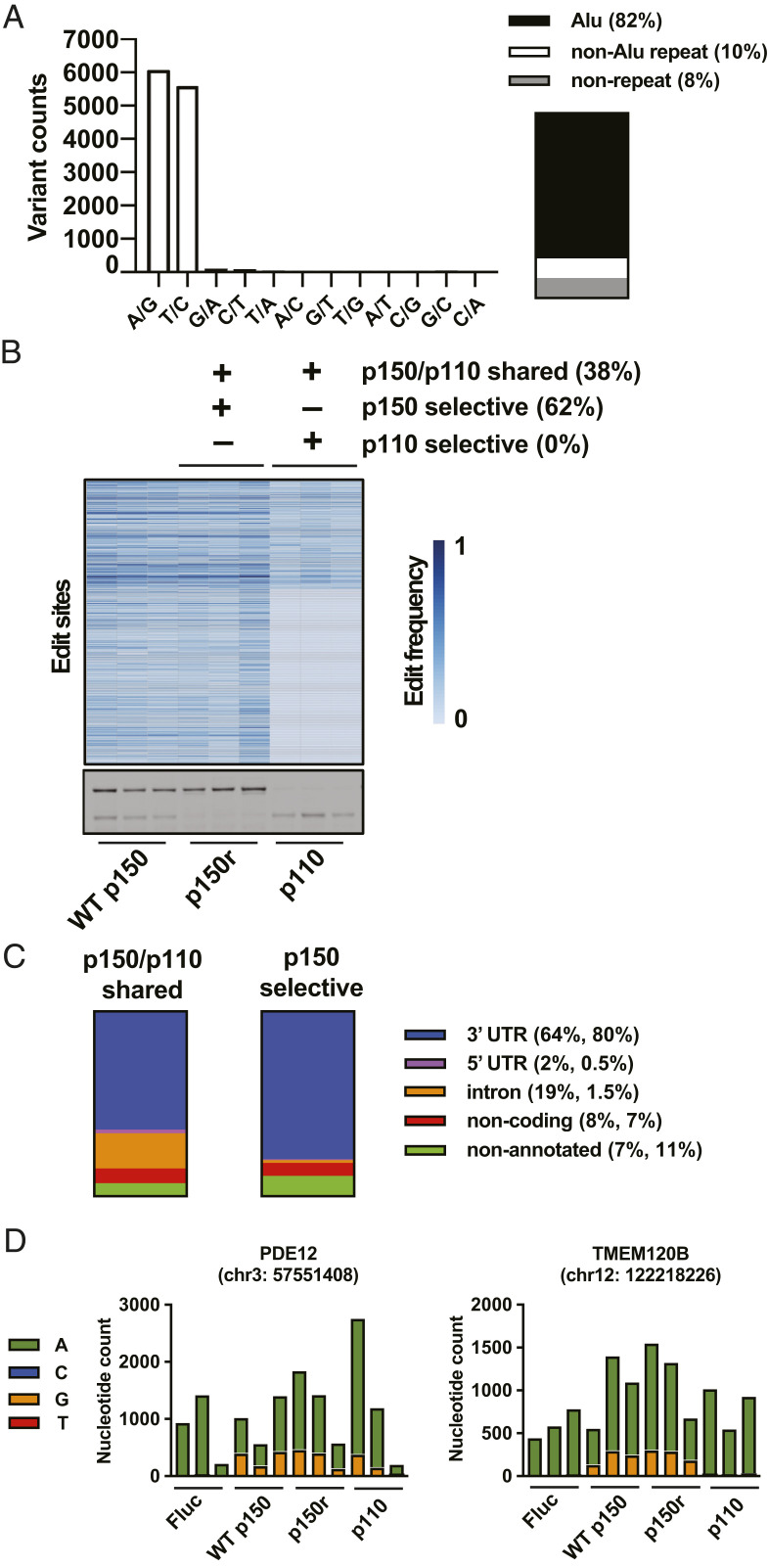
Following total RNA extraction, library preparation, and sequencing, alignment and variant identification were done using STAR and GATK. Following identification of total variants, additional filters were applied to identify putative ADAR1-edit sites (SI Appendix, Fig. S3). As validation of the editing analysis, 96% of the 12,173 potential ADAR1-edit sites are A-to-G or T-to-C reference-read mismatches (Dataset S1). 82% of edit sites are located in Alu repeat elements, 10% in non-Alu repeat elements, and 8% in nonrepetitive sequences (Fig. 4A).
Fig. 4.
The A-to-I editome includes p150-selective and p150/p110-shared edit sites. (A) Histogram shows counts of the 12 possible variants after filtering for ADAR1-edit sites. The components chart shows proportions of ADAR1-edit sites that are located in Alu repeat sequences, non-Alu repeat sequences, and nonrepetitive sequences. (B) Potential ADAR1-edit sites are shown classified as p150-selective edit sites (edited in all replicates of the p150r group and not edited in all replicates of the p110 group) and p150/p110 shared edit sites (edited in all replicates of both p150r and p110 groups); p110-selective edit sites were not identified using the current filtering criteria. The heatmap shows, grouped together, the edit frequencies of p150/p110 shared and p150-selective edit sites. (C) The components charts show proportions of p150-selective and p150/p110 shared edit sites that are located in different genomic annotations: 3′ UTR, 5′ UTR, intron, noncoding region, and nonannotated (intergenic) region. Of note, further analysis of sites located in nonannotated intergenic regions reveals that about 25% of these sites are located within 1 kb downstream of an annotated 3′ UTR (Dataset S1). (D) The amplicon sequencing data are displayed using bar graphs, which show, for each of the 12 samples, nucleotide counts (based on counting reads from unique molecular identifiers) from an edit site in PDE12 that is shared by p150 and p110, and another edit site in TMEM120B that is edited selectively by p150.
To identify sites that are selectively edited by p150 or p110 and sites that can be edited by both p150 and p110, the set of ADAR1-edit sites was classified based on whether certain sites are detected as edited or not in the p150r and p110 groups. A site is considered edited (present) if each replicate in a group has the site detected as edited. A site is considered not edited (absent) if each replicate in a group does not have the site detected as edited. Of the sites that were classified, 62% are p150-selective edit sites (present in the p150r group and absent in the p110 group), and 38% are p150/p110 shared edit sites (present in both the p150r and p110 groups) (Fig. 4B). Of note, with the current filtering criteria, p110-selective sites were not detected, but relaxing the filtering criteria does result in detection of putative p110-selective edits. However, amplicon sequencing, as will be discussed below, showed that these sites were also edited by p150. For p150-selective edit sites, 0.5% are located in 5′ UTR, 80% in 3′ UTR, 7% in noncoding RNA, 11% in nonannotated RNA, and 1.5% in introns; and for p150/p110 shared edit sites, 2% are located in 5′ UTR, 64% in 3′ UTR, 8% in noncoding RNA, 7% in nonannotated RNA, and 19% in introns (Fig. 4C) (Datasets S2 and S3). Genes containing exclusively p150-selective edit sites comprise 52% of edited genes, and genes containing exclusively p150/p110 shared edit sites comprise 33% of edited genes; 15% of genes contain both p150-selective and p150/p110 shared edit sites (Datasets S4 and S5). As validation of the editing classification, selected sites were analyzed by amplicon sequencing, and unique molecular tags were incorporated during the reverse transcription step to obtain accurate counts of each nucleotide at the selected sites (Fig. 4D). Collectively, the editing analysis reveals that the p150-selective editome is remarkably vast, and that a large number of sites can also be edited by both p150 and p110.
Discussion
ADAR1 is expressed as two distinct isoforms, a full-length p150 and smaller p110 (5). We found that expression of both isoforms is linked because of how the canonical p150 mRNA is organized. The p110 start codon is in an optimal Kozak context, and the large distance and lack of in-frame start codons between the p150 and p110 start codons suggest the p150 mRNA is optimized to express p150 with p110 and no other isoforms. Introducing synonymous mutations between the p150 and p110 start codons that increase ribosome activity in an alternate reading frame while reducing ribosome activity in the correct reading frame retains p150 expression while suppressing p110 expression. This system allows for the ability to examine the in vivo A-to-I editome when either p150 or p110 is independently the dominant editing isoform.
With the current stable transduction system and analysis workflow, we find that p150 is the dominant editing isoform—more than half of the sites are selectively edited by p150. This could be attributed to the ability of p150 to bind a broader range of RNA molecules compared to p110 as a result of its unique N-terminal domain, which is known to bind both RNA and DNA in Z-conformation with high affinity (35–37). From a clinical standpoint, the P193A mutation found in Aicardi-Goutières syndrome patients is located specifically in the Z-alpha domain specific to p150, raising the possibility that the P193A mutation may reduce the overall number of sites edited by p150 (9, 38).
Our finding of an expansive p150-selective editome, when taken in context with prior studies that report p150 as essential for enhancing A-to-I edit frequencies during the interferon response, suggest that p150 not only enhances the edit frequency per edit site, but also allows for an increase in the total number of edited sites that are otherwise not edited by p110 alone during the interferon response. Our isoform-specific editome analysis also uncovered the characteristics of p150-selective edit sites, which are primarily found in the 3′ UTR (80%) but minimally present in introns (1.5%). Prior studies have shown that A-to-I edits are mostly present in Alu elements within introns and 3′ UTRs (6, 7), and our findings suggest that p150 plays a nonredundant role in editing Alu repeats in 3′ UTRs. It is critical for future studies to examine the functional significance of p150-selective editing in the 3′ UTR since p150 KO mice and human cells exhibit embryonic lethality and autoinflammation, respectively (10, 17).
In addition to selectively editing a large number of sites, p150 also shares with p110 a significant proportion of edit sites. Compared to p150-selective edit sites, over a log-fold more p150/p110 shared edit sites are located in introns, likely because of the contribution of nuclear-localized p110 and a subset of nuclear p150. A separate study in HeLa cells also concluded that, in the absence of p150, the A-to-I editing is more prevalent in intronic regions (39). Motif analysis of the sequences surrounding p150-selective and p150/p110 shared edit sites reveals no significant difference. There is a preference for U preceding the edited adenosine and G following the edited adenosine for both classifications of edit sites.
One of the implications of leaky p110 translation on the interferon-inducible p150 exon 1A mRNA variant is that p110 can also be an interferon-inducible protein, much like p150. Prior studies have established a role for p150 in preventing endogenous RNAs from activating viral RNA-sensing pattern recognition receptors (PRRs), including PKR and MDA5 (10, 11, 17, 40). It is postulated that p150-edited endogenous RNAs can evade detection by PRRs. We examined if leaky p110 translation from p150 is required to suppress PKR activation. We find, at least for 293T cells, that p150 alone (p150r samples) is necessary and sufficient to suppress PKR activation in the context of interferon signaling, suggesting that p110-mediated editing contributes minimally to suppressing the sensing of immunogenic RNAs by PKR (SI Appendix, Fig. S4). The roles of p150 and p110 editing in regulating activation of other PRRs, such as MDA5 and OAS, need to be further investigated.
Our finding of interferon-inducible p110 expression also provides insights on results from a prior study in which A-to-I editing analysis was performed on WT, ADAR1 KO, and ADAR1 p150 KO cells with and without interferon treatment (10). In that study, edit frequencies of p150-dependent sites and ADAR1-dependent sites were increased following interferon treatment. The edit frequencies of p150-dependent sites were increased because p150 expression increases with interferon treatment. We can now imagine, knowing that p110 is also up-regulated following interferon treatment, that p110 up-regulation may contribute alongside p150 to increased editing frequencies of the p150/p110 shared edit sites following interferon treatment.
Of note, what we call p150/p110 shared edit sites are different from the theoretically possible p150/p110 synergistic edit sites, in which presence of both p150 and p110 is required for editing to occur. We did not find such synergistic sites in the current study. Furthermore, while p110-selective edit sites (adenosines that are edited by p110 but not p150) are also theoretically possible, they were not identified using the current filtering requirements for classification. The filtering algorithm requires p110- and p150-selective edit sites to have a read depth ≥50 in each sample, and the A-to-G mismatch frequency must be ≥0.1 in edited sites and =0 in nonedited sites. With different filtering criteria, such as read depth ≥5 rather than ≥50, potential p110-selective sites can be identified, but amplicon sequencing, which gives higher coverage at each site compared to bulk RNA sequencing, showed that these potential p110-selective sites had A-to-G mismatch frequencies >0 in the p150r group. Therefore, we conclude that increasing the read depth is important to improve the sensitivity of identifying A-to-G mismatches, thereby reducing the false negative rate in terms of classifying a site as not edited.
In conclusion, our study establishes the p150 and p110 coexpression phenomenon on the canonical p150-encoding mRNA that results from leaky ribosome scanning and translation initiation at the p110 start codon. This means that p110 expression goes together with p150 expression, and that p110 can be interferon-inducible alongside the canonical interferon-stimulated p150. We were able to suppress leaky expression of p110 using mutations that preserve the p150 coding sequence, allowing us to examine the p150 and p110 A-to-I editomes independently. The results reveal how expansive p150 is as an editing isoform, in that it selectively edits more than half of the sites but also shares many edit sites with p110. Taken together, one can imagine that the extensive p150-selective editome in combination with the interferon inducibility of p110 may together contribute to optimal editing during the interferon response. Our findings and system of analysis also set the stage for future studies that can be aimed at understanding the potentially diverse consequences of having p110 linked to p150 in expression. In particular, knocking in the p150r mutations or similar mutations into genomic DNA will allow an investigation into how endogenous expression of p150 and p110 plays a role during the interferon response and in other cellular contexts.
Materials and Methods
Cell Culture and Antibodies.
Human embryonic kidney (HEK) 293T cells (ATCC ACS-4500) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (Millipore Sigma TMS-013-B) and MEM Non-Essential Amino Acids Solution (GIBCO 11140076). Cells were seeded for experiments at densities of 6 × 104 and 4 × 105 cells per well for 24-well plates and 6-well plates, respectively. Human interferonβ 1a (PBL Assay Science 11415-1) was added to culture media at 1 nM concentration for 24 h. Cells were maintained at 37 °C and 5% CO2. Antibodies used for immunoblots include mouse anti-ADAR1 (Santa Cruz Biotechnology D-8), mouse monoclonal anti-beta-actin (Millipore Sigma A5441), horseradish peroxidase (HRP) mouse anti-beta actin antibody (Abcam ab49900), HRP goat anti-mouse secondary antibody (Abcam ab97023), and IRDye 800CW goat anti-mouse secondary antibody.
CRISPR Knockout and Single-Cell Cloning.
Guide RNAs targeting ADAR1 exons 1B, 1A, 1C, and their respective promoters were designed using the genomic sequences surrounding the exons based on the hg19 human reference genome. The CRISPR knockout method was based on a previously published genome engineering protocol (41). Briefly, CRISPR oligonucleotides were annealed and cloned into an expression vector (PX458) that encodes both Cas9 and GFP, allowing for single-cell selection by fluorescent-aided cell sorting (FACS). Plasmids were transfected into cells using Lipofectamine 2000 Transfection Reagent, following manufacturer guidelines. At 48 h following transfection, GFP-positive cells were sorted using the BD FACSAria II flow cytometer at The Rockefeller University Flow Cytometry Resource Center. After 2 wk, single-cell clones were moved from 96-well flat-bottom plates into 6-well plates for expansion. Next, single-cell clones were screened by genomic DNA PCR to look for homozygous deletion of the targeted exon (SI Appendix, Fig. S1). All PCR reactions were set up using the KOD Hot Start DNA Polymerase kit (Millipore Sigma 71086). CRISPR oligonucleotide sequences are listed in SI Appendix, Table S1.
Immunoblot and Quantification.
Cells were lysed using 2× Sample Buffer with 400 mM dithiothreitol (DTT), passed through a 26-G needle, boiled for 10 min, and centrifuged at 10,000 × g for 10 min. Cell lysates were loaded into 4 to 12% Bis-Tris gels and run at 130 V in 1× MOPS buffer for 2 h at room temperature. Next, proteins were transferred from gels onto nitrocellulose membranes at 300 mA for 2 h at 4 °C. The 10× transfer buffer was diluted using water and methanol to create 1× transfer buffer with 20% methanol. Following transfer, membranes were blocked for 1 h at room temperature either with 5% milk in tris-buffered saline (TBS) with Tween or LI-COR Odyssey blocking buffer. Following blocking, membranes were incubated with primary antibody overnight at 4 °C, followed by three 5 min washes, incubation with secondary antibody for 1 h at room temperature, and another set of three 5 min washes. For chemiluminescence, membranes were incubated for 5 min using SuperSignal West Pico Chemiluminescent Substrate. For fluorescence, membranes were imaged using the LI-COR Odyssey machine. Quantification of immunoblot bands was done in ImageJ.
RNA Extraction and In Vitro Transcription.
Total RNA from cells was extracted using phenol-chloroform extraction. Briefly, cells were lysed using TRIzol Reagent for 5 min at room temperature, vortexed with chloroform, and incubated for 3 min at room temperature. The mixture was centrifuged for 15 min at 12,000 × g and the aqueous portion was removed for RNA extraction using the Zymo Research Direct-zol RNA MiniPrep kit. Reverse transcription of RNA into cDNA and Sanger sequencing were performed as previously described (10). In vitro transcription of RNA was performed using the Promega T7 RiboMAX in vitro transcription kit.
Lentivirus Transduction.
ADAR1 mutations were introduced into the WT sequence using primers that contain the desired mutations along with the p150 expression plasmid as a template. Overlap PCR was used to assemble the fragments with mutations to be cloned back into the expression plasmid, which was packaged into lentivirus via transfection of WT 293T cells with plasmids encoding vesicular stomatitis virus glycoprotein (VSVG), Gag-Pol polyprotein, and the expression sequences of interest, as previously described (10). Primer sequences used to generate ADAR1 mutant constructs are listed in SI Appendix, Table S2.
High-Throughput RNA Sequencing and Analysis.
Libraries were prepared using the TruSeq Stranded Total RNA kit (Illumina), with two modifications to enrich for larger fragment sizes: 2-min fragmentation time at the 94 °C step; volume reduction of AMPure XP Beads (Beckman Coulter) to 60% of the standard amount. RNA libraries were pooled, diluted, and sequenced using the 150-base paired-end sequencing option on the Illumina NextSeq 500 High Output and NovaSeq S1 flow cells. Base calling data stored in BCL files were converted into FASTQ files and demultiplexed using bcl2fastq Conversion Software at The Rockefeller University Genomics Resource Center. The NextSeq and NovaSeq sequencing results were merged and aligned to hg19 using STAR 2.5.4b with 2-pass mapping. The alignment algorithm allows up to three mismatches for each 22-nucleotide region and considers the mean insert sizes of the RNA library fragments when determining if a read pair is concordant. Unique mappings were selected for variant calling. For each sample, Picard 2.18.1 was used to calculate total read counts, Phred quality scores, and alignment percentages. Aligned SAM files were converted into BAM files using SAMtools 1.9. Mutect2 (GATK 4.0.8) was used to identify reference-read mismatches for each of the 12 samples (four groups in biological triplicates: firefly luciferase, WT p150, p150r, and p110). Genomic positions with a single mismatch were considered for editing analysis. Genomic positions with a total read count of ≥5 in all 12 samples were selected for downstream analysis. A mismatch site was called for a group if the mismatched nucleotide read count at that site is ≥2 in each of the biological triplicates. For each mismatch, a mean mismatch frequency was calculated based on read counts from the biological triplicates: ΣG/ΣA+C+G+T. To create a list of putative ADAR1-edit sites, mismatch sites were selected from any one of the three groups: WT p150, p150r, or p110, with the requirement that a selected site must have a mean mismatch frequency greater than two SDs above the mean mismatch frequency of that site in the firefly luciferase (fluc) group. The mismatches were annotated using ANNOVAR (parsed from RefGene). Finally, mismatch sites found in the dbSNP138 database were excluded from further analysis. To subcategorize the list of putative ADAR1-edit sites, mismatches were selected that have total read counts of ≥50 in all 12 samples, and that are A-to-G following addition of annotated genomic strand information. A site is considered edited in a group if the mismatch frequency is ≥0.1 and not edited in a group if the mismatch frequency = 0.
Amplicon Sequencing and Analysis.
Amplicons were made from total RNA using gene-specific antisense primers with unique molecular identifiers (10-nucleotide degenerate sequences) on each primer. Reverse transcription was done using SuperScript III reverse transcriptase, following manufacturer guidelines. The first round of PCR was done using gene-specific sense primers paired with a common primer that binds outside the unique molecular identifier sequence, and the second round of PCR was done using sets of primers that add on indexes for sample multiplexing and complete the Illumina adapter sequences. Primer sequences used to build the amplicons are listed in SI Appendix, Table S2. Sequencing was done using the MiSeq Nano format at The Rockefeller University Genomics Resource Center, and data analysis was done using dms_tools2, developed by the Jesse Bloom group. The bcsubamp program was used with adaptations to the arguments to fit the design of the amplicons.
Supplementary Material
Acknowledgments
We thank Joseph Luna, Kathryn Rozen-Gagnon, Alison Ashbrook, Lauren Aguado, and Jesse Bloom for advice and discussions; Shira Weingarten-Gabbay for sharing reagents and critical reading of the manuscript; James Sun for help with programming; and The Rockefeller University Flow Cytometry Resource Center and Genomics Resource Center. This study was supported by NIH grant R01AI091707 (to C.M.R.) and NIH grant R01AI151029 (to B.R.R.). H.C. was funded by NIH/NIAID F32AI114211 and Searle Scholars Program (SSP-2020-114). A.A. was funded by NIH/NIAID 5F32AI126892. T.S. was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under Award T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD–PhD Program. B.H. is a Ruth Kirschstein NIH Predoctoral Fellow (F30CA236239-01).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021757118/-/DCSupplemental.
Data Availability
Sequencing data have been deposited in Classification of ADAR1 p150 and p110 edit sites (PRJNA590956). Datasets/analyses are included as supplementary files.
References
- 1.Bass B. L., Weintraub H., An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55, 1089–1098 (1988). [DOI] [PubMed] [Google Scholar]
- 2.O’Connell M. A., Keller W., Purification and properties of double-stranded RNA-specific adenosine deaminase from calf thymus. Proc. Natl. Acad. Sci. U.S.A. 91, 10596–10600 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner R. W., Smith J. E., Cooperman B. S., Nishikura K., A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. U.S.A. 86, 2647–2651 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hough R. F., Bass B. L., Purification of the Xenopus laevis double-stranded RNA adenosine deaminase. J. Biol. Chem. 269, 9933–9939 (1994). [PubMed] [Google Scholar]
- 5.Patterson J. B., Samuel C. E., Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 15, 5376–5388 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan M. H., et al., Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenberg E., Levanon E. Y., A-to-I RNA editing–Immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Liddicoat B. J., et al., RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice G. I., et al., Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung H., et al., Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad S., et al., Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797–810.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., et al., Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. eLife 6, 1–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu L. D., Öhman M., ADAR1 editing and its role in cancer. Genes (Basel) 10, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fumagalli D., et al., Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 13, 277–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishizuka J. J., et al., Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gannon H. S., et al., Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells. Nat. Commun. 9, 5450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pestal K., et al., Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43, 933–944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakurai M., et al., ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 24, 534–543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weier H. U. G., George C. X., Greulich K. M., Samuel C. E., The interferon-inducible, double-stranded RNA-specific adenosine deaminase gene (DSRAD) maps to human chromosome 1q21.1-21.2. Genomics 30, 372–375 (1995). [DOI] [PubMed] [Google Scholar]
- 20.George C. X., Samuel C. E., Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. U.S.A. 96, 4621–4626 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George C. X., Samuel C. E., Characterization of the 5′-flanking region of the human RNA-specific adenosine deaminase ADAR1 gene and identification of an interferon-inducible ADAR1 promoter. Gene 229, 203–213 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Zeng Y., Murray J. M., Nishikura K., Genomic organization and chromosomal location of the human dsRNA adenosine deaminase gene: The enzyme for glutamate-activated ion channel RNA editing. J. Mol. Biol. 254, 184–195 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Liu Y., George C. X., Patterson J. B., Samuel C. E., Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem. 272, 4419–4428 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Herbert A., et al., A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. U.S.A. 94, 8421–8426 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulsen H., Nilsson J., Damgaard C. K., Egebjerg J., Kjems J., CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 21, 7862–7871 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barraud P., Banerjee S., Mohamed W. I., Jantsch M. F., Allain F. H. T., A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1. Proc. Natl. Acad. Sci. U.S.A. 111, E1852–E1861 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strehblow A., Hallegger M., Jantsch M. F., Nucleocytoplasmic distribution of human RNA- editing enzyme ADAR1 is modulated by double- stranded RNA-binding domains, a Leucine-rich export signal, and a putative dimerization domain. Mol. Biol. Cell 13, 3822–3835 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chilibeck K. A., et al., FRET analysis of in vivo dimerization by RNA-editing enzymes. J. Biol. Chem. 281, 16530–16535 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Cho D. S. C., et al., Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 278, 17093–17102 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Valente L., Nishikura K., RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 282, 16054–16061 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weingarten-Gabbay S., et al., Comparative genetics: Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, aad4939 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Kozak M., Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 12, 857–872 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochetov A. V., et al., uORFs, reinitiation and alternative translation start sites in human mRNAs. FEBS Lett. 582, 1293–1297 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Vattem K. M., Wek R. C., Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schade M., et al., The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA. Proc. Natl. Acad. Sci. U.S.A. 96, 12465–12470 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz T., Rould M. A., Lowenhaupt K., Herbert A., Rich A., Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284, 1841–1845 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Placido D., Brown B. A. 2nd, Lowenhaupt K., Rich A., Athanasiadis A., A left-handed RNA double helix bound by the Z α domain of the RNA-editing enzyme ADAR1. Structure 15, 395–404 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbert A., Mendelian disease caused by variants affecting recognition of Z-DNA and Z-RNA by the Zα domain of the double-stranded RNA editing enzyme ADAR. Eur. J. Hum. Genet. 28, 114–117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaller C. K., Donohue R. C., Nersisyan S., Brodsky L., Cattaneo R., Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150. PLoS Biol. 16, e2006577 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George C. X., Ramaswami G., Li J. B., Samuel C. E., Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J. Biol. Chem. 291, 6158–6168 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran F. A., et al., Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in Classification of ADAR1 p150 and p110 edit sites (PRJNA590956). Datasets/analyses are included as supplementary files.