Abstract
At the Bundeswehr Hospitals of Hamburg and Westerstede, patients repatriated from subtropical war and crisis zones of Northern Africa and the Middle East were medically treated, including microbiological assessment. Within a six-year interval, 16 Acinetobacter spp. strains, including 14 Acinetobacter baumannii (Ab) isolates with resistance against carbapenems and origins in Afghanistan (n = 4), Iraq (n = 2), Libya (n = 2), and Syria (n = 8) were collected. While clonal relationships of Libyan and Syrian strains had been assessed by superficial next generation sequencing (NGS) and “DiversiLab” repetitive elements sequence-based (rep-)PCR so far, this study provides core genome-based sequence typing and thus more detailed epidemiological information. In detail, sequencing allowed a definitive species identification and comparison with international outbreak-associated Ab strains by core genome multi locus sequence typing (cgMLST) and the identification of MLST lineages, as well as the identification of known resistance genes. The sequence analysis allowed for the confirmation of outbreak-associated clonal clusters among the Syrian and Afghan Ab isolates, indicating likely transmission events. The identified acquired carbapenem resistance genes comprised blaOXA-23, blaOXA-58, blaNDM-1, and blaGES-11, next to other intrinsic and acquired, partly mobile resistance-associated genes. Eleven out of 14 Ab isolates clustered with the previously described international clonal lineages IC1 (4 Afghan strains), IC2 (6 Syrian strains), and IC7 (1 Syrian strain). Identified Pasteur sequence types of the 14 Ab strains comprised ST2 (Syrian), ST25 (Libyan), ST32 (Iraqi), ST81 (Afghan), ST85 (Libyan), and ST1112 (Syrian), respectively. In conclusion, the study revealed a broad spectrum of resistance genes in Ab isolated from war-injured patients from Northern Africa and the Middle East, thereby broadening the scarcely available data on locally abundant clonal lineages and resistance mechanisms.
Keywords: Acinetobacter baumannii, war injury, Libya, Syria, Iraq, Afghanistan, epidemiology, sequence typing, carbapenem resistance, resistance gene, crisis zone
1. Introduction
During the last two decades, military conflicts have affected many regions of Northern Africa, the Middle East and neighboring countries, including Afghanistan, Iraq, Libya, and Syria. As previously summarized [1,2], war-associated wounds are prone to colonization or infection with multidrug-resistant Gram-negative bacteria including Acinetobacter baumannii (Ab).
The epidemiology of war trauma-associated infections, nosocomial transmission as well as deployment-associated colonization with A. baumannii has been best studied during the recent military interventions of US-American and British Forces in Iraq [3,4,5,6,7,8,9,10,11,12,13]. Epidemiological data also exist from civilian Iraqi health care facilities [14,15] as well as from the field of environmental biology [16]. As early as in 2006, blaOXA-58-like and blaPER-like genes were identified via sequencing sequenced in Iraqi A. baumannii strains [4]. Six years later, identified carbapenemase genes comprised of blaOXA-40, blaOXA-23, blaOXA-58, and ISAbal located upstream of the intrinsic blaOXA-51 on plasmids and/or the chromosome [10]. In addition, blaOXA-40-like genes were detected in Acinetobacter strains isolated in the Iraqi Kurdistan region. In 2008, multidrug-resistant European clone II-associated isolates were described to be linked with local outbreaks in soldiers deployed to Iraq [6]. From the Iraqi Kurdistan region, the Pasteur sequence types [17] ST2, ST136, ST94, ST623, ST792, and ST793 were reported. Although skin colonization had been discussed as a predictor of later traumatic A. baumannii-associated wound infections [5], a considerable proportion of 27% infections did not show respective clonal links [8]. Interestingly, there was even a decline in A. baumannii colonization in US troops during post-deployment screening from 21% to 4% between 2005 and 2009 [9].
Similar experience with multidrug resistant A. baumannii strains leading to trauma-associated wound infections were reported from the military conflicts and civil-war-haunted settings in Afghanistan [4,7,9,18,19,20,21], Syria [22,23,24,25,26], and Libya [27,28,29,30,31,32]. While blaNDM-1 was identified in carbapenem-resistant A. baumannii strains from Syria [23,24], alternative beta-lactam resistance mechanisms like blaOXA-94 (a blaOXA-51 variant) as well as the elements ISAba13, ISAba14, and ISAba17 were also detected in strains of the ST85 clone [26]. In addition, gyrA-gene- and parC-gene-based resistance to fluoroquinolones was reported from those strains isolated from Syrian civil war victims [26]. In Libyan patients, A. baumannii strains carrying the carbapenemase-encoding gens blaNDM-1, blaOXA-23-like, and blaOXA-40like were detected as well as high abundance of sequence type ST2 [27,29,30].
In the course of a six-year-interval, 16 Acinetobacter spp. isolates with reduced susceptibility towards carbapenems and origins in Afghanistan (n = 4), Iraq (n = 2), Libya (n = 2), and Syria (n = 8) were collected at the Bundeswehr Hospitals Hamburg and Westerstede, Germany. At those sites, patients repatriated from subtropical war and crisis zones of Northern Africa and the Middle East were medically treated and assessed. Superficial epidemiological and resistance information about the Libyan and Syrian strains had previously been provided by low-coverage next generation sequencing (NGS) and “DiversiLab” repetitive elements sequence-based (rep-)PCR [33], however, a detailed analysis of clonal relationship and resistance mechanisms was still missing.
In order to provide this information, whole genome sequencing as well as analysis based on core genome multi locus sequence typing (cgMLST) [34] were performed in the present study. By performing these analyses, we aimed at providing additional, more detailed epidemiological information on the international distribution of resistant Acinetobacter clones, as well as on underlying molecular resistance mechanisms.
2. Results
2.1. Confirmation on Species Level and Clustering with International Outbreak Strains Based on Core Genome Analysis
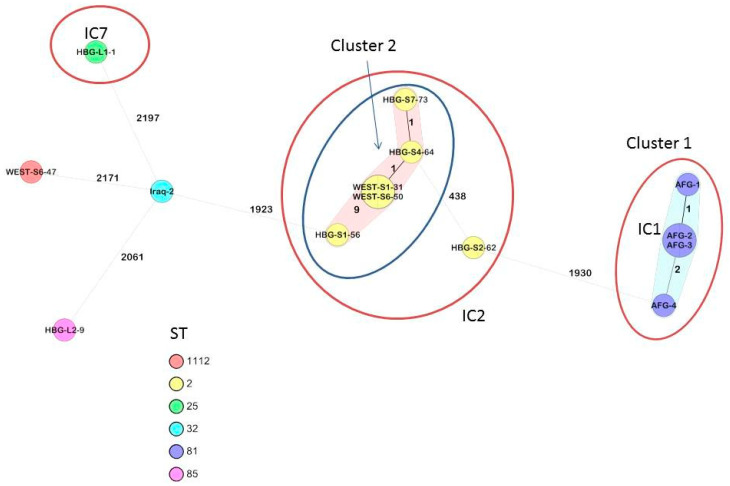
From the 16 Acinetobacter spp. isolates assessed, 14 were confirmed as A. baumannii by the gyrB multiplex PCR and presence of blaOXA-51-like. The remaining two strains were identified as A. dijkshooriniae (sample name Iraq-1) and A. radioresistens (sample name WEST-S5-44) using the Jspecies Tetra correlation search [35] and matrix-assisted laser-desorption-ionization time-of-flight mass spectrometry (MALDI TOF MS) (Table 1). Details on the sequence-based identification of these two Acinetobacter spp. on species level are provided in the Appendix A Table A1. All 14 A. baumannii isolates were subjected to cgMLST analysis, which showed that the isolates clustered with the previously identified international clonal lineages IC1 (the four Afghan strains), IC2 (six out of seven Syrian strains), IC7 (one out of two Libyan strains) [36], as shown in Figure 1, while for three isolates (one from Iraq, one from Syria, one from Libya), no matching IC could be identified.
Table 1.
Analysis of antimicrobial resistance determinants, ordered by strain, MLST type, and the international clonal lineage of the assessed Acinetobacter spp. isolates. N.a. = not applicable, ST = Sequence type, IC = International clone. Clusters are indicated in the same colors as in Figure 1.
| Sample, Species, Country of Origin, Year of Isolation |
MLST | Clonal Lineage |
Antibiotic Resistance Determinants | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| STox | STpas | Sulfonamide | Phenicol | Beta-Lactam | Aminoglycoside | Macrolide | Tetracycline | Trimethoprim | Fluoroquinolone and Aminoglycoside |
||
| Iraq-2, A. baumannii, Iraq, 2010 |
1627 | 32 | n.a. | sul2-like | blaADC-25-like, blaOXA-100, blaOXA-58 | aadB-like | |||||
| AFG-1, A. baumannii, Afghanistan, 2008 |
498 | 81 | IC1 | sul2 | blaADC-25-like, blaOXA-23, blaOXA-69 |
aadB-like, aph(3′)-Ia |
|||||
| AFG-2, A. baumannii, Afghanistan, 2008 |
498 | 81 | IC1 | sul2 | blaADC-25-like, blaOXA-23, blaOXA-69 |
aadB-like, aph(3′)-Ia |
|||||
| AFG-3, A. baumannii, Afghanistan, 2008 |
498 | 81 | IC1 | sul2 | blaADC-25-like, blaOXA-23, blaOXA-69 | aadB-like, aph(3′)-Ia, aph(3′)-VIa-like | |||||
| AFG-4, A. baumannii, Afghanistan, 2008 |
498 | 81 | IC1 | sul2 | blaADC-25-like, blaOXA-23, blaOXA-69 |
aadB-like, aph(3′)-Ia |
|||||
| HBG-L1-1, A. baumannii, Libya, 2011 |
440 | 25 | IC7 | sul2 | blaADC-25-like, blaOXA-23, blaOXA-64 | aadB-like, aph(3′)-Ic, strA, strB | |||||
| HBG-L2-9, A. baumannii, Libya, 2011 |
1089 | 85 | n.a. | sul2 | floR-like | blaADC-25-like,blaNDM-1, blaOXA-94 |
aadB-like, aph(3′)-VIa-like |
mph(E), msr(E) | |||
| WEST-S6-47, A. baumannii, Syria, 2013 |
2271 | 1112 | n.a. | sul1 | cmlA1-like | blaADC-25-like, blaGES-11, blaOXA-715 | aadA2, aadB, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | dfrA7 | aac(6′)Ib-cr-like | ||
| WEST-S1-31, A. baumannii, Syria, 2013 |
218 | 2 | IC2 | sul2 | blaADC-25-like, blaOXA-23, blaOXA-66, blaTEM-1D |
aph(3′)-Ic, armA,
strA, strB |
mph(E), msr(E) | tet(B)-like | |||
| WEST-S6-50, A. baumannii, Syria, 2013 |
218 | 2 | IC2 | sul2 | blaADC-25-like, blaOXA-23, blaOXA-66, blaTEM-1D |
aph(3′)-Ic, armA,
strA, strB |
mph(E), msr(E) | tet(B)-like | |||
| HBG-S4-64, A. baumannii, Syria, 2013 |
218 | 2 | IC2 | blaADC-25-like, blaOXA-23, blaOXA-66, blaTEM-1D |
aph(3′)-Ic, armA,
strA, strB |
mph(E), msr(E) | tet(B)-like | ||||
| HBG-S1-56, A. baumannii, Syria, 2013 |
195 | 2 | IC2 | blaADC-25-like, blaOXA-23, blaOXA-66, blaTEM-1D |
aph(3′)-Ic,
strA, strB |
tet(B)-like | |||||
| HBG-S7-73, A. baumannii, Syria, 2013 |
218 | 2 | IC2 | blaADC-25-like, blaOXA-23, blaOXA-66, blaTEM-1D |
aph(3′)-Ic, armA,
strA, strB |
mph(E), msr(E) | tet(B)-like | ||||
| HBG-S2-62, A. baumannii, Syria, 2013 |
1114 | 2 | IC2 | sul1,sul2 | cmlA1-like | blaADC-25-like, blaGES-11, blaOXA-23, blaOXA-66, blaTEM-1D | aadA2, aadB-like, aph(3′)-VIa-like, strA-like, strB-like, aacA4-like | dfrA7 | aac(6′)Ib-cr-like | ||
| WEST-S5-44, A. radioresistens, 2013 |
n.a. | n.a. | n.a. | blaOXA-815 | |||||||
| Iraq-1, A. dijkshoorniae, 2010 |
1605 | 1141 | n.a. | bla OXA-819 | |||||||
Figure 1.
Minimum spanning tree of the A. baumannii strains based on 2390 target alleles (core genome). Isolate numbers are within the nodes, and the numbers between the nodes indicate the number of alleles that were different. Isolates are colored based on their Pasteur sequence type.
Of note, cgMLST (Figure 1) confirmed the close clonal relationship of the four IC1 strains which had been isolated in the course of a small local outbreak in Afghanistan, suggesting a common source of infection. Regarding the Syrian A. baumannii strains of the IC2 cluster comprising isolates from two different Bundeswehr Hospitals (WEST for “Bundeswehr Hospital Westerstede” and HBG for “Bundeswehr Hospital Hamburg”), the clonal relationship that had already been suggested by rep-PCR and superficial sequencing (for comparison, also see Figure 2 in reference [33]) could be confirmed by cgMLST, making transmission events prior to hospital admission likely.
All isolates were typed based on two 7-loci multi-locus sequence typing (MLST) schemes, i.e., the Oxford scheme and the Pasteur scheme (Table 1) [37,38]. Notably, six of the A. baumannii were Pasteur ST-2 which corresponds to IC2, but were subdivided into four Oxford STs, while four isolates were Pasteur ST-81 (Oxford ST-498) which is a single locus variant of ST-1 and corresponds to IC1. The other isolates, including the non-A. baumannii, were represented by single STs.
2.2. Identified Molecular Resistance Mechanisms and Comparison with Phenotypic Resistance Testing
Table 1 summarizes the analysis of antimicrobial resistance determinants, ordered by strain, MLST type and international clonal lineage. Identified molecular resistance determinants of the carbapenem-susceptible Iraqi A. dijkshooriniae strain and the Syrian A. radioresistens strain are shown as well.
In most cases, isolates that were related carried the same resistomes. The most frequently detected resistance mechanisms among the assessed isolates comprised beta-lactamases as well as sulfonamide and aminoglycoside resistance genes. This is reflected by observed phenotypic resistance as indicated in the Appendix A Table A2, indicating high resistance rates against these substance groups, in particular for the isolates WEST-S6-47 and HBG-S2-62 which were resistant to amikacin, gentamicin, and tobramycin. These two isolates also possessed the same array of aminoglycoside modifying enzymes, as well as being the only isolates with dfrA7, aac(6′)Ib-cr-like, cmlA1-like, and blaGES-11, suggesting a shared plasmid or resistance island (Table 1). All A. baumannii were carbapenem resistant and this was mediated through possession of blaOXA-23 in the Afghan strains, by bla OXA-23 and/or blaGES-11 in the Syrian isolates, by blaOXA-58 in the Iraqi isolate, and blaNDM-1 in the Libyan isolate. Fluoroquinolone resistance was observed for all A. baumannii isolates (Appendix A Table A2). Analysis of the gyrA and parC genes revealed that the resistant isolates had a Ser83-Leu substitution in GyrA, while all but one (Iraq-2) also harbored a Ser80-Leu substitution in ParC. Resistance determinants to sulfonamides were present in all isolates resistant to co-trimoxazole despite a dfr gene being present in only two isolates, suggesting other mechanisms such as efflux might be playing a role, while tetB-like genes were detected in four isolates. All isolates remained susceptible to colistin.
All A. baumannii were in possession of the intrinsic blaOXA-51-like, the variants of which correlated with their Pasteur STs and IC clonal designation (Table 1). ISAba1 was not associated with this gene. The A. dijkshoorniae possessed blaOXA-819, which is similar to the instrinic blaOXA-213-like from the closely related A. pittii, while the A. radioresistens had blaOXA-815 which is a blaOXA-23-like that is intrinsic to this species. Furthermore, all A. baumannii isolates carried their intrinsic blaADC.
3. Discussion
The assessment was conducted to expand the knowledge about the micro-epidemiology of A. baumannii isolates with resistance against carbapenems from patients from Northern Africa and the Middle East, thus extending the results of a previous study published in 2018 [33]. A total of 16 Acinetobacter spp. strains including 14 A. baumannii from the strain collection of the Department of Microbiology and Hospital Hygiene, Bundeswehr Hospital Hamburg, were included in the study. Therefore, in addition to the previous study, clinical isolates from patients from Afghanistan and Iraq, which had been first introduced in a technical evaluation scheme on diagnostic identification of Acinetobacter spp. by fluorescence in situ hybridization (FISH) [39], could be included in the analysis. In comparison to the previous epidemiological assessment based on rep-PCR and superficial sequencing [33], cgMLST- and MLST-based typing applying the Pasteur and Oxford schemes [37,38] provided considerably more details, allowing origin-specific assignments to international clones and MLST-based sequence types. The comparably small size of the collection of those rare strains made a complete assessment of all available isolates, even of the non-A. baumannii strains, possible without any economic restrictions. Of note, all assessed A. baumannii isolates could be assigned to MLST sequence types of the Oxford and Pasteur schemes [37,38].
The cgMLST approach confirmed two nosocomial transmission clusters. Cluster 1 comprised the four Afghan isolates, which were isolated during a small local outbreak, while five out of seven Syrian A. baumannii strains which were isolated in 2013 at the Bundeswehr Hospitals Westerstede and Hamburg formed cluster 2. The remaining five isolates were singletons. The latter clustering as observed with the cgMLST-typing confirmed likely transmission events between Syrian patients prior to admission at the Bundeswehr Hospitals Westerstede and Hamburg, which had already been suspected based on the results of rep-PCR and superficial sequencing [33]. Those transmission events might either have occurred in medical facilities in the patients′ home country but also, as previously discussed for Syrian and Ukrainian war-injured patients treated at German military hospitals [33,40,41], during the evacuation flights out of the patients’ crisis-struck home countries. Of note, two strains isolated from Syrian patients at the military hospital in Westerstede (“WEST”-strains) showed completely identical sequences and other isolates of this cluster, which were isolated at the Bundeswehr Hospital Hamburg (“HBG”-strains) showed only one, two, and nine allele differences, respectively (Figure 1). Such very close genetic relationship makes recent transmission events highly likely.
When comparing the cluster 2 from Figure 1 of the results chapter with the epidemiology at the Bundeswehr hospitals as recently described [33], the resulting likely nosocomial transmission events were as follows. All patients had been admitted to the German Bundeswehr Hospitals at the same day in 2013. Focusing on the Bundeswehr Hospital Westerstede, the strain WEST-S1-31, which had been isolated at day 21 after admission from the anus of a patient, was most likely nosocomially acquired from another patient, from which strain WEST-S6-50 had been isolated at day 1 after admission from a wound. Focusing on the cluster-related strains HBG-S1-56, HBG-S4-64 and HBG-S7-73 isolated at the Bundeswehr Hospital Hamburg, the first two isolates had already been isolated at day 2 after admission from a deep wound and the perineal skin of the patients, respectively, so nosocomial transmission had most likely occurred prior to hospital admission. The strain HBG-S7-73, however, had been isolated at day 9 from the pharynx of a patient. The sequence homology as indicated in Figure 1 makes nosocomial transmission from the patient carrying the strain HBG-S4-64 at the perineal skin highly likely.
As an interesting side-effect, cgMLST confirmed the clonal relationships as suggested by rep-PCR in the recent assessment [33], thus additionally confirming the basic reliability of this comparably cheap and easy-to-perform typing approach for local outbreak scenarios, in spite of the undeniably higher resolution of genome-based typing [34,40,42]. A comparison with other A. baumannii isolates from Ukrainian conflict zones that we have investigated did not show any clonality to those in the current study (data not shown) [40].
When looking at the detected beta-lactam resistance genes, the quantitatively most relevant one was blaOXA-23, which could be identified in strains from all assessed geographic regions, and which is also frequent in resistant A. baumannii in Europe and beyond [43]. Other beta-lactamase genes with relevance for the observed carbapenem resistance comprised blaOXA-58, the class A beta-lactamase gene blaGES-11, and blaNDM-1, respectively. It is interesting to note that isolate WEST-S6-47 had the lowest carbapenem-MICs of the A. baumannii in this study and it only had the acquired blaGES-11. In addition, and especially when looking for the resistome beyond just beta-lactamases, considerable diversity could be seen even in spite of an otherwise low number of allelic differences between some isolates within the observed clonal clusters, which is suggestive for the abundance of plasmids and mobile genetic elements. Despite the high-level of resistance seen in these isolates, colistin still retained activity.
Any comparisons with previous reports from the geographic regions of interest are limited by a paucity of data from respective war and crisis zones. Further, the relatively low number of available isolates is, of course, insufficient to provide a comprehensive overview on locally abundant resistant A. baumannii clones as well as their molecular resistance determinants, an admitted limitation of the study. Nevertheless, the blaOXA-58 gene as detected in the Iraqi A. baumannii strain is one of the genes mediating carbapenem resistance which had been reported already in 2006 and thus early after the beginning of the local conflict [4]. In a similar way, the occurrence of blaNDM-1 and blaOXA-23 in A. baumannii isolated in Libya had been described before [27,29,30], making those detections not further surprising. In so far, several of the reported findings are not completely new but just complement previously available epidemiological information.
4. Materials and Methods
4.1. Patient Isolates
All Acinetobacter strains with reduced susceptibility towards carbapenems and origin from North African and Middle Eastern war and crisis zones available at the laboratory of the Bundeswehr Hospital Hamburg were included in the assessment. There were no exclusion criteria. In detail, the strains Iraq-1 and Iraq-2 were isolated from wounds of injured US-American soldiers in the Iraqi conflict and kindly provided by the US Landstuhl Regional Medical Center (LRMC) as described elsewhere [39]. The strains AFG-1, AFG-2, AFG-3, and AFG-4 isolated from casualties of the Afghanistan conflict were shipped via the German field hospital in Mazar-e Sharif as previously detailed as well [39]. No patient specific data were provided for those strains for security reasons. The origin of the strains from patients injured in the conflicts in Libya and Syria and treated at the Bundeswehr Hospitals Hamburg and Westerstede (HBG-L1 (1), HBG-L2 (9) from Libya; WEST-S1 (31), WEST-S5 (44), WEST-S6 (47), WEST-S6 (50), HBG-S1 (56), HBG-S2 (62), HBG-S4 (64), HBG-S7 (73) from Syria) was described in detail elsewhere [33] next to epidemiological links and a preliminary molecular resistance analysis based on superficial NGS and rep-PCR typing. No environmental isolates were available. A. baumannii were identified using MALDI-TOF-MS and gyrB multiplex PCR [44,45].
4.2. DNA Extraction and Whole Genome Sequencing
DNA extraction and whole genome sequencing was performed exactly as described recently [40]. In detail, MagAttract HMW DNA Kits (Qiagen, Hilden, Germany) were used to extract the genomic DNA of the isolates according to the manufacturer’s instructions for whole genome sequencing (WGS). Nextera XT library prep kits (Illumina GmbH, Munich, Germany) were applied for the preparation of sequencing libraries for 250 bp paired-end sequencing runs on an Illumina MiSeq sequencer. The Velvet assembler integrated in the Ridom SeqSphere+ v.7.0.4. software allowed the de novo assembling of the obtained reads. Any raw sequencing reads from the project were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena/, last accessed on 10 March 2021) with the Accession numbers PRJEB42650.
4.3. Molecular Epidemiology and Determination of Antibiotic Resistance Genes
All isolates were initially screened for the presence of common carbapenemases by PCR [46,47]. Using the pubMLST website (https://pubmlst.org/abaumannii/, last accessed on 10 March 2021)), the genome assemblies of all isolates were used to identify sequence types (STs) according to the Oxford and the Pasteur 7-loci MLST schemes [17,37,38].
Based on a validated core genome multi-locus sequence typing (cgMLST) scheme [34] using the Ridom SeqSphere+ v. 7.0.4 software (Ridom GmbH, Münster, Germany), the isolates were further analyzed. The clonal relatedness was visualized using the same software by generating a minimum spanning tree including 2390 target alleles, also including an in-house library with the established international clones.
Finally, the beta-lactamase database (http://www.bldb.eu/, last accessed on 10 March 2021)) as well as the webtool Resfinder [48,49] were applied for the identification of resistance genes.
4.4. Phenotypic Resistance Testing and Spectrometry-Based Discrimination
A preliminary discrimination of the strains to the A baumannii complex (ABC) level had been performed applying MALDI TOF MS as described previously [33]. In detail, a Shimadzu/Kratos “AXIMA Assurance” MALDI TOF MS (Shimadzu Deutschland GmbH, Duisburg, Germany) and the “IVD-mode VitekMS-ID” database version 3.2.0.-6 (bioMérieux, Marcy-l′Étoile, France) had been used. Regarding phenotypic resistance testing, colistin susceptibility was assessed applying the microbroth dilution assay MICRONAUT-S (MERLIN Diagnostika GmbH, Bornheim, Germany). All other tested substances were assessed applying AST-N248 cards in a VITEK-II automated device (bioMérieux).
4.5. Ethical Clearance
Ethical clearance for the molecular typing of the Acinetobacter strains was obtained from the ethics committee of the medical association of Hamburg (WF-042/15) in line with National German laws, without the need for informed consent.
5. Conclusions
In conclusion, this assessment provided epidemiological information on carbapenem-resistant A. baumannii strains as isolated from patients from Northern African and Middle Eastern war and crisis zones and deposited at the Bundeswehr Hospital of Hamburg, Germany. Respective epidemiological assessments, further analyzing the molecular background of the distribution and spread of antimicrobial resistance in such difficult-to-access regions in the world, are neglected so far, as technological resources for molecular surveillance purposes are scarce in politically unstable areas. Certainly, our current small assessment of more or less randomly isolated strains collected over several years cannot replace continuous surveillance programs but may provide a glimpse on information which is otherwise difficult to obtain and usually relies on small cross-sectional assessments [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30]. Considering the ongoing problem of infections due to carbapenem resistant A. baumannii isolates worldwide, the provided data may serve as a piece in the puzzle of global antimicrobial resistance without diminishing the need for larger future studies on the local epidemiology of carbapenem resistance in Northern African and Middle Eastern A. baumannii isolates in order to amend and broaden this preliminary information.
Appendix A
Table A1.
Results of querying genomes with the TCS function of JSpeciesWS.
| Isolate WEST-S5-44 | ||||||||
| Pos. | Species | Strain | Domain | Phylum | Class | Order | Family | Z-Score |
| 1 | Acinetobacter radioresistens SH164 | SH164 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99921 |
| 2 | Acinetobacter radioresistens NIPH 2130 | NIPH 2130 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99869 |
| 3 | Acinetobacter radioresistens DSM 6976 = NBRC 102413 = CIP 103788 | CIP 103788 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99862 |
| 4 | Acinetobacter radioresistens SK82 | SK82 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99842 |
| 5 | Acinetobacter radioresistens WC-A-157 | WC-A-157 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99811 |
| 6 | Acinetobacter sp. 1461402 | 1461402 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99798 |
| 7 | Acinetobacter sp. 230853 | 230853 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99788 |
| 8 | Acinetobacter sp. 1239920 | 1239920 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99774 |
| 9 | Acinetobacter sp. 263903-1 | 263903-1 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99763 |
| 10 | Acinetobacter sp. 272263 | 272263 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99756 |
| Isolate Iraq-1 | ||||||||
| Pos. | Species | Strain | Domain | Phylum | Class | Order | Family | Z-Score |
| 1 | Acinetobacter lactucae ABBL098 | ABBL098 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99922 |
| 2 | Acinetobacter pittii ABBL016 | ABBL016 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99922 |
| 3 | Acinetobacter lactucae NRRL B-41902 | NRRL B-41902 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.999 |
| 4 | Acinetobacter sp. 1542444 | 1542444 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99894 |
| 5 | Acinetobacter pittii ABBL148 | ABBL148 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99894 |
| 6 | Acinetobacter pittii null | null | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99892 |
| 7 | Acinetobacter pittii ABBL126 | ABBL126 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99891 |
| 8 | Acinetobacter pittii ABBL019 | ABBL019 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.9989 |
| 9 | Acinetobacter pittii ABBL033 | ABBL033 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.9989 |
| 10 | Acinetobacter pittii ANC 4050 | ANC 4050 | Bacteria | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | 0.99885 |
Table A2.
Phenotypic resistance testing of the Acinetobacter spp. isolates based on micro-broth dilution (applied for colistin) and automated resistance testing using a VITEK-II device (BioMéreux, Nürtingen, Germany; applied for all other assessed antimicrobial substances). MICs (minimum inhibitory concentrations) in µg/mL are shown in brackets. Interpretation was performed according to EUCAST NAK (European Committee on Antimicrobial Susceptibility Testing “Nationales Antibiotia-Sensitivitätstest-Kommittee”). S = susceptible. I = Susceptible at increased dosage. R = Resistant. N.d. = not defined.
| Strain Number | Species | Imipenem | Meropenem | Amikacin | Gentamicin | Tobramycin | Ciprofloxacin | Cotrimoxazole | Colistin |
|---|---|---|---|---|---|---|---|---|---|
| Iraq-2 | A. baumannii | R (8) | I (8) | S (≤2) | R (8) | S (4) | R (≥4) | R (160) | S (0.0625) |
| AFG-1 | A. baumannii | R (≥16) | R (≥16) | S (≤2) | S (4) | S (2) | R (≥4) | R (≥320) | S (0.5) |
| AFG-2 | A. baumannii | R (≥16) | R (≥16) | S (≤2) | S (4) | S (≤1) | R (≥4) | R (≥320) | S (0.25) |
| AFG-3 | A. baumannii | R (8) | I (8) | S (8) | S (4) | S (2) | R (≥4) | R (≥320) | S (2) |
| AFG-4 | A. baumannii | R (8) | R (≥16) | S (≤2) | S (4) | S (≤1) | R (≥4) | R (≥320) | S (1) |
| HBG-L1-1 | A. baumannii | R (≥16) | R (≥16) | S (≤2) | R (≥16) | R (≥16) | R (≥4) | R (≥320) | S (0.5) |
| HBG-L2-9 | A. baumannii | R (≥16) | I (8) | S (≤2) | R (8) | R (8) | R (≥4) | R (≥320) | S (0.5) |
| WEST-S6-47 | A. baumannii | I (1) | I (4) | R (≥64) | R (8) | R (8) | R (≥4) | R (≥320) | S (1) |
| WEST-S1-31 | A. baumannii | R (≥16) | R (≥16) | S (4) | R (≥16) | R (≥16) | R (≥4) | R (≥320) | S (0.5) |
| WEST-S6-50 | A. baumannii | R (≥16) | R (≥16) | S (4) | R (≥16) | R (≥16) | R (≥4) | R (≥320) | S (1) |
| HBG-S4-64 | A. baumannii | R (≥16) | R (≥16) | S (8) | R (≥16) | R (≥16) | R (≥4) | S (≤20) | S (0.25) |
| HBG-S1-56 | A. baumannii | R (≥16) | R (≥16) | S (≤2) | S (4) | S (≤1) | R (≥4) | S (≤20) | S (0.125) |
| HBG-S7-73 | A. baumannii | R (≥16) | R (≥16) | S (4) | R (≥16) | R (≥16) | R (≥4) | S (≤20) | S (0.25) |
| HBG-S2-62 | A. baumannii | R (≥16) | R (≥16) | R (16) | R (≥16) | R (≥16) | R (≥4) | R (≥320) | S (1) |
| WEST-S5-44 | A. radioresistens | S (≤0.25) | S (≤0.25) | S (≤2) | S (≤1) | S (≤1) | I (≤0.25) | S (≤20) | S (0.5) |
| Iraq-1 | A. dijkshoorniae | S (≤0.25) | I (4) | S (≤2) | S (≤1) | S (≤1) | I (0.5) | S (≤20) | S (0.125) |
Author Contributions
Conceptualization, P.G.H., R.M.H., B.K., H.F., and U.L.; methodology, P.G.H.; software, P.G.H.; validation, P.G.H.; formal analysis, P.G.H., P.W.; investigation, P.G.H., P.W.; resources, H.F., P.G.H., B.K., A.P., U.L.; data curation, P.G.H.; writing—original draft preparation, P.G.H., H.F.; writing—review and editing, P.G.H., R.M.H., B.K., P.W., A.P., H.F., U.L.; visualization, P.G.H.; project administration, H.F. All authors have read and agreed to the published version of the manuscript.
Funding
We acknowledge support by the Open Access Publication Funds of the University of Göttingen.
Institutional Review Board Statement
Ethical clearance for the molecular typing of the Acinetobacter strains was obtained from the ethics committee of the medical association of Hamburg (WF-042/15, provided on 27 October 2015) in line with National German laws. The study was conducted according to the guidelines of the Declaration of Helsinki.
Data Availability Statement
All relevant data are provided in the manuscript, its figure, its table, and its Appendix A. Raw sequence data are deposited and available as stated in the methods chapter. Further raw data can be made available on reasonable request.
Conflicts of Interest
The sponsors did not have any role in the collection, analysis, or interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Davis K.A., Moran K.A., McAllister C.K., Gray P.J. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 2005;11:1218–1224. doi: 10.3201/1108.050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Shea M.K. Acinetobacter in modern warfare. Int. J. Antimicrob. Agents. 2012;39:363–375. doi: 10.1016/j.ijantimicag.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Turton J.F., Kaufmann M.E., Gill M.J., Pike R., Scott P.T., Fishbain J., Craft D., Deye G., Riddell S., Lindler L.E., et al. Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J. Clin. Microbiol. 2006;44:2630–2634. doi: 10.1128/JCM.00547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hujer K.M., Hujer A.M., Hulten E.A., Bajaksouzian S., Adams J.M., Donskey C.J., Ecker D.J., Massire C., Eshoo M.W., Sampath R., et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 2006;50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffith M.E., Lazarus D.R., Mann P.B., Boger J.A., Hospenthal D.R., Murray C.K. Acinetobacter skin carriage among US army soldiers deployed in Iraq. Infect. Control Hosp. Epidemiol. 2007;28:720–722. doi: 10.1086/518966. [DOI] [PubMed] [Google Scholar]
- 6.Whitman T.J., Qasba S.S., Timpone J.G., Babel B.S., Kasper M.R., English J.F., Sanders J.W., Hujer K.M., Hujer A.M., Endimiani A., et al. Occupational transmission of Acinetobacter baumannii from a United States serviceman wounded in Iraq to a health care worker. Clin. Infect. Dis. 2008;47:439–443. doi: 10.1086/589247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray C.K., Yun H.C., Griffith M.E., Thompson B., Crouch H.K., Monson L.S., Aldous W.K., Mende K., Hospenthal D.R. Recovery of multidrug-resistant bacteria from combat personnel evacuated from Iraq and Afghanistan at a single military treatment facility. Mil. Med. 2009;174:598–604. doi: 10.7205/MILMED-D-03-8008. [DOI] [PubMed] [Google Scholar]
- 8.Sensenig R.A., Murray C.K., Mende K., Wolf S.E., Chung K.K., Hospenthal D.R., Yun H.C. Longitudinal characterization of Acinetobacter baumannii-calcoaceticus complex, Klebsiella pneumoniae, and methicillin-resistant Staphylococcus aureus colonizing and infecting combat casualties. Am. J. Infect. Control. 2012;40:183–185. doi: 10.1016/j.ajic.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 9.Hospenthal D.R., Crouch H.K., English J.F., Leach F., Pool J., Conger N.G., Whitman T.J., Wortmann G.W., Robertson J.L., Murray C.K. Multidrug-resistant bacterial colonization of combat-injured personnel at admission to medical centers after evacuation from Afghanistan and Iraq. J. Trauma. 2011;71(Suppl. 1):S52–S57. doi: 10.1097/TA.0b013e31822118fb. [DOI] [PubMed] [Google Scholar]
- 10.Huang X.Z., Chahine M.A., Frye J.G., Cash D.M., Lesho E.P., Craft D.W., Lindler L.E., Nikolich M.P. Molecular analysis of imipenem-resistant Acinetobacter baumannii isolated from US service members wounded in Iraq, 2003–2008. Epidemiol. Infect. 2012;140:2302–2307. doi: 10.1017/S0950268811002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell W.R., Li P., Whitman T.J., Blyth D.M., Schnaubelt E.R., Mende K., Tribble D.R. Multi-Drug-Resistant Gram-Negative Infections in Deployment-Related Trauma Patients. Surg. Infect. 2017;18:357–367. doi: 10.1089/sur.2017.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Kadmy I.M.S., Ali A.N.M., Salman I.M.A., Khazaal S.S. Molecular characterization of Acinetobacter baumannii isolated from Iraqi hospital environment. New Microbes New Infect. 2017;21:51–57. doi: 10.1016/j.nmni.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weintrob A.C., Murray C.K., Xu J., Krauss M., Bradley W., Warkentien T.E., Lloyd B.A., Tribble D.R. Early Infections Complicating the Care of Combat Casualties from Iraq and Afghanistan. Surg. Infect. 2018;19:286–297. doi: 10.1089/sur.2017.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganjo A.R., Maghdid D.M., Mansoor I.Y., Kok D.J., Severin J.A., Verbrugh H.A., Kreft D., Fatah M.H., Alnakshabandi A.A., Dlnya A., et al. OXA-Carbapenemases Present in Clinical Acinetobacter baumannii-calcoaceticus Complex Isolates from Patients in Kurdistan Region, Iraq. Microb. Drug. Resist. 2016;22:627–637. doi: 10.1089/mdr.2015.0060. [DOI] [PubMed] [Google Scholar]
- 15.Fily F., Ronat J.B., Malou N., Kanapathipillai R., Seguin C., Hussein N., Fakhri R.M., Langendorf C. Post-traumatic osteomyelitis in Middle East war-wounded civilians: Resistance to first-line antibiotics in selected bacteria over the decade 2006–2016. BMC Infect. Dis. 2019;19:103. doi: 10.1186/s12879-019-3741-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassan P.A., Khider A.K. Correlation of biofilm formation and antibiotic resistance among clinical and soil isolates of Acinetobacter baumannii in Iraq. Acta Microbiol. Immunol. Hung. 2019;13:1–10. doi: 10.1556/030.66.2019.026. [DOI] [PubMed] [Google Scholar]
- 17.Gaiarsa S., Batisti Biffignandi G., Esposito E.P., Castelli M., Jolley K.A., Brisse S., Sassera D., Zarrilli R. Comparative Analysis of the Two Acinetobacter baumannii Multilocus Sequence Typing (MLST) Schemes. Front. Microbiol. 2019;10:930. doi: 10.3389/fmicb.2019.00930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun H.C., Branstetter J.G., Murray C.K. Osteomyelitis in military personnel wounded in Iraq and Afghanistan. J. Trauma. 2008;64(Suppl. 2):S163–S168. doi: 10.1097/TA.0b013e318160868c. [DOI] [PubMed] [Google Scholar]
- 19.Calhoun J.H., Murray C.K., Manring M.M. Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin. Orthop. Relat. Res. 2008;466:1356–1362. doi: 10.1007/s11999-008-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuart T.L., Mulvey M., Simor A.E., Tien H.C., Battad A., Taylor G., Vayalumkal J.V., Weir C., Ofner M., Gravel D., et al. Acinetobacter baumannii in casualties returning from Afghanistan. Can. J. Infect. Control. 2007;22:152–154. [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention (CDC) Acinetobacter baumannii infections among patients at military medical facilities treating injured U.S. service members, 2002–2004. MMWR Morb. Mortal. Wkly. Rep. 2004;53:1063–1066. [PubMed] [Google Scholar]
- 22.Hamzeh A.R., Al Najjar M., Mahfoud M. Prevalence of antibiotic resistance among Acinetobacter baumannii isolates from Aleppo, Syria. Am. J. Infect. Control. 2012;40:776–777. doi: 10.1016/j.ajic.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Rafei R., Dabboussi F., Hamze M., Eveillard M., Lemarié C., Mallat H., Rolain J.M., Joly-Guillou M.L., Kempf M. First report of blaNDM-1-producing Acinetobacter baumannii isolated in Lebanon from civilians wounded during the Syrian war. Int. J. Infect. Dis. 2014;21:21–23. doi: 10.1016/j.ijid.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Heydari F., Mammina C., Koksal F. NDM-1-producing Acinetobacter baumannii ST85 now in Turkey, including one isolate from a Syrian refugee. J. Med. Microbiol. 2015;64:1027–1029. doi: 10.1099/jmm.0.000132. [DOI] [PubMed] [Google Scholar]
- 25.Angeletti S., Ceccarelli G., Vita S., Dicuonzo G., Lopalco M., Dedej E., Blasi A., Antonelli F., Conti A., De Cesaris M., et al. Sanitary Bureau of Asylum Seekers Center of Castelnuovo di Porto. Unusual microorganisms and antimicrobial resistances in a group of Syrian migrants: Sentinel surveillance data from an asylum seekers centre in Italy. Travel Med. Infect. Dis. 2016;14:115–122. doi: 10.1016/j.tmaid.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Salloum T., Tannous E., Alousi S., Arabaghian H., Rafei R., Hamze M., Tokajian S. Genomic mapping of ST85 blaNDM-1 and blaOXA-94 producing Acinetobacter baumannii isolates from Syrian Civil War Victims. Int. J. Infect. Dis. 2018;74:100–108. doi: 10.1016/j.ijid.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Hammerum A.M., Larsen A.R., Hansen F., Justesen U.S., Friis-Møller A., Lemming L.E., Fuursted K., Littauer P., Schønning K., Gahrn-Hansen B., et al. Patients transferred from Libya to Denmark carried OXA-48-producing Klebsiella pneumoniae, NDM-1-producing Acinetobacter baumannii and meticillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents. 2012;40:191–192. doi: 10.1016/j.ijantimicag.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Dau A.A., Tloba S., Daw M.A. Characterization of wound infections among patients injured during the 2011 Libyan conflict. East. Mediterr. Health J. 2013;19:356–361. doi: 10.26719/2013.19.4.356. [DOI] [PubMed] [Google Scholar]
- 29.Mathlouthi N., Areig Z., Al Bayssari C., Bakour S., Ali El Salabi A., Ben Gwierif S., Zorgani A.A., Ben Slama K., Chouchani C., Rolain J.M. Emergence of Carbapenem-Resistant Pseudomonas aeruginosa and Acinetobacter baumannii Clinical Isolates Collected from Some Libyan Hospitals. Microb. Drug. Resist. 2015;21:335–341. doi: 10.1089/mdr.2014.0235. [DOI] [PubMed] [Google Scholar]
- 30.Mathlouthi N., El Salabi A.A., Ben Jomàa-Jemili M., Bakour S., Al-Bayssari C., Zorgani A.A., Kraiema A., Elahmer O., Okdah L., Rolain J.M., et al. Early detection of metallo-β-lactamase NDM-1- and OXA-23 carbapenemase-producing Acinetobacter baumannii in Libyan hospitals. Int. J. Antimicrob. Agents. 2016;48:46–50. doi: 10.1016/j.ijantimicag.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Kraiem A.G., Zorgani A., Elahmer O., El Salabi A.A., Ghenghesh K.S. Carbapenem-resistant gram-negative bacilli in Tripoli, Libya. Am. J. Infect. Control. 2016;44:1192–1194. doi: 10.1016/j.ajic.2016.04.245. [DOI] [PubMed] [Google Scholar]
- 32.Kieffer N., Ahmed M.O., Elramalli A.K., Daw M.A., Poirel L., Álvarez R., Nordmann P. Colistin-resistant carbapenemase-producing isolates among Klebsiella spp. and Acinetobacter baumannii in Tripoli, Libya. J. Glob. Antimicrob. Resist. 2018;13:37–39. doi: 10.1016/j.jgar.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 33.Frickmann H., Köller T., Hagen R.M., Ebert K.P., Müller M., Wenzel W., Gatzer R., Schotte U., Binder A., Skusa R., et al. Molecular Epidemiology of Multidrug-Resistant Bacteria Isolated from Libyan and Syrian Patients with War Injuries in Two Bundeswehr Hospitals in Germany. Eur. J. Microbiol. Immunol. 2018;8:1–11. doi: 10.1556/1886.2018.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins P.G., Prior K., Harmsen D., Seifert H. Development and evaluation of a core genome multilocus typing scheme for whole-genome sequence-based typing of Acinetobacter baumannii. PLoS ONE. 2017;12:e0179228. doi: 10.1371/journal.pone.0179228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarrilli R., Pournaras S., Giannouli M., Tsakris A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents. 2013;41:11–19. doi: 10.1016/j.ijantimicag.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Wisplinghoff H., Hippler C., Bartual S.G., Haefs C., Stefanik D., Higgins P.G., Seifert H. Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic species 13TU isolates using a multilocus sequencing typing scheme. Clin. Microbiol. Infect. 2008;14:708–715. doi: 10.1111/j.1469-0691.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- 38.Diancourt L., Passet V., Nemec A., Dijkshoorn L., Brisse S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE. 2010;5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frickmann H., Essig A., Hagen R.M., Riecker M., Jerke K., Ellison D., Poppert S. Rapid identification of Acinetobacter spp. by fluorescence in situ hybridization (FISH) from colony and blood culture material. Eur. J. Microbiol. Immunol. 2011;1:289–296. doi: 10.1556/EuJMI.1.2011.4.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins P.G., Hagen R.M., Podbielski A., Frickmann H., Warnke P. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii Isolated from War-Injured Patients from the Eastern Ukraine. Antibiotics. 2020;9:579. doi: 10.3390/antibiotics9090579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granzer H., Hagen R.M., Warnke P., Bock W., Baumann T., Schwarz N.G., Podbielski A., Frickmann H., Koeller T. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter Baumannii Complex Isolates from Patients that were Injured During the Eastern Ukrainian Conflict. Eur. J. Microbiol. Immunol. 2016;6:109–117. doi: 10.1556/1886.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruppitsch W., Pietzka A., Prior K., Bletz S., Fernandez H.L., Allerberger F., Harmsen D., Mellmann A. Defining and Evaluating a Core Genome Multilocus Sequence Typing Scheme for Whole-Genome Sequence-Based Typing of Listeria monocytogenes. J. Clin. Microbiol. 2015;53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nowak J., Zander E., Stefanik D., Higgins P.G., Roca I., Vila J., McConnell M.J., Cisneros J.M., Seifert H., MagicBullet Working Group WP4 High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 2017;72:3277–3282. doi: 10.1093/jac/dkx322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higgins P.G., Wisplinghoff H., Krut O., Seifert H. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin. Microbiol. Infect. 2007;13:1199–1201. doi: 10.1111/j.1469-0691.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- 45.Higgins P.G., Lehmann M., Wisplinghoff H., Seifert H. gyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J. Clin. Microbiol. 2010;48:4592–4594. doi: 10.1128/JCM.01765-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Higgins P.G., Pérez-Llarena F.J., Zander E., Fernández A., Bou G., Seifert H. OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013;57:2121–2126. doi: 10.1128/AAC.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerezales M., Biniossek L., Gerson S., Xanthopoulou K., Wille J., Wohlfarth E., Kaase M., Seifert H., Higgins P.G. Novel multiplex PCRs for detection of the most prevalent carbapenemase genes in Gram-negative bacteria within Germany. J. Med. Microbiol. 2021 doi: 10.1099/jmm.0.001310. [DOI] [PubMed] [Google Scholar]
- 48.Kleinheinz K.A., Joensen K.G., Larsen M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zankari E. Comparison of the web tools ARG-ANNOT and ResFinder for detection of resistance genes in bacteria. Antimicrob. Agents Chemother. 2014;58:4986. doi: 10.1128/AAC.02620-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are provided in the manuscript, its figure, its table, and its Appendix A. Raw sequence data are deposited and available as stated in the methods chapter. Further raw data can be made available on reasonable request.