Abstract
The recent report of distinct Xanthomonas lineages of Xanthomonas arboricola pv. juglandis and Xanthomonas euroxanthea within the same walnut tree revealed that this consortium of walnut-associated Xanthomonas includes both pathogenic and nonpathogenic strains. As the implications of this co-colonization are still poorly understood, in order to unveil niche-specific adaptations, the genomes of three X. euroxanthea strains (CPBF 367, CPBF 424T, and CPBF 426) and of an X. arboricola pv. juglandis strain (CPBF 427) isolated from a single walnut tree in Loures (Portugal) were sequenced with two different technologies, Illumina and Nanopore, to provide consistent single scaffold chromosomal sequences. General genomic features showed that CPBF 427 has a genome similar to other X. arboricola pv. juglandis strains, regarding its size, number, and content of CDSs, while X. euroxanthea strains show a reduction regarding these features comparatively to X. arboricola pv. juglandis strains. Whole genome comparisons revealed remarkable genomic differences between X. arboricola pv. juglandis and X. euroxanthea strains, which translates into different pathogenicity and virulence features, namely regarding type 3 secretion system and its effectors and other secretory systems, chemotaxis-related proteins, and extracellular enzymes. Altogether, the distinct genomic repertoire of X. euroxanthea may be particularly useful to address pathogenicity emergence and evolution in walnut-associated Xanthomonas.
Keywords: walnut bacterial blight, comparative genomics, Xanthomonas arboricola pv. juglandis, Xanthomonas euroxanthea, pathogenicity
1. Introduction
Xanthomonas is a genus of gammaproteobacteria [1], which include numerous species acknowledged as important plant-associated bacteria with the capacity to cause disease in a wide range of plant species, including important agricultural crops [2,3]. At the current date, the genus comprises 32 species (with validly published and correct names) [4], some of which are subdivided into distinct pathogenicity groups, also known as pathovars, according to their high degree of host specificity, disease symptoms, and infection mechanisms [5,6,7,8]. Within this genus, Xanthomonas arboricola [9] is responsible for severe diseases in important stone fruits and nut trees [10]. Pathogenic strains of X. arboricola belonging to the pathovar juglandis [11] cause serious diseases on walnut, namely walnut bacterial blight (WBB), brown apical necrosis (BAN), and vertical oozing canker (VOC), altogether responsible for major drops in walnut production and extended damages in walnut nurseries, leading to major economic losses [12,13,14].
Compared to other X. arboricola pathovars, X. arboricola pv. juglandis is characterized by a larger genetic diversity recorded throughout geographically distinct walnut cultivation regions [15,16,17,18,19]. Genotyping and population studies of walnut-associated xanthomonads identified a high diversity of X. arboricola pv. juglandis pathogenic strains responsible for walnut bacterial diseases [13] and nonpathogenic strains of X. arboricola shown to be asymptomatic on walnut [20]. Based on a large body of evidence, carefully reviewed by Büttner and Bonas (2010) [21] and more recently by An et al. (2020) [22], it is currently acknowledged that Xanthomonas pathogenesis is dependent on an array of pathogenicity and virulence factors, including type 3 secretion system (T3SS) and type 3 effectors (T3E), but also other type secretion systems, adhesins, extracellular polysaccharides (EPS), lipopolysaccharides (LPS), and plant cell wall hydrolytic enzymes, among others. Furthermore, it has been suggested that these genetic determinants of pathogenicity and virulence are under the control of tight transcriptional and post-transcriptional regulatory mechanisms that allow infective bacteria to adhere, invade the plant host tissues, and to multiply and overcome the plant defense mechanisms [21].
Comparative genomics of pathogenic and nonpathogenic strains of X. arboricola has been instrumental to unveil genetic determinants of pathogenicity and virulence [23,24,25] and to provide insights into evolutionary events linked to pathoadaptation, as emphasized by Cesbron et al. (2015) [24]. Still, in order to gain more insight regarding adaptations of nonpathogenic strains to new environments, comparative genomics of pathogenic and nonpathogenic strains isolated from the same host is required [22]. Although T3SS and T3E have been pointed out as major players of pathogenicity in X. arboricola, the different repertoires of virulence-associated genes between pathogenic and nonpathogenic strains of X. arboricola suggests that pathogenicity and virulence are determined by a complex network of genetic determinants, which is not fully understood [24].
Most of the X. arboricola whole genomes currently available are scattered in multiple contigs assembled from millions of short-reads (typically < 300 bp) obtained by sequencing-by-synthesis technologies (such as Illumina) incapable of resolving repetitive domains and impairing accurate comparisons across wide genomic domains [26]. More recently, single-molecule-sequencing technologies (such as Oxford Nanopore Technologies (ONT)) have emerged. These, are capable of producing long reads (up to several thousand bp) that may span over entire repetitive regions, achieving a complete structural resolution of genetic elements, regardless the higher error rates (above 10% of mismatches and indels, compared to ~1% of mismatches for short reads) that blur the sequence resolution [26,27]. Hybrid assembling approaches, capable to conciliate the accuracy of Illumina short-reads and the gap-free long-read sequences obtained by ONT, result in single chromosome scaffolds with high confidence per base call, and particularly suitable for high accuracy comparative genomics to unveil genetic and structural variants [28,29].
The recent description of pathogenic and nonpathogenic strains isolated from walnut belonging to the new species Xanthomonas euroxanthea [30] raises further questions regarding new pathoadaptations to this host [19,31,32]. Furthermore, the fact that these X. euroxanthea strains were isolated from the same walnut host tree, together with characteristic X. arboricola pv. juglandis strains (CPBF 427, CPBF 1521) [19,30,33,34], suggest a sympatric lifestyle that may contribute to unveil events of genetic recombination and trade-offs related to pathogenicity and virulence of Xanthomonas in walnut. The present work discloses a comprehensive comparative genomics study between the complete genome sequences of three X. euroxanthea strains (CPBF 367, CPBF 424T, CPBF 426) and one X. arboricola pv. juglandis strain (CPBF 427) isolated from the same diseased walnut tree. To better understand the ecology and evolution of these two walnut-associated Xanthomonas species, emphasis has been given to the genetic determinants of pathogenicity and virulence and putative niche-specific adaptations.
2. Materials and Methods
2.1. Xanthomonas Strains Used in This Study and Genome Sequencing
Four isolates (CPBF 367, CPBF 424T, CPBF 426, and CPBF 427) obtained from a single walnut tree host located in the region of Loures, Portugal [19], with distinct genotypes and pathogenicity in walnut (while CPBF 367 was nonpathogenic, CPBF 424T and CPBF 427 were pathogenic) [30], were chosen for genome sequencing. A previous MLSA of acnB, fyuA, gyrB, and rpoD genes grouped the strains CPBF 427 along with other X. arboricola pv. juglandis strains [19], whereas CPBF 367, CPBF 424T, and CPBF 426 were recently described as belonging to the new species X. euroxanthea [30].
Bacteria culture, DNA extraction for sequencing, library preparation, and genome sequencing and annotation were carried out as previously described by Teixeira et al. [32,34]. Briefly, bacteria were recovered on culture medium M2 at 28 °C and 100 rpm for 48 h. DNA was extracted using an E.Z.N.A. bacterial DNA purification kit (Omega Bio-tek, Norcross, GA, USA) and sequenced with Illumina and Oxford Nanopore Technologies (ONT) MinION platforms. Illumina sequencing was outsourced to GATC Biotech, AG (Konstanz, Germany), using an Illumina HiSeq instrument with a standard 2 × 150 bp paired-end library protocol, while ONT sequencing was performed on a MinION sequencer using an R9.4.1 flow cell. Reads were base called and demultiplexed using Guppy v3.4.1 (high accuracy base-calling mode) and assembled de novo following a hybrid Nanopore-Illumina approach using Unicycler v. 0.4.8 [35] and annotated with PGAP v2020-03-30.build4489 [36]. The complete genome sequences of Xanthomonas strains CPBF 367, CPBF 424T, CPBF 426, and CPBF 427 have been deposited in the European Nucleotide Archive (ENA). Illumina reads, ONT reads, and the assembled genomes are respectively available under the following accession numbers: ERX2780809, ERX4296808, and GCA_903989455 for CPBF 367; ERR2767968, ERX4911540, and GCA_905187425 for CPBF 424T; ERX2780811, ERX4296809, and GCA_903989465 for CPBF 426; and ERX2780812, ERX4296810, and GCA_903989475 for CPBF 427.
2.2. Average Nucleotide Identity
The average nucleotide identity (ANI), based on BLASTn, was carried out with OrthoANI v1.40 [37] to determine the genetic distance between each of the four bacterial genomes (CPBF 367, CPBF 424T, CPBF 426, and CPBF 427) sequenced within the framework of this study and 40 genomes of Xanthomonas spp., including 29 genomes of X. arboricola and eight distinct pathovars (Table S1), all available at the NCBI genome database.
2.3. Comparative Genome Analysis
A total of 1149 genes from BUSCO (v. 4.0.6, database xanthomonadales_odb10) [38,39], present in all the 44 mentioned Xanthomonas genomes, were aligned with TCoffee (v11.00.8cbe486, mcoffee mode) [40] to generate a tree, using RAxML (raxmlGUI 2.0, model GTR+FO) [41,42] with 500 bootstrap replicates. The corresponding phylogram was visually represented with the R package “ggtree” [43]. In order to unveil strain-specific features and pools of genes shared between the CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, a pairwise comparison of genomes based on SRVs [44,45] was performed in the EDGAR platform (EDGAR v. 2.3. [45,46] and represented in a Venn diagram.
2.4. Homologous of Pathogenicity and Virulence-Associated Proteins Inferred by tBLASTn Analysis
The genome sequences of the four Xanthomonas strains used in this study were scrutinized for the presence of protein homologs by tBLASTn analysis (tBLASTn v. 2.10.1 [47]) against a created database of protein sequences previously reported to be involved in the pathogenesis and virulence of Xanthomonas (Table S3). To ensure that only closely-related orthologous proteins were selected, the tBLASTn cut-offs criteria to identify protein homologs were ≥ 40% identity and ≥ 75% query sequence length. The search for homologs was performed using as query sequences proteins of xanthan biosynthesis identified by Lee et al. [48] and Vorhölter et al. [49], a list of proteins of the flagellar system from X. campestris pv. vesicatoria 85-10 [50] and from X. fragariae LMG 25863 (AJRZ00000000.1, NCBI database), as well as proteins of the rpf gene cluster for regulation of pathogenicity factors in X. campestris, [51] and X. fragariae LMG 25863, AJRZ00000000.1, NCBI database). Homologs of chemotaxis and methyl-accepting chemotaxis proteins, proteins involved in the biosynthesis of quorum sensing signals and non-fimbrial adhesins, were identified using as query the protein sequences previously used for X. arboricola genomes by Garita-Cambronero et al. [25]. The presence or absence of homologs associated with components of the different secretion systems were also predicted, using as query sequences proteins of the type II secretion system (T2SS) [52,53] and related hemicellulolytic, cellulolytic, and pectolytic enzymes, lipases, and proteases [25]; proteins of the type IV secretion system and type IV pilus [25,54]; and proteins of the type VI secretion system [55].
3. Results
3.1. General Features of X. euroxanthea and X. arboricola pv. juglandis Genome Assemblies
The complete genome sequences obtained by hybrid assemblies of Illumina and Nanopore reads for the four studied strains (CPBF 367, CPBF 424T, CPBF 426, and CPBF 427), allowed a single chromosomal scaffold to be achieved for all four strains and revealed the presence of one plasmid in strains CPBF 367 and CPBF 426 (Table 1). When comparing the genome properties, while some features are similar between the four genomes, namely a G + C content around 65%, a number of coding genes proportional to genome size, and the number of rRNA operons and 5S tRNA genes, some other features are clearly distinct between the three X. euroxanthea strains (CPBF 367, CPBF 424T, and CPBF 426) and the X. arboricola pv. juglandis strain CPBF 427. These differences are particularly underlined by a higher genome size for the X. arboricola pv. juglandis strain CPBF 427 (5.23 Mb), in comparison with the slightly smaller genomes of X. euroxanthea strains CPBF 367, CPBF 424T, and CPBF 426, i.e., less than 5.0 Mb, the higher number of RNA genes, non-coding RNAs (ncRNA), and pseudogenes observed for strain CPBF 427 comparatively with the three X. euroxanthea strains (Table 1).
Table 1.
General genomic features of the four strains analyzed in this study.
| General Features | Xanthomonas euroxanthea Strains | Xanthomonas arboricola pv. juglandis Strain | ||
|---|---|---|---|---|
| CPBF 367 | CPBF 424T | CPBF 426 | CPBF 427 | |
| Genome size (bp) | 4,968,459 | 4,900,930 | 4,900,648 | 5,228,174 |
| Contigs | 2 | 1 | 2 | 1 |
| N50 (bp) | 4,923,218 | 4,900,930 | 4,883,254 | 5,228,174 |
| G+C content (%) | 65.81 | 65.88 | 65.85 | 65.38 |
| Plasmids | 1 | 0 | 1 | 0 |
| Total genes | 4157 | 4119 | 4143 | 4465 |
| Total CDSs | 4077 | 4040 | 4066 | 4367 |
| Coding genes | 4012 | 3993 | 4003 | 4237 |
| RNA genes | 80 | 79 | 77 | 98 |
| rRNA (5S, 16S, 23S) | 2, 2, 2 | 2, 2, 2 | 2, 2, 2 | 2, 2, 2 |
| ncRNA | 18 | 17 | 18 | 38 |
| tRNA | 56 | 56 | 53 | 54 |
| Pseudogenes | 65 | 47 | 63 | 130 |
| ENA/GenBank accession number | GCA_903989455 | CGA_905187425 | GCA_903989465 | GCA_903989475 |
| Reference | [32] | This study | [32] | [34] |
3.2. Genomic Distance Assessed by ANI and Phylogenetic Analysis
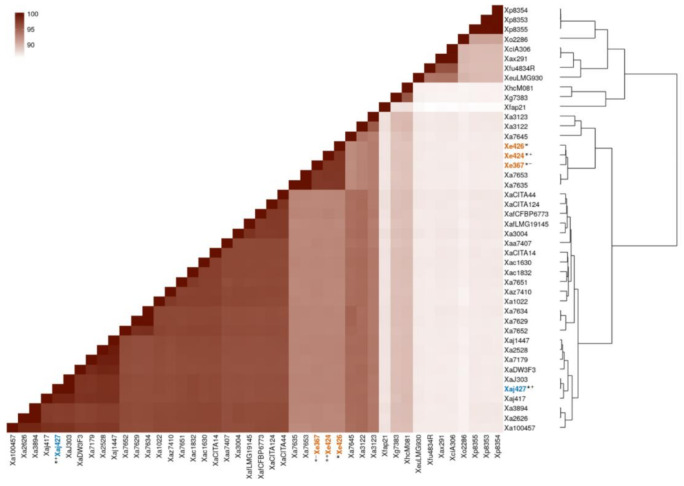
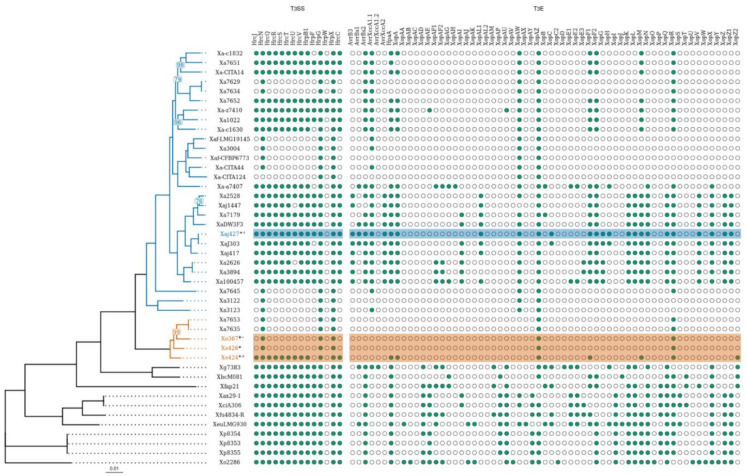
The ANI values determined for a sampling of 44 Xanthomonas strains, including the four walnut-associated Xanthomonas isolates characterized in the current study, assigned strains CPBF 367, CPBF 424T, and CPBF 426 to the recently described new species X. euroxanthea [30], and strain CPBF 427 as a member of X. arboricola pv. juglandis. In fact, the high ANI similarity values of ≥ 97.9% shared between strains CPBF 367, CPBF 424T, and CPBF 426 and the ANI values of ≤ 93.6 and ≤ 89.4% observed between these three strains and X. arboricola and CPBF 427 or other Xanthomonas species strains, respectively (Figure 1, Table S2), are clearly above and below the high stringent threshold of > 95% to separate different species [56]. Interestingly, two nonpathogenic X. arboricola strains (CFBP 7635 and CFBP 7653), previously described [20], share ANI values of ≥ 97.8% with the three X. euroxanthea strains (CPBF 367, CPBF 424T, and CPBF 426), strongly suggesting that these two strains are misclassified and likely belong to X. euroxanthea. The phylogenetic tree obtained for the Xanthomonas 44 strains using the 1149 single-copy orthologous genes from BUSCO placed X. euroxanthea strains (CPBF 367, CPBF 424T, and CPBF 426) and CFBP 7635 and CFBP 7653 in a cluster well separated from all the other xanthomonads considered in the analysis, including the CPBF 427 and other X. arboricola pv. juglandis strains with which they share the same plant host (Figure 2).
Figure 1.
Heatmap representing average nucleotide identity (ANI) (disclosed in Table S2) and the respective distance tree cladogram, determined for 44 Xanthomonas genomes including the four genomes disclosed in this study, indicated with (*) and highlighted in orange (CPBF 367, CPBF 424T, CPBF 426) or blue (CPBF 427). Pathogenic and nonpathogenic on walnut are marked with (+) or (−), respectively. The color scale ranging from white to dark red depicts lower to higher similarity values, respectively. The strain names refer to the code field from Table S1.
Figure 2.
Maximum likelihood tree based on concatenated sequences of the 1149 core genes of 44 Xanthomonas genomes, including the Xanthomonas euroxanthea strains (branches in orange) and Xanthomonas arboricola strains (branches in blue). Phylogenetic relations were inferred using RaxML and the phylogram represented with the R package “ggtree”. Supporting values from 500 bootstrap replicates are indicated near nodes. Dot plot scheme represents the presence/absence scheme for type 3 secretion system (T3SS) and effectors (T3E) putative homologs; ●, present; ○, not present; considering a tBLASTn hit with a query coverage threshold ≥ 75%, and sequence identity with ≥ 40% cut-off. Results for genomes disclosed in this study are marked with (*); also, X. euroxanthea strains CPBF 367, CPBF 424T, and CPBF 426 are highlighted in orange and for X. arboricola pv. juglandis CPBF 427 in blue. Pathogenic and nonpathogenic on walnut are marked with (+) or (−), respectively. The strain names refer to the code field from Table S1. Best BLAST results and accession numbers of sequences used as query are disclosed in Table S3.
3.3. Genetic Patrimony Retrieved from the X. euroxanthea and X. arboricola pv. juglandis Strains Isolated from a Single Walnut Host Tree
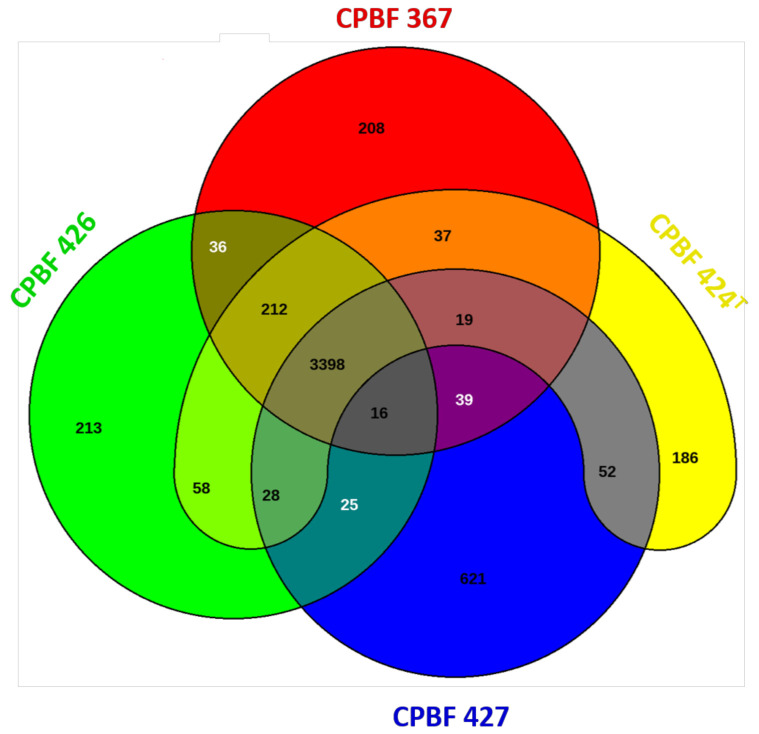
To disclose the gene pool of xanthomonads found in a single walnut tree host, the total number of CDSs corresponding to non-redundant genes retrieved from the four strains studied (CPBF 367, CPBF 424T, CPBF 426, and CPBF 427) was determined. The results highlighted in a Venn diagram showed that the core genome, i.e., the set of genes common to the four strains (Figure 3). The remaining CDSs, which corresponds to the accessory genome, represent the differential gene content of these strains; that is, they include the strain-specific CDSs and genes shared by two or more strains. When focusing on genomic sub-sets, 212 CDSs are shared exclusively by X. euroxanthea strains (CPBF 367, CPBF 424T, and CPBF 426), and 52 CDSs are shared exclusively by the two pathogenic strains (CPBF 424T, and CPBF 427). Among the genes shared by X. euroxanthea, a great number of regulator proteins and hypothetical proteins were found, with the latter constituting 24% of the total genes shared by X. euroxanthea. Regarding strain-specific genomic contents, 621 strain-specific CDSs were retrieved for X. arboricola pv. juglandis, while for X. euroxanthea strains, 208 unique CDSs were identified for CPBF 367, 213 unique CDSs for CPBF 426, and 186 unique CDSs for CPBF 424T. From these singletons, 60%, 52%, and 45% were assigned as hypothetical proteins for CPBF 367, CPBF 424T, and CPBF 426, respectively.
Figure 3.
Venn diagram highlighting the genomic patrimony from CPBF 367, CPBF 424T, CPBF 426, and CPBF 427 genomes. The core genome is given by the interception of the four strains and corresponds to the number of orthologous CDSs shared (3398 CDSs). Strain specific CDSs are also represented in the periphery of the diagram (208 for CPBF 367, 213 for CPBF 426, 186 for CPBF 424T, and 621 for CPBF 427). The remaining combinations represent the number of orthologous shared between two to three genomes. The strain names refer to the code field from Table S1.
3.4. Pathogenic and Virulence-Related Factor Prediction
In addition to the functional analysis, the profile of Xanthomonas pathogenicity and virulence factors characterized in previous studies was determined, unravelling notable differences between the consortium strains isolated from the same walnut host (Figures S1–S6, Table S3). All four xanthomonad strains were found to share numerous genes associated with pathogenesis, namely genes for the biosynthesis of the xanthan polysaccharide biosynthesis (operon gumBCDEFGHIJKLMN), lipopolysaccharide biosynthesis, the flagellar system, the regulatory rpf cluster of pathogenicity factor synthesis, the xps gene of T2SS (xpsD, E, F, G, H, I, J, K, L, and M), homologs of the type IV pilus (T4P), several T4SS genes (virB1, virB2, virB3, virB4, virB6, virB8, virB9, virB10, virB11, and virD4), as well as genes from the T6SS (Table S3). Regarding the T2SS, the xpsN gene was only present in the X. euroxanthea strains (CPBF 367, CPBF 424T, and CPBF 426) (Table 2, Figure S1), and five T4P-related genes (pilY1, pilX, pilW, pilV, and fimT) were found to be exclusively present in the pathogenic CPBF 424T (Table 2, Figure S2). Concerning the presence of non-fimbrial adhesins, it is noticeable that the X. euroxanthea CPBF 424T and CPBF 426 strains did not harbor homologous genes for fhaB1 and fhaB2, which encode filamentous hemagglutinin-related proteins, regardless of the fact that all four strains share at least three homologs to non-fimbrial adhesins (Table 2, Figure S3). Additionally, no major differences were observed between the four strains studied regarding genes encoding proteins associated with Xanthomonas sensing and chemotaxis mechanisms, with the exception of a methyl-accepting chemotaxis protein and a chemotaxis protein of which homologs were present in all three X. euroxanthea strains but not in X. arboricola pv. juglandis strain CPBF 427 (Table 2, Figure S4). The main differences between X. arboricola pv. juglandis and X. euroxanthea strains were observed in the profile of pectolytic enzymes associated with T2SS (Table 2, Figure S5). Homologs for a pectate lyase E and a pectinesterase were only identified in the genomes of the three X. euroxanthea strains. On the contrary, homologs of a degenerated pectate lyase, an endoglucanase, a rhamnogalacturonase B, and a polygalacturonase were present in CPBF 427 and not in X. euroxanthea strains. Furthermore, homologs of xylosidase/arabinosidase (xylB) were found in the two X. arboricola pv. juglandis strains CPBF 427 and in the pathogenic X. euroxanthea strain CPBF 424T.
Table 2.
Comparison of the occurrence of homologs associated with chemotaxis, non-fimbrial adhesins, T2SS, extracellular enzymes, and T4P, between Xanthomonas euroxanthea strains (CPBF 367, CPBF 424T, and CPBF 426) and the Xanthomonas arboricola pv. juglandis (Xaj) strain (CPBF 427).
| X. euroxanthea | Xaj | |||||
|---|---|---|---|---|---|---|
| Protein/Gene Name | Label | GenBank Accession Number | CPBF 367 | CPBF 424T | CPBF 426 | CPBF 427 |
| Chemotaxis-related proteins: | ||||||
| Methyl-accepting chemotaxis protein | XCV1938 | CAJ23615.1 | ● * | ● | ● | ○ |
| Chemotaxis protein | XAC3768 | AAM38611.1 | ● | ● | ● | ○ |
| Non-fimbrial adhesins: | ||||||
| Filamentous hemagglutinin-related protein (fhaB1) | FhaB1 | CAJ23537.1 | ● | ○ | ○ | ● |
| Filamentous hemagglutinin-related protein (fhaB2) | FhaB2 | CAJ23538.1 | ● | ○ | ○ | ● |
| Type II secretion system (T2SS): | ||||||
| General secretion pathway protein XpsN (xpsN) | XpsN | WP_011035909.1 | ● | ● | ● | ○ |
| Extracellular enzymes: | ||||||
| Xylosidase/arabinosidase (xylB) | XylB.1 | WP_011036375.1 | ○ | ● | ○ | ● |
| Xylosidase/arabinosidase (xsa) | Xsa.1 | WP_011037540.1 | ○ | ○ | ○ | ● |
| Xylosidase/arabinosidase (xylB) | XylB.2 | WP_011039174.1 | ○ | ● | ○ | ● |
| Endoglucanase (bcsZ) | bcsZ | AAM38359.1 | ○ | ○ | ○ | ● |
| Polygalacturonase (pglA) | pglA | WP_011037410.1 | ○ | ○ | ○ | ● |
| Pectate lyase E (pelA) | pelA | WP_011035380.1 | ● | ● | ● | ○ |
| Degenerated pectate lyase (pel) | pel | AAM37225.1 | ○ | ○ | ○ | ● |
| Pectinesterase | XCC0121 | WP_011035379.1 | ● | ● | ● | ○ |
| Type IV pilus (T4P): | ||||||
| PilY1 protein (pilY1) | pilY1 | WP_011051753.1 | ○ | ● | ○ | ○ |
| PilX protein (pilX) | pilX | WP_011051754.1 | ○ | ● | ○ | ○ |
| PilW protein (pilW) | XAC2667 | WP_040107776.1 | ○ | ● | ○ | ○ |
| Pre-pilin leader sequence (pilV) | pilV | WP_011051756.1 | ○ | ● | ○ | ○ |
| Pre-pilin like leader sequence (fimT) | fimT | WP_011051757.1 | ○ | ● | ○ | ○ |
* ●, present; ○, not present.
3.5. Type 3 Secretion System and Its Effectors
The presence of homologous proteins for the type 3 secretion system and its effectors were evaluated in CPBF 367, CPBF 424T, CPBF 426, CPBF 427, and other Xanthomonas genomes, including known pathogenic and nonpathogenic X. arboricola strains (Figure 2 and Figure S6). For both T3SS and T3E proteins, X. euroxanthea CPBF 367 and CPBF 426 presented the same profiles as nonpathogenic X. arboricola strains CFBP 7653 and CFBP 7635 and a reduced number of homologs than pathogenic X. euroxanthea strains CPBF 424T. Particularly for T3SS, X. euroxanthea strains CPBF 367 and CPBF 426 revealed a similar profile to nonpathogenic CFBP 7653, CFBP 7635, CFBP 7645, CFBP 7634, CFBP 7629, CITA 124, and CITA 44 with the presence of only HrpG, HrpX, and HrcN. Conversely, pathogenic X. euroxanthea strain CPBF 424T demonstrated a more complete profile that includes HrpG, HrpX, HrcN, HrcJ, HrcQ, HrcR, HrcS, HrcT, HrcV, HrpB1, HrcC, and HrcU, similarly to X. arboricola pathogenic strains including CPBF 427 (strain isolated from the same walnut tree host, at the same sampling event) and nonpathogenic X. arboricola strains CFBP 7652, CFBP 7651, and CITA 14 (Figure 2 and Figure S6). Regardless of the arsenal of T3SS proteins observed for CPBF 424T, this strain includes fewer T3 effector proteins (XopAZ, XopR, HpaA, XopM, XopF1, XopA, XopZ2), similarly to nonpathogenic X. arboricola strains CFBP 7651, CFBP 7652, CITA 14. CPBF 367, and CPBF 426, which only possess XopAZ and XopR, as observed for nonpathogenic CFBP 7653 and CFBP 7635.
4. Discussion
The occurrence of distinct Xanthomonas populations colonizing the same host plant has been previously documented [57]. In walnut and stone fruit trees, besides the presence of X. arboricola strains belonging to pathovar juglandis and pruni, characteristic of these two host species, respectively, the isolation of distinct yellow-pigmented xanthomonads has been reported, mostly represented by nonpathogenic lineages that do not form a phylogenetically coherent group with the pathogenic strains of X. arboricola pathovars [20,58,59]. This raises the need to understand the role played in the pathosystems by these bacteria characterized by distinct genotypes.
In this study, four Xanthomonas strains (CPBF 367, CPBF 424T, CPBF 426, CPBF 427) isolated from the same walnut tree were sequenced to provide an in-depth characterization of these co-colonizing strains to disclose differential genomic contents putatively related to pathogenicity, virulence, and other specific niche adaptations. ANI and a core-genome phylogenetic analysis disclosed the presence of two different Xanthomonas species in one disease walnut tree, with the confirmation that CPBF 427 belongs to X. arboricola pv. juglandis, whilst the other three strains, CPBF 367, CPBF 424T, and CPBF 426, were already assigned to the recently described species X. euroxanthea [30]. Moreover, two of the atypical strains described by Essakhi et al. [20] as X. arboricola, CFBP 7635 and CFBP 7653, were now confirmed to belong to X. euroxanthea. The role of nonpathogenic strains in Xanthomonas evolution and its potential for pathogenicity emergence is often neglected due to their unvalued direct agro-economic impact. However, a recent genomics study on nonpathogenic strains reinforces our lack of knowledge regarding the lifestyle of these strains. In fact, a nonpathogenic isolate from citrus (LMG 8993) was revealed to belong to X. arboricola species, being placed phylogenetically closest to nonpathogenic X. arboricola isolates from walnut (CFBP 7634 and CFBP 7651), than with other nonpathogenic citrus isolates [60]. Currently, the ecological, evolutionary, and pathogenicity implications of this co-colonization are not understood, but it is hardly refutable that this knowledge is needed for the improvement of efficient phytosanitary practices and the design of appropriate management strategies [61].
The genome size of CPBF 427 (5.23 Mbp) is roughly equal to the genome size reported for other sequenced X. arboricola pv. juglandis genomes, namely Xaj 417 [62], NCPPB 1447 [9], J303 [63], DW3F3, [64], and CFBP 2528T and CFBP 7179 [24] or CFSAN033077 and CFSAN033080 [65]. The genomes of X. euroxanthea strains CPBF 367, CPBF 424T, and CPBF 426 were smaller (ranging from ≈ 4.90 to 4.97 Mbp), presenting values closer to the nonpathogenic or avirulent strains of X. arboricola, i.e., with uncertain pathogenicity or belonging to non-juglandis, non-pruni, and non-corylina pathovars, with genome sizes inferior to 5 Mb [66,67,68]. Despite the presence of one plasmid in strains CPBF 367 and CPBF 426, all the virulence-related homologs were chromosomal. The genetic patrimony of the studied strains illustrated by a Venn diagram encompasses genes associated with basic biological aspects of the Xanthomonas genus, as phenotypic traits [69]. Among these, it was possible to discern 212 genes shared exclusively by X. euroxanthea strains. The analysis of these gene sets specific to X. euroxanthea or X. arboricola pv. juglandis suggests the presence of genes encoding for proteins associated with biochemical functions that may confer selective advantages, such as adaptation to different niches, pathogenicity, or colonization of a new host [69]. Some genetic determinants of virulence were shown to be group-specific or even strain-specific. For example, X. euroxanthea strains evidenced an exclusive presence of the xpsN gene, which encodes the XpsN protein of the type II secretion system. Proteins of the xps system were shown to be associated with virulence of Xanthomonas species such as X. campestris, X. oryzae, and X. euvesicatoria [7]. Furthermore, the X. euroxanthea pathogenic strain CPBF 424T harbors a set of genes that encodes proteins associated with type IV pilus (PilY1, PilX, PilW, PilV, FimT), some of which are considered primary structures of the T4P pilin subunits. Indeed, T4P could play an important role in the pathogenesis of various species of Xanthomonas, and in some cases it is thought that this system has a role in plant colonization [54,70]. Comparative studies between pathogenic X. arboricola pv. juglandis and nonpathogenic X. arboricola strains have shown a differential repertoire of genes encoding chemotaxis-related proteins and proteins related to type I, II, and IV secretion systems [24]. Furthermore, a differential repertoire of non-fimbrial adhesins involved in different functions related to bacterial attachment to the host surface were found in all strains, whereas homologs of non-fimbrial adhesins fhaB probably associated with the bacteria colonization were only identified in pathogenic strains [24]. In the same way, homologs of proteins involved in the biogenesis of type IV pilus were observed, but the absence of PilA, PilX, and/or PilV proteins in the genomes of pathogenic and nonpathogenic strains may point to the absence of bacterial surface filaments in all strains [24]. Moreover, differences can be pinpointed between pathogenic and nonpathogenic X. euroxanthea strains, particularly, the five type IV pilus proteins exclusively present in CPBF 424T, and the xylosidases (XylB.1 and XylB.2) only present in CPBF 424T and also in CPBF 427. It was also possible to identify homologs encoding for proteins specific to X. arboricola pv. juglandis strain CPBF 427 that are missing in X. euroxanthea, such as the extracellular enzymes xylosidase (Xsa.1), endoglucanase, polygalacturonase, and degenerate pectate lyase.
Furthermore, major genomic differences between the strains analyzed in this study were observed for T3SS and related T3E homologs. In Xanthomonas spp., T3SS is crucial for translocating effector proteins that have a key role in bacterial proliferation in host tissues and the development of disease symptoms [21]. The majority of pathogenic strains from the 44 analyzed genomes, including CPBF 427 and X. euroxanthea CPBF 424T, displayed a T3SS profile comprised of most homologous genes for highly conserved T3SS of the Hrp2 family [23,24,25,58,71]. Interestingly, a similar pattern was spotted for nonpathogenic X. arboricola strains CFBP 7652, CFBP 7651, and CITA 14 and for the pathogenic X. euroxanthea strain CPBF 424T, with the exception for T3SS homologs HrpF and HrpW, which were slightly below the 75% query coverage threshold used to filter for the most conserved homologs. Conversely, nonpathogenic X. euroxanthea CPBF 367 and CPBF 426, and nonpathogenic X. arboricola CFBP 7653, CFBP 7645, CFBP 7635, CFBP 7634, CFBP 7629, CITA 44, and CITA 124 lacked most of the genes coding for the macromolecular structure of T3SS, as well as the hrpF gene, involved in the translocation of T3E [24], despite harboring regulators genes of T3SS, as hrpX and hrpG [72,73]. Interestingly CPBF 424T lacks homologs for several pathogenicity genes thought to be essential for X. arboricola pv. juglandis strains. In fact, CPBF 424T harbors homologs for seven known effectors, i.e., less than the nine to ten T3Es found in nonpathogenic X. arboricola CFBP 7651, CFBP 7652, and CITA 14. Only two T3Es were identified in nonpathogenic X. euroxanthea stains (CPBF 367 and CPBF 426), suggesting that X. euroxanthea may also make use of other virulence and pathogenicity-related proteins to trigger infection. Still, the intricate mechanism for successful pathogenicity of X. euroxanthea CPBF 424T can only be disclosed with further investigation and dedicated functional assays.
Cesbron et al. [24] and Garita-Cambronero et al. [71] started to elucidate the mechanisms associated with the emergence of X. arboricola pv. juglandis and X. arboricola pv. pruni pathogenic strains. This was achieved by comparative genomics of X. arboricola strains differing on their pathogenicity, including the nonpathogenic X. arboricola strains isolated from walnut and evaluated in this study (CFBP 7634 and CFBP 7651), and X. arboricola strains (CFBP 14, CFBP 44, and CFBP 124) isolated from Prunus. Regardless of the contributions of these comparative genomics studies in highlighting the importance of T3SS and T3E genes, dedicated functional studies are still required to identify essential genes for successful infection. Furthermore, these nonpathogenic strains are particularly valuable to address questions regarding pathogenicity evolution in Xanthomonas.
5. Conclusions
The extensive genomic comparison of four walnut-associated strains isolated from the same walnut specimen and belonging to two species (X. arboricola pv. juglandis and X. euroxanthea), including pathogenic (CPBF 427 and CPBF 424T) and nonpathogenic strains (CPBF 367 and CPBF 426), provides insights about niche-specific adaptations that could inform on the role played by each of these strains in the co-colonization of walnut. Comprehensive genomics analysis, which also includes previously reported nonpathogenic X. arboricola strains, shows that two of these strains, CFBP 7635 and CFBP 7653, belong to X. euroxanthea species. The data gathered suggest a pattern of homologous genes putatively associated with pathogenicity, virulence, and niche-specific adaptations that need to be addressed in future functional studies to determine their importance in walnut diseases caused by Xanthomonas.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/9/3/624/s1, Figure S1. (a) Scheme representing presence/absence of type 2 secretion system (T2SS) putative homologs in 44 Xanthomonas spp. genomes; (b) BLAST identity (%) and length (i.e., % of query coverage) for type 2 secretion system (T2SS) putative homologs in CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, Figure S2. (a) Scheme representing presence/absence of type 4 pilus (T4P) putative homologs in 44 Xanthomonas spp. genomes; (b) BLAST identity (%) and length (i.e., % of query coverage for type 4 pilus T4P) putative homologs in CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, Figure S3. (a) Scheme representing presence/absence of non-fimbrial adhesin putative homologs in 44 Xanthomonas spp. genomes; (b) BLAST identity (%) and length (i.e., % of query coverage) of non-fimbrial adhesins putative homologs in CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, Figure S4. (a) Scheme representing presence/absence of chemotaxis-related protein putative homologs in 44 Xanthomonas spp. genomes; (b) BLAST identity (%) and length (i.e., % of query coverage) of chemotaxis-related protein putative homologs in CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, Figure S5. (a) Scheme representing presence/absence of extracellular enzyme putative homologs in 44 Xanthomonas spp. genomes; (b) BLAST identity (%) and length (i.e., % of query coverage) of extracellular enzyme putative homologs in CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, Figure S6. (a) BLAST identity (%) and length (i.e., % of query coverage) of type 3 secretion system (T3SS) putative homologs in CPBF 367, CPBF 424T, CPBF 426, and CPBF 427; Figure S6. (b) BLAST identity (%) and length (i.e., % of query coverage) of type 3 effector (T3E) putative homologs in CPBF 367, CPBF 424T, CPBF 426. and CPBF 427. Table S1: List of Xanthomonas spp. genomes used in this study, Table S2: Average nucleotide identity results for the genomes of the 44 strains analyzed in this study, Table S3: Best BLAST hit results of putative homologs.
Author Contributions
Conceptualization, C.F., J.F.P., and F.T.; data curation, M.T. and N.A.F.; formal analysis, C.F., L.M., M.T., J.F.P., N.A.F., and F.T.; funding acquisition, N.A.F., and F.T.; investigation, C.F., L.M., and M.T.; project administration, F.T.; resources, J.B., J.F.P., N.A.F., and F.T.; software, M.T., J.B., and N.A.F.; supervision, N.A.F. and F.T.; validation, C.F., L.M., M.T., N.A.F., and F.T.; visualization, C.F., L.M., and M.T.; writing—original draft, C.F., L.M., J.F.P., and F.T.; Writing—review and editing, C.F., L.M., M.T., J.B., J.F.P., N.A.F., and F.T. All authors have read and agreed to the published version of the manuscript.
Funding
Funding for this work was provided by the EU Horizon 2020 Research and Innovation program (grant agreement 668981 and 857251), European Structural and Investment Funds (ESIFs) through COMPETE 2020, National Funds through FCT (project EVOXANT-PTDC/BIA-EVF/3635/2014-POCI-01-0145-FEDER-016600), and the Operational Thematic Program for Competitiveness and Internationalization (POCI) under the PORTUGAL 2020 Partnership Agreement through the European Regional Development Fund (FEDER) project PORBIOTA—POCI-01-0145-FEDER-022127. C.F. and L.M. were supported by fellowships from FCT (SFRH/BD/95913/2013 and SFRH/BD/137079/2018, respectively). This article is based upon work from COST Action CA16107 EuroXanth, supported by COST (European Cooperation in Science and Technology). C.F. was granted a Short-Term Scientific Mission by this COST Action to perform genomic assemblies in Wädenswil (CH). The EDGAR platform is financially supported by the BMBF grant FKZ031A533 within the de.NBI network. The APC was funded COST Action CA16107 EuroXanth.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The assembled genome sequences have been deposited in the European Nucleotide Archive (ENA) under the accession numbers GCA_903989455, CGA_905187425, GCA_903989465, and GCA_903989475 for CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, respectively.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dowson D.W. On the systematic position and generic names of the Gram negative bacterial plant pathogens. Zentralblatt Fur Bakteriol. Parasitenkd. Und Infekt. 2. 1939;100:177–193. [Google Scholar]
- 2.Leyns F., De Cleene M., Swings J.G., De Ley J. The host range of the genus Xanthomonas. Bot. Rev. 1984;50:308–356. doi: 10.1007/BF02862635. [DOI] [Google Scholar]
- 3.Swings J.G., Civerolo E.L. Xanthomonas. Chapman & Hall; London, UK: 1993. [Google Scholar]
- 4.Parte A.C., Carbasse J.S., Meier-Kolthoff J.P., Reimer L.C., Göker M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. [(accessed on 2 February 2021)]; doi: 10.1099/ijsem.0.004332. Available online: https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed]
- 5.Young J.M., Dye D.W., Bradbury J.F., Panagopoulos C.G., Robbs C.F. A proposed nomenclature and classification for plant pathogenic bacteria. J. Agric. Res. 1978;21:153–177. doi: 10.1080/00288233.1978.10427397. [DOI] [Google Scholar]
- 6.Vauterin L., Rademaker J., Swings J. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology. 2000;90:677–682. doi: 10.1094/PHYTO.2000.90.7.677. [DOI] [PubMed] [Google Scholar]
- 7.Ryan R.P., Vorhölter F.J., Potnis N., Jones J.B., Van Sluys M.A., Bogdanove A.J., Dow J.M. Pathogenomics of Xanthomonas: Understanding bacterium-plant interactions. Nat. Rev. Microbiol. 2011;9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 8.Meyer D.F., Bogdanove A.J. Genomics-driven advances in Xanthomonas biology. In: Jackson R., editor. Genomics-Driven Advances in Xanthomonas Biology. Caister Academic Press; Norfolk, UK: 2009. pp. 147–161. [Google Scholar]
- 9.Vauterin L., Hoste B., Kersters K., Swings J. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 1995;45:472–489. doi: 10.1099/00207713-45-3-472. [DOI] [Google Scholar]
- 10.Lamichhane J.R. Xanthomonas arboricola diseases of stone fruit, almond, and walnut trees: Progress toward understanding and management. Plant Dis. 2014;98:1600–1610. doi: 10.1094/PDIS-08-14-0831-FE. [DOI] [PubMed] [Google Scholar]
- 11.Pierce N.B. Walnut bacteriosis. Bot. Gaz. 1901;31:272–273. doi: 10.1086/328100. [DOI] [Google Scholar]
- 12.Smith R.E., Smith C.O., Ramsey H.J. Walnut Culture in California: Walnut Blight. California Agricultural Experiments Station Publications; Berkeley, CA, USA: 1912. [Google Scholar]
- 13.Hajri A., Meyer D., Delort F., Guillaumès J., Brin C., Manceau C. Identification of a genetic lineage within Xanthomonas arboricola pv. juglandis as the causal agent of vertical oozing canker of Persian (English) walnut in France. Plant Pathol. 2010;59:1014–1022. doi: 10.1111/j.1365-3059.2010.02362.x. [DOI] [Google Scholar]
- 14.Belisario A., Maccaroni M., Corazza L., Balmas V., Valier A. Occurrence and etiology of brown apical necrosis on Persian (English) walnut fruit. Plant Dis. 2002;86:599–602. doi: 10.1094/PDIS.2002.86.6.599. [DOI] [PubMed] [Google Scholar]
- 15.Ivanović Ž., Popović T., Janse J., Kojić M., Stanković S., Gavrilović V., Fira D. Molecular assessment of genetic diversity of Xanthomonas arboricola pv. juglandis strains from Serbia by various DNA fingerprinting techniques. Eur. J. Plant Pathol. 2014;141:133–145. doi: 10.1007/s10658-014-0531-5. [DOI] [Google Scholar]
- 16.Kaluzna M., Pulawska J., Waleron M., Sobiczewski P. The genetic characterization of Xanthomonas arboricola pv. juglandis, the causal agent of walnut blight in Poland. Plant Pathol. 2014;63:1404–1416. doi: 10.1111/ppa.12211. [DOI] [Google Scholar]
- 17.Fischer-Le Saux M., Bonneau S., Essakhi S., Manceau C., Jacques M.-A. Aggressive Emerging Pathovars of Xanthomonas arboricola Represent Widespread Epidemic Clones Distinct from Poorly Pathogenic Strains, as Revealed by Multilocus Sequence Typing. Appl. Environ. Microbiol. 2015;81:4651–4668. doi: 10.1128/AEM.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovanardi D., Bonneau S., Gironde S., Fischer-Le Saux M., Manceau C., Stefani E. Morphological and genotypic features of Xanthomonas arboricola pv. juglandis populations from walnut groves in Romagna region, Italy. Eur. J. Plant Pathol. 2016;145:1–16. doi: 10.1007/s10658-015-0809-2. [DOI] [Google Scholar]
- 19.Fernandes C., Albuquerque P., Mariz-Ponte N., Cruz L., Tavares F. Comprehensive diversity assessment of walnut-associated xanthomonads reveal the occurrence of distinct Xanthomonas arboricola lineages and of a new species (Xanthomonas euroxanthea) within the same tree. Plant Pathol. 2021:1–16. doi: 10.1111/ppa.13355. [DOI] [Google Scholar]
- 20.Essakhi S., Cesbron S., Fischer-Le Saux M., Bonneau S., Jacques M.-A., Manceau C. Phylogenetic and Variable-Number Tandem-Repeat Analyses Identify Nonpathogenic Xanthomonas arboricola Lineages Lacking the Canonical Type III Secretion System. Appl. Environ. Microbiol. 2015;81:5395–5410. doi: 10.1128/AEM.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Büttner D., Bonas U. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 2010;34:107–133. doi: 10.1111/j.1574-6976.2009.00192.x. [DOI] [PubMed] [Google Scholar]
- 22.An S.Q., Potnis N., Dow M., Vorhölter F.J., He Y.Q., Becker A., Teper D., Li Y., Wang N., Bleris L., et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2019;44:1–32. doi: 10.1093/femsre/fuz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajri A., Pothier J.F., Le Saux M.F., Bonneau S., Poussier S., Boureau T., Duffy B., Manceau C. Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant-pathogenic Xanthomonas arboricola. Appl. Environ. Microbiol. 2011;78:371–384. doi: 10.1128/AEM.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cesbron S., Briand M., Essakhi S., Gironde S., Boureau T., Manceau C., Fischer-Le Saux M., Jacques M.-A. Comparative Genomics of Pathogenic and Nonpathogenic Strains of Xanthomonas arboricola Unveil Molecular and Evolutionary Events Linked to Pathoadaptation. Front. Plant Sci. 2015;6:1126. doi: 10.3389/fpls.2015.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garita-Cambronero J., Palacio-Bielsa A., Cubero J. Xanthomonas arboricola pv. pruni, causal agent of bacterial spot of stone fruits and almond: Its genomic and phenotypic characteristics in the X. arboricola species context. Mol. Plant Pathol. 2018;19:2053–2065. doi: 10.1111/mpp.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kchouk M., Gibrat J.-F., Elloumi M. Generations of Sequencing Technologies: From First to Next Generation. Biol. Med. 2017;9:1–8. doi: 10.4172/0974-8369.1000395. [DOI] [Google Scholar]
- 27.Lu H., Giordano F., Ning Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinforma. 2016;14:265–279. doi: 10.1016/j.gpb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oxford Nanopore Technologies Large Insights into Microorganisms. [(accessed on 28 January 2021)]; Available online: https://Nanoporetech.com/publication.
- 29.Goldstein S., Beka L., Graf J., Klassen J.L. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genom. 2019;20:23. doi: 10.1186/s12864-018-5381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martins L., Fernandes C., Blom J., Dia N.C., Pothier J.F., Tavares F. Xanthomonas euroxanthea sp. nov., a new xanthomonad species including pathogenic and non-pathogenic strains of walnut. Int. J. Syst. Evol. Microbiol. 2020;70:6024–6031. doi: 10.1099/ijsem.0.004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandes C., Blom J., Pothier J.F., Tavares F. High-Quality Draft Genome Sequence of Xanthomonas sp. Strain CPBF 424, a Walnut-Pathogenic Strain with Atypical Features. Microbiol. Resour. Announc. 2018;7 doi: 10.1128/MRA.00921-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teixeira M., Martins L., Fernandes C., Chaves C., Pinto J., Tavares F., Fonseca N.A. Complete Genome Sequences of Walnut-Associated Xanthomonas euroxanthea Strains CPBF 367 and CPBF 426 Obtained by Illumina/Nanopore Hybrid Assembly. Microbiol. Resour. Announc. 2020;9:e00902–20. doi: 10.1128/MRA.00902-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandes C., Blom J., Pothier J.F., Tavares F. High-Quality Draft Genome Sequence of Xanthomonas arboricola pv. juglandis CPBF 1521, Isolated from Leaves of a Symptomatic Walnut Tree in Portugal without a Past of Phytosanitary Treatment. Microbiol. Resour. Announc. 2018;7:e00887–18. doi: 10.1128/MRA.00887-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teixeira M., Fernandes C., Chaves C., Pinto J., Tavares F., Fonseca N.A. Complete Genome Sequence Obtained by Nanopore and Illumina Hybrid Assembly of Xanthomonas arboricola pv. juglandis CPBF 427, Isolated from Buds of a Walnut Tree. Microbiol. Resour. Announc. 2021;10:e00085–21. doi: 10.1128/MRA.00085-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee I., Kim Y.O., Park S.C., Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 38.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 39.Waterhouse R.M., Seppey M., Simao F.A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E.V., Zdobnov E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Notredame C., Higgins D.G., Heringa J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 41.Edler D., Klein J., Antonelli A., Silvestro D. RaxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021;12:373–377. doi: 10.1111/2041-210X.13512. [DOI] [Google Scholar]
- 42.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 43.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T. ggtree: An r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 44.Lerat E., Daubin V., Moran N.A. From Gene Trees to Organismal Phylogeny in Prokaryotes:The Case of the γ-Proteobacteria. PLoS Biol. 2003;1:e19. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blom J., Albaum S.P., Doppmeier D., Pühler A., Vorhölter F.J., Zakrzewski M., Goesmann A. EDGAR: A software framework for the comparative analysis of prokaryotic genomes. BMC Bioinform. 2009;10:1–14. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blom J., Kreis J., Spänig S., Juhre T., Bertelli C., Ernst C., Goesmann A. EDGAR 2.0: An enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016;44:W22–W28. doi: 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 48.Lee B.M., Park Y.J., Park D.S., Kang H.W., Kim J.G., Song E.S., Park I.C., Yoon U.H., Hahn J.H., Koo B.S., et al. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 2005;33:577–586. doi: 10.1093/nar/gki206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vorhölter F.J., Schneiker S., Goesmann A., Krause L., Bekel T., Kaiser O., Linke B., Patschkowski T., Rückert C., Schmid J., et al. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 2008;134:33–45. doi: 10.1016/j.jbiotec.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 50.Darrasse A., Carrère S., Barbe V., Boureau T., Arrieta-Ortiz M.L., Bonneau S., Briand M., Brin C., Cociancich S., Durand K., et al. Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genom. 2013;14:1–30. doi: 10.1186/1471-2164-14-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crossman L., Dow J.M. Biofilm formation and dispersal in Xanthomonas campestris. Microbes Infect. 2004;6:623–629. doi: 10.1016/j.micinf.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Filloux A. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta Mol. Cell Res. 2004;1694:163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Szczesny R., Jordan M., Schramm C., Schulz S., Cogez V., Bonas U., Büttner D. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria. New Phytol. 2010;187:983–1002. doi: 10.1111/j.1469-8137.2010.03312.x. [DOI] [PubMed] [Google Scholar]
- 54.Dunger G., Llontop E., Guzzo C.R., Farah C.S. The Xanthomonas type IV pilus. Curr. Opin. Microbiol. 2016;30:88–97. doi: 10.1016/j.mib.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Shrivastava S., Mande S.S. Identification and functional characterization of gene components of type VI secretion system in bacterial genomes. PLoS ONE. 2008;3:e2955. doi: 10.1371/journal.pone.0002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacques M.A., Guimbaud J.F., Briand M., Indiana A., Darrasse A. Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. Volume 2. Wiley Blackwell; Hoboken, NJ, USA: 2016. Flagellar Motility and Fitness in Xanthomonads; pp. 1265–1273. [Google Scholar]
- 58.Garita-Cambronero J., Palacio-Bielsa A., López M.M., Cubero J. Comparative genomic and phenotypic characterization of pathogenic and non-pathogenic strains of Xanthomonas arboricola reveals insights into the infection process of bacterial spot disease of stone fruits. PLoS ONE. 2016;11:e0161977. doi: 10.1371/journal.pone.0161977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacques M.-A., Arlat M., Boulanger A., Boureau T., Carrère S., Cesbron S., Chen N.W.G., Cociancich S., Darrasse A., Denancé N., et al. Using Ecology, Physiology, and Genomics to Understand Host Specificity in Xanthomonas. Annu. Rev. Phytopathol. 2016;54:163–187. doi: 10.1146/annurev-phyto-080615-100147. [DOI] [PubMed] [Google Scholar]
- 60.Bansal K., Kumar S., Patil P.B. Phylogenomic Insights into Diversity and Evolution of Nonpathogenic Xanthomonas Strains Associated with Citrus. mSphere. 2020;5:e00087-20. doi: 10.1128/mSphere.00087-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quibod I.L., Perez-Quintero A., Booher N.J., Dossa G.S., Grande G., Szurek B., Vera Cruz C., Bogdanove A.J., Oliva R. Effector Diversification Contributes to Xanthomonas oryzae pv. Oryzae Phenotypic Adaptation in a Semi-Isolated Environment. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep34137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pereira U.P., Gouran H., Nascimento R., Adaskaveg J.E., Goulart L.R., Dandekar A.M. Complete genome sequence of Xanthomonas arboricola pv. juglandis 417, a copper-resistant strain isolated from Juglans regia L. Genome. Genome Announc. 2015;3:e01126-15. doi: 10.1128/genomeA.01126-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Retamales J., Segovia C., Alvarado R., Nuñez P., Santander J. Draft Genome Sequence of Xanthomonas arboricola pv. juglandis J303, Isolated from Infected Walnut Trees in Southern Chile. Genome Announc. 2017;5:e01085–17. doi: 10.1128/genomeA.01085-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fu B., Chen Q., Wei M., Zhu J., Zou L., Li G., Wang L. Complete Genome Sequence of Xanthomonas arboricola pv. juglandis Strain DW3F3, Isolated from a Juglans regia L. Bacterial Blighted Fruitlet. Genome Announc. 2018;6:e00023–18. doi: 10.1128/genomeA.00023-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Higuera G., González-Escalona N., Véliz C., Vera F., Romero J. Draft Genome Sequences of Four Xanthomonas arboricola pv. juglandis Strains Associated with Walnut Blight in Chile: TABLE 1. Genome Announc. 2015;3:e01160–15. doi: 10.1128/genomeA.01160-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garita-Cambronero J., Palacio-Bielsa A., López M.M., Cubero J. Draft genome sequence for virulent and avirulent strains of Xanthomonas arboricola isolated from Prunus spp. in Spain. Stand. Genom. Sci. 2016;11:1–10. doi: 10.1186/s40793-016-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ignatov A.N., Kyrova E.I., Vinogradova S.V., Kamionskaya A.M., Schaad N.W., Luster D.G. Draft genome sequence of Xanthomonas arboricola strain 3004, a causal agent of bacterial disease on barley. Genome Announc. 2015;3:1572–1586. doi: 10.1128/genomeA.01572-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harrison J., Grant M.R., Studholme D.J. Draft genome sequences of two strains of Xanthomonas arboricola pv. celebensis isolated from banana plants. Genome Announc. 2016;4:e01705-15. doi: 10.1128/genomeA.01705-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Medini D., Donati C., Tettelin H., Masignani V., Rappuoli R. The microbial pan-genome. Curr. Opin. Genet. Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Hersemann L., Wibberg D., Blom J., Goesmann A., Widmer F., Vorhölter F.J., Kölliker R. Comparative genomics of host adaptive traits in Xanthomonas translucens pv. graminis. BMC Genom. 2017;18:35. doi: 10.1186/s12864-016-3422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garita-Cambronero J., Palacio-Bielsa A., López M.M., Cubero J. Pan-genomic analysis permits differentiation of virulent and non-virulent strains of Xanthomonas arboricola that cohabit Prunus spp. and elucidate bacterial virulence factors. Front. Microbiol. 2017;8:1–17. doi: 10.3389/fmicb.2017.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo Y., Figueiredo F., Jones J., Wang N. HrpG and HrpX Play Global Roles in Coordinating Different Virulence Traits of Xanthomonas axonopodis pv. citri. Mol. Plant Microbe Interact. 2011;24:649–661. doi: 10.1094/MPMI-09-10-0209. [DOI] [PubMed] [Google Scholar]
- 73.Jacobs J.M., Pesce C., Lefeuvre P., Koebnik R. Comparative genomics of a cannabis pathogen reveals insight into the evolution of pathogenicity in Xanthomonas. Front. Plant. Sci. 2015;6:431. doi: 10.3389/fpls.2015.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled genome sequences have been deposited in the European Nucleotide Archive (ENA) under the accession numbers GCA_903989455, CGA_905187425, GCA_903989465, and GCA_903989475 for CPBF 367, CPBF 424T, CPBF 426, and CPBF 427, respectively.