Abstract
Nuclear pore complexes (NPCs) are the central mediators of nucleocytoplasmic transport. Increasing evidence shows that many cancer cells have increased numbers of NPCs and become addicted to the nuclear transport machinery. How reducing NPC numbers affects the physiology of normal and cancer cells and if it could be exploited for cancer therapies has not been investigated. We report that inhibition of NPC formation, a process mostly restricted to proliferating cells, causes selective cancer cell death, prevents tumor growth and induces tumor regression. While cancer cells die in response to NPC assembly inhibition, normal cells undergo a reversible cell cycle arrest that allows them to survive. Mechanistically, reducing NPC numbers results in multiple alterations contributing to cancer cell death including abnormalities in nuclear transport, catastrophic alterations in gene expression, and the selective accumulation of DNA damage. Our findings uncover the NPC formation process as a novel targetable pathway in cancer cells.
Keywords: Nuclear pore complex, nucleoporins, nucleocytoplasmic transport, assembly, cancer target
INTRODUCTION
Nuclear pore complexes (NPCs), the multiprotein channels that connect the nucleus with the cytoplasm, are the sole gateway to the genome (1). Together with nuclear transport receptors, NPCs control nucleocytoplasmic molecule exchange. Recent studies have shown that many cancer cells, in particular multidrug resistance cells and cells from aggressive tumors, have increased numbers of NPCs, higher nucleocytoplasmic transport rates, and become addicted to the nuclear transport system, suggesting that the nuclear transport machinery is a vulnerability of several cancers (2–7). Consistent with this idea, inhibitors of nucleocytoplasmic transport have been found to be highly effective at inducing cancer cell death and are the subject of multiple clinical trials (8,9).
Because NPCs are essential cellular structures, directly targeting these channels has historically been viewed as an unfeasible therapeutic strategy. Notably, we and others previously uncovered that non-dividing cells strongly downregulate the expression of key NPC components and maintain their assembled NPCs for years, maybe even decades, suggesting the formation of NPCs would be mostly restricted to cell proliferation (10,11). These findings put forth the exciting possibility that preventing the assembly of new NPCs would decrease the number of these structures only in cells that are actively dividing. Moreover, cells that divide faster, require more nuclear pores, or have a stronger dependency on the nuclear transport process, such as transformed cells, would be expected to be more sensitive to the reduction in NPC numbers. But whether reducing NPC numbers can indeed be exploited to differentially induce cancer cell death, and how it affects the physiology of normal cells has not been investigated. Here, we provide evidence for the selective requirement of NPC formation in proliferating cells and uncover that inhibition of nuclear pore formation induces cancer cell death while it triggers a reversible cell cycle arrest in normal cells that allows them to survive a block in NPC production. We also show that reducing NPC numbers is sufficient to prevent the growth of melanoma and colorectal xenograft tumors and to induce tumor regression. Consistent with their multifunctional nature, we demonstrate that suppression of NPC assembly impacts multiple cellular processes associated with transformation resulting in cancer-specific cell death. The restricted nature of nuclear pore assembly, the selective sensitivity of cancer cells to the inhibition of this process, and the simultaneous alterations of multiple cellular processes that contribute to cancer cell death, indicate that blocking nuclear pore formation represents a promising strategy for anti-neoplastic therapies.
RESULTS
NUP160 and NUP93 scaffold nucleoporins are required for NPC assembly
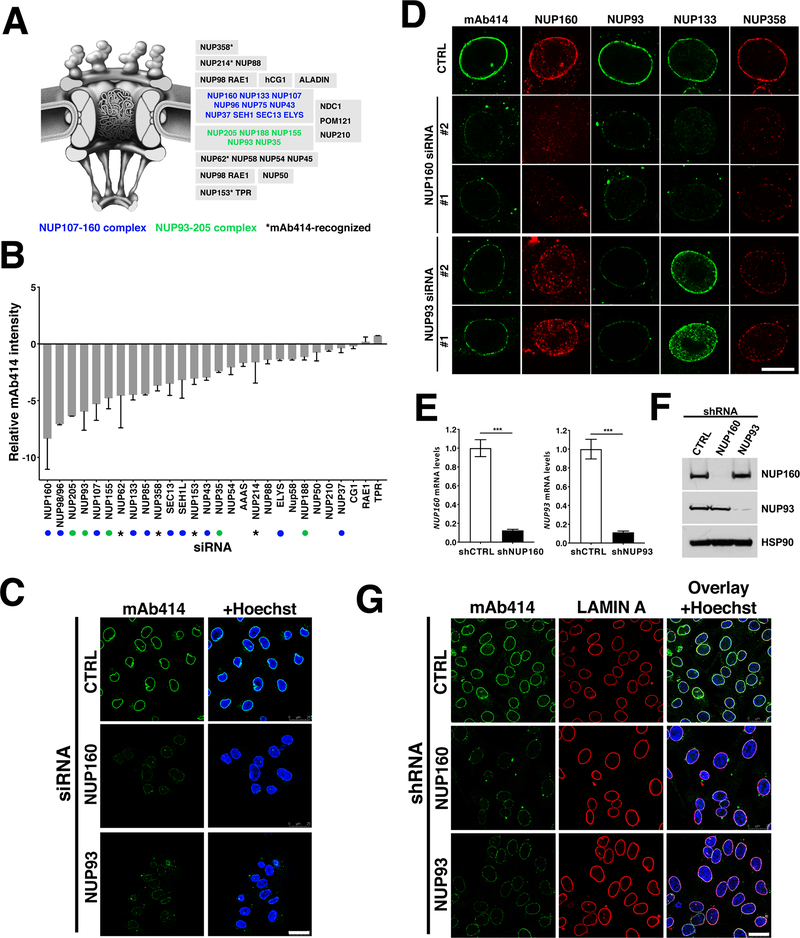
As a first approach to investigate if NPC assembly represents a vulnerability of cancer cells, we performed an siRNA screen against most nucleoporins to identify the most critical components for NPC formation. For this, malignant human melanoma A375 (BRAF V600E) cells were transfected with siRNA pools against 28 of the 32 nucleoporins (Fig. 1A), and NPCs at the nuclear envelope were quantified by immunofluorescence using the mAb414 NPC antibody recognizing 4 different nucleoporins (NUP62, NUP153, NUP214 and NUP358). Consistent with previously described functions of scaffold nucleoporins (12–19), depletion of components from the NUP107–160 and NUP93–205 scaffold subcomplexes most strongly inhibited NPC assembly (Fig. 1B). Nuclear pore complex biogenesis is a highly ordered process that involves the stepwise recruitment of different components, with the NUP107–160 and NUP93–205 complexes being required at different stages of assembly (20). The NUP107-NUP160 complex comes early during assembly and is required for the subsequent recruitment of the NUP93-NUP205 complex to the nuclear envelope (20). The NUP93-NUP205 complex is critical for the later recruitment of the central channel nucleoporins required for the initiation of nucleocytoplasmic transport (20). To inhibit NPC formation at different stages of the process we chose one nucleoporin from each complex, NUP160 and NUP93. Downregulation of these nucleoporins using the pool of 4 siRNAs from our screen (Fig. 1C) or individual siRNAs (Fig. 1D) strongly decreased the NPC signal at the nuclear envelope. Consistent with the previously reported order of assembly, depletion of NUP160 resulted in the loss of all tested NPC components from the nuclear envelope (Fig. 1D). On the other hand, downregulation of NUP93 did not entirely block the recruitment of the early scaffold nucleoporin NUP133 to chromatin and the nuclear envelope but inhibited the association of the later nucleoporins recognized by mAb414, preventing the formation of mature/functional NPCs (Fig. 1D).
Figure 1. NUP93 and NUP160 nucleoporins are critical for NPC assembly.
A, Schematic illustration of nuclear pore complexes. Subcomplexes are shown in gray boxes. Blue and green nucleoporins are part of NPC scaffold subcomplexes. Asterisk shows the four different FXFG-repeat-containing nucleoporins recognized by the mAb414 antibody. B, Nucleoporins were downregulated in A375 cells using specific siRNA pools (four siRNAs per target) and NPC intensity at the nuclear envelope was quantified 48 hours post-transfection using the mAb414 antibody. 500–2000 cells were analyzed for each well and siRNA treatment was done in duplicate wells. Green and blue dots indicate scaffold nucleoporins and asterisks indicate mAb414-recognized nucleoporins as shown in (A). C, NUP93 and NUP160 were downregulated for 72 hours using the specific siRNA pools described in (B) and NPCs were analyzed by immunofluorescence with the mAb414 antibody. Scale bar 25 μm. D, A375 cells treated with control siRNAs or two different siRNAs against NUP93 and NUP160 for 72 hours were stained against different NPC components. Images show the nuclear cross-sections. Scale bar, 10 μm. E, F, A375 cells stably expressing doxycycline-inducible control shRNAs or shRNAs against NUP93 and NUP160 were treated with doxycycline for 72 hours and the expression levels of both nucleoporins was analyzed by qPCR (E) and western blot (F). G, Immunofluorescence analysis of LAMIN A and mAb414 in A375 cells after 72 hours of Control, NUP160, or NUP93 shRNA induction. Scale bar 25 μm. Data are mean ± s.d. Experiments are representative of 3–5 independent repeats.
NPC assembly is linked to cell proliferation and critical for the survival of multiple cancer cells
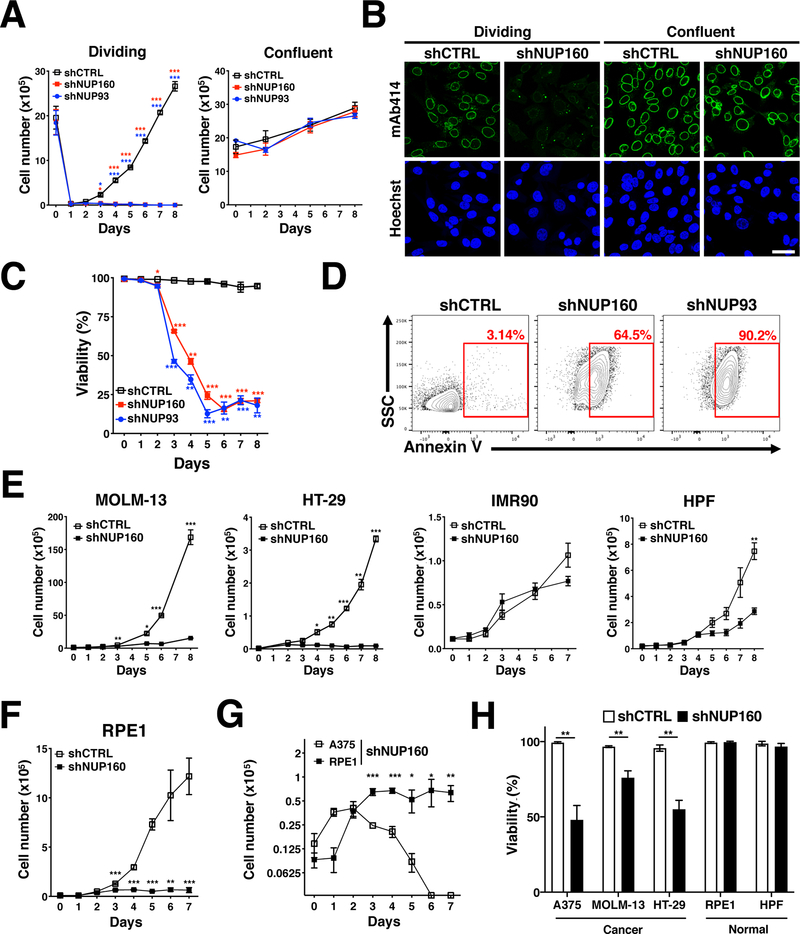
The strong downregulation in the expression of critical assembly nucleoporins during cell cycle exit and the low turnover of NPCs in post-mitotic cells suggest that the formation of these structures is mostly restricted to actively dividing cells (10,11). To test this hypothesis, we analyzed the consequence of blocking NPC assembly in non-proliferating and proliferating cells. We performed these experiments with A375 melanoma cells, which have been found to be sensitive to alterations in nucleocytoplasmic transport (2,21), and used the depletion of the essential nucleoporins NUP160 and NUP93 to inhibit NPC assembly. For this, we generated stable A375 cell lines expressing doxycycline-inducible control shRNA or shRNAs against NUP160 and NUP93 that allowed us to temporally control the depletion of these nucleoporins. Treatment of these cells with doxycycline led to a strong depletion of NUP160 and NUP93 and resulted in inhibition of NPC assembly comparable to siRNA downregulation (Fig. 1E–G and Supplementary Fig. 1A). A375 cells were allowed to grow to confluency before being treated with doxycycline for 72 hours to eliminate the synthesis of new nucleoporins. Cells were then either replated at 1:10 dilution (T=0) to allow cell proliferation or maintained in a confluent non-dividing state, both in the continuous presence of doxycycline. We found that blocking NPC assembly by depletion of these essential nucleoporins strongly reduced the number of dividing cells without significantly affecting non-dividing/confluent cells (Fig. 2A). Analysis of RNA expression and NPC staining indicated that, even though nucleoporin expression was strongly downregulated in both conditions (Supplementary Fig. 1B), blocking nuclear pore formation only decreased NPC numbers in proliferating cells (Fig. 2B). The same results were observed when growth was slowed down by decreasing the serum concentration in the culture media (Supplementary Fig. 1C). Cells grown in low serum, which decreased their proliferation, showed a lower inhibition of NPC assembly compared to cells grown in high serum media (Supplementary Fig. 1D). To further confirm that NPC formation is only required during cell proliferation, the expression of NUP160 was eliminated in proliferating myoblasts or differentiated post-mitotic myotubes and NPC levels at the nuclear envelope were analyzed 3 or 10 days after shRNA induction respectively. Eliminating the expression of the essential nucleoporin NUP160 in proliferating myoblasts significantly reduced the number of NPCs in these cells, while the number of NPCs in post-mitotic myotubes remained unaffected (Supplementary Fig. 1E). These findings further indicate that NPC formation is only required during cell proliferation. To investigate if inhibition of NPC assembly is cytostatic or results in cell death (cytotoxic), proliferating A375 cells depleted of NUP160 or NUP93, were stained with trypan blue or the apoptotic marker Annexin V and the percentage of viable cells and apoptotic/necrotic cells was quantified. Figure 2C and D show that blocking the formation of NPCs results in decreased cell viability and a significant increase in the levels of Annexin V.
Figure 2. NPC assembly is required for cancer cell survival.
A, A375 cells were treated with doxycycline to induce Control, NUP160, or NUP93 shRNA for 72 hours and at T=0 either replated at 1:10 dilution to induce cell proliferation or maintained in a confluent state, both in the continuous presence of doxycycline. The number of viable cells was counted over time. B, Dividing and confluent A375 cells expressing inducible Control or NUP160 shRNAs were treated with doxycycline to inhibit NPC formation. Dividing and confluent cells were fixed and stained with the NPC antibody mAb414 after 3 or 6 days of knockdown respectively. Images show a representative immunofluorescence of 3 independent experiments. Scale bar 25 μm. C, The viability of A375 proliferating cells expressing Control, NUP160 or NUP93 shRNAs was determined at the indicated time points by automatic counting with the Trypan blue exclusion dye method. stained cells. Graph shows the percentage of cells excluding Trypan Blue (live cells). D, Flow cytometry analysis of Annexin V staining in A375 cells after 6 days of Control, NUP160, or NUP93 shRNA induction. Red rectangles show percentage of Annexin V positive cells. E, MOLM-13, HT-29, IMR90, and HPF cells stably expressing inducible Control or NUP160 shRNAs were treated with doxycycline and the number of viable cells was measured over time. F, RPE1 cells stably expressing inducible Control or NUP160 shRNAs were treated with doxycycline and the number of viable cells was measured over time. G, A375 and RPE1 cells stably expressing inducible Control or NUP160 shRNAs were treated with doxycycline and the number of viable cells was measured over time. H, Viability was determined by automated counting of Trypan blue inclusion/exclusion of A375, RPE1, MOLM-13, HT-29 and HPF cells after 4 days of Control or NUP160 shRNA induction. Data are mean ± s.d. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 by multiple unpaired Student’s t tests with Holm-Sidak method to correct for multiple comparisons in (A, C, E, F, G, H). Experiments are representative of 3 independent repeats.
Previous studies have identified that cancer cells from different tumor types including melanoma (2,21), colorectal cancer (7,22), lung cancer (3), leukemia and lymphomas (9,23) have high sensitivity to nuclear transport inhibitors. The susceptibility of cancer cells to nuclear transport inhibitors suggests that they could also be more sensitive to inhibition of NPC formation than normal cells. In agreement with this prediction, blocking NPC formation in MOLM-13 (Acute myeloid leukemia) and HT-29 (colorectal cancer) cells rapidly reduced the number of proliferating cancer cells, while normal proliferating IMR90 and HPF primary fibroblasts showed a much smaller reduction in cell growth after several days of inhibition (Fig. 2E). Because IMR90 and HPF fibroblasts grow slower than most cancer cells analyzed (doubling time: ~35–50 vs 18–20 hours respectively (Supplementary Fig. 1F), we examined the effect of inhibiting NPC assembly in normal RPE1 retina pigment epithelial cells (doubling time: ~20 hours) to account for the differences in growth rates. Even though the inhibition of NPC assembly also reduced the number of proliferating RPE1 cells (Fig. 2F), we noticed that unlike A375 cancer cells that undergo rapid cell death after ~3–4 days of NPC formation inhibition, these normal cells stopped proliferating without dying (Fig. 2G, H and Supplementary Movie). The same differential cell death response to inhibition of NPC assembly was observed between the HT-29 and MOLM-13 cancer cells and normal HPF cells (Fig. 2H), indicating that the selective sensitivity to the reduction in NPC numbers is a common feature of several cancer cells. Altogether, these findings suggest that differently from cancer cells that die in response to inhibition of NPC assembly, normal cells might undergo cell cycle arrest.
Inhibition of NPC assembly leads to a reversible cell cycle arrest in normal cells but triggers death in cancer cells
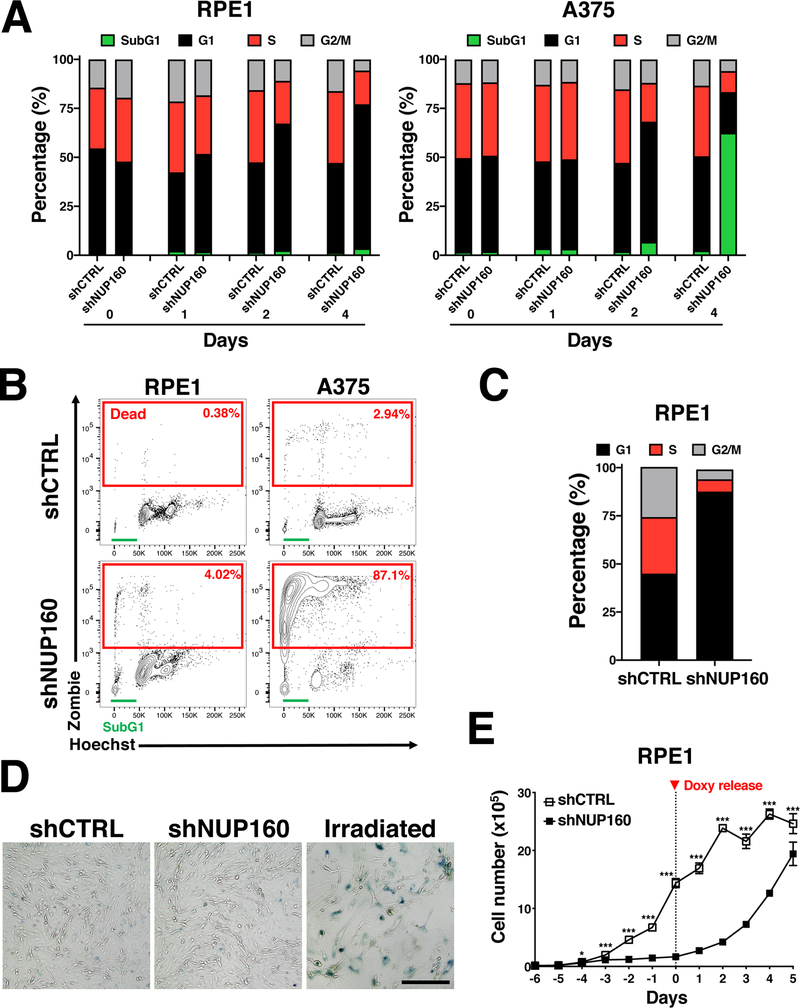
To further investigate the response of normal and cancer cells to inhibition of NPC assembly we performed cell cycle analyses on NUP160-depleted A375 and RPE1 cells at different times after inducing knockdown. We identified that by day 4 of NPC assembly inhibition, the majority of normal RPE1 cells were arrested in the G1 phase of the cell cycle (Fig. 3A) and showed no significant cell death (≤2%). On the other hand, when NPC formation was blocked in melanoma A375 cells ≥50% of the cells were labeled with the Zombie cell death marker, and the remaining Zombie-negative cells (considered live cells) displayed a massive expansion of the SubG1 population which represents dying cells (Fig. 3A, and Supplementary Fig. 2A). A similar cell death response was observed when inhibition of NPC formation was performed by depletion of NUP93 (Supplementary Fig. 2B). After 7 days of NPC inhibition the small number of A375 cells remaining were almost all (≥87%) stained with the Zombie cell death marker, while RPE1 cells showed no significant cell death and remained arrested in the G1 phase (Fig. 3B, C and Supplementary Fig. 2A). Like RPE1 cells, normal HPF primary fibroblasts also arrested in G1 after NPC assembly was inhibited (Supplementary Fig. 2C). Cell cycle-arrested RPE1 cells showed no staining for β-galactosidase (Fig. 3D), indicating they did not become senescent and suggesting a reversible cell cycle arrest. To investigate if reestablishing NPC assembly could restore the growth of RPE1 cells, doxycycline was eliminated from the culture media after 6 days of treatment and cell number was measured over time. We observed that RPE1 cells grew back when the expression of NUP160 was reestablished (Fig. 3E). However, no increase in A375 cell number was observed under these conditions (Supplementary Fig. 2D). To determine if other normal cells respond similarly to inhibition of NPC formation, we depleted NUP160 in the immortalized human melanocyte cell line H3A, normally used as a control for the A375 melanoma cells (24), and in H9 human embryonic stem cells (ESCs) (25), which are most closely related to cancer cells. For these experiments, H3A stably expressing control or NUP160 shRNAs were treated with doxycycline to reduce NUP160 levels, while control or NUP160 siRNAs were used to inhibit NPC assembly in H9 ESCs. Both treatments resulted in strong downregulation of NUP160 expression levels, decreased NPC numbers and increased p21 (CDKN1A) expression, a marker of G1/S cell cycle arrest (26,27) (Supplementary Fig 3A and B). Release from NPC inhibition in H3A cells was done by removing doxycycline from the media on day 6 while the release in ESCs was caused by the standard progressive decrease of siRNA-induced knockdown due to the dilution of siRNA levels over time (average duration of >70–80% siRNA knockdown is ~5–7 days according to manufacturer) (Supplementary Fig. 3C). In the same way as RPE1 cells, inhibition of NPC assembly in these cell types resulted in a strong growth arrest that was relieved when NUP160 expression was restored, and these cells regained their ability to proliferate with no major impact on cell survival (Supplementary Fig. 3D and E).
Figure 3. Selective sensitivity of melanoma A375 cells to inhibition of NPC formation.
A, Flow cytometry analysis of cell cycle in RPE1 and A375 cells at the indicated times after induction of Control or NUP160 shRNAs. The percentage of cells in each phase of the cell cycle is indicated. Data are the mean result representative of one of three independent experiments. B, Flow cytometry analysis of viability (Zombie) and cell cycle (Hoechst) in RPE1 and A375 cells at 7 days of Control or NUP160 shRNA treatment. The percentage of non-viable cells, positively stained with the viability dye, is indicated for each condition in red rectangles. The sub-G1 phase of the cell cycle is indicated by a green line. C, Flow cytometry analysis of cell cycle in RPE1 cells at 7 days of Control or NUP160 shRNA treatment. The percentage of live cells in each phase of the cell cycle is indicated. D, β-Galactosidase staining of RPE1 cells after 4 days of Control or NUP160 shRNA induction. RPE1 cells irradiated with 10 Gy and cultured for 7 days were used as positive controls for senescence. Images are representative of 2 independent experiments. E, RPE1 cells expressing inducible or Control or NUP160 shRNAs were treated with doxycycline (doxy) for 6 days. Cell were then incubated with doxycycline-free media (T= 0) to shut down shRNA expression. The number of viable cells before and after doxy release was counted over time by trypan blue exclusion. Data are mean ± s.d. * P ≤ 0.05, *** P ≤ 0.001 by multiple unpaired Student’s t tests with Holm-Sidak method to correct for multiple comparisons in (E). Unless noted, experiments are representative of a minimum of 3 independent repeats.
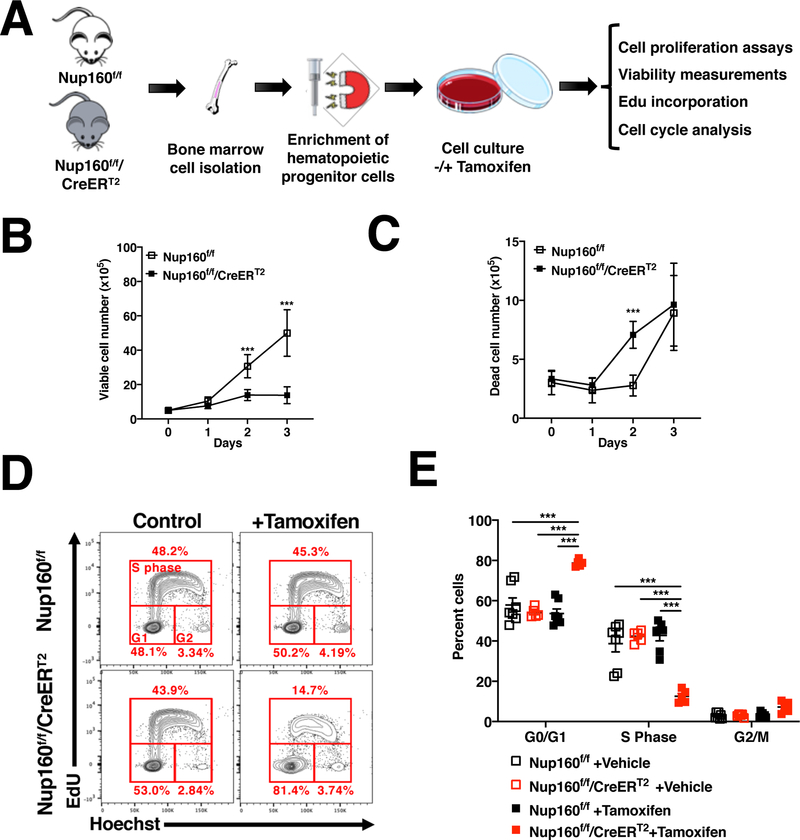
To determine if primary cells also arrest in response to inhibition of NPC formation we generated a conditional Nup160 knockout mouse line in which exon 4 is flanked by loxP sites (Nup160f/f) and crossed it with a mouse line constitutively expressing a tamoxifen-regulated Cre recombinase (R26-CreERT2) (28), which allows temporal control of Nup160 ablation. Hematopoietic progenitors isolated from these mice were treated with tamoxifen to eliminate Nup160 and inhibit NPC assembly, and cell proliferation, survival and cell cycle progression was analyzed over time (Fig. 4A and Supplementary 4A and B). As shown in Figure 4B and C, knocking out Nup160 in primary hematopoietic progenitors was associated with decreased cell numbers but did not strongly increase cell death (<10% dead cells in the population), supporting the idea that primary hematopoietic cells also arrest in response to inhibition of NPC assembly. To confirm this, cell cycle and EdU incorporation analyses were performed in control and Nup160 knockout cells at 72 hours of tamoxifen treatment. Consistent with a cell cycle arrest, Nup160-depleted cells, but not control cells, showed a significant decrease in the number of EdU-positive cells and a strong increase in the G1 population (Fig. 4D and E). Altogether, our data indicate that when NPC formation is inhibited, normal cells reversibly arrest in G1 while transformed cells undergo massive cell death. These findings confirm the selective sensitivity of cancer cells to alterations in nuclear pore complex formation.
Figure 4. Primary hematopoietic cells arrest in response to inhibition of NPC assembly.
A, Schematic illustration of experimental approach to evaluate the effect of Nup160 knockout in mouse primary hematopoietic cells. B, C, Hematopoietic progenitors isolated from control (Nup160f/f) or Nup160 inducible knockout mice (Nup160f/f/CreERT2) were treated with DMSO vehicle or 4-hydroxytamoxifen to knockout Nup160 and the number of viable (B) and dead (C) cells was quantified over time by trypan blue exclusion. D, Flow cytometry analysis of cell cycle and EdU incorporation in Nup160f/f and Nup160f/f/CreERT2 hematopoietic progenitor cells after 72 hours of tamoxifen treatment followed by incubation with EdU for 2 hours. E, Quantification of EdU incorporation and cell cycle stages from (D). Each data point represents cells isolated from an individual animal (n = 5–7). Data are mean ± s.e.m. * P ≤ 0.05, *** P ≤ 0.001 by multiple unpaired Student’s t tests with Holm-Sidak method to correct for multiple comparisons in (B, C) or by two-way analysis of variance (ANOVA, E). Graphs are representative of 3 independent experiments
Blocking nuclear pore formation similarly reduces the number of NPCs in normal cells and cancer cells
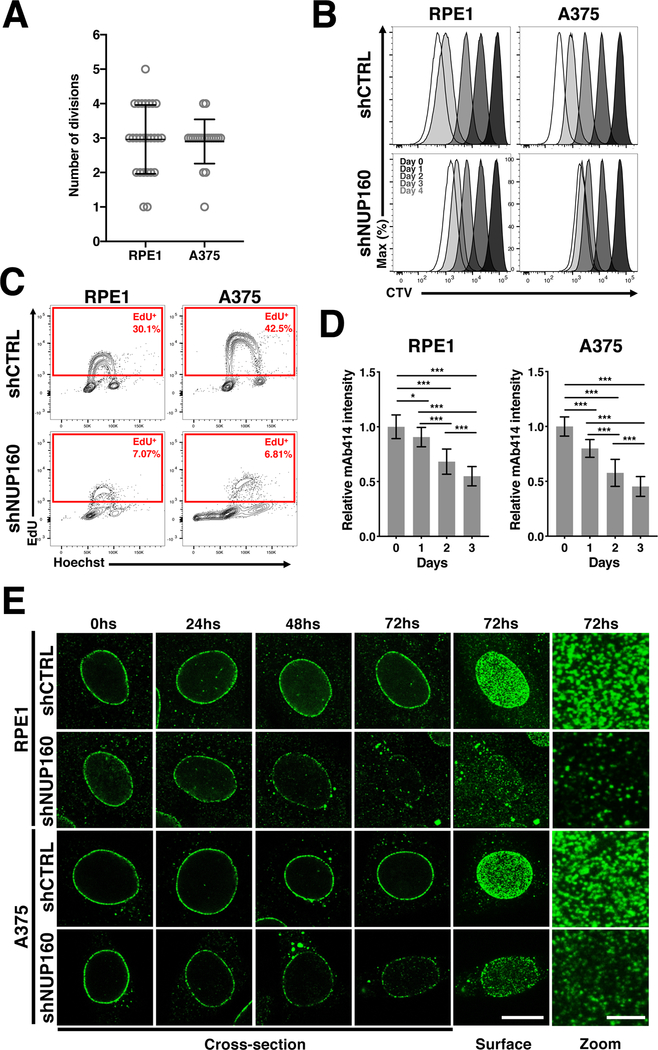
Proliferating cells have two mechanisms of NPC formation. One occurs at the end of mitosis when nuclear envelopes reform around the daughters’ genomes (post-mitotic NPC assembly), and the other one takes place during interphase when cells double their number of NPCs to prepare for the next cell division (interphase NPC assembly) (29). Even though both assembly processes result in the formation of the same structure, they occur through fundamentally different mechanisms (20,29). During mitotic nuclear envelope breakdown, the NPCs of the mother cell are disassembled into stable subcomplexes that are reused for the formation of new nuclear pores during post-mitotic NPC assembly. If all nucleoporins are recycled and distributed evenly between daughter cells, each cell is expected to receive enough components to assemble half of the nuclear pores that the mother cell had when entering M phase. Inhibiting interphase NPC assembly, for example by blocking the expression of new NPC components, should not prevent cells from assembling nuclear pores using pre-existing components at the end of mitosis but would preclude cells from forming new NPCs during interphase when their numbers need to double. This would lead to a progressive reduction in the number of NPCs with each cell division. Because cancer cells die in response to NPC assembly inhibition, one possibility is that when NUP160 expression is inhibited, these cells continue to divide therefore diluting their NPC numbers to levels that are incompatible with cell survival. To test this hypothesis A375 and RPE1 cells were allowed to reach confluency and then treated with doxycycline for 72 hours to eliminate NUP160 as described before. Cells were then diluted in presence of doxycycline to maintain the inhibition of NPC formation during cell proliferation and the number of times each cell divided before either dying or arresting was analyzed by live imaging. Notably, most A375 and RPE1 cells divided 3 times before undergoing cell death or cell cycle arrest respectively (Fig. 5A). If anything, it was more common to see cells that divided more than 3 times in the RPE1 population than in the A375 (Fig. 5A). To confirm this using a different synchronization and quantification method, cells were arrested in G1/S using thymidine in the presence of doxycycline to reduce NUP160 expression, labeled with CTV, which allows us to quantify the number of times cells divide (30), and released in the presence of doxycycline to maintain the inhibition of NPC formation during cell division. Consistent with our imaging assays, we found that most NUP160-depleted cells divided a maximum of 3 times, while control cells kept dividing until the CTV dye was undetectable (Fig. 5B). Also, EdU incorporation assays on day 4 after inhibition of NPC assembly showed a similar reduction in the percentage of actively proliferating RPE1 and A375 cells, further indicating that A375 melanoma cells do not divide more than normal cells when NPC formation is inhibited (Fig. 5C and Supplementary Fig. 5A). Even more, when cells were analyzed 16 hours after the EdU pulse, none of the SubG1 cells showed EdU incorporation, confirming that dying cells are not undergoing cell division at this stage (Supplementary Fig. 5B).
Figure 5. Blocking NPC assembly similarly affects cell division and NPC numbers in melanoma and epithelial cells.
A, The number of cell divisions that RPE1 and A375 undergo after induction of NUP160 shRNA before dying or arresting was quantified using live cell imaging. Circles represent individual cells (n = 20–26 cells). B, RPE1 and A375 cells were labeled with the cell tracking dye CTV and cell division number after inhibition of NPC assembly with NUP160 shRNAs (dilution of CTV signal) was determined by flow cytometry. The CTV dilution across time is indicated, with each peak representing a cell division. C, Flow cytometry analysis of EdU incorporation and cell cycle (Hoechst) in cells treated with Control or NUP160 shRNAs for 4 days and with EdU for 2 hours before analysis. The rectangles indicate the percentage of proliferating, EdU positive, cells. D, Quantification of NPC signal (n = 20–40 cells) at the nuclear envelope in proliferating RPE1 and A375 cells at the indicated times after Control or NUP160 shRNA induction with doxycycline. E, Representative images of NPC staining (nuclei cross-section or surface) in RPE1 and A375 cells at different times after Control or NUP160 shRNA induction. Surface scale bar, 10 μm. Zoom scale bar 2.5 μm. Data are mean ± s.d. * P ≤ 0.05, *** P ≤ 0.001 by two-way analysis of variance (ANOVA, D). Unless otherwise stated, experiments are representative of a minimum of 3 independent repeats.
Another possibility is that cancer cells are more dependent on interphase NPC assembly and that inhibition of this process results in a stronger dilution of nuclear pores with each cell division. To investigate this, we quantified the NPC signal at the nuclear envelope of A375 and RPE1 cells at different times after induction of control or NUP160 shRNAs. Consistent with a progressive dilution of NPC numbers, downregulation of NUP160 expression resulted in a gradual decrease of nuclear pore signal in both A375 and RPE1 cells (Fig. 5D and E). Notably, the treatment resulted in the same reduction of NPCs relative to controls in both cell types, with RPE1 cells having slightly less NPCs at the end of treatment consistent with their lower starting number of NPCs compared to A375 cells (Supplementary Fig. 5C). Altogether these findings indicate that the death of cancer cells in response to the inhibition of NPC formation is not due to a higher dilution of NPCs at the nuclear envelope but to an increased dependency on nuclear pores, and provides additional evidence for the selective susceptibility of cancer cells to alterations in the nuclear transport machinery.
Reducing NPC numbers results in multiple cellular alterations contributing to cancer cell death
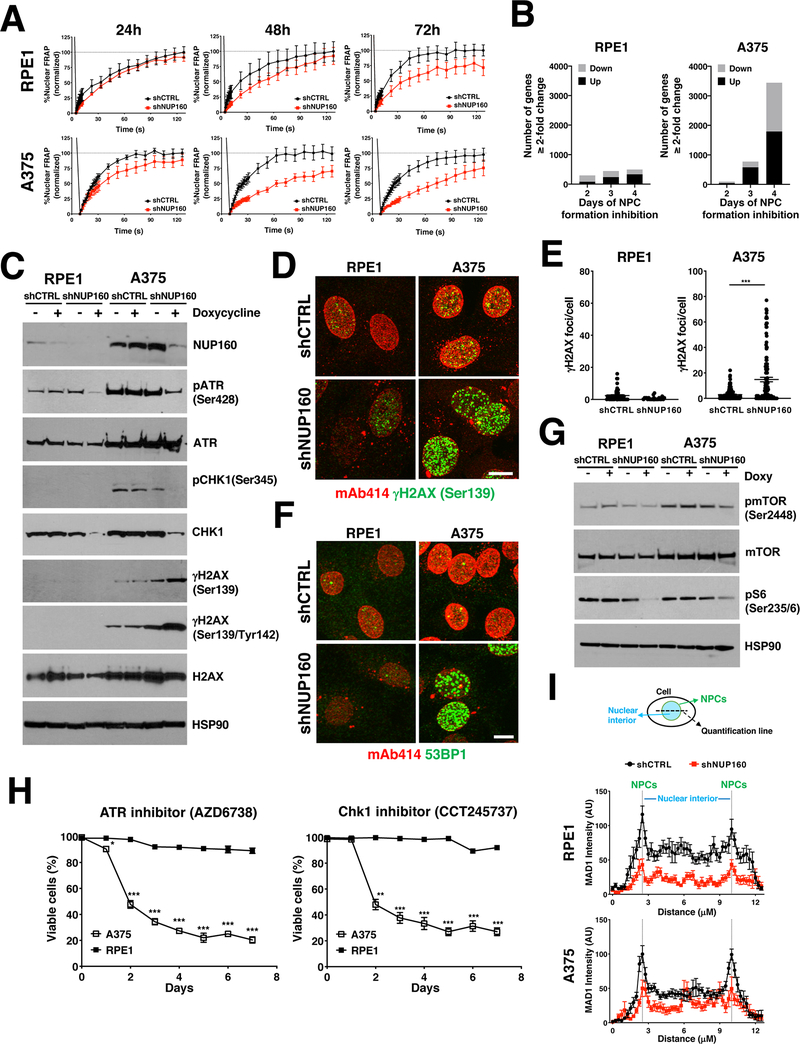
In recent years it has become evident that NPCs play critical roles in many cellular processes. Thus, reducing their numbers is likely to result in multiple alterations contributing to the decreased survival of cancer cells. Consistent with their canonical role in regulating nucleocytoplasmic molecule exchange, using two different transport reporters we found that inhibiting the formation of NPCs was associated with a gradual decline in nuclear import efficiency in both cell types (Fig. 6A, Supplementary Fig. 6A and B). For RPE1 cells, nuclear import rates recovered when cells were released from NPC inhibition and cell proliferation was restored (Supplementary Fig. 6B). RNAseq analyses of gene expression at different times after the inhibition of NPC assembly showed a progressive deregulation of gene activity that paralleled the alterations of nucleocytoplasmic transport (Fig. 6B). Even though the reduction in NPC numbers was associated with decreased nuclear import rates and the deregulation of genes in both cell types, A375 cells showed much larger alterations in these processes with a massive deregulation of gene activity by day 4 of NUP160 knockdown, supporting the higher dependency of these cells on the nuclear transport machinery. Because NPCs also play a key role in maintaining the nuclear permeability barrier, it is also a possibility that NUP160 depletion could lead to nuclear permeability alterations and the leaking of cytoplasmic and nuclear molecules that could allow normal cells to partially compensate the nuclear transport reduction. To investigate this possibility, we performed two complementary approaches. In the first one (Supplementary Fig. 7A and B), we investigated the diffusion of two inert fluorescent proteins of different sizes, Dronpa (~26kDa) and 2x-GFP (~52kDa), into the nucleus of control and NUP160-depleted RPE1 cells using FRAP as previously described (31,32). In the second approach (Supplementary Fig. 7C), we analyzed nuclear permeability in vitro using a combination of two fluorescent dextrans of different sizes above the NPC passive diffusion limit of 40–60kDa in plasma membrane-permeabilized cells (10,33). Using these methods, we found that depletion of NUP160 was not associated with alterations in nuclear permeability (Supplementary Fig. 7A–C), consistent with the NUP107–160 complex being required at the very early stages of NPC assembly before the nucleo-cytoplasmic connection is established (34).
Figure 6. Reduction in NPC number results in multiple cellular alterations.
A, Fluorescence recovery after photobleaching (FRAP) analysis showing nuclear import of the NES-Tomato-NLS reporter in RPE1 and A375 cells after treatment for the indicated number of days with Control or NUP160 siRNAs. The data are expressed as percentage of FRAP relative to maximum recovery of Control (n = 8–16 cells). B, RNAseq analysis of genes significantly changed ≥ 2-fold (q-value < 0.05) in RPE1 and A375 cells treated with NUP160 shRNAs for the indicated number of days compared to Control cells. C, Western blot analysis of DNA damage response proteins in RPE1 and A375 cells untreated or treated with doxycycline for 72 hours to induce Control or NUP160 shRNAs. D, Immunofluorescence analysis of NPCs (mAb414) and γH2AX (Ser139) in RPE1 and A375 cells after 96 hours days of treatment with Control or NUP160 shRNAs. Images show the maximum projection of entire nuclei. Scale bar, 10 μm. E, Quantification of images from (D) (n = 59–79 cells). F, Immunofluorescence analysis of NPCs (mAb414) and 53BP1 in RPE1 and A375 cells after 96 hours days of treatment with Control or NUP160 shRNAs. Representative images show the maximum projection of entire nuclei. Scale bar, 10 μm. G, Western blot analysis of mTOR signaling proteins in RPE1 and A375 cells untreated or treated with doxycycline for 3 days to induce Control or NUP160 shRNA. H, Viability was determined by automated counting of Trypan blue inclusion/exclusion of proliferating A375 and RPE1 cells grown with 10 μM of ATR or CHK1 inhibitors for the indicated time points. I, Top panel shows schematic illustration of method used to quantify MAD1 association with nuclear pores. Bottom panel shows quantification of MAD1 immunofluorescent images of RPE1 and A375 cells (n = 12–14 cells) 72 hours after induction of Control or NUP160 shRNAs with doxycycline. Dashed lines indicate edge of nuclear periphery/NPCs. Data are mean ± s.e.m (A, E) or mean ± s.d. (H, I). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 by unpaired Student’s t test (E) or multiple unpaired Student’s t tests with Holm-Sidak method to correct for multiple comparisons (H). Unless otherwise stated, experiments are representative of a minimum of 3 independent repeats.
In addition to regulating nucleocytoplasmic transport, NPCs have been found to play important roles in DNA damage response (DDR) (35–39). These findings suggest that the reduction in NPC numbers could lead to an altered DDR and increased accumulation of DNA damage. Alterations in the DNA repair machinery and higher levels of genome damage are a hallmark of cancer cells (40). Due to the increased mutation rates and fast proliferation, cancer cells generally show higher levels of DNA damage and replication stress and are more sensitive than normal cells to inhibition of the mechanisms that modulate these processes (40,41). Consistent with cancer cells having more DNA damage than normal cells, A375 cells showed higher endogenous levels of total and phosphorylated (p) ATR (Ser428) and CHK1 (Ser345), two key components of DDR, as well as increased basal levels of the DNA damage marker γH2AX compared to RPE1 cells (Fig. 6C and Supplementary Fig. 8A). Remarkably, we found that the reduction in NPC number by depletion of NUP160 or NUP93 led to decreased levels of pATR, pChk1 and total Chk1 in both cell types but increased the levels of γH2AX only in A375 cells (Fig. 6C and Supplementary Fig. 8B). NPC formation inhibition resulted in more A375 cells with γH2AX foci that were larger in size and intensity when compared to control treated cells (Fig. 6D, E, and Supplementary Fig. 8C and D). Similarly, blocking NPC assembly increased the fraction of cells with large 53BP1 foci in the A375 population, but not in RPE1 cells (Fig. 6F and Supplementary Fig. 8E). Notably, many γH2AX and 53BP1 foci observed in NUP160 knockdown cells were localized to the nuclear periphery (Supplementary Fig. 8F), supporting the previously described local role for NPCs in the resolution of DNA damage at the nuclear envelope (42,43). Even though RPE1 cells do not show increased DNA damage when NPC assembly is inhibited, these cells show decreased nuclear import rates and transcriptional alterations (Fig. 6A and B). It is a possibility that when these cells are released from NPC assembly inhibition, re-entering S-phase with these alterations could lead to DNA damage and genome instability. To test this possibility, we analyzed DNA damage in RPE1 and H3A cells 6 days after restoring NUP160 expression as described in Fig. 3E. We found that after regaining cell proliferation, these cells showed no increase in γH2AX staining compared to control cells (Supplementary Fig. 8G).
In normal cells, activation of the DNA damage checkpoint results in mTOR inhibition, which leads to reduced protein translation and G1/S cell cycle arrest (44). In many cancer cells, mTOR activity is deregulated and not inhibited by DNA damage (44,45). In fact, sustained mTOR activity in these cells has been found to promote CHK1 gene expression and to be required for cell survival in the presence of increased DNA damage (45). Consistent with a role for mTOR in alleviating DNA damage accumulation in cancer cells, pharmacological inhibition of this signaling pathway leads to decreased CHK1 protein levels (45,46), accumulation of γH2AX (45,47), increased levels and activity of 53BP1 (47), and reduced viability in several cancer cell types including A375 cells (48). mTOR regulation of CHK1 gene expression occurs through the activation of S6 kinase (45). Because we identified that the reduction in NPC numbers in A375 and RPE1 cells is associated with decreased CHK1 protein (Fig. 6C and Supplementary 8A) and mRNA levels (Supplementary Fig. 9A), as well as with the accumulation of γH2AX and 53BP1 foci (Fig. 6C–F), we investigated if blocking NPC assembly affected mTOR signaling by analyzing the phosphorylation levels of mTORC1 and its downstream effector S6. While mTOR phosphorylation on Ser2448 was not strongly affected by NUP160 downregulation, we found that reducing NPC numbers was associated with a strong reduction in S6 phosphorylation (Ser235/6) (Fig. 6G). These findings suggest that inhibiting NPC assembly prevents cells from properly signaling through mTOR, which leads to decreased CHK1 expression. Our findings indicate that inhibition of NPC assembly affects ATR and CHK1 signaling in both RPE1 and A375 cells, but only induces cell death in the latter. If decreased activity of CHK1 and/or ATR selectively induce the death of A375 when NPC numbers are reduced, we could expect that these cells should be more sensitive to inhibition of this DDR pathway. To test this hypothesis A375 cells and RPE1 cells were incubated with the ATR inhibitor AZD6738 or CHK1 inhibitor CCT245737 and cell viability was quantified over 7 days. Figure 6H shows that both inhibitors selectively induced cell death in A375 cells but had minimal effect on RPE1 cell survival. Moreover, we determined that depletion of NUP160 in A375 cells did not further increase the sensitivity of these cells to ATR or CHK1 inhibition, suggesting that NPCs and ATR/Chk1 are part of the same pathway (Supplementary Fig. 9B). Together, these findings suggest that inhibition of NPC assembly induces the death of A375 cells, at least in part, through the modulation of ATR/CHK1 signaling.
MAD1 is a critical component of the spindle assembly checkpoint (SAC) (49) and its displacement from NPCs was previously shown to lead to mitotic timing defects, mitotic checkpoint alterations, and genomic instability (50,51). Notably, we found that the reduction of NPC numbers was associated with the loss of Mad1 from the nuclear periphery of interphase cells (Fig. 6I and Supplementary Fig. 9C). In addition to its mitotic roles, MAD1 was recently found to work with MAD2 and ATR to facilitate DNA damage repair at the nuclear periphery during interphase (52), which suggests that loss of MAD1 from NPCs might contribute to the increased DNA damage associated with the reduction in NPC number. Altogether, our findings indicate that inhibition of NPC assembly and the consequent reduction in the number of these channels results in multiple cellular alterations that contribute to the selective death of cancer cells.
Inhibition of NPC assembly prevents tumor growth and induces tumor regression
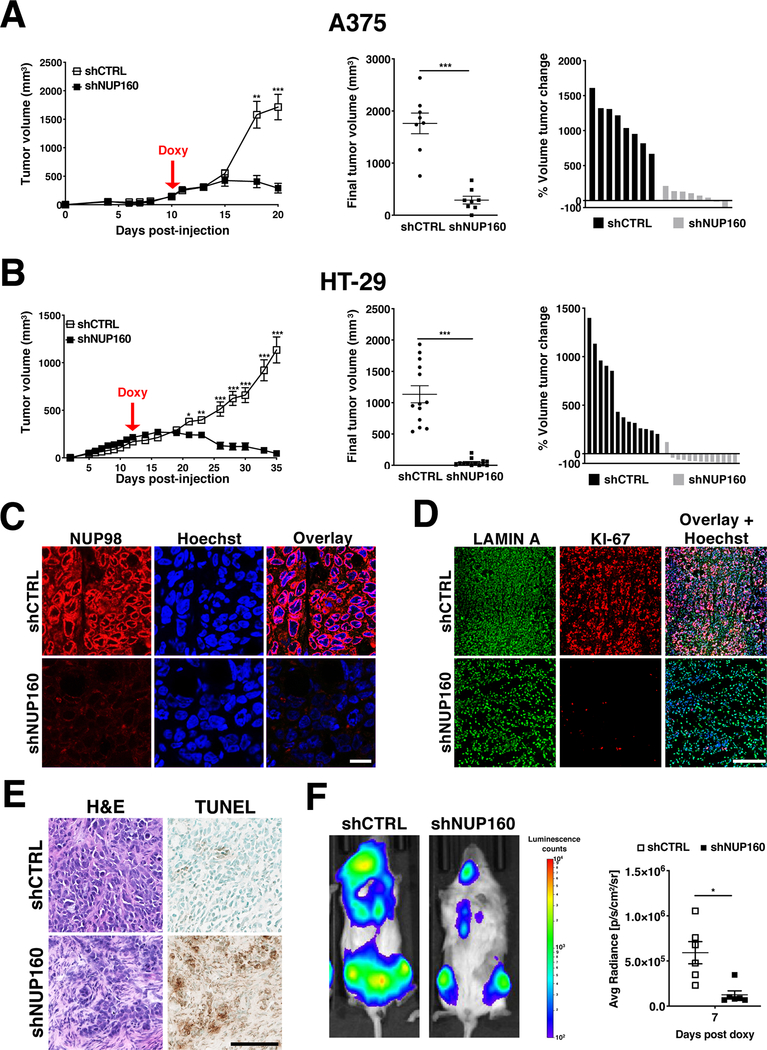
The higher dependency of cancer cells on the nuclear transport machinery and their differential cell death response to inhibition of NPC assembly compared to normal cells suggest that targeting this process could represent a promising strategy for anti-cancer therapies. To investigate if inhibition of NPC formation is sufficient to affect tumor growth in vivo, A375 and HT-29 cells carrying inducible control or NUP160 shRNAs were used for xenograft studies. Cells were injected subcutaneously in NOD-SCID immunodeficient mice and tumors were allowed to grow to 100–125 mm3 (~9–14 days) before shRNA induction. Control or NUP160 shRNAs were induced by doxycycline and tumor volume was measured over time. As shown in Figures 7A & B inhibition of NPC formation in established tumors by depletion of the essential nucleoporin NUP160 prevented tumor growth and led to significant tumor regression. Similarly, depletion of NUP93 was recently described to delay the growth of breast cancer tumors in vivo (53). To confirm that NUP160 depletion in vivo prevents NPC formation, tumors were collected at the end of treatment, sectioned and stained with antibodies against the NPC component NUP98 (Fig. 7C, Supplementary Fig. 10A). NUP160-depleted tumors showed drastically reduced levels of the cell proliferation marker KI-67, increased necrosis, and higher TUNEL staining indicative of apoptotic cell death (Fig. 7D and E, Supplementary Fig. 10B and C). To investigate if inhibition of NPC assembly could also be exploited for the treatment of liquid tumors, we performed xenograft studies using MOLM-13 acute myeloid leukemia cells. To follow the growth of leukemia cells in vivo, MOLM-13 cells carrying control or NUP160 shRNAs were transduced with lentivirus expressing luciferase and selected to generate stable cell lines. Cells were transplanted into NOD-SCID immunocompromised mice and animals were followed by in vivo bioluminescent imaging. Similar to A375 and HT-29 cells, inhibition of NPC assembly in MOLM-13 cells strongly inhibited tumor growth (Fig. 7F, Supplementary Fig. 10D). Altogether, these findings demonstrate that inhibition of NPC formation could represent an effective treatment for solid and liquid tumors.
Figure 7. Inhibition of NPC assembly prevents tumor growth and causes tumor regression.
A, B, Tumor volume analyses in A375 and HT-29 xenografts (n = 8 or 13) before and after induction of Control and NUP160 shRNAs with doxycycline (left) and at the final time point (center). Each symbol represents an individual tumor. Percent change of final tumor volume to the start of doxycycline treatment (right). C, Representative images of NUP98 staining in sections from Control or NUP160 shRNA HT-29 tumors from (B) at the final experimental time point (Day 35). Scale bar, 25 μm D, Representative images of LAMIN A and KI-67 staining in sections from Control or NUP160 shRNA HT-29 tumors from (B) at the final time point (Day 35). Scale bar, 250 μm. E, Representative images of Hematoxylin & Eosin (H&E) and Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining of sections from HT-29 Control or NUP160 shRNA tumors from (B) at the final time point (Day 35). Scale bar 100 μm. F, Representative bioluminescence images (left) and quantification (right) of injected with MOLM-13-luciferase cells expressing Control or NUP160 shRNAs (n = 6 mice). Data are mean ± s.e.m. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 by unpaired Student’s t test (A, B middle, F) or multiple unpaired Student’s t tests with Holm-Sidak method to correct for multiple comparisons (A, B left). Images (C, D, E) are representative of 2 independent experiments.
DISCUSSION
Nuclear pore complexes are essential cellular channels and the only gateway into the genome. Besides controlling the transport of molecules across the nuclear envelope these structures perform multiple transport-independent functions that are critical for cell homeostasis. The formation of NPCs is tightly regulated in cells and alterations in the numbers and function of these structures have been proposed to contribute to malignant transformation and cancer development. Here we demonstrate that NPC assembly is predominantly required in proliferating cells, and that inhibiting this process selectively affects the survival of cancer cells. We found that while reducing the number of NPCs leads to a reversible cell cycle arrest in several normal cells, it induces the rapid death of cancer cells, providing evidence of how normal and transformed cells differentially respond to the reduction of nuclear pore numbers. The selective response of cancer cells to the inhibition of NPC assembly exposes the potential targetability of this process for cancer therapies.
Alterations in the nuclear transport machinery have long been observed in cancer cells (54). Recent evidence showing that many cancer cells become addicted to nuclear transport has fueled the idea that the nuclear transport machinery represents a promising target for cancer (3,6,7). Consistent with this idea, inhibitors of the main nuclear export factor CRM1/XPO1 have been found to be highly effective in inducing cancer cell death and the focus of more than 40 different clinical trials encompassing both solid tumors and hematologic malignancies (8). Different from nucleocytoplasmic transport, our findings show that the formation of NPCs is primarily required in proliferating cells. As a consequence, blocking NPC assembly might achieve the same result as inhibition of nuclear transport but without affecting non-dividing cells such as neurons and muscle. Our data also indicates that while cancer cells undergo rapid cell death in response to inhibition of NPC assembly, normal cells undergo a reversible cell cycle arrest and recuperate their ability to divide once NPC formation is restored. Notably, we observed that cancer cells die even though the dilution of NPC numbers is not stronger than in normal cells, indicating that they need more NPCs to survive and further confirming the increased dependency of cancer cells on the nuclear transport machinery. This selective sensitivity of cancer cells to the inhibition of NPC formation suggests that blocking this process could be exploited for cancer treatments. In support of this idea, we present evidence that blocking NPC formation inhibits tumor growth and promotes tumor regression. Collectively, our data expose the process of nuclear pore formation as a novel susceptibility of cancer cells and puts forth the exciting possibility that reducing the number of these structures might represent a viable strategy for cancer therapies. The fact that many different cancer cells are highly sensitive to nuclear transport alterations suggest that a therapy based on inhibition of nuclear pore complex formation could be exploited for the treatment of multiple cancer types. Finally, we found that besides reducing nuclear transport rates, the inhibition of NPC assembly results in multiple cellular alterations that contribute to cancer cell death, including alterations in gene expression regulation, DNA damage repair, cell signaling and cell cycle progression. Replication stress is another likely contributor to the decreased survival in NPC-depleted cancer cells. Consistent with the previously described role of NPCs in DNA replication (55,56), we identified that dilution of NPC numbers results in decreased ATR signaling, a key responder to replication stress, and increased γH2AX levels in cancer cells, a marker of stalled replication forks and double-strand breaks (57). Because cancer cells normally have higher levels of replication stress and aberrant responses to DNA damage, the ATR signaling pathway becomes critical for their survival. Consequently, the decreased activity of the ATR-mediated response that results from lowering NPC numbers is likely to selectively impact the survival of cancer cells. The simultaneous alterations of multiple cellular processes that contribute to cell death is a strong therapeutic advantage that would make the emergence of resistance to the inhibition of NPC formation more challenging for cancer cells.
In this work we inhibited NPC assembly by depletion of essential nucleoporins. Therapeutically, this could be achieved by indirectly modulating the transcription, translation or degradation of individual nucleoporins. Directly targeting nucleoporins, on the other hand, could represent a more difficult technical challenge. Most of these proteins have a very limited set of domains, generally lacking functional or catalytic domains that could be targeted by small molecules (1,58). Moreover, the crystal structure for most structural components of the mammalian NPC have not been solved yet (58), limiting the ability to develop structure-based rationally designed inhibitors. As an alternative approach, NPC formation could be inhibited by modulating the activity of cellular factors that regulate the assembly process. This could also avoid the undesired phenotypes that could result from the tissue-specific and NPC-independent functions described for several nucleoporins.
METHODS
Cell culture
A375 and HT-29 cells were obtained from ATCC. HPF cells (Catalog #3310) were obtained from ScienCell. RPE1 cells engineered to knockout puromycin were generated and kindly provided by Dr. Andrew Holland (John Hopkins University). Primary human myoblasts, IMR-90, MOLM-13, Hermes 3A (H3A) and H9 cells were kindly provided by Drs. Alessandra Sacco, Pier Lorenzo Puri, Anirunda Deshpande, Ze’ev Ronai and Evan Snyder respectively (Sanford Burnham Prebys Medical Discovery Institute). Cell lines not obtained from ATCC have been authenticated by the SBP Genomics Core Facility, which utilizes the GenePrint10 (Promega) system for short tandem repeat (STR) analysis on genomic DNA. All cells were cultured at a humidified 37 °C with 5% CO2 in media supplemented with 10% heat-inactivated fetal bovine serum (Sigma-Aldrich) and 1 unit/mL penicillin-streptomycin (GE Healthcare Life Sciences) unless otherwise noted. Cells were routinely tested for mycoplasma contamination (MycoAlert™ Mycoplasma Detection Kit LT07–118). A375 and HPF cells were cultured in DMEM medium (Mediatech). HT-29, MOLM-13, and H3A cells were cultured in RPMI-1640 medium (GE Healthcare Bio Science, Hyclone). IMR-90 cells were cultured in EMEM medium (Quality Biological) with 20% FBS and no penicillin-streptomycin. RPE1 cells were cultured in DMEM/F-12 with GlutaMAX (Gibco). Primary human myoblasts were cultured on plates coated with collagen (Corning, Rat Collagen I) in proliferation media (Ham’s F-10 Nutrient Mix with 15% FBS) or differentiation media (DMEM with 2% Horse Serum, Gibco) and no penicillin-streptomycin. H9 cells were cultured in complete mTesR1 medium (STEMCELL Technologies) with no penicillin-streptomycin on Matrigel (Cell Applications)-coated tissue culture plates. For low serum experiments, cells were grown in complete media with 0.6% FBS. For irradiation-induced senescence, cells were irradiated with 10 Gy using an RS2000 (Rad Source Technologies). For inhibitor treatments, cells were grown with CHK1 inhibitor CCT245737 (Selleck Chemicals) or ATR inhibitor VE-821 (R&D Systems) or AZD6738 (Cayman Chemical Company) for the specified times and counted directly or imaged using a Celigo Imaging Cytometer (Nexcelom Biosciences).
Proliferation and viability assays
Cells were plated at the indicated amounts and counted daily. To count cells, conditioned media was collected, adherent cells were washed once with PBS, and incubated with trypsin-EDTA (0.25%, Life Technologies) for 5 minutes. Cells in trypsin were pooled with conditioned media and spun down. Cells were resuspended in PBS and counted 50% in Trypan Blue Solution, 0.4% (Life Technologies) with a TC20 cell counter (Bio-Rad Laboratories). Doubling time was calculated by dividing relative change in cell number by time when cells were in log phase growth.
Immunofluorescence microscopy
Cells were seeded in polymer coverslip 8-well chamber slides (IBIDI) and grown for the indicated times. All subsequent steps were performed at room temperature. Cells were fixed in 2–4% PFA (methanol-free) in PBS for 5 minutes and blocked using IF buffer (1x PBS, 10 mg/ml BSA, 0.02% SDS, 0.1% Triton X-100) for 30 minutes. Cells were then incubated with primary antibody in IF buffer for 1 hour at room temperature (RT) or overnight at 4°C and washed with IF buffer. Cells were then incubated with secondary antibody in IF buffer for 1 hour at RT and washed with IF buffer. Cells were then incubated with 1 μg/mL Hoechst 33342 (Life Technologies) in PBS for 5 minutes and washed with PBS before imaging.
Live imaging
Cells were seeded with doxycycline in polymer coverslip 8-well μSlide dishes (IBIDI) and grown in a humidified 37°C with 5% CO2 stage top incubator (H301, Okolab) controlled by the Oko-Touch (Okolab). Cells were imaged on a Leica DMi8 microscope at 10 X magnification one frame per 20 minutes for 5 days. 20–26 cells were manually tracked, and number/time of cell divisions were recorded.
Flow cytometry
Analytical cytometry was performed in the Sanford Burnham Prebys Flow Cytometry Core using a BD LSRFortessa™ (BD Biosciences) or a BD LSRFortessa™ X-20 (BD Biosciences). For proliferation, live cells were labeled with 5 μM CTV (Life Technologies) for 10 minutes at 37°C, washed with cold DMEM medium supplemented with 0.5% FBS and 10 mM HEPES, and cultured in complete medium at 37°C. CTV dilutions were analyzed at the indicated times by flow cytometry. For viability staining, live cells were stained with Zombie NIR™ Fixable Viability dye (BioLegend) in PBS at 1:500 for 10 minutes. For cell cycle staining, cells were fixed with 70% cold ethanol in PBS for 1 hour at −20°C and stained with 2.5 μg/mL Hoechst (Invitrogen) in PBS with 2% FBS. For EdU staining, cells were incubated with 10 μM EdU for the indicated times and Click-iT™ Plus EdU Alexa Fluor 647 Flow Cytometry Assay (C10634, Invitrogen) was used according to manufacturer’s directions. For Annexin V staining, live cells were stained with 2.5 μL Alexa Fluor 488 Annexin V (Life Technologies A13201) in 100 μL1X Binding Buffer (140 mM NaCl, 10 mM HEPES, 2.5 mM CaCl2) for 15 minutes before flow cytometry.
Hematopoietic progenitor cell isolation and culture
Nup160 exon 4 floxed (fl) mice were generated by Cyagen US Inc in the C57BL/6N background using Turboknockout technology. Nup160fl/f animals were crossed with R26-CreERT2 mice (Stock No. 008463, The Jackson Laboratory) to generate Nup160fl/fl-CreERT2 mice. Lineage negative hematopoietic progenitor cells were isolated from mouse bone marrow (BM) of these animals. BM cells were flushed from femurs and tibia into PBS, 1.2% FBS and passed through a 40 μm nylon mesh to obtain a single cell suspension. After red blood cells lysis, Lineage negative hematopoietic progenitor cells were isolated by negative selection using EasySep™ Mouse Hematopoietic Progenitor Cell Isolation Kit (STEMCELL) according to manufacturer’s instructions. Briefly, BM cells were incubated first with rat serum and biotinylated antibodies directed against non-hematopoietic stem cells and non-progenitor cells (CD5, CD11b, CD19, CD45R/B220, Ly6G/C(Gr-1), TER119), and then with streptavidin-coated magnetic particles. Labeled cells are captured using a magnet and the Lin negative cells were collected. The efficacy of enrichment was verified by flow cytometry. Purified Lin negative cells were cultured in 6-well plates at 0.2*106 cells/ml in DMEM, 15%FBS, 20ng/ml SCF, 10ng/ml IL6 and 6ng/ml IL3 for 24 hours at 37°C and subsequently treated with DMSO vehicle or 4-hydroxytamoxifen (0.5 μM, Sigma) for 72 hours. Cells were counted daily with Trypan Blue.
Fluorescence recovery after photobleaching (FRAP) and Dronpa activation
Cells were transfected with the plasmids pcDNA6.2/C-Lumio expressing NES-Tomato-NLS, 2x-GFP and 2x-GFP-NLS (59), or Dronpa (Addgene # 54682) using Lipofectamine RNAiMAX (Life Technologies), according to manufacturer’s instructions. 24–72 hours post-transfection, cells were imaged on a Leica SP8 confocal microscope. For FRAP, nuclear signal was photobleached with 50% power 488 nm (GFP) or 552 nm (Tomato) laser for 3 seconds, transport of fluorescent signal was recorded for 2 minutes using the LAS X software. For Dronpa diffusion, entire cell Dronpa signal was inactivated with 50% power 488 nm laser for 3 seconds, a 10 μm2 area of cytoplasm was activated using 25% power 405 nm laser for 100 ns, diffusion of Dronpa signal out of the cytoplasm and into the nucleus was recorded for 12 seconds using LAS X software. A total of 7–9 nuclei per group were analyzed. Data is normalized by calculating the difference between the nuclear and cytoplasmic background-subtracted signal and normalizing to the average maximal recovery of the control.
In vitro nuclear permeability assay
RPE1 cells transfected with control or NUP160-specific siRNAs were seeded on 8-well μSlide IBIDI dishes to achieve 60–80% confluency at 96hs post-transfection. At 96 hours of knockdown cells were washed with PBS twice and placed on ice. Cells were equilibrated with permeabilization buffer (20mM Tris-HCL pH7.5, 110mM potassium acetate, 5mM magnesium chloride, 0.25M sucrose) for 5 minutes before being permeabilized in permeabilization buffer plus 20μg/ml digitonin (Millipore CHR-103() for 4–7 minutes. Cells were washed 3 times with assay buffer (20mM Tris-HCL pH7.5, 110mM potassium acetate, 5mM sodium chloride, 2mM magnesium chloride, 0.25M sucrose) for 1, 5, and 10 minutes. A mixture of 65–85kDa (Sigma T-1162) and 500kDa (Sigma FD-500S) fluorescent dextrans, 0.02mg/ml each, plus Hoechst 5mg/ml were added and cells were imaged on a Leica SP8 confocal microscope. The intranuclear signal for the 65–85kDa and 500 kDa dextran were quantified using Image J and normalized. For each image, the intranuclear dextran intensity was normalized to the extracellular signal and graphed as a relative intensity of 65–85kDa/500kDa.
Mice and tumor xenograft models
All mice were housed under 12-hour light/12-hour dark cycles, fed ad libitum, and all experiments started between 6 and 12 weeks of age. Immune compromised animals were bred and housed in sterile caging and maintained under pathogen-free conditions. Doxycycline hyclate (Alfa Aesar) was administered in drinking water at 0.5 mg/ml for the indicated time. The tumor injections and measurements were performed by the Sanford Burnham Prebys Animal Facility Core. In 100 μL 50% Matrigel (Cell Applications) and 50% DMEM, for each condition 2 × 106 cells were injected subcutaneously into both the right and left flank. Length and width were measured, and volume was calculated using the formula (length x width2)/2 as previously described(3). When average tumor size reached 100–125mm3 doxycycline was administered. For MOLM-13, 2 × 106 cells were injected via tail vein. To measure bioluminescence 250 μL luciferin (PerkinElmer, 15 mg/mL) was IP injected and radiance was measured by Xenogen IVIS Spectrum imager. When luciferase signal reached 25,000 average radiance (p/s/cm2/sr) doxycycline was administered. All animals were housed at the Sanford Burnham Prebys Medical Discovery Institute and all experiments were approved by the Institutional Animal Care and Use Committee and were performed in accordance with institutional guidelines and regulations.
Tumor collection and histology
On the final day of the experiment mice were euthanized by isoflurane overdose and tumors were measured and frozen in OCT (Tissue-Tek) using isopentane equilibrated to liquid nitrogen. Tumors were cut in 10 μm frozen sections using a cryostat (Leica Biosystems). Frozen sections were thawed and processed for immunofluorescence or histology. H&E staining and TUNEL assay were performed by the Sanford Burnham Prebys Histology Core.
Data collection and analysis
Microsoft Excel was used to record and calculate data. GraphPad Prism 8 software v8.2 (GraphPad Software, Inc.) was used to prepare graphs and to perform statistical analyses. Outcomes were compared using a two-tailed unpaired Student’s t test when comparing two groups. Multiple t tests with corrections for multiple comparisons or two-way ANOVA were used when comparing greater than two groups. Microscopy data were collected with a DMi8 Leica SP8 confocal microscope and analyzed using the Leica Application Suite X software v3.1.5.16308 (Leica Microsystems) or ImageJ v2.0.0-rc-54/1.51h (NIH). Relative mAb414 intensity was calculated on single Z section images in ImageJ. For each cell a polygon was drawn around the nuclear periphery, mean mAb414 fluorescence was calculated, and values were expressed relative to control. γH2AX foci were calculated on max projection images in ImageJ by finding maxima on after minimum threshold. MAD1 line intensities were extracted from single Z section images in ImageJ. Flow cytometry data were collected using the BD FACSDIVA™ Software (BD Biosciences) and analyzed using FlowJo software v10.0.8r1 (Tree Star, Inc.) and cell cycle distribution was calculated with the Dean-Jet-Fox model. qPCR data were collected in a CFX384 Real-Time PCR Detection System (Bio-Rad). Celigo images were acquired with Celigo Software Version 2.1 (Nexcelom Biosciences).
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Reducing nuclear pore complex (NPC) numbers in cancer cells induces death, prevents tumor growth and results in tumor regression. Conversely, normal cells undergo a reversible cell cycle arrest in response to inhibition of NPC assembly. These findings expose the potential of targeting NPC formation in cancer.
ACKNOWLEDGMENTS
M.A.D. was supported by a Pew Biomedical Science Scholar Award and Research Scholar Grant RSG-17-148-01- CCG from the American Cancer Society. This work was also supported by the National Institutes of Health (awards RO1AR065083, RO1AR065083-S1 and R01AI148668). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was additionally supported by the NCI Cancer Center grant P30 CA030199, which supports the animal, flow cytometry, genomics, and bioinformatics cores at the SBP La Jolla campus. We thank Drs. S. Heynen-Genel and P. Aza-Blanc for assistance with the siRNA screen. We also thank Drs. A. Holland, A. Sacco, P.L. Puri, A. Deshpande, Z. Ronai and E. Snyder for providing cell lines used in this work.
Financial support: This work was supported by a Research Scholar Grant RSG-17-148-01-CCG from the American Cancer Society and by the National Institutes of Health (awards RO1AR065083 and R01AI148668). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was additionally supported by the NCI Cancer Center grant P30 CA030199, which supports the Animal, Flow Cytometry, Genomics, Bioinformatics and High Content Screening cores, and the Conrad Prebys Center for Chemical Genomics at SBP.
Footnotes
The authors declare no conflict of interest. M.A.D has a related patent (PCT/US2018/041461) pending.
REFERENCES
- 1.Beck M, Hurt E. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 2017;18(2):73–89 doi 10.1038/nrm.2016.147. [DOI] [PubMed] [Google Scholar]
- 2.Pathria G, Wagner C, Wagner SN. Inhibition of CRM1-mediated nucleocytoplasmic transport: triggering human melanoma cell apoptosis by perturbing multiple cellular pathways. J Invest Dermatol 2012;132(12):2780–90 doi 10.1038/jid.2012.233. [DOI] [PubMed] [Google Scholar]
- 3.Kim J, McMillan E, Kim HS, Venkateswaran N, Makkar G, Rodriguez-Canales J, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature 2016;538(7623):114–7 doi 10.1038/nature19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewin JM, Lwaleed BA, Cooper AJ, Birch BR. The direct effect of nuclear pores on nuclear chemotherapeutic concentration in multidrug resistant bladder cancer: the nuclear sparing phenomenon. J Urol 2007;177(4):1526–30 doi 10.1016/j.juro.2006.11.048. [DOI] [PubMed] [Google Scholar]
- 5.Kuusisto HV, Wagstaff KM, Alvisi G, Roth DM, Jans DA. Global enhancement of nuclear localization-dependent nuclear transport in transformed cells. FASEB J 2012;26(3):1181–93 doi 10.1096/fj.11-191585. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Bravo V, Pippa R, Song WM, Carceles-Cordon M, Dominguez-Andres A, Fujiwara N, et al. Nuclear Pores Promote Lethal Prostate Cancer by Increasing POM121-Driven E2F1, MYC, and AR Nuclear Import. Cell 2018;174(5):1200–15 e20 doi 10.1016/j.cell.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vecchione L, Gambino V, Raaijmakers J, Schlicker A, Fumagalli A, Russo M, et al. A Vulnerability of a Subset of Colon Cancers with Potential Clinical Utility. Cell 2016;165(2):317–30 doi 10.1016/j.cell.2016.02.059. [DOI] [PubMed] [Google Scholar]
- 8.Jans DA, Martin AJ, Wagstaff KM. Inhibitors of nuclear transport. Curr Opin Cell Biol 2019;58:50–60 doi 10.1016/j.ceb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Taylor J, Sendino M, Gorelick AN, Pastore A, Chang MT, Penson AV, et al. Altered Nuclear Export Signal Recognition as a Driver of Oncogenesis. Cancer Discov 2019;9(10):1452–67 doi 10.1158/2159-8290.CD-19-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009;136(2):284–95 doi S0092–8674(08)01512–2 [pii] 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013;154(5):971–82 doi 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, et al. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 2003;11(4):853–64. [DOI] [PubMed] [Google Scholar]
- 13.Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 2003;113(2):195–206. [DOI] [PubMed] [Google Scholar]
- 14.Sachdev R, Sieverding C, Flotenmeyer M, Antonin W. The C-terminal domain of Nup93 is essential for assembly of the structural backbone of nuclear pore complexes. Mol Biol Cell 2012;23(4):740–9 doi 10.1091/mbc.E11-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, et al. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell 1997;8(10):2017–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A 2003;100(3):981–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 2010;141(6):1030–41 doi S0092–8674(10)00490–3 [pii] 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, et al. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J 2005;24(20):3519–31 doi 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Angelo MA, Anderson DJ, Richard E, Hetzer MW. Nuclear pores form de novo from both sides of the nuclear envelope. Science 2006;312(5772):440–3 doi 312/5772/440 [pii] 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 20.Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol 2010;191(1):15–22 doi 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salas Fragomeni RA, Chung HW, Landesman Y, Senapedis W, Saint-Martin JR, Tsao H, et al. CRM1 and BRAF inhibition synergize and induce tumor regression in BRAF-mutant melanoma. Mol Cancer Ther 2013;12(7):1171–9 doi 10.1158/1535-7163.MCT-12-1171. [DOI] [PubMed] [Google Scholar]
- 22.Niu M, Chong Y, Han Y, Liu X. Novel reversible selective inhibitor of nuclear export shows that CRM1 is a target in colorectal cancer cells. Cancer Biol Ther 2015;16(7):1110–8 doi 10.1080/15384047.2015.1047569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hing ZA, Fung HY, Ranganathan P, Mitchell S, El-Gamal D, Woyach JA, et al. Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies. Leukemia 2016;30(12):2364–72 doi 10.1038/leu.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y, Sessions EH, Zhang F, Ban F, Placencio-Hickok V, Ma CT, et al. Identification and characterization of small molecule inhibitors of the ubiquitin ligases Siah1/2 in melanoma and prostate cancer cells. Cancer Lett 2019;449:145–62 doi 10.1016/j.canlet.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 2000;227(2):271–8 doi 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 26.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 1995;377(6549):552–7 doi 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 27.Collier AE, Spandau DF, Wek RC. Translational control of a human CDKN1A mRNA splice variant regulates the fate of UVB-irradiated human keratinocytes. Mol Biol Cell 2018;29(1):29–41 doi 10.1091/mbc.E17-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, et al. Restoration of p53 function leads to tumour regression in vivo. Nature 2007;445(7128):661–5 doi 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 29.Otsuka S, Ellenberg J. Mechanisms of nuclear pore complex assembly - two different ways of building one molecular machine. FEBS Lett 2018;592(4):475–88 doi 10.1002/1873-3468.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borlido J, Sakuma S, Raices M, Carrette F, Tinoco R, Bradley LM, et al. Nuclear pore complex-mediated modulation of TCR signaling is required for naive CD4(+) T cell homeostasis. Nat Immunol 2018;19(6):594–605 doi 10.1038/s41590-018-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dultz E, Huet S, Ellenberg J. Formation of the nuclear envelope permeability barrier studied by sequential photoswitching and flux analysis. Biophys J 2009;97(7):1891–7 doi 10.1016/j.bpj.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardarelli F, Serresi M, Bizzarri R, Giacca M, Beltram F. In vivo study of HIV-1 Tat arginine-rich motif unveils its transport properties. Mol Ther 2007;15(7):1313–22 doi 10.1038/sj.mt.6300172. [DOI] [PubMed] [Google Scholar]
- 33.Roehrig S, Tabbert A, Ferrando-May E. In vitro measurement of nuclear permeability changes in apoptosis. Anal Biochem 2003;318(2):244–53 doi 10.1016/s0003-2697(03)00242-2. [DOI] [PubMed] [Google Scholar]
- 34.Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, et al. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol 2008;180(5):857–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackay DR, Howa AC, Werner TL, Ullman KS. Nup153 and Nup50 promote recruitment of 53BP1 to DNA repair foci by antagonizing BRCA1-dependent events. J Cell Sci 2017;130(19):3347–59 doi 10.1242/jcs.203513. [DOI] [PubMed] [Google Scholar]
- 36.Duheron V, Nilles N, Pecenko S, Martinelli V, Fahrenkrog B. Localisation of Nup153 and SENP1 to nuclear pore complexes is required for 53BP1-mediated DNA double-strand break repair. J Cell Sci 2017;130(14):2306–16 doi 10.1242/jcs.198390. [DOI] [PubMed] [Google Scholar]
- 37.Palancade B, Liu X, Garcia-Rubio M, Aguilera A, Zhao X, Doye V. Nucleoporins prevent DNA damage accumulation by modulating Ulp1-dependent sumoylation processes. Molecular biology of the cell 2007;18(8):2912–23 doi 10.1091/mbc.E07-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemaitre C, Fischer B, Kalousi A, Hoffbeck AS, Guirouilh-Barbat J, Shahar OD, et al. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene 2012;31(45):4803–9 doi 10.1038/onc.2011.638. [DOI] [PubMed] [Google Scholar]
- 39.Chung DK, Chan JN, Strecker J, Zhang W, Ebrahimi-Ardebili S, Lu T, et al. Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat Commun 2015;6:7742 doi 10.1038/ncomms8742. [DOI] [PubMed] [Google Scholar]
- 40.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer 2012;12(12):801–17 doi 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 41.Brown JS, O’Carrigan B, Jackson SP, Yap TA. Targeting DNA Repair in Cancer: Beyond PARP Inhibitors. Cancer Discov 2017;7(1):20–37 doi 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, et al. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol 2015;17(11):1401–11 doi 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 2008;322(5901):597–602 doi 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamm N, Rogers S, Cesare AJ. The mTOR pathway: Implications for DNA replication. Prog Biophys Mol Biol 2019;147:17–25 doi 10.1016/j.pbiomolbio.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Zhou X, Liu W, Hu X, Dorrance A, Garzon R, Houghton PJ, et al. Regulation of CHK1 by mTOR contributes to the evasion of DNA damage barrier of cancer cells. Sci Rep 2017;7(1):1535 doi 10.1038/s41598-017-01729-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koppenhafer SL, Goss KL, Terry WW, Gordon DJ. mTORC1/2 and Protein Translation Regulate Levels of CHK1 and the Sensitivity to CHK1 Inhibitors in Ewing Sarcoma Cells. Mol Cancer Ther 2018;17(12):2676–88 doi 10.1158/1535-7163.MCT-18-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandhakavi S, Kim YM, Ro SH, Xie H, Onsongo G, Jun CB, et al. Quantitative nuclear proteomics identifies mTOR regulation of DNA damage response. Mol Cell Proteomics 2010;9(2):403–14 doi 10.1074/mcp.M900326-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He H, Nan X, Liu S, Zhang L, Yang Z, Wu Y, et al. Anticancer effects of combinational treatment with BRAF(V600E) siRNA and PI3K pathway inhibitors in melanoma cell lines harboring BRAF(V600E). Oncol Lett 2018;16(1):632–42 doi 10.3892/ol.2018.8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol 2012;22(22):R966–80 doi 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, et al. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 2014;156(5):1017–31 doi 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lussi YC, Shumaker DK, Shimi T, Fahrenkrog B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus 2010;1(1):71–84 doi 10.4161/nucl.1.1.10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence KS, Chau T, Engebrecht J. DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity. PLoS Genet 2015;11(4):e1005150 doi 10.1371/journal.pgen.1005150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bersini S, Lytle NK, Schulte R, Huang L, Wahl GM, Hetzer MW. Nup93 regulates breast tumor growth by modulating cell proliferation and actin cytoskeleton remodeling. Life Sci Alliance 2020;3(1) doi 10.26508/lsa.201900623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nat Rev Cancer 2004;4(2):106–17. [DOI] [PubMed] [Google Scholar]
- 55.Su XA, Dion V, Gasser SM, Freudenreich CH. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev 2015;29(10):1006–17 doi 10.1101/gad.256404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aguilera P, Whalen J, Minguet C, Churikov D, Freudenreich C, Simon MN, et al. The nuclear pore complex prevents sister chromatid recombination during replicative senescence. Nat Commun 2020;11(1):160 doi 10.1038/s41467-019-13979-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang BJ, Adhikary G, Eckert RL, Lu AL. Chk1 inhibition as a novel therapeutic strategy in melanoma. Oncotarget 2018;9(54):30450–64 doi 10.18632/oncotarget.25765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin DH, Hoelz A. The Structure of the Nuclear Pore Complex (An Update). Annu Rev Biochem 2019;88:725–83 doi 10.1146/annurev-biochem-062917-011901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Developmental Cell 2012;22(2):446–58 doi 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.