Abstract
Gene therapy promises to revolutionize biomedicine and personalized medicine by modulating or compensating the expression of abnormal genes. The biggest obstacle for clinical application is the lack of an effective, non-immunogenic delivery system. We show that bovine colostrum exosomes and polyethyleneimine matrix (EPM) delivers short interfering RNA (siRNA) or plasmid DNA (pDNA) for effective gene therapy. KRAS, a therapeutic focus for many cancers, was targeted by EPM-delivered KRAS siRNA (siKRAS) and inhibited lung tumor growth (>70%) and reduced KRAS expression (50%−80%). Aberrant p53 is another therapeutic focus for many cancers. EPM-mediated introduction of wild-type (WT) p53 pDNA (pcDNA-p53) resulted in p53 expression in p53-null H1299 cells in culture subcutaneous lung tumor, and tissues of p53-knockout mice. Additionally, chemo-sensitizing effects of paclitaxel were restored by exogenous WT p53 in lung cancer cells. Together, this novel EPM technology represents an effective ‘platform’ for delivery of therapeutic nucleic acids to treat human disease.
1. Introduction
Gene therapy has revolutionized biomedical research and has become an indispensable research tool. Gene therapy is the therapeutic delivery of genetic material into cells to compensate for abnormal genes by either turning off genes that produce faulty proteins or introducing genes to produce a beneficial protein to treat disease. Gene therapy could be a viable treatment option for a wide range of diseases including cancer, cystic fibrosis, heart disease, diabetes, neurodegenerative disorders, infectious and inflammatory diseases [1]. Nucleic acids that have been widely exploited for gene therapy include DNA and mRNA macromolecules for overexpression of genes and smaller siRNA, miRNA, and antisense oligonucleotides (ASO) for gene knockdown (collectively referred to as biologics) [2, 3]. The largest limitation for gene therapy is effective delivery to the cell. Nucleic acids in their native form have poor cellular uptake, and are prone to degradation and clearance and therefore require a vector for cellular delivery. Several viral and non-viral vector-mediated gene delivery systems have thus been developed [4–6].
Viruses are highly efficient at nucleic acid transfer and thus were an initial choice for gene delivery, with nearly 70% of clinical studies having used viral vectors [7, 8]. A limitation of viral vectors is immune rejection, however, there has been an extensive drive towards producing non-pathogenic viruses for gene therapy. Several modified viruses such as retroviruses, lentiviruses, adenoviruses, adeno-associated viruses (AAVs), and others are widely used in this respect [9]. With more than two-dozen viral-based gene therapy drugs currently in clinical trials, this field continues to grow with new generations of viral vectors. It is also noteworthy that many of the virus-based gene therapy successes are achieved in ex-vivo, in situ, or for localized tissue delivery and thus have advantages such as no immunologic response, no risk to germ-line cells, and lower renal clearance [4, 10]. However, many diseases will require systemic delivery of viral vectors for gene therapy, which is not feasible at the moment. Although, virus-based vectors have shown considerable promise in preclinical and early clinical studies for gene delivery their success is greatly hampered by concerns of systemic inflammatory responses, immunogenicity, and insufficient gene expression [6, 11, 12]. An array of physical and chemical non-viral methods has been used to address the limitations of viral vectors, particularly concerning safety, limited cargo capacity, and cost. Physical methods of gene delivery include gene gun, electroporation, hydrodynamic delivery, sonoporation, and magnetofection [13, 14]. However, these techniques are more suitable for pre-clinical studies and have so far failed to show clinical translatability. Chemical gene carriers generally have defined structures, designed functions, non-immunogenicity, and the potential for large-scale production [15]. Other advantages of these carriers include the protection of their cargo from enzymatic degradation and immune recognition [16]. Chemical vectors used in gene therapy have taken advantage of the negative charge of nucleic acids by complexing them electrostatically with cationic materials. Cationic lipids, cationic polymers, or lipidpolymer hybrids have been developed for gene delivery purposes by complexing with DNA, oligodeoxynucleotides, and various forms of synthetic RNA [17, 18]. A mixture of cationic lipids, neutral lipids, and nucleic acid spontaneously assemble into lipoplexes or liposomes, in which nucleic acid is entrapped within the lipid bilayers. Cationic polymers, such as polyethyleneimine (PEI), poly(l-lysine), polyamidoamine, and polybeta-amino esters complex with nucleic acids via ionic interaction and aid in the condensation of DNA or RNA into nanoparticles. Cationic systems have been shown to primarily accumulate in the lungs, liver, and spleen [19]. This accumulation can be explained by the fact that therapeutic doses require high concentrations of cationic particles that have a tendency to readily aggregate at increasing concentrations [20]. Therefore, the nucleic acid-complexes must demonstrate stability under physiological conditions but with no aggregation. One of the main limitations of current non-viral systems is the failure to deliver a high dose to the target site in vivo.
A new frontier in drug delivery is the use of cell secretory nano-vesicles, especially exosomes (30–100 nm), which have clinical value based on biocompatibility and natural trafficking ability to deliver endogenous payloads [21–23]. Exosomes have been isolated from a wide range of donor cells including, tumor cells, macrophages, dendritic cells (DCs), and mesenchymal stem cells (MSCs). Selecting the source of donor cells and engineering exosomes to integrate specific cargo at the target site are key factors determining their clinical application. Recent reports suggest that exosomes encapsulate and protect exogenous oligonucleotides (such as siRNA and miRNA) for delivery to target cells [24, 25]. Early phase clinical trials with DCs and MSCs exosomes indicate the safety of exosomes and to some extent the feasibility of clinical scale exosome production [26, 27]. There have also been reports of rapid large-scale purification of cell culture-derived therapeutic exosomes [28, 29] but cost-effectiveness remains elusive.
Our laboratory has identified bovine milk exosomes as an effective delivery vehicle for small molecule drugs [30–33] and siRNA [34]. To develop a nano ‘platform’ for the targeted delivery of a wide range of macromolecules, including RNA and DNA, we formulated the novel EPM technology. Our approach utilizes ionic interactions between positively charged PEI, negatively charged nucleic acids, and exosomes to create a polyplex containing all three moieties while retaining a nanostructure that naturally traffics cargo for systemic delivery.
In this report, we demonstrate the novel EPM approach for gene delivery using PEI (60 kDa) in combination with bovine colostrum exosomes. To achieve tumor targeting, exosomes are functionalized with folic acid (FA). Thus, FA-functionalization of exosomes, followed by interaction with PEI, complex formation with siRNA or pDNA, and recovery of the complex by precipitation results in the final formulation (Fig. 1a). EPM as the transfection vector for siRNA renders enhanced gene silencing and tumor targeting. Our results indicate significant silencing of oncogenic genes and tumor growth inhibition with siRNA, and effective gene transfer for overexpression of green fluorescence protein (GFP) and p53 protein with pDNAs when delivered by EPM. As a transfection vector, PEI alone was only slightly effective in gene silencing in vitro, but showed cytotoxicity. Analysis of liver and kidney function enzymes and hematopoietic parameters confirm the lack of systemic toxicity of EPM mediated gene delivery.
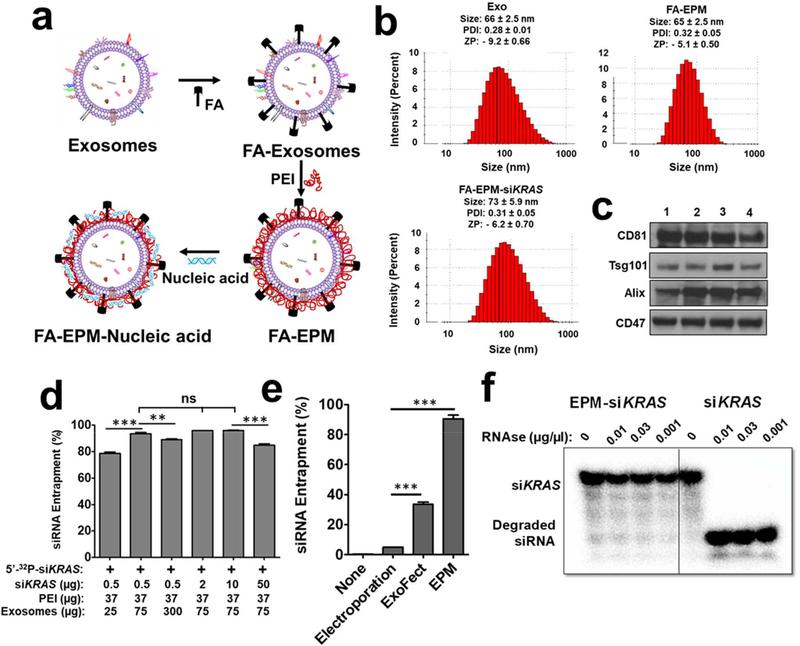
Figure 1.
(a) Graphical representation of folic acid (FA) covalently attached to the exosome-polyethyleneimine matrix (EPM) as a transfection vector for delivery of nucleic acid. (b) Size, polydispersity index (PDI), and zeta potential (ZP) of exosomes, FA-EPM, and FA-EPM-siKRAS, analyzed by Zetasizer. Data represent mean ± SD from 3 preparations. (c) Surface-bound hallmark protein markers in bovine colostrum exosomes, as analyzed by western blot; exosomes isolated from different batches (1 – 4) of bovine colostrum powder were analyzed. (d) The entrapment efficiency of EPM with siKRAS; 5’−32P-labeled siKRAS was included as a tracer. Entrapment efficiency was calculated by the distribution of the radioactivity in the EPM-siKRAS recovered by precipitation with ExoQuick™ and in the supernatant, followed by radioactivity quantitation by Packard InstantImager. (e) The relative entrapment efficiency of the EPM versus the conventional methods involving electroporation and Exo-Fect™. Test siRNA was entrapped by EPM, electroporation, and Exo-Fect™; radiolabeled siRNA was included as a tracer. Entrapment efficiency was determined as described in Fig. 1d legend. (f) Susceptibility of 5’−32P-labeled siKRAS entrapped in EPM to enzymatic degradation. Following a brief incubation of EPM with the radiolabeled siKRAS, the reaction mixture was incubated with indicated concentrations of RNase A for 30 min. The reaction products were resolved by polyacrylamide gel electrophoresis after adding heparin to dissociate the siKRAS from PEI, and radioactive products were detected by Packard InstantImager. Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparison test (d) and using student’s t-test (e) to compare between the indicated groups. **, p < 0.01; *** p < 0.001.
2. Materials and Methods.
2.1. Isolation of exosomes from colostrum powder:
Exosomes were isolated from colostrum powder. Briefly, colostrum powder was rehydrated in deionized water achieving a final concentration of 5% w/v, and exosomes were isolated by sequential centrifugations (13,000 x g, 30 min; 65,000 x g, 60 min; and 135,000 x g, 2 h, as described [33]. The pellet was suspended in PBS and exosomal protein concentration was measured by standard BCA kit. The exosome suspension was stored at 6 mg/ml at −80°C. Exosomes from raw bovine milk were isolated, as described [33].
2.2. FA-functionalization of exosomes:
To stabilize the interaction of folic acid (FA) with exosomal proteins in vivo, we attached FA covalently by using activated FA. Activated FA was prepared using standard EDC (1-ethyl-3-(−3-dimethyl aminopropyl) carbodiimide hydrochloride) and NHS (N-hydroxysuccinimide esters). The degree of functionalization was achieved by varying FA concentration, and FA loading was determined by releasing the FA from the formulation in the presence of NaOH, followed by the recovery of the exosomes. The FA and exosomal proteins were measured by spectrophotometry and BCA assay, respectively, and percent FA loading was calculated.
2.3. siRNA:
siRNA-6 was purchased from Dharmacon (Lafayette, CO), with and without manufacturer’s propriety ON-TARGET plus modifications in the phosphate backbone. For siKRAS and scrambled siKRAS (siSCR), 3’ overhangs of UU and TT, respectively, were added to the targeting sequence to increase the stability of the molecule. siKRAS in the initial testing was also purchased from Millipore-Sigma (Woodlands, Tx) and Integrated DNA Technologies (Coralville, IA). siSurvivin and siSCR were custom synthesized from Millipore-Sigma (Woodlands, Tx). The sequences are provided below.
siKRAS Sense 5’ GGA GGG CUU UCU UUG UGU AUU 3’
Antisense 3’ UU CCU CCC GAA AGA AAC ACA U 5’
siSCR Sense 5’ GUU GGA GCU AGU GGC GUA GTT 3’
Antisense 3’ TT CAA CCU CGA UGA CCG CAU C 5’
siSurvivin Sense 5’ GGA CCA CCG CAU CUC UAC A 3’
Antisense 3’ CCU GGU GGC GUA GAG AUG U 5’
2.3.1. Loading of siRNA in the EPM:
Entrapment of siRNAs in the EPM was achieved by brief incubation of siRNA with exosomes in the presence of PEI. In a typical reaction, exosomes (25 μg – 300 μg) were mixed with 0.005% – 0.1% (7 – 150 μg) PEI (MW 800 Da – 600 kDa) in 150 μl PBS. After 5 min incubation at 23°C, siRNA (0.1 – 10 μg corresponding to 10 – 1,000 pmoles) was added and the incubation was continued for 15 min. The resulting EPM-siRNA or FA-EPM-siRNA complex was collected by precipitation with ExoQuickTC reagent (System Biosciences, Palo Alto, CA) on ice for 60 min, followed by centrifugation at 13,000×g for 10 min. The pellet was suspended in PBS and used for transfection studies. Alternatively, the EPM-siRNA or FA-EPM-siRNA complex was purified by ultrafiltration with 300,000 molecular weight-cutoff spin filters for analysis of size, zeta potential, and polydispersity index (PDI) by Zetasizer (Malvern, Westborough, MA) and transmission electron microscopy (TEM) (Make, City, state).
2.3.2. Preparation of 32P-labeled siRNA:
siKRAS and siVEGF were 5′−32P labeled with T4 polynucleotide kinase and [γ−32P]ATP (>6,000 Ci/mmol) using a molar excess of the siRNA. Any residual ATP was removed using Illustra ProbeQuant G-50 Micro Column (GE Healthcare), and the labeled siRNA was purified by a small RNA isolation kit as described [34]. The purity of the labeled siRNA was determined by polyacrylamide gel electrophoresis with detection by Packard InstantImager.
2.3.3. Electroporation:
Colostrum exosomes were loaded with siRNA by electroporation using the Gene Pulser Xcell (Bio-Rad, Hercules, CA) electroporation system as described [34]. Briefly, Exosomes in electroporation buffer (Bio-Rad, Hercules, CA), were mixed with non-radioactive siKRAS (0.5 – 10 μg) in combination with a 32P-labeled siRNA tracer. The mixture was electroporated at 400 V and 125 μF capacitance for a pulse time of 10–15 ms. Samples were kept on ice for at least 10 min pre- and post-electroporation pulse. Samples were then centrifuged at 7,000×g for 5 min to remove any aggregates. The resulting siRNA-loaded exosomes were harvested by precipitation with ExoQuick™ and the siRNA entrapment was determined.
2.3.4. Chemical transfection:
Colostrum exosomes were loaded with siRNA using a chemical transfecting reagent, Exo-Fect™ (System Biosciences, Palo Alto, CA, USA). Briefly, exosomes (75 – 300 μg in 50 μl) were incubated with Exo-Fect and non-radioactive siRNA (0.5 – 10 μg) and a 32P-labeled siRNA tracer at 37 °C for 10 min. The resulting exosome complex was collected and the siRNA entrapment was determined.
2.3.5. Entrapment and loading efficiency of siRNA:
To determine the entrapment and loading of siRNA in the EPM, we included P-labeled siRNA as a tracer along with non-radioactive. Entrapment efficiency was calculated by the distribution of the radioactivity present in the EPM-siKRAS recovered by precipitation with ExoQuick™ and in the supernatant, followed by radioactivity quantitation by Packard InstantImager. Percent entrapment and loading efficiencies of siRNA were calculated as follows:
2.4. emGFP (Emerald Green Fluorescent Protein) and p53 plasmids:
Both emGFP and p53 plasmids were constructed using the mammalian expression vector pcDNA™ 3.3 TOPO™-TA (ThermoFisher Scientific, Waltham, MA). The plasmid constructions details are provided in the supplementary materials. Both plasmids were maintained in bacterial culture, grown in the presence of ampicillin to ensure plasmid maintenance. For transfection, plasmids were isolated using the PureLink® HiPure Plasmid Maxiprep Kit (Thermo Fisher Scientific, Waltham, MA).
2.4.1. Loading of emGFP, and p53 plasmids in the EPM:
emGFP and p53 plasmids were entrapped in the EPM essentially the same way as described for siKRAS, except that different concentration of exosomes and nucleic acids were used.
2.5. Cell culture:
Human lung cancer cells, H1299, A549, and H522, and human pancreatic cancer cells, Panc-1 and MiaPaCa-2 were cultured in DMEM Glutamax (Gibco, Waltham, MA) supplemented with 10% FBS (or 2.5% horse serum for MiaPaCa-2) and antibiotics were incubated at 37 °C in 5% CO2. Human breast cancer cells, MDA-MB-231 were cultured in L-15 (Gibco, Waltham, MA) supplemented with 10% FBS and antibiotics at 37 °C without CO2.
2.6. In vitro transfection of EPM-siRNA and pDNA:
In vitro gene knock-down experiments were performed in H1299, A549, Panc-1, MiaPaCa-2, and MDA-MB-231. Cells were seeded in 24-well plates at a density of 75 × 103 cells/well and treated with either siKRAS or siSurvivin at 0.01 – 20 μg entrapped by EPM for up to 48 h. Additional reactions with siRNA, PEI-siRNA, Exo-siRNA, and exosomes alone were also included. The treated cells were analyzed by western blot to determine target gene silencing.
Studies related to the delivery of pemGFP and pcDNA-p53 were performed similarly as described for siRNA. H1299, A549, and Panc-1 were treated with EPM-pcDNA-p53 using 0.15 – 15 μg pDNA. Additional reactions included PEI-pcDNA-p53, Exo-pcDNA-p53, and exosomes alone as controls. emGFP expression studies were performed with the A549 cells. Cells treated with the EPM-pemGFP formulations were analyzed by fluorescent microscopy using BioTek Lionheart FX (Winooski, VT) and western blot to determine gene expression levels.
2.7. qRT-PCR for target gene expression:
One-Step SYBR green qRT-PCR Kit (Quanta Biosciences, Gaithersburg, MD) was used to perform cDNA synthesis and PCR amplification simultaneously from 100 ng of total RNA according to the manufacturer’s instructions and as described [35]. Relative gene expression was assessed using the differences in normalized Ct (ΔΔCt) method after normalization to β-actin [36]. Fold changes were calculated by 2−ΔΔCt. Experiments were performed in triplicate.
2.8. cell viability assay:
Cell viability following treatment with different formulations was measured by MTT assay, trypan blue assay and colony formation assay as described previously [37]. Experimental details are provided in Supplementary materials.
2.9. Western blot analysis:
Whole cell lysates were analyzed for proteins of interest by western blot as described previously [34]. Experimental details are provided in Supplementary materials.
2.10. Confocal imaging:
For the analysis of Texas Red (TR) siRNA uptake in vitro, images were acquired with a Nikon confocal microscope as described previously [37]. Experimental details are provided in Supplementary materials.
2.11. Biodistribution of exosomes and EPM
2.11.1. Preparation of AF750-labeled exosomes, with and without FA-functionalization:
We covalently attached FA and near-infrared dye AF750 to exosomes by using standard stable amide chemistry. This was achieved in a two-step process: In the first step, exosomes (10 μg/μl) were mixed with 100 μl of NaHCO3 buffer (1 M, pH 8.4) followed by mixing with 100 μl of freshly prepared activated FA-NHS (N-hydroxysuccinimidyl) ester (25 mg/ml in 0.1 N NaOH). Following incubation for 1 h at room temperature, the unbound FA was removed by ultrafiltration using 300,000 molecular weight cut-off spin filters (Sartorius, Goettingen, Germany). In the second step, AF750 NHS-ester was attached to exosomes or FA-exosomes as described for FA, except that it was dissolved in PBS.
2.11.2. Preparation of AF750-labeled EPM, with and without FA-functionalization:
In a typical reaction, AF750-labeled exosomes (150 μg), with and without FA-functionalization, were mixed with PEI (MW 60K) (0.025%) in 150 μl PBS. After incubation at room temperature for 20 min, the AF750-labeled EPM, with and without FA-functionalization, were recovered by precipitation with ExoQuick™ and the pellet was reconstituted in PBS.
2.11.3. Biodistribution study in tumor-bearing mice:
Female athymic nude (nu/nu) mice (5–6 weeks old) were maintained according to the Institutional Animal Care and Use Committee (IACUC) guidelines. Lung tumor xenografts were produced by subcutaneously injecting human lung A549 cells (2.5 × 106). Once tumors grew to >200 mm3, animals were randomized (n = 3) and administered a single intravenous dose of Exo-AF750, EPM-AF750, FA-Exo-AF750, or FA-EPM-AF750; one group of animals remained untreated as a control. The dose of AF750 in each treatment was maintained the same (20 – 50 μg per mouse) as determined by measurement of the fluorescence by spectrofluorometry. The animals were euthanized and various organs and tumor tissues were imaged ex vivo using Advanced Molecular Imager AMI 1000 (Spectral Instruments Imaging, Tucson, AZ). The relative intensities were measured and compared with untreated animals.
2.12. Application of EPM-siKRAS in vivo:
All experiments relating to animal models were carried out as per the IACUC guidelines. For the subcutaneous lung tumor xenograft model, A549 cells (2.5 × 106), in serum-free media, were mixed with Matrigel matrix and subcutaneously injected into the left flank of female athymic nude (nu/nu) mice (5–6-week old). Animals received purified AIN-93M diet and water ad libitum. Once the average tumor size reached about 100 mm3, mice were randomized into 3 groups (n = 10–12). Animals were treated intravenously three times a week with vehicle (PBS) or EPM-siKRAS (15 μg/dose). An additional group was treated with EPM-siSCR (scrambled siKRAS; 15 μg/dose). Tumor size was measured using Vernier caliper and animal weights, diet intake, and overall animal health were monitored weekly. After 31 days of treatment, the animals were euthanized and select tissues were collected for further analysis.
For the orthotopic lung tumor inhibition study, groups of female NOD-Scid mice were inoculated with Bioware® Brite A549-Red-Fluc cells (2 ×106 cells) via intrathoracic injection in 50 μl of Matrigel mixed with serum-free media (1:1; v/v) via intrathoracic injection using 30-gauge needles [38, 39]. After 10 days when the luminescence intensity reached approximately 6 × 106 photons, animals were randomized (n = 10–12) and treated intravenously three times a week (15 μg/dose) with FA-EPM, EPM-siKRAS, and EPM-siKRAS; a group of animals received vehicle (PBS) treatment. Animals received the purified AIN-93M diet and water ad libitum. For luciferase expression, mice were imaged twice a week, 15 min post intraperitoneal injection of luciferin (120 mg/kg), and the luciferase signals were detected by using Advanced Molecular Imager, AMI1000. When the average luminescence intensity in the vehicle treatment reached approximately 20 × 108 photons, all animals were euthanized and various tissues were collected and imaged ex vivo. Lung, liver, and tumor tissues were collected and stored at −80 °C for marker analysis.
2.13. Application of EPM-pcDNA-p53 in vivo:
For the p53-knockout model, following acclimation, female p53-knockout (Trp53tm1Tyj/J) mutant mice (Jackson Laboratories, Bar Harbor, ME) were randomized into 2 groups. Animals received EPM-pcDNA-p53 (10 μg/dose) (n=4) or vehicle (n=2) intravenously daily. Animals were euthanized after five days. At euthanasia, organs such as the lung, liver, kidneys, and spleen were collected and analyzed for p53 expression levels by western blot and RT-PCR.
For the subcutaneous xenografts study, A549 cells (2.5 × 106) were administered into female athymic nude (nu/nu) mice as described above. Animals received three intravenous doses a week either vehicle (PBS) or EPM-pcDNA-pP53 (10 μg/dose). After 10 days of treatment, animals were euthanized by CO2 asphyxiation and select tissues and the tumor were collected and analyzed for p53 expression levels by western blot.
2.14. Assessment of systemic toxicity:
Following acclimation, wild-type female C57Bl/6 mice (5–6-week old) were randomized (n = 5) into four groups and treated intravenously with vehicle (PBS), EPM, exosomes, and a PEI-exosomes three times a week for four weeks. EPM was prepared by incubation of colostrum exosomes (150 μg) and PEI (MW 60K; 37 μg) in 150 μl PBS, followed by precipitation of the exosomes and PEI complex (EPM) with ExoQuick™. The dose of exosomes in the exosome alone group was the same as used for the EPM formulation. Animals were euthanized by CO2 asphyxiation and blood was collected. Hematological parameters were analyzed by Abbott Laboratories (Santa Clara, CA). Serum was used to analyze the liver and kidney function enzymes. Electrolyte analysis was done by using the ion-selective electrode while other biochemical parameters were analyzed spectrophotometrically by using AU640 Chemistry Immuno Analyzer (Beckman Coulter, Inc., Brea, CA).
2.15. Statistical analysis:
Statistical analysis was performed using Graphpad Prism 8.0 (La Jola, CA). Data are presented as means ± SD of at least 3–4 replicates. Student unpaired t-test, one-way ANOVA followed by Tukey’s multiple comparison test, and two-way ANOVA followed by Bonferroni posttests were used to compare treatment groups. Details of specific tests performed are provided in the figure legends. Data with two replicates were not analyzed statistically. All western blot data were quantitated by ImageJ software and averages were compared between treatment and control using the student’s t-test. In each assay, a p-value of <0.05 was considered to be statistically significant.
3. Results
3.1. Characterization of EPM formulations
Exosomes were harvested from standardized bovine colostrum powder as yield is higher from this source compared to fresh raw milk (0.8 – 1.0 g/L of liquid containing 5% colostrum powder compared with 0.08 – 0.1 g/L milk). Colostrum powder was rehydrated, and exosomes were isolated by differential centrifugation, as described [33]. The average isolated exosomes were physically homogenous with particle size distribution peaking at 66 ± 2.5 nm, polydispersity index (PDI) of 0.28 ± 0.01, and zeta potential of −9.2 ± 0.66 mV, as determined by Zetasizer (Fig. 1b and Supplementary Table I); similar sizes were observed with transmission electron microscopy (TEM) (Supplementary Fig. 1a). In addition to hallmark protein markers CD81, Tsg101, and Alix, colostrum exosomes exhibited CD47, a known anti-phagocytic protein (Fig. 1c). The size of exosome was slightly increased to 77 ± 1.53 nm after FA conjugation (Supplementary Table I).
The size and PDI of exosomes, FA-EPM, and FA-EPM-siKRAS remained essentially unaltered (Fig. 1b and Supplementary Table I), which was supported by TEM analysis (Supplementary Fig. 1a). However, the zeta potential of the FA-EPM (−5.1 ± 0.50) was somewhat higher than the exosomes alone (−9.2 ± 0.66) which may be due to the cationic nature of the polymer (Supplementary Table I). Next, we examined the effect of exosomes and siRNA concentrations on PEI entrapment (MW 60 kDa, 37 μg, or 0.025% w/v). 5’−32P-labeled siKRAS was included as a tracer to determine the entrapment efficiency of the siRNA. 75 μg exosomes per 150 μl reaction volume resulted in the optimal entrapment of the siKRAS while siKRAS between 0.5 – 10 μg achieved similar entrapment efficiency (≈95%). Increasing the siRNA cargo to 50 μg somewhat decreased the entrapment efficiency (≈85%) (Fig. 1d and Supplementary Fig. 1b, 1c). Screening PEI of different MWs (0.8 – 60 kDa) revealed that the highest siKRAS entrapment (90% - 95%) was achieved with ≥37 μg of PEI (60 kDa) (Supplementary Fig. 1d, 1e). Similar conclusions were drawn with siVEGF (Supplementary Fig. 1f). The highest entrapment efficiency (>90%) was observed when exosomes: PEI: polynucleotide were 75:37:2 on a weight (μg) basis; the entrapment efficiency was essentially unaffected up to 10 μg polynucleotide (Figure 1d and Supplementary Figure 1f). The highest transfection efficiency was found when exosomes: PEI was 75:37, irrespective of the amount of polynucleotide in the range of 0.1 – 4 μg (Figure 3c and 3e and Supplementary Figure 3e). We found that nearly 70% of the initial PEI was not part of the matrix (Supplementary Fig. 1g) and was removed before cell treatment, reducing cytotoxicity concerns of unbound high molecular weight PEI. The EPM resulted in significantly (p<0.001) higher entrapment of siRNA (>90%) compared with the conventional approaches – electroporation (<5%) and the chemical transfection, Exo-Fect™ (about 35%) (Fig. 1e). siKRAS entrapped with the EPM was found to be safeguarded effectively based on lack of enzymatic degradation of the 5’−32P-labeled siKRAS used as a tracer (Fig. 1f).
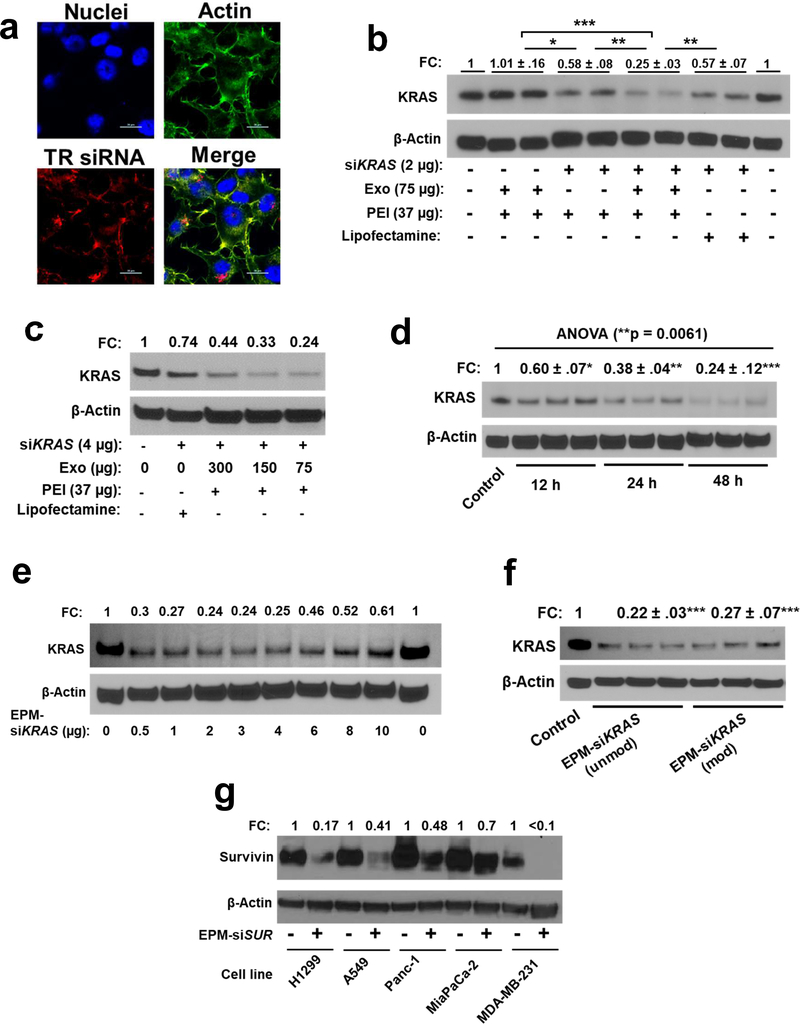
Figure 3.
(a) Uptake of Texas Red (TR)-labelled control siRNA entrapped in the exosome-PEI matrix (EPM) in human H1299 cells. Cells were treated with EPM-TR siRNA for 24 h, nuclei, and actin were stained with DAPI and phalloidin 488, respectively, and analyzed by confocal microscopy (scalebar indicate 20 μm). (b) Effect of siKRAS entrapped in EPM, PEI alone, and lipofectamine, respectively, on the modulation of the target protein in A549 cells. (c-f) Effect of siKRAS entrapped in EPM on the modulation of the target protein in A549 cells. The effect of different parameters such as (c) varying amounts of exosomes in EPM, (d) treatment time, (e) varying amounts of siKRAS, and (f) siKRAS carrying no modifications (Unmod) or modifications (Mod) were analyzed. (g) Effect of siSurvivin (siSUR) entrapped in EPM on the modulation of the target protein in indicated human cancer cell lines. (b-g) cells were treated with the various formulations along with vehicle control for 48 h and in the presence of EPM containing 37 μg PEI and 75 μg Exo, unless specifically indicated otherwise, and protein lysates were analyzed by western blot. Reactions were performed in triplicates and data represent mean ± SD (n = 3; blots shown from only one or two replicates for presentation purpose). Fold change (FC) was calculated and statistical analysis was performed by student’s t-test and one-way ANOVA followed by Tukey’s multiple comparison test to compare between the indicated groups; *, p < 0.05; **, p < 0.01; *** p < 0.001.
3.2. Biodistribution
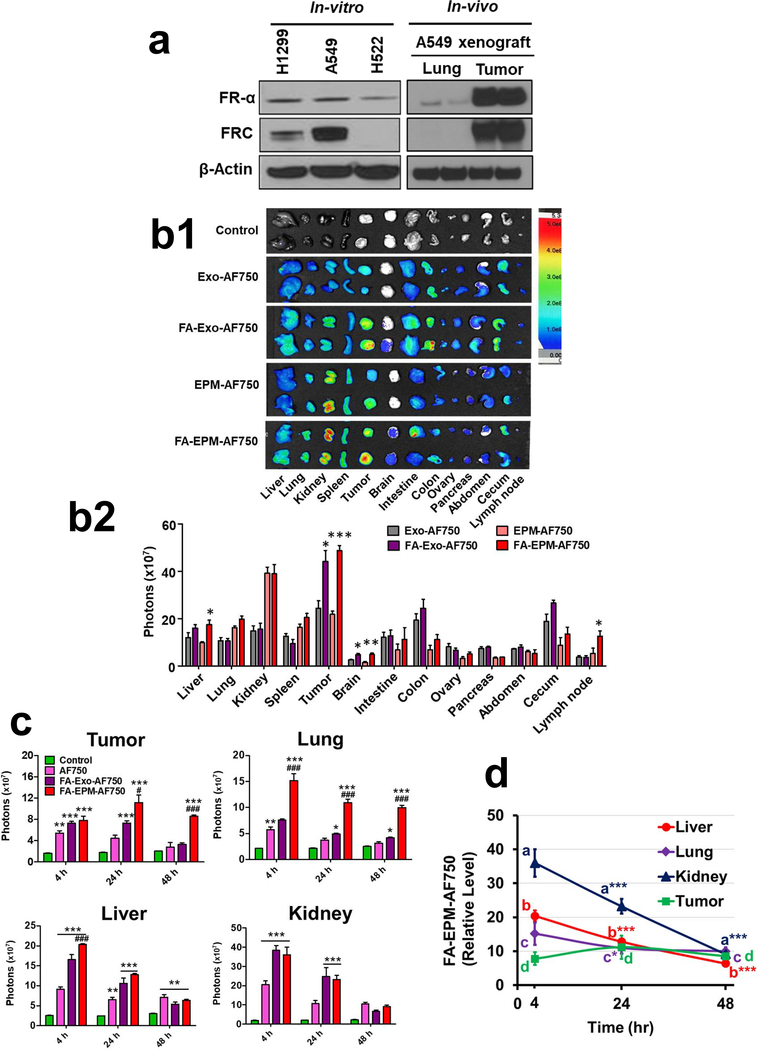
The use of the FA-functionalization of exosomes was prompted by the overexpression of the folate receptor-α (FR-α) in a broad range of cancerous cells, including lung cancer cell lines A549 and H1299. Analysis of whole-cell lysates indicated the two aggressive cell lines A549 and H1299 express higher FR-α compared to H522 cells (Fig. 2a). A549 xenograft tumors expressed nearly 100-fold higher FR-α compared to normal mouse lung tissue (Fig. 2a), suggesting folate receptor targeting as a viable approach to achieve tumor-specific delivery. We explored tumor cell targeting using Alexa Fluor™-750 (AF750)-labeled exosomes and EPM delivery, with and without FA-functionalization. Ex vivo imaging of tumors after single intravenous injection (20 μg AF750/500 μg exosomes) indicated all test formulations localized to tumors as well as a variety of other tissues. However, fluorescent signals from FA-Exo-AF750 and FA-EPM-AF750 treatment resulted in significantly (p<0.01) higher tumor accumulation of exosomes compared with non-functionalized Exo-AF750 and EPM-AF750, respectively (Fig. 2b1, b2, 2c, and Supplementary Fig. 2a), indicating tumor-targeted delivery. We observed that the total signals in the FA-functionalized formulations were higher compared to non-functionalized formulations; however, both the FA-Exo-AF750 and FA-EPA-AF750 showed higher tumor accumulation based on the mean signals and fraction of the total signals (Supplementary Tables II and IIA). A time-course biodistribution of FA-Exo-AF750 and FA-EPM-AF750 revealed a continuous decline in fluorescent signals in the kidney and liver, indicating presumable clearance of the formulations. The decline in the lung was more modest and no decline was observed in the tumor after 48 h of the FA-EPM-AF750 treatment (Fig. 2c, 2d, and Supplementary Fig. 2b, 2c, 2d). Higher accumulation was also detected in orthotopic mammary tumors with FA-EPM-AF750 compared with FA-Exo-AF750 (Supplementary Fig. 2e). FA-Exo-750 and FA-EPM-AF750 also resulted in significantly (p<0.01) enhanced signals in the brain and lymph nodes (Fig. 2b1, b2), demonstrating the ability of these formulations to reach the sites difficult to target. Notably, FA-EPM-AF750 accumulated more in both tumor and lung tissue, especially after 24 and 48 h, than AF750 alone (Fig. 2c).
Figure 2.
(a). Detection of folate receptors in highly aggressive (H1299 and A549) and less aggressive (H522) lung cancer cell lines and in A549 subcutaneous lung tumor xenografted in nude mice. Cells and tissue protein lysates were analyzed by western blot. FR-α, folate receptor α. (b) Tissue distribution of bovine colostrum exosomes and EPM, with and without FA-functionalization of exosomes. Exosomes were tagged with fluorescent dye Alexa Fluor™ 750 (AF750). A549 subcutaneous lung tumor-bearing nude mice (n=3) were treated with single intravenous doses of indicating formulations and animals were euthanized after 4 h and various tissues, including tumor, were imaged ex vivo using advanced molecular imager (AMI1000) (b1). Fluorescence intensity was measured, and the tissue distribution of exosomes is presented as relative fluorescence intensity in panel b2. Background values from untreated animals are subtracted from all groups and not shown. Statistical analysis was performed using the student’s t-test (b2) =Fluorescent intensity represents mean photons per second that leave a square centimeter of tissue and radiate into a solid angle of one steradian (photons/s/cm^2/sr). (c, d) Time-dependent tissue distribution of FA-functionalized EPM and FA-exosomes in A549 subcutaneous lung tumor-bearing nude mice. Exosomes were tagged with AF750. Animals were treated with a single dose of different formulations intravenously; AF750 alone was included as a free dye control. Groups of animals (n = 3 each) were euthanized after 4 h, 24 h, and 48 h, and different tissues, including the tumors, were imaged ex vivo using an AMI1000 animal imager, and the fluorescence intensity was measured. Statistical analysis was performed by two-way ANOVA followed by Bonferroni posttests to compare between the indicated groups (c). (d) Represent accumulation/clearance of FA-EPM-AF750 in indicated tissues at different time points. Data represent mean ± SD. Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparison test to compare between the indicated groups. A comparison of groups to control is represented by asterisks whereas a comparison of FA-Exo-AF750 and FA-EPM-AF750 is shown by the # sign. * #, p < 0.05; **, ##, p < 0.01; ***, ### p < 0.001.
3.3. In vitro gene silencing with EPM-siRNA
To assess cellular uptake, we treated human cancer cells with reference Texas Red (TR)-labeled siRNA (EPM-TR-siRNA), and to assess gene silencing ability we treated human cancer cells with EPM-siKRAS. Confocal microscopy confirmed the effective internalization of the poly complex resulting in red fluorescent signals in H1299 (Fig. 3a, Supplementary Fig. 3a), and MiaPaCa-2 pancreatic (Supplementary Fig. 3b) cancer cells. Nine different commercially available KRAS siRNA (siR-1 through siR-9) sequences were screened to identify the sequence with the highest KRAS knockdown when delivered by EPM in A549 cells (Supplementary Fig. 3c); the sequence siR-6 eliciting the highest knockdown was used for all subsequent studies. We demonstrate efficient gene knock-down with 2 μg of siRNA; EPM-siKRAS reduced expression by 4-fold while PEI-siKRAS and siKRAS were delivered by lipofectamine, an industry-standard transfecting agent, each only reduced expression by 1.7-fold (Fig. 3b). The MW of PEI played a significant role in defining the transfection efficiency of the EPM, with maximal transfection efficacy observed with MW 60 kDa or higher, followed by MW 5 kDa; MW lower than 5 kDa was ineffective (Supplementary Fig. 3d). In all subsequent studies, we used PEI MW 60 kDa, unless specifically indicated otherwise. Further, we show that 37 μg PEI (Supplementary Fig. 3e) along with 75 μg exosomes resulted in the highest gene knockdown in the EPM formulation Fig. 3c). This optimal formulation was used to determine the time-dependent effect on gene expression, with the highest knock-down occurring at 48 h (Fig. 3d). Also, as little as 0.1 – 0.5 μg of siKRAS was sufficient to knock-down 60%−70% of the target gene (Fig. 3e and Supplementary Fig. 3f). We note that KRAS gene silencing was equally efficient with unmodified (EPM-siKRAS (unmod)) and modified (EPM-siKRAS (mod)) carrying manufacturer’s propriety phosphate backbone modifications for higher specificity and efficiency (Fig. 3f), indicating that EPM presumably protected the siRNA leading to similar transfection efficiencies. Thus, the use of EPM could eliminate the need for chemical modifications which may affect the properties of siRNA resulting in undesired effects. We also compared the gene-silencing efficiency of EPM-siKRAS using exosomes from colostrum powder versus mature milk. KRAS expression was reduced with both sources of exosomes, but the effect was greater with colostrum exosomes (Supplementary Fig. 3g). A second siRNA targeting survivin in EPM formulation resulted in a 30%−80% knock-down of the target gene in lung, pancreatic, and breast cancer cells (Fig. 3g), indicating the versatility of EPM to deliver different siRNAs across various cancer types.
3.4. Anti-tumor effect of gene-knock down with EPM-siRNA
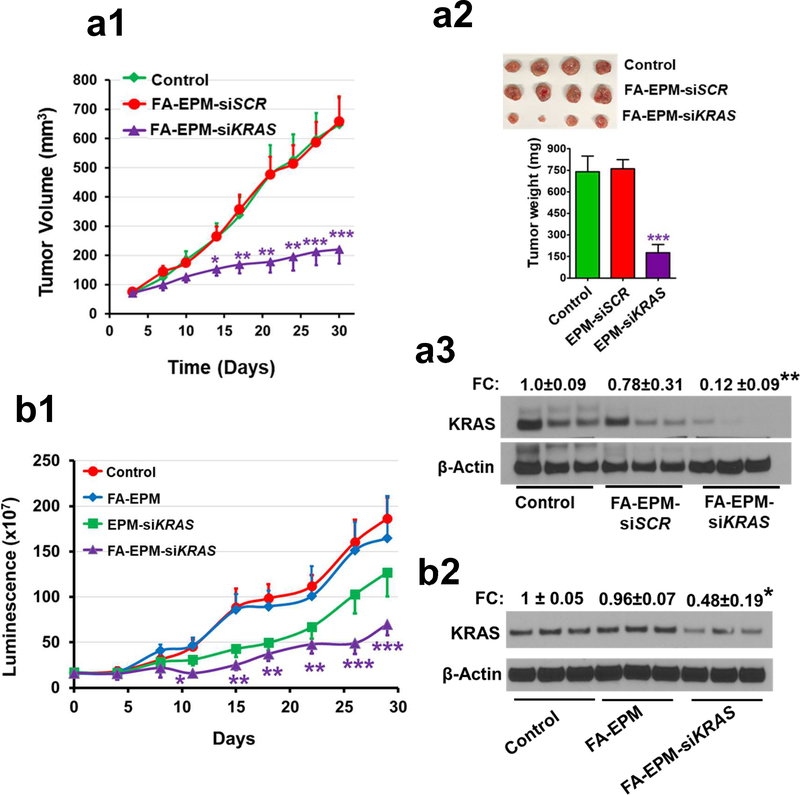
To confirm the therapeutic potential of EPM-siRNA in vivo, we administered FA-EPM-siKRAS (loaded with 15 μg siKRAS) intravenously to subcutaneous A549 tumor-bearing mice. A significant decrease in tumor volume (67%; p<0.001) and weight (76%; p<0.001) correlated with >85% knockdown of KRAS protein (p<0.01) levels in tumors treated with FA-EPM-siKRAS (Fig. 4a1, a2, a3) compared to vehicle control. These data provided evidence of effective, targeted EPM-mediated delivery of siRNA in vivo with therapeutic value. Next, we investigated the therapeutic activity of FA-EPM-siKRAS (loaded with 15 μg siKRAS) against orthotopic A549-Red-FLuc tumors in NOD-Scid mice. Bioluminescent imaging revealed a significant reduction in tumor growth rate by FA-EPM-siKRAS (62%, p<0.001) compared to FA-EPM and vehicle treatments. EPM-siKRAS also slowed down the tumor growth but the effect was statistically insignificant; FA-EPM did not affect tumor growth (Fig. 4b1). Analysis of cancerous lung tissues revealed a significant knockdown of KRAS protein with FA-EPM-siKRAS (52%, p<0.05), whereas, EPM-siKRAS and FA-EPM paralleled the KRAS levels in the untreated control (Fig. 4b2), thus validating the importance of FA-functionalization to achieve tumor-specific gene knockdown for effective therapy.
Figure 4.
(a) Effect of siKRAS entrapped in EPM on the growth of subcutaneous lung tumors growth (a1), weight (a2), and modulation of the target gene in the tumor tissues (a3) in female nude mice. Animals were subcutaneously inoculated with human A549 cells (2.5 × 106 cells) and when tumors grew to 80–100 mm3, animals were randomized (n = 8–10 each) and treated intravenously three times a week (Monday, Wednesday, and Friday) with siKRAS or siSCR (scrambled of siKRAS) entrapped in folic acid (FA)-functionalized EPM. Tumor growth was measured with a caliper weekly; at the end of the study, tumors were weighed and analyzed for the expression of the target protein by western blot. (b) Effect of siKRAS entrapped in EPM on the growth of orthotopic lung tumors (b1) and modulation of the target gene in the tumor tissues (b2) in female NOD Scid mice. Animals were inoculated with Bioware® Brite A549 Red-FLuc lung cancer cells (2 × 106 cells) in 50 μl Matrigel via intrathoracic injection. Ten days later, animals were imaged, randomized, and treated with siKRAS entrapped in EPM, with and without FA-functionalization of exosomes. Two additional groups received FA-EPM or vehicle as controls (n=10 each group). Animals were given luciferin substrate (80 mg/kg, i.p.), 15 min before imaging. Tumor growth was measured in live animals using AMI1000 Imager; at the termination of the study, tumors from randomly selected animals (n=3) were analyzed for the expression of the target protein; data represent mean ± SD. FC, fold change. Statistical analysis was performed by student’s t-test; *, p < 0.05; **, p < 0.01; *** p < 0.001.
3.5. Knock-in of the exogenous gene with EPM-pDNA in vitro
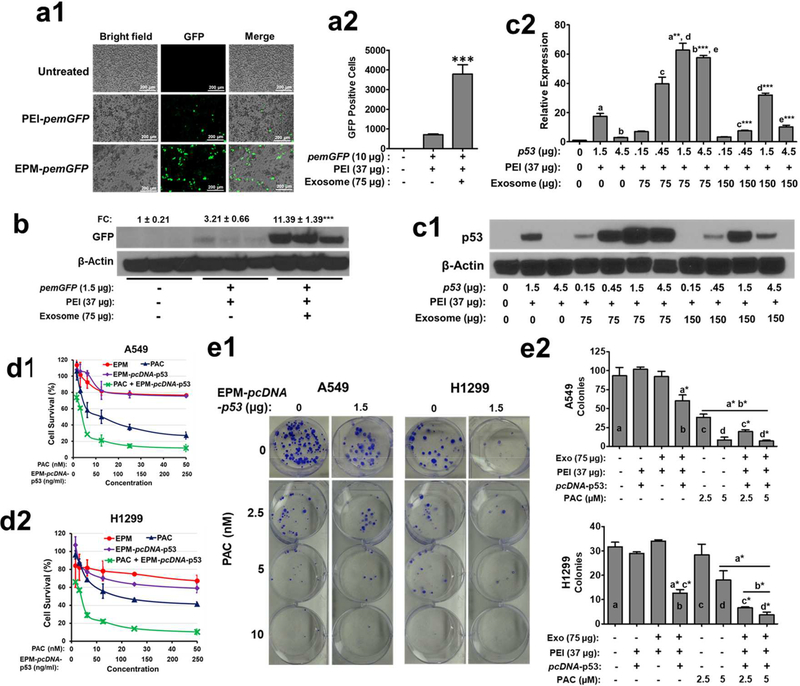
After a successful demonstration of the gene knock-down ability of EPM-siRNA formulations, we investigated the potential of EPM to deliver pDNA to knock-in exogenous genes. Our in vitro findings indicate successful cellular uptake and expression of GFP with EPM-pemGFP in A549 cells (Figs. 5a1, a2, Fig. 5b and Supplementary Fig. 4a); the highest expression was found after 48 h of transfection (Supplementary Fig. 4b). Exogenous gene expression tended to decrease with increasing amounts of exosomes used in the EPM-pcDNA-p53 formulation (Fig. 5c1, c2). We noted that p53 expression was detectable with as little as 0.15 μg pcDNA-pP53 entrapped by the EPM with the highest expression at 1.5 μg pcDNA-p53, but the expression levels declined (or plateaued) when higher amounts (4.5 μg) of the pDNA were used (Figs. 5c1, 5c2). Similar knock-in efficiency of EPM-pcDNA-p53 was also observed in Panc-1 pancreatic cancer cells (Supplementary Fig. 4c). Formulations lacking exosomes, i.e., PEI-pcDNA-p53, was significantly less effective than EPM-pcDNA-p53 (Supplementary Fig. 4c). Treatment with paclitaxel (PAC) of knock-in of WT p53 using EPM in p53-null H1299 cells resulted in enhanced cytotoxicity (Fig. 5d2) and reduced colony formation (Fig. 5e1, 5e2); no significant effects were observed when WT p53 was delivered via PEI alone (Supplementary Fig. 4d). Furthermore, we found chemo-sensitizing effects by exogenous p53 when co-treated with the chemotherapeutic drug, PAC in lung cancer cells (Fig. 5d1, 5d2, 5e1, 5e2). The effect on cell viability was confirmed by trypan blue assay (Supplementary Fig. 4e). These findings provide proof-of-concept for application of EPM loaded with pDNA to deliver exogenous genetic material to cells to compensate for loss-of-function mutations.
Figure 5.
(a). Introduction of pemGFP (emerald green fluorescence protein) plasmid DNA (10 μg) entrapped in the exosome-PEI matrix (EPM) in A549 cells. Cells were treated with EPM-pemGFP for 48 h and GFP signals were detected by fluorescent microscope (a1) and relative emGFP positive cells following treatment with the different formulations (a2). Data represent mean ± SD (n = 3). (b) Introduction of pemGFP entrapped in EPM in A549 cells. Cells were treated with EPM-pemGFP for 48 h and protein lysates were analyzed by western blot. (c) Introduction of wild type (WT) p53 plasmid DNA (pcDNA-p53) entrapped in EPM in H1299 cells. Cells were treated with EPM-pcDNA-p53 carrying different amounts of exosomes and pcDNA, for 48 h. Protein lysates were analyzed by western blot (c1) and blots were quantitated using ImageJ software (c2). Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparison test to compare between the indicated groups. The asterisk followed by letters shows a significant difference from the mean values represented by the same letters at p≤0.05. (d) Antiproliferative activity of paclitaxel (PAC) and EPM-pcDNA-p53, alone and in combination, against A549 (d1) and H1299 (d2) cells. pcDNA-p53 (1000 ng) was entrapped with the EPM. A549 and H1299 cells were plated in 96-well microtiter plates at an initial density of 3 × 103 cells per well and treated with different concentrations of paclitaxel (PAC) and EPM-pcDNA-p53, alone and in combination. Antiproliferative activity was determined by the MTT assay after 72 h. (e) Effect of PAC and EPM-pcDNA-p53, alone and in combination, on colony-forming in A549 and H1299 cells. Five hundred cells were plated in 6-well plates and treated with indicated concentrations of PAC and EPM-pcDNA-p53 (1.5 μg/well), alone and in combination. After 10 days, colonies were washed with PBS and stained with crystal violet. Colonies (e1) were counted and inhibition of colony formation was calculated (e2). Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparison test to compare between the indicated groups. The asterisk followed by letters shows a significant difference from the mean values represented by the same letters at p≤0.05.
3.6. Exogenous gene expression in vivo with EPM-pDNA
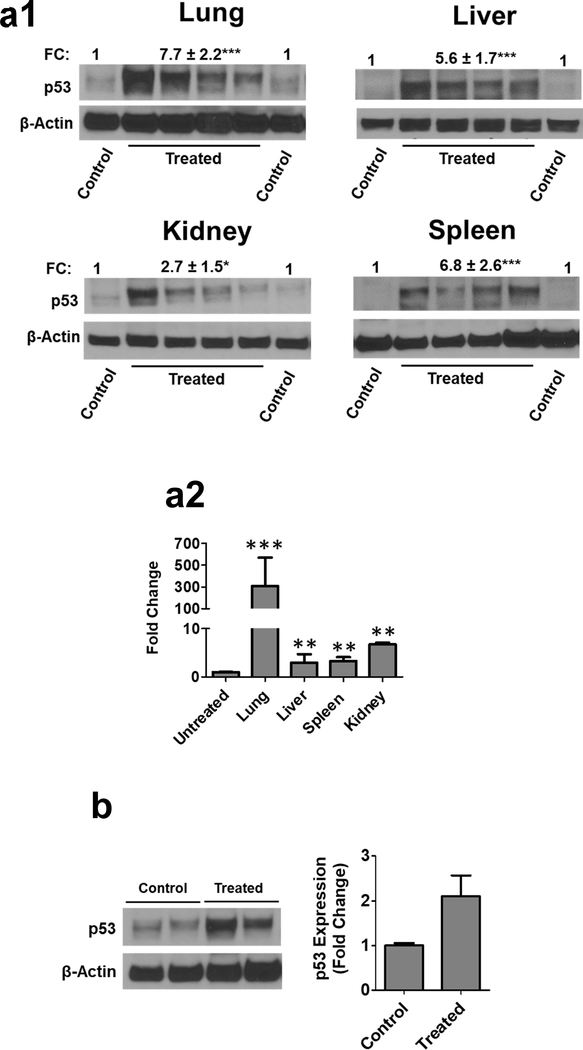
Finally, to demonstrate the ability of EPM-pDNA to knock-in genes in vivo, we administered EPM-pcDNA-p53 (10 μg) to p53-knockout (Trp53tm1Tyj/J) mutant mice. As p53 mutations are largely associated with tumors in the lung, colon, breast, and brain, we examined lung and several other tissues for p53 mRNA and protein expression levels. p53 protein and mRNA expression were detectable in the lung, liver, kidney, and spleen tissues (Fig. 6a1). This effect was most pronounced in the lung (Fig. 6a2), a common location of p53 mutations resulting in cancer. The expression of exogenous p53 in the tumor was also evident in A549 subcutaneous tumor xenografts following treatment with FA-EPM-pcDNA-p53 (10 μg) (Fig. 6b). Together, these experiments demonstrate the potential of EPM-mediated gene delivery to achieve exogenous gene expression in vivo.
Figure 6.
Introduction of wild-type (WT) p53 plasmid DNA (pcDNA) entrapped EPM in female p53-null mice. Animals were treated with EPM-pcDNA-p53 intravenously (10 μg pcDNA-p53/mouse) for 6 days. 24 h after the last dose, animals were euthanized, and different tissues were analyzed by western blot for p53 expression (a1) and RT-PCR (a2). Relative gene expression was assessed using the differences in normalized Ct (ΔΔCt) method after normalization to β-actin. Tissues were analyzed individually (n=4), and data represent mean ± SD. (b) Introduction of WT pcDNA-p53 entrapped in EPM in subcutaneous A549 lung tumors of female nude mice. Animals were treated with folic acid (FA)-functionalized EPM-pcDNA-p53 (n= 4) (10 μg pcDNA-p53/mouse) or vehicle (n = 2) intravenously for 10 days. 24 h after the last dose, animals were euthanized, and tumor tissues were analyzed by western blot for p53 expression. FC, fold change. All blots were quantitated using ImageJ software. Data represent mean ± SD (n = 3). Student’s t-test was used for statistical analysis; **, p < 0.01; ***, p < 0.001.
3.7. Systemic toxicity
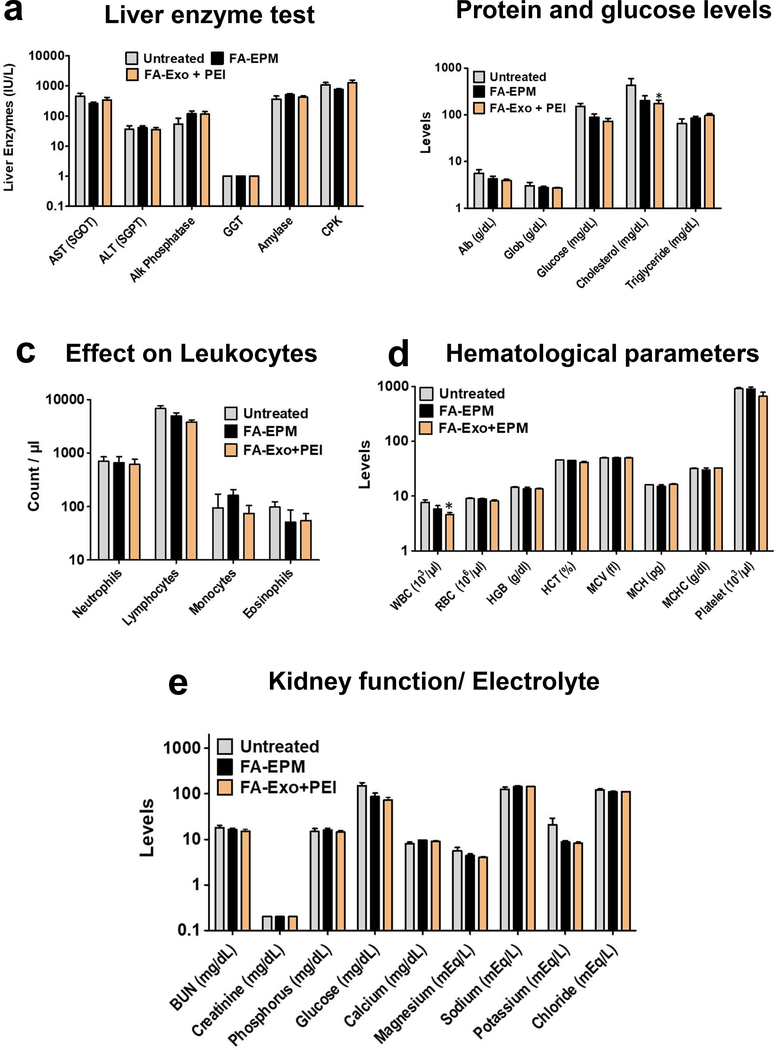
To evaluate systemic toxicity of EPM, WT female C57BL/6 mice were treated with intravenous injections of FA-EPM and FA-Exo + PEI (which would include FA-EPM and free PEI); the untreated group served as control. No signs of gross toxicity were observed with any of the formulations compared with control, based on the physical wellness of the animals, diet intake, and body weight gain (Supplementary Fig. 5). Analysis of liver and kidney function enzymes, hematopoietic, and biochemical parameters (Figure 7) indicated that test formulations were well tolerated during the 28-day treatment.
Figure 7.
Effect of FA-EPM and mixture of FA-exosomes and polyethyleneimine (PEI) on liver and kidney function enzymes, hematopoietic parameters and biochemical parameters in wild-type female C57BL/6 mice. Groups of animals (n= 5) were treated intravenously three doses a week for 4 weeks with FA-EPM and FA-exosomes + PEI (which contains FA-Exo and free PEI without precipitation step); untreated group served as control. EPM was prepared by mixing exosomes and PEI, followed by precipitation of EPM to remove unbound PEI. Animals were euthanized 24 h after the last dose and blood and plasma was analyzed for (a) liver enzyme test, (b) protein and glucose levels, (c) Effect on Leukocytes, (d) Hematological parameters and (e) Electrolyte levels and kidney function tests. Data represent mean ± SD. Student’s t-test was used for statistical analysis. *, p<0.05; **, p < 0.01.
4. Discussion
Gene therapy seeks to treat diseases that were previously considered either treatable but not curable or untreatable with conventional medication by introducing therapeutic nucleic acids into target cells. The development of an ideal delivery system for gene therapy that can effectively carry exogenous nucleic acid cargo, with minimal to no side effects, has been a great challenge to clinical applications. Several viral and non-viral vectors have been developed for gene therapy over the past two decades. However, clinical success has been very limited and hindered by the challenges associated with these delivery systems. Therefore, the need to develop safe and effective delivery systems is still ongoing. The use of cell secretory nano-vesicles, especially exosomes (30–100 nm) for nucleic acid delivery has brought tremendous excitement to the field of gene therapy, presumably due to favorable pharmacokinetics, biocompatibility, minimal or no inherent toxicity, long plasma half-life, and its intrinsic ability to cross biological barriers [21–23]. While some researchers engineered parent cells in vitro to secrete exosomes with the payload (such as siRNA, and mRNA) for delivery to target cells, others have manipulated exosomes post-secretion by physical or chemical methods to encapsulate the desired cargo [24, 25, 40]. Our group has utilized bovine milk exosomes to effectively deliver small molecule drugs [30, 32, 33] and siRNA [34].
Intending to develop a nano ‘platform’ for the targeted delivery of a wide range of macromolecules, including RNA and DNA, we formulated a matrix that utilizes ionic interactions between positively charged PEI (60 kDa), negatively charged nucleic acids, and exosomes to create a polyplex containing all three moieties, retaining a nanostructure. PEI has been largely used as a benchmark transfection reagent in in vitro studies for gene delivery between 0.8 – 25 kDa molecular weight (MW). The use of high MW PEI at an effective concentration is associated with in vivo toxicity and is of major concern [41]. On the other hand, low MW PEI is less toxic but is also less effective as a transfection reagent [42]. Although there is no report on the excretion of high MW PEI, it has been shown by PET imaging and the ICP-MS analysis that the PEI of MW 25,000 was accumulated in the liver and spleen at the first 24 h, and then excreted slowly from the body through the fecal route within 30 days [43]. Despite these limitations, PEI is considered a gold standard for gene delivery [44] due to its simple and robust transfection ability, extremely high stability, and cost-effectiveness. Additionally, PEI carries amino groups that facilitate chemical modifications for conjugation of targeting groups. Researchers have harnessed these positive attributes of PEI by complexing with lipid nanoparticles [45], inorganic nanoparticles [46], and other polymeric materials [47]. PEI has also been used in conjugation with viral vectors to enhance viral transduction [48]. Some early phase clinical trials used locally administered PEI-based nanoparticles to treat bladder cancer [49], intraperitoneally for ovarian cancer [50], and intratumorally for pancreatic cancer [51], and these nanoparticles were well tolerated and exhibited good safety profiles. These reports provide a precedent for the use of PEI in humans.
We show that exosomes harvested from standardized bovine colostrum powder were physically homogenous with particle size, PDI, and zeta potential, similar to raw milk-derived exosomes [33]. The use of colostrum powder minimizes the potential variability in the exosome population that could occur due to seasonal variation in the quality of raw milk. Colostrum exosomes, in addition to hallmark protein markers such as CD81, Tsg101, and Alix, abundantly expressed anti-phagocytic protein marker CD47. Although we did not examine the role CD47 in this report, CD47 on exosomes have been demonstrated to suppress their clearance by monocytes and increase exosomes half-life in the circulation [25].
The size and physical properties of EPM nanoparticles are comparable to exosomes alone. Importantly, EPM nanoparticles achieved several fold higher transfection efficiency of siRNA compared to electroporation and/or chemical-based transfection. While studies are limited comparing siRNA transfection efficiencies, Kamerkar et. al [25] reported similar siRNA loading efficiency for liposomes and electroporation of exosomes. Although electroporation of exosomes with siRNA has been widely used, researchers noted adverse morphological changes and aggregation of exosomes [52] and siRNA [53]. Given these concerns, EPM mediated delivery could be a successful alternative.
Our in vitro studies indicated that the EPM-entrapped siRNA was well protected from enzymatic degradation resulting in an effective knockdown of target protein expression. Using both unmodified and modified nucleotides for preparations of siKRAS sequences we have shown that EPM not only entraps both moieties with the same efficiency, but also results in similar gene silencing efficacy. These results affirm that EPM stabilizes both modified and unmodified nucleic acid sequences against nuclease degradation, a subject of concern. Thus, the use of EPM could eliminate the need for chemical modifications which may unfavorably affect the properties of siRNA such as its sensitivity to ribonucleases, recognition by the RNAi system, alter the localization of siRNA and leads to mild to severe toxicity [54, 55]. Successful gene silencing with multiple siRNAs against several cancer types and the ability to knock-in exogenous plasmid DNAs via EPM indicate the robustness of this nano-matrix. The significant inhibition of tumor growth in mouse models and exogenous expression of p53 in p53-knockout mutant mice endorses the clinical feasibility of this approach. Milk and colostrum exosomes have shown anti-proliferative effect on different cancer cells both in vitro and in vivo, although the effects were statistically insignificant [33]. EPM complex included in our present cell culture (Figure 2b, 5d, 5e) and in vivo (Figure 4b) studies shown no significant effect on the tumor cell growth or gene knockdown. The endogenous payload of mRNA and miRNA content (about 65 ng/100 μg exosomes) is substantially lower than the exogenous siRNA delivered (1– 2 μg in vitro and 10–15 μg in vivo) and we do not anticipate any adverse effect. In fact, milk/colostrum exosome have shown a modest anti-proliferative and anti-inflammatory effects [33]. These findings thus demonstrate the versatility of the EPM approach by its ability to entrap different siRNAs and pDNAs and provide proof-of-concept for application of EPM to deliver exogenous genetic material to cells to silence target genes or to compensate for loss-of-function mutations.
Therapeutics can be made more effective by delivering the drug to the target site thus reducing off-target effects and potential adverse side effects. Tumor-targeted delivery approaches have exploited several cell surface receptors that are overexpressed in cancer cells [56, 57]. Folate receptors, especially FRα, is one such target that is well studied in the field of nanoparticle-based drug delivery systems to improve the efficacy of anticancer agents by increasing their concentration in tumor cells relative to normal cells [58]. Since the normal tissues have much lower expression and 100–200 times lower binding affinity to FA [59], off-target toxicity is expected to be minimal. Pawar et al. [59] showed FA- functionalization significantly improved drug efficacy and its respective pharmacokinetics profile. Reports of FA decorated non-viral vectors for delivery of antisense oligodeoxyribonucleotides [60], pDNA [61] and siRNA [62, 63] are also available in the literature, indicating the expansion of folate targeting approach for gene therapy. We achieved lung tumor-targeted delivery of EPM by covalent conjugation of FA with amino groups of exosomal surface proteins. Biodistribution studies in tumor-bearing mice confirmed the ability of FA-functionalized EPM formulations to achieve higher tumor accumulation. Our findings substantiate the importance of the FA-functionalization of EPM to achieve tumor-specific gene knock-down resulting in enhanced anti-tumor activity. These findings are in corroboration with Zheng et al [64], who reported efficient suppression of colorectal cancer tumor xenograft with the FA-displaying exosomes loaded with survivin siRNA. In this study folate-displaying exosomes were shown to mediate cytosolic delivery of siRNA by avoiding endosome trapping [64].
Importantly, EPM formulations were also found to reach the sites difficult to target including the brain and lymph nodes after systemic administration, indicating EPM to be an effective delivery vehicle with ability to cross physiological barriers, while protecting cargo molecules.
Since the focus of this study is to develop a platform technology for gene delivery, it is essential to determine the acute and chronic toxicity of the EPM vector. EPM demonstrated no untoward side effects in our sub-chronic (28-day) in vivo study. These data indicate that toxicity concerns associated with the use of high molecular weight PEI can be reduced to a large extent when using the EPM-nucleic acid complex as it was harvested by precipitation leaving behind the bulk of the unbound PEI. This was supported by results showing no change in hematopoietic parameters in the EPM treated group. These data corroborated the report of low MW PEI in which hematological analyses show that all parameters were within the normal ranges when the mice were treated with PEI@UCNPs administered by the i.p., i.v., and i.g. injections [43]. Additionally, milk exosomes were previously shown to have no immunological or cross-species reactions [30, 33]. However, the possible impact of PEI in the EPM on immunotoxicity needs to be investigated in greater detail. Further, longer-term studies examining the safety of EPM formulation are required to advance our understanding and to determine the clinical suitability of this platform gene delivery technology.
The EPM based approach has the potential to make a significant impact in the field of gene therapy as it possesses several advantages over both viral and non-viral transfection methods. Viral vector-mediated gene delivery has seen the most success with several FDA-approved treatments in recent years [65, 66] and many more are currently under clinical evaluation. Apart from prohibitively high cost, ranging between $300,000 to $1.5 million for the currently approved therapies [67], concerns of adverse effects and safety are major challenges of viral vectors. It is interesting to note that exosome-entrapped viruses are currently being studied to overcome physical and biochemical barriers and to reduce off-target toxicities [68, 69] associated with viral vectors. Many non-viral vectors especially those using PEI, lipid, and PLGA-based nanoparticles are being developed for safe and effective gene delivery given the limitations of the current viral-mediated methods. However, many of these nanoparticle-based gene therapies failed to meet their endpoints in clinical trials. So far only one lipid-based nanoparticle therapy has been approved by the FDA in August of 2018 [70].
Such low success rates are associated with inherent limitations of synthetic nanoparticles such as rapid degradation and clearance in circulation, short half-life, low uptake by target cells, and the toxic effect that arose by immune response stimulation causing failure in clinical settings [71]. Compared to viral and other non-viral methods, including cell culture-derived exosomes, bovine colostrum exosomes represent a biocompatible, scalable, and cost-effective delivery vehicle for clinical applications.
In conclusion, this novel EPM approach for gene therapy can load and protect a diverse range of nucleic acids and can safely elicit a biological response. FA-functionalized EPM formulations can further enhance the specificity of the therapeutic delivery. EPM-mediated nucleic acid delivery represents a new technology that opens avenues in the field of gene therapy to treat cancer and other degenerative diseases and has potential applications for vaccine development and anti-viral therapeutics.
Supplementary Material
Highlights.
Exosomes and PEI matrix (EPM) represent a new exosomal-based nano delivery ‘platform’.
EPM is a biocompatible means for nucleic acid delivery.
EPM provides high nucleic acid entrapment and protection from enzymatic degradation.
EPM enhances the specificity of delivery by the addition of a targeting ligand.
Intravenous dosing of EPM formulations was well tolerated with no adverse response.
Acknowledgments
This work was supported by funds from 3P Biotechnologies, Inc. and, in part, from the NCI SBIR grant R44-CA-221487 and Agnes Brown Duggan Endowment (to R.C.G.). Drs. Ashish Agrawal and Al-Hassan Kyakulaga are thankfully acknowledged for establishing the FA-functionalization of exosomes and performing confocal microscopy analysis, respectively.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Competing Interests and Financial Disclosures
Dr. Ramesh C. Gupta holds positions both at the University of Louisville and 3P Biotechnologies.
The authors have filed an international patent application (PCT) based on the results reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Anguela XM, High KA, Entering the Modern Era of Gene Therapy, Annu Rev Med, 70 (2019) 273–288 [DOI] [PubMed] [Google Scholar]
- [2].Sridharan K, Gogtay NJ, Therapeutic nucleic acids: current clinical status, Br J Clin Pharmacol, 82 (2016) 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Del Pozo-Rodriguez A, Rodriguez-Gascon A, Rodriguez-Castejon J, Vicente-Pascual M, Gomez-Aguado I, Battaglia LS, Solinis MA, Gene Therapy, Adv Biochem Eng Biotechnol, 171 (2020) 321–368. [DOI] [PubMed] [Google Scholar]
- [4].Mali S, Delivery systems for gene therapy, Indian J Hum Genet, 19 (2013) 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nayerossadat N, Maedeh T, Ali PA, Viral and nonviral delivery systems for gene delivery, Adv Biomed Res, 1 (2012) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG, Non-viral vectors for gene-based therapy, Nature Reviews Genetics, 15 (2014) 541. [DOI] [PubMed] [Google Scholar]
- [7].Mancheno-Corvo P, Martin-Duque P, Viral gene therapy, Clin Transl Oncol, 8 (2006) 858–867. [DOI] [PubMed] [Google Scholar]
- [8].Chen YH, Keiser MS, Davidson BL, Viral Vectors for Gene Transfer, Curr Protoc Mouse Biol, 8 (2018) e58. [DOI] [PubMed] [Google Scholar]
- [9].Lundstrom K, Viral Vectors in Gene Therapy, Diseases, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Herrero MJ, Sabater L, Guenechea G, Sendra L, Montilla AI, Abargues R, Navarro V, Alino SF, DNA delivery to ‘ex vivo’ human liver segments, Gene Ther, 19 (2012) 504–512. [DOI] [PubMed] [Google Scholar]
- [11].Hartman ZC, Appledorn DM, Amalfitano A, Adenovirus vector induced innate immune responses: impact upon efficacy and toxicity in gene therapy and vaccine applications, Virus Res, 132 (2008) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sakurai H, Kawabata K, Sakurai F, Nakagawa S, Mizuguchi H, Innate immune response induced by gene delivery vectors, Int J Pharm, 354 (2008) 9–15. [DOI] [PubMed] [Google Scholar]
- [13].Mellott AJ, Forrest ML, Detamore MS, Physical non-viral gene delivery methods for tissue engineering, Ann Biomed Eng, 41 (2013) 446–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Du X, Wang J, Zhou Q, Zhang L, Wang S, Zhang Z, Yao C, Advanced physical techniques for gene delivery based on membrane perforation, Drug Deliv, 25 (2018) 1516–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Che J, Xue Y, Feng J, Bai G, Yuan W, Comparison of Biological Responses of Polymers Based on Imine and Disulfide Backbones for siRNA Delivery, ACS applied materials & interfaces, 10 (2018) 5196–5202. [DOI] [PubMed] [Google Scholar]
- [16].Xin Y, Huang M, Guo WW, Huang Q, Zhang LZ, Jiang G, Nano-based delivery of RNAi in cancer therapy, Molecular cancer, 16 (2017) 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jin L, Zeng X, Liu M, Deng Y, He N, Current progress in gene delivery technology based on chemical methods and nano-carriers, Theranostics, 4 (2014) 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sung YK, Kim SW, Recent advances in the development of gene delivery systems, Biomater Res, 23 (2019) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Blanco E, Shen H, Ferrari M, Principles of nanoparticle design for overcoming biological barriers to drug delivery, Nat Biotechnol, 33 (2015) 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sprouse D, Reineke TM, Davis ME, Polymeric Delivery Vehicles for Exogenous Nucleic Acid Delivery,, Reference Module in Materials Science and Materials Engineering, Elsevier, 2016. [Google Scholar]
- [21].Darband SG, Mirza-Aghazadeh-Attari M, Kaviani M, Mihanfar A, Sadighparvar S, Yousefi B, Majidinia M, Exosomes: natural nanoparticles as bio shuttles for RNAi delivery, J Control Release, 289 (2018) 158–170. [DOI] [PubMed] [Google Scholar]
- [22].Jiang XC, Gao JQ, Exosomes as novel bio-carriers for gene and drug delivery, Int J Pharm, 521 (2017) 167–175. [DOI] [PubMed] [Google Scholar]
- [23].Shahabipour F, Barati N, Johnston TP, Derosa G, Maffioli P, Sahebkar A, Exosomes: Nanoparticulate tools for RNA interference and drug delivery, J Cell Physiol, 232 (2017) 1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ, Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes, Nat Biotechnol, 29 (2011) 341–345. [DOI] [PubMed] [Google Scholar]
- [25].Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R, Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer, Nature, 546 (2017) 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, Li G, Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer, Mol Ther, 16 (2008) 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mendt M, Rezvani K, Shpall E, Mesenchymal stem cell-derived exosomes for clinical use, Bone Marrow Transplant, 54 (2019) 789–792. [DOI] [PubMed] [Google Scholar]
- [28].Chen YS, Lin EY, Chiou TW, Harn HJ, Exosomes in clinical trial and their production in compliance with good manufacturing practice, Ci Ji Yi Xue Za Zhi, 32 (2019) 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee JH, Ha DH, Go HK, Youn J, Kim HK, Jin RC, Miller RB, Kim DH, Cho BS, Yi YW, Reproducible Large-Scale Isolation of Exosomes from Adipose Tissue-Derived Mesenchymal Stem/Stromal Cells and Their Application in Acute Kidney Injury, Int J Mol Sci, 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Agrawal AK, Aqil F, Jeyabalan J, Spencer WA, Beck J, Gachuki BW, Alhakeem SS, Oben K, Munagala R, Bondada S, Gupta RC, Milk-derived exosomes for oral delivery of paclitaxel, Nanomedicine, 13 (2017) 1627–1636. [DOI] [PubMed] [Google Scholar]
- [31].Aqil F, Jeyabalan J, Agrawal AK, Kyakulaga AH, Munagala R, Parker L, Gupta RC, Exosomal delivery of berry anthocyanidins for the management of ovarian cancer, Food Funct, 8 (2017) 4100–4107. [DOI] [PubMed] [Google Scholar]
- [32].Munagala R, Aqil F, Jeyabalan J, Agrawal AK, Mudd AM, Kyakulaga AH, Singh IP, Vadhanam MV, Gupta RC, Exosomal formulation of anthocyanidins against multiple cancer types, Cancer Lett, 393 (2017) 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Munagala R, Aqil F, Jeyabalan J, Gupta RC, Bovine milk-derived exosomes for drug delivery, Cancer Lett, 371 (2016) 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aqil F, Munagala R, Jeyabalan J, Agrawal AK, Kyakulaga AH, Wilcher SA, Gupta RC, Milk exosomes - Natural nanoparticles for siRNA delivery, Cancer Lett, 449 (2019) 186–195. [DOI] [PubMed] [Google Scholar]
- [35].Munagala R, Aqil F, Vadhanam MV, Gupta RC, MicroRNA ‘signature’ during estrogen-mediated mammary carcinogenesis and its reversal by ellagic acid intervention, Cancer Lett, 339 (2013) 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pfaffl MW, A new mathematical model for relative quantification in real-time RT-PCR, Nucleic Acids Res, 29 (2001) e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kyakulaga AH, Aqil F, Munagala R, Gupta RC, Synergistic combinations of paclitaxel and withaferin A against human non-small cell lung cancer cells, Oncotarget, 11 (2020) 1399–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fan TW, Lane AN, Higashi RM, Yan J, Stable isotope resolved metabolomics of lung cancer in a SCID mouse model, Metabolomics : Official journal of the Metabolomic Society, 7 (2011) 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhong W, Hansen R, Li B, Cai Y, Salvador C, Moore GD, Yan J, Effect of yeast-derived beta-glucan in conjunction with bevacizumab for the treatment of human lung adenocarcinoma in subcutaneous and orthotopic xenograft models, Journal of immunotherapy, 32 (2009) 703–712. [DOI] [PubMed] [Google Scholar]
- [40].Mathiyalagan P, Sahoo S, Exosomes-Based Gene Therapy for MicroRNA Delivery, Methods Mol Biol, 1521 (2017) 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cao W, Mishina M, Amoah S, Mboko WP, Bohannon C, McCoy J, Mittal SK, Gangappa S, Sambhara S, Nasal delivery of H5N1 avian influenza vaccine formulated with GenJet or in vivo-jetPEI((R)) induces enhanced serological, cellular and protective immune responses, Drug Deliv, 25 (2018) 773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chollet P, Favrot MC, Hurbin A, Coll JL, Side-effects of a systemic injection of linear polyethylenimine-DNA complexes, J Gene Med, 4 (2002) 84–91. [DOI] [PubMed] [Google Scholar]
- [43].Yu J, Yin W, Peng T, Chang YN, Zu Y, Li J, He X, Ma X, Gu Z, Zhao Y, Biodistribution, excretion, and toxicity of polyethyleneimine modified NaYF4:Yb,Er upconversion nanoparticles in mice via different administration routes, Nanoscale, 9 (2017) 4497–4507. [DOI] [PubMed] [Google Scholar]
- [44].Lungwitz U, Breunig M, Blunk T, Gopferich A, Polyethylenimine-based non-viral gene delivery systems, Eur J Pharm Biopharm, 60 (2005) 247–266. [DOI] [PubMed] [Google Scholar]
- [45].Sun W, Wang Y, Cai M, Lin L, Chen X, Cao Z, Zhu K, Shuai X, Codelivery of sorafenib and GPC3 siRNA with PEI-modified liposomes for hepatoma therapy, Biomater Sci, 5 (2017) 2468–2479. [DOI] [PubMed] [Google Scholar]
- [46].Babaei M, Eshghi H, Abnous K, Rahimizadeh M, Ramezani M, Promising gene delivery system based on polyethylenimine-modified silica nanoparticles, Cancer Gene Ther, 24 (2017) 156–164. [DOI] [PubMed] [Google Scholar]
- [47].Nam JP, Nah JW, Target gene delivery from targeting ligand conjugated chitosan-PEI copolymer for cancer therapy, Carbohydr Polym, 135 (2016) 153–161. [DOI] [PubMed] [Google Scholar]
- [48].Fan G, Fan M, Wang Q, Jiang J, Wan Y, Gong T, Zhang Z, Sun X, Bio-inspired polymer envelopes around adenoviral vectors to reduce immunogenicity and improve in vivo kinetics, Acta Biomater, 30 (2016) 94–105. [DOI] [PubMed] [Google Scholar]
- [49].Gofrit ON, Benjamin S, Halachmi S, Leibovitch I, Dotan Z, Lamm DL, Ehrlich N, Yutkin V, Ben-Am M, Hochberg A, DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer, J Urol, 191 (2014) 1697–1702. [DOI] [PubMed] [Google Scholar]
- [50].Thaker PH, Brady WE, Lankes HA, Odunsi K, Bradley WH, Moore KN, Muller CY, Anwer K, Schilder RJ, Alvarez RD, Fracasso PM, A phase I trial of intraperitoneal GEN-1, an IL-12 plasmid formulated with PEG-PEI-cholesterol lipopolymer, administered with pegylated liposomal doxorubicin in patients with recurrent or persistent epithelial ovarian, fallopian tube or primary peritoneal cancers: An NRG Oncology/Gynecologic Oncology Group study, Gynecol Oncol, 147 (2017) 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Buscail L, Bournet B, Vernejoul F, Cambois G, Lulka H, Hanoun N, Dufresne M, Meulle A, Vignolle-Vidoni A, Ligat L, Saint-Laurent N, Pont F, Dejean S, Gayral M, Martins F, Torrisani J, Barbey O, Gross F, Guimbaud R, Otal P, Lopez F, Tiraby G, Cordelier P, First-in-man phase 1 clinical trial of gene therapy for advanced pancreatic cancer: safety, biodistribution, and preliminary clinical findings, Mol Ther, 23 (2015) 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Johnsen KB, Gudbergsson JM, Skov MN, Christiansen G, Gurevich L, Moos T, Duroux M, Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes, Cytotechnology, 68 (2016) 2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kooijmans SAA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJA, Schiffelers RM, Raemdonck K, Vader P, Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles, J Control Release, 172 (2013) 229–238. [DOI] [PubMed] [Google Scholar]
- [54].Garber K, Alnylam terminates revusiran program, stock plunges, Nat Biotechnol, 34 (2016) 1213–1214. [DOI] [PubMed] [Google Scholar]
- [55].lannitti T, Morales-Medina JC, Palmieri B, Phosphorothioate oligonucleotides: effectiveness and toxicity, Curr Drug Targets, 15 (2014) 663–673. [DOI] [PubMed] [Google Scholar]
- [56].Chen S, Zhao X, Chen J, Kuznetsova L, Wong SS, Ojima I, Mechanism-based tumor-targeting drug delivery system. Validation of efficient vitamin receptor-mediated endocytosis and drug release, Bioconjug Chem, 21 (2010) 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ojima I, Zuniga ES, Berger WT, Seitz JD, Tumor-targeting drug delivery of new-generation taxoids, Future Med Chem, 4 (2012) 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Narmani A, Rezvani M, Farhood B, Darkhor P, Mohammadnejad J, Amini B, Refahi S, Abdi Goushbolagh N, Folic acid functionalized nanoparticles as pharmaceutical carriers in drug delivery systems, Drug Dev Res, 80 (2019) 404–424. [DOI] [PubMed] [Google Scholar]
- [59].Pawar H, Surapaneni SK, Tikoo K, Singh C, Burman R, Gill MS, Suresh S, Folic acid functionalized long-circulating co-encapsulated docetaxel and curcumin solid lipid nanoparticles: In vitro evaluation, pharmacokinetic and biodistribution in rats, Drug Deliv, 23 (2016) 1453–1468. [DOI] [PubMed] [Google Scholar]
- [60].Jhaveri MS, Rait AS, Chung KN, Trepel JB, Chang EH, Antisense oligonucleotides targeted to the human alpha folate receptor inhibit breast cancer cell growth and sensitize the cells to doxorubicin treatment, Mol Cancer Ther, 3 (2004) 1505–1512. [PubMed] [Google Scholar]
- [61].Liang B, He ML, Chan CY, Chen YC, Li XP, Li Y, Zheng D, Lin MC, Kung HF, Shuai XT, Peng Y, The use of folate-PEG-grafted-hybranched-PEI nonviral vector for the inhibition of glioma growth in the rat, Biomaterials, 30 (2009) 4014–4020. [DOI] [PubMed] [Google Scholar]
- [62].Xu L, Yeudall WA, Yang H, Folic acid-decorated polyamidoamine dendrimer exhibits high tumor uptake and sustained highly localized retention in solid tumors: Its utility for local siRNA delivery, Acta Biomater, 57 (2017) 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang Y, Sun G, Gong Y, Zhang Y, Liang X, Yang L, Functionalized Folate-Modified Graphene Oxide/PEI siRNA Nanocomplexes for Targeted Ovarian Cancer Gene Therapy, Nanoscale Res Lett, 15 (2020) 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zheng Z, Li Z, Xu C, Guo B, Guo P, Folate-displaying exosome mediated cytosolic delivery of siRNA avoiding endosome trapping, J Control Release, 311–312 (2019) 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Geyer MB, First CAR to Pass the Road Test: Tisagenlecleucel’s Drive to FDA Approval, Clin Cancer Res, 25 (2019) 1133–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ma CC, Wang ZL, Xu T, He ZY, Wei YQ, The approved gene therapy drugs worldwide: from 1998 to 2019, Biotechnol Adv, 40 (2020) 107502. [DOI] [PubMed] [Google Scholar]
- [67].Darrow JJ, Luxturna: FDA documents reveal the value of a costly gene therapy, Drug Discov Today, 24 (2019) 949–954. [DOI] [PubMed] [Google Scholar]
- [68].Wassmer SJ, Carvalho LS, Gyorgy B, Vandenberghe LH, Maguire CA, Exosome-associated AAV2 vector mediates robust gene delivery into the murine retina upon intravitreal injection, Sci Rep, 7 (2017) 45329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Orefice NS, Souchet B, Braudeau J, Alves S, Piguet F, Collaud F, Ronzitti G, Tada S, Hantraye P, Mingozzi F, Duconge F, Cartier N, Real-Time Monitoring of Exosome Enveloped-AAV Spreading by Endomicroscopy Approach: A New Tool for Gene Delivery in the Brain, Mol Ther Methods Clin Dev, 14 (2019) 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Anselmo AC, Mitragotri S, Nanoparticles in the clinic: An update, Bioeng Transl Med, 4 (2019) e10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen J, Guo Z, Tian H, Chen X, Production and clinical development of nanoparticles for gene delivery, Mol Ther Methods Clin Dev, 3 (2016) 16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.