Abstract
Atopic dermatitis (AD) is a common chronic relapsing inflammatory disease. There is substantial evidence suggesting that noncoding RNAs have indispensable roles in the pathogenesis of AD. Exosomal transfer RNA-derived fragments (tRFs) have been identified as potential biomarkers for various disorders. However, the role of tRFs in AD has remained to be elucidated, which was thus the aim of the present study. Plasma samples from 23 pediatric patients with AD and 23 healthy controls were collected. Exosomes were successfully isolated from plasma according to the manufacturer's protocol. Small RNA sequencing was performed in 3 patients with AD and 3 controls, and 135 significantly differentially expressed plasma exosomal tRFs were identified, including 36 upregulated and 99 downregulated tRFs. Using the miRanda and RNAhybrid databases, 58,227 target genes of these 135 differentially expressed tRFs were predicted. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses suggested that these target genes of tRFs are involved in multiple functions and pathways associated with AD. Downregulation of tRF-28-QSZ34KRQ590K and tRF-33-Q99P9P9NH57SD3 were validated in 20 patients with AD and 20 controls by reverse transcription-quantitative PCR and tRF-28-QSZ34KRQ590K exhibited significance in the receiver operating characteristic curve analysis. The present study was the first to provide a tRF profile in AD and implied that plasma exosomal tRF-28-QSZ34KRQ590K may be a potential biomarker for pediatric patients with AD. The present study enhanced the understanding of the pathogenesis of AD and provided a novel marker for the diagnosis and targeted treatment of AD.
Keywords: atopic dermatitis, biomarker, plasma, exosome, sequence, transfer RNA-derived fragments
Introduction
Atopic dermatitis (AD) is a common dermatological disease with prevalence rates of 15-30% in children and 2-10% in adults (1). The combination of genetic, immune and environmental stimuli appears to contribute to the manifestation of AD. Hereditary susceptibility is considered a primary factor. To elucidate the genetic factors of AD, multiple genetic studies have been performed and a series of susceptibility genes/loci have been identified, which indicate two major biological pathways responsible for the etiology of AD, including skin epithelial dysfunction and innate/adaptive immune response dysregulation, which affect each other (2). However, its exact pathogenesis has remained largely elusive.
Exosomes are thought to have important roles in intercellular communication and have been proven to be potential markers for numerous diseases, such as tumours (3) and renal disease (4). Along with the identification of RNA in exosomes in 2007 and the development of high-throughput techniques for nucleic acid analyses, there have been a growing number of studies on RNA sequences in exosomes (5). One class of small noncoding (snc)RNAs, transfer RNA-derived fragments (tRFs), which are generated from transfer RNA (tRNA), have been suggested to have roles in the regulation of gene expression, cell proliferation, RNA processing, modulation of the DNA damage response, priming of viral reverse transcriptase, neurodegeneration and tumour suppression (6). tRFs have been implicated in various infections, such as human T-cell leukaemia virus type 1(7) and HIV-1-infected cells (8). Different tRFs have been identified in cellular proliferation and tRF signatures have been described in myelodysplastic syndromes (9) and various cancer types, such as prostate cancer (10) and squamous cell carcinoma (11). These studies highlighted the potential role of tRFs in disease progression. A database of tRFs has been developed where each unique tRF has been given a name following the progress of high-throughput sequencing and analyses of small tRNA fragments (12). According to their mapped positions on the primary or mature tRNA transcript, tRFs may be classified into various types, such as tRNA halves (tiRNA/tiR) (13), tRF-1, tRF-3, tRF-5(14), and endogenous tRFs (i-tRFs) (15).
However, to the best of our knowledge, no previous study has characterized tRFs in AD. In the present study, the possible roles of tRFs in AD were explored, which may provide novel information regarding the pathogenesis of AD.
Materials and methods
Ethics statement and human specimens
This study was approved by the ethics committee of Shanghai Skin Disease Hospital (Shanghai, China) and was conducted according to the principles of the Declaration of Helsinki. A total of 23 pediatric patients with AD meeting the Hanifin-Rajka diagnostic criteria (16) and 23 healthy controls were enrolled from Shanghai Skin Disease Hospital (Shanghai, China) between December 1, 2017 and October 1, 2018 in the present study. All controls were evaluated by experienced doctors and were required to have no atopic disease history in individuals and their immediate family members over three generations. All patients and controls were required to have no history of malignancy or other systemic autoimmune diseases. None of the patients were treated with systemic drugs in the two weeks prior to examination. The disease severity in patients was evaluated using the objective SCORing Atopic Dermatitis (SCORAD) index (17), according to which patients were divided into the mild (0-24 points), moderate (25-50 points) and severe (51-103 points) groups. Written informed consent forms were signed by all of the subjects and their guardians. There was no significant difference between the two groups in terms of sex or age. The clinical features of the AD and control groups are presented in Table I.
Table I.
Clinical features of subjects in the AD and control groups.
| Item | AD group (n=23) | Control group (n=23) |
|---|---|---|
| Age (years) | 9.5 (3-13) | 9.0 (6-12) |
| Sex (female/male) | 12/11 | 10/13 |
| SCORAD | - | |
| Mild | 7 | - |
| Moderate | 10 | - |
| Severe | 6 | - |
Values are expressed as mean (range) or n. As the control group, subjects with no atopic disease history in individuals and three generations of immediate family members were used. As an indicator of AD severity, the SCORAD system was used as follows: 0~24, mild; 25-50, moderate; 51-103, severe. SCORAD, SCORing atopic dermatitis; AD, atopic dermatitis.
Plasma from patients with AD and healthy controls
Peripheral venous blood was collected into EDTA anticoagulant tubes, stored at 4˚C and further processed within 1 h. The blood was centrifuged at 2,500 x g at 4˚C for 10 min and the supernatant was collected and stored at -80˚C.
Exosome isolation and transmission electron microscopy (TEM) examination
Using the ExoQuick Plasma Prep and Exosome Precipitation kit (cat. no. EXOQ5TM-1, System Biosciences LLC) according to the manufacturer's protocol, exosomes were isolated from plasma. The following protocol were followed: i) Purified thrombin was diluted to a concentration of [611 U/ml] in PBS; ii) 4 µl of [611 U/ml] thrombin per 0.5 ml plasma was added to a final concentration of 5 U/ml; iii) the mixture was then incubated at room temperature for 5 min while mixing; iv) centrifugation was performed in a standard microfuge at 4,500 x g for 5 min at room temperature; v) the visible fibrin pellet at the bottom of the tubes was transferred to a new clean tube; vi) the serum-like supernatant was then treated with ExoQuick to precipitate exosomes for 30-60 min at 5˚C; vii) 5-10 µl of the exosome supernatant was absorbed and added to a copper net, precipitated for 1 min, and the floating liquid was removed from the edge with a filter paper at room temperature; viii) the sample was rinsed with PBS at room temperature; ix) 10 µl phosphotungstic acid was dropped on a copper net, precipitated for 1 min, before the floating liquid was absorbed with filter paper at room temperature; and x) this was fried at room temperature for 2 min, before TEM was performed (JEM-1200EX, Japan Electronics Corporation).
RNA extraction, small RNA library preparation and sequencing
Total RNA from plasma exosomes was extracted with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, purified RNAs were sent to Yingbio for constructing small RNA libraries and performing small RNA sequencing analysis. In brief, the small RNA was bound with 3'- and 5'-adapters and complementary (c)DNA constructs were created by reverse transcription (RT) followed by PCR. The small RNA fragments (15-40 nt) were excised and purified and the purified libraries were quantified and validated. Small RNA sequencing was performed on an Illumina HiSeq 2500 (Illumina, Inc.) (18).
Data analysis
Low-quality reads and short reads (<15 nt) of the raw sequencing data were filtered out. All small RNA clean reads were aligned to the piwi-interacting RNA (piRNA) database (http://pirnabank.ibab.ac.in/), miRbase database (http://www.mirbase.org/), National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), tRF database (tRFdb; http://genome.bioch.virginia.edu/trfdb/), Genomic tRNA database (http://gtrnadb.ucsc.edu/) and MintBase (https://mintbase.io/) to identify known sncRNAs. The clean data were analysed to detect the expression of sncRNAs between patients and controls, the criteria for differentially expressed small RNAs were as follows: Log2FC, >1 or <-1; P<0.05. MiRanda (http://www.microrna.org/microrna/home.do; selection criteria, score ≥150 and energy <-20) and RNAhybrid (https://omictools.com/rnahybrid-tool; selection criteria, energy <-25) were used to predict the target mRNAs of the screened small RNAs with significant differences. The overlapping genes of these two databases were taken as the final result of the target gene prediction. Gene Ontology (GO) assignment (http://www.geneontology.org/) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg) were used to analyse the functions and pathways of all target genes.
RT-quantitative (q)PCR
A total of 100 ng of RNA was reverse-transcribed to cDNA using the NEBNext® Multiplex Small RNA Library Prep Kit (NEB; cat. no. E7560S) according to the manufacturer's protocol. First, 1 µl of RT primer and 100 ng RNA was mixed, and then added to RNase-Free H2O to make up 12 µl. The mixture was incubated at 65˚C for 5 min. Subsequently, 4 µl of buffer (5X), 2 µl of dNTP Mix, 1 µl of protector RNase Inhibitor, 1 µl of transcriptase, and 12 µl of the mixture from the previous step, were added, followed by incubation for 1 h at 42˚C, then at 70˚C for 5 min. The obtained cDNA was cryopreserved. Next, 5 µl of 2X Master Mix (Roche Diagnostics, 0.3 µl of PCR specific primer forward, and 0.3 µl of PCR specific primer reverse was mixed, and added to ddH2O to make up 9 µl. Then, 9 µl mixture was added to each well of the 384-PCR plate, along with 1 µl of the corresponding cDNA. qPCR was performed on an ABI Q6 detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95˚C for 10 min; followed by 95˚C for 15 sec, then to 60˚C for 60 sec, for 45 cycles. U6 was selected as the internal control. The PCR specific primer sequences were as follows: tRF-33-Q99P9P9NH57SD3 forward 5'-GCAGGCTTCTGTAGTGTAGTGGTTA-3' and reverse, 5'-AGTGCGTGTCGTGGAGTCG-3'; tRF-28-QSZ34KRQ590K forward, 5'-AGGCTCGTTGGTCTAGGGGTA-3' and reverse, 5'-AGTGCGTGTCGTGGAGTCG-3'; and U6 forward, 5'-CGATACAGAGAAGATTAGCATGGC-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'. The 2-ΔΔCq method was used to quantify the tRFs (19).
Statistical analysis
Spearman's nonparametric correlation analysis was used to analyse the correlation between the expression of tRFs and disease severity using SPSS 23 (IBM Corp.). The predictive value of biomarkers regarding the probability of AD was determined by receiver operating characteristic (ROC) curve analysis, which was performed using MedCalc software (version 10.4.7.0; MedCalc). The area under the ROC curve (AUC) was calculated to evaluate the diagnostic potential of tRFs for AD. The optimal diagnostic score of the signature of each tRF was evaluated at the cut-off value with the largest Youden index. P<0.05 was considered to indicate statistical significance. All the experimental data were from at least three independent experiments.
Results
Characterization of the isolated plasma exosomes by TEM
There was no significant difference between the two groups in terms of sex or age. The clinical features of the AD and control groups are presented in Table I. To obtain the purified exosomes, plasma samples from pediatric patients with AD and individuals from the control group were subjected to centrifugation. TEM revealed that the plasma exosomes were spherical particles with a diameter of 100-150 nm and a complete membrane structure (Fig. S1), which indicated the successful isolation of exosomes from human plasma.
Sequencing of tRFs from plasma exosomes
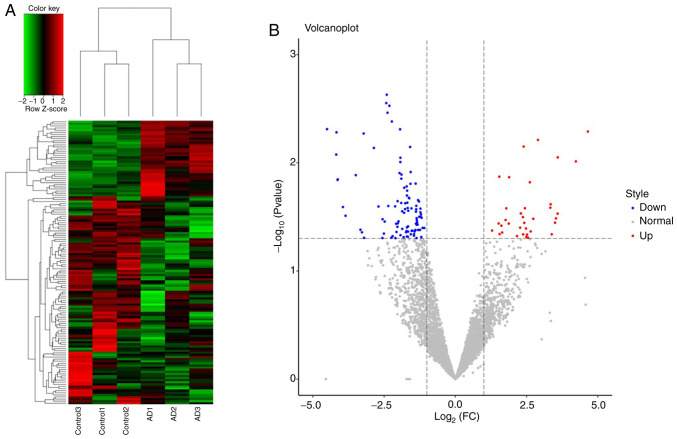
A total of 4,637 tRFs were identified between 3 patients with AD and 3 controls. Of these tRFs, 135 were significantly differentially expressed, including 36 upregulated and 99 downregulated tRFs (Fig. 1); the details are provided in Table SI. A total of 6 types of tRFs were identified, namely tRF-1, tRF-3, tRF-5, 3'-halves, 5'-halves and i-tRF (Table SII).
Figure 1.
(A) Hierarchical clustering of differentially expressed tRFs between the AD (n=3) and control groups (n=3). The colour scale represents high (red) to medium (black) to low (green) relative expression. (B) Volcano plot displaying the differentially expressed tRFs displaying significantly upregulated tRFs (red), significantly downregulated tRFs (blue) and nonsignificant tRFs (grey). AD, atopic dermatitis; tRFs, transfer RNA-derived fragments; FC, fold change.
Target gene prediction and functional analysis of differentially expressed tRFs
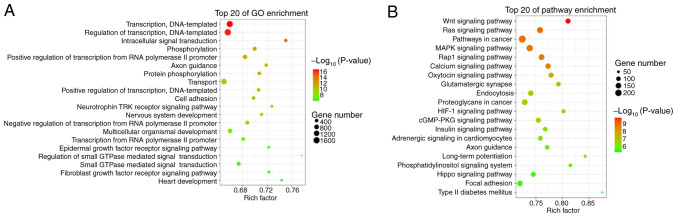
Analyses with the miRanda and RNAhybrid databases identified 58,227 target genes associated with 135 differentially expressed tRFs. GO analysis revealed that these target genes were functionally enriched in transcription, DNA templating, regulation of transcription, intracellular signal transduction, transport and epidermal growth factor receptor signalling pathway (Fig. 2A). KEGG pathway analysis revealed that these target genes were enriched in the Wnt signalling pathway, Ras signalling pathway, MARK signalling pathway, Rap1 signalling pathway, calcium signalling pathway, endocytosis and hypoxia-inducible factor (HIF)-1 signalling pathway (Fig. 2B).
Figure 2.
Bubble graphs of (A) GO and (B) KEGG pathway enrichment analyses each displaying 20 identified top pathways for target genes of differentially expressed tRFs (larger circles represent a larger number of genes). The colour scale represents low (green) to high (red) P-values. tRFs, transfer RNA-derived fragments; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Validation of differentially expressed tRFs
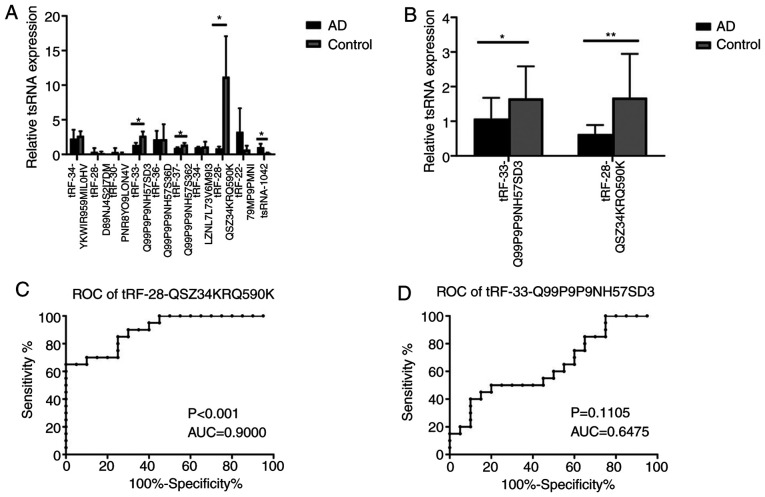
To further confirm the tRFs, 10 differentially expressed tRFs, including 6 downregulated tRFs (tRF-34-YKWIR959MIL0HV, tRF-34-LZNL7L73V6M9I3, tRF-28-QSZ34KRQ590K, tRF-33-Q99P9P9NH57SD3, tRF-36-Q99P9P9NH57S36D and tRF-37-Q99P9P9NH57S362) and 4 upregulated tRFs (tRF-28-D89NJ4S2I7DM, tRF-30-PNR8YO9LON4V, tRF-22-79MP9PMNI and tsRNA-1042), were subjected to RT-qPCR analysis in plasma exosomes from 3 patients with AD and 3 controls. A total of four tRFs, namely tRF-28-QSZ34KRQ590K, tRF-33-Q99P9P9NH57SD3, tRF-37-Q99P9P9NH57S362 and tsRNA-1042, exhibited differential expression (P=0.037, 0.030, 0.047 and 0.041 respectively; Fig. 3A). Of these, tRF-28-QSZ34KRQ590K and tRF-33-Q99P9P9NH57SD3 were further selected to be validated by RT-qPCR in 20 patients with AD and 20 controls. The results indicated that the expression of both of the tRFs exhibited significant differences between the AD and control groups (P<0.001 and 0.048, respectively; Fig. 3B).
Figure 3.
Verification of tRFs and ROC curve analysis. (A) Verification of 10 tRFs in 3 subjects with AD and 3 controls by RT-qPCR. (B) Verification of 2 tRFs in 20 patients with AD and 20 controls by RT-qPCR. *P<0.05; **P<0.01. (C and D) ROC curve analysis of (C) tRF-28-QSZ34KRQ590K and (D) tRF-33-Q99P9P9NH57SD3 for the differentiation of subjects with AD from the controls. tRFs, transfer RNA-derived fragments; AD, atopic dermatitis; RT-qPCR, reverse transcription-quantitative PCR; ROC, receiver operating characteristic; AUC, area under the ROC curve.
ROC curve analyses for tRF-28-QSZ34KRQ590K and tRF-33-Q99P9P9NH57SD3
ROC curve analysis was used to determine the diagnostic accuracy of plasma exosomal tRF-28-QSZ34KRQ590K and tRF-33-Q99P9P9NH57SD3. The ΔCq value of each sample was used for ROC curve analysis. and the AUCs of the two tRFs were 0.900 and 0.6475, respectively (P<0.001 and P=0.1105; Fig. 3C and D, respectively), which revealed that plasma exosomal tRF-28-QSZ34KRQ590K may serve as a biomarker for AD.
Correlation of tRF-28-QSZ34KRQ590K with AD severity
According to the SCORAD, the patients with AD were divided into three groups (Table I). There was no correlation between the expression of tRFs and disease severity (P=0.842, data not shown).
Discussion
Exosomes are thought to have important roles in intercellular communication and have been proven to be potential markers for numerous diseases. The discovery of tRFs in exosomes and their importance in gene regulation have substantially promoted an increase in their study in recent years. They are closely related to the occurrence of numerous human diseases and have the potential to become novel biomarkers for diseases (20). In the present study, the first analysis of tRFs in plasma exosomes from patients with AD was performed. Exosomes were isolated successfully from plasma with combined centrifugation and were then further identified by TEM. A total of 135 differentially expressed plasma exosomal tRFs were identified in 3 subjects with AD and 3 controls. GO and KEGG pathway enrichment analyses revealed a series of the possible functions and pathways of the target genes of these 135 differentially expressed tRFs. The downregulation of plasma exosomal tRF-28-QSZ34KRQ590K and tRF-33-Q99P9P9NH57SD3 was further verified by RT-qPCR in 20 subjects with AD and 20 controls. The ROC curve analysis indicated that only tRF-28-QSZ34KRQ590K may be a potential biomarker for AD.
tRFs have multiple biological functions, including acting as signalling molecules in cell proliferation and stress responses and as regulators of gene expression, and have been considered to be involved in RNA processing, suppression of translation, neurodegeneration and modulation of the DNA damage response (20). Identifying the entire repertoire of the roles of tRFs is particularly important to better understand their functions. In the present study, a total of 135 differentially expressed plasma exosomal tRFs were identified in a small sample, including 36 upregulated and 99 downregulated tRFs, which contained 6 types of tRFs, namely tRF-1, tRF-3, tRF-5, 3'-halves, 5'-halves and i-tRF. tRFs represent a heterogeneous class of sncRNAs and different sncRNAs are still being identified. In addition, there are limitations concerning tRF identification by current methods; in particular, certain tRNA modifications may block reverse transcriptase and modified tRFs may escape detection (21). Thus, it is possible that additional classes of tRFs are uncovered in the future. The present study was the first to identify differentially expressed plasma exosomal tRFs in AD, which provided a preliminary outline on tRFs in AD, and additional functional studies are required to further understand their mechanisms of action.
The different subclasses of tRFs have been indicated to be involved in several cellular functions that are also implicated in cancer, infection and neurodegenerative disorders (21). In the present study, two downregulated differentially expressed plasma exosomal tRFs, tRF-28-QSZ34KRQ590K and tRF-33-Q99P9P9NH57SD3, were verified in a larger sample by RT-qPCR. Growing evidence has indicated that tRFs regulate gene expression, epigenetic inheritance, apoptosis and RNA degradation and stability, although their detailed biological functions have not been fully elucidated. Downregulated tRF-28-QSZ34KRQ590K was demonstrated to be a significant marker for AD in the ROC curve analysis. Studies have revealed that tRFs may serve as noninvasive diagnostic biomarkers for various diseases (20). Therefore, tRF-28-QSZ34KRQ590K may also be a diagnostic biomarker for AD in pediatric patients, which may provide a novel source for the early diagnosis and targeted treatment of AD in the future. However, no correlation between tRF-28-QSZ34KRQ590K and the severity of AD was obtained (P>0.05). Thus, further studies are required to clarify the potential role of tRF-28-QSZ34KRQ590K in AD.
Analysis with the miRanda and RNAhybrid databases retrieved 58,227 target genes associated with the 135 differentially expressed tRFs. GO analysis indicated functional enrichment of these target genes in transcription, DNA templating, regulation of transcription, intracellular signal transduction and epidermal growth factor receptor signalling pathways. Epidermal growth factor receptor signalling has been previously reported to have a protective role in AD and to modulate IL-17 responses in the skin (22). KEGG pathway analysis suggested that these target genes were enriched in pathways including the Wnt, Ras, MAPK, Rap1, calcium and HIF-1 signalling pathways. The Rap1 signalling pathway (23) and MAPK signalling pathway (24) have been reported to be associated with inflammation in previous studies and the inflammatory response has also been suggested to have roles in the pathogenesis of AD (25). Therefore, the present study implied that plasma exosome tRFs may participate in the pathogenesis of AD by being involved in the inflammatory response and the epidermal growth factor receptor signalling pathway. However, further functional studies are required to better understand the pathogenesis of AD.
tRF-28-QSZ34KRQ590K demonstrated a significant diagnostic value for AD in the ROC curve analysis. There were 28 target genes of tRF-28-QSZ34KRQ590K, including tachykinin receptor 1, RANBP2-like and GRIP domain-containing 1, non-SMC condensin I complex subunit H (NCAPH), ectodysplasin-A receptor, contactin-associated protein family member 5, cytochrome P450 family (CYP)27 subfamily C member 1, WD repeat domain 33, POTE ankyrin domain family member F, insulin receptor substrate 1, thyroid hormone receptor interactor 12, S-antigen visual arrestin, ATPase plasma membrane Ca2+ transporting 2, neurexophilin and PC-esterase domain family member 3, Rho GTPase activating protein 31, p21 (RAC1)-activated kinase 2, midnolin, unc-13 homolog A, potassium channel tetramerization domain containing 15, endogenous retrovirus group V member 1 envelope, zinc finger and SCAN domain containing 18, KIAA0930 (also known as C22orf9, LSC3), SET binding factor 1, forkhead box K1, AVL9 cell migration-associated, FKBP prolyl isomerase family member 6, X-ray repair cross complementing 2, phosphatase and actin regulator 1 and mitochondrial calcium uniporter regulator 1. A previous study reported on a pediatric patient with AD, mental retardation, autistic features, epilepsy, developmental delay and abnormal immunological results, who carried a 7.9 Mb de novo deletion of chromosome 22q13.2/qter, a region containing the NCAPH2, SH3 and multiple ankyrin repeat domains 3 and CYP2D6 genes (26). These genes are associated with the T-cell immune response and the inflammatory response has also been suggested to have roles in the pathogenesis of AD. Thus, tRF-28-QSZ34KRQ590K may be involved in the pathogenesis of AD by affecting the inflammatory response. Further studies are required to uncover the role of tRF-28-QSZ34KRQ590K in the pathogenesis of AD. At present, it remains undetermined whether tRF-28-QSZ34KRQ590K is specifically expressed in AD, although it has not been reported to be expressed in any other diseases, to the best of our knowledge. However, tRF-28-QSZ34KRQ590K exhibited a significant diagnostic value in the ROC curve analysis, which suggested that tRF-28-QSZ34KRQ590K may be a potential biomarker for pediatric patients with AD, although further studies are required to confirm the role of this tRF in AD.
In conclusion, in the present study, 135 differentially expressed tRFs in the plasma exosomes of pediatric patients with AD were identified, and the significantly downregulated tRF-28-QSZ34KRQ590K and tRF-33-Q99P9P9NH57SD3 were further verified. tRF-28-QSZ34KRQ590K exhibited significance in the ROC curve analysis. Thus, tRF-28-QSZ34KRQ590K may be a potential biomarker for AD in pediatric patients. To the best of our knowledge, the present study was the first to report on the roles of tRFs in AD. It provided novel biological information for uncovering the pathogenesis of AD and a novel potential source for the early diagnosis and targeted treatment of AD in the future. However, further studies are required to better understand the roles of tRFs in AD.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: This work was supported by the National Natural Science Foundation of China (grant nos. 81602744 and 81573063).
Availability of data and materials
The raw data of small RNA sequencing of plasma exosomes has been uploaded to the sequence read archive (http://www.ncbi.nlm.nih.gov/bioproject/609458; submission ID: SUB7071247; BioProject accession no. PRJNA609458). The raw data will be publicly available on March 1, 2021.
Authors' contributions
LM and LJ involved in drafting the manuscript and revising it critically for important intellectual content, and have made substantial contributions in the conception and design of the current study. JC, HR, ZG, YW and FW were involved in the acquisition of data, analysis and interpretation of data. LY was responsible for the conception and design of the current study and manuscript revision. LM and LY confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the ethics committee of Shanghai Skin Disease Hospital (Shanghai, China) and all subjects and their guardians provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Abrams EM, Sicherer S. Cutaneous sensitization to peanut in children with atopic dermatitis: A window to prevention of peanut allergy. JAMA Dermatol. 2018;155:13–14. doi: 10.1001/jamadermatol.2018.3722. [DOI] [PubMed] [Google Scholar]
- 2.Bin L, Leung DY. Genetic and epigenetic studies of atopic dermatitis. Allergy Asthma Clin Immunol. 2016;12(52) doi: 10.1186/s13223-016-0158-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J Neural Transm (Vienna) 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 4.Miranda KC, Bond DT, McKee M, Skog J, Paunescu TG, Da Silva N, Brown D, Russo LM. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 6.Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G, Grassi A, Zanovello P, Bronte V, Ciminale V, D'Agostino DM. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: A role for a tRNA fragment as a primer for reverse transcriptase. J Virol. 2014;88:3612–3622. doi: 10.1128/JVI.02823-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: Evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Bosompem A, Mohan S, Erdogan B, Ye F, Vickers KC, Sheng Q, Zhao S, Li CI, Su PF, et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genomics. 2015;16(727) doi: 10.1186/s12864-015-1929-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, Bangma CH, Litman T, Visakorpi T, Jenster G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 11.Victoria Martinez B, Dhahbi JM, Nunez Lopez YO, Lamperska K, Golusinski P, Luczewski L, Kolenda T, Atamna H, Spindler SR, Golusinski W, Masternak MM. Circulating small non-coding RNA signature in head and neck squamous cell carcinoma. Oncotarget. 2015;6:19246–19263. doi: 10.18632/oncotarget.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141–D145. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajka G, Hanifin JM. Diagnostic features of atopic dermatitis. Acta Dermato-venereologica. 1980;60:44–47. [Google Scholar]
- 17.The SCORAD index. Consensus report of the European task force on atopic dermatitis. Dermatology. 1993;186:23–31. doi: 10.1159/000247298. Severity scoring of atopic dermatitis. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Liu S, Wang JH, Zou J, Zeng H, Zhao H, Zhang B, He Y, Shi J, Yoshida S, Zhou Y. Differential expressions of microRNAs and transfer RNA-derived Small RNAs: Potential targets of choroidal neovascularization. Curr Eye Res. 2019;44:1226–1235. doi: 10.1080/02713683.2019.1625407. [DOI] [PubMed] [Google Scholar]
- 19.Mazzei M, Vascellari M, Zanardello C, Melchiotti E, Vannini S, Forzan M, Marchetti V, Albanese F, Abramo F. Quantitative real time polymerase chain reaction (qRT-PCR) and RNAscope in situ hybridization (RNA-ISH) as effective tools to diagnose feline herpesvirus-1-associated dermatitis. Vet Dermatol. 2019;30:e491–e147. doi: 10.1111/vde.12787. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Yu X, Zhu L, Li T, Yan Z, Guo J. Transfer RNA-derived fragments and tRNA halves: Biogenesis, biological functions and their roles in diseases. J Mol Med. 2018;96:1167–1176. doi: 10.1007/s00109-018-1693-y. [DOI] [PubMed] [Google Scholar]
- 21.Soares AR, Santos M. doi: 10.1002/wrna.1423. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA: 8, 2017 doi: 10.1002/wrna.1423. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Xiao C, Gibson AM, Bass SA, Khurana Hershey GK. EGFR signaling blunts allergen-induced IL-6 production and Th17 responses in the skin and attenuates development and relapse of atopic dermatitis. J Immunol. 2014;192:859–866. doi: 10.4049/jimmunol.1301062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu A, Chen H, Xu C, Zhou J, Chen S, Shi Y, Xu J, Gan J, Zhang J. miR-203a is involved in HBx-induced inflammation by targeting Rap1a. Exp Cell Res. 2016;349:191–197. doi: 10.1016/j.yexcr.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: A 10-year update. Physiol Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 25.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Sakashita M, Yamada T, Fujieda S, Tanaka S, Doi S, et al. Genome-wide association study identifies eight new susceptibility loci for atopic dermatitis in the Japanese population. Nat Genet. 2012;44:1222–1226. doi: 10.1038/ng.2438. [DOI] [PubMed] [Google Scholar]
- 26.Chen CP, Lin SP, Chern SR, Tsai FJ, Wu PC, Lee CC, Chen YT, Chen WL, Wang W. A de novo 7.9 Mb deletion in 22q13.2→qter in a boy with autistic features, epilepsy, developmental delay, atopic dermatitis and abnormal immunological findings. Eur J Med Genet. 2010;53:329–332. doi: 10.1016/j.ejmg.2010.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of small RNA sequencing of plasma exosomes has been uploaded to the sequence read archive (http://www.ncbi.nlm.nih.gov/bioproject/609458; submission ID: SUB7071247; BioProject accession no. PRJNA609458). The raw data will be publicly available on March 1, 2021.