Abstract
A cysteine proteinase, papain-like proteinase (PL1pro), of the human coronavirus 229E (HCoV) regulates the expression of the replicase polyproteins, pp1a and ppa1ab, by cleavage between Gly111 and Asn112, far upstream of its own catalytic residue Cys1054. In this report, using bioinformatics tools, we predict that, unlike its distant cellular homologues, HCoV PL1pro and its coronaviral relatives have a poorly conserved Zn2+ finger connecting the left and right hand domains of a papain-like fold. Optical emission spectrometry has been used to confirm the presence of Zn2+ in a purified and proteolytically active form of the HCoV PL1pro fused with theEscherichia coli maltose-binding protein. In denaturation/renaturation experiments using the recombinant protein, its activity was shown to be strongly dependent upon Zn2+, which could be partly substituted by Co2+ during renaturation. The reconstituted, Zn2+-containing PL1pro was not sensitive to 1,10-phenanthroline, and the Zn2+-depleted protein was not reactivated by adding Zn2+ after renaturation. Consistent with the proposed essential structural role of Zn2+, PL1pro was selectively inactivated by mutations in the Zn2+ finger, including replacements of any of four conserved Cys residues predicted to co-ordinate Zn2+. The unique domain organization of HCoV PL1pro provides a potential framework for regulatory processes and may be indicative of a nonproteolytic activity of this enzyme.
Proteolytic enzymes control a large variety of processes in cellular organisms and viruses. Some proteases can completely digest most proteins, while others are highly selective, cleaving only one bond in a protein or a set of proteins (1). Mechanistically, four classes of proteases have been recognized, and they are named in accordance with the chemical nature of their catalytic site, namely cysteine proteases, serine proteases, aspartate proteases, and metalloproteases (Zn2+) (2). Each protease class can be further divided into protein families that are united by a common origin and a common structural fold (3). Recently, however, it has become evident that more complex relationships exist among proteases. Thus, proteolytic enzymes employing different catalytic mechanisms within the framework of the same structural fold have been identified (showing that protease families can extend across the borders separating classes (4, 5, 6)) and proteases that are decorated with determinants of nonproteolytic activity, for example, polynucleotide-binding cysteine proteases (7, 8, 9) and Zn2+-binding nonmetalloproteases (see below) have also been described. The proteases that exhibit these unusual features are very often encoded by positive sense RNA viruses. Many of these viruses rely upon proteolytic enzymes to regulate viral and cellular gene expression and virion morphogenesis (10, 11, 12).
The majority of RNA viral proteinases are recognized as being very distantly related to either cellular chymotrypsin-like or papain-like proteases (CLpro and PLpro, respectively).1 RNA viral CLpro enzymes have been found that exhibit three different catalytic triads, the canonical Ser-His-Asp and two variations not found in cellular proteins, Cys-His-Asp and Cys-His-Glu (5, 13, 14, 15, 16). Some RNA viral CLpro enzymes, with the canonical or Cys-His-Asp catalytic triads (e.g. the hepatitis C virus NS3 and picornavirus 2A proteases, respectively) employ a structural Zn2+ bound by two loops of the two β-barrel fold (17, 18, 19, 20, 21, 22). Until very recently, the relationship between cellular and RNA viral PLpros had been primarily based on the results of sequence (23, 24, 25, 26) and predicted secondary structure (27) analyses. A recent x-ray analysis of the foot-and-mouth virus (FMDV) Lpro (leader protease), one of the most well characterized viral PLpros (28), has proven the distant relationship between viral and cellular PLpros by showing that the FMDV Lpro adopts a compact version of the PLpro fold (29). Like its cellular homologues, the FMDV Lpro employs the catalytic Cys-His dyad residues, which are assisted by unique Asn and Asp residues replacing, respectively, Gln forming the oxyanion hole as well as Asn hydrogen-bonded to the catalytic His in cellular enzymes. Most probably, similar or other replacements could be found in the catalytic center of the other RNA viral PLpros (26) (see also below). It is reasonable to think that the structural diversity of RNA viral proteinases is related to the extremely high mutation rate of RNA viruses (30). As a result, many RNA viral proteinases have diverged to the point where sequence similarity between homologues can barely be detected.
One of the groups of RNA viral PLpro enzymes includes seven ortologous and paralogous coronaviral proteinases. Three coronaviruses (murine hepatitis virus (MHV) (31, 32), human coronavirus 229E (HCoV) (33), and porcine transmissible gastroenteritis virus (TGEV) (34)) encode two divergent copies of PLpro (PL1pro and PL2pro), while avian infectious bronchitis virus (IBV) (23, 26, 35) encodes only one PLpro. These enzymes are produced as part of large replicative precursor proteins (pp1a and pp1ab), with pp1ab being expressed by a mechanism involving (−1) ribosomal frameshifting (36). The PL1pros of MHV (37, 38, 39, 40) and HCoV (41) and the PLpro of IBV (42) cleave one or two sites upstream of the protease domain. HCoV PL1pro, which is the focus of the work reported here, was previously mapped between the Gly861 and Gln1285 residues of pp1a/pp1ab. This protease releases an N-terminal protein, p9, from the pp1a/pp1ab precursors via the cleavage of the Gly111–Asn112 bond. The conserved residues Cys1054 and His1205, which were predicted to be the catalytic dyad, were found to be indispensable for proteolytic activity of the HCoV PL1pro (41). In this paper, we predict and provide experimental evidence to show that coronaviral PLpros comprise a unique group of enzymes that rely upon a structural Zn2+ within the framework of a papain-like fold.
EXPERIMENTAL PROCEDURES
Computer Sequence and Structure Analyses
Amino acid sequences were derived from Swiss-Prot and GenbankTM data bases, and protein structures were derived from the Brookhaven data base (Protein Data Bank, Brookhaven National Laboratory, Upton, NY). Sequence alignments were produced using a family of Clustal programs (43, 44) and the Macaw workbench (45). Nonredundant sequence data bases were searched with single sequences (46), blocks (47), and Hidden Markov Models trained on multiple sequence alignments (48). These alignments were also sent as input for the PhD (49) or DSC (50) programs to predict secondary structure. The 123D program (51) was used for the fold prediction by threading. The structure superimposition and three-dimensional protein homology modeling (52) were done and evaluated with the help of the Quanta 97 and Whatif 4.99 (53) packages. Analysis of the structure-based alignments was assisted by the CORE package (54, 55). Ribbons 2.81 (56) was used to display protein structures, and DSSP (57) was used to calculate secondary structure.
Plasmids
Plasmid pT7-IRES-Pap, encoding amino acids (aa) 1–1315 of pp1a/pp1ab, has been described previously (41). DNA was amplified from pT7-IRES-Pap by standard polymerase chain reaction procedures using oligonucleotides I (5′-ACTGCCATGGGTGGTATTTTGGCAGTAATA-3′) and II (5′-TTATCACTTGGTAGAAAGCTACATTGTC-3′). The polymerase chain reaction product was cloned into the XmnI site of pMal-c2 (New England Biolabs, Schwalbach, Germany), resulting in pMal-PL1, and the construction was verified by sequencing. Plasmid pMal-PL1 encodes an isopropyl-1-thio-β-d-galactopyranoside-inducible fusion protein, MBP-PL1, consisting of three parts: the Escherichia coli maltose-binding protein (MBP), a linker of three artificial aa, and the HCoV PL1pro domain consisting of aa 861–1285 of pp1a/pp1ab (the so-called “minimal” trans-active protease) (41).
Site-directed Mutagenesis
A modified protocol of site-directed mutagenesis (58) was used. Two partially complementary oligonucleotides carrying the desired mutation were used in a polymerase chain reaction with plasmid DNA as template. The parental DNA was digested with DpnI, and the polymerase chain reaction products were used to transform E. coli Top 10F′ cells. In vivo recombined plasmid DNA was isolated from individual clones and sequenced. Then DNA fragments from the parental plasmids were replaced with appropriate DNA fragments from the mutated plasmids.
Bacterial Expression and Purification of Recombinant Proteins
Plasmids pMal-c2 and pMal-PL1 were used to transformE. coli TB1 cells. Single colonies were inoculated into LB medium supplemented with 100 μm ZnOAc and incubated at 37 °C until an A 600 of 0.5 was reached. Expression of the recombinant protein was induced with 1 mmisopropyl-1-thio-β-d-galactopyranoside. After incubation at 25 °C for 10 h, the cells were harvested and resuspended in 3 ml of buffer A (20 mm Tris-HCl, pH 7.5, 200 mm NaCl, 1 mm β-mercaptoethanol)/g of cell pellet. Cells were broken with a French pressure cell (SLM Instruments Inc.), and the lysate was clarified by centrifugation at 5000 ×g for 30 min. The clarified lysate was loaded on a column containing amylose and washed with 10 column volumes of buffer A, and MBP-containing proteins were eluted using buffer A containing 10 mm maltose. The protein concentration was determined and adjusted to 1 or 2 mg/ml. The bacterially expressed recombinant proteins were analyzed by SDS-polyacrylamide gel electrophoresis. Protein preparations were stored at −70 °C.
In Vitro Trans-cleavage Assay
The proteolytic activity ofin vitro synthesized proteins expressed from pT7-IRES-Pap and its derivatives or the activity of bacterially expressed MBP-PL1 was assayed using in vitro synthesized, [35S]Met-labeled substrate, representing aa 1–956 of pp1a/pp1ab (41). The proteolytic reaction was done at 30 °C by incubation of 7 μl of the reticulocyte lysate substrate with either 20 μl of in vitro synthesized enzyme for 3 h (41) or 1 μg (0.5 μl) of recombinant protein for 1 h. The reaction products were immunoprecipitated with the polyclonal rabbit antiserum IS1720 (specific for aa 41–250 of pp1a/pp1ab) and analyzed by 10–17.5% gradient SDS-polyacrylamide gel electrophoresis (41). Radioactively labeled polypeptides were visualized by autoradiography.
Denaturation and Renaturation of MBP-PL1
Two 1-ml aliquots (I and II) of purified MBP-PL1 (1 mg/ml) were mixed with 9 volumes of buffer A containing 8.0 m urea. After incubation at 20 °C for 4 h, aliquot I was adjusted with EDTA to a final concentration of 10 mm, and aliquot II was adjusted with ZnOAc to a final concentration of 300 μm. The aliquots were subsequently dialyzed twice against 500 volumes of buffer A containing 1 mm EDTA (aliquot I) or 100 μmZnOAc (aliquot II) at 4 °C for 16 h. The dialyzed material was then concentrated 10-fold using Centricon-10 membranes (Amicon). The EDTA-treated sample (aliquot I) was subjected to a second round of denaturation as described above and divided into three aliquots (Ia, Ib, and Ic). Aliquot Ia was adjusted with EDTA to a final concentration of 10 mm, aliquot Ib was adjusted with ZnOAc to a final concentration of 300 μm, and aliquot Ic was adjusted with CoOAc to a final concentration of 300 μm. The aliquots were dialyzed against 500 volumes of buffer A containing 1 mm EDTA (aliquot Ia), 100 μm ZnOAc (aliquot Ib), or 100 μm CoOAc (aliquot Ic).
Determination of Metal Content
The recombinant protein solutions were analyzed for the presence of Ca2+, Co2+, Cu2+, Fe2+, Mg2+, Mn2+, Ni2+, Pb2+, and Zn2+ by ICP-OES on a JY30 Plus spectrometer (Jobin Yvon, France). Prior to analysis, all samples were treated twice with Chelex-100 resin by mixing in suspension at 4 °C for 30 min.
RESULTS
Coronaviral PL Proteases Are Predicted to Have the α + β Structural Organization Found in Cellular Papain-like Proteinases
In this study, we have employed bioinformatics tools to gain insights into the structure and function of coronaviral PLpros. The predictions derived from these analyses have then been tested experimentally. Previously, this same strategy has proven very efficient in identifying and characterizing numerous RNA viral enzymes (for a review, see Ref. 59), including coronaviral PLpros (37,40, 41, 42).
First, we extended previous analyses (26, 31, 33, 34) of the primary structure of the coronavirus PL protein family using multiple sequence comparison tools (see “Experimental Procedures”). A set of multiple alignments of the conserved PL domains, consisting of aa 202–216, was generated, and an overall alignment was constructed (Fig. 1 , A and C). A number of different approaches were then used to search the data bases for similarities (see “Experimental Procedures”). Only several matches of marginal statistical significance were detected with cysteine proteases, namely between the coronavirus PLpros and the Lpro of different strains of FMDV (26) and also proteases of the ubiquitin isopeptidase T family (60). These weak similarities were structurally reasonable, since in the matches (putative) catalytic Cys or His residues were aligned (data not shown). This failure to identify a pronounced similarity between coronaviral and the other PLpros was, perhaps, not surprising given that pairwise similarity within the coronavirus PLpro family itself is very low (13–32% identical residues). Thus, to check for the existence of more remote structural similarities, which were not evident at the amino acid sequence level (61), the predicted secondary structure of the coronavirus PLpros was threaded (62, 63) through the Protein Data Bank. Even then, no high scores were returned. However, consistent with the above observations and a previous analysis (26), papain was among the three top hits (Z score was 3.16) with the catalytic cysteines of papain and the coronavirus PLpros being matched (data not shown). It is also noteworthy that coronaviral PLpros were predicted to belong to the α + β structural class, which also includes cellular PLpros. Thus, we were encouraged to elaborate our model by producing a secondary structure-based alignment of seven coronaviral and 11 cellular PLpros, whose tertiary structures have been solved. This alignment includes 10 ungapped blocks encompassing the majority of the (predicted) structural elements of the two protein sets and has interblock spacing that is similar in the proteases of both cellular and viral origin (Fig. 1 A). The interfamily amino acid conservation is most evident in blocks II, IV, VI, and VIII, which contain both catalytic and substrate-binding pocket residues of the cellular enzymes and four out of eight absolutely conserved residues of the viral enzymes (three in block II and one in block VIII). Three other invariant residues are located between blocks V and VI and described below. The eighth coronaviral invariant residue, Asp, is located in block IX. In an alignment slightly different from that shown in Fig. 1 A, this residue would correspond to a highly conserved Asn, which is hydrogen-bonded to catalytic His residue of cellular enzymes. However, this alternative alignment was not favored by the MACAW-assisted analysis or by the results of a site-directed mutagenesis study (see below). In this respect, it is worth noting that the conserved Asn of cellular PLpros does not play the catalytic role (64) and is substituted by Lys in stem bromelain, one of the plant PLpros (65). Importantly, in the alignment shown in Fig. 1 A, the coronavirus PL residues match the majority (77 out of 91) of cellular PL residues with a low space variation (the so-called core residues (55)) and only a minority (32 out of 87) of structurally less conserved residues (Fig. 1, A and B). Taken together, all of the above observations strongly imply that the coronaviral PLpros may adopt a variant of the α + β fold conserved in cellular PLpros.
Figure 1.
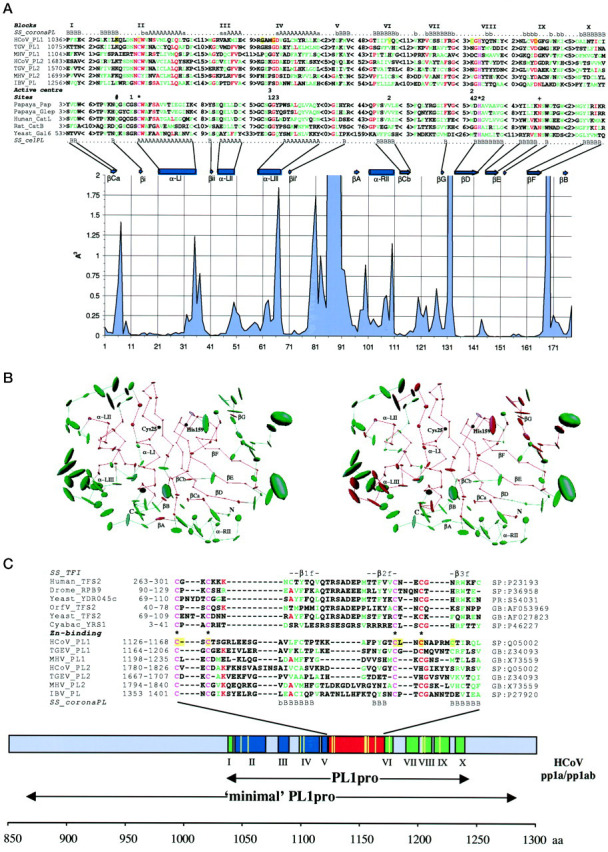
Fig. 1. Coronaviral and cellular PLpros: structural similarities and unique features.A, secondary structure-based sequence alignment of coronaviral and cellular PLpros. The primary structures of HCoV PL1pro and its coronaviral relatives (for accession numbers see C) were aligned using ClustalW program (43) in a stepwise manner and manually corrected with the ClustalX program (44) and the MACAW workbench (45). The main portion of this alignment is presented as 10 ungapped blocks. Only blocks II and III were statistically significant (p < 10−20 and p = 1.5−13, respectively), and blocks IV and VII, excluding MHV PL1pro, were conditionally significant, using a searching space between blocks III and VIII and between blocks IV and VIII, respectively. The validity of block VIII was previously confirmed by site-directed mutagenesis of conserved His for MHV and HCoV PL1pros and IBV PLpro (37, 41, 42). The secondary structures predicted by the PhD program are shown at the top (SS_coronaPL; A anda represent α-helix, and B and brepresent β-strand, predictions in capital letters have a reliability >5 and predictions inlowercase letters have a reliability of 5 and less (49)). The validity of this prediction was confirmed when similar secondary structure profiles were also returned for (i) the same alignment using the DSC program (50) and (ii) two automatically generated alignments containing either PL1pros or PL2pros encoded by HCoV and TGEV (not shown). The secondary structure profile of coronaviral PLpros was aligned with secondary structure elements conserved in the tertiary structure of 11 cellular PLpros (SS_celPL) (Protein Data Bank accession numbers: 1ppn, Papaya_Pap, papain (77);1gec, Papaya_Glep, glycyl endopeptidase (78); 1ppo, caricain (79);1yac, chymopapain (80); 1mem, cathepsin K (81); 1cjl, Human_CatL, cathepsin L (82); 1cte, Rat_Catb, cathepsin B (83); 2aim, trypanosoma cruzain (84); 2act, actinidin (85); 1gcb, yeast Gal6/bleomycin hydrolase (86)). The secondary structure alignment guided a sequence alignment of coronaviral and cellular proteases. A register of the alignment within each block was (arbitrarily) selected to maximize interfamily sequence similarity, although two or more poorly discriminated alignments were produced for all blocks except blocks II and VIII. When the three-dimensional structures of coronaviral PLpros become available, this alignment may need to be locally adjusted. For cellular PLpros, only a representative set of five sequences is shown. Coloring of the alignment of 12 sequences indicates the following:pink, invariant residues; red, residues conserved in >50% of the sequences; green, group of similar residues. The alignments of coronaviral and cellular PLpros highlight the active site residues of cellular proteases (66, 78). *, principal catalytic; +, “accessory” catalytic; 1, 2,3, and 4, substrate-binding pocket subsites S1, S2, S3, and S4, respectively; #, oxyanion hole-forming residue.Beneath the alignments, a plot displaying the positional structural variability (55) of cellular PLpros is shown.Above the plot, the positions of conserved secondary structure elements of cellular PLpros (66) as well as four conserved hydrogen-forming elements consisting of one residue (not marked) in the primary structure are displayed. Vertical axis, space variability at a position of the alignment; horizontal axis, numeration in the structural alignment containing only aligned residues. B, core structural residues of the cellular PLpros and residues conserved in cellular and coronaviral PLpros. Using the CORE package (55), a structural alignment of 11 cellular PLpros was converted into an average PL structure. It is characterized by the mean position of each C-α atom common in the family. The size of the ellipsoid around each of these atoms is proportional to the volume of atom variance. The two identical average PL structures, consisting of 178 atoms, are displayed in the “standard” papain orientation (66) featuring left-hand and right-hand domains as well as the interdomain active site cleft with the two catalytic residues of papain, Cys25 and His159. Conserved secondary structure elements of cellular PLpros are also marked. These structures are colored ingreen and red as follows. The left structure, the half of C-α atoms plus two atoms having the lowest space variance (91 atoms) are colored in red (core), and the remaining atoms are in green (noncore). The right structure, 109 atoms, whose residues were aligned with coronaviral PL residues in Fig. 1A, are shown in red (interfamily conserved residues), and the remaining atoms are in green. Note that the cellular PL core residues and the interfamily conserved residues are mainly from the same pool. C, A unique Zn2+finger connects the two domains of the PL fold of coronaviral PLpros. A region of the coronaviral PLpros between blocks V and VI was aligned as specified in Fig. 1A. Using the secondary structures predicted for the PLpros (SS_coronaPL) (50) and derived from the NMR structure (69) of the TFIIS Zn2+ ribbon (SS_TFI), an alignment of Zn2+ fingers of coronaviral PLpros and TFIIS was generated. The positions of these sequences in the corresponding proteins are given on the left, and accession numbers in the sequence data bases are shown on the right.Coloring of the alignment is as detailed for A. Residues involved in Zn2+ binding in TFIIS (69) are marked. A bar depicts the region of HCoV pp1a/pp1ab characterized in this study with the conserved blocks (Fig. 1A) shown. These blocks are organized in three groups colored differently.Blue, left-hand α-helix domain; green, right-hand β-sheet domain without counterparts of βA- and βB-strands; red, Zn2+ finger domain.Beneath the bar, the positions of the PL1pro domain, which is conserved among coronaviruses, and the HCoV minimal PL1pro domain determined by deletion analysis (41) are shown. The positions of mutations (Ref. 41 and Table II) are depicted withyellow vertical lines in thebar and yellow amino acid background in the alignments in A and C.
Coronaviral PL Proteases Contain a Unique Zn2+ Finger Connecting the Two Domains of a Papain-like Fold
In the alignment displayed in Fig. 1 A, three structural elements of the cellular PLpros, βA, βB, and α-RII, which are spatially juxtaposed in front of the substrate-binding pocket and comprise a peripheral part of the right-hand domain (Fig. 1 B), were not aligned with coronaviral sequences. Two of these elements, βA and α-RII, form part of a long, poorly conserved structure found in the middle of the cellular PLpros (66). The corresponding region between blocks V and VI in the coronaviral PLpros was highly diverged, but, most conspicuously, it was found to contain a unique conserved sequence pattern characteristic of Zn2+ fingers, CX 1–2CX 22–31CX 1–2-[CH], where X is any aa (Fig. 1 C).
When the predicted HCoV PL1pro Zn2+ finger was compared with the structural data base of Zn2+ fingers (67, 68), the nucleic acid binding domain of human transcriptional elongation factor TFIIS, which has a three-stranded, antiparallel, β-sheet fold (zinc ribbon) (69), was selected as being the most similar. Both proteins are enriched with β-strands, belong to the C4 class, have a similar spacing between the pair of the C2 halves, and have a variable spacing separating cysteines within the C2 elements (or cysteine and histidine in the CH element of related proteins) (Fig. 1 C). The zinc ribbon architecture is apparently conserved in a wide variety of Zn2+ fingers (69, 70) and was shown to tolerate a large size difference in the loop structure (71). These observations strongly suggest that the specific features of the coronaviral PLpro Zn2+ fingers, which are evident from the alignment shown in Fig. 1 C, may also be compatible with this architecture.
From the comparative sequence analyses outlined above, we predicted that coronaviral PLpros are composed of the two domains of the papain-like fold which, unlike their known cellular relatives, are connected by a Zn2+ finger.
Purified PL1pro Fused with E. coli Maltose-binding Protein Is Proteolytically Active
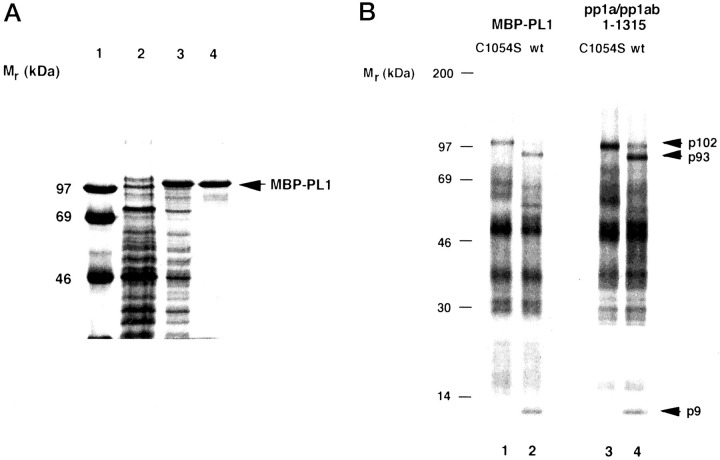
To address the enzymatic and predicted Zn2+ binding properties of coronaviral PLpros experimentally, we decided to study a recombinant, purified HCoV PL1pro that was amenable to genetic and biochemical manipulations. The minimal HCoV PL1pro domain, which had been shown to be active when synthesized in a reticulocyte lysate (41), was expressed in E. coli as the fusion protein MBP-PL1 (Fig. 2 A, lanes 2 and 3). MBP-PL1 was partly purified by amylose affinity chromatography (Fig. 2 A, lane 4) and assayed for proteolytic activity. The recombinant MBP-PL1, but not a derivative carrying a replacement of the catalytic Cys1054 by serine, was proteolytically active (Fig. 2 B,lanes 1 and 2). The proteolytic activity of recombinant MBP-PL1 was identical to that of in vitro synthesized HCoV PL1pro (Fig. 2 B,lanes 3 and 4; Ref. 41).
Figure 2.
Expression and purification of proteolytically activeHCoVPL1pro fused with the E. colimaltose-binding protein.A, purification of the fusion protein. MBP-PL1 was purified by affinity chromatography on amylose column from lysates of E. coli transformed with TB1[pMal-PL1] as described under “Experimental Procedures.” Aliquots taken from different stages of the purification were analyzed by 12.5% SDS-polyacrylamide gel electrophoresis. Lane 1, molecular mass markers; lane 2, noninduced bacterial lysate; lane 3, isopropyl-1-thio-β-d-galactopyranoside-induced bacterial lysate; lane 4, protein after amylose affinity chromatography. The position of MBP-PL1 is indicated. B, proteolytic activity of MBP-PL1. The trans-cleavage assay usingin vitro generated [35S]Met-labeled substrate was used to monitor proteolytic activity of purified MBP-PL1 (lanes 1 and 2) and in vitro generated, nonlabeled polypeptide containing PL1pro (pp1a/pp1ab-(1–1315)) and its mutated derivative (lanes 3 and 4). After immunoprecipitation of the cleavage reaction with IS 1720, proteins were separated by 10–17.5% gradient SDS-polyacrylamide gel electrophoresis, and labeled polypeptides were visualized by autoradiography. The positions of molecular mass markers, the substrate (p102), and cleavage products (p93 and p9) are indicated. The source of enzyme was as follows: MBP-PL1 C1054S (lane 1), MBP-PL1 (lane 2), in vitro produced PL1pro C1054S (lane 3), in vitro produced PL1pro (lane 4).
HCoV PL1pro Is a Zn2+-binding Enzyme
The partly purified MBP-PL1 was analyzed for eight metals by ICP-OES. No other metals except Zn2+ and Fe2+ were detected in significant amounts. The Zn2+/Fe2+ ratio was approximately 3–4.5/1 (Table I ). MBP, expressed and purified under identical conditions, bound only background amounts of the metal ions, confirming that it is the coronaviral portion of the fusion protein that binds Zn2+. It was calculated that HCoV PL1pro binds Zn2+ in equimolar amounts.
Table I.
Metal content in MBP-PL1
| Sample | Batch | Zinc | Iron |
|---|---|---|---|
| μg/ml | μg/ml | ||
| Buffer | NDa | ND | |
| MBP (2 mg/ml)b | 1 | 25 | 26 |
| 2 | ND | ND | |
| MBP-PL1 (2 mg/ml)b | 1 | 546 | 120 |
| 2 | 554 | 184 |
ND, not detectable.
Total protein concentration, using gel densitometry.
Zn2+ Is an Essential Structural Cofactor of Proteolytically Active HCoV PL1pro
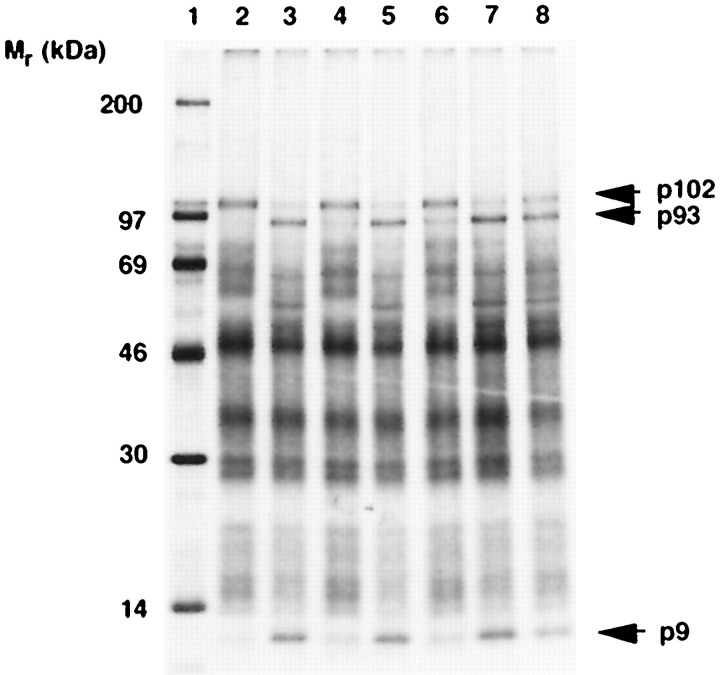
In a series of preliminary experiments, we observed that if ZnOAc was not added to the bacterial growth media, purified MBP-PL1 was proteolytically less active and there was a decreased amount of Zn2+ and increased amount of Fe2+ bound to the protein (data not shown). This observation indicated that Zn2+ might be an essential co-factor of PL1pro. To test this hypothesis, MBP-PL1 was denatured in the presence of 8 m urea and then renatured by dialysis against buffer A supplemented with either ZnOAc or EDTA (72). MBP-PL1 renatured in the presence of Zn2+, but not EDTA, was proteolytically active (Fig. 3 ,lanes 4 and 5). Furthermore, the Zn2+-depleted MBP-PL1 (apoenzyme) could be reactivated after a second round of denaturation and renaturation in the presence of ZnOAc or, to a lesser degree, CoOAc (Fig. 3, lanes 6–8). Importantly, Fe2+ could not substitute for Zn2+, the addition of 100 μm ZnOAc to the apoenzyme did not restore activity, and the reconstituted Zn2+-PL1pro was not inhibited by 10 mm1,10-phenanthroline (data not shown). These results strongly suggest that Zn2+ is tightly bound to the PL1pro and that coordination of the Zn2+ has to occur during folding of the protein. This also implies that Zn2+ plays an essential structural rather than catalytic role.
Figure 3.
Effect of Zn2+ on the proteolytic activity of MBP-PL1. The trans-cleavage assay was used to monitor proteolytic activity of MBP-PL1 subjected to one or two cycles of denaturation in the presence of 8 m urea and renaturation in the presence of EDTA, ZnOAc, or CoOAc. The substrate and cleavage products are indicated as in Fig. 2. Lane 1, molecular markers. The source of enzyme was as follows: buffer A (lane 2), MBP-PL1 (not treated) (lane 3), denatured MBP-PL1 renatured in the presence of EDTA (apoenzyme) (lane 4), denatured MBP-PL1 renatured in the presence of ZnOAc (lane 5), denatured apoenzyme MBP-PL1 renatured in the presence of EDTA (lane 6), denatured apoenzyme MBP-PL1 renatured in the presence of ZnOAc (lane 7), denatured apoenzyme MBP-PL1 renatured in the presence of CoOAc (lane 8).
Four Cysteine Residues Implicated in Binding Zn2+ Are Crucial for Proteolytic Activity of HCoV PL1pro
In our model of the HCoV PL1pro, the Zn2+ is predicted to be tetrahedrally coordinated by Cys1126, Cys1128, Cys1154, and Cys1157 (Fig. 1 C). If this is correct, and in view of the strong dependence of MBP-PL1 activity on Zn2+ (Fig. 3), these four cysteine residues should be indispensable for proteolytic activity. We tested this prediction by assaying the proteolytic activity of in vitrosynthesized PL1pro mutants. Consistent with our prediction, mutants with a replacement of any of the putative Zn2+-coordinating cysteine residues were proteolytically inactive (TableII ). Furthermore, PL1pro was found to be inactivated by two other mutations in the vicinity of the Zn2+-coordinating Cys residues, an insertion of Val between aa 1126 and 1127 or a deletion of Leu1155 (Table II and Fig. 1 A). The effect of the above mutations was highly selective, since two different mutations at Cys1163, a nonconserved residue of the Zn2+ finger, did not inactivate the enzyme (Table II). Also, six other highly conserved residues, Lys1048, Gly1099, Gly1102, Val1175, Cys1203, and Asp1218, were probed outside the Zn2+ finger. The majority of these residues were predicted to be involved in the formation of the active site, as can be seen from the alignment of the coronaviral and cellular PLpros (Fig. 1 A). Each of these residues (with the exception of Lys1048) was replaced by a number of amino acids found in the same position in other PLpro(s) or by other amino acids (Fig. 1 A and Table II). In contrast to the four conserved Cys residues of the Zn2+ finger, the majority of these residues tolerated at least one replacement. The exception, Lys1048, which was aligned with the oxyanion hole-forming Gln15 of papain, did not tolerate a Glu substitution, the only tested mutation. These results are fully compatible with our alignment of coronaviral and cellular PLpros (Fig. 1) and indicate that the four conserved Cys residues of the Zn2+ finger are as equally important for the proteolytic activity of the HCoV PL1pro as the previously identified catalytic Cys1054 and His1205residues (41).
Table II.
Proteolytic activity of HCoV PL1pro mutants
| Region of HCoV PL1pro | Replacements in pp1a/pp1ab (from → to)a | PLpro whose sequence at this position matches the HCoV PL1 mutant | Proteolytic activityb | Reference |
|---|---|---|---|---|
| Zinc finger | C1126D, H | No | This work | |
| V1126/27ins | HCoV PL2 | No | This work | |
| C1128A, D, P | No | This work | ||
| C1154A, H, D | No | This work | ||
| L1155del | MHV PL1/HCoV PL2 | No | This work | |
| C1157A, D, P | No | This work | ||
| H | HCoV PL2/TGV PL2 | No | ||
| C1163A | IBV PL | Yes | This work | |
| D | Yes | |||
| Outside of zinc finger | K1048E | IBV PL | No | This work |
| C1054S, A, G | No | 41 | ||
| G1099P | MHV PL1 | Yes | This work | |
| A | No | |||
| G1102S | Yes | This work | ||
| A | MHV PL2/IBV PL | Yes | ||
| V1175H | MHV PL1 | No | This work | |
| P | No | |||
| N | Yes | |||
| T | TGV PL2 | Yes | ||
| C1203D | MHV PL1 | Yes | This work | |
| A | No | |||
| H1205A, G | No | 41 | ||
| D1218N, Q, E, | Yes | This work | ||
| H, K, A | Yes |
V1126/27ins, insertion of Val between aa 1126 and 1127; L1155del, deletion of L1155.
Proteolytic activity was monitored using SDS-polyacrylamide gel electrophoresis of the products of reactions catalyzed by anin vitro produced PL1pro or its derivatives in the trans-cleavage and monomolecular assays (41).
DISCUSSION
In this report, we describe a unique cysteine proteinase, dependent on Zn2+ and encoded by an RNA virus. A combination of bioinformatics, biochemical, and molecular-genetic techniques has been used to obtain evidence that strongly supports a structural, rather than a catalytic, role of Zn2+ in the HCoV PL1pro. To characterize the HCoV PL1pro, we have expressed the enzyme in E. coli as a fusion protein that was shown to be proteolytically active in a trans-cleavage assay. The ability of PL1pro to cleave a cognate substrate seems to be completely dependent on additional (viral) cofactor(s), since we have been unable to develop a peptide-based assay for this protease to date. These putative cofactor(s) may reside in the approximately 900-aa region that separates the catalytic domain of PL1pro and its cleavage site. This region is known to modulate PL1pro-mediated processing in MHV (37,73).
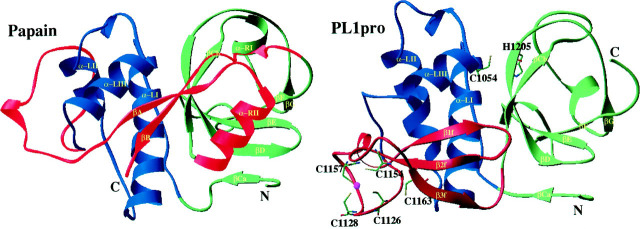
The most intriguing aspect of the HCoV PL1pro structure is, without doubt, the zinc finger. We have, first of all, delineated this structure on theoretical grounds and subsequently used a recombinant PL1pro to characterize the Zn2+ binding properties by spectroscopic analysis as well as the structure-function relationships by mutational analysis. The experimental results have validated the functional importance of the HCoV PL1pro Zn2+ finger, although we acknowledge that it remains to be shown directly that its four conserved Cys residues bind Zn2+. In the first analysis, the Zn2+ finger was predicted to adopt a β-sheet topology, most probably of the zinc ribbon type, and be incorporated into a papain-like fold. As an extension of this analysis, a possible tertiary organization of PL1pro is presented in Fig. 4 as a ribbon model that was built by homology modeling (52) using the alignments shown in Fig. 1. We believe that this model is useful for rationalizing the results presented here (see below) and for the development of more precise mutagenesis analyses. At the same time, we recognize that the model most probably deviates significantly from the coordinates of the actual structure. This lack of robustness is due to the fact that none of the current methods of homology modeling (52) can rigorously predict the structure of proteins, when modeling is based upon two separate templates of a marginal similarity, as was the case for PL1pro. Despite these reservations about the model, the compatibility of the Zn2+ribbon and the PL fold is supported by two observations: (i) the distance between the N and C termini of the Zn2+ ribbon fits between the two “acceptor” residues of the core PL domains in papain and (ii) the Zn2+ ribbon spatially replaces the C-terminal βB-strand in papain, and, accordingly, a counterpart of this strand is not conserved in the coronaviral PLpros (Fig. 1 A). The interdomain junction role of the Zn2+finger is also compatible with the observation that many RNA viral PLpros, although not all (26, 74), have considerably smaller sizes than coronaviral PLpros and may contain only a short interdomain loop in place of the Zn2+finger.2 A unique β-sheet topology of the interdomain junction has most recently been also described for the FMDV Lpro, the only RNA viral PLpro whose three-dimensional structure was solved (29).
Figure 4.
A crude structural model ofHCoVPL1pro.The PL1pro model (right; pp1a/1ab residues 1033–1242) was generated using the structures of papain (1ppn; shown left) and human TFIIS (1tfi) as templates for building the core and a loop library for constructing the variable regions by using Quanta 97 and Whatif 4.99. The model was partly refined but not minimized. Secondary structure was calculated according to Ref. 57 and, for some positions in PL1pro, from the predicted secondary structure. The structures of both proteases are displayed (56) in the standard papain orientation (66) and split into three domains. These are coloredaccording to a scheme given in Fig. 1C as follows.Blue, left-hand α-helix domain; green, right-hand β-sheet domain without counterparts of βA- and βB-strands; red, Zn2+ finger domain in PL1pro and the interdomain loop along with βA- and βB-strands and α-RII-helix in papain. Cysteine residues of the Zn2+finger as well as the catalytic dyad residues of PL1pro that have been probed by site-directed mutagenesis (Table II) are shown in theball-and-stick model.
The Zn2+ finger occupies one of the most pronouncedly diverged regions of coronaviral PLpros and seems to form a separate domain (Figs. 1, A and C, and Fig. 4). Surprisingly, the proteolytic activity of HCoV PL1pro was abolished by any of three Zn2+ finger mutations that mimicked the wild-type sequences of HCoV PL1pro relatives (V1126/27ins, L1155del, and C1157H in Table II). To reconcile these findings, one must propose that, in the evolution of the HCoV PL1pro homologues, such mutations have been suppressed by other replacements in the Zn2+finger or the catalytic domains. If the changes were limited to the Zn2+ finger, this would imply that the Zn2+finger is either involved in substrate binding or controls movement of the catalytic domains. Both options seem possible given the interdomain position of the Zn2+ finger and its likely proximity to the active site (Fig. 4). If the catalytic domains accepted mutations as well, this would be indicative of an interaction between these domains and the Zn2+ finger. Again, the current model does not preclude an interaction between the Zn2+ finger and α-LI helix, which is capped by the catalytic Cys1054 (Fig. 4). It is conceivable that the PL1pro Zn2+ finger interacts with both the catalytic domains and substrate.
HCoV PL1pro and, by implication, the other coronaviral PLpros extend the list of nonmetal proteinases dependent on Zn2+ and encoded by RNA viruses. Recently, a Zn2+ has been identified as a structural component in the picornavirus 2A and hepatitis C virus NS3 proteases (17, 18, 19, 20, 21, 22). In these enzymes, Zn2+ is uniquely coordinated by residues that have been naturally engineered in two loops of the CLpro fold. In both the chymotrypsin-like or papain-like Zn2+-binding viral proteases, Zn2+ is located at the site opposite to the active center. The recurrent emergence of Zn2+ dependence in the two independent lineages of RNA viral nonmetal proteinases could be linked to their biology (12). For example, in the coronaviral PLpros, the prominent position of the Zn2+ finger makes it an excellent candidate for mediating external signals that might modulate proteolytic activity inside cells. Through the Zn2+ finger, the PLpros might interact with proteins or polynucleotides and, in turn, affect processes regulated by the PLpro partner (75).
Among the positive sense RNA viruses, coronaviruses have the largest genomes (close to 30 kilobases) and probably use the most sophisticated mechanisms of RNA replication and transcription (76). As is the case for poliovirus (7), coronaviral PLpros might turn out to be important regulators at these levels. It may not be entirely coincidental that the Zn2+ finger used as the template in our HCoV PL1pro model employs a variant of the general architecture motif of RNA polymerases (70). The lack of an infectious DNA clone for any coronavirus remains a major technical obstacle in our attempts to elucidate the possible roles of PLpros in the viral replicative cycle. However, regardless of the role(s) played by PLpros, the integrity of its unusual domain organization could be targeted by specific drugs for the control of coronaviral infections.
Acknowledgments
We thank B. Schelle for excellent technical assistance, P. Schramel for ICP-OES analyses, M. Gerstein and R. Altman for help with the CORE package, K. Miaskiewicz for the administration of computer resources, and S. Burt and J. Maizel for commenting on the manuscript.
Footnotes
The abbreviations used are: CLpro, chymotrypsin-like protease; PLpro, papain-like proteinase; aa, amino acid(s); ICP-OES, inductively coupled plasma optical emission spectrometry; MBP, maltose-binding protein; FMDV, foot-and-mouth disease virus; HCoV, human coronavirus 229E; MHV, murine hepatitis virus; TGEV, porcine transmissible gastroenteritis virus; IBV, avian infectious bronchitis coronavirus; CoOAc, cobalt acetate; ZnOAc, zinc acetate.
A. E. Gorbalenya, unpublished observations.
REFERENCES
- 1.Neurath H. Science. 1984;224:350–357. doi: 10.1126/science.6369538. [DOI] [PubMed] [Google Scholar]
- 2.Hartley B.S. Annu. Rev. Biochem. 1960;29:45–72. doi: 10.1146/annurev.bi.29.070160.000401. [DOI] [PubMed] [Google Scholar]
- 3.Rawlings N.D., Barrett A.J. Biochem. J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbalenya A.E., Blinov V.M., Donchenko A.P. FEBS Lett. 1986;194:253–257. doi: 10.1016/0014-5793(86)80095-3. [DOI] [PubMed] [Google Scholar]
- 5.Bazan J.F., Fletterick R.J. Proc. Natl. Acad. Sci. U. S. A. 1988;85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ollis D.L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S.M., Harel M., Remington S.J., Silman I., Schrag J., Sussman J.L., Verschueren K.H.G., Goldman A. Prot. Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 7.Andino R., Rieckhof G.E., Baltimore D. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-j. [DOI] [PubMed] [Google Scholar]
- 8.Xu H.E., Johnson S.A. J. Biol. Chem. 1994;269:21177–21183. [PubMed] [Google Scholar]
- 9.Mangel W.F., Toledo D.L., Ding J., Sweet R.M., McGrath W.J. Trends Biochem. Sci. 1997;22:393–398. doi: 10.1016/s0968-0004(97)01123-7. [DOI] [PubMed] [Google Scholar]
- 10.Kräusslich H.G., Wimmer E. Annu. Rev. Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty W.G., Semler B.L. Microbiol. Rev. 1993;57:781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorbalenya A.E., Snijder E.J. Perspect. Drug Discov. Des. 1996;6:64–86. doi: 10.1007/BF02174046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya A.E., Donchenko A.P., Blinov V.M., Koonin E.V. FEBS Lett. 1989;243:103–114. doi: 10.1016/0014-5793(89)80109-7. [DOI] [PubMed] [Google Scholar]
- 14.Choi H.-K., Tong L., Minor W., Dumas P., Boege U., Rossmann M.G., Wengler G. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 15.Allaire M., Chernaia M.M., Malcolm B.A., James M.N.G. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 16.Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Sisson W., Villafranca J.E., Janson C.A., McElroy H.E., Gribskov C.L., Worland S. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 17.Yu S.F., Lloyd R.E. Virology. 1992;186:725–735. doi: 10.1016/0042-6822(92)90039-r. [DOI] [PubMed] [Google Scholar]
- 18.De Francesco R., Urbani A., Nardi M.C., Tomei L., Steinkuhler C., Tramontano A. Biochemistry. 1996;35:13282–13287. doi: 10.1021/bi9616458. [DOI] [PubMed] [Google Scholar]
- 19.Love R.A., Parge H.E., Wickersham J.A., Hostomsky Z., Habuka N., Moomaw E.W., Adachi T., Hostomska Z. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 20.Kim J.L., Morgenstern K.A., Lin C., Fox T., Dwyer M.D., Landro J.A., Chambers S.P., Markland W., Lepre C.A., O'Malley E.T., Harbeson S.L., Rice C.M., Murcko M.A., Caron P.R., Thomson J.A. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 21.Sommergruber W., Seipelt J., Fessl F., Skern T., Liebig H.D., Casari G. Virology. 1997;234:203–214. doi: 10.1006/viro.1997.8595. [DOI] [PubMed] [Google Scholar]
- 22.Stempniak M., Hostomska Z., Nodes B.R., Hostomsky Z. J. Virol. 1997;71:2881–2886. doi: 10.1128/jvi.71.4.2881-2886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya A.E., Koonin E.V., Donchenko A.P., Blinov V.M. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy W.R., Strauss J.H. J. Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh C.-S., Carrington J.C. Virology. 1989;173:692–699. doi: 10.1016/0042-6822(89)90582-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorbalenya A.E., Koonin E.V., Lai M.M.C. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skern T., Fita I., Guarne A. J. Gen. Virol. 1998;79:301–307. doi: 10.1099/0022-1317-79-2-301. [DOI] [PubMed] [Google Scholar]
- 28.Ryan M.D., Flint M. J. Gen. Virol. 1997;78:699–723. doi: 10.1099/0022-1317-78-4-699. [DOI] [PubMed] [Google Scholar]
- 29.Guarne A., Tormo J., Kirchweger R., Pfistermueller D., Fita I., Skern T. EMBO J. 1998;17:7469–7479. doi: 10.1093/emboj/17.24.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holland J., Domingo E. Virus Genes. 1998;16:13–21. doi: 10.1023/a:1007989407305. [DOI] [PubMed] [Google Scholar]
- 31.Lee H.J., Shieh C.K., Gorbalenya A.E., Koonin E.V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M.M. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonilla P.J., Gorbalenya A.E., Weiss S.R. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herold J., Raabe T., Schelle-Prinz B., Siddell S.G. Virology. 1993;195:680–691. doi: 10.1006/viro.1993.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eleouet J.F., Rasschaert D., Lambert P., Levy L., Vende P., Laude H. Virology. 1995;206:817–822. doi: 10.1006/viro.1995.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boursnell M.E.G., Brown T.D.K., Foulds I.J., Green P.F., Tomley F.M., Binns M.M. J. Gen. Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 36.Brierley I. J. Gen. Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 37.Baker S.C., Yokomori K., Dong S., Carlisle R., Gorbalenya A.E., Koonin E.V., Lai M.M. J. Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong S., Baker S.C. Virology. 1994;204:541–549. doi: 10.1006/viro.1994.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes S.A., Bonilla P.J., Weiss S.R. J. Virol. 1995;69:809–813. doi: 10.1128/jvi.69.2.809-813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonilla P.J., Hughes S.A., Weiss S.R. J. Virol. 1997;71:900–909. doi: 10.1128/jvi.71.2.900-909.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herold J., Gorbalenya A.E., Thiel V., Schelle B., Siddell S.G. J. Virol. 1998;72:910–918. doi: 10.1128/jvi.72.2.910-918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim K.P., Liu D.X. Virology. 1998;245:303–312. doi: 10.1006/viro.1998.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson J.D., Higgins D.G., Gibson T.J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuler G.D., Altschul S.F., Lipman D.J. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 46.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henikoff S., Pietrokovski S., Henikoff J.G. Nucleic Acids Res. 1998;26:309–312. doi: 10.1093/nar/26.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eddy S.R. Curr. Opin. Struct. Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 49.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 50.King R.D., Saqi M., Sayle R., Sternberg M.J. Comput. Appl. Biosci. 1997;13:473–474. doi: 10.1093/bioinformatics/13.4.473. [DOI] [PubMed] [Google Scholar]
- 51.Alexandrov N.N., Nussinov R., Zimmer R.M. In: Pacific Symposium on Biocomputing, 1996. Hunter L., Klein T.E., editors. World Scientific; Singapore: 1996. pp. 53–72. [PubMed] [Google Scholar]
- 52.Sanchez R., Sali A. Curr. Opin. Struct. Biol. 1997;7:206–214. doi: 10.1016/s0959-440x(97)80027-9. [DOI] [PubMed] [Google Scholar]
- 53.Vriend G., Sander C. J. Appl. Crystallogr. 1993;26:47–60. [Google Scholar]
- 54.Altman R.B., Hughes C., Gerstein M.B. J. Mol. Graph. 1995;13:142–152. doi: 10.1016/0263-7855(95)00002-n. [DOI] [PubMed] [Google Scholar]
- 55.Gerstein M., Altman R.B. Comput. Appl. Biosci. 1995;11:633–644. doi: 10.1093/bioinformatics/11.6.633. [DOI] [PubMed] [Google Scholar]
- 56.Carson M. Methods Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 57.Kabsch W., Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 58.Herold J., Siddell S., Ziebuhr J. Methods Enzymol. 1996;275:68–89. doi: 10.1016/S0076-6879(96)75007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorbalenya A.E., Koonin E.V. Sov. Sci. Rev. D. Physicochem. Biol. 1993;11:1–84. [Google Scholar]
- 60.Wilkinson K.D. Annu. Rev. Nutr. 1995;15:161–189. doi: 10.1146/annurev.nu.15.070195.001113. [DOI] [PubMed] [Google Scholar]
- 61.Aurora R., Rose G.D. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2818–2823. doi: 10.1073/pnas.95.6.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexandrov N.N., Fischer D. Proteins. 1996;25:354–365. doi: 10.1002/(SICI)1097-0134(199607)25:3<354::AID-PROT7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 63.Bowie J.U., Luthy R., Eisenberg D. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 64.Vernet T., Tessier D.C., Chatellier J., Plouffe C., Lee T.S., Thomas D.Y., Storer A.C., Menard R. J. Biol. Chem. 1995;270:16645–16652. doi: 10.1074/jbc.270.28.16645. [DOI] [PubMed] [Google Scholar]
- 65.Ritonja A., Rowan A.D., Buttle D.J., Rawlings N.D., Turk V., Barrett A.J. FEBS Lett. 1989;247:419–424. doi: 10.1016/0014-5793(89)81383-3. [DOI] [PubMed] [Google Scholar]
- 66.Kamphuis I.G., Drenth J., Baker E.N. J. Mol. Biol. 1985;182:317–329. doi: 10.1016/0022-2836(85)90348-1. [DOI] [PubMed] [Google Scholar]
- 67.Schwabe J.W., Klug A. Nat. Struct. Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- 68.Hubbard T.J.P., Murzin A.G., Brenner S.E., Chothia C. Nucleic Acids Res. 1997;25:236–239. doi: 10.1093/nar/25.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian X., Gozani S.N., Yoon H., Jeon C.J., Agarwal K., Weiss M.A. Biochemistry. 1993;32:9944–9959. doi: 10.1021/bi00089a010. [DOI] [PubMed] [Google Scholar]
- 70.Wang B., Jones D.N., Kaine B.P., Weiss M.A. Structure. 1998;6:555–569. doi: 10.1016/s0969-2126(98)00058-6. [DOI] [PubMed] [Google Scholar]
- 71.Zhu W., Zeng Q., Colangelo C.M., Lewis L.M., Summers M.F., Scott R.A. Nat. Struct. Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]
- 72.Sommergruber W., Casari G., Fessl F., Seipelt J., Skern T. Virology. 1994;204:815–818. doi: 10.1006/viro.1994.1599. [DOI] [PubMed] [Google Scholar]
- 73.Bonilla P.J., Hughes S.A., Pinon J.D., Weiss S.R. Virology. 1995;209:489–497. doi: 10.1006/viro.1995.1281. [DOI] [PubMed] [Google Scholar]
- 74.Liu X., Ropp S.L., Jackson R.J., Frey T.K. J. Virol. 1998;72:4463–4466. doi: 10.1128/jvi.72.5.4463-4466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berg J.M., Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 76.Lai M.M.C., Cavanagh D. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harris G., Pickersgill R., Howlin B., Moss D. Acta Crystallogr. B. 1992;48:67–75. doi: 10.1107/s0108768191006663. [DOI] [PubMed] [Google Scholar]
- 78.O'Hara B.P., Hemmings A.M., Buttle D.J., Pearl L.H. Biochemistry. 1995;34:13190–13195. doi: 10.1021/bi00040a034. [DOI] [PubMed] [Google Scholar]
- 79.Pickersgill R.W., Rizkallah P., Harris G.W., Goodenough P.W. Acta Crystallogr. Sec. B. 1991;47:766–771. [Google Scholar]
- 80.Maes D., Bouckaert J., Poortmans F., Wyns L., Looze Y. Biochemistry. 1996;35:16292–16298. doi: 10.1021/bi961491w. [DOI] [PubMed] [Google Scholar]
- 81.McGrath M.E., Klaus J.L., Barnes M.G., Bromme D. Nat. Struct. Biol. 1997;4:105–109. doi: 10.1038/nsb0297-105. [DOI] [PubMed] [Google Scholar]
- 82.Coulombe R., Grochulski P., Sivaraman J., Menard R., Mort J.S., Cygler M. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 83.Jia Z., Hasnain S., Hirama T., Lee X., Mort J.S., To R., Huber C.P. J. Biol. Chem. 1995;270:5527–5533. doi: 10.1074/jbc.270.10.5527. [DOI] [PubMed] [Google Scholar]
- 84.McGrath M.E., Eakin A.E., Engel J.C., McKerrow J.H., Craik C.S., Fletterick R.J. J. Mol. Biol. 1995;247:251–259. doi: 10.1006/jmbi.1994.0137. [DOI] [PubMed] [Google Scholar]
- 85.Baker E.N. J. Mol. Biol. 1980;141:441–484. doi: 10.1016/0022-2836(80)90255-7. [DOI] [PubMed] [Google Scholar]
- 86.Joshua-Tor L., Xu H.E., Johnston S.A., Rees D.C. Science. 1995;269:945–950. doi: 10.1126/science.7638617. [DOI] [PubMed] [Google Scholar]