Abstract
The invasion of glioblastoma is a complex process based on the interactions of tumor cells and the extracellular matrix (ECM). Tumors that are engineered using biomaterials are more physiologically relevant than a two-dimensional cell culture system. Matrix metalloproteinases (MMPs) and the plasminogen activator generated by tumor cells regulate a tumor’s invasive behavior. In this study, microtumors were fabricated by encapsulating U87 glioma cells in type I collagen and then glioma cell migration in the collagen hydrogels was investigated. Crosslinking of collagen with 8S-StarPEG increased the hydrogel viscosity and reduced the tumor cell migration speed in the hydrogels. The higher migration speed corresponded to the increased gene expression of MMP-2, MMP-9, urokinase plasminogen activator (uPA), and tissue plasminogen activator (tPA) in glioma cells grown in non-crosslinked collagen hydrogels. Inhibitors of these molecules hindered U87 and A172 cell migration in collagen hydrogels. Aprotinin and tranexamic acid did not inhibit U87 and A172 migration on the culture dish. This study demonstrated the differential effect of pharmacologic molecules on tumor cell motility in either a two-dimensional (2D) or three-dimensional (3D) culture environment.
Keywords: Glioblastoma, migration, collagen, hydrogel, inhibition
Introduction
Glioblastoma is the most common type of brain tumor, and often causes poor prognosis and a high mortality rate. The malignancy of glioblastoma is mainly the result of the highly invasive behavior of glioma cells. Although surgery is the major therapeutic method, a complete resection is difficult since the tumor is usually infiltrative (Cohen and Colman, 2015; Jhaveri et al., 2016; Ranger et al., 2009; Sherman et al., 2011). Glioblastoma research has typically been based on tumor cells on cell culture dishes. Multicellular tumor spheres have been generated to mimic in vivo tumors because the two-dimensional (2D) cell culture method does not emulate the microenvironment of tumor tissue in vivo. However, the tumor sphere generated by the hanging drop method or centrifugation cannot efficiently create microtumors with extracellular matrix (ECM) components. The invasion of glioblastoma is a complex process based on the interactions between tumor cells and the extracellular matrix (Huijbers et al., 2010; Ma et al., 2012; Veeravalli and Rao, 2012). Biomaterial scaffolds now show promise in forming three-dimensional (3D) cancer models for in vitro and in vivo studies. Tumors engineered using biomaterials have shown that they are more physiologically relevant than the traditional 2D cell culture system (DelNero et al., 2013; Jang et al., 2017; Long et al., 2014).
In the normal brain, the ECM contains high levels of glycosaminoglycan (GAG) hyaluronan, and proteoglycans (PGs), and low levels of fibrous proteins such as collagens, fibronectin, and laminin (Ruoslahti, 1996; Zimmermann and Dours-Zimmermann, 2008). Fibrous proteins are the major components of basement membranes of the brain’s vasculature system (Berczi and Szentiványi, 2009). The aggressive glioblastoma alters the ECM microenvironment by generating collagens (I and IV), fibronectin, and laminin (Mahesparan et al., 2003). The increased generation of collagen by the glioblastoma can modify the mechanical properties of the ECM, thereby modulating the motility of the tumor cell. Previous studies have demonstrated that glioma cells have responded to the stiffness of the ECM and changed the cell motility. In a 2D cell culture, stiffness of the polymeric substrate affected the morphology and motility of the glioma cell (Ulrich et al., 2009). A 3D cell culture model provides a more relevant microenvironment than the 2D cell culture system in order to study the physical behavior of tumor cells. Collagen, a type of natural biomaterial, is one component of the glioblastoma ECM. A 3D matrix generated by collagen hydrogel can mimic human tissue in a tumor-invasion study.
A number of signaling pathways facilitate the invasion of a glioblastoma. Matrix metalloproteinases (MMPs) and a plasminogen activator generated by the tumor cells are involved in the progression and invasive behavior of tumors (Chen et al., 2013; Guan et al., 2015; Lei et al., 2015; Musumeci et al., 2015; Song et al., 2009; Tabouret et al., 2013; Wang et al., 2015; Wang et al., 2014b). MMPs are proteolytic endopeptidases responsible for cancer progression. MMP-2 and MMP-9 are highly expressed in glioblastomas in comparison with normal brain tissue. The elevation of MMP-2 and MMP-9 levels in tumor cells is correlated with an increased grade of glioblastoma malignancy. Previous studies showed that MMP-2 and MMP-9 regulate cellular proliferation, motility, invasion, and angiogenesis of glioblastomas (Lei et al., 2015; Musumeci et al., 2015; Rao, 2003; Song et al., 2009; Tabouret et al., 2013). Binding of the plasminogen activator and its receptor activates the proteolytic cascades that breakdown the ECM and result in tumor cell migration and invasion (Blasi and Carmeliet, 2002; Rao, 2003). A previous study showed that the downregulation of urokinase plasminogen activator (uPA) in cultured glioma cells inhibited the PI3k/Akt signaling pathway, thereby inhibiting cell migration (Chandrasekar et al., 2003). An in vitro 3D model based on collagen hydrogels that can mimic the in vivo environment provides an effective microenvironment to facilitate the analysis of the complex process of tumor cell interaction with the ECM. The role of the plasminogen activator in determining glioma cell migration in 3D matrices is not yet clear. Direct comparison of the effect of MMPs and plasminogen activator inhibitors (PAIs) on glioma cell migration in 2D and 3D environment has not been reported.
Previous studies have investigated glioma cell migration in collagen matrices and shown that the change in matrix stiffness altered tumor cell motility (Kaufman et al., 2005; Ulrich et al., 2010). However, these studies are not sufficient to fully interpret the effect of the mechanical properties of the ECM on glioma cell migration. The viscosity of collagen hydrogel can be altered by crosslinking, and the response of tumor cells to different mechanical properties when they are grown in the crosslinked collagen hydrogels can be studied. In this work, we investigated glioma cell migration in crosslinked collagen hydrogels to provide insight into the effect of ECM viscosity on cell motility. We also applied MMP-2 and MMP-9 inhibitors as well as plasminogen activators to glioma cells cultured on both a 2D culture dish and a 3D collagen hydrogel to study their regulatory effect on tumor cell migration.
Materials and Methods
Generation of microtumors and tumor cell culture
To generate microtumors, U87 glioma cells (50,000) were mixed with a collagen solution (3 mg/ml, 100 µl) or an 8arm PEG Succinimidyl Glutarate (hexaglycerol) (8S-StarPEG, JenKem Technology, Plano, TX)-crosslinked collagen solution (100 µl). The final concentration of 8S-StarPEG was 0.1 mM. This mixture was transferred into a syringe with a needle (25G1) and directly injected into the cell culture medium on a cell culture plate using a syringe pump (flow rate, 0.1 µl/min) (Berndt et al., 2016). Then the cells were cultured in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (100 IU/ml penicillin and streptomycin) at 37°C with 5% CO2. After fabrication, the microsphere size was analyzed and quantified based on images taken at various times (6 hours, day 1, day 3, day 5, and day 7).
Tumor cell migration from microtumors in collagen hydrogels
After the microtumors were cultured in medium for seven days, they were transferred in collagen hydrogel (3 mg/ml) or 8S-StarPEG-crosslinked collagen hydrogel (3 mg/ml). The final concentration of the 8S-StarPEG in collagen hydrogel was 1 mM. The collagen hydrogel was generated as reported in our previous research (Seyedhassantehrani et al., 2016). The distance of cell migration from the microtumors was measured and quantified.
Cell viability and proliferation assays of microtumors
After the microtumor cells were cultured in collagen hydrogels for seven days, their viability was analyzed using a LIVE/DEAD® cell vitality assay kit (Life Technologies Corporation, Grand Island, NY). To perform the assay, the microtumors were incubated with the phosphate-buffered saline (PBS) including EthD-1 stock solution and calcein AM stock solution for 30 minutes at room temperature. Then the cells labeled with green (indicating live cells) or red (indicating dead cells) fluorescent dye were counted under a fluorescent microscope (Axiovert 200M, Carl Zeiss, Inc.).
The proliferation of glioma cells in the microspheres was studied by monitoring their metabolic activity using AlamarBlue® assay (Pierce Biotechnology, Rockford, IL). The microtumors were incubated with a cell culture medium containing 10% (v/v) AlamarBlue® reagent for four hours at day 3 and day 7 of cell culturing. Then the solution was taken from the cell culture well for the assay of absorbance at wavelengths of 570 nm and 600 nm using a microplate reader (Synergy™ Mx Monochromator-Based Multi-Mode Microplate Reader, Winooski, VT).
Porosity of collagen hydrogel and tumor cell-mediated collagen hydrogel degradation
To study the degradation of collagen hydrogels encapsulating U87 cells, the non-crosslinked hydrogels or crosslinked hydrogels (1 mM 8S-StarPEG) were fabricated (Seyedhassantehrani et al., 2016) and incubated with 5% CO2 at 37°C overnight. U87 glioma cells (100,000 cells) were seeded in 500 µl collagen hydrogel or collagen hydrogel crosslinked with 8S-StarPEG (0.5 mM and 1 mM). To study the digestion of the hydrogels by the tumor cells, the hydrogels were freeze-dried after being generated after 6 hours, 5 days, and 7 days. The weight of the dry materials was then measured and quantified.
Rheological testing
Rheological testing was conducted to check the viscosity of non-crosslinked or crosslinked (8S-StarPEG, 1 mM) collagen hydrogel. Rheological and viscometric measurements were conducted on a TA instruments ARES rheometer using the ASTM D4473 test method with 1200% strain at a frequency of 2 rad/s for 180 minutes. Parallel-plate geometry (50 mm diameter) at 0.9 mm gap height was used on a 3000 µL crosslinked and non-crosslinked collagen hydrogel concentration after 24 hours of gelation. The temperature was set at 37°C. The real-time plot results were obtained using TA Orchestrator software. G’ indicates the storage modulus that describes the elastic properties, and G” indicates the loss modulus that describes the viscous properties.
Migration of tumor cells in collagen hydrogels
To study the migration of glioma cells in the hydrogels, U87 cells or A172 cells (20,000 cells) were seeded in 200 µl collagen hydrogel and 8S-StarPEG (1 mM)-crosslinked collagen hydrogel. The U87 or A172 glioma cells were seeded in a 24-well plate (5,000 cells/well), and tumor cell migration on the cell culture dish was studied. After the cells were cultured for 48 hours, a time-lapse microscope (Zeiss Axio Observer microscope) placed in a plastic incubator with 5% CO2 at 37°C was utilized to record tumor cell migration. The time-lapse images of cell migration were recorded using ZEN 2011 imaging microscope software. Images were captured using a digital camera (AxioCam MRm Rev.3 with FireWire). Images were taken every 5 minutes for 3 hours, and then analyzed to quantify the cell migration distance and speed using NIH ImageJ software (National Institutes of Health, Bethesda, MD). The distance represents the full-length cell migration, while the cell migration speed was calculated from the distance during 3 hours of recording.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The qRT-PCR technique was performed to study the gene expression of MMP-2, MMP-9, uPA, tissue plasminogen activator (tPA), and PAI in tumor cells grown in collagen hydrogels. U87 or A172 glioma cells (100,000 cells) were seeded in 500 µl control and crosslinked collagen hydrogels or on a cell culture dish. After culturing for 3 days, the total RNA extraction for the cultured cells was performed using a Direct-zol™ RNA MiniPrep Plus (Zymo Research, Irvine, CA). A NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA) was used to measure the amount of RNA. The cDNA was generated using a High-Capacity cDNA Reverse Transcription Kit (Life Technologies Corporation, Grand Island, NY). Power SYBR® Master Mix was used to perform the qRT-PCR using the Applied Biosystems™ QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA). Conditions for the qRT-PCR were set as 95°C for 10 minutes, and then 40 cycles at 95°C for 15 seconds followed by 60°C for 60 seconds. Gene transcription was normalized in relation to transcription of the housekeeping human glyceraldehyde 3-phosphate dehydrogenase (GAPDH). To calculate the relative gene expression of each target gene, the 2−ΔΔCt value was calculated and presented. Primers used in the qRT-PCR procedure are listed in Table 1.
Table 1.
Primers for qRT-PCR
| Gene | Oligonucleotide (5’–3’) |
|---|---|
| MMP-2 | Forward: CCACTGCCTTCGATACAC |
| Reverse: GAGCCACTCTCTGGAATCTTAAA | |
| MMP-9 | Forward: TGGGCTACGTGACCTATGACAT |
| Reverse: GCCCAGCCCACCTCCACTCCTC | |
| tPA | Forward: GGCTGTGGACAGAAGGATGT |
| Reverse: GCTTTTGAGGAGTCGGGTGT | |
| uPA | Forward: CACGCAAGGGGAGATGAA |
| Reverse: ACAGCATTTTGGTGGTGACTT | |
| PAI | Forward: TGCTGGTGAATGCCCTCTACT |
| Reverse: CGGTCATTCCCAGGTTCTCTA | |
| GAPDH | Forward: CGAGATCCCTCCAAAATCAA |
| Reverse: TTCACACCCATGACGAACAT |
Inhibition of tumor cell migration in collagen hydrogels
The U87 or A172 glioma cells were cultured on cell culture dishes (5,000 cells/well of 24-well plate) or in collagen hydrogels (20,000 cells/200 µl hydrogel). To investigate the effect of the MMP-2 inhibitor (ARP 100, R&D System, Minneapolis, MN), MMP-9 inhibitor (CAS 1177749–58-4, Sigma), tranexamic acid (Santa Cruz Biotechnology, Inc., Dallas, TX), and aprotinin (Sigma-Aldrich, St. Louis, MO) on tumor cell migration on a cell culture dish and in non-crosslinked collagen hydrogel, these inhibitors were added to the cell culture medium or incorporated into the collagen hydrogels. After 48 hours, the migration of tumor cells on cell culture dishes or in the collagen hydrogels was recorded. The concentration of MMP-2 inhibitor, MMP-9 inhibitor, tranexamic acid, and aprotinin in the cell culture medium or in the collagen hydrogels was 100 µM, 25 µM, 3 mM, and 40 µg/ml, respectively.
Statistical analysis
Statistical analysis was conducted using a two-tailed Student’s t-test. A p-value of 0.05 was considered to be statistically significant. Data are expressed as means ± standard deviation.
Results
Migration of tumor cell into collagen hydrogels from microtumors
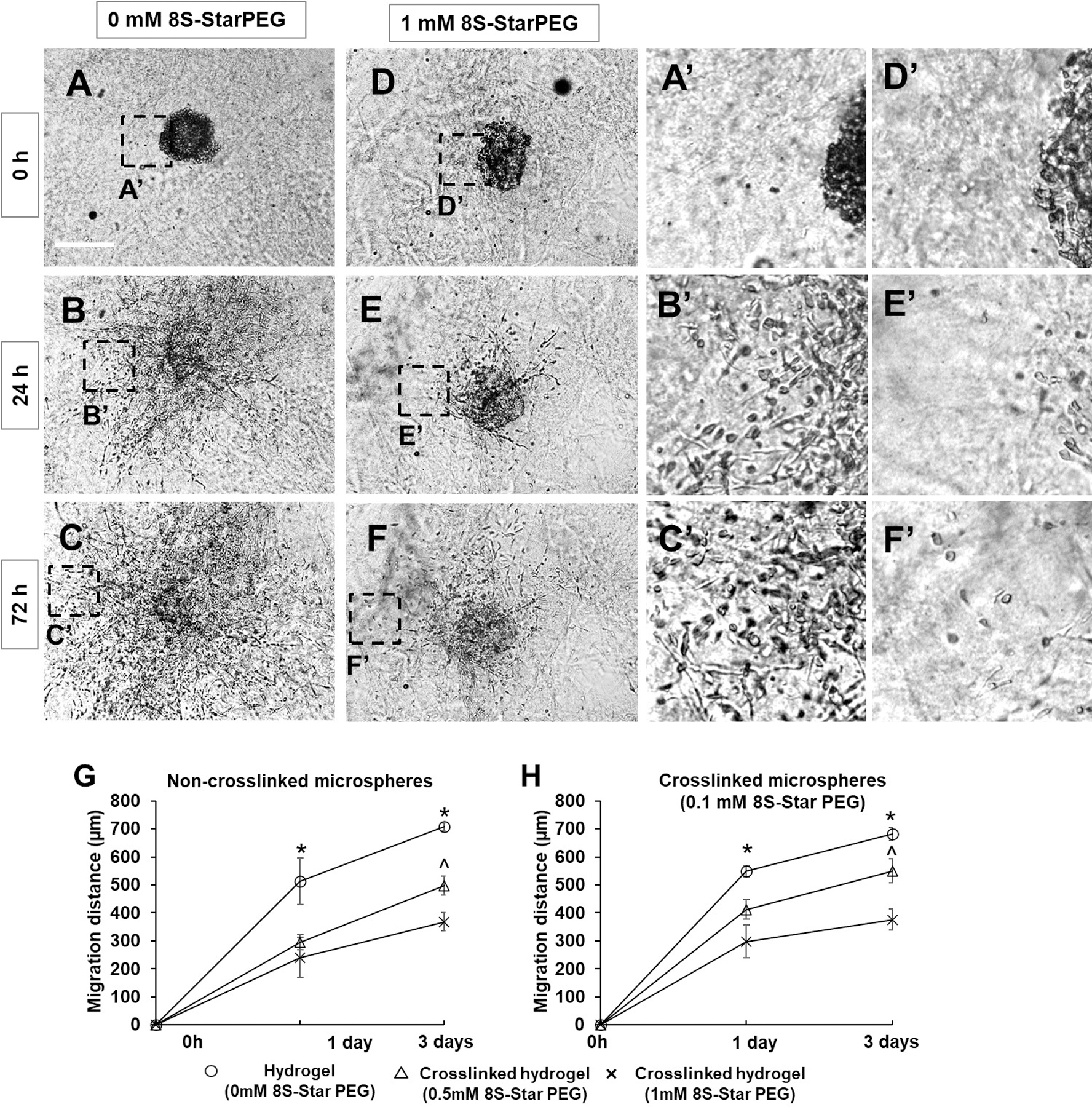
The microtumors were generated by encapsulating U87 glioma cells in non-crosslinked collagen or crosslinked collagen (8S-StarPEG, 0.1 mM). The size of the non-crosslinked and crosslinked microspheres encapsulating glioma cells were 773.9 ± 27.0 µm and 726 ± 27.0 µm, respectively, after they were cultured for 6 hours. The size of these microtumors decreased significantly after they were cultured for 3 days. The size of the non-crosslinked and crosslinked microtumors continued to decrease significantly to 330.5 ± 10.5 µm (p < 0.01) and 315.7 ± 39.9 µm (p < 0.01), respectively, after further culturing for another 4 days (Figure 1). Cells showed extended multiple processes in the microtumors. However, the size of crosslinked and non-crosslinked collagen microspheres was not significantly different at each time point.
Figure 1.
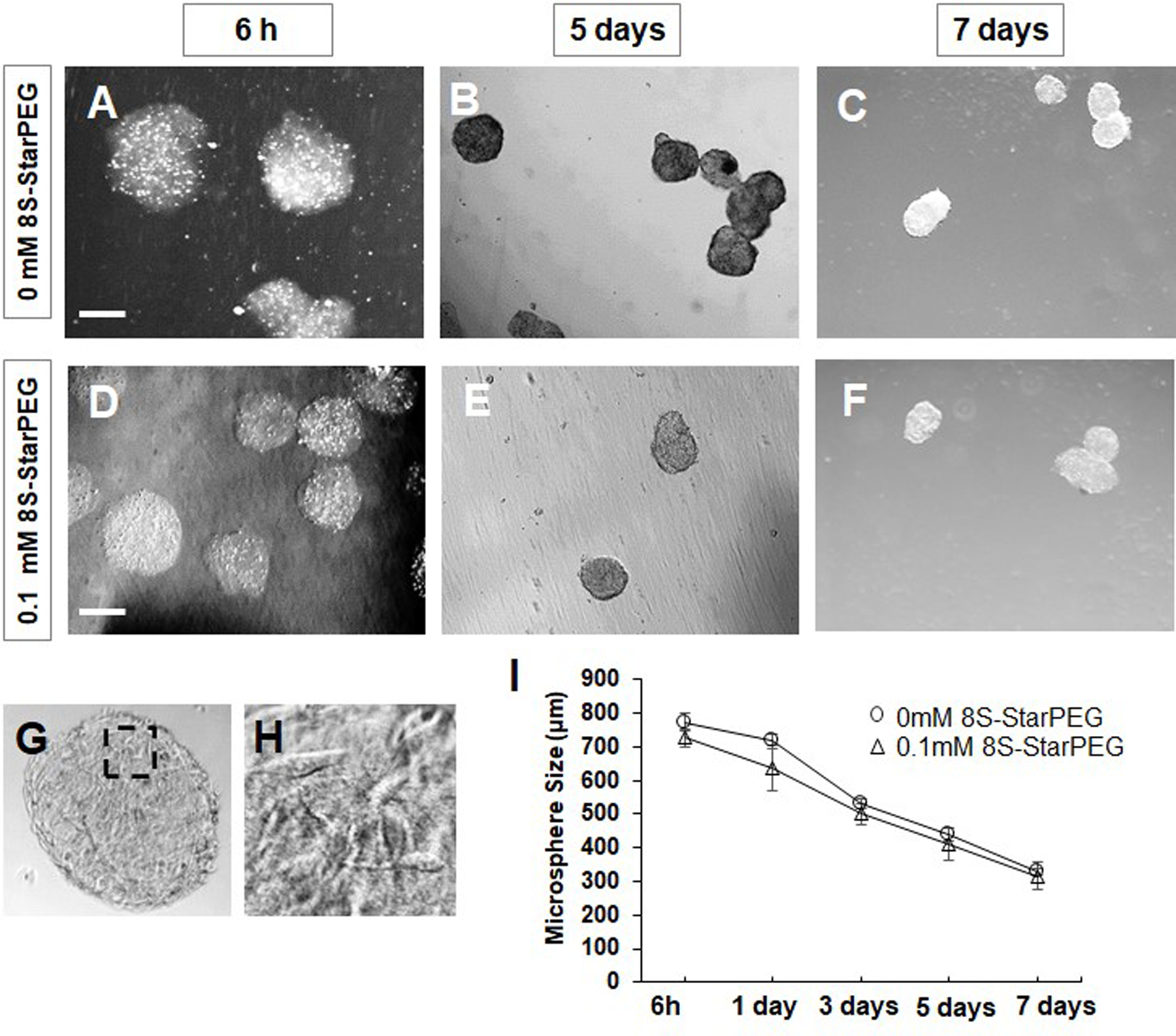
Fabrication of microtumors of U87 glioma cells. (A–C) Non-crosslinked microtumors and (D–F) Microtumors crosslinked with 8S-StarPEG and cultured for 7 days in cell culture medium. Microtumor size decreased gradually. Scale bar: 400 µm. (G) Dense tumor cells seen in microtumors after 7 days. (H) Magnified image of inset indicated in (G). (I) Quantification of microtumor size showing decrease when cultured in cell culture medium.
Tumor cells migrated into the collagen hydrogels from these microtumors after they were seeded in the hydrogels (Figures 2A–2F). The migration distance of tumor cells in the non-crosslinked collagen hydrogel was significantly higher than in that of the 8S-StarPEG (1 mM)-crosslinked collagen hydrogel after culturing for 3 days.
Figure 2.
Migration of glioma cells from microtumors grown in collagen hydrogels. (A–C) Images of tumor cell migration in non-crosslinked collagen hydrogel. (D–F) Images of tumor cell migration in crosslinked collagen hydrogel. Microtumors formed by mixing tumor cells with non-crosslinked collagen solution. Scale bar: 400 μm. (A’–F’) Magnified images of insets indicated in (A–F). (G, H) Quantification of glioma cell migration from microtumors formed by non-crosslinked collagen or crosslinked collagen. *p < 0.05, compared with cells in hydrogels crosslinked with 0.5 mM and 1 mM 8S-StarPEG at same time point. ^p < 0.05, compared with cells in hydrogels crosslinked with 1 mM 8S-StarPEG at same time point.
Cell viability of tumor cells encapsulated in microspheres
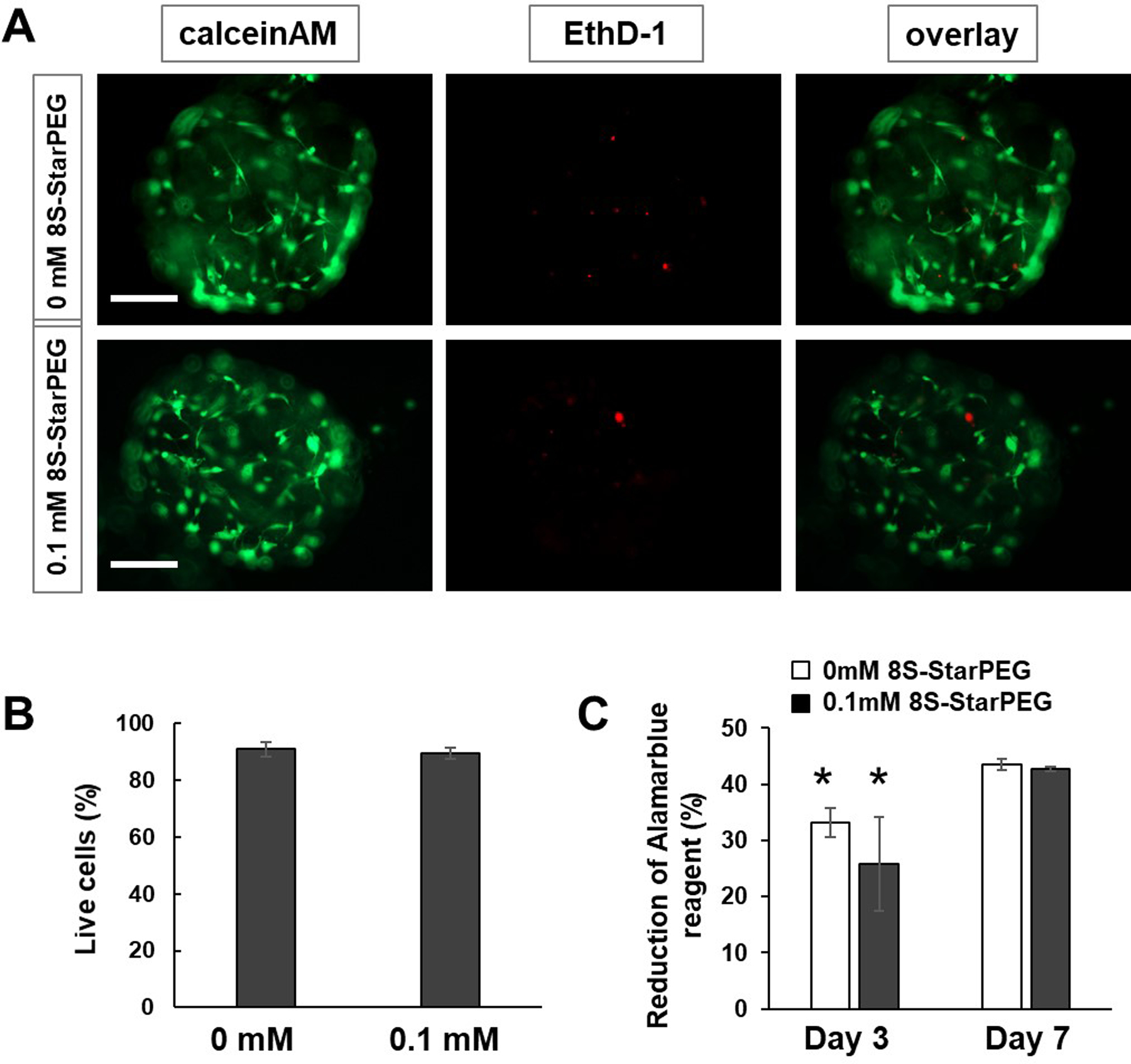
The cell viability of U87 cells encapsulated in the collagen microspheres was determined using a LIVE/DEAD® cell vitality assay kit and viewed under a fluorescent microscope. After the cells were cultured for 7 days, most of them survived in the non-crosslinked microspheres and crosslinked microspheres (Figures 3A). The tumor cells showed typical processes in microtumors. Quantification of live and dead cells in the microspheres showed that the ratio of live cells to total cells was 90.8 ± 2.4% in the non-crosslinked microspheres, and 89.5 ± 2.0% in the 0.1 mM 8S-StarPEG-crosslinked microspheres (Figure 3B).
Figure 3.
Cell viability assay. (A) LIVE/DEAD® cell viability assay of microtumors. Scale bar: 250 μm. (B) Quantification of live tumor cells. (C) Measurement of cell viability using AlamarBlue® assay for microtumors. *p < 0.05, compared with result at day 7.
The AlamarBlue® assay showed the metabolic activity of U87 cells in collagen microspheres (Figure 3C). After culturing for three days, the reduction of AlamarBlue® reagent for tumor cells in non-crosslinked collagen microspheres and 0.1 mM 8S-StarPEG-crosslinked collagen microspheres was 33.1 ± 2.6% and 25.8 ± 8.3%, respectively. The reduction value increased to 43.5 ± 0.9% and 42.8 ± 0.4% (p < 0.01), respectively, after culturing for 7 days. Results indicated tumor cell proliferation in the microspheres.
Characterization of crosslinked collagen hydrogels
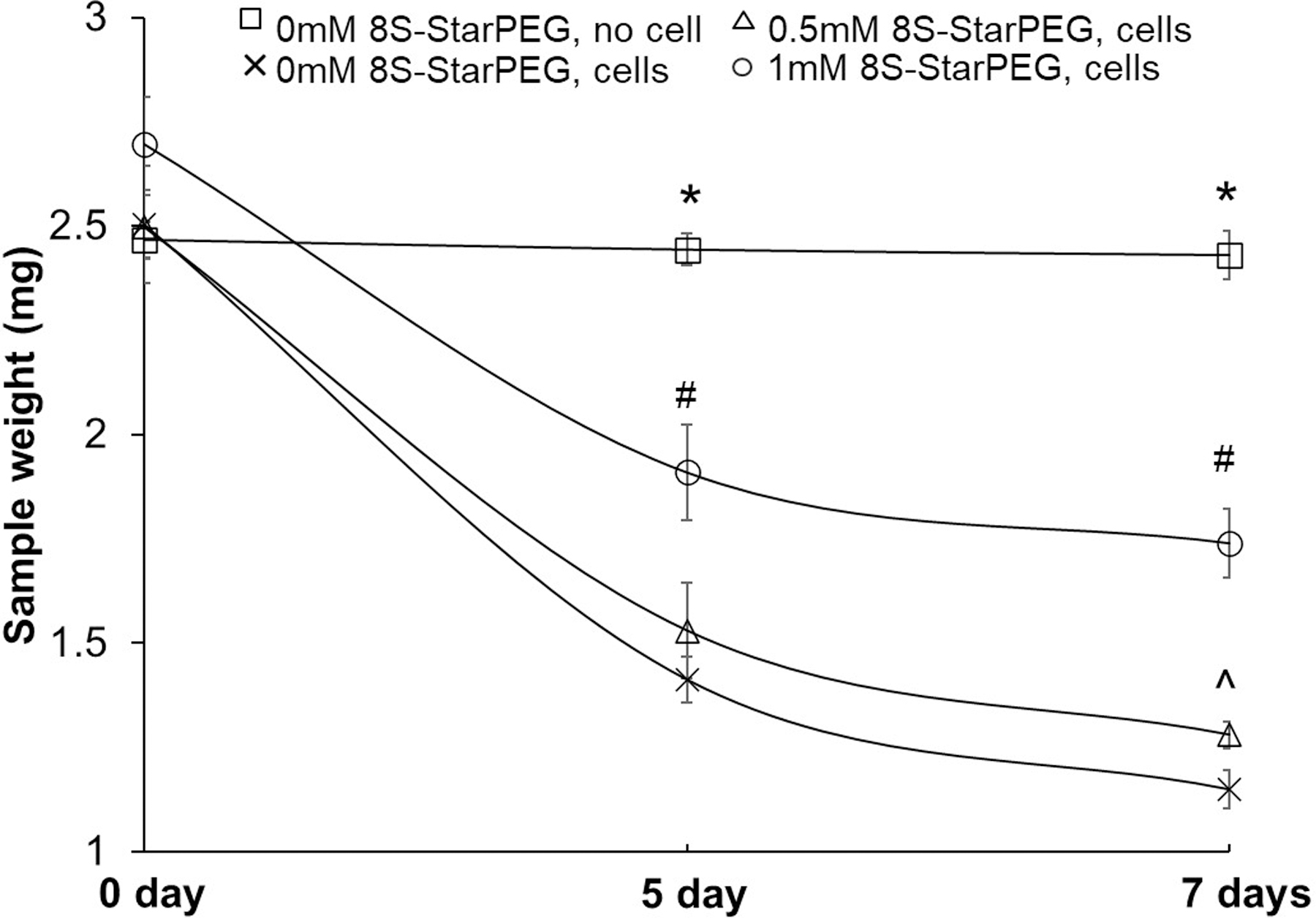
The purpose of this study was to determine the efficacy of crosslinking for reduction of the tumor cell-mediated degradation of matrices. Crosslinking of collagen hydrogels reduced the tumor cell-mediated collagen degradation. After the U87 tumor cells were cultured in collagen hydrogels for seven days, the weight of collagen in the hydrogel decreased (Figure 4). The collagen weight (1.74 ± 0.1 mg) in collagen hydrogel crosslinked with 1 mM 8S-StarPEG was significantly higher than that in the collagen hydrogel crosslinked with 0.5 mM 8S-StarPEG (1.28 ± 0.0 mg, p < 0.05) and non-crosslinked hydrogel (1.15 ± 0.0, p < 0.05).
Figure 4.
Cross-linking collagen with 8S-StarPEG decreased rate of glioma cell-mediated collagen hydrogel degradation. Glioma cells grown in collagen hydrogels cross-linked with different amounts of 8S-StarPEG. *p < 0.05, compared with collagen hydrogels with tumor cell growth at same time point. #p < 0.05, compared with non-crosslinked collagen hydrogels and hydrogels crosslinked with 0.5 mM 8S-StarPEG at same time point. ^p < 0.05, compared with non-crosslinked collagen hydrogels at same time point.
Rheological testing showed that G” was higher in the crosslinked collagen than in non-crosslinked collagen, indicating that viscosity of the crosslinked collagen hydrogel was higher than that of the non-crosslinked collagen hydrogel. Dynamic viscosity (η) is calculated as shear stress divided by shear rate and is used to measure the collagen hydrogel’s resistance to flow when an external force is applied. In this study, the dynamic viscosity of crosslinked collagen hydrogel (0.43 poise) was higher than that of the non-crosslinked collagen hydrogel (0.15 poise) (Figure 5).
Figure 5.
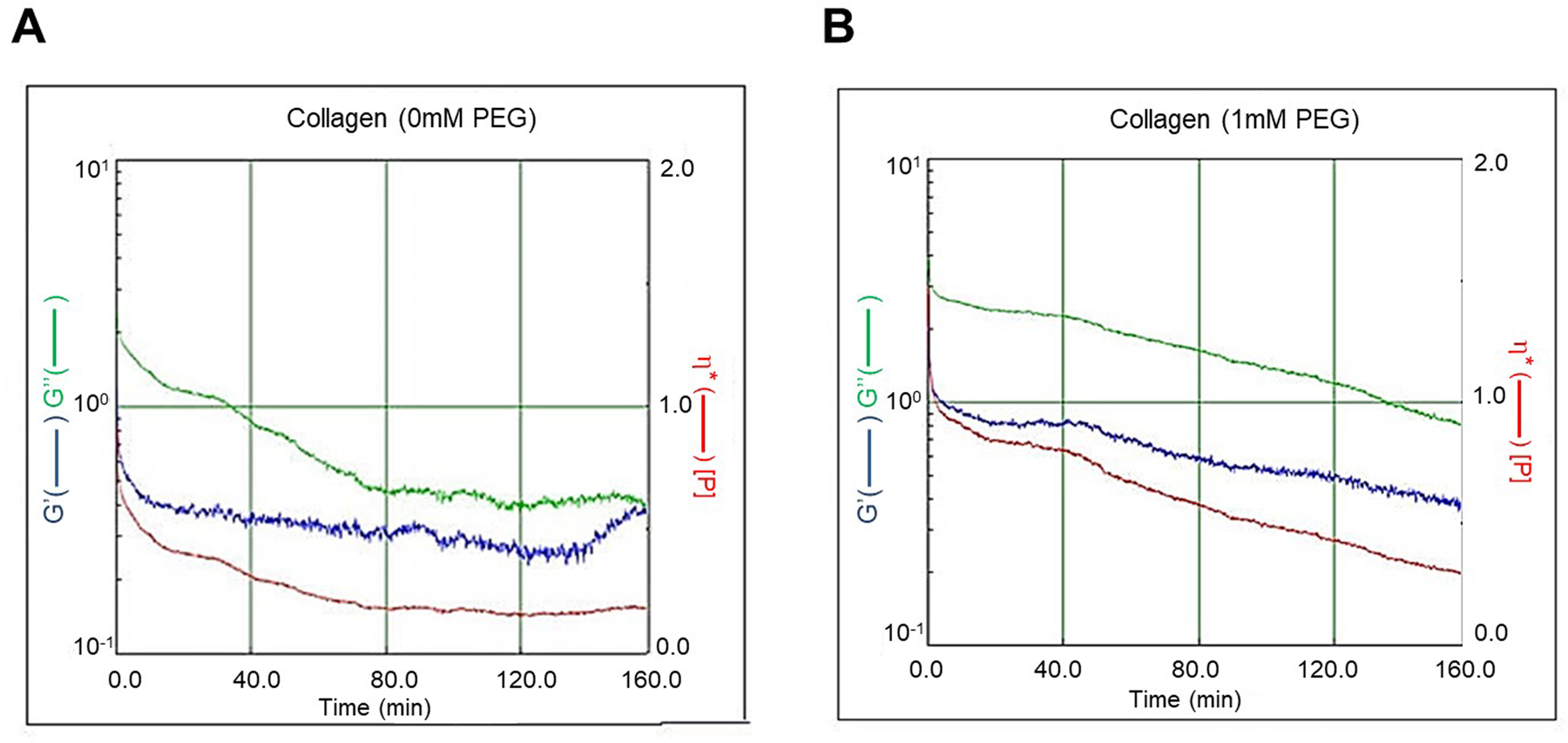
Rheological testing showing viscosity of crosslinked (8S-StarPEG, 1 mM) collagen hydrogel higher than that of non-crosslinked hydrogel.
Crosslinking of collagen hydrogel decreased tumor cell migration speed in hydrogels
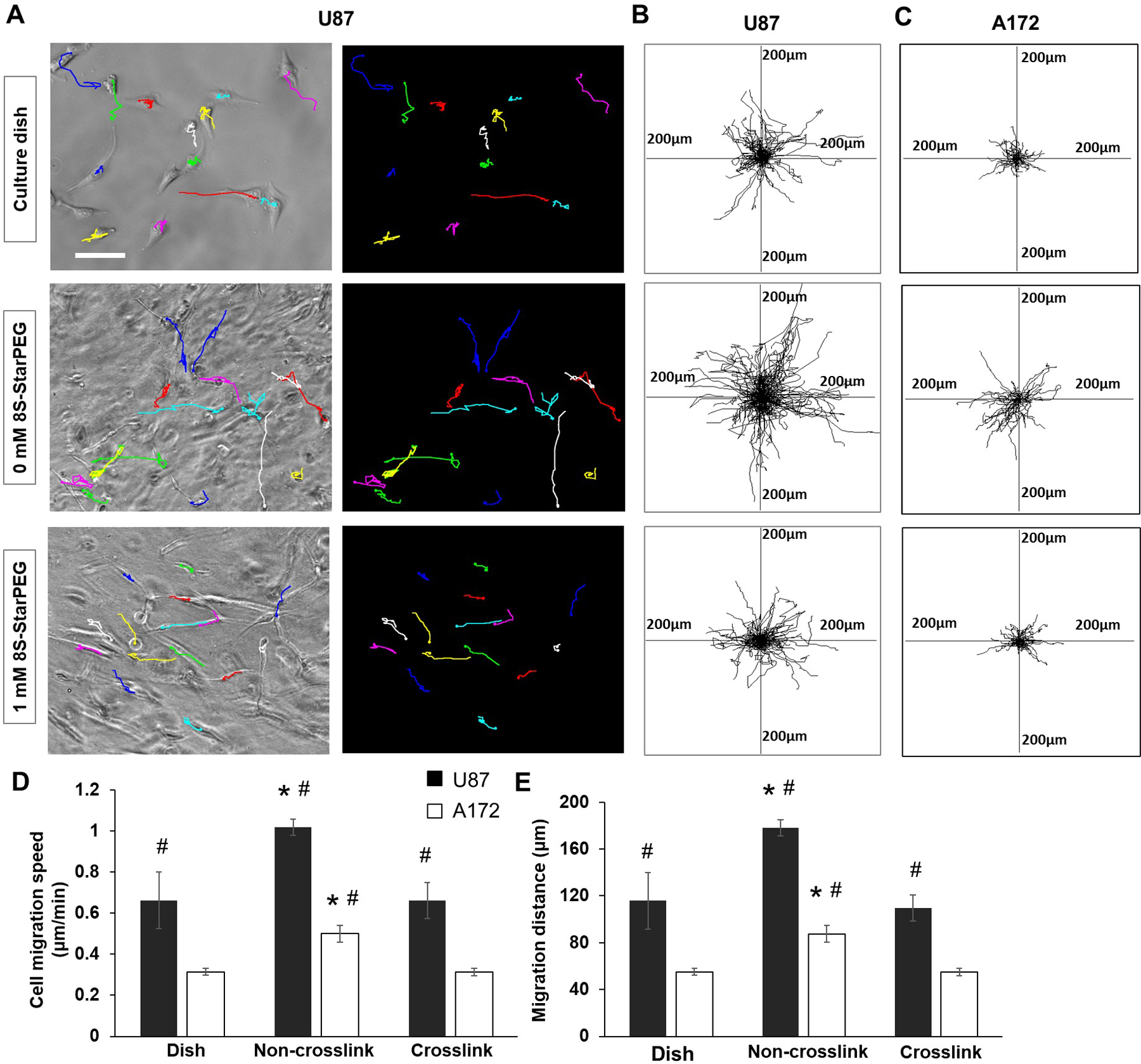
In this study, we investigated the migration of U87 and A172 glioma cells on cell culture dish and in collagen hydrogels. Figure 6A shows the cell migration pathways highlighted with lines of different colors. The tumor cell migration distance in non-crosslinked collagen hydrogel (see supplemental video 2) was longer than that on the cell culture dish (see supplemental video 1) and in the crosslinked collagen hydrogel (see supplemental video 3).
Figure 6.
Migration of glioma cells in hydrogels. (A) U87 cells migrated on cell culture dish or in hydrogels. Cell migration tracks presented as lines in graph. Scale bar: 100 µm. (B) and (C) U87 and A172 glioma cell migration tracks for 3 hours. Center of frame represents cell initial location of migration at t = 0 min. (D) Tumor cell migration speed. (E) Tumor cell migration distance. *p < 0.05, compared with tumor cell migration on cell culture dish or in crosslinked collagen hydrogel. #p < 0.01, compared with corresponding A172 glioma cell migration.
Each frame of Figures 6B and 6C shows the superimposed migration tracks of U87 and A172 glioma cells, respectively. The positions of all cells at initial locations are centered at (0, 0) in the frame. Each line in the frame demonstrates the single-cell dislocation over a three-hour period.
The migration speed and distance of tumor cells were quantified. The migration speed of U87 (1.01 ± 0.03 µm/min) and A172 cells (0.499 ± 0.0.04 µm/min) in the collagen hydrogel was significantly greater than that on the cell culture dish (U87, 0.66 ± 0.13 µm/min; A172, 0.31 ± 0.01 µm/min; p < 0.01) and in the 8S-StarPEG-crosslinked collagen hydrogel (U87, 0.66 ± 0.08 µm/min; A172, 0.31 ± 0.01 µm/min p < 0.01) (Figure 6D). Similar to the cell migration speed, the migration distance of U87 and A172 cells in collagen hydrogel was significantly greater than that on the cell culture dish and in the crosslinked collagen hydrogel (Figure 6E). The migration speed of U87 cells was significantly higher than that of A172 cells on the cell culture dish or in collagen hydrogels (p < 0.01).
Investigation of gene expression of tumor cells in hydrogels
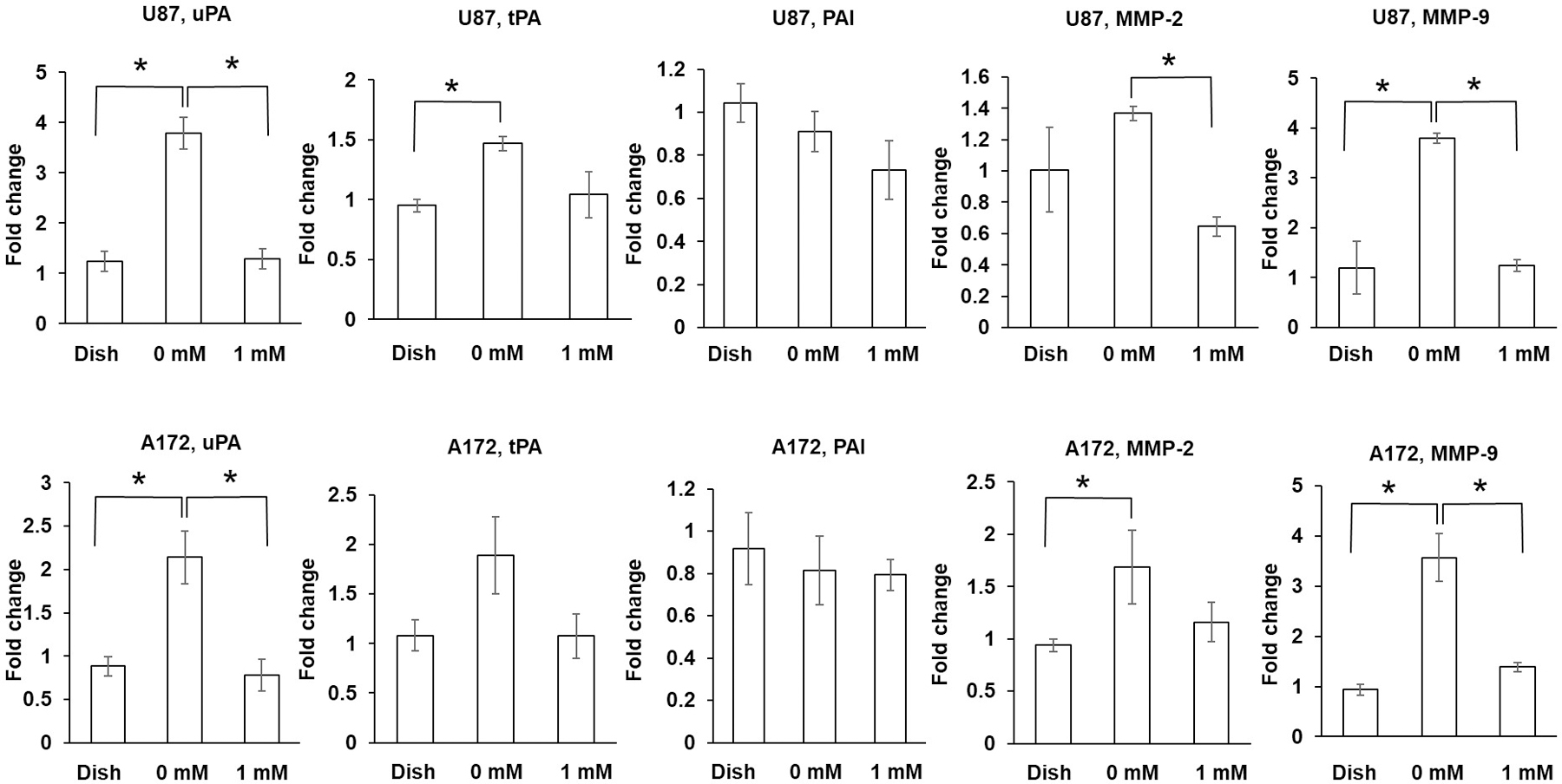
The expression of uPA and MMP9 of U87 and A172 glioma cells in non-crosslinked collagen hydrogel was significantly higher than that of cells on the culture dish or in crosslinked collagen hydrogel (p < 0.01). The MMP-2 expression of U87 glioma cells in non-crosslinked collagen hydrogel was significantly higher than that of cells in crosslinked collagen hydrogel (p < 0.01). The MMP-2 expression of A172 glioma cells in non-crosslinked collagen hydrogel was significantly higher than that of cells on the culture dish (p < 0.05). The expression of tPA of U87 glioma cells in non-crosslinked collagen hydrogel was significantly higher than that of cells on the culture dish (p < 0.05). (Figure 7).
Figure 7.
Expression of uPA, tPA, PAI, MMP-2, and MMP-9 by glioma cells grown on cell culture dish or in hydrogel. Expressed as fold change compared with gene expression by U87 and A172 glioma cells on cell culture dish. Each target gene normalized to GAPDH. *p < 0.05.
Investigation of regulation of tumor cell migration in collagen hydrogel
A pharmacological approach was used to investigate the effect of MMP-2, MMP-9, and plasminogen activators on tumor cell migration (Figure 8). After the U87 cells grown on a cell culture dish or in non-crosslinked collagen hydrogels were treated with MMP-2 or MMP-9 inhibitors (ARP 100, 100µM; CAS 1177749-58-4, 25 µM), their migration speed was reduced significantly compared with that of control groups (p < 0.01). When U87 cells grown on a cell culture dish were treated with tranexamic acid (3 mM) and aprotinin (40 µg/ml), the cell migration speeds were 0.66 ± 0.20 µm/min and 0.65 ± 0.19 µm/min, respectively, which were not significantly different than that of the control study on a cell culture dish. When U87 cells grown in collagen hydrogel were treated with tranexamic acid and aprotinin, the tumor cell migration speeds were 0.55 ± 0.08 µm/min and 0.69 ± 0.05 µm/min, respectively, which were significantly lower than that of the control study in collagen hydrogels. The treatment of MMP-2, MMP-9, uPA, and tPA inhibitors to A172 cells in collagen hydrogels significantly reduced the cell migration speed compared with that of non-treated cells. The treatment of inhibitors to the A172 cells grown on a cell culture dish did not significantly decrease the cell migration speed compared with that of non-treated cells.
Figure 8.
Effect of inhibiting MMP-2, MMP-9, uPA, and tPA on glioma cell migration. (A) Track graphs showing effects of MMP-2, MMP-9, uPA, and tPA inhibitors on U87 cell migration. Each graph axis represents 200 µm. Inhibitors of MMP-2 (ARP 100, 100 µM) and MMP-9 (CAS 1177749-58-4, 25 µM) significantly decreased U87 cell migration on cell culture dish or in collagen hydrogels. Aprotinin (40 µg/ml) and tranexamic acid (3 mM) inhibited U87 migration in collagen hydrogel but not on cell culture dish. (B) and (C) Quantification of tumor cell migration after U87 and A172 glioma cells treated with inhibitors respectively. *p < 0.05, compared with tumor cell migration of corresponding group without inhibitor treatment. (D) and (E) Quantification of tumor cell migration distance after U87 and A172 glioma cells treated with inhibitors, respectively. *p < 0.05, compared with tumor cell migration of corresponding group without inhibitor treatment.
Discussion
Engineered 3D microtumors are cellular aggregates and considered to be more accurate models for reflecting the structural complexity and physical properties of the glioblastoma in vivo than 2D cell cultures (Friedrich et al., 2007; Kunz-Schughart et al., 2004). Microtumors were generated using the tumor cell aggregation method such as the hanging drop method (Del Duca et al., 2004), spontaneous aggregation of tumor spheroids (Deisboeck et al., 2001; Singh et al., 2003), aggregation in spinner flasks (Wartenburg et al., 2001), or rotary cell culture systems (Mazzoleni et al., 2009; Unsworth and Lelkes, 1998). Ultra-low attachment (ULA) 96-well round-bottomed plates were used to generate suspension cultures of reproducibly sized, single spheroids in each well (Vinci et al., 2012). Additionally, microtumors were also fabricated by encapsulating tumor cells using synthetic polymers (Long et al., 2014) or natural materials (Jang et al., 2017). The generated microtumors using the ECM are a valued model for studying the role of 3D cell-cell and cell-ECM interactions.
In this study, microtumors were fabricated by self-assembly of a collagen solution with glioma cells in a cell culture medium. The microspheres encapsulating the tumor cells formed solid microtumors after they were cultured in the cell culture medium. Collagen type I is a biocompatible and biodegradable natural material that provides a permissive microenvironment for cell growth. In a previous study, we demonstrated that the astrocytes encapsulated in collagen microspheres showed high cell viability (Berndt et al., 2016). Glioma cells also showed high cell viability in collagen microspheres. When collagen microspheres were incubated in a cell culture medium for ten days, their morphology was stable. However, cells encapsulated in the collagen microspheres can condense the microspheres. We found that the size of microspheres encapsulating either astrocytes (Berndt et al., 2016) or glioma cells were reduced after culturing in the medium. Crosslinking of collagen microspheres with 8S-StarPEG reduced the degradation rate of collagen microspheres against collagenase digestion. In this study, microtumors were generated by crosslinking the collagen with 8S-StarPEG. The control and crosslinked microtumors showed a similar size reduction after culturing for seven days. The crosslinking of the collagen hydrogel was characterized by digesting the crosslinked collagen hydrogels with tumor cells. It was observed that the tumor cells can degrade the collagen hydrogels, and crosslinking decreased the tumor cell-mediated collagen degradation.
The fibril structure of collagen hydrogels provides scaffolds for tumor cell migration. A previous study showed that the integrin-dependent cell-matrix interaction formed by fibroblasts grown in 3D matrices is different than that on 2D substrata (Cukierman et al., 2001) It was suggested that the reciprocal interaction between cells and the 3D collagen matrix provides a mechanically compliant environment to the cell (Rhee, 2009). In this study, a higher migration speed of glioma cells in collagen hydrogels compared with those on the cell culture dish was observed. The different molecular and mechanical interactions between the cells and the matrix may cause different cell motility of glioma cells in collagen hydrogels and on a cell culture dish.
A previous study showed that ECM rigidity can alter the cell motility of glioma cells. Glioma cells can respond to the stiffness of the substrate and show decreased stress fiber formation, focal adhesion, and cell motility when they are cultured on a soft substrate compared to a rigid material (Ulrich et al., 2009). The biophysical property of the 3D model generated by biomaterial scaffolds mimics the in vivo environment in the investigation of tumor cell interaction with the ECM. Glioma cells modify the normal brain tissue by producing ECM molecules such as collagen and thereby increase the rigidity of the microenvironment (Ulrich et al., 2009). As a ligand of tumor cell receptors, collagen can mediate tumor cell growth and differentiation (Leitinger, 2011). Collagen can also act as a scaffold and provide adhesion sites for tumor cell invasion. The changes in tumor ECM rigidity can alter cell motility (Thomas and DiMilla, 2000). The migration of glioma cells grown in collagen gels was used to study tumor cell invasion. The high ECM stiffness enhanced the expression of Rho, which mediates cell motility (Paszek et al., 2005). It was reported that the invasion rate of glioma cells was enhanced when they were grown in collagen matrices with a higher collagen concentration because of the higher stiffness of the matrices (Kaufman et al., 2005). However, when the elasticity of the collagen ECM was increased by incorporating agarose, the glioma cell invasion decreased (Ulrich et al., 2010). The invasion study was also performed on other tumor cell types. The cell motility and invasion of both MV3 and HT1080 cells were delayed when the collagen density was increased (Haeger et al., 2014).
The protease-independent cell migration depends on deformability of the tumor cell body and ECM molecules (Beadle et al., 2008; Ilina et al., 2011; Wolf et al., 2007; Wyckoff et al., 2006). In this study, the migration of U87 from microtumors in collagen hydrogels was tested. Crosslinking of collagen hydrogels decreased the tumor cell migration distance, and crosslinking increased the viscosity of the collagen hydrogels. Time-lapse imaging showed that crosslinking increased the viscosity of collagen hydrogels and decreased the tumor cell migration speed. These studies suggest that the decreased flexibility of crosslinked collagen fibers restricts cell locomotion and causes a reduction in the tumor cell migration speed.
TPA, uPA, and their receptors regulate the cell-extracellular matrix interaction and cell migration. The binding of plasminogen activator and its receptor can induce cell adhesion and migration (Rao, 2003). When the uPA gene expression was downregulated in glioma cells, the tumor cell motility decreased. The PI3k/Akt signaling pathway was inhibited when uPA was downregulated. This study suggested that the PI3k/Akt pathway regulates uPA-induced cell migration (Chandrasekar et al., 2003). A previous study demonstrated the inhibition of myeloma cell migration with aprotinin, which is a competitive serine protease inhibitor that forms stable complexes with plasmin. The treatment of myeloma cells with aprotinin decreased the osteoblast-mediated myeloma cell invasion into Matrigel or type I collagen hydrogel (Hecht et al., 2007). Tranexamic acid competitively inhibits the activation of plasminogen to plasmin by interfering with the binding of plasminogen to the cell surface. An in vivo study showed that treatment of tranexamic acid to male C57BL/6 mice resulted in a significant reduction of ischemia-reperfusion-elicited intravascular firm adherence and transmigration of neutrophils (Reichel et al., 2011). Another in vivo study reported that the administration of tranexamic acid by the rat reduced smooth muscle cell migration four days after a balloon catheter injury to the rat carotid artery (Jackson et al., 1993). Plasminogen activator activity can also regulate astrocyte migration (Faber-Elman et al., 1995). We found that the migration of astrocytes in fibrin hydrogel does so in a plasminogen activator activity-dependent manner (Berndt et al., 2016). When aprotinin is added to the fibrin gel, the astrocyte-induced fibrinolysis slowed down, and the astrocyte migration was completely inhibited. However, in this study, the migration speed of U87 or A172 cells was decreased only 30% to 50% when treated with the same amount of aprotinin.
The proteolytic and non-proteolytic mechanisms are both involved in the in vivo tumor cell invasion process (Wolf and Friedl, 2011). MMPs and serine proteases such as plasminogen activator contribute to degradation of the ECM during the proteolytic process. The gene expression level of these molecules varies in response to different extracellular microenvironments (Egeblad et al., 2010; Kessenbrock et al., 2010). In this study, we observed higher MMP-2, MMP-9, tPA, and uPA expression of U87 and A172 glioma cells in non-crosslinked collagen hydrogels compared with that of cells grown on culture dishes or in crosslinked collagen hydrogels. When glioma cells in the collagen hydrogels were treated with inhibitors of the molecules, the cell migration speed was decreased compared with non-treated cells in the hydrogels. Results indicate that these molecules play an important role in the proteolytic movement of U87 and A172 glioma cells in collagen hydrogels. We also observed a lower MMP-9 expression in crosslinked collagen hydrogels, which is consistent with previous observations that reported a decreased MMP-9 expression in stiffer hydrogels (Pedron and Harley, 2013; Wang et al., 2014a). The higher expression of MMPs enhanced the ECM degradation process, thereby leading to the higher migration speed in non-crosslinked hydrogel. When tumor cells are cultured on a cell culture dish, which is a non-deformable and non-degradable substrate, the change in cell morphological alone leads to cell migration. In this study, the inhibitors of MMP-2, MMP-9, uPA, and tPA did not inhibit A172 cell migration, and uPA and tPA did not inhibit U87 cell migration on the culture dish. The non-proteolytic mechanism contributes to the glioma cell migration on the culture dish. However, we also observed different responses of U87 and A172 cells grown on a cell culture dish to inhibitors of MMP-2 and MMP-9. Unlike the A172 cells, the migration speed of U87 cells grown on a cell culture dish was reduced when they were treated with MMP-2 or MMP-9 inhibitors. This finding indicates that the migration of U87 is also regulated by MMP-2 and MMP-9 and intrinsic molecular pathways. Further studies will be needed to explore the interaction of MMPs and signaling molecules, and different responses of the A172 glioma cell to MMP-2 and MMP-9 inhibitors. The U87 and A172 cells are established cell lines and broadly used glioma cell types for investigating glioma biological behavior. Although they are well-characterized, the limitation of these cell lines is that the cells cannot represent the highly invasive behavior of primary glioma cells. This research provides a preliminary data for further study of tumor cell invasion using primary glioma cells.
Supplementary Material
Supplemental video 1. U87 cell migration on cell culture dish.
Supplemental video 3. U87 cell migration in 8S-StarPEG crosslinked collagen hydrogel.
Supplemental video 2. U87 cell migration in collagen hydrogel.
Acknowledgements
The start-up funding of Li Yao, Wichita State University; the National Institute of General Medical Sciences (P20 GM103418) of the National Institutes of Health; Wichita Medical Research and Education Foundation (WMREF) for their support of this study; Flossie E West Memorial Trust.
Contract grant sponsors:
1. Li Yao’s start-up funding at Wichita State University.
2. The National Institute of General Medical Sciences (P20 GM103418) of the National Institutes of Health.
3. Wichita Medical Research and Education Foundation (WMREF) and Flossie E. West Memorial Trust.
REFERENCES
- Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. 2008. The role of myosin II in glioma invasion of the brain. Molecular Biology of the Cell 19(8):3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berczi I, Szentiványi A. 2009. Foreword: Neurogenic inflammation coming of age. NeuroImmune Biology. 8:vii–xiv. [Google Scholar]
- Berndt M, Li Y, Seyedhassantehrani N, Yao L. 2016. Fabrication and characterization of microspheres encapsulating astrocytes for neural regeneration. ACS Biomaterials Science & Engineering 3(7):1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi F, Carmeliet P. 2002. uPAR: A versatile signalling orchestrator. Nature Reviews Molecular Cell Biology 3(12):932. [DOI] [PubMed] [Google Scholar]
- Chandrasekar N, Mohanam S, Gujrati M, Olivero WC, Dinh DH, Rao JS. 2003. Downregulation of uPA inhibits migration and PI3k/Akt signaling in glioblastoma cells. Oncogene 22(3):392. [DOI] [PubMed] [Google Scholar]
- Chen S-H, Hung W-C, Wang P, Paul C, Konstantopoulos K. 2013. Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Scientific Reports 3:1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Colman H. 2015. Glioma biology and molecular markers. In Current understanding and treatment of gliomas: Springer. pp. 15–30. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. 2001. Taking cell-matrix adhesions to the third dimension. Science 294(5547):1708–1712. [DOI] [PubMed] [Google Scholar]
- Deisboeck T, Berens M, Kansal A, Torquato S, Stemmer‐Rachamimov A, Chiocca E. 2001. Pattern of self-organization in tumour systems: complex growth dynamics in a novel brain tumour spheroid model. Cell Proliferation 34(2):115–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Duca D, Werbowetski T, Del Maestro RF. 2004. Spheroid preparation from hanging drops: Characterization of a model of brain tumor invasion. Journal of Neuro-Oncology 67(3):295–303. [DOI] [PubMed] [Google Scholar]
- DelNero P, Song YH, Fischbach C. 2013. Microengineered tumor models: Insights & opportunities from a physical sciences-oncology perspective. Biomedical Microdevices 15(4):583–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Nakasone ES, Werb Z. 2010. Tumors as organs: Complex tissues that interface with the entire organism. Developmental Cell 18(6):884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FaberElman A, Miskin R, Schwartz M. 1995. Components of the plasminogen activator system in astrocytes are modulated by tumor necrosis factor-alpha and interleukin-1 beta through similar signal transduction pathways. Journal of Neurochemistry 65(4):1524–1535. [DOI] [PubMed] [Google Scholar]
- Friedrich J, Ebner R, Kunz-Schughart LA. 2007. Experimental anti-tumor therapy in 3-D: Spheroids—Old hat or new challenge? International Journal of Radiation Biology 83(11–12):849–871. [DOI] [PubMed] [Google Scholar]
- Guan P-P, Yu X, Guo J-J, Wang Y, Wang T, Li J-Y, Konstantopoulos K, Wang Z-Y, Wang P. 2015. By activating matrix metalloproteinase-7, shear stress promotes chondrosarcoma cell motility, invasion and lung colonization. Oncotarget 6(11):9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeger A, Krause M, Wolf K, Friedl P. 2014. Cell jamming: Collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochimica et Biophysica Acta (BBA)-General Subjects 1840(8):2386–2395. [DOI] [PubMed] [Google Scholar]
- Hecht M, Heider U, Kaiser M, Von Metzler I, Sterz J, Sezer O. 2007. Osteoblasts promote migration and invasion of myeloma cells through upregulation of matrix metalloproteinases, urokinase plasminogen activator, hepatocyte growth factor and activation of p38 MAPK. British Journal of Haematology 138(4):446–458. [DOI] [PubMed] [Google Scholar]
- Huijbers IJ, Iravani M, Popov S, Robertson D, Al-Sarraj S, Jones C, Isacke CM. 2010. A role for fibrillar collagen deposition and the collagen internalization receptor Endo180 in glioma invasion. PLoS One 5(3):e9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilina O, Bakker G-J, Vasaturo A, Hoffman RM, Friedl P. 2011. Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and-independent collective cancer cell invasion. Physical Biology 8(1):015010. [DOI] [PubMed] [Google Scholar]
- Jackson CL, Raines EW, Ross R, Reidy MA. 1993. Role of endogenous platelet-derived growth factor in arterial smooth muscle cell migration after balloon catheter injury. Arteriosclerosis, Thrombosis, and Vascular Biology 13(8):1218–1226. [DOI] [PubMed] [Google Scholar]
- Jang M, Koh I, Lee SJ, Cheong J-H, Kim P. 2017. Droplet-based microtumor model to assess cell-ECM interactions and drug resistance of gastric cancer cells. Scientific Reports 7:41541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri N, Chen TC, Hofman FM. 2016. Tumor vasculature and glioma stem cells: Contributions to glioma progression. Cancer Letters 380(2):545–551. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Brangwynne C, Kasza K, Filippidi E, Gordon VD, Deisboeck T, Weitz D. 2005. Glioma expansion in collagen I matrices: Analyzing collagen concentration-dependent growth and motility patterns. Biophysical Journal 89(1):635–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z. 2010. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141(1):52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz-Schughart LA, Freyer JP, Hofstaedter F, Ebner R. 2004. The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. Journal of Biomolecular Screening 9(4):273–285. [DOI] [PubMed] [Google Scholar]
- Lei X, Chang L, Ye W, Jiang C, Zhang Z. 2015. Raf kinase inhibitor protein (RKIP) inhibits the cell migration and invasion in human glioma cell lines in vitro. International Journal of Clinical and Experimental Pathology 8(11):14214. [PMC free article] [PubMed] [Google Scholar]
- Leitinger B 2011. Transmembrane collagen receptors. Annual Review of Cell and Developmental Biology 27:265–290. [DOI] [PubMed] [Google Scholar]
- Long TJ, Sprenger CC, Plymate SR, Ratner BD. 2014. Prostate cancer xenografts engineered from 3D precision-porous poly (2-hydroxyethyl methacrylate) hydrogels as models for tumorigenesis and dormancy escape. Biomaterials 35(28):8164–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Cui W, He S-m, Duan Y-h, Heng L-j, Wang L, Gao G-d. 2012. Human U87 astrocytoma cell invasion induced by interaction of βig-h3 with integrin α5β1 involves calpain-2. PLoS One 7(5):e37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahesparan R, Read T-A, Lund-Johansen M, Skaftnesmo K, Bjerkvig R, Engebraaten O. 2003. Expression of extracellular matrix components in a highly infiltrative in vivo glioma model. Acta Neuropathologica 105(1):49–57. [DOI] [PubMed] [Google Scholar]
- Mazzoleni G, Di Lorenzo D, Steimberg N. 2009. Modelling tissues in 3D: The next future of pharmaco-toxicology and food research? Genes & Nutrition 4(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci G, Magro G, Cardile V, Coco M, Marzagalli R, Castrogiovanni P, Imbesi R, Graziano ACE, Barone F, Di Rosa M. 2015. Characterization of matrix metalloproteinase-2 and-9, ADAM-10 and N-cadherin expression in human glioblastoma multiforme. Cell and Tissue Research 362(1):45–60. [DOI] [PubMed] [Google Scholar]
- Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D. 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell 8(3):241–254. [DOI] [PubMed] [Google Scholar]
- Pedron S, Harley B. 2013. Impact of the biophysical features of a 3D gelatin microenvironment on glioblastoma malignancy. Journal of Biomedical Materials Research Part A 101(12):3404–3415. [DOI] [PubMed] [Google Scholar]
- Ranger A, McDonald W, Bauman GS, Del Maestro R. 2009. Effects of surgical excision and radiation on medulloblastoma cell invasiveness. Canadian Journal of Neurological Sciences 36(5):631–637. [DOI] [PubMed] [Google Scholar]
- Rao JS. 2003. Molecular mechanisms of glioma invasiveness: The role of proteases. Nature Reviews Cancer 3(7):489. [DOI] [PubMed] [Google Scholar]
- Reichel CA, Lerchenberger M, Uhl B, Rehberg M, Berberich N, Zahler S, Wymann MP, Krombach F. 2011. Plasmin inhibitors prevent leukocyte accumulation and remodeling events in the postischemic microvasculature. PLoS One 6(2):e17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S 2009. Fibroblasts in three dimensional matrices: cell migration and matrix remodeling. Experimental & Molecular Medicine 41(12):858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruoslahti E 1996. Brain extracellular matrix. Glycobiology 6(5):489–492. [DOI] [PubMed] [Google Scholar]
- Seyedhassantehrani N, Li Y, Yao L. 2016. Dynamic behaviors of astrocytes in chemically modified fibrin and collagen hydrogels. Integrative Biology 8(5):624–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman JH, Hoes K, Marcus J, Komotar RJ, Brennan CW, Gutin PH. 2011. Neurosurgery for brain tumors: Update on recent technical advances. Current Neurology and Neuroscience Reports 11(3):313–319. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. 2003. Identification of a cancer stem cell in human brain tumors. Cancer Research 63(18):5821–5828. [PubMed] [Google Scholar]
- Song H, Li Y, Lee J, Schwartz AL, Bu G. 2009. Low-density lipoprotein receptor-related protein 1 promotes cancer cell migration and invasion by inducing the expression of matrix metalloproteinases 2 and 9. Cancer Research 69(3):879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabouret E, Boudouresque F, Barrie M, Matta M, Boucard C, Loundou A, Carpentier A, Sanson M, Metellus P, Figarella-Branger D. 2013. Association of matrix metalloproteinase 2 plasma level with response and survival in patients treated with bevacizumab for recurrent high-grade glioma. Neuro-Oncology 16(3):392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, DiMilla P. 2000. Spreading and motility of human glioblastoma cells on sheets of silicone rubber depend on substratum compliance. Medical and Biological Engineering and Computing 38(3):360–370. [DOI] [PubMed] [Google Scholar]
- Ulrich TA, de Juan Pardo EM, Kumar S. 2009. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Research 69(10):4167–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich TA, Jain A, Tanner K, MacKay JL, Kumar S. 2010. Probing cellular mechanobiology in three-dimensional culture with collagen–agarose matrices. Biomaterials 31(7):1875–1884. [DOI] [PubMed] [Google Scholar]
- Unsworth BR, Lelkes PI. 1998. Growing tissues in microgravity. Nature Medicine 4(8):901. [DOI] [PubMed] [Google Scholar]
- Veeravalli KK, Rao JS. 2012. MMP-9 and uPAR regulated glioma cell migration. Cell Adhesion & Migration 6(6):509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinci M, Gowan S, Boxall F, Patterson L, Zimmermann M, Lomas C, Mendiola M, Hardisson D, Eccles SA. 2012. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biology 10(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Tong X, Yang F. 2014a. Bioengineered 3D brain tumor model to elucidate the effects of matrix stiffness on glioblastoma cell behavior using PEG-based hydrogels. Molecular Pharmaceutics 11(7):2115–2125. [DOI] [PubMed] [Google Scholar]
- Wang P, Chen S, Hung W, Paul C, Zhu F, Guan P, Huso D, Kontrogianni-Konstantopoulos A, Konstantopoulos K. 2015. Fluid shear promotes chondrosarcoma cell invasion by activating matrix metalloproteinase 12 via IGF-2 and VEGF signaling pathways. Oncogene 34(35):4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Guan P-P, Wang T, Yu X, Guo J-J, Konstantopoulos K, Wang Z-Y. 2014b. Interleukin-1β and cyclic AMP mediate the invasion of sheared chondrosarcoma cells via a matrix metalloproteinase-1-dependent mechanism. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1843(5):923–933. [DOI] [PubMed] [Google Scholar]
- Wartenberg M, Dönmez F, Ling FC, Acker H, Hescheler J, Sauer H. 2001. Tumor-induced angiogenesis studied in confrontation cultures of multicellular tumor spheroids and embryoid bodies grown from pluripotent embryonic stem cells. The FASEB Journal 15(6):995–1005. [DOI] [PubMed] [Google Scholar]
- Wolf K, Friedl P. 2011. Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends in Cell Biology 21(12):736–744. [DOI] [PubMed] [Google Scholar]
- Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. 2007. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nature Cell Biology 9(8):893. [DOI] [PubMed] [Google Scholar]
- Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. 2006. ROCK-and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Current Biology 16(15):1515–1523. [DOI] [PubMed] [Google Scholar]
- Zimmermann DR, Dours-Zimmermann MT. 2008. Extracellular matrix of the central nervous system: From neglect to challenge. Histochemistry and Cell Biology 130(4):635–653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental video 1. U87 cell migration on cell culture dish.
Supplemental video 3. U87 cell migration in 8S-StarPEG crosslinked collagen hydrogel.
Supplemental video 2. U87 cell migration in collagen hydrogel.


