Abstract
Nurr1/NR4A2 is an orphan nuclear receptor transcription factor implicated as a drug target for neurological disorders including Alzheimer’s and Parkinson’s diseases. Previous studies identified small molecule NR4A nuclear receptor modulators, but it remains unclear if these ligands affect transcription via direct binding to Nurr1. We assessed twelve ligands reported to affect NR4A activity for Nurr1-dependent and Nurr1-independent transcriptional effects and the ability to bind the Nurr1 ligand-binding domain (LBD). Protein NMR structural footprinting data show that amodiaquine, chloroquine, and cytosporone B bind the Nurr1 LBD; ligands that do not bind include C-DIM12, celastrol, camptothecin, IP7e, isoalantolactone, TMPA, and three high-throughput screening hit derivatives. Importantly, ligands that modulate Nurr1 transcription also show Nurr1-independent effects on transcription in a cell type-specific manner, indicating care should be taken when interpreting the functional response of these ligands in transcriptional assays. These findings should help focus medicinal chemistry efforts that desire to optimize Nurr1-binding ligands.
Graphical Abstract
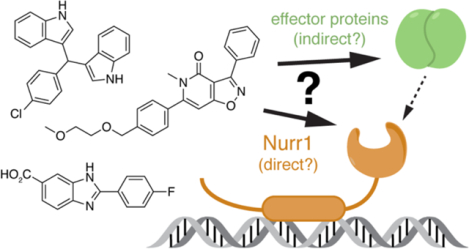
Introduction
The NR4A nuclear receptor transcription factors regulate important physiological processes including cellular homeostasis, metabolic regulation, apoptosis, and cell differentiation1. The three members of the NR4A family—NR4A1 (Nur77), NR4A2 (Nurr1), and NR4A3 (NOR1)—regulate the transcription of target genes through binding to specific genomic DNA response element sequences. Nuclear receptors are generally classified as ligand-dependent transcription factors. However, the NR4As have an unconventional putative orthosteric ligand-binding pocket compared to other nuclear receptors. Crystal structures of the Nur77 and Nurr1 ligand-binding domains (LBDs) show a collapsed orthosteric pocket that is filled with residues containing bulky hydrophobic sidechains suggesting these receptors may function independent of binding ligand within an orthosteric pocket2, 3. Furthermore, contributing to their status as orphan receptors, it remains unclear if the NR4As are regulated by binding physiological or endogenous ligands, although unsaturated fatty acids4–6, prostaglandins7, and dopamine metabolites8 have been shown to interaction with the Nur77 and/or Nurr1 LBDs.
Regulating Nurr1 activity with small molecule ligands is implicated to provide a therapeutic benefit in several Nurr1-related diseases including neurological disorders, such as Alzheimer’s and Parkinson’s diseases, inflammation, autoimmunity, cancer, and multiple sclerosis9–11. To develop therapies to treat these diseases, several groups have initiated studies to discover small molecules that modulate Nurr1 transcription. Among these studies, ligands with a 4-amino-7-chloroquinoline scaffold—including amodiaquine, chloroquine, and glafenine—were identified in a high-throughput screen as ligands that increase Nurr1 transcription in human neuroblastoma SK-N-BE(2)-C cells12. Amodiaquine improves behavioral alterations in a Parkinson’s disease animal model12 and improves neuropathology and memory impairment in an Alzheimer’s disease animal model13. NMR structural footprinting studies revealed the Nurr1 LBD binding epitopes of these ligands are similar to the binding epitopes of unsaturated fatty acids4, 5, 12.
Amodiaquine is the most potent and efficacious Nurr1 agonist of the 4-amino-7-chloroquinoline ligands, but these ligands are not Nurr1 specific; they also target other proteins including apelin receptor14 and are capable of antiviral15, 16 and antimalarial17 activity. However, knowledge that the 4-amino-7-chloroquinoline scaffold can directly bind to the Nurr1 LBD opens the path to future structure-activity relationship (SAR) studies to develop more potent and efficacious ligands with better specificity for Nurr1 over other molecular targets. Screening efforts have identified other classes of Nurr1-activating ligands with poorly defined mechanisms of action, and ligands reported to influence Nur77 activity represent another potential source to discover Nurr1 ligands since evolutionarily related nuclear receptors (e.g. ERα and ERβ) often display broad specificity for similar ligands.
It would be useful to know if the Nurr1- and Nur77-modulating ligands reported in the literature affect Nurr1 transcription through direct binding to Nurr1. We tested twelve ligands reported to affect the activity of Nurr1 or Nur77 for Nurr1-dependent transcription using cellular reporter assays and for direct binding to the Nurr1 LBD using a protein NMR structural footprinting assay that provides information on ligand binding epitopes. We found that most of the ligands display cell type-specific transcriptional activities through Nurr1-dependent and Nurr1-independent mechanisms. Furthermore, using protein NMR we found that only three of the ligands directly bind to the Nurr1 LBD: amodiaquine, chloroquine, and cytosporone B. These findings should be of interest to medicinal chemists that want to focus on the discovery and optimization of Nurr1-binding ligands as opposed to ligands that affect Nurr1 activity through other indirect mechanisms.
Results
NR4A ligand selection and properties
The twelve ligands we selected represent most, if not all, of the chemical scaffolds reported to modulate Nurr1- or Nur77-dependent transcription or other activities (Figure 1). Of these, nine were available from commercial sources and three others (SR10098, SR24237, and SR10658; compounds 1, 2, and 3, respectively) were synthesized in-house.
Figure. 1.
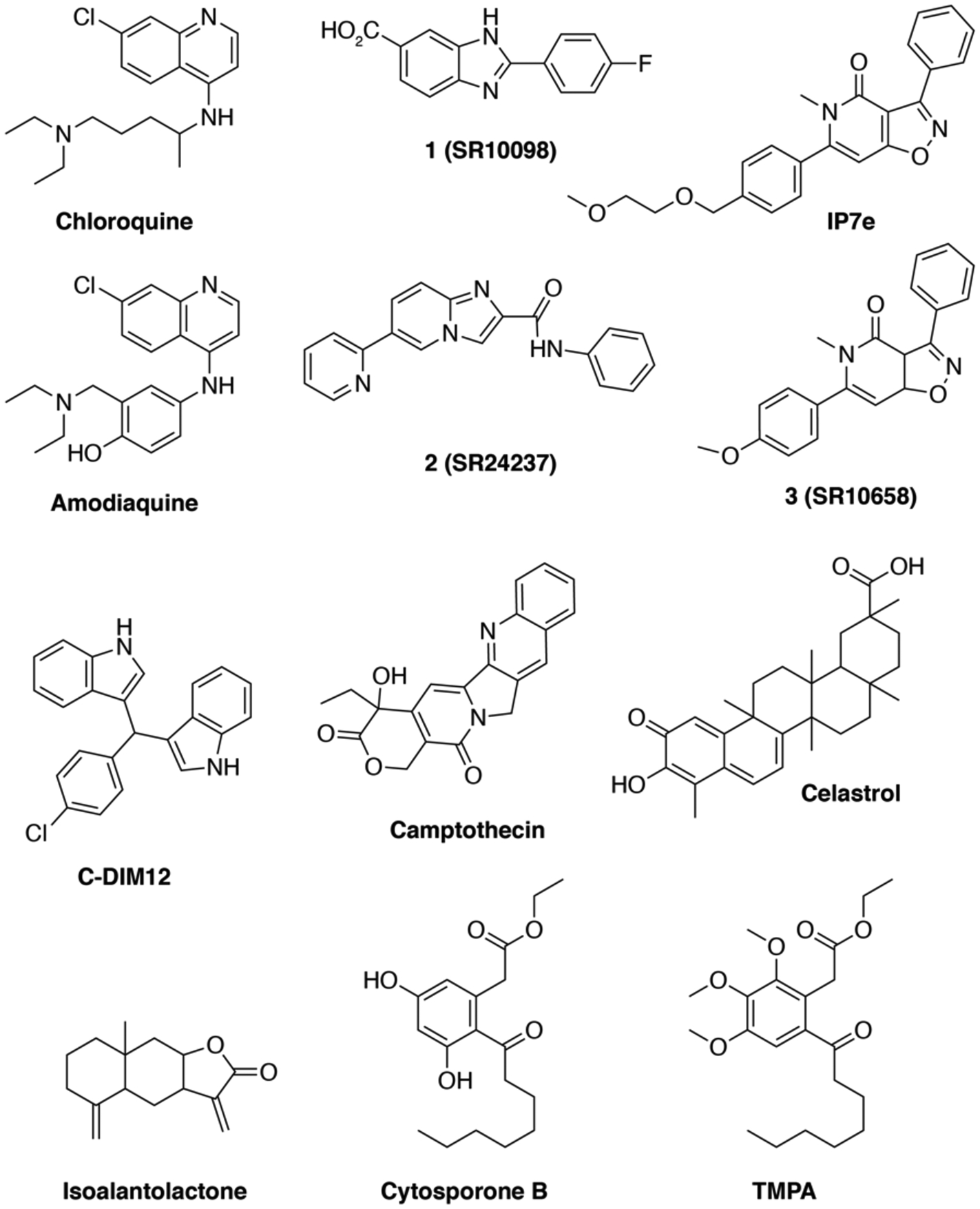
Chemical structures of the twelve NR4A ligands characterized in this study.
Amodiaquine and chloroquine are antimalarial ligands that contain a 4-amino-7-chloroquinoline scaffold and were identified in a screen using SK-N-BE(2)-C cells transfected with full-length Nurr1 and a luciferase reporter plasmid containing four copies of the NGFI-B response element (NBRE) motif12. These ligands are activators of Nurr1 transcription with micromolar potency that directly bind to the Nurr1 LBD at low nanomolar potency.
Four ligands were hits or optimized from a hit identified in different high-throughput screening (HTS) campaigns as ligands that increase Nurr1-dependent transcription with low nanomolar potency using a NBRE-based luciferase reporter assay. The benzimidazole SR10098 (compound 12 in18; full-length Nurr1 EC50 = 24 nM) and the isoxazolo-pyridinone SR10658 (compound 7a in19; full-length Nurr1 EC50 = 4.1 nM) were discovered in a screen using the MN9D dopaminergic cellular model; IP7e (full-length Nurr1 EC50 = 3.9 nM) is an analog of SR10658 reported to have improved solubility for in vivo studies and displayed in vivo efficacy in the experimental autoimmune encephalomyelitis mice (EAE) model of multiple sclerosis (MS)20. The imidazopyridine SR24237 (compound 3 in21; full-length Nurr1 EC50 = ~1–240 nM) is an optimized ligand from a hit discovered in two screens using Chinese hamster ovary (CHO) and a mouse neuronal N2A cell lines; an analog of SR24237 displays neuroprotective and anti-inflammatory activity.
Two additional ligands were previously reported as Nurr1 modulators. C-DIM12, or 1,1-bis(3’-indolyl)-1-(p-chlorophenyl)methane, was reported as a synthetic Nurr1 activator with micromolar potency that increases transcription in a Gal4-Nurr1 fusion assay, affects the expression of dopaminergic genes in pancreatic cells, keratinocyte epidermal cells, and primary neurons, and displays in vivo efficacy in models of Parkinson’s disease22–25. Camptothecin, an antitumor chemotherapeutic agent and cyclooxygenase-2 inhibitor, is a natural product identified in a high-throughput screen using a NBRE-based luciferase reporter assay in HEK293T cells as an inhibitor of Nurr1 transcription at 200 nM that triggers an antitumor response by reducing Foxp3+ T regulatory cells and inducing IFNγ+ T helper 1 cells indicating Nurr1 may be a target for cancer immunotherapy26.
We also tested four ligands reported to modulate the activity of Nur77. Celastrol is a natural product discovered in surface plasmon resonance screen that binds the Nur77 LBD with high affinity (Kd = 290 nM), inhibits Nur77 transcription at 500 nM in a reporter assay in HEK293T cells, and influences Nur77 activity through multiple mechanisms27. Isoalantolactone is a natural product discovered in a screen using a Gal4-Nur77 fusion reporter assay in MiaPaCa-2 cells as an inhibitor of Nur77 transcription at 5 μg/ml and activator of AMPKα in 3T3-L1 cells leading to a cascade of events that highlights a role for targeting Nur77 in protection against metabolic disorders and obesity28. Cytosporone B (CsnB) is a natural product identified as a Nur77 agonist that binds to the Nur77 LBD (LBD Kd = 1.5 μM) and activates Nur77 transcription (EC50 = ~0.1–0.3 nM)29. CsnB was also shown to activate transcription in a Gal4-Nurr1 fusion assay in BGC-823 human gastric carcinoma cells. Later work using another chemical screen identified an analog of CsnB, ethyl 2-[2,3,4-trimethoxy-6-(1-octanoyl)phenyl]acetate (TMPA), which showed low micromolar affinity for the Nur77 LBD30. Unlike CsnB, TMPA does not function as a canonical agonist of Nur77 transcription; rather, it inhibits the interaction between Nur77 and Liver kinase B1 (LKB1), and it also enhances phosphorylation of AMPKα. Another CsnB analog that was not commercially available, PDNPA, inhibits the interaction between Nur77 and the MAP kinase p38α31, suggesting inhibition of Nur77-kinase interactions may be a general mode of action for these ligands.
Cellular assays to characterize ligand-mediated transcription of Nurr1
Nurr1 regulates gene expression as a monomer through binding to NGFI-B response element (NBRE) motifs32, which are present in the majority of well characterized Nurr1 genomic binding sites including the promoter region of the tyrosine hydroxylase (TH)33, 34 and topoisomerase IIβ (topIIβ)35. Most of the ligands in Figure 1 were discovered or validated using a monomeric NBRE luciferase (NBRE-luc) reporter plasmid. Another motif termed the Nur response element (NurRE) was identified in the promoter of the pro-opiomelanocortin (POMC) gene36 that enables binding of Nur77 or Nurr1 dimers37, 38. Furthermore, Nurr1 can form a heterodimer with retinoid X receptor (RXR) on a DR5 motif that contains two consensus nuclear receptor binding elements separated by a 5 nucleotide linker39. To the best of our knowledge, no Nurr1 target genes have been reported to contain DR5 motifs. However, RXR ligands can activate transcription of Nurr1 monomers on NBRE motifs40.
Given these observations, we assessed the effect of the ligands on Nurr1-mediated transcription by cotransfecting full-length Nurr1 with a luciferase reporter plasmid containing three repeats of the NBRE or NurRE response element (NBRE3-luc and NurRE3-luc, respectively). Because RXR influences transcription of Nurr1 monomers, we also tested the effect of cotransfection of full-length Nurr1 and full-length RXRα together. We performed the assays in three cell lines including HEK293T, a kidney embryonic cell line commonly used to assess general nuclear receptor activity, as well as two cell lines relevant to Nurr1 functions in neurons: PC12, a rat pheochromocytoma cell line exhibiting neuronal-like characteristics; and SK-N-BE(2)-C, a neuroblastoma cell line displaying moderate levels of tyrosine hydroxylase activity and dopamine-b-hydroxylase activity. We used four ligand concentrations in the assays, which differ among all the ligands; based on previously reported cellular potencies in the original reports of each ligand as well as initial studies that included higher concentrations that for some ligands gave bell shaped response curves or a significant decrease in luciferase activity, indicative of colloidal aggregation and/or toxicity.
We also performed two control assays to determine the specificity of the ligands and assess Nurr1-dependent and Nurr1-independent effects on transcription using the same ligand concentrations tested in the NBRE and NurRE luciferase assays. First, we used a VP16-Gal4 control assay to test for toxicity and overall effects on general transcription; cells are transfected with a reporter plasmid containing five copies of the yeast Gal4 upstream activation sequence (UAS) followed by the firefly luciferase gene (UAS-luc) along with an expression plasmid containing the yeast Gal4 DNA-binding domain fused to the herpes simplex virus protein VP16 activation domain. The Gal4-VP16 fusion protein displays constitutively high luciferase activity; ligands that show decreased activity may either display cytotoxicity or inhibit general transcription in a Nurr1-independent manner, whereas ligands that show increased activity activate general transcription in a Nurr1-indepenent manner. Second, we also tested the ligands using the CellTiter-Glo luminescence cell viability assay as a more direct measure of cytotoxicity. For each assay type (reporter or control), we displayed the same y-axis range corresponding to the fold change in luciferase activity normalized to vehicle-treated cells in order to directly compare the magnitude of the activity changes between different ligands in the same assay.
Characterization of ligands in the cellular assays
For the 4-amino-7-chloroquinoline scaffold ligands, amodiaquine (Figure 2A) and to a lesser extent chloroquine (Figure 2B) increased activity of the NBRE-luc (Nurr1 and Nurr1-RXRα) and NurRE-luc (Nurr1) reporters in SK-N-BE(2)-C cells, consistent with previously published data12. However, decreases in luciferase signal were observed in HEK293T and PC12 cells, respectively. Both ligands increased activity of the Gal4-VP16/UAS-luc control assay in SK-N-BE(2)-C cells at concentrations up to ~15 μM and abruptly decreased activity at 125 μM without showing some cytotoxicity activity in the CellTiter-Glo assay. Taken together, these data indicate amodiaquine and chloroquine affect general transcription through Nurr1-independent mechanisms.
Figure. 2.
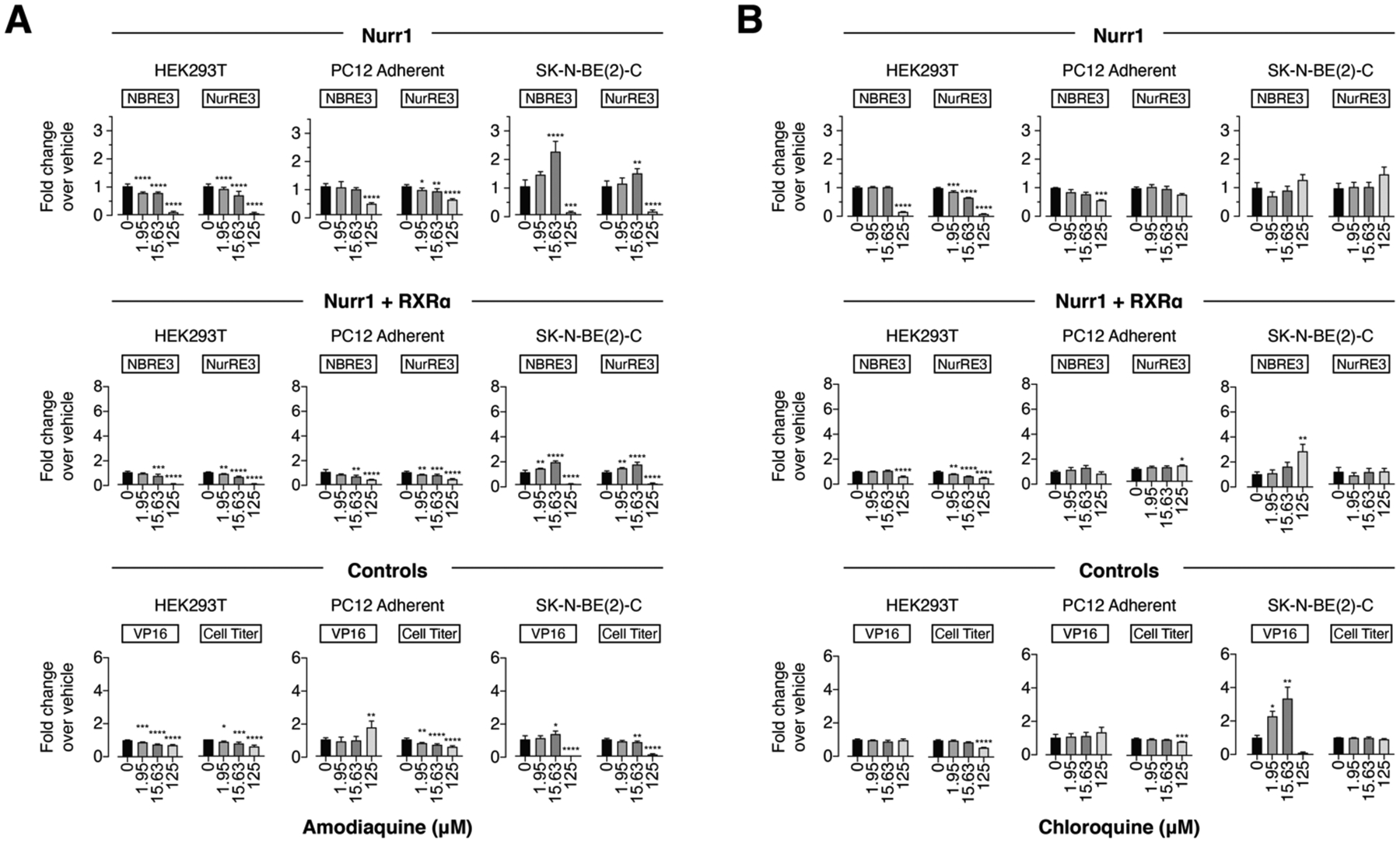
Effect of the 4-amino-7-chloroquinoline derivatives (A) amodiaquine and (B) chloroquine on transcription of full-length Nurr1 or Nurr1-RXRα using NBRE3-luc or NurRE3-luc reporters in HEK293T, PC12, and SK-N-BE(2)-C cells; as well as control assays including a constitutively active Gal4-VP16/UAS-luc assay and a CellTiter-Glo toxicity assay. Data represent mean ± s.e.m. (n=4); *P< 0.05, **P< 0.01, ***P< 0.001, ****P ≤ 0.0001, one-way ANOVA with multiple comparisons to vehicle control in each condition.
For the HTS hits, SR10098 (Figure 3A) slightly increased luciferase activity of both reporters under certain conditions in HEK293T cells (Nurr1 and Nurr1-RXRα), SK-N-BE(2)-C cells (Nurr1, NBRE-luc and NurRE-luc; Nurr1-RXRα, NurRE-luc), and PC12 cells (Nurr1-RXRα, NurRE-luc). SR24237 (Figure 3B) and SR10658 (Figure 3C) showed slight to moderate dose-responsive increased activity in most conditions. IP7e (Figure 3D) only showed increased activity at the highest concentration tested (100 nM) in most conditions. However, all four of these HTS ligands increased the activity of the Gal4-VP16/UAS-luc control assay in the same cell lines and conditions that showed activation in the NBRE-luc and NurRE-luc assays without showing any significant cytotoxicity activity in the CellTiter-Glo assay, indicating they affect general transcription via Nurr1-independent mechanisms.
Figure 3.
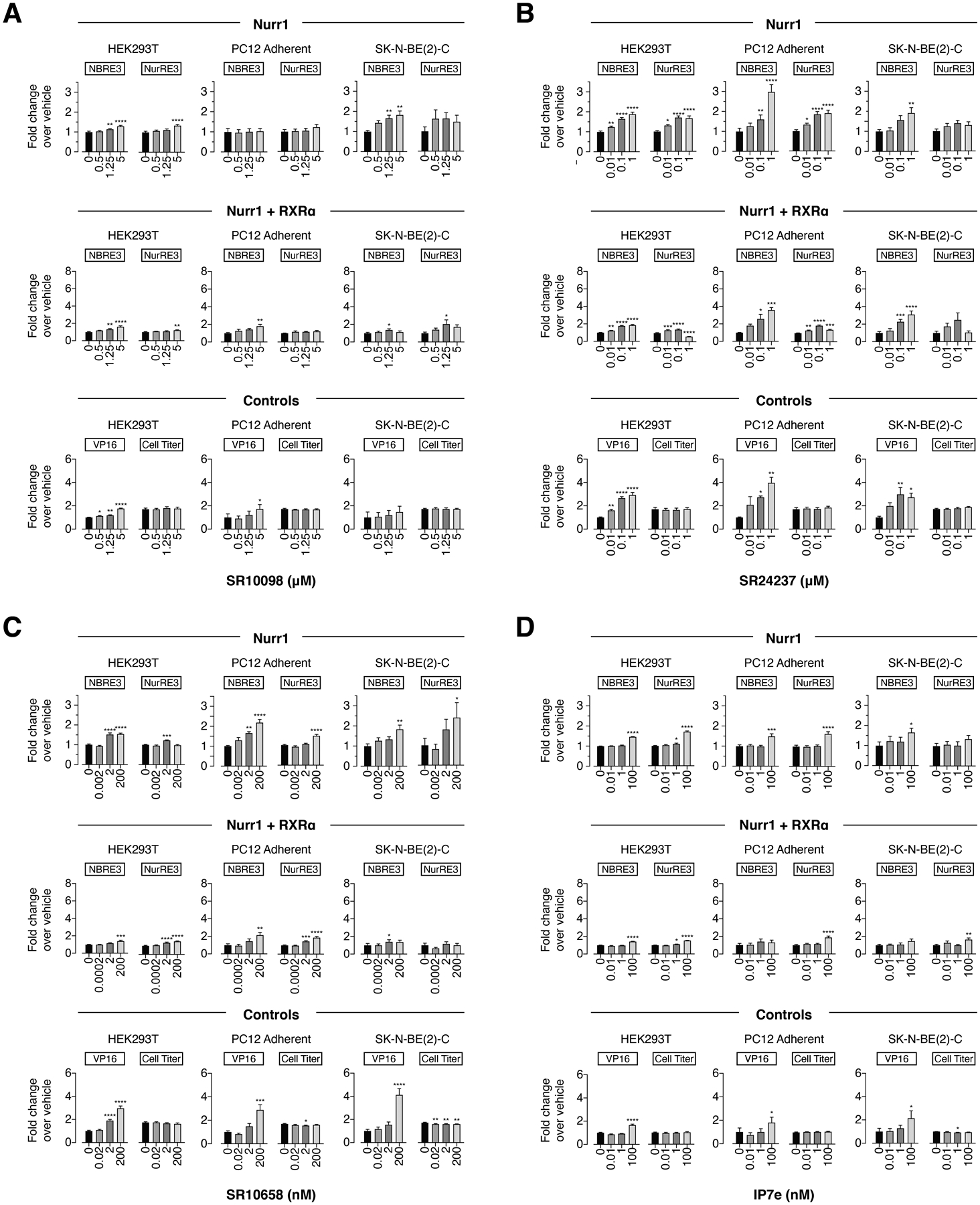
Effect of the Nurr1 HTS derived ligands (A) SR10098, (B) SR24237, (C) SR10658, and (D) IP7e on transcription of full-length Nurr1 or Nurr1-RXRα using NBRE3-luc or NurRE3-luc reporters in HEK293T, PC12, and SK-N-BE(2)-C cells; as well as control assays including a constitutively active Gal4-VP16/UAS-luc assay and a CellTiter-Glo toxicity assay. Data represent mean ± s.e.m. (n=4); *P< 0.05, **P< 0.01, ***P< 0.001, ****P ≤ 0.0001, one-way ANOVA with multiple comparisons to vehicle control in each condition.
For the Nurr1 modulators, C-DIM12 (Figure 4A) showed decreased activity in HEK293T and SK-N-BE(2)-C cells in most conditions (Nurr1 and Nurr1-RXRα, NBRE-luc and NurRE-luc) except in the Nurr1-RXRα/NBRE3-luc assay where there was an increase and decrease in activity, and relatively no effect in PC12 cells except for Nurr1-RXRα/NBRE3-luc. Camptothecin (Figure 4B) showed decreased activity in most conditions except in PC12 cells (Nurr1, NurRE-luc). For the most part, both ligands also showed decreased activity of the Gal4-VP16/UAS-luc control assay and/or CellTiter-Glo activity in the same cell lines and conditions that showed decreased activity in the NBRE-luc and NurRE-luc assays, indicating that Nurr1-independent and cytotoxic mechanisms contribute to activity of these ligands.
Figure 4.
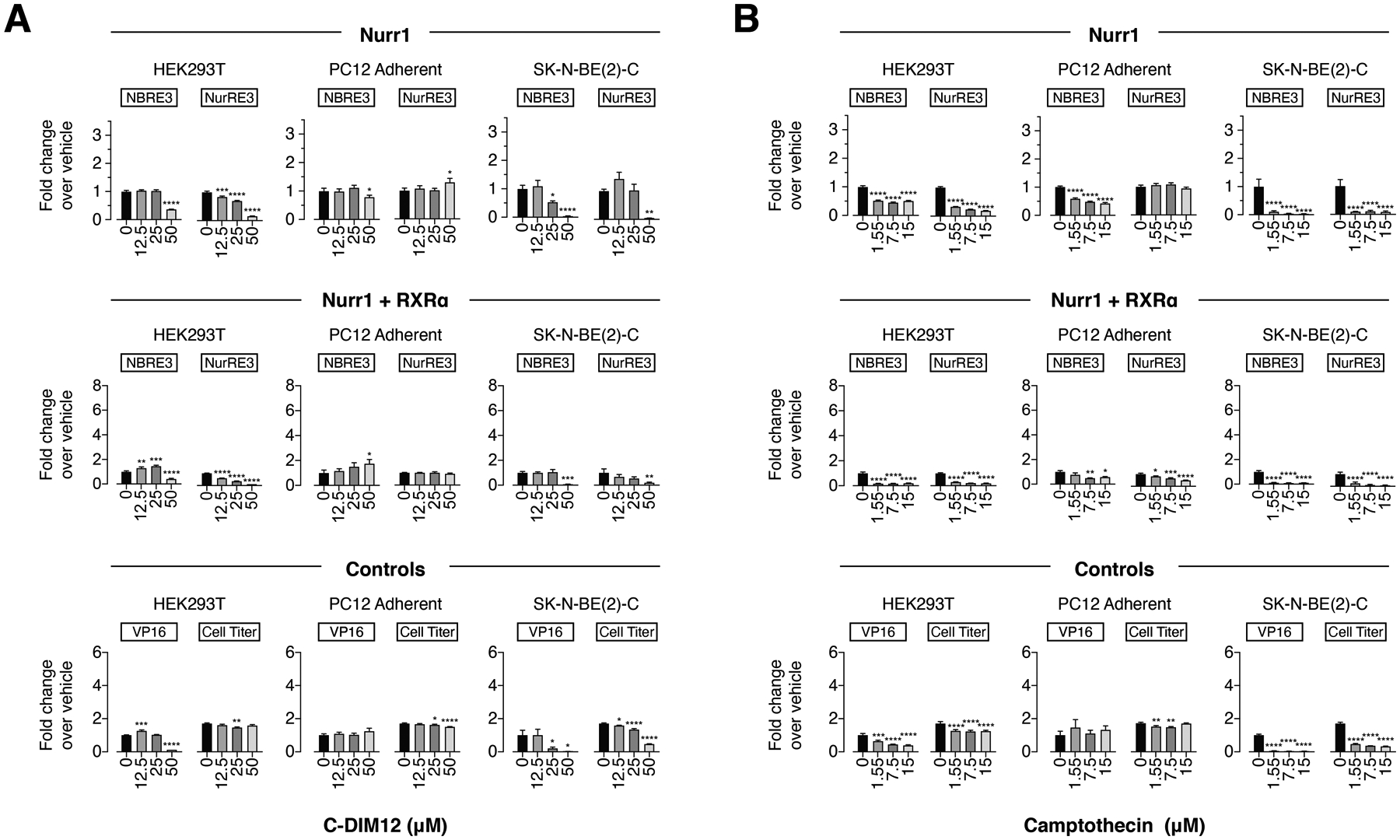
Effect of the other Nurr1 modulators (A) C-DIM12 and (B) camptothecin on transcription of full-length Nurr1 or Nurr1-RXRα using NBRE3-luc or NurRE3-luc reporters in HEK293T, PC12, and SK-N-BE(2)-C cells; as well as control assays including a constitutively active Gal4-VP16/UAS-luc assay and a CellTiter-Glo toxicity assay. Data represent mean ± s.e.m. (n=4); *P< 0.05, **P< 0.01, ***P< 0.001, ****P ≤ 0.0001, one-way ANOVA with multiple comparisons to vehicle control in each condition.
Finally, for the Nur77 modulators, Celastrol (Figure 5A) showed increased or decreased activity in a cell type-specific and reporter-specific manner, affecting the Nurr1 and Nurr1-RXRα assays similarly. Isoalantolactone (Figure 5B) showed decreased activity in HEK293T and SK-N-BE(2)-C cells (Nurr1 and Nurr1-RXRα, NBRE-luc and NurRE-luc), and a slight decrease in activity for Nurr1/NurRE-luc in PC12 cells under any condition. Cytosporone B (Figure 5C) showed decreased activity for some conditions at the highest 1–2 concentrations tested (25 μM and/or 100 μM), whereas TMPA (Figure 5D) showed relatively no activity under all conditions. Notably, all four ligands showed decreased activity of the Gal4-VP16/UAS-luc control assay and/or CellTiter-Glo activity in the same cell lines and conditions that showed decreased activity in the NBRE-luc and NurRE-luc assays, indicating that Nurr1-independent and cytotoxic mechanisms contribute to activity of these ligands.
Figure 5.
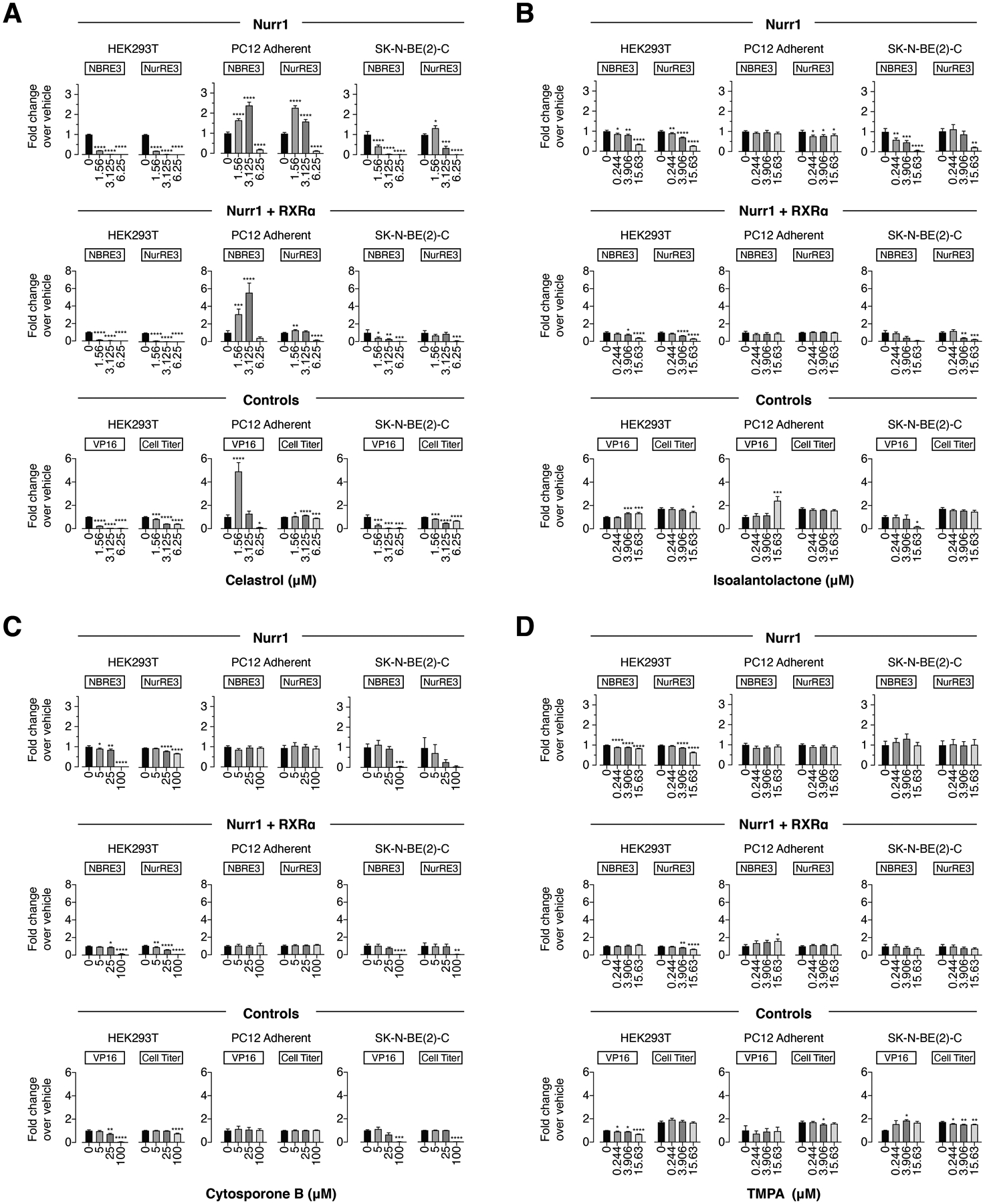
Effect of the Nur77 modulators on transcription of (A) celastrol, (B) isoalantolactone, (C) cytosporone B, and (D) TMPA on full-length Nurr1 or Nurr1-RXRα using NBRE3-luc or NurRE3-luc reporters in HEK293T, PC12, and SK-N-BE(2)-C cells; as well as control assays including a constitutively active Gal4-VP16/UAS-luc assay and a CellTiter-Glo toxicity assay. Data represent mean ± s.e.m. (n=4); *P< 0.05, **P< 0.01, ***P< 0.001, ****P ≤ 0.0001, one-way ANOVA with multiple comparisons to vehicle control in each condition.
Binding analysis by protein NMR structural footprinting
The notion that all of the ligands tested in the cellular assays show Nurr1-independent mechanisms on general transcription raise a question as to whether they physically interact with and bind to the Nurr1 LBD as opposed to binding upstream effector proteins such as kinases that in turn regulate Nurr1 transcription through downstream cellular functions. We therefore used a protein NMR spectroscopy structural footprinting assay to determine if the NR4A ligands directly bind to the Nurr1 LBD. We collected 2D [1H,15N]-TROSY-HSQC of 15N-labeled Nurr1 LBD in the presence of vehicle control or 2 molar equivalents of ligand (Figure 6).
Figure 6.
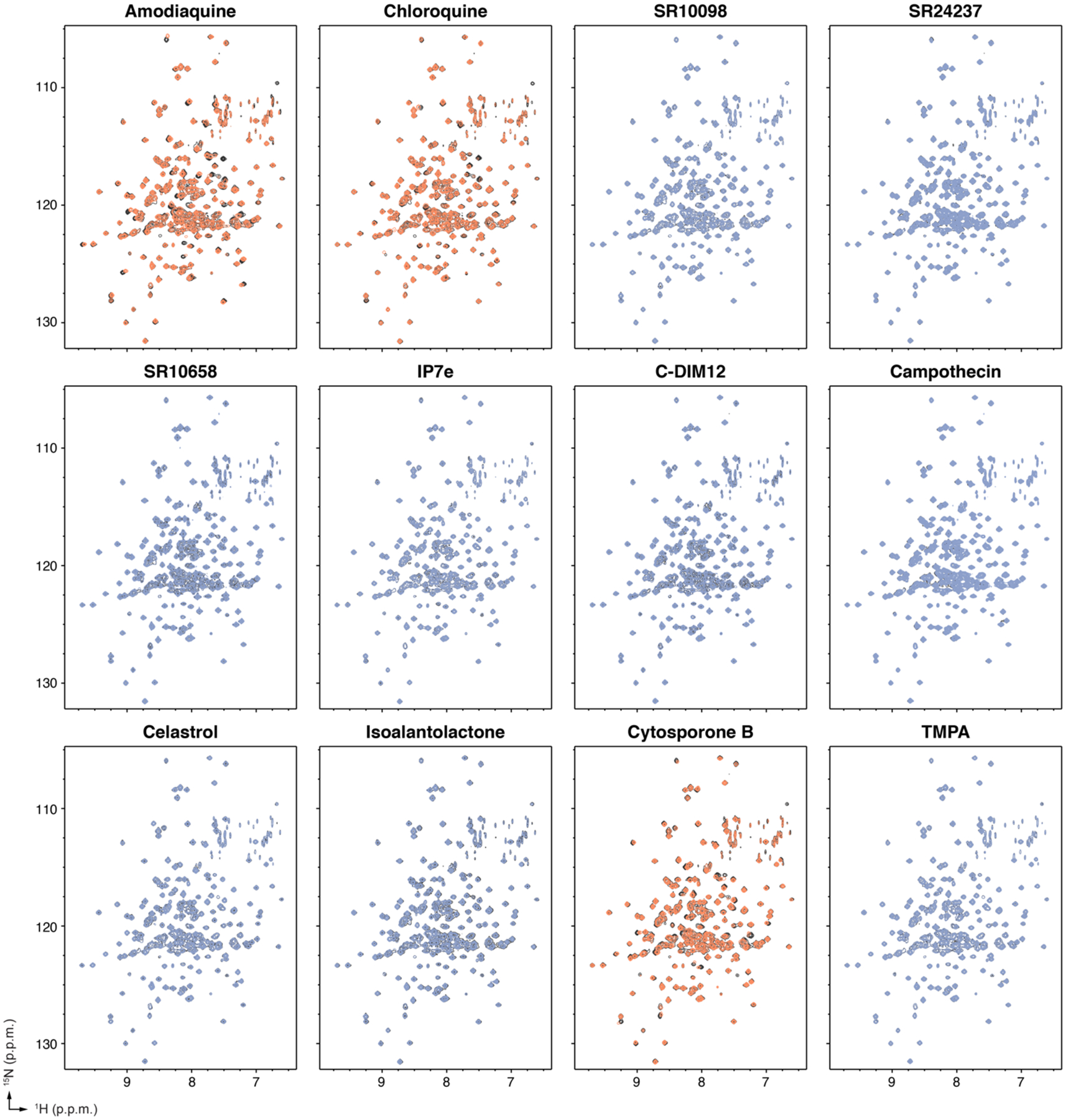
Protein NMR spectroscopy ligand footprinting data. 2D [1H,15N]-TROSY-HSQC data (700 MHz, 298K) of 15N-labeled Nurr1 LBD in the presence of vehicle control (black spectra) or 2X ligand reveals ligands that do not bind to the Nurr1 LBD (blue spectra) and ligands that bind to the Nurr1 LBD (orange spectra).
To provide a direct comparison of the relative residue-specific binding effects caused by the twelve ligands, we calculated a mean and standard deviation of the CSP changes (changes in chemical environment caused by structural changes upon binding) and peak intensity changes (NMR line broadening indicative of changes in protein dynamics and/or exchange effects due to residues involved in ligand binding) for all residues and ligands grouped together (Figure 7). Then for each ligand we plotted the per-residue NMR chemical shift perturbation (CSP) changes (Figure 8A) and peak intensity changes (Figure 8B).
Figure 7.
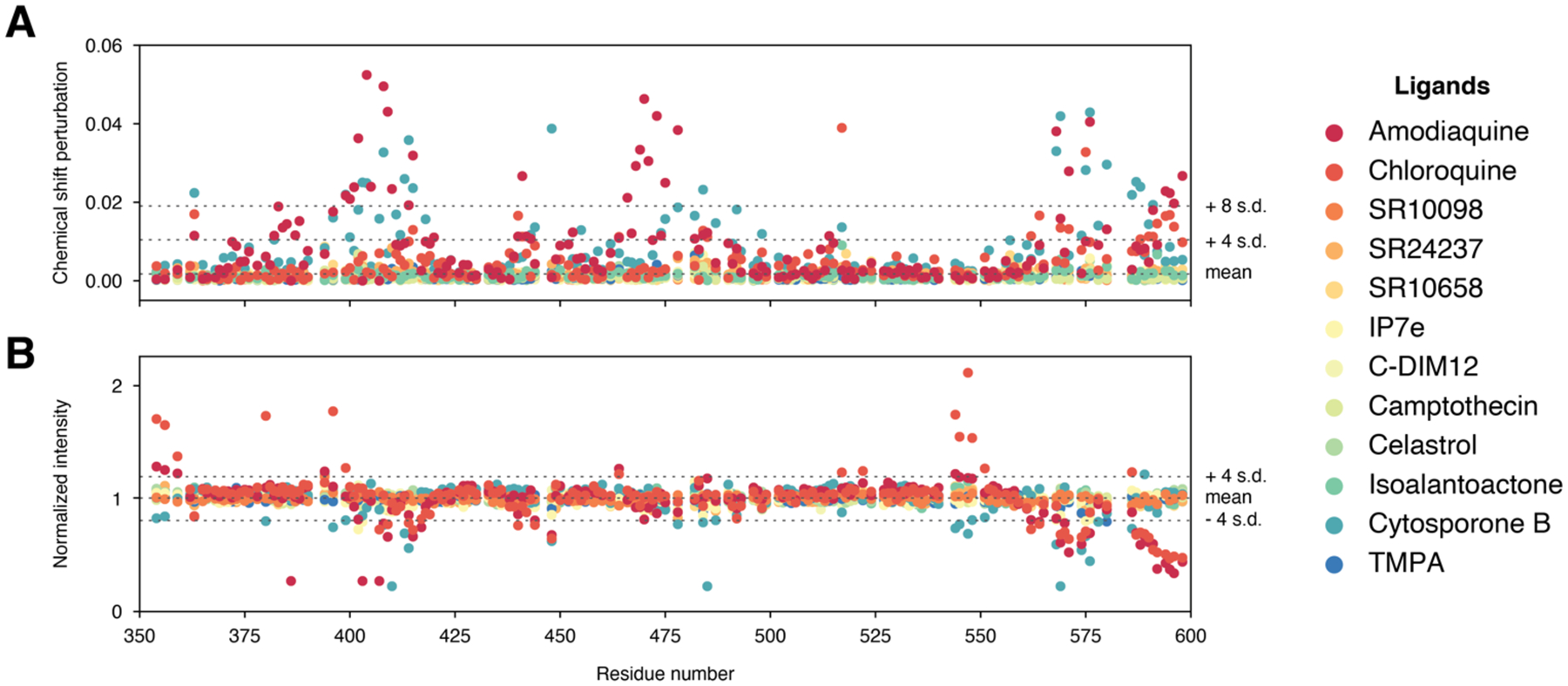
Grouped quantitative analysis of the 2D NMR ligand footprinting data. (A) NMR chemical shift perturbations and (B) peak intensities were used determine the group mean and standard deviation cutoffs to highlight significant changes in the per-ligand quantitative analysis.
Figure 8.
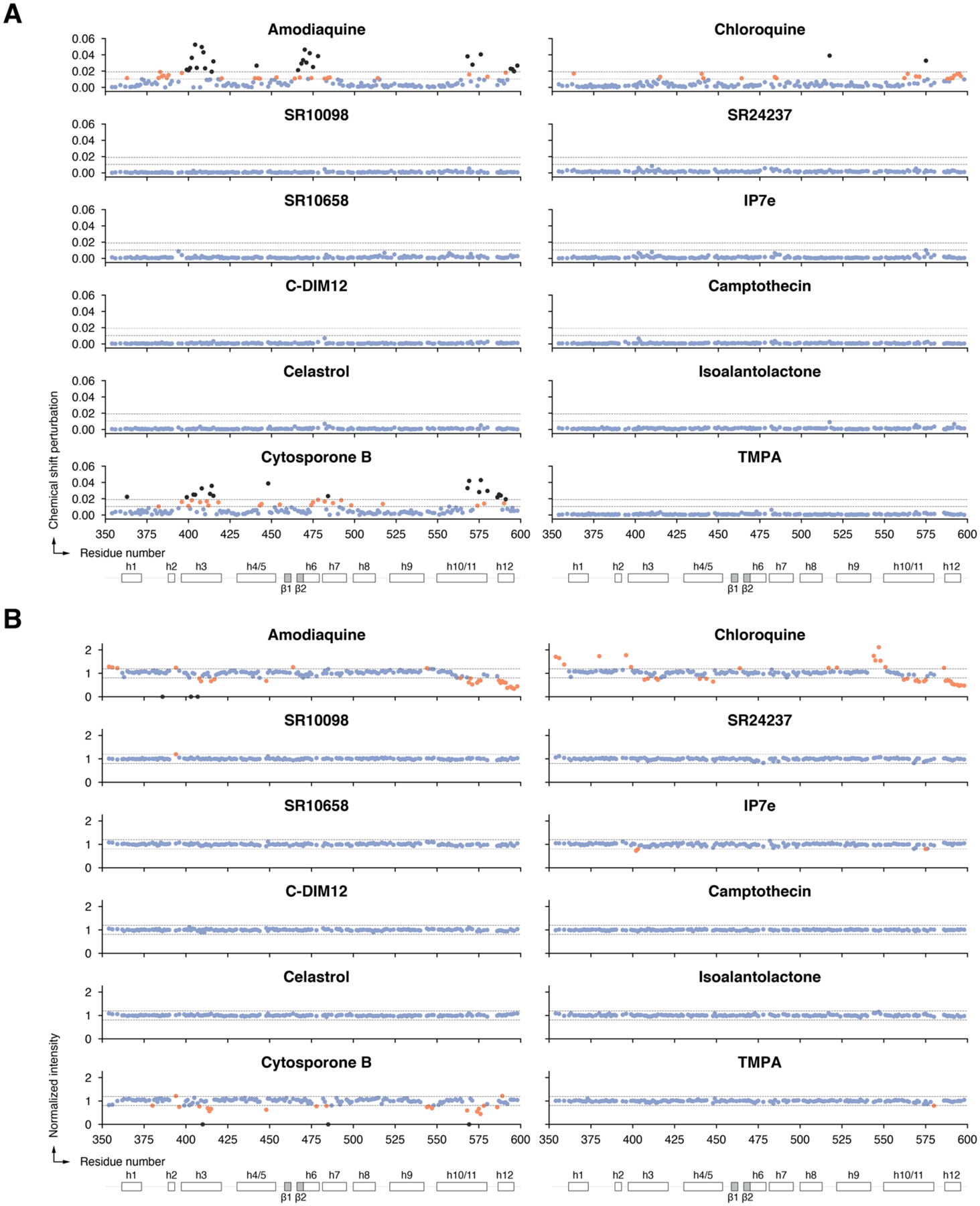
Per-ligand quantitative analysis of the 2D NMR ligand footprinting data, including (A) differential NMR chemical shift perturbation analysis and (B) NMR peak intensity analysis. Blue circles, residues with no significant effect; residues with a CSP or peak intensity change ≥ mean ± 4 s.d. or 8 s.d. are colored orange or black, respectively; residues with peak intensities that decrease or disappear into the noise are colored orange or black, respectively.
Consistent with previous data4, 12, addition of amodiaquine and to a lesser degree chloroquine, which is less potent than amodiaquine12, showed select NMR CSP and peak intensity changes for Nurr1 LBD residues within helix 3, helix 6, helix 10/11 and helix 12, indicating they directly bind to the Nurr1 LBD. Of the other ten NR4A ligands, only cytosporone B showed NMR structural footprinting results indicating direct binding to the Nurr1 LBD. In contrast, SR10098, SR24237, SR10658, IP7e, C-DIM12, camptothecin, celastrol, isoalantolactone, and TMPA showed no evidence of binding to the Nurr1 LBD. The profile of cytosporone B-induced CSP and peak intensity changes are similar to the changes caused by amodiaquine, indicating they likely share a common binding site within the Nurr1 LBD.
To visualize the NMR-detected binding effects, we mapped the NMR structural footprinting changes for amodiaquine, chloroquine, and cytosporone B, onto the crystal structure of Nurr1 LBD2 (Figure 9A) and compared the binding epitopes to the crystal structure of TMPA-bound Nur77 LBD30, PDNPA-bound Nur77 LBD31, and the Nur77 modeled binding sites of cytosporone B29 and celastrol27 relative to the canonical ligand-binding pocket (Figure 9B). Amodiaquine, chloroquine, and cytosporone B show similar binding epitopes on the Nurr1 LBD, which most of the NMR-detected changes occurring for residues within or near the canonical orthosteric-ligand binding pocket. For Nur77, the crystallized TMPA binding poses and modeled celastrol and cytosporone B binding poses are all surface exposed.
Figure 9.
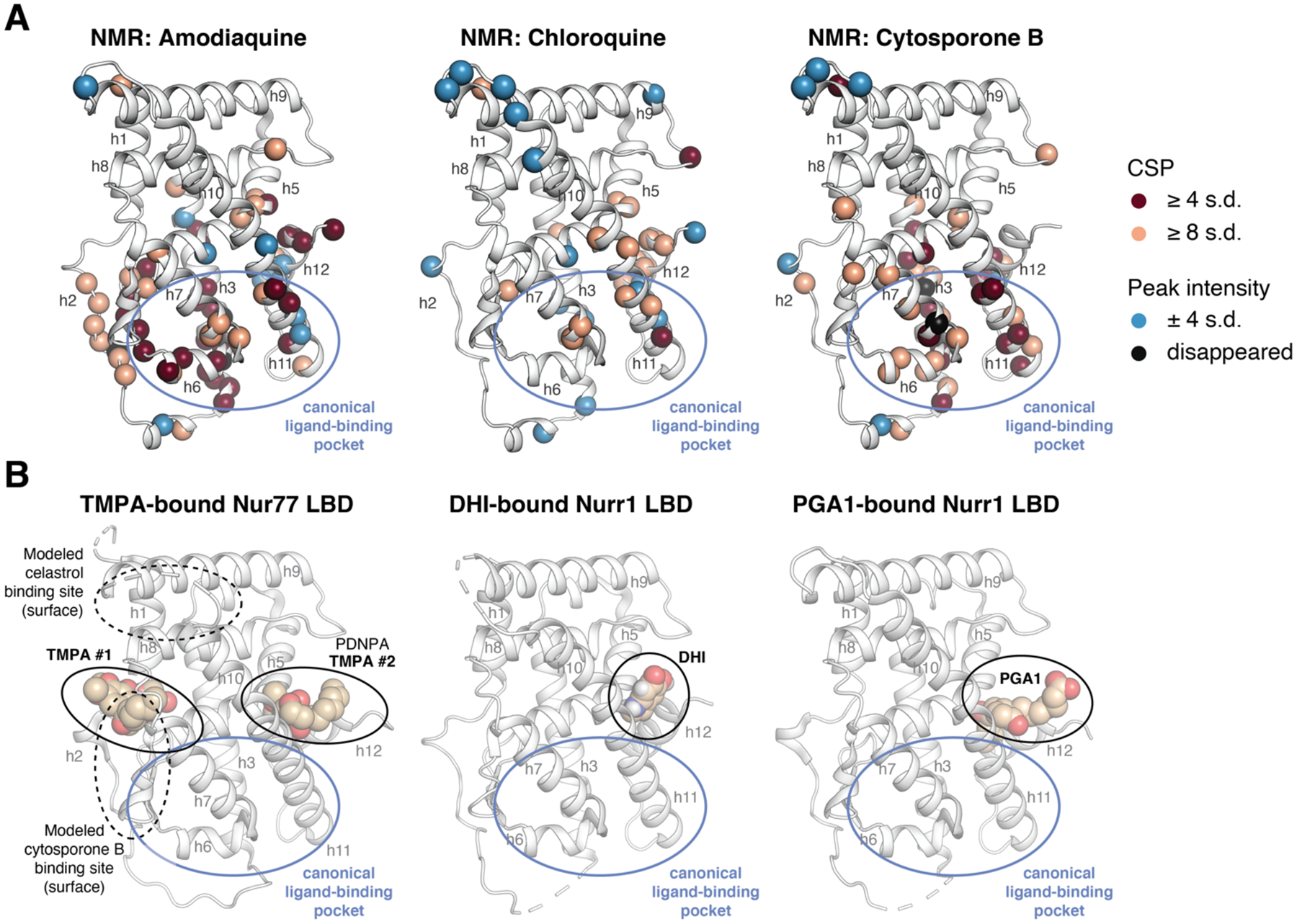
Comparison of NMR structural footprinting data to ligand-bound NR4A LBD crystal structures. (A) NMR mapped onto the Nurr1 LBD crystal structure (PDB 1OVL, chain B). The inset legend shows the coloring scheme for residues with significant NMR CSP or peak intensity values. (B) Crystal structures of TMPA-bound Nur77 LBD (PDB 3V3Q) with other modeled and crystallized Nur77 ligand binding sites reported in the literature highlighted, as well as Nurr1 LBD crystal structures bound to 5,6-dihydroxyindole (DHI; PDB 6DDA, chain A) and prostaglandin A1 (PGA1; PDB 5Y41, chain A).
Although there is some overlap between the surface exposed Nur77 interaction sites and the Nurr1 NMR-detected binding epitopes, there are also differences. For example, cytosporone B and TMPA are derived from the same scaffold, and the modeled cytosporone B interaction site agrees with one of the two crystallized TMPA binding modes—a solvent exposed surface in the Nur77 LBD. However, the NMR-detected cytosporone B binding epitope on Nurr1 is different, suggesting the interaction occurs within the canonical ligand-binding pocket similar to amodiaquine, chloroquine, and unsaturated fatty acids4, 5, 12. These data suggest that cytosporone B likely binds differently to Nurr1 and Nur77, but another explanation could be that the solution NMR structural footprinting analysis picks up on binding events or structural changes that are not apparent in solid state crystallography studies.
Characterization of binding ligands for NR4A specificity
Finally, we performed cellular luciferase reporter assays using full dose responses for amodiaquine, chloroquine, and cytosporone B to obtain insight into the activity of these three Nurr1-binding compounds against all three NR4A receptors. We cotransfected HEK293T, PC12, and SK-N-BE(2)-C cells with full-length Nurr1, Nur77, NOR-1, or empty control plasmids along with the NBRE3-luc or NurRE3-luc reporter plasmids in the absence and presence of full-length RXRα (Figures S1–S3). Notably, the ligands all show complicated dose response curves due to a combination of cellular toxicity issues (Figures 2–5, Cell-Titer Glo data) and NR4A-independent non-specific transcriptional effects (Figures 2–5, VP16-Gal4 control data). Furthermore, the ligand dose response profiles look nearly identical for cells transfected with Nurr1, Nur77, NOR-1, or an empty control plasmid—the latter of which, when considered with the Cell-Titer Glo and VP16-Gal4 control data, indicates the luciferase reporter dose response profiles are not NR4A-dependent. We therefore did not attempt to fit these data to extract EC50/IC50 values given that the potency and efficacy responses in the dose response experiments appear to be dominated by NR4A-independent contributions; examples could include binding to an off-target (non-NR4A) protein, cell death, fluorescent assay artifacts, etc.
Discussion
Defining if and how ligands bind to Nurr1 is critical not only for understanding Nurr1 function and regulation but also in prioritizing and directing medicinal chemistry efforts on Nurr1-binding ligands. Relative to other nuclear receptors, crystal structures have not revealed a well-defined ligand-binding pocket in the Nurr1 LBD. Solution-state structural studies indicate the Nurr1 ligand-binding pocket is dynamic and solvent accessible, indicating the absence of a pocket captured in Nurr1 LBD crystal structures represents a collapsed conformation4. The lack of a well-defined Nurr1 ligand binding pocket has arguably stunted efforts to discover and design Nurr1-binding ligands. Several ligands have been reported in the literature to interact with the Nurr1 LBD. Unsaturated fatty acids, amodiaquine, and chloroquine appear to bind to the Nurr1 orthosteric ligand-binding pocket4, 5, 12, whereas the endogenous dopamine metabolite 5,6-dihydroxyindole (DHI) and prostaglanding A1 (PGA1) covalently bind to noncanonical site via covalent attachment to a surface-exposed cysteine residue on helix 11 in the Nurr1 LBD8.
Our studies here show that most of the NR4A ligands we studied that influence the cellular functions of Nurr1 do not appear to function through direct binding to Nurr1. These findings indicate that these ligands may exert their effects on Nurr1 activity through binding to upstream effector proteins such as kinases, which could then affect Nurr1 cellular activity via downstream effects. Several other observations support this idea.
First, most if not all of the NR4A ligands show cell type-specific functions. It is possible this is due to the availability of different transcriptional coregulator proteins present within different cell types that could be recruited to Nurr1 in a ligand-dependent manner. However, given that most of the NR4A ligands do not directly bind Nurr1, it is possible that cell type-specific expression of effector proteins upstream of the NR4As or other non-NR4A proteins also contributes to the cell type-specific activities of the ligands. Cell type-specific Nurr1 activity has been highlighted previously in neuronal cell lines, suggesting that endogenous factors expressed in specific neuronal cell lines influence Nurr1 activation41. Cell type-specific dependence may apply to other types including bladder cancer cells and other human cancer cells where Nurr1 activity has been found to be critical for survival25, 42, 43.
Second, some of these NR4A ligands are polypharmacology modulators. As one example, C-DIM12 and related analogs were reported as Nurr1 activators in pancreatic cancer cells22, bladder cancer cells25, and neuronal cells23, 24; a Nurr1 inhibitor in glioblastoma cells44; and activates other nuclear receptors in various cell types including Nur7745, 46, COUP-TF147 and PPARγ48–50. Indeed, previous studies suggest that the mechanisms of action of these C-DIM ligands may occur independent of nuclear receptor binding or via nuclear receptor-independent mechanisms through affecting kinase activity46, 51–56. In silico ligand docking studies suggested that C-DIM12 may bind to the Nurr1 LBD coregulator binding surface24. However, our protein NMR structural footprinting data clearly show C-DIM12 does not directly bind to the Nurr1 LBD. Further selectivity profiling such as chemoproteomic methods is warranted to determine the molecular target of C-DIM12 and related analogs.
Related to polypharmacology, some of the other NR4A ligands that we profiled that do not directly bind to the Nurr1 LBD but activate Nurr1 transcription have been shown to function through targets other than the NR4As, which in principle could affect Nurr1 activity through downstream functions of the targets. Celastrol, which contains a reactive quinone methide moiety enabling covalent attachment to cysteine residues57, including direct binding and inhibition of c-Myc-Max heterodimers58, cancerous inhibitor of protein phosphatase 2A (CIP2A)59, STAT360, SHOC-2 to inhibit ERK signaling61, IKKα and IKKβ62, and HSP90-chaperone interactions63–65; directly binds nearly 70 protein targets in a proteome microarray assay61; and affects other cellular signaling pathways including protein phosphatase 2A-Akt, AMPK, and WNT/β-catenin66. Isoalantolactone activates AMPKα28 and inhibits STAT367 and IKKβ68. Camptothecin inhibits topoisomerase I69. Given that the other Nurr1 activators identified in HTS screens or derived from HTS hits (SR10098, SR10658 and the related analog IP7e, and SR24237) do not directly bind to the Nurr1 LBD, these ligands likely target other effector proteins that influence Nurr1 activity or other general transcriptional machinery since some showed activity in the VP16-Gal4 assay. This concept of ligands affecting Nurr1 activity via binding to upstream effectors of Nurr1 is supported by a study showing that kinase inhibitors can activate and inhibit Nurr1 transcription70. Thus, kinases could act as upstream effector proteins on downstream Nurr1 activities.
Amodiaquine and chloroquine were two of the three ligands in our panel of twelve NR4A ligands that physically bind to the Nurr1 LBD and activated Nurr1 transcription in a screen using SK-N-BE(2)-C neuronal cells12. However, we also found that amodiaquine and chloroquine activated transcription in the VP16-Gal4 assay in SK-N-BE(2)-C cells, indicating they also have Nurr1-independent effects on transcription. Amodiaquine is well known as an antimalarial drug mostly used against strains of Plasmodium falciparum71, but it also inhibits human histamine N-methyltransferase72 and several human cytochrome P450 enzymes73. Chloroquine, which is also an antimalarial drug, is a chemokine receptor CXCR4 antagonist74 and used as anticancer agent capable of inhibiting autophagy by disrupting the fusion of autophagosomes with lysosomes75, 76. Future work on this 4-amino-7-chloroquinoline scaffold may result in the development of direct Nurr1-binding ligands with better specificity towards Nurr1 and reduced general effects on transcription. In fact, the observation that amodiaquine shows more efficacious activity over chloroquine suggests there is the possibility to develop structure-activity relationship (SAR) on this scaffold77.
Our NMR studies show that cytosporone B, which was the first identified Nur77 agonist and shown to activate a Gal4-Nurr1 LBD fusion construct in BGC-823 human gastric cancer cells29 directly binds to the Nurr1 LBD; however, the related analog TMPA that binds to the Nur77 LBD30 does not bind to the Nurr1 LBD. We found that cytosporone B did not activate Nurr1 transcription in HEK293T cells or the PC12 and SK-N-BE(2)-C neuronal cell lines, but it did decrease NBRE3/NurRE-luc activity as well as the activity in the Gal4-VP16/UAS-luc and CellTiter-Glo assays indicating it also displays Nurr1-independent and cytotoxic effects on transcription. Future studies are needed to detail whether cytosporone B can affect Nurr1 transcription through direct binding in other cell types to determine if it represents a potential starting point for future medicinal chemistry efforts on Nurr1-binding ligands.
In conclusion, our studies emphasize the need for determining whether ligands that affect Nurr1 activity, or NR4A activity more broadly, indeed bind directly to the LBD or function through other cellular mechanisms such as effector proteins that function upstream of the NR4As. The fact that amodiaquine and chloroquine show NR4A-independent effects in luciferase assays since they show activity in VP16-Gal4 constitutive control and CellTiter-Glo cell viability assays indicates caution should be taken when interpreting the functional response of 4-amino-7-chloroquinoline ligands in Nurr1 reporter assays. The discovery of these ligands as Nurr1 agonists was performed using luciferase reporter assays normalized by an internal β-galactosidase internal control plasmid (reporter activity over control activity)12. A more recent study used a Renilla luciferase plasmid to normalize Nurr1 luciferase reporter data for amodiaquine and chloroquine, which produced a profile suggesting activation of Nurr1 with a very steep dose response transition (hill slope >> 1)78. Internal control plasmids, which can be used to account for well-to-well differences in transfection efficiency and/or cell number in multiwell assay plates, typically contain a general promoter (e.g., SV40) for constitutive expression. However, their use is implicit on the premise that ligand treatment does not influences cell viability, which is the case for amodiaquine and chloroquine. Ligand concentration-dependent cell death will cause a decrease in β-galactosidase or Renilla luciferase internal control activity79–83. Normalization of luciferase reporter activity by the internal control will artifactually influence the dose response curves, often causing a steep dose-responsive increase in normalized luciferase activity. For amodiaquine and chloroquine, normalization of luciferase reporter data to an internal control would mask the real impacts on Nurr1-mediated transcription. Thus, both the luciferase reporter activity and internal control activity should be carefully inspected to ensure that ligand concentration-dependent profiles modulate the reporter and not the internal control. Future studies employing biochemical, biophysical, or structure-based screening might lead to new direct binding Nurr1/NR4A ligand scaffolds that do not show notable NR4A-independent functional effects, which would open new possibilities for developing Nurr1-binding ligands. Furthermore, chemoproteomic studies could aid in the identification of the molecular target(s) of the ligands that impact Nurr1/NR4A activity but do not bind to their LBDs.
Experimental Section
General
Nine of the twelve ligands were purchased from commercial vendors: amodiaquine (Xenotech Llc), chloroquine (Chem Impex Intl Inc), IP7e (Tocris), C-DIM12 (Sigma-Aldrich), camptothecin (Cayman), celastrol (Cayman), isoalantolactone (Indofine), cytosporone B (Tocris), and TMPA (EMD Millipore). Three ligands were synthesized in-house using previously described methods including SR1009818, SR1065819, and SR2423721. The purity of all synthesized and commercially purchased is > 95%. For synthesized ligands, purity was confirmed using reverse-phase analytical HPLC (Shimadzu Prominence; 5->95 CH3CN/H2O, 3mL/min, YMC Pack-Pro C18 50×4.6mm ID, S-5μm) and identity was confirmed by 1H NMR (Bruker 600 MHz NMR spectrometer) and mass analysis (Thermo Scientific Ultimate 3000/LCQ Fleet system ESI mass spectrometer). For commercially purchased ligands, purity was confirmed in vendor certificate of analysis (CoA) sheets. Ligands were suspended, according to vendor recommendations when applicable, in water (chloroquine), ethanol-d6 (amodiaquine), or DMSO-d6 (all other ligands).
Spectral characterization of synthesized compounds
1 (SR10098).
1H NMR (DMSO-D6, 600 MHz), δ(p.p.m.): 8.42–8.4 (m, 1H), 8.23–8.19 (m, 2H), 8.16 (dd, 1H), 7.79 (dd, 1H), 7.48–7.43 (m, 2H). MS (ESI): Expected mass for C14H9FN2O2 (M + H)+: 256.06 Da, observed mass: 256.79 Da.
2 (SR24237).
1H NMR (DMSO-D6, 600 MHz), δ(p.p.m.): 10.37 (s, 1H), 9.49 (s, 1H), 8.79–8.77 (m, 1H), 8.69 (d, 1H), 8.18 (dd, 1H), 8.09 (dt, 1H), 8.03 (dt, 1H), 7.98–7.96 (m, 2H), 7.83 (dt, 1H), 7.49 (ddd, 1H), 7.43–7.39 (m, 1H), 7.18–7.15 (m, 1H). MS (ESI): Expected mass for C19H14N4O (M + H)+: 314.12 Da, observed mass: 314.80 Da.
3 (SR10658).
1H NMR (DMSO-D6, 600 MHz), δ(p.p.m.): 8.17–8.15 (m, 2H), 7.53–7.44 (m, 5H), 7.06–7.03 (m, 2H), 6.74 (s, 1H). MS (ESI): Expected mass for C26H22N2O3 (M + H)+: 334.13 Da, observed mass: 332.87 Da.
Cell lines
All cell lines were obtained from ATCC, including HEK293T (#CRL-11268), PC12 (#CRL-1721.1), and SK-N-BE(2)-C (#CRL-2268) and cultured according to ATCC guidelines. HEK293T cells were grown at 37°C, 5% CO2 in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and 100 units/mL of Penicillin, 100 μg/mL of Streptomycin and 0.292 mg/mL of Glutamine (Gibco) until 90 to 95% confluence prior to subculture or use. SK-N-BE(2)-C were maintained at 37°C, 5% CO2 in a media containing 1:1 mixture of EMEM (ATCC) and F12 medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) until 90 to 95% confluence prior to subculture or use. PC12 cells were grown at 37°C, 5% CO2 in FK-12 medium (Gibco) supplemented with 15% horse serum (Sigma) and 2.5% fetal bovine serum (Gibco) until 90 to 95% confluence prior to subculture or use.
Plasmids
DNA encoding the Nurr1 LBD (NR4A2; residues 353–598) was cloned into the pET-46 expression vector using the Ek/LIC system (EMD Chemicals/Novagen) as tobacco etch virus (TEV) protease-cleavable N-terminal 6x-polyhistidine tag fusion protein. Full-length human Nurr1 and Nur77 were cloned into the mammalian expression vector pcDNA3.1; full-length NOR-1, cloned into the mammalian expression vector pEGFP, was kindly provided by Dr. R. Brandes (Goethe-Universität). The 3xNBRE- and 3xNurRE-luciferase reporters were generated by cloning three copies of the NBRE DNA response element (5’-GAGTTTTAAAAGGTCATGCTCAATT TGTC-3’) and 3 copies of the NurRE POMC promoter DNA response element (5’-GATCGTGATATTTACCTCCAAATGCCA-3’) respectively into the pGL3 promoter luciferase reporter vector (Promega). The pBIND Gal4-VP16 plasmid, pCMV-SPORT6 full-length RXRα, and pGL4.35–5xUAS luciferase reporter plasmid were kindly provided by Dr. P. Griffin (Scripps Research). All plasmids were confirmed by Sanger sequencing (Genewiz) prior to use.
Cellular transcription assays
HEK293T (#CRL-11268), PC12 (#CRL-1721.1), and SK-N-BE(2)-C (#CRL-2268) cells were seeded in 10-cm petri dish at 1.5 million cells. The following day, cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) and Opti-MEM with full-length Nurr1, Nur77, NOR-1, or empty vector control (pcDNA3.1 or pEGFP) expression plasmid without (2 μg) or with (1 μg) contransfection of full-length RXRα plasmid (1μg)—along with 3xNBRE3 or 3xNurRE-luciferase reporter plasmid (6 μg) to a total of 8 μg total DNA and incubated for 18 h. For Gal4-VP16 transactivation, cells were transfected the same way but with a Gal4-VP16 expression plasmid (2 μg) and 5xUAS-luciferase reporter plasmid (Upstream Activation Sequence; 2μg). Cells were transferred to a white 384-well plates (Thermo Fisher Scientific) at 10,000 cells/well in 20 μL and incubated for 4 h. Ligands were prepared in dose response dilutions using vehicle control: water (chloroquine), ethanol (amodiaquine), or DMSO (all others). Ligand in dose response format or the respective vehicle control were added to the cells (20 μL) in quadruplicate. Cells were then incubated for 18 h and harvested for luciferase activity quantified using Britelite Plus (Perkin Elmer; 20 μL) on a Synergy Neo plate reader (Biotek). Data were plotted as mean ± s.e.m in GraphPad Prism and are generally representative of ≥ 2 independent experiments. One-way ANOVA analysis was used for statistical analysis of the data using multiple comparisons (vehicle control compared to each ligand-treated condition).
Cell viability assays
HEK293T, PC12 or SK-N-BE(2)-C cells were seeded in 10-cm petri dish at 1.5M cells. The following day, Cells were transferred to a white 384-well plates (Thermo Fisher Scientific) at 10,000 cells/well in 20 μL and incubated for 4 h. Ligands (or vehicle control) were added (20 μL) in quadruplicate, cells incubated for 18 h and harvested for cell viability quantitation using CellTiter-Glo (Promega; 20 μL) on a Synergy Neo plate reader (Biotek). Data were plotted as mean ± s.e.m in GraphPad Prism and are generally representative of ≥ 2 independent experiments. One-way ANOVA analysis was used for statistical analysis of the data using multiple comparisons (vehicle control compared to each ligand-treated condition).
Expression and purification of 15N-labeled Nurr1 LBD
Recombinant 15N-labeled Nurr1 LBD (NR4A2; residues 353 to 598) was expressed and purified as previously described5. Briefly, the protein was expressed in Escherichia coli BL21(DE3) cells (Life Technologies) using a pET-46 tobacco etch virus (TEV) protease-cleavable N-terminal hexahistidine tag fusion protein in M9 media supplemented with 15NH4Cl (Cambridge Isotope Labs, Inc.). Nurr1 LBD was eluted against a 500 mM imidazole gradient through a Ni-NTA column, followed by overnight dialysis against a buffer without imidazole for TEV protease His tag cleavage at 4°C. The next morning, the sample is loaded onto the Ni-NTA column for contaminants and tag removal. The flow through containing the purified protein was collected, concentrated and ran through a S75 size exclusion column (GE healthcare) in NMR buffer (20 mM KPO4 pH 7.4, 50 mM KCl, and 0.5 mM EDTA). The corresponding protein peak is collected and stored at −80°C. All the ligands were dissolved in either water, DMSO-d6, or ethanol-d6 for NMR experiments.
Protein NMR spectroscopy structural footprinting
Data were collected on a Bruker 700 MHz NMR spectrometer equipped with a QCI cryoprobe at 298 K. For each ligand titration, 2D [1H,15N]-TROSY-HSQC were acquired at 298°K using 200 μM 15N-labeled Nurr1 LBD in NMR buffer containing 10% D2O in the presence of 400 μM ligand or vehicle control as follows: amodiaquine, 0.8% ethanol-d6; chloroquine, water; SR10098, 0.8% DMSO-d6; SR24237, 1.6% DMSO-d6; SR10658, 0.8% DMSO-d6; IP7e, 0.4% DMSO-d6; C-DIM12, 1.6% DMSO-d6; camptothecin, 2.4% DMSO-d6; celastrol, 2% DMSO-d6; isoalantolactone, 2% DMSO-d6; cytosporone B, 1.6% DMSO-d6; TMPA, 2% DMSO-d6. Data were processed using NMRFx84 and analyzed using NMRViewJ85. Chemical shift perturbation (CSP) analysis was performed by transfer of Nurr1 LBD NMR chemical shift assignments86 that we previously validated and reported5 from the vehicle for each ligand (described above) to the 2X ligand spectra using the minimal NMR chemical shift method87. Peaks were identified to have broadened to zero if there was no confident peak in proximity to the vehicle peak. CSP values were calculated using the equation where α is the ratio of the 15N and 1H gyromagnetic ratios (−27130000 and 267520000 rad s−1 T−1, respectively). The average CSP and the standard deviation (s.d.) in the CSPs and peak intensities were calculated for titrations after rejecting outliers more than 2 s.d. from the mean. Peaks that displayed changes in CSP or peak intensity greater than ≥ 4 s.d. above (CSP and peak intensity) or below (peak intensity) the average were noted as significant.
Supplementary Material
Funding Sources
This work was supported in part by National Institutes of Health (NIH) grants R01GM114420 (D.J.K.) and S10OD021550.
Abbreviations Used
- CSP
chemical shift perturbation
- DHI
5,6-dihydroxyindole
- EAE
experimental autoimmune encephalomyelitis
- LBD
ligand-binding domain
- NBRE
NGFI-B response element
- NurRE
Nur response element
- PGA1
prostaglandin A1
- RXR
retinoid X receptor
Footnotes
Supporting information availability
HPLC data for in-house synthesized compounds
Certificate of Analysis (CoA) data for purchased compounds
Figures S1–S3
Molecular Formula Strings
References
- 1.Herring JA; Elison WS; Tessem JS Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization Across Tissues. Cells 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z; Benoit G; Liu J; Prasad S; Aarnisalo P; Liu X; Xu H; Walker NP; Perlmann T Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 2003, 423, 555–560. [DOI] [PubMed] [Google Scholar]
- 3.Flaig R; Greschik H; Peluso-Iltis C; Moras D Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain. J Biol Chem 2005, 280, 19250–19258. [DOI] [PubMed] [Google Scholar]
- 4.de Vera IMS; Munoz-Tello P; Zheng J; Dharmarajan V; Marciano DP; Matta-Camacho E; Giri PK; Shang J; Hughes TS; Rance M; Griffin PR; Kojetin DJ Defining a Canonical Ligand-Binding Pocket in the Orphan Nuclear Receptor Nurr1. Structure 2019, 27, 66–77 e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vera IM; Giri PK; Munoz-Tello P; Brust R; Fuhrmann J; Matta-Camacho E; Shang J; Campbell S; Wilson HD; Granados J; Gardner WJ Jr.; Creamer TP; Solt LA; Kojetin DJ Identification of a Binding Site for Unsaturated Fatty Acids in the Orphan Nuclear Receptor Nurr1. ACS Chem Biol 2016, 11, 1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinayavekhin N; Saghatelian A Discovery of a protein-metabolite interaction between unsaturated fatty acids and the nuclear receptor Nur77 using a metabolomics approach. J Am Chem Soc 2011, 133, 17168–17171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajan S; Jang Y; Kim CH; Kim W; Toh HT; Jeon J; Song B; Serra A; Lescar J; Yoo JY; Beldar S; Ye H; Kang C; Liu XW; Feitosa M; Kim Y; Hwang D; Goh G; Lim KL; Park HM; Lee CH; Oh SF; Petsko GA; Yoon HS; Kim KS PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function. Nat Chem Biol 2020, 16, 876–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruning JM; Wang Y; Oltrabella F; Tian B; Kholodar SA; Liu H; Bhattacharya P; Guo S; Holton JM; Fletterick RJ; Jacobson MP; England PM Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite. Cell Chem Biol 2019, 26, 674–685 e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safe S; Jin UH; Morpurgo B; Abudayyeh A; Singh M; Tjalkens RB Nuclear receptor 4A (NR4A) family - orphans no more. J Steroid Biochem Mol Biol 2016, 157, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekiya T; Kashiwagi I; Inoue N; Morita R; Hori S; Waldmann H; Rudensky AY; Ichinose H; Metzger D; Chambon P; Yoshimura A The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun 2011, 2, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiya T; Kashiwagi I; Yoshida R; Fukaya T; Morita R; Kimura A; Ichinose H; Metzger D; Chambon P; Yoshimura A Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol 2013, 14, 230–237. [DOI] [PubMed] [Google Scholar]
- 12.Kim CH; Han BS; Moon J; Kim DJ; Shin J; Rajan S; Nguyen QT; Sohn M; Kim WG; Han M; Jeong I; Kim KS; Lee EH; Tu Y; Naffin-Olivos JL; Park CH; Ringe D; Yoon HS; Petsko GA; Kim KS Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc Natl Acad Sci U S A 2015, 112, 8756–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon M; Jung ES; Jeon SG; Cha MY; Jang Y; Kim W; Lopes C; Mook-Jung I; Kim KS Nurr1 (NR4A2) regulates Alzheimer’s disease-related pathogenesis and cognitive function in the 5XFAD mouse model. Aging Cell 2019, 18, e12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McAnally D; Siddiquee K; Gomaa A; Szabo A; Vasile S; Maloney PR; Divlianska DB; Peddibhotla S; Morfa CJ; Hershberger P; Falter R; Williamson R; Terry DB; Farjo R; Pinkerton AB; Qi X; Quigley J; Boulton ME; Grant MB; Smith LH Repurposing antimalarial aminoquinolines and related compounds for treatment of retinal neovascularization. PLoS One 2018, 13, e0202436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Y; Pham HT; Xu H; Quan Y; Mesplede T Antimalarial drugs and their metabolites are potent Zika virus inhibitors. J Med Virol 2019, 91, 1182–1190. [DOI] [PubMed] [Google Scholar]
- 16.Sakurai Y; Sakakibara N; Toyama M; Baba M; Davey RA Novel amodiaquine derivatives potently inhibit Ebola virus infection. Antiviral Res 2018, 160, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan DJ Jr.; Gluzman IY; Russell DG; Goldberg DE On the molecular mechanism of chloroquine’s antimalarial action. Proc Natl Acad Sci U S A 1996, 93, 11865–11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubois C; Hengerer B; Mattes H Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem 2006, 1, 955–958. [DOI] [PubMed] [Google Scholar]
- 19.Hintermann S; Chiesi M; von Krosigk U; Mathe D; Felber R; Hengerer B Identification of a series of highly potent activators of the Nurr1 signaling pathway. Bioorg Med Chem Lett 2007, 17, 193–196. [DOI] [PubMed] [Google Scholar]
- 20.Montarolo F; Raffaele C; Perga S; Martire S; Finardi A; Furlan R; Hintermann S; Bertolotto A Effects of isoxazolo-pyridinone 7e, a potent activator of the Nurr1 signaling pathway, on experimental autoimmune encephalomyelitis in mice. PLoS One 2014, 9, e108791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lesuisse D; Malanda A; Peyronel JF; Evanno Y; Lardenois P; De-Peretti D; Abecassis PY; Barneoud P; Brunel P; Burgevin MC; Cegarra C; Auger F; Dommergue A; Lafon C; Even L; Tsi J; Luc TPH; Almario A; Olivier A; Castel MN; Taupin V; Rooney T; Vige X Development of a novel NURR1/NOT agonist from hit to lead and candidate for the potential treatment of Parkinson’s disease. Bioorg Med Chem Lett 2019, 29, 929–932. [DOI] [PubMed] [Google Scholar]
- 22.Li X; Lee SO; Safe S Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3’-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem Pharmacol 2012, 83, 1445–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond SL; Safe S; Tjalkens RB A novel synthetic activator of Nurr1 induces dopaminergic gene expression and protects against 6-hydroxydopamine neurotoxicity in vitro. Neurosci Lett 2015, 607, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond SL; Popichak KA; Li X; Hunt LG; Richman EH; Damale PU; Chong EKP; Backos DS; Safe S; Tjalkens RB The Nurr1 Ligand,1,1-bis(3’-Indolyl)-1-(p-Chlorophenyl)Methane, Modulates Glial Reactivity and Is Neuroprotective in MPTP-Induced Parkinsonism. J Pharmacol Exp Ther 2018, 365, 636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inamoto T; Papineni S; Chintharlapalli S; Cho SD; Safe S; Kamat AM 1,1-Bis(3’-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol Cancer Ther 2008, 7, 3825–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hibino S; Chikuma S; Kondo T; Ito M; Nakatsukasa H; Omata-Mise S; Yoshimura A Inhibition of Nr4a Receptors Enhances Antitumor Immunity by Breaking Treg-Mediated Immune Tolerance. Cancer Res 2018, 78, 3027–3040. [DOI] [PubMed] [Google Scholar]
- 27.Hu M; Luo Q; Alitongbieke G; Chong S; Xu C; Xie L; Chen X; Zhang D; Zhou Y; Wang Z; Ye X; Cai L; Zhang F; Chen H; Jiang F; Fang H; Yang S; Liu J; Diaz-Meco MT; Su Y; Zhou H; Moscat J; Lin X; Zhang XK Celastrol-Induced Nur77 Interaction with TRAF2 Alleviates Inflammation by Promoting Mitochondrial Ubiquitination and Autophagy. Mol Cell 2017, 66, 141–153 e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung YS; Lee HS; Cho HR; Kim KJ; Kim JH; Safe S; Lee SO Dual targeting of Nur77 and AMPKalpha by isoalantolactone inhibits adipogenesis in vitro and decreases body fat mass in vivo. Int J Obes 2019, 43, 952–962. [DOI] [PubMed] [Google Scholar]
- 29.Zhan Y; Du X; Chen H; Liu J; Zhao B; Huang D; Li G; Xu Q; Zhang M; Weimer BC; Chen D; Cheng Z; Zhang L; Li Q; Li S; Zheng Z; Song S; Huang Y; Ye Z; Su W; Lin SC; Shen Y; Wu Q Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat Chem Biol 2008, 4, 548–556. [DOI] [PubMed] [Google Scholar]
- 30.Zhan YY; Chen Y; Zhang Q; Zhuang JJ; Tian M; Chen HZ; Zhang LR; Zhang HK; He JP; Wang WJ; Wu R; Wang Y; Shi C; Yang K; Li AZ; Xin YZ; Li TY; Yang JY; Zheng ZH; Yu CD; Lin SC; Chang C; Huang PQ; Lin T; Wu Q The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat Chem Biol 2012, 8, 897–904. [DOI] [PubMed] [Google Scholar]
- 31.Li L; Liu Y; Chen HZ; Li FW; Wu JF; Zhang HK; He JP; Xing YZ; Chen Y; Wang WJ; Tian XY; Li AZ; Zhang Q; Huang PQ; Han J; Lin T; Wu Q Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation. Nat Chem Biol 2015, 11, 339–346. [DOI] [PubMed] [Google Scholar]
- 32.Law SW; Conneely OM; DeMayo FJ; O’Malley BW Identification of a new brain-specific transcription factor, NURR1. Mol Endocrinol 1992, 6, 2129–2135. [DOI] [PubMed] [Google Scholar]
- 33.Sakurada K; Ohshima-Sakurada M; Palmer TD; Gage FH Nurr1, an orphan nuclear receptor, is a transcriptional activator of endogenous tyrosine hydroxylase in neural progenitor cells derived from the adult brain. Development 1999, 126, 4017–4026. [DOI] [PubMed] [Google Scholar]
- 34.Kim KS; Kim CH; Hwang DY; Seo H; Chung S; Hong SJ; Lim JK; Anderson T; Isacson O Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J Neurochem 2003, 85, 622–634. [DOI] [PubMed] [Google Scholar]
- 35.Heng X; Jin G; Zhang X; Yang D; Zhu M; Fu S; Li X; Le W Nurr1 regulates Top IIbeta and functions in axon genesis of mesencephalic dopaminergic neurons. Mol Neurodegener 2012, 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philips A; Lesage S; Gingras R; Maira MH; Gauthier Y; Hugo P; Drouin J Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol 1997, 17, 5946–5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maira M; Martens C; Philips A; Drouin J Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol 1999, 19, 7549–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang L; Dai S; Li J; Liang X; Qu L; Chen X; Guo M; Chen Z; Chen L; Wei H; Chen Y Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements. J Biol Chem 2019, 294, 19795–19803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perlmann T; Jansson L A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev 1995, 9, 769–782. [DOI] [PubMed] [Google Scholar]
- 40.Forman BM; Umesono K; Chen J; Evans RM Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 1995, 81, 541–550. [DOI] [PubMed] [Google Scholar]
- 41.Castro DS; Arvidsson M; Bondesson Bolin M; Perlmann T Activity of the Nurr1 carboxyl-terminal domain depends on cell type and integrity of the activation function 2. J Biol Chem 1999, 274, 37483–37490. [DOI] [PubMed] [Google Scholar]
- 42.Guo J; Zu G; Zhou T; Xing J; Wang Z Clinicopathological significance of orphan nuclear receptor Nurr1 expression in gastric cancer. Clin Transl Oncol 2015, 17, 788–794. [DOI] [PubMed] [Google Scholar]
- 43.Wang J; Yang J; Li BB; He ZW High cytoplasmic expression of the orphan nuclear receptor NR4A2 predicts poor survival in nasopharyngeal carcinoma. Asian Pac J Cancer Prev 2013, 14, 2805–2809. [DOI] [PubMed] [Google Scholar]
- 44.Karki K; Li X; Jin UH; Mohankumar K; Zarei M; Michelhaugh SK; Mittal S; Tjalkens R; Safe S Nuclear receptor 4A2 (NR4A2) is a druggable target for glioblastomas. J Neurooncol 2020, 146, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chintharlapalli S; Burghardt R; Papineni S; Ramaiah S; Yoon K; Safe S Activation of Nur77 by selected 1,1-Bis(3’-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J Biol Chem 2005, 280, 24903–24914. [DOI] [PubMed] [Google Scholar]
- 46.Cho SD; Yoon K; Chintharlapalli S; Abdelrahim M; Lei P; Hamilton S; Khan S; Ramaiah SK; Safe S Nur77 agonists induce proapoptotic genes and responses in colon cancer cells through nuclear receptor-dependent and nuclear receptor-independent pathways. Cancer Res 2007, 67, 674–683. [DOI] [PubMed] [Google Scholar]
- 47.Yoon K; Chen CC; Orr AA; Barreto PN; Tamamis P; Safe S Activation of COUP-TFI by a Novel Diindolylmethane Derivative. Cells 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chintharlapalli S; Smith R 3rd; Samudio I; Zhang W; Safe S 1,1-Bis(3’-indolyl)-1-(p-substitutedphenyl)methanes induce peroxisome proliferator-activated receptor gamma-mediated growth inhibition, transactivation, and differentiation markers in colon cancer cells. Cancer Res 2004, 64, 5994–6001. [DOI] [PubMed] [Google Scholar]
- 49.Qin C; Morrow D; Stewart J; Spencer K; Porter W; Smith R 3rd; Phillips T; Abdelrahim M; Samudio I; Safe S A new class of peroxisome proliferator-activated receptor gamma (PPARgamma) agonists that inhibit growth of breast cancer cells: 1,1-Bis(3’-indolyl)-1-(p-substituted phenyl)methanes. Mol Cancer Ther 2004, 3, 247–260. [PubMed] [Google Scholar]
- 50.Tjalkens RB; Liu X; Mohl B; Wright T; Moreno JA; Carbone DL; Safe S The peroxisome proliferator-activated receptor-gamma agonist 1,1-bis(3’-indolyl)-1-(p-trifluoromethylphenyl)methane suppresses manganese-induced production of nitric oxide in astrocytes and inhibits apoptosis in cocultured PC12 cells. J Neurosci Res 2008, 86, 618–629. [DOI] [PubMed] [Google Scholar]
- 51.Lei P; Abdelrahim M; Cho SD; Liu X; Safe S Structure-dependent activation of endoplasmic reticulum stress-mediated apoptosis in pancreatic cancer by 1,1-bis(3’-indoly)-1-(p-substituted phenyl)methanes. Mol Cancer Ther 2008, 7, 3363–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lei P; Abdelrahim M; Cho SD; Liu S; Chintharlapalli S; Safe S 1,1-Bis(3’-indolyl)-1-(p-substituted phenyl)methanes inhibit colon cancer cell and tumor growth through activation of c-jun N-terminal kinase. Carcinogenesis 2008, 29, 1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldberg AA; Draz H; Montes-Grajales D; Olivero-Verbel J; Safe SH; Sanderson JT 3,3’-Diindolylmethane (DIM) and its ring-substituted halogenated analogs (ring-DIMs) induce differential mechanisms of survival and death in androgen-dependent and -independent prostate cancer cells. Genes Cancer 2015, 6, 265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho SD; Lei P; Abdelrahim M; Yoon K; Liu S; Guo J; Papineni S; Chintharlapalli S; Safe S 1,1-bis(3’-indolyl)-1-(p-methoxyphenyl)methane activates Nur77-independent proapoptotic responses in colon cancer cells. Mol Carcinog 2008, 47, 252–263. [DOI] [PubMed] [Google Scholar]
- 55.Chintharlapalli S; Papineni S; Safe S 1,1-bis(3’-indolyl)-1-(p-substitutedphenyl)methanes inhibit growth, induce apoptosis, and decrease the androgen receptor in LNCaP prostate cancer cells through peroxisome proliferator-activated receptor gamma-independent pathways. Mol Pharmacol 2007, 71, 558–569. [DOI] [PubMed] [Google Scholar]
- 56.Lei P; Abdelrahim M; Safe S 1,1-Bis(3’-indolyl)-1-(p-substituted phenyl)methanes inhibit ovarian cancer cell growth through peroxisome proliferator-activated receptor-dependent and independent pathways. Mol Cancer Ther 2006, 5, 2324–2336. [DOI] [PubMed] [Google Scholar]
- 57.Salminen A; Lehtonen M; Paimela T; Kaarniranta K Celastrol: Molecular targets of Thunder God Vine. Biochem Biophys Res Commun 2010, 394, 439–442. [DOI] [PubMed] [Google Scholar]
- 58.Wang H; Teriete P; Hu A; Raveendra-Panickar D; Pendelton K; Lazo JS; Eiseman J; Holien T; Misund K; Oliynyk G; Arsenian-Henriksson M; Cosford ND; Sundan A; Prochownik EV Direct inhibition of c-Myc-Max heterodimers by celastrol and celastrol-inspired triterpenoids. Oncotarget 2015, 6, 32380–32395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z; Ma L; Wen ZS; Hu Z; Wu FQ; Li W; Liu J; Zhou GB Cancerous inhibitor of PP2A is targeted by natural compound celastrol for degradation in non-small-cell lung cancer. Carcinogenesis 2014, 35, 905–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye S; Luo W; Khan ZA; Wu G; Xuan L; Shan P; Lin K; Chen T; Wang J; Hu X; Wang S; Huang W; Liang G Celastrol Attenuates Angiotensin II-Induced Cardiac Remodeling by Targeting STAT3. Circ Res 2020, 126, 1007–1023. [DOI] [PubMed] [Google Scholar]
- 61.Xiao-Pei H; Ji-Kuai C; Xue W; Dong YF; Yan L; Xiao-Fang Z; Ya-Min P; Wen-Jun C; Jiang-Bo Z Systematic identification of Celastrol-binding proteins reveals that Shoc2 is inhibited by Celastrol. Biosci Rep 2018, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JH; Koo TH; Yoon H; Jung HS; Jin HZ; Lee K; Hong YS; Lee JJ Inhibition of NF-kappa B activation through targeting I kappa B kinase by celastrol, a quinone methide triterpenoid. Biochem Pharmacol 2006, 72, 1311–1321. [DOI] [PubMed] [Google Scholar]
- 63.Chadli A; Felts SJ; Wang Q; Sullivan WP; Botuyan MV; Fauq A; Ramirez-Alvarado M; Mer G Celastrol inhibits Hsp90 chaperoning of steroid receptors by inducing fibrillization of the Co-chaperone p23. J Biol Chem 2010, 285, 4224–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang T; Hamza A; Cao X; Wang B; Yu S; Zhan CG; Sun D A novel Hsp90 inhibitor to disrupt Hsp90/Cdc37 complex against pancreatic cancer cells. Mol Cancer Ther 2008, 7, 162–170. [DOI] [PubMed] [Google Scholar]
- 65.Zhang T; Li Y; Yu Y; Zou P; Jiang Y; Sun D Characterization of celastrol to inhibit hsp90 and cdc37 interaction. J Biol Chem 2009, 284, 35381–35389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen SR; Dai Y; Zhao J; Lin L; Wang Y; Wang Y A Mechanistic Overview of Triptolide and Celastrol, Natural Products from Tripterygium wilfordii Hook F. Front Pharmacol 2018, 9, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen W; Li P; Liu Y; Yang Y; Ye X; Zhang F; Huang H Isoalantolactone induces apoptosis through ROS-mediated ER stress and inhibition of STAT3 in prostate cancer cells. J Exp Clin Cancer Res 2018, 37, 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xing JS; Wang X; Lan YL; Lou JC; Ma B; Zhu T; Zhang H; Wang D; Yu Z; Yuan Z; Li XY; Zhang B Isoalantolactone inhibits IKKbeta kinase activity to interrupt the NF-kappaB/COX-2-mediated signaling cascade and induces apoptosis regulated by the mitochondrial translocation of cofilin in glioblastoma. Cancer Med 2019, 8, 1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li F; Jiang T; Li Q; Ling X Camptothecin (CPT) and its derivatives are known to target topoisomerase I (Top1) as their mechanism of action: did we miss something in CPT analogue molecular targets for treating human disease such as cancer? Am J Cancer Res 2017, 7, 2350–2394. [PMC free article] [PubMed] [Google Scholar]
- 70.Williams KL; Wells CI; Moore JT Identification of Kinase Inhibitors that Regulate Nuclear Receptor Nurr1 (NR4A2) Cellular Activity. 2019, bioRxiv:420976. biorxiv.org e-Print archive. (accessed Nov 4, 2020). [Google Scholar]
- 71.Mariga ST; Gil JP; Sisowath C; Wernsdorfer WH; Bjorkman A Synergism between amodiaquine and its major metabolite, desethylamodiaquine, against Plasmodium falciparum in vitro. Antimicrob Agents Chemother 2004, 48, 4089–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokoyama A; Mori S; Takahashi HK; Kanke T; Wake H; Nishibori M Effect of amodiaquine, a histamine N-methyltransferase inhibitor, on, Propionibacterium acnes and lipopolysaccharide-induced hepatitis in mice. Eur J Pharmacol 2007, 558, 179–184. [DOI] [PubMed] [Google Scholar]
- 73.Wennerholm A; Nordmark A; Pihlsgard M; Mahindi M; Bertilsson L; Gustafsson LL Amodiaquine, its desethylated metabolite, or both, inhibit the metabolism of debrisoquine (CYP2D6) and losartan (CYP2C9) in vivo. Eur J Clin Pharmacol 2006, 62, 539–546. [DOI] [PubMed] [Google Scholar]
- 74.Kim J; Yip ML; Shen X; Li H; Hsin LY; Labarge S; Heinrich EL; Lee W; Lu J; Vaidehi N Identification of anti-malarial compounds as novel antagonists to chemokine receptor CXCR4 in pancreatic cancer cells. PLoS One 2012, 7, e31004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jia L; Wang J; Wu T; Wu J; Ling J; Cheng B In vitro and in vivo antitumor effects of chloroquine on oral squamous cell carcinoma. Mol Med Rep 2017, 16, 5779–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mauthe M; Orhon I; Rocchi C; Zhou X; Luhr M; Hijlkema KJ; Coppes RP; Engedal N; Mari M; Reggiori F Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim CH; Leblanc P; Kim KS 4-amino-7-chloroquinoline derivatives for treating Parkinson’s disease: implications for drug discovery. Expert Opin Drug Discov 2016, 11, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Willems S; Whitney K; Ni X; Chaikuad A; Knapp S; Heering J; Merk D The orphan nuclear receptor Nurr1 is responsive to non-steroidal anti-inflammatory drugs. Commun Chem 2020, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shifera AS; Hardin JA Factors modulating expression of Renilla luciferase from control plasmids used in luciferase reporter gene assays. Anal Biochem 2010, 396, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang D; Atlasi SS; Patel KK; Zhuang Z; Heaney AP False responses of Renilla luciferase reporter control to nuclear receptor TR4. Mol Cell Biochem 2017, 430, 139–147. [DOI] [PubMed] [Google Scholar]
- 81.Ibrahim NM; Marinovic AC; Price SR; Young LG; Frohlich O Pitfall of an internal control plasmid: response of Renilla luciferase (pRL-TK) plasmid to dihydrotestosterone and dexamethasone. Biotechniques 2000, 29, 782–784. [DOI] [PubMed] [Google Scholar]
- 82.Zhang X; Chen HZ; Rovin BH Unexpected sensitivity of synthetic Renilla luciferase control vectors to treatment with a cyclopentenone prostaglandin. Biotechniques 2003, 35, 1144–1146, 1148. [DOI] [PubMed] [Google Scholar]
- 83.Dougherty DC; Sanders MM Comparison of the responsiveness of the pGL3 and pGL4 luciferase reporter vectors to steroid hormones. Biotechniques 2005, 39, 203–207. [DOI] [PubMed] [Google Scholar]
- 84.Norris M; Fetler B; Marchant J; Johnson BA NMRFx Processor: a cross-platform NMR data processing program. J Biomol NMR 2016, 65, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson BA Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 2004, 278, 313–352. [DOI] [PubMed] [Google Scholar]
- 86.Michiels P; Atkins K; Ludwig C; Whittaker S; van Dongen M; Gunther U Assignment of the orphan nuclear receptor Nurr1 by NMR. Biomol NMR Assign 2010, 4, 101–105. [DOI] [PubMed] [Google Scholar]
- 87.Williamson MP Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc 2013, 73, 1–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


