Abstract
Carotenoids are important isoprenoids produced in the plastids of photosynthetic organisms that play key roles in photoprotection and antioxidative processes. β-Carotene is generated from lycopene by lycopene β-cyclase (LCYB). Previously, we demonstrated that the introduction of the Daucus carota (carrot) DcLCYB1 gene into tobacco (cv. Xanthi) resulted in increased levels of abscisic acid (ABA) and especially gibberellins (GAs), resulting in increased plant yield. In order to understand this phenomenon prior to exporting this genetic strategy to crops, we generated tobacco (Nicotiana tabacum cv. Petit Havana) mutants that exhibited a wide range of LCYB expression. Transplastomic plants expressing DcLCYB1 at high levels showed a wild-type-like growth, even though their pigment content was increased and their leaf GA1 content was reduced. RNA interference (RNAi) NtLCYB lines showed different reductions in NtLCYB transcript abundance, correlating with reduced pigment content and plant variegation. Photosynthesis (leaf absorptance, Fv/Fm, and light-saturated capacity of linear electron transport) and plant growth were impaired. Remarkably, drastic changes in phytohormone content also occurred in the RNAi lines. However, external application of phytohormones was not sufficient to rescue these phenotypes, suggesting that altered photosynthetic efficiency might be another important factor explaining their reduced biomass. These results show that LCYB expression influences plant biomass by different mechanisms and suggests thresholds for LCYB expression levels that might be beneficial or detrimental for plant growth.
Keywords: β-Carotene, biomass, carotenoids, lycopene β-cyclase, Nicotiana tabacum cv. Petit Havana, photosynthesis, phytohormones, RNAi, transplastomic
A lycopene β-cyclase expression threshold influences plant biomass, development, and metabolism by modulating pigment and phytohormone content in transplastomic and RNAi tobacco lines.
Introduction
Carotenoids are 40-carbon isoprenoids with polyene chains containing up to 15 conjugated double bonds (Hirschberg, 2001). In plants, carotenoids have many important functions, such as chlorophyll protection from UV light, and multiple antioxidant activities, including the ability to scavenge reactive oxygen species (ROS), such as singlet oxygen, and peroxyl radicals (Krinsky, 1989). In addition, carotenoids play a critical role in light absorption processes such as photosynthetic light harvesting via singlet state energy transfer (Frank and Cogdell, 1993; Dall’Osto et al., 2007b; Andrade-Souza et al., 2011). As membrane-bound compounds, they provide membrane stabilization (Frank and Cogdell, 1993, 1996; Demmig-Adams et al., 1996; Havaux, 1998). Furthermore, carotenoids are structural constituents of the photosynthetic machinery, forming pigment–protein complexes in photosystems (PS) I and II and their respective light harvesting complexes (LHCs) as well as in the cytochrome b6f complex (Dall’Osto et al., 2007b, 2014). Moreover, they are precursors of the apocarotenoid phytohormone abscisic acid (ABA), which is involved in developmental processes and adaptive stress responses to environmental stimuli such as drought-induced stomatal closure in plants (Nambara and Marion-Poll, 2005; Taylor et al., 2005; Shi et al., 2015), and of strigolactones (SLs), involved in plant development (Brewer et al., 2013; Al-Babili and Bouwmeester, 2015). One of the most important carotenoids, for human health and nutrition is β-carotene, the precursor of vitamin A (Olson, 1996). β-Carotene deficiency leads to blindness, xerophthalmia, and premature death in humans (Giuliano et al., 2003).
Lycopene-β-cyclase (LCYB) catalyses the last step of β-carotene synthesis, and therefore plays a key role in this pathway. Overexpression of LCYB leads to increased tolerance to abiotic stresses (e.g. drought and salt) in tomato (D’Ambrosio et al., 2004), Arabidopsis (Chen et al., 2011), and sweet potato (Kang et al., 2018). Additionally, transgenic tomatoes (Solanum lycopersicum) expressing the tobacco LCYB gene showed tolerance to the bleaching herbicide 2-(4-chlorophenylthio) triethylamine (CPTA), a β-cyclase inhibitor (Ralley et al., 2016). Moreover, increased β-carotene, violaxanthin, lutein, and zeaxanthin content in plants was shown to improve tolerance to abiotic stresses such as high light, UV irradiation, and salt stress by scavenging ROS (Davison et al., 2002; Götz et al., 2002; Han et al., 2008; Shi et al., 2015; Kang et al., 2018). In tobacco, overexpression of NtLCYB led to increased expression of genes for phytoene synthase (PSY), phytoene desaturase (PDS), ζ-carotene desaturase (ZDS), zeaxanthin epoxidase (ZEP), violaxanthin de-epoxidase (VDE), and neoxanthin synthase (NXS), increased accumulation of β-carotene, violaxanthin, lutein, and neoxanthin, and enhanced tolerance to salt and drought stress, whereas transgenic NtLCYB RNA interference (RNAi) lines either showed an albino phenotype in leaves or did not survive beyond the early developmental stages (Shi et al., 2015).
Recently, a major growth advantage was reported in tobacco lines expressing the carrot DcLCYB1 (Moreno et al., 2016). This was shown under fully controlled (constant and fluctuating light regimes) and non-controlled climate conditions (Moreno et al., 2020). Increased levels of photoprotective molecules such as β-carotene, lutein/zeaxanthin, and violaxanthin enhanced photoprotection and contributed to the increased photosynthetic efficiency, especially under fluctuating light conditions. However, unexpectedly, transgenic DcLCYB1 lines also showed increased transcript levels of key genes involved in several metabolic pathways related to isoprenoid metabolism, resulting in increased biosynthesis of ABA and especially of different gibberellins (GAs; Fig. 1A). The increased GA/ABA ratio altered plant development and architecture, for example reducing shading of mature leaves. Thereby, induction of leaf senescence was delayed, and mature leaves maintained a high photosynthetic capacity. This ultimately resulted in higher fitness and yield in these tobacco lines (Moreno et al., 2020).
Fig. 1.
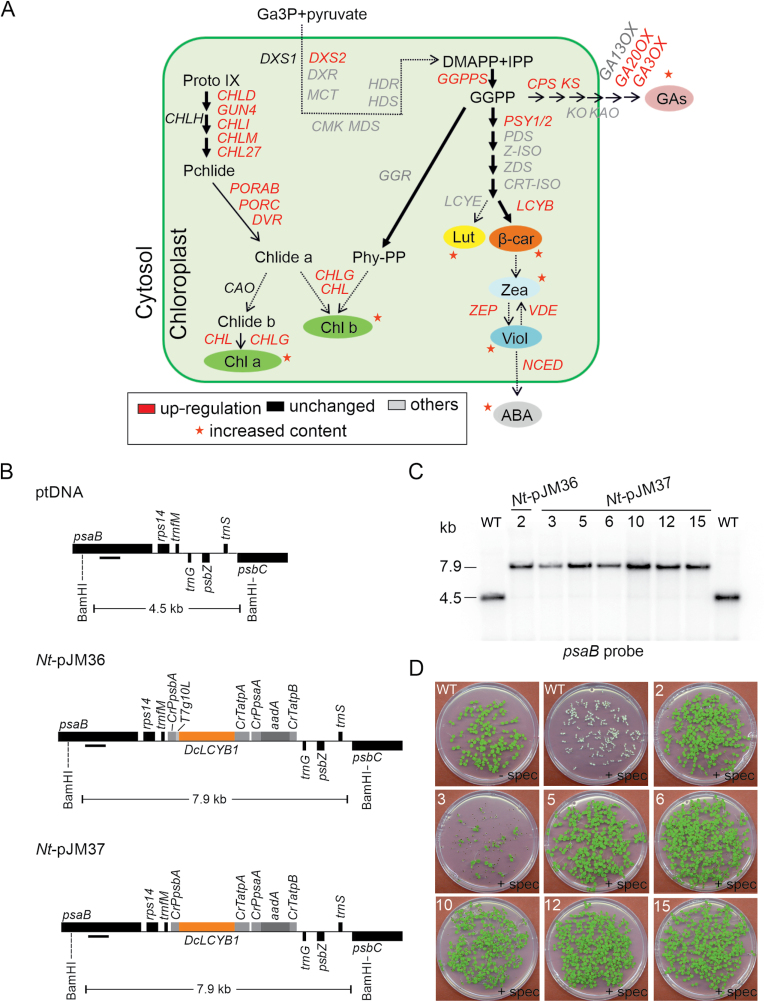
Introduction of the Daucus carota lycopene β-cyclase1 (DcLCYB1) gene into the plastid genome by stable transformation. (A) Schematic representation for isoprenoid-derived pathways (e.g. carotenoids, GA, ABA) of the changes caused upon expression of DcLCYB1 in nuclear tobacco lines. Up-regulated genes are marked in red; genes shown in black were not changed; and genes shown in grey were not measured. Red stars indicate increased metabolite content. ABA, abscisic acid; β-car, β-carotene; Chl, chlorophyll; DMAPP, dimethylallyl diphosphate; GA, gibberellins; IPP, isopentenyl diphosphate; Lut, lutein; Pchlide, protochlorophillidae; Phy-PP, phytyl diphosphate; Viol, violaxanthin; Zea, zeaxanthin. Gene abbreviation, protein name, and a brief description of each gene measured by qPCR can be found in Moreno et al. (2020). (B) Physical map of the plastid genome region (ptDNA) used as a site for integration, and of the transgenic loci in the generated transplastomic tobacco lines (Nt-pJM37, Nt-pJM36). Native plastid genes are shown in black; introduced elements are shown in pale grey and orange. Genes above the line are transcribed from left to right; genes below the line are transcribed right to left. Promoter (P) and terminator (T) sequence from Chlamydomonas reinhardtii (Cr) are shown in pale grey. T7g10L is the gene 10 leader sequence from the T7 phage, which has been shown to be highly efficient at facilitating high-level translation in green plastids (Oey et al., 2009; Elghabi et al., 2011). The aadA gene encodes the AMINOGLYCOSIDE 3′-ADENYLYLTRANSFERASE enzyme, which confers resistance to the aminoglycoside-type antibiotics spectinomycin and streptomycin and as such serves as a selectable marker for transformed chloroplasts. The black box under psaB indicates the probe used for RFLP analysis. (C) RFLP analysis of transplastomic plants. Total DNA from wild-type and transformed lines was digested with BamHI producing fragments of the sizes indicated in (A). A single, independently isolated transplastomic line is shown for Nt-pJM36; six lines are shown for Nt-pJM37. (D) Inheritance assay of transplastomic DcLCYB1 plants. Germination of the T1 generation of plastid transformants in the presence of spectinomycin (+spec) revealed the homoplasmic stage for the integrated selectable marker gene aadA. Numbering of the lines refers to the nomenclature use in (B). spec, spectinomycin; WT, wild type.
The unexpected contribution of altered GA and ABA levels to the growth phenotype raises the question of whether or not similar effects have been overlooked so far in other mutants with altered carotenoid metabolism (Qin et al., 2007; Li et al., 2009; Cazzaniga et al., 2012; Avendaño-Vázquez et al., 2014; Kromdijk et al., 2016). For example, impaired growth of mutants with reduced carotenoid accumulation due to repression or inactivation of enzymes involved in carotenoid biosynthesis (Pogson et al., 1998; Dall’Osto et al., 2007a, 2013; Qin et al., 2007; Li et al., 2009; Cazzaniga et al., 2012; Fiore et al., 2012) has been solely attributed to defects in photosynthesis and impaired protection against oxidative stress, in line with the well-established functions of carotenoids. Possible additive effects of altered carotenoid biosynthesis on the synthesis of other isoprenoids, especially phytohormones, have not been considered so far. Therefore, a broader analysis is needed of the consequences of altered carotenoid biosynthesis for phytohormones, especially GAs, ABA, and strigolactones, whose synthesis is closely related to carotenoid synthesis.
In light of the reported growth benefit in tobacco (Moreno et al., 2020), it is of interest to understand how a single gene transformation can trigger molecular and physiological responses that are reflected in higher yield. The higher DcLCYB1 expression in the nuclear tobacco lines was reflected in a 2-fold increase in β-carotene content. In addition, higher DcLCYB1 expression triggered a co-expression of key genes in carotenoid- and carotenoid-related pathways, which resulted, for instance, in higher chlorophyll and GA content, two important isoprenoid pathways influencing plant growth and development. Higher β-carotene was reflected in higher xanthophyll production, and therefore enhanced photoprotection, and ABA content. These lines of evidence suggest that DcLCYB1 expression can induce various molecular pathways (Fig. 1A), at genetic and metabolic levels, that stimulate plant growth, development, photosynthesis, and yield. These findings bring into question the role of LCYB expression levels in growth regulation. Following this idea, higher LCYB expression levels might induce (stronger induction than in the nuclear DcLCYB1 lines) the expression of key genes of carotenoid, ABA, GA, and chlorophyll biosynthesis pathways (Fig. 1A), and therefore, higher increases (compared with the nuclear lines) in β-carotene, ABA, GA, and chlorophyll contents. By contrast, a reduction in LCYB expression should trigger a down-regulation of key genes of carotenoid, ABA, GA, and chlorophyll biosynthesis pathways (Fig. 1A), and therefore, reductions in β-carotene, ABA, GA, and chlorophyll contents. In both cases, higher or reduced LCYB expression should lead to enhanced or reduced plant biomass, respectively. Based on the previous evidence we combined both cases and formulate the following hypothesis: LCYB expression regulates plant growth by influencing carotenoid (β-carotene and xanthophylls) and hormone (ABA and GA) contents through an activation of key genes of carotenoid and hormone biosynthesis pathways.
To test our hypothesis, we designed two genetic strategies to further increase (more than in the nuclear lines) and reduce LCYB expression in tobacco plants. One possible way to achieve very high transgene expression is via chloroplast transformation. Transformation of plastid DNA has enormous advantages over nuclear transformation (Bock, 2007; Clarke and Daniell, 2011). For instance, high transgene expression levels of more than 70% of soluble leaf protein can be achieved due to the large chloroplast number per cell, high ploidy per chloroplast, and high translation capacity of this compartment (Oey et al., 2009). While expression levels of carotenoid biosynthesis enzymes in the range of several percent of soluble leaf protein is certainly not required to manipulate carotenoid biosynthesis, the appropriate choice of expression elements for chloroplast transformation allows the generation of mutants covering a wide range of transgene expression levels. As another advantage of chloroplast transformation, gene dispersal in the environment is largely abolished due to maternal inheritance of chloroplasts, which excludes plastid genes and transgenes from pollen transmission (Ruf et al., 2007). Finally, transgene insertion via the homologous recombination process avoids position effects and gene silencing of the transgene, making plastid transformation ideal for crop improvement and metabolic engineering. Previously, plastid transformation approaches aiming to increase isoprenoid content succeeded in tobacco and tomato (Apel and Bock, 2009; Kumar et al., 2012; Lu et al., 2013). Tomato lines expressing the daffodil (Narcissus pseudonarcissus) LCYB gene from the plastid genome (transplastomic lines) showed increased β-carotene content in the fruit (changed from red to orange), but not in the leaves (Apel and Bock, 2009). In addition, β-carotene content measured in leaves of tomatoes and tobacco lines transformed with a bacterial LCYB gene (Erwinia herbicola) remained unchanged (Wurbs et al., 2007; Apel and Bock, 2009). In both tomato and tobacco transplastomic lines, higher LCYB activity was related to the higher tolerance to the herbicide CPTA. In order to reduce LCYB expression we used RNAi in order to obtain a wide range of transgene silencing and distinguish between harmful and non-harmful (for plant growth and development) reductions in the expression of the gene.
Here, by generating both transplastomic tobacco lines strongly expressing DcLCYB1 and RNAi mutants repressing the tobacco LCYB enzyme to different residual levels, we will be able to conclude if LCYB expression is tightly related to growth regulation. Following our recently published results for nuclear DcLCYB1 lines (Moreno et al., 2020), we would expect that high DcLCYB1 expression levels in our transplastomic lines could trigger a co-expression of key genes of carotenoid and carotenoid-related pathways, thus causing an increase in the content of key isoprenoids (e.g. β-carotene, xanthophylls, chlorophylls, GA, and ABA) and ultimately leading to higher growth and biomass. By contrast, reduced NtLCYB expression in our RNAi lines was hypothesized to trigger reduced expression of key genes of carotenoid and carotenoid-related pathways, thus causing a reduction in the content of key isoprenoids (e.g. β-carotene, xanthophylls, chlorophylls, GA, and ABA) and ultimately leading to reduced growth and biomass. If this is true, the reduced plant biomass might be due to these additional disturbances and impaired photosynthetic performance, which is directly related to the structural function of carotenoid in the photosynthetic apparatus. Hence, gene expression levels, pigment and hormone contents, biomass parameters, photosynthetic efficiency, chloroplast structure, and primary and secondary metabolism were evaluated in these lines. We show that plastid DcLCYB1 expression leads to increased LCYB activity (indirectly measured as increased CPTA tolerance) and pigment content (β-carotene and violaxanthin), but not necessarily to increased biomass and photosynthesis, whereas reduced NtLCYB expression negatively affects plant physiology and development, primary and secondary metabolism, photosynthetic efficiency, and ultimately plant biomass.
Materials and methods
Plant material and growth conditions
Tobacco (N. tabacum cv. Petit Havana) wild type, RNAi, and transplastomic lines were raised from seeds germinated in Petri dishes containing Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 30 g l−1 sucrose. Kanamycin (100 μg ml−1) was used for selection of nuclear-transgenic plants, and spectinomycin and streptomycin (500 μg ml−1) were used for selection of transplastomic plants. The leaf tissue for all the experiments (qPCR and pigment, metabolite, and hormone quantification) were taken from the fourth fully developed (bottom to the top) leaf from 6-week-old tobacco plants. For photosynthetic measurements, seedlings were transferred 14 d after germination to a soil–vermiculite mixture (2:1) and grown in a controlled-environment chamber at 350 μmol photons m−2 s−1 light intensity (16 h day, 22 °C, 75% relative humidity). For growth-related measurements (plant height, leaf surface, flower and leaf numbers), sampling, and seed production, plants were grown under greenhouse conditions.
Transplastomic vector design and transformation
The complete coding sequence of the carrot DcLCYB1 gene was synthesized by Thermo Fisher Scientific, and further amplified with primers designed to introduce 5′- and 3′-EcoRV restriction sites for incorporation into plastid expression vectors. The expression vector pDK325, containing a Chlamydomonas reinhardtii (Cr) CrpsbA promoter (CrPpsbA) coupled to the T7 phage gene10 leader (T7g10L) (Svab and Maliga, 1991) sequence upstream of the EcoRV site, and a CratpA terminator (CrTatpA), was used for creation of Nt-pJM36. The expression vector pDK326, containing the CrPpsbA and 5′-untranslated region and the CrTatpA, was used for creation of Nt-pJM37 vector. Both vectors included a chimeric aadA gene, including aadA and plastid expression elements (Svab and Maliga, 1993), for selection of transformed plants. Plant transformation was undertaken using the previously described biolistic protocols (Svab and Maliga, 1993; Ruf and Bock, 2011). Transformed plants were selected by growth on spectinomycin-containing medium (500 µg ml−1), and underwent several rounds of regeneration to obtain homoplasmic plants. Spontaneous spectinomycin-resistant lines were eliminated by double selection on medium containing both spectinomycin and streptomycin (500 µg ml−1 each) (Svab and Maliga, 1991; Bock, 2001). The homoplasmic state was confirmed by restriction fragment length polymorphism (RFLP) analysis. Briefly, total DNA was isolated by the CTAB method (Doyle and Doyle, 1990) from wild-type and transplastomic plants. BamHI-digested samples were separated by agarose gel electrophoresis, transferred to Hybond XL membranes by capillary blotting, and hybridized using an α-[32P]dCTP-labelled probe targeting the psaB gene fragment (Fig. 1C). Homoplasmic transplastomic lines were rooted on hormone-free medium and subsequently transferred to the greenhouse for seed production.
RNAi silencing vector design and transformation
Using the pENTR™ directional TOPO® Cloning Kit (Thermo Fisher Scientific), the PCR-derived NtLCYB fragment was cloned into pENTR/SD/D-TOPO® according to the manufacturer’s protocol. The PCR product (used for GatewayTM cloning) was amplified with a forward primer containing CACC at its 5′ end (5′-CACCTTGTTGGATTGCCTCGACGCC-3′) to match the overhang in the cloning vector (GTGG; reverse primer 5′-CTCCACTTCTGCCAATATGCC-3′). This entry vector was then used to perform a Gateway recombination reaction generating the final expression vector pK7GWIWG2 (I) with the NtLCYB RNAi regions in sense and antisense orientation. The Gateway® LR cloning (Thermo Fisher Scientific) reaction was carried out following the manufacturer’s instructions. Transformation of the NtLCYB RNAi constructs into tobacco (N. tabacum cv. Petit Havana) was undertaken by Agrobacterium tumefaciens-mediated gene transfer using bacterial strain C58C1:pGV2260 (Rosahl et al., 1987).
Physiological measurements and plant biomass experiment
T1 generation homoplasmic DcLCYB1 and T1 heterozygous NtLCYB RNAi seeds were used for physiological and biomass measurements as previously described (Moreno et al., 2020). T2 generation homoplasmic DcLCYB1 and T3 heterozygous NtLCYB RNAi seeds were used for germination (n=20), root length (n=6), and biomass quantification (n=6) experiments (10-day-old tobacco plants) in Petri dishes (repeated three times).
RNA isolation, cDNA synthesis, and qPCR experiments
For transplastomic and RNAi lines, the fourth leaf from the bottom of the plants and the leaf with the most severe phenotype were sampled. Leaf tissue was frozen immediately in liquid nitrogen and ground to a fine powder. RNA extraction, cDNA synthesis, and qPCR experiments were performed as previously described (Moreno et al., 2020), using the primers described therein. Three biological and three technical replicates per line were analysed. The relative transcript levels of each gene were determined using the formula (1+E)−ΔΔCtwhere E is the binding efficiency of the primers (Pfaffl, 2001). All the primers used in this study were previously assessed and tested for expression analysis using at least two references genes (Schmidt and Delaney, 2010; Albus et al., 2012; Kromdijk et al., 2016; Moreno et al., 2016; Armarego-Marriott et al., 2019; Moreno et al., 2020). In addition, expression analysis were performed following the same parameters and conditions as in Moreno et al. (2020), in which three different reference genes (ACTIN, UBIQUITIN, PDF2) were used. Therefore, in our current work only Actin transcript levels were measured as reference. Previously published primers that were used in this work can be found in Supplementary Table S1.
UHPLC analysis of pigments
Plant pigments were extracted and subsequently analysed by Acquity UPLC™ H-Class System (Waters, Milford, MA, USA) equipped with an autosampler, quaternary solvent manager and a photodiode-array detection detector as described in Moreno et al. (2020). Five biological replicates per line were measured and data were analysed with Empower software (v. 3, Waters, Manchester, UK).
ABA and GA measurements
Extraction and purification of ABA and GAs were performed using 15 mg (dry weight) leaf tissue following the procedure described in Turečková et al. (2009) and Urbanová et al. (2013). All data were processed using MassLynx™ software (v. 4.2, Waters, USA). Hormone levels were calculated on the basis of the standard isotope dilution method (Rittenberg and Foster, 1940).
Hormone and inhibitor treatments
Hormone and inhibitor treatments were performed as previously described (Moreno et al., 2020) but using liquid MS (Murashige and Skoog, 1962), supplemented with 1% sucrose, and 24-wells plates. Six tobacco seedlings per line were used in each treatment (n=6). Tobacco seedlings grown in agar MS medium (10-day-old) were transferred to liquid MS medium (with agitation) and treated for 7 d with hormones (GA3, GA4, ABA, 1 µM; GA3/ABA, 1 µM/0.66 µM; and GA4/ABA, 1 µM/0.66 µM) and inhibitors (paclobutrazol/PBZ, 1 µM). Six biological replicates for each transgenic line and wild type were grown in liquid MS medium in the 24-well plate to evaluate the effect of each hormone and hormone inhibitor treatment, and therefore each set of transgenic lines (transplastomic or RNAi lines) was compared separately.
Extraction and phase separation for GC and LC analyses
Tobacco samples (fourth fully developed leaf from 6-week-old plants) were extracted using methyl tertiary butyl ether buffer as previously described (Salem et al., 2016). After removal of the lipid phase, aliquots of 200 μl and 300 μl of the polar phase were transferred to new 1.5 ml Eppendorf tubes for GC-MS and LC-MS analysis, respectively. Samples were evaporated to dryness (speed-vac concentrator, Thermo Fisher Scientific) without heating.
Analysis of primary metabolites from the methanol–water phase by GC-MS
Samples were derivatized and analysed as previously described (Lisec et al., 2006; Caldana et al., 2013). Mass chromatograms were processed and peak areas integrated using the software Xcalibur (v. 4.0, Thermo Fisher Scientific). Peak annotation was performed by matching the retention index relative to the fatty acid methylesters and mass spectra against an in-house reference library (Cuadros-Inostroza et al., 2009). Results reported as log2 fold change were plotted as a heatmap using the pheatmap package (Kolde, 2019). The primary metabolite-reporting list following the recommendations described in Fernie et al. (2011) is provided in Supplementary Dataset S1.
Analysis of secondary metabolites from the methanol–water phase by UPLC-MS
Samples were re-suspended in 200 μl methanol–water (1:1, v/v) and analysed as previously described (Giavalisco et al., 2011). Data processing and statistical analysis was performed as previously described for primary metabolites. Peaks were annotated based on an accurate mass and elution profile of secondary metabolites, previously characterized in Nicotiana attenuata (Li et al., 2016). The secondary metabolite-reporting list following the recommendations described by Fernie et al. (2011) is provided in Supplementary Dataset S2.
Quantification of apocarotenoids using UHPLC-QQQ-MS/MS
Analysis of apocarotenoids from tobacco leaves was performed on a Vanquish™ Flex UHPLC System with an ACQUITY UPLC BEH C18 column (100×2.1 mm×1.7 mm) coupled with a QQQ-MS (TSQ Altis™ Triple Quadrupole Mass Spectrometer, Thermo Scientific) with a heated-electrospray ionization source according to the protocol modified from Mi et al. (2018). Briefly, tobacco leaves were harvested, lyophilized, and powdered. Approximate 25 mg tissue powder spiked with internal standards mixture (including D3-β-ionone, D3-β-apo-11-carotenal, D3-3-OH-β-apo-13-carotenone, D3-β-apo-13-carotenone, D3-β-apo-15-carotenal, D3-β-apo-14′-carotenal, D3-β-apo-12′-carotenal, D3-β-apo-10′-carotenal, and D3-β-apo-8′-carotenal; 2.5 ng of each standard) was extracted with methanol containing 0.1% butylated hydroxytoluene twice in an ultrasound bath, followed by centrifugation. The supernatant was collected and dried under vacuum. The residue was re-dissolved in 150 μl of acetonitrile–water (90:10, v/v) and filtered through a 0.22 µm filter for LC-MS analysis (Mi et al., 2018). Apocarotenoid profiling was performed by using UHPLC-QQQ-MS/MS in selective reaction monitoring (SRM) mode. The SRM transition list is shown in Supplementary Dataset S3. Apocarotenoid standards (Buchem BV, Apeldoorn, Netherlands) including β-cyclocitral, 3-OH-β-ionone, β-ionone, 3-OH-β-apo-11-carotenal, β-apo-11-carotenal, 3-OH-β-apo-13-carotenone, β-apo-13-carotenone, 3-OH-β-apo-15-carotenal, β-apo-15-carotenal, β-apo-14′-carotenal, 3-OH-β-apo-12′-carotenal, β-apo-12′-carotenal, 3-OH-β-apo-10′-carotenal, β-apo-10′-carotenal, 3-OH-β-apo-8′-carotenal, and β-apo-8′-carotenal were used to validate the identification of apocarotenoids from tobacco leaves using UHPLC-MS.
Photosynthesis measurements
Measurements of chlorophyll content and the Chl a/b ratio (fourth leaf from bottom to the top) were undertaken with a Jasco V-630 photometer (Jasco GmbH, Groß-Umstadt, Germany) in 80% (v/v) acetone (Porra et al., 1989). Chl a fluorescence of intact plants was measured using a DUAL-PAM-100 instrument (Heinz Walz GmbH, Effeltrich, Germany) after 30 min of dark adaptation. Light intensity was increased stepwise from 0 to 2500 µE m−2 s−1, with a measuring time of 150 s per step under light-limited conditions and of 60 s under light-saturated conditions. Light-response curves of non-photochemical quenching (qN; Krause and Weis, 1991), the redox state of the PSII acceptor side (qL; Kramer et al., 2004), and of the donor-side limitation of PSI (Y(ND); Schreiber and Klughammer, 2016) were determined. Linear electron transport was corrected for leaf absorptance, which was calculated from leaf transmittance and reflectance spectra as 100% minus transmittance (%) minus reflectance (%). Spectra were measured between 400 and 700 nm wavelengths using an integrating sphere attached to a photometer (V650, Jasco Inc.). The spectral bandwidth was set to 1 nm, and the scanning speed was 200 nm min−1.
Microscopy techniques
Leaf samples at similar developmental stages were fixed for a period of 1 h in a solution containing 45% ethanol, 5% glacial acetic acid, and 5% formaldehyde. The samples were subjected to ethanol dehydration series for observing leaf anatomy. Subsequently the samples were infiltrated with 1% Hardener I in Technovit 7100 and incubated for up to 24 h after a short 2 h infiltration with Technovit 7100. The embedding was carried out using 15 parts of infiltration solution with 1 part of Hardener II in molds and polymerized at room temperature overnight. Sectioning was undertaken using a rotary microtome (Leica RM2265) and sections of 5 µm were prepared and stained with 0.05% toluidine blue and visualized and recorded using an Olympus light microscope (BX51).
For chloroplasts counting, leaves were selected and sampled using the same procedure as for the leaf cross sections (see above). To each leaf piece 1.5 ml fixation solution (TissuePrep Buffered 10% Formalin; Electron Microscopy Sciences; Hatfield, PA, USA) was added and incubated for 1.5 h under vacuum. Subsequently, the fixation solution was replaced by fresh solution and the tubes were constantly inverted for 2 h. Then, samples were incubated at 4 °C overnight and afterwards heated for 3 h at 60 °C. Images of fixed leaf cells were taken using an Olympus light microscope (BX51). Chloroplasts of 20 cells per line were counted.
For chloroplast ultrastructural analysis, leaf tissue (of the fourth leaf of the transplastomic and the most variegated of the RNAi lines) was fixed in 2.5% glutaraldehyde supplemented with 0.2 M sodium cacodylate for a period of 8 h. The samples were then incubated with 2% osmium tetroxide for a period of 4 h. The samples were rinsed and taken through dehydration steps and embedded in Epon resin using standard protocols (Austin and Staehelin, 2011). Sections of 1–2 µm were cut by diamond knife, and stained (2% uranyl acetate and lead citrate) prior to imaging using a Zeiss EM 912 Omega transmission electron microscope.
Statistical analysis
Three main experiments with transgenic plants (transplastomic and RNAi lines) growing under greenhouse or fully controlled conditions (phytochamber) were performed in this work. Transgenic lines were compared in two groups (transplastomic and RNAi lines) due to the high number of lines and biological replicates. Thus, transplastomic and RNAi lines were always grown and compared in two different groups with their own wild-type control. The same leaf tissue (fourth leaf bottom to top) from 6-week-old tobacco was used to perform all the molecular analysis. The first experiment included a set of tobacco plants used for molecular analysis such as qPCR (n=3 and three technical replicates), pigment quantification via UPLC (n=5), and hormone (n=5 and three technical replicates) quantification. In addition, for primary and secondary metabolites (n=6) statistical analysis was performed in R (R Core Team, 2018) using an unpaired Wilcoxon test with the default parameters within the function compare_means of ggpubr package (Kassambara, 2018). The second main experiment consisted of tobacco plants used for physiological parameters and biomass quantification (n=5) of tobacco plants grown in the greenhouse. Due to the high number of transgenic lines and biological replicates, the experiments were performed with 3 d of difference, starting with the transplastomic lines and then the RNAi lines (each transgenic group with their own wild type). The third main experiment consisted of a set of tobacco plants used for photosynthetic analysis (n=3–8). Plants were grown in a controlled-environment chamber in a consecutive manner due to space constrains (first transplastomic and then RNAi lines, each group with its own wild-type control). In a fourth experiment, tobacco seedlings grown in solid MS medium were used. Germination rate (n=20), root length (n=6), and biomass (n=6) experiments were performed in 10-day-old tobacco seedlings. These experiments were repeated three times in different Petri dishes. For the last (fifth) experiment, corresponding to the hormone and hormone inhibitors experiments, 10-day-old tobacco seedlings growing on half-strength solid MS medium were transferred to a 24-well plate. Each row consisted of six wild type, and six replicates of the three transplastomic or RNAi lines, and therefore the plants within a plate were always compared with each other (these plants came from the same Petri dish where they grew for 10 d). Due to the experimental design for the five different data sets of plants, an unpaired, two-tailed Student’s t-test was performed to compare each set of transgenic lines with the wild type (unless otherwise stated). GraphPad Prism 5.0 software was used to prepare the figures and perform the t-tests.
Results
Generation of homoplasmic Daucus carota lycopene-β-cyclase1 expressing lines
The full-length carrot DcLCYB1 gene was inserted into the pDK325 and pDK326 vectors to generate the pJM36 and pJM37 chloroplast transformation vectors, respectively (Fig. 1B). The pJM36 and pJM37 vectors contained the Chlamydomonas reinhardtii (Cr) PsbA promoter (CrPpsbA) and CrAtpA terminator (CrTatpA), flanking the synthetic DcLCYB1 gene, and a chimeric spectinomycin resistance gene (aadA, an aminoglycoside adenylyltransferase) as selectable marker. In addition, the CrPpsbA in the pJM36 vector was fused to the strong translation initiation signals derived from the gene10 leader of coliphage T7 (T7g10L) (Kuroda and Maliga, 2001). Both constructs were introduced into the genome of Nicotiana tabacum cv. Petit Havana by biolistic chloroplast transformation (Svab and Maliga, 1993; Ruf et al., 2001; Ruf and Bock, 2011), and transplastomic plants were selected on spectinomycin-containing medium (see Materials and methods). Resistance to spectinomycin can arise from expression of the aadA marker gene, but also from spontaneous mutations proximal to the tRNA binding region of the 16S rRNA. As these spontaneous mutants are still sensitive to streptomycin, which, however, is detoxified by aadA (Svab and Maliga, 1991), mutant lines isolated on spectinomycin were further tested for streptomycin resistance.
Interestingly, only two aadA-containing positive (spectinomycin and streptomycin resistant plants) and 20 spontaneous mutant (plants resistant to spectinomycin but sensitive to streptomycin) (Svab and Maliga, 1991) lines were obtained with the pJM36 vector (T7g10L; Supplementary Table S2). By contrast, 23 positive transformants and only 18 spontaneous mutants were obtained with the pJM37/LCYB1 vector (Supplementary Table S2). The high number of spontaneous mutants relative to true mutants (Supplementary Table S2) suggests the possibility of a toxic effect attributed to the transformation with the pJM36/LCYB1 vector (Svab and Maliga, 1991). Moreover, one of the positive pJM36/LCYB1 lines died during the tissue culture process, supporting the idea of a toxic effect caused by the extremely high expression of the transgene. Due to this, we performed further analysis with only one pJM36/LCYB1 line. From the obtained lines, seven putative transformants (one transformed with pJM36 and six with pJM37) were purified to homoplasmy by passing them through additional regeneration cycles under antibiotic selection. To confirm chloroplast transformation, proper integration of the transgene via homologous recombination and homoplasmy of the transplastomic lines, RFLP analysis using Southern blotting was performed (Fig. 1C). BamHI digestion of the wild type DNA generated a single band of 4.5 kb (Fig. 1C), while for the seven transgenic lines a band of 7.9 kb (consistent with expected length following the insertion of the transgene and the respective plastid expression elements), but no 4.5 kb wild type band, was obtained (Fig. 1C). In addition, and as an ultimate support for the homoplasmic state, a large-scale inheritance test was conducted, involving germination of T1 seeds on spectinomycin-containing medium (Fig. 1D) (Bock, 2001). No appearance of antibiotic-sensitive seedlings, as could be observed for the wild type, was observed in any of the analysed lines (Fig. 1D), suggesting a homoplasmic state in the DcLCYB1 transplastomic lines. Homoplasmic T1Nt-pJM37/LCYB1 plants were indistinguishable from the wild-type control at the seedling stage, indicating that integration and expression of the transgene was phenotypically neutral at early stages of development. By contrast, it seems that very high LCYB1 expression in the Nt-pJM36/LCYB1 plants caused a toxic effect (probably causing an imbalance in phytohormone content, see Discussion) that impaired plant development impeding the generation of more transplastomic lines.
Increased DcLCYB1 expression results in increased pigment content in transplastomic DcLCYB1 lines
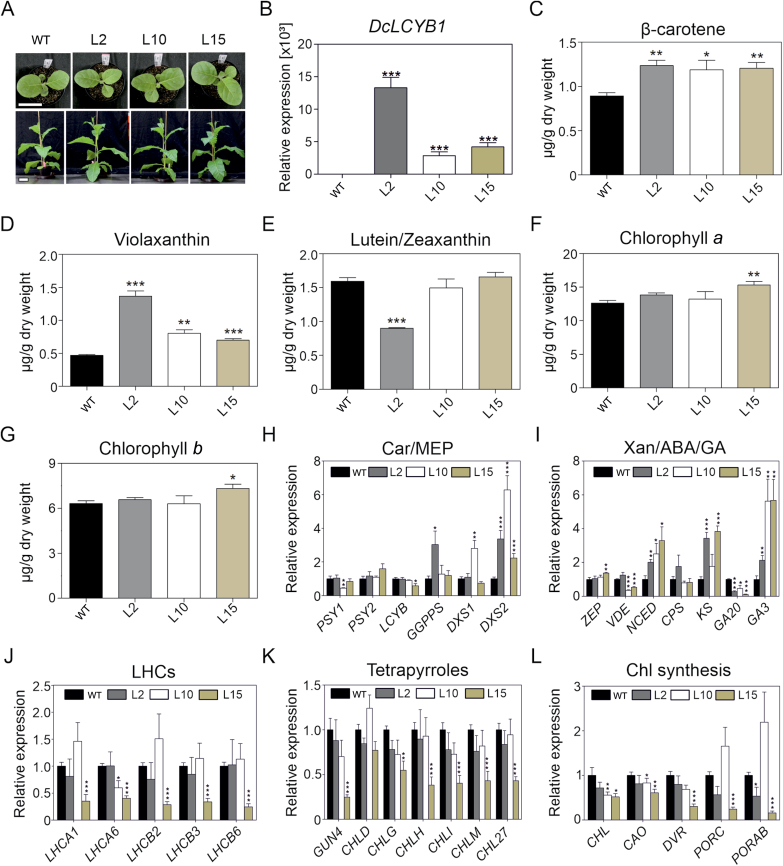
From the seven confirmed homoplasmic lines, we choose the lines L2 (pJM36), L10, and L15 (pJM37) to perform further molecular analysis. All three lines showed a phenotype similar to the wild type after 2 and 4 weeks of growth under greenhouse conditions (Fig. 2A). Line L2 was transformed with the pJM36 vector containing the expression element T7g10L (Fig. 1A), and therefore DcLCYB1 expression was much higher (~12 000-fold relative to the wild type) than that of line L10 and L15, which were transformed with the pJM37 vector without the T7g10L expression element (~5000-fold higher relative to the wild type; Fig. 2B). Despite the differences in expression level, all three lines had an increase of approximately 30% in β-carotene content (Fig. 2C). This correlated with the increased tolerance to CPTA, which specifically inhibits LCYB activity (Schuetz and Baldwin, 1958; Tao et al., 2004), observed in the transplastomic lines subjected to CPTA treatment (Supplementary Fig. S1). Wild-type plants showed chlorosis/leaf variegation and stunted growth, while lines L10 and L15 only showed moderate variegation and line L2 remained completely green after 8 d of CPTA treatment (Supplementary Fig. S1C). Enhanced CPTA tolerance indirectly suggests increased LCYB activity in the transgenic lines. In addition, violaxanthin content was increased in all lines, about 190%, 82%, and 48% for line L2, L10, and L15, respectively (Fig. 2D). Additionally, line L2, which has the highest DcLCYB1 expression, showed a decrease to 56% of the wild type in lutein/zeaxanthin content (Fig. 2E). Moreover, chlorophyll a and b contents were significantly increased in line L15 (21% and 16%, respectively; Fig. 2F, G).
Fig. 2.
Highly increased DcLCYB1 expression causes no changes in phenotype but increases pigment content and affects gene expression in tobacco plants. (A) Two-week-old (upper) and 4-week-old (lower) transplastomic and wild-type plants. Scale bar: 10 cm. (B) DcLCYB1 expression levels measured by qPCR. (C–G) Pigment content (carotenoids and chlorophylls) measured by UPLC. (H–L) Gene expression analysis of carotenoid and carotenoid-related pathways. The expression of Actin as a stable reference gene was used for normalization. Increases, decreases and/or no change in all three lines for the majority of the genes involved in carotenoid, MEP, chlorophyll, GA, LHC, and tetrapyrrole pathways were observed. Columns and bars represent the means and SEM for the qPCR (three biological replicates and three technical replicates) and UPLC (five biological replicates) experiments. Unpaired two-tailed Student’s t-test was performed to compare transgenic lines with the wild type. *P<0.05, **P<0.001, ***P<0.0001. ABA, abscisic acid; Car, carotenoids; Chl, chlorophyll; GA, gibberellins; L2, pJM36-2; L10, pJM37-10; L15, pJM37-15; LHC, light harvesting complex; MEP, 2-C-methyl-D-erythritol 4-phosphate; WT, wild type; Xan, xanthophylls.
Recently, it was shown that expression of DcLCYB1 in tobacco (cv. Xanthi) leads to a general increase in transcripts of carotenoid and carotenoid-related genes (Moreno et al., 2020). However, such a general positive impact on carotenoid and carotenoid-related pathways was not observed in the transplastomic lines (Fig. 2H–L; Supplementary Fig. S2A, B). Transcript accumulation of the majority of the genes analysed did not increase (it was unchanged or reduced), possibly due to a negative impact of extremely high DcLCYB1 expression (feedback regulation) on another isoprenoid pathway. However, some changes were consistently significant in all lines: steady state accumulation of deoxyxylulose 5-phosphate synthase2 (DXS2) transcripts was highly increased, 3.4-, 6.3-, and 2.4-fold in lines L2, L10, and L15, respectively (Fig. 2H). Additionally, 9-cis-epoxycarotenoid dioxygenase (NCED) transcript accumulation was increased, 2-, 2.5-, and 3.3-fold, respectively, in the three lines (Fig. 2I). Transcript accumulation of genes encoding key enzymes involved in GA biosynthesis, gibberellin 20- and 3-oxidases (GA20ox and GA3ox), was reduced (3.6-, 2.1-, and 9.1-fold in L2, L10, L15, respectively) and increased (2.1-, 5.6-, and 5.7-fold in L2, L10, L15, respectively) in all the lines, respectively (Fig. 2I). Furthermore, the majority of transcripts of tetrapyrrole and chlorophyll biosynthesis genes and genes encoding LHC subunits were decreased in line L15 (Fig. 2J–L). Transcript abundance of genes encoding subunits of PSI and PSII and the cytochrome b6f (cyt b6f) complex were not consistently changed across all lines (Supplementary Fig. S2A, B).
Reduction in NtLCYB expression leads to a decrease in pigment content and negatively impacts gene expression in carotenoid and carotenoid-related pathways
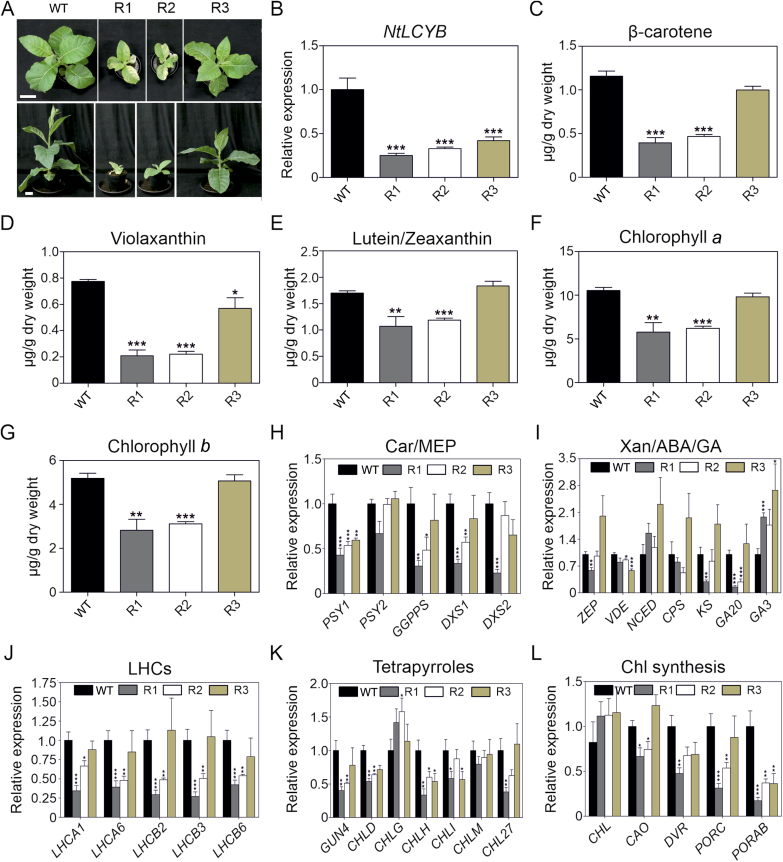
Reduction in NtLCYB gene expression was accomplished through silencing using RNAi. Reduced NtLCYB expression levels resulted in a variegated phenotype for approximately half of the transformants (10) growing in sugar-supplemented medium (Supplementary Fig. S3A). These lines were able to later grow photoautotrophically in soil under greenhouse conditions (Supplementary Fig. S3B). The other half presented a white/pale green phenotype when grown in synthetic medium and were not able to survive after transfer to soil (Fig. S3A, C). These phenotypes suggest the importance of proper β-carotene levels for plant viability. The T1 generation of NtLCYB RNAi lines was obtained and three independent transgenic lines with different strengths of variegation (R1>R2>R3) were chosen to perform further molecular analysis. Lines R1, R2, and R3, showing 25%, 33%, and 42% residual expression of NtLCYB gene, showed delayed development compared with the wild type (Fig. 3A, B). The relative fitness and residual NtLCYB expression level are reflected in the reduction of β-carotene content, which decreased ~65% for the line with the strongest phenotype (R1) and ~15% for the line with the weakest phenotype (R3; Fig. 3C). Furthermore, violaxanthin content was significantly reduced in all lines (Fig. 3D). In addition, lutein/zeaxanthin and chlorophyll a and b content were decreased in R1 and R2 but remain comparable to those of the wild type in R3 (Fig. 3E–G).
Fig. 3.
Reduced NtLCYB expression causes decreases in plant growth, pigment content, and gene expression in tobacco plants. (A) Side and top view from 4-week-old RNAi and wild type plants. Scale bar: 10 cm. (B) NtLCYB expression levels measured by qPCR. (C–G) Pigment content (carotenoids and chlorophylls) measured by UPLC. (H–L) Gene expression analysis of carotenoid and carotenoid-related pathways measured by qPCR. The expression of Actin as a stable reference gene was used for normalization. A general decrease in transcript abundance of all three lines for the majority of the genes involved in carotenoid, MEP, chlorophyll, and GA pathways was observed. Decrease in transcript accumulation of genes encoding LHC subunits and enzymes involved in tetrapyrrole biosynthesis was observed. Columns and bars represent the means and SEM for the qPCR (three biological replicates and three technical replicates) and UPLC (five biological replicates). Unpaired two-tailed Student’s t-test was performed to compare transgenic lines with the wild type. Letters represent statistical differences among the lines. *P<0.05, **P<0.001, ***P<0.0001. ABA, abscisic acid; Car, carotenoids; Chl, chlorophyll; GA, gibberellins; LHCs, light harvesting complex; MEP, 2-C-methyl-D-erythritol 4-phosphate; WT, wild type; Xan, xanthophylls.
In order to understand the reductions in carotenoid and chlorophyll content, we performed gene expression analysis to measure transcript abundance of genes encoding enzymes involved in carotenoid and chlorophyll biosynthesis. Carotenoid, chlorophyll, and GA pathways share a common biosynthetic precursor, geranyl geranyl diphosphate (GGPP), which is produced via the MEP pathway. For this reason, key genes of the MEP and GA pathways were also analysed. Expression of genes with products involved in the production of GGPP such as GGPP synthase (GGPPS), DXS1, and DXS2 were strongly reduced in R1 and R2 (Fig. 3H). Moreover, PSY1 transcript was reduced ~50% for all lines while PSY2 transcript remain unchanged.
Transcript abundance of other carotenoid biosynthetic genes such as ZEP and VDE, which encode enzymes involved in the xanthophyll cycle, were not consistently reduced in the RNAi lines (ZEP was reduced in R1 while VDE was reduced in R2 and R3; Fig. 3I). In addition, transcript of NCED, which encodes the enzyme catalysing the first step of abscisic acid biosynthesis, was unchanged in the RNAi lines (Fig. 3I). CPS and KS (with products involved in the early stage of the GA pathway) transcripts remain mostly constant in all the lines (significant decrease of KS in R1; Fig. 3I). Interestingly, GA20ox expression was decreased in R1 and R2 (Fig. 3I). It was previously reported that the expression of GA20ox has a strong impact on plant height, with higher expression leading to larger plants and lower expression resulting in dwarf phenotypes (Davière and Achard, 2013). Therefore, these changes may partially account for the phenotypes observed in line R1 and R2 compared with the wild type (Coles et al., 1999; Vidal et al., 2001). Moreover, GA3-OXIDASE is encoded by GA3ox acting downstream of the GA20ox converting inactive GA into bioactive GA such as GA1 and GA4 (Gallego-Giraldo et al., 2008; Hedden and Thomas, 2012). In our RNAi lines, GA3ox was increased in all transgenic lines (Fig. 3I), perhaps as a response to counteract the GA deficit.
The accumulation of transcripts encoding five LHCs from PSI and PSII (LHCA1, LHCA6, LHCB2, LHCB3, and LHCB6) was reduced in the two strongest RNAi lines while in R3 LHC transcript accumulation was not changed (Fig. 3J). In addition, the transcript accumulation of genes encoding enzymes involved in tetrapyrrole biosynthesis was decreased in R1 (GUN4, CHLD, CHLH, CHLI, CHL27), R2 (GUN4, CHLD, CHLH), and R3 (CHLH, CHLI; Fig. 3K). Moreover, accumulation of the transcripts of genes encoding enzymes involved in chlorophyll biosynthesis such as CAO (R1 and R2), DVR (R1), PORC (R1 and R2), and PORAB (R1, R2 and R3) was reduced (Fig. 3K). Strikingly, the majority of transcripts encoding subunits of PSI, PSII, and cyt b6f were decreased in R1 (19 out of 20), R2 (13 out of 20), and R3 (seven genes) lines (Supplementary Fig. S2C, D). These results indicate a relationship between the reduction of the NtLCYB transcript, the strength of the phenotype, and the reduction in the different key photosynthesis and photosynthesis-related genes.
Altered LCYB expression influences plant biomass in tobacco plants
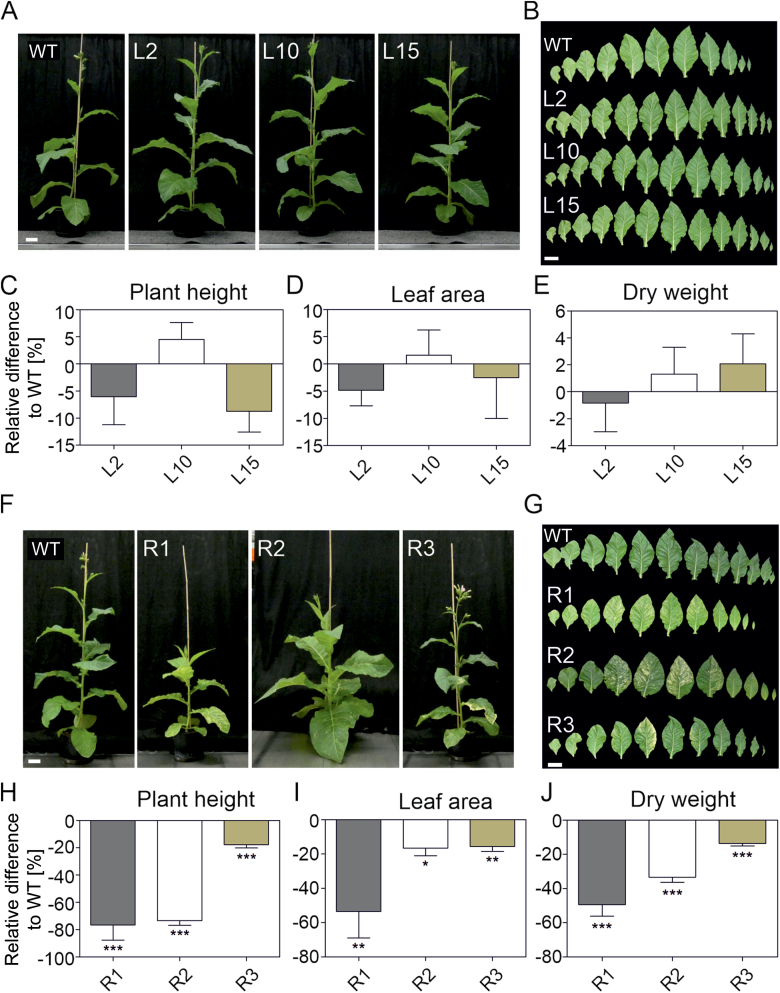
As mentioned earlier, transplastomic lines expressing the DcLCYB1 gene were phenotypically indistinguishable from the wild type (Fig. 2A), while 4-week-old LCYB RNAi lines showed leaf variegation, delayed development, and reduced growth (Fig. 3A). Moreover, at the end of the tobacco life cycle transplastomic lines were similar in height to the wild type (Supplementary Fig. S4A). By contrast, at the end of the plant life cycle (3-month-old plants), R2 and R3 plants had a comparable height to wild type plants while R1 remained dwarfed and produced fewer flowers (Supplementary Fig. S4B; Supplementary Table S3).
In order to analyse the relationship between LCYB transcript/β-carotene production and plant biomass, we performed physiological and biomass measurements with transplastomic DcLCYB1 and NtLCYB RNAi lines containing increased and reduced LCYB transcript levels (Fig. 4). Transplastomic plants visibly resembled the wild type (Fig. 4A, B) and showed no differences in plant height or leaf area through different time points of their development (Fig. 4C–E; Supplementary Fig. S5A–D). By contrast, the RNAi lines showed a variegated-plant phenotype and delayed development (Fig. 4F), most pronounced in R1, then R2 and R3 (Fig. 4G–J). Interestingly, the variegated phenotype not only appeared in the leaves but also in stem and capsules of the two strongly affected RNAi lines (R1 and R2; Supplementary Fig. S4C, D). Physiological parameters such as plant height, leaf area, and plant biomass were drastically reduced in all three RNAi lines, with more pronounced reductions in the mutants with the stronger reductions, R1 and R2 (Fig. 4H–J). Furthermore, quantification of plant height, leaf area, leaf number, and internodal space through development supports our observation of the different phenotypic strength of each line (Supplementary Fig. S5E–H). This phenotype was also observed in the T3 generation of tobacco lines, in which R1 was even more affected than the parental R1 plants, probably due to homozygosity in this line (Supplementary Fig. S6A–C).
Fig. 4.
Plant productivity of transplastomic DcLCYB1 and NtLCYB RNAi lines. (A) Side view of 5-week-old wild type and transplastomic tobacco plants. (B) Leaf series of 5-week-old wild type and transplastomic lines. (C–E) Plant height, leaf area, and dry weight biomass of 4-week-old transplastomic plants. (F) Side view of 5-week-old wild type and RNAi tobacco plants. (G) Leaf series of 5-week-old wild type and RNAi lines. (H–J) Plant height, leaf area, and dry weight biomass of 4-week-old RNAi plants. Columns and bars represent the means and SEM of five biological replicates. Unpaired two-tailed Student’s t-test was performed to compare transgenic lines with the wild type. *P<0.05, **P<0.001, ***P<0.0001. Scale bar: 10 cm. WT, wild type.
Altered LCYB expression influence hormone metabolism and apocarotenoid synthesis in transgenic tobacco plants
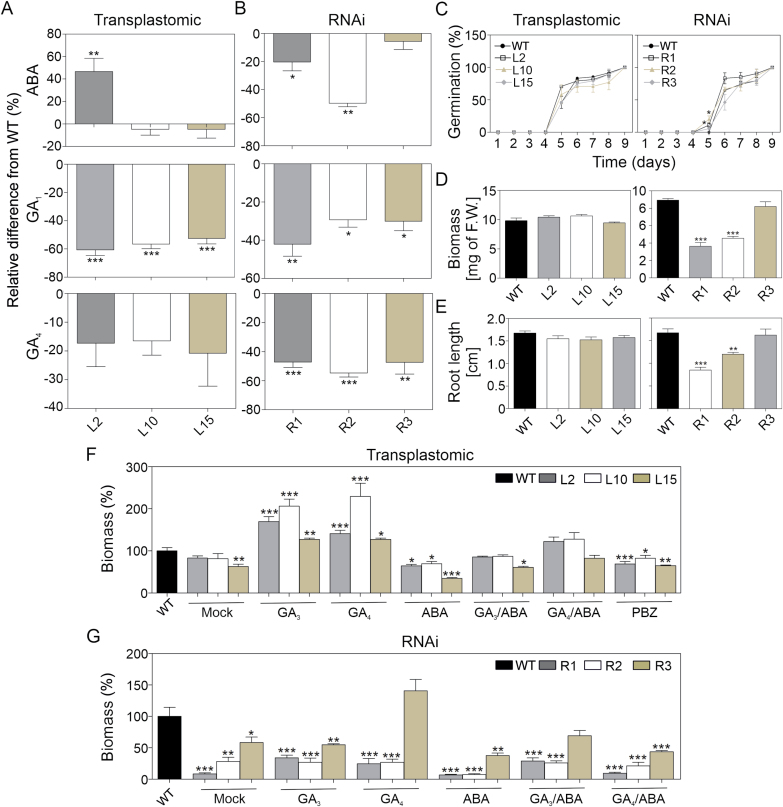
Hormone content was altered in both transplastomic and RNAi lines. Interestingly, ABA content was increased only in L2, the line with the highest DcLCYB1 expression levels (Fig. 5A). GA1 was reduced ~60% in all three lines while GA4 remain unchanged (Fig. 5A). ABA (R1 and R2, ~20–40%), GA1 (~30%), and GA4 (~40–55%) contents were strongly reduced in the RNAi lines (Fig. 5B). Due to the changes observed in GA and ABA content in transplastomic DcLCYB1 and LCYB RNAi lines, we measured ABA and GA metabolic intermediaries to gain further insights into GA and ABA metabolism. As expected, various GA and ABA metabolic intermediaries were affected in transplastomic (Supplementary Fig. S7) and RNAi lines (Supplementary Fig. S8). These results suggest that LCYB expression can modify hormone content, and therefore influence plant development and physiology. For instance, unchanged or slightly increased ABA levels in the transplastomic lines did not influence seed germination or plant biomass at the seedling stage (Fig. 5C–E) as was previously reported for increased ABA content (Garciarrubio et al., 1997). By contrast, reduced ABA levels in R1 and R2 were reflected in early seed germination, reduced plant biomass, and shorter primary root length in tobacco seedlings compared with the wild type (Fig. 5C–E). To further dissect the hormonal effect on our plants we designed an experiment in which 10-day-old tobacco seedlings grown on agar MS medium were transferred to liquid MS medium in 24-well plates, and were subjected to hormone and inhibitor treatments for 7 d. Interestingly, results from hormone and inhibitor treatments shed light on how these tobacco lines can achieve higher/lower biomass (Fig. 5F, G). Transplastomic L2 and L10 lines treated with water (mock) showed the same plant biomass and architecture as the wild type, while the biomass of L15 was slightly lower (Fig. 5F; Supplementary Fig. S9A). GA3 and GA4 treatments resulted in increased biomass in all lines and the wild type, while ABA treatment resulted in decreased biomass. Under treatment with paclobutrazol, a specific inhibitor of GA synthesis, biomass of the transplastomic lines was also reduced, relative to the wild type (Fig. 5F). In addition, combinations of GA3 and GA4 with ABA did not alter plant biomass in the transplastomic lines. Water-treated (mock) RNAi lines showed reduced biomass compared with the wild type and confirmed the observed phenotype strength observed under greenhouse conditions (Figs 5G, 3A; Supplementary Fig. S9B). Interestingly, in the most affected RNAi lines, R1 and R2, neither GA3 or GA4 nor combinations of GA3 and GA4 with ABA treatments could rescue the phenotype to the wild-type level (Fig. 5G). In addition, RNAi lines showed reduced biomass after ABA treatment. Taken together these results suggest a slightly and profoundly altered hormonal balance in the transplastomic and RNAi lines, respectively.
Fig. 5.
Hormone metabolism and its effect on plant physiology. (A) Abscisic acid and gibberellin content in transplastomic lines. (B) Abscisic acid and gibberellin (GA1 and GA4) content in RNAi lines. Columns and bars represent the means and SEM of five biological replicates and three technical replicates. (C) Seed germination in DcLCYB1 transplastomic and NtLCYB RNAi lines (three independent plates, n=20). (D) Biomass of 10-day-old DcLCYB1 transplastomic and NtLCYB RNAi lines (three independent plates, n=6). (E) Root length of 10-day-old DcLCYB1 transplastomic and NtLCYB RNAi lines (three independent plates, n=6). (F) Hormone and inhibitor treatments in 10-day-old DcLCYB1 transplastomic lines. (G) Hormone and inhibitor treatments in 10-day-old NtLCYB RNAi lines. Tobacco seedlings grown in agar MS medium (10-day-old) were transferred to liquid MS medium (with agitation) and treated for 7 d with hormones (GA3, GA4, ABA, 1 µM; GA3/ABA, 1 µM/0.66 µM; GA4/ABA, 1 µM/0.66 µM) and inhibitors (paclobutrazol/PBZ, 1 µM). Biomass was calculated as percentage for each treatment normalized to wild type as 100% in each respective treatment (n=6 for each transgenic line and wild type). Unpaired two-tailed Student’s t-test was performed to compare transgenic lines with the wild type (*P<0.05, **P<0.001, ***P<0.0001). ABA, abscisic acid, GA, gibberellins, GA3, gibberellin A3; GA4, gibberellin A4; PBZ, paclobutrazol; WT, wild type.
β-Carotene-derived apocarotenoids (e.g. β-cyclocitral and zaxinone) play roles as growth (promoting) regulators in Arabidopsis, rice, and tomato (Dickinson et al., 2019; Wang et al., 2019); because carotenoid levels were altered in the DcLCYB1 and LCYB RNAi lines (Figs 2C–E, 3C–E), we profiled apocarotenoid accumulation to determine if some of these molecules were also altered and thus may contribute to the growth phenotype observed in our tobacco lines. Due to the similar changes in carotenoid content obtained in transplastomic lines, we chose only one transplastomic line (L15) (Fig. 2C–E) for profiling, as well as R1, R2, and R3 RNAi lines because they showed different reductions in carotenoid content (Fig. 3C–E). Using a UHPLC-MS method (Mi et al., 2018) we quantified different non-hydroxylated and hydroxylated apocarotenoids (Supplementary Fig. S10A, B). Despite increases in β-carotene and violaxanthin in the transplastomic lines, apocarotenoid levels remain the same as those in the wild type (Supplementary Fig. S10A, B). From the RNAi lines, only R1 (the most affected line at the phenotypic and molecular level) showed significant changes in apocarotenoid content including increases in β-apo-8′-, β-apo-10′-, β-apo-12′-, and 3-OH-β-apo-8′-carotenal. In addition, other apocarotenoids, such as β-apo-11-, β-apo-15-, 3-OH-β-apo-10′-, 3-OH-β-apo-11-, 3-OH-β-apo-14′-, and 3-OH-β-apo-15-carotenal, showed slight but significant reductions (Supplementary Fig. S10A, B). Although β-cyclocitral (β-cc) did not show a significant reduction, 3-OH-β-apo-13-carotenone (zaxinone), another growth (promoting) regulator in rice (Wang et al., 2019), was strongly reduced in the most affected line (~30%, P<0.005). These results suggest that (i) strong reductions in carotenoid accumulation cause strong changes in apocarotenoid content, and (ii) zaxinone might be contributing to the observed growth phenotype in the RNAi lines.
Chloroplast structure and photosynthetic analysis in DcLCYB1 transplastomic and NtLCYB RNAi lines
In order to understand how phenotypic changes were present at a microscopic level (e.g. leaf structure), we performed light microscopy on sectioned leaf tissue of transplastomic and RNAi lines (Supplementary Fig. S11). We selected two transplastomic lines, one with the strongest LCYB1 expression (L2) and another with moderate expression (L15), and in the case of the RNAi lines in terms of phenotype (yellow tissue/green tissue). As R1 and R3 exhibit a strong and moderate variegated-leaf phenotype, respectively, we decided to examine both green and yellow tissue independently.
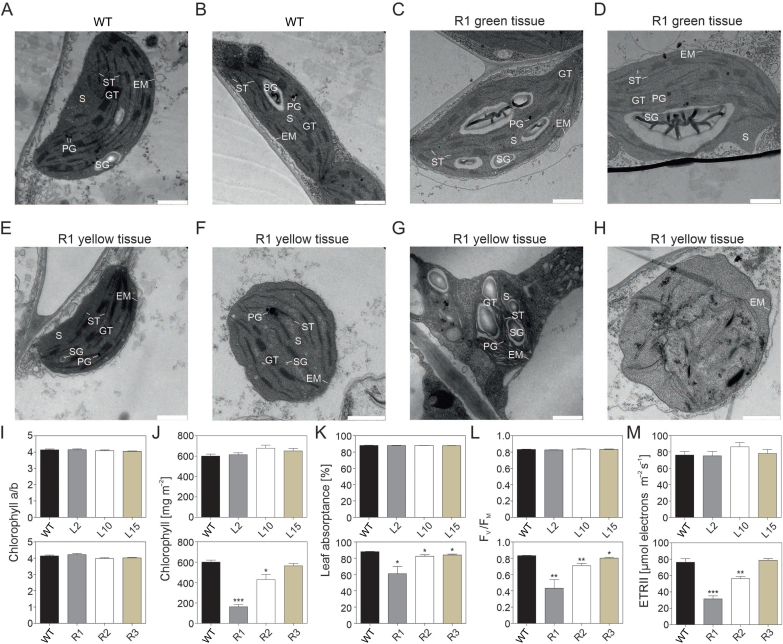
In the transplastomic lines and in the RNAi line R3, no changes in the arrangement of cell layers in leaf cross sections were observed compared with the wild type (Supplementary Fig. S11A–C, F, G). By contrast, in yellow tissue of the strong variegated R1 line, palisade cells were smaller with reduced spacing between the cells (Supplementary Fig. S11E). Furthermore, a significantly increased number of chloroplasts was observed in the transplastomic lines (P<0.05) and decreased number in the RNAi lines (P<0.0001; Supplementary Table S4). To gain further insight into chloroplast ultrastructure, we performed transmission electron microscopy of wild type, R1 (strong phenotype), and R3 (mild phenotype) leaves. We observed that wild-type chloroplasts were organized side by side against the cell contour or densely packed in some regions of the cells. The green sectors of R1 leaves did not have a significantly different number of chloroplasts (16 per 1000 μm2) from the wild type (19 per 1000 μm2) and they had similarly positioned chloroplasts (Supplementary Fig. S12A–D; Supplementary Table S4). Chloroplast structure and organization was similar in R1 and R3, but the mild R3 line showed alleviated effects compared with the strongest R1 line. Green sectors of R1 and R3 leaves exhibited wild-type-like chloroplasts (Fig. 6C, D; Supplementary Fig. S12A–D, G–J). Yellow sectors of the R1 and R3 leaves showed four different types of chloroplast shapes: wild-type-like (30% and 52%, respectively), round (41% and 39%, respectively; chloroplasts not present close to the cell contour unlike the wild type), chloroplasts with several starch granules and an impaired thylakoid structure (16% and 6%, respectively), and chloroplasts with completely altered structure (14% and 4%, respectively; Fig. 6E–H; Supplementary Fig. S12E, F, K–N). By contrast, yellow sectors of R1 leaves had only a very few chloroplasts, which were either not present close to the cell contour, unlike the wild type, or unstructured (Supplementary Fig. S12E, F).
Fig. 6.
Chloroplast ultrastructure and photosynthetic analysis of DcLCYB1 transplastomic and NtLCYB RNAi lines. Transmission electron microscopy images of chloroplasts of wild type plants and green and yellow sectors of 4-week-old R1 lines. (A, B) Wild type chloroplasts. (C, D) Chloroplasts in the green tissue of R1 leaves are similar to those of the wild type, but differences in starch granule number, apparent size, and shape can be observed. (E–H) A wide range of phenotypes were observed in the yellow sectors of R1 leaves (wild type-like, 23.3%; round, altered chloroplast morphology with impaired thylakoid structure, 5.6%; and unstructured and round chloroplasts 71.1%). (I–M) Photosynthetic parameters in transplastomic (upper panel) and RNAi (bottom panel) lines: (I) Chlorophyll a/b ratio; (J) chlorophyll content; (K) leaf absorptance; (L) maximum quantum efficiency of PSII in the dark-adapted state (Fv/Fm); and (M) ETRII, linear electron flux capacity, which was corrected for leaf absorptance. Transplastomic and RNAi lines were 4 weeks old. Columns and bars represent the means and SEM of three to eight biological replicates. Unpaired two-tailed Student’s t-test was performed to compare transgenic lines with the wild type. *P<0.05, **P<0.001, ***P<0.0001. EM, envelope membranes; GT, grana thylakoids; PG, plastoglobules; S, stroma; SG, starch granule; ST, stroma thylakoids; WT, wild type. Scale bar: 1000 nm.
In order to better understand the impact of altered leaf and chloroplast structure on plant growth, we measured several photosynthetic parameters. Measurements were always performed on the youngest fully expanded leaves, to avoid effects of leaf shading or from the onset of leaf senescence. In transplastomic plants in their vegetative state (before flower production), the chlorophyll a/b ratio, chlorophyll content (a and b), leaf absorptance, maximum quantum efficiency of PSII in the dark-adapted state (Fv/Fm), and the light-saturated capacity of linear electron transport (ETRII) did not change compared with the wild type (Fig. 6I–M). Accordingly, light response curves of linear electron transport (Supplementary Fig. S13A), photoprotective non-photochemical quenching (qN, Supplementary Fig. S13B), the redox state of the PSII acceptor side (qL; Supplementary Fig. S13C), and the donor-side limitation of PSI (Y(ND); Supplementary Fig. S13D) also did not reveal significant differences between the wild type and the transplastomic lines. By contrast, the RNAi lines showed reduced chlorophyll content (to 27% of the wild-type levels in R1 and to 71% in R2), leaf absorptance (to 69% in R1, to 94% in R2, and to 95% in R3), Fv/Fm (to 52% in R1, to 85% in R2, and to 96% in R3) and ETRII (to 29% in R1 and to 69% in R2; Fig. 6I–M). The light response curves of linear electron transport confirmed the strongly reduced electron transport capacity (Supplementary Fig. S13E). In the most strongly affected line, R1, induction of non-photochemical quenching was somewhat impaired (Supplementary Fig. S13F), in line with reduced contents of the xanthophyll cycle pigments violaxanthin and zeaxanthin (Fig. 3D, E). The redox state of the PSII acceptor side (Supplementary Fig. S13G) was less affected, but PSI became more rapidly oxidized in limited light (Supplementary Fig. S13H), in line with the strongly decreased capacity of linear electron transport.
Primary/secondary metabolites are altered in transplastomic DcLCYB1 and NtLCYB RNAi lines
To further investigate causes for the phenotype observed in the transgenic lines, metabolite levels were determined (see Materials and methods). A total of 72 primary (by GC-MS) and 31 secondary (by LC-MS) metabolites were measured. We observed 65 significant changes in DcLCYB1 transplastomic and NtLCYB RNAi lines (Fig. 7). In the transplastomic lines only a few metabolites changed significantly (e.g. proline and serine for line L2). Additionally, three and four O-acyl sugars in line L2 and line L15 were decreased, respectively. However, other metabolites exhibited only a few or very moderate changes (Fig. 7). By contrast, the RNAi lines, especially the strongest variegated line, R1, exhibited extensive significant changes. Most of the amino acids were increased; for instance asparagine content increased by ~50-fold and glutamine content by ~8-fold in R1. The only amino acids that were decreased in R1 were GABA (~5-fold decrease) and tryptophan (~3-fold decrease). Furthermore, the abundance of most measured sugars was affected. Erythritol (~4-fold, ~3-fold, and ~2-fold increase for R1, R2, and R3, respectively) and galacturonate (~34-fold, ~3-, and ~11-fold increased for R1, R2, and R3, respectively) were increased in all three RNAi lines (Fig. 7). In the strongest variegated line, increased abundance of five sugars (e.g. galactonate ~2-fold increase) and decreased abundance of six sugars (e.g. myo-inositol ~6-fold decrease) was observed. Moreover, metabolites involved in the TCA cycle were reduced in R1 (e.g. fumarate and pyruvate with ~5- and ~3-fold decrease, respectively). Additionally, O-acyl sugars were significantly reduced in the R1 line (e.g. O-AS45 class IV ~3-fold reduction) and exhibited a similar trend for R3 (non-significant changes). Three phenylpropanoids were reduced (e.g. monohydrated N,N-dicaffeoyl-spermidine ~7-fold decrease) and two were increased (e.g. 4-O-trans-caffeoyl-D-quinic acid ~2-fold increase) in R1. Furthermore, allantoin and uric acid were increased ~25- and ~70-fold, whereas maleic acid and spermidine-based phenolamide #7 were ~14- and 10-fold decreased in R1 (Fig. 7). Altogether, the results suggest mild metabolic changes (primary and secondary) for transplastomic DcLCYB1 lines but very pronounced metabolic changes in the line with the highest reduction in NtLCYB expression.
Fig. 7.
Metabolite content in transplastomic DcLCYB1 and NtLCYB RNAi lines. Heat map of metabolite content for transplastomic and RNAi lines. Only metabolites with significant changes for at least one line are shown. Results are reported as log2-fold changes relative to the wild type. Statistical analysis was performed in R (R Core Team, 2018) using an unpaired Wilcoxon test with the default parameters within the function compare_means of the ggpubr package (Kassambara, 2018). WT, wild type.
Discussion
Increased plant yield and photosynthetic efficiency from nuclear DcLCYB1 expression was recently reported in tobacco (cv. Xanthi) (Moreno et al., 2020). An increased GA/ABA ratio played a key role in the higher growth phenotype in the nuclear DcLCYB1 tobacco lines (Fig. 8C). In an attempt to fully understand this phenomenon, we generated transgenic lines with high and reduced LCYB expression levels. Here, we took advantage of the high expression capacity granted by the transformation of plastid DNA and we generated transplastomic DcLCYB1 tobaccos with very high DcLCYB1 expression levels. High LCYB expression levels favored the β-branch of the carotenoid pathway, resulting in increased β-carotene and violaxanthin content. Interestingly, differences in DcLCYB1 transcript levels within the transplastomic lines are not reflected in different levels of β-carotene accumulation. This has also been observed in our previous tobacco nuclear lines where L14 (~900-fold), L15 (~100-fold), and L16 (~60-fold) showed different DcLCYB1 transcript levels but a similar β-carotene content increase of 2-fold (Moreno et al., 2020). In carrot, this phenomena was observed in different tissues (leaf and root; Moreno et al., 2013), where higher DcLCYB1 expression in lines L6, L8, and L9 (2-, 4-, and 8-fold, respectively, compared with the wild type) was reflected in ~3-fold increase in β-carotene content. In carrot roots, different DcLCYB1 transcript levels (1.25-, 1.6-, and 2.3-fold) was reflected in ~2-fold increase in β-carotene content. Altogether, these lines of evidence suggest that despite higher DcLCYB1 transcript accumulation in the different transgenic lines, β-carotene accumulates at the same level in tobacco and carrot. This points towards the existence of an upper threshold in LCYB transcript levels and the accumulation of β-carotene.
Fig. 8.
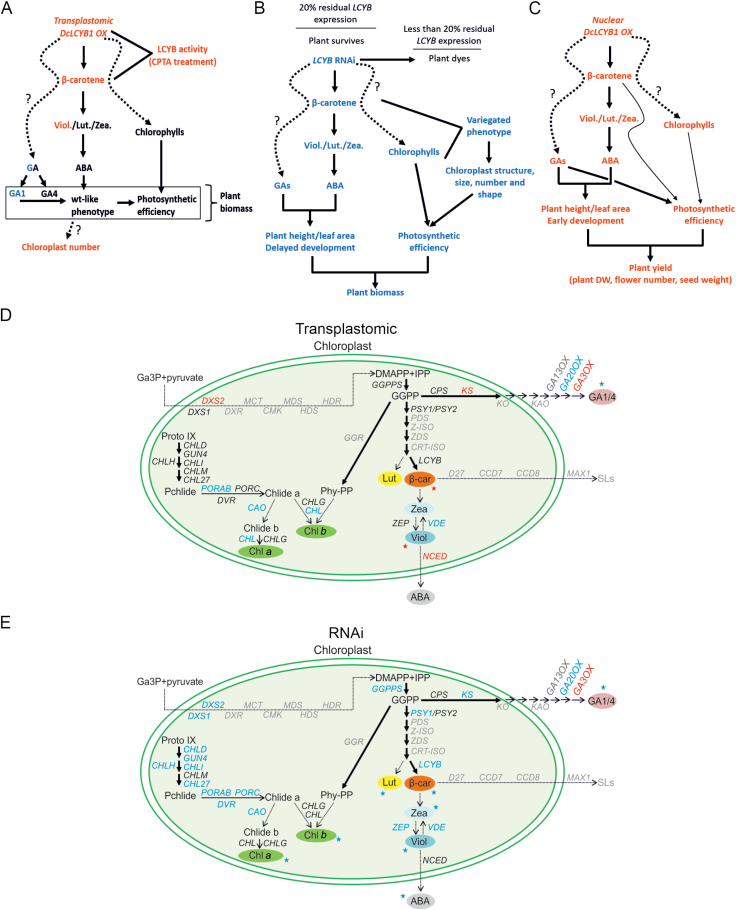
Proposed models for altered plant yield in transplastomic DcLCYB1 and NtLCYB RNAi lines. (A) Proposed model for transplastomic lines. (B) Proposed model for RNAi lines. (C) Model for nuclear DcLCYB1-expressing lines (model was built from pigment, hormone, photosynthetic, and physiological data in Moreno et al. (2020)). In (A–C) the relations of physiological parameters (e.g. biomass, photosynthetic efficiency), gene expression, pigment content, and hormone content are shown (red font: increase; blue font: decrease; black font: no change). (D, E) Molecular response triggered by increased and reduced LCYB expression in transplastomic and RNAi lines, respectively. Changes occurring in at least two lines are shown. Increases are marked in blue, decreases in red. Genes shown in black were not changed and genes shown in grey were not measured. ABA, abscisic acid; β-car, β-carotene; Chl, chlorophyll; DMAPP, dimethylallyl diphosphate; GA, gibberellins; IPP, isopentenyl diphosphate; Lut, lutein; OX, overexpressor; Pchlide, protochlorophillidae; Phy-PP, phytyl diphosphate; SLs, strigolactones; Viol, violaxanthin; Zea, zeaxanthin. Gene abbreviation, protein name, and a brief description of each gene measured by qPCR can be found in Moreno et al. (2020).
Despite the extremely high DcLCYB1 expression, transplastomic lines showed a wild type-like phenotype (Fig. 2A; Supplementary Fig. S1A–D). One possibility to explain the wild type-like biomass phenotype observed in the transplastomic lines lies in the connection between several isoprenoid plastid pathways. Carotenoid, chlorophyll, and GA biosynthesis pathways take place in the chloroplast and all share the same precursor, GGPP (Bouvier et al., 2005; Pulido et al., 2012). A positive co-expression of key genes encoding enzymes involved in the MEP, chlorophyll, carotenoid, and GA pathways accompanied by increased carotenoid, chlorophylls, and ABA/GA contents was observed in nuclear DcLCYB1 tobaccos (Moreno et al., 2020). By contrast, in our transplastomic lines we observed an altered/not consistent accumulation of transcripts encoding enzymes involved in MEP, tetrapyrrole, chlorophyll, LHCs, carotenoid, and ABA/GA biosynthesis pathways (Fig. 2H–L). For instance, GA20ox and GA3ox are key regulators of GA biosynthesis (Hedden and Thomas, 2012; Gupta and Chakrabarty, 2013). Increased GA20ox expression levels resulted in larger plants, higher GA content, biomass, and photosynthetic efficiency (Biemelt et al., 2004; Voorend et al., 2016). Gene expression of GA20ox was decreased in the transplastomic lines, whereas GA3ox expression was increased. This expression pattern is in line with the reduced GA1 and unchanged GA4 content in the transplastomic lines and might explain the observed wild type-like phenotype, in terms of biomass and photosynthetic efficiency (Figs 4A, B, 6I–M; Supplementary Figs S13A–D, S14A). Moreover, a reduced GA1 content might suggest a reduction in the GGPP pool as previously shown for carotenoid and chlorophyll contents in plants with modifications (GGPPS and LCYB overexpressors or the expression of an astaxanthin pathway) in the isoprenoid or carotenoid metabolic flux (Wurbs et al., 2007; Apel and Bock, 2009; Tata et al., 2016; Lu et al., 2017). Additionally, unchanged/moderately increased ABA content (L2) was not reflected in any developmental phenotype (Figs 4C, E, 5C, E). However, an ABA catabolite, phaseic acid (PA), has been shown to be a signaling molecule that fine-tunes plant physiology (e.g. seed germination, plant growth), environmental adaptation, and development (Weng et al., 2016). That PA functions as a phytohormone (PA is recognized by a subset of ABA receptors) suggest that chemically related metabolites generated from plant hormones might additionally have signaling functions (Weng et al., 2016), and therefore that it is important to analyse global changes in hormone metabolism caused by genetic manipulation of their precursors (e.g. ABA and GA). In our transplastomic lines, PA is reduced in L10 and L15 (Supplementary Fig. S7A) indicating that although ABA content is not drastically changed, ABA function might be influenced by the reduction in PA. In contrast, in our previously published nuclear DcLCYB1 lines, the content of ABA and its catabolites (PA, neoPA, and ABA-glucose ester) was increased in all the lines. This evidence might suggest that together the lower β-carotene accumulation (30% more than in the wild type) and altered gene expression in the transplastomic lines (Fig. 2) in comparison with the nuclear DcLCYB1 lines, affected ABA catabolism in a negative manner (reduced PA). GA metabolism was also affected in the transplastomic lines. For instance, reduction in GA20ox expression levels was correlated with GA53 accumulation and reduction of downstream-hydroxylated precursors (GA19 and GA20) of GA1 (Supplementary Fig. S7E, F), thus explaining the reduced GA1 content in our transgenic lines (Fig. 5A). Interestingly, hydroxylated and non-hydroxylated GA29 and GA51 contents were increased in the transplastomic lines pointing towards an increased GA2ox expression; however, we did not analyse the expression of this gene since it is not involved in the generation of bioactive GAs. Exogenous GA3 and GA4 application increased plant biomass in the transplastomic lines, while ABA treatment reduced biomass accumulation as previously described (Nagel et al., 1994; Ma et al., 2008). As previously reported, an appropriate GA/ABA ratio increases plant biomass in tobacco cv. Xanthi (Moreno et al., 2020). Exogenous GA/ABA application (as previously reported) did not increase plant biomass in the transplastomic lines, probably due to the preexistent imbalance in ABA and GA metabolism in the transplastomic lines (Fig. 5A; Supplementary Fig. S7A–C). Taken together, our results suggest that very high DcLCYB1 expression levels (i) shift the metabolic flux towards the β-branch of the carotenoid pathway, (ii) cause an altered expression pattern in isoprenoid genes of different plastid isoprenoid biosynthesis pathways, and (iii) (subsequently) influence phytohormone metabolism. In addition, increases in carotenoid content are not sufficiently high to alter the synthesis of apocarotenoids that have previously been reported to be involved in promoting growth (e.g. β-cc and zaxinone; Supplementary Fig. S10), which is in line with the wild type-like observed phenotype in the transplastomic lines.
Another possible explanation for the wild type-like phenotype observed in the transplastomic lines (Fig. 8A), could be related to a secondary effect from the plastid transformation. In transplastomic overexpressors, mRNA degradation or side products with lower molecular mass can accumulate and interfere with the accumulation of a desired metabolite (Wurbs et al., 2007; Fernández-San Millán et al., 2018). For instance, plastid expression of LCYB genes from Phycomyces blakesleeanus (LPP) and Erwinia (LCE) did not influence carotenoid and chlorophyll content or plant growth in tobacco cv. Petit Havana (Wurbs et al., 2007). However, LCE did strongly increased CPTA tolerance, suggesting strong LCYB activity in these lines. In tomato, however, LCE fruits produced a 4-fold increase in β-carotene content. Interestingly, in tomato leaves, β-carotene remain unchanged while reductions (lutein) and increases (antheraxanthin, zeaxanthin, and violaxanthin) of several carotenoids were observed. The unsuccessful expression of the LPP was attributed to the observed instability of the transcript in the plastid. This may have occurred in our transplastomic lines, which accumulated lower levels of β-carotene compared with nuclear lines (Moreno et al., 2020), despite higher steady state transcript accumulation of LYCB. However, in our transplastomic lines, the 30% increase in β-carotene (and ~50–200% in violaxanthin) content and high tolerance to CPTA suggest increased LCYB activity arguing against protein instability or enzymatic inactivity of the protein. In fact, to date, the 30% increase in β-carotene in our transplastomic lines is the highest reported in leaves of transplastomic LCYB-expressing tobacco and tomato lines (Wurbs et al., 2007; Apel and Bock, 2009). Moreover, violaxanthin increase in leaves is higher in our transplastomic tobaccos than in leaves of transplastomic tomatoes (Wurbs et al., 2007; Apel and Bock, 2009). This suggests that the DcLCYB1 gene might confer an extra advantage for carotenoid production (due to different regulation or less susceptibility to feedback inhibition) compared with other LCYBs from daffodil and Erwinia. This might be explained by the vast β-carotene accumulation in leaves, but especially in carrot roots (Just et al., 2007, 2009). Interestingly, nuclear DcLCYB1 overexpression in carrot increased 3-fold the β-carotene content in leaves (Moreno et al., 2013), which is, to date, the highest increase in β-carotene content in leaves compared with previously reported nuclear and transplastomic transformations (Rosati et al., 2000; Wurbs et al., 2007; Apel and Bock, 2009; Ji et al., 2009; Shi et al., 2015). However, if we compared the transplastomic strategy with the nuclear transformation approach, it seems that the nuclear transformation approach is more efficient in terms of β-carotene accumulation. In our nuclear DcLCYB1 lines (cv. Xanthi) (Moreno et al., 2020), β-carotene accumulation is higher than in the transplastomic DcLCYB1 lines (cv. Petit Havana, this work). In addition, fruits of transplastomic tomato plants expressing the daffodil LCYB produced less β-carotene than the fruits of the nuclear LCYB overexpressors (D’Ambrosio et al., 2004; Apel and Bock, 2009). These lines of evidence show that in tobacco and tomato the nuclear strategy worked better than the transplastomic approach in terms of β-carotene accumulation. Although in the particular case of the LCYB expression in tobacco and tomato the nuclear strategy was proven to be more efficient, the insertion of an astaxanthin pathway by a transplastomic approach resulted in pink/orange tobacco plants (Lu et al., 2017), due to an extremely high astaxanthin accumulation. This suggests that strategies to manipulate carotenoid genes and enhance carotenoid content in plants have to be evaluated case by case.
By contrast, the reduced NtLCYB expression caused leaf variegation in the RNAi lines, as reported for other mutants in Arabidopsis, Brassica napus, and rice, with defects in carotenoid biosynthesis (Aluru et al., 2001, 2009; Fang et al., 2008; Zhao et al., 2020). Here, by choosing three lines with different phenotype strength and gene expression we were able to dissect the importance of β-carotene for plant growth and development. Although strong LCYB RNAi lines (R1 and R2) showed strong plant variegation and perhaps accumulation of secondary effects not related to the LCYB silencing, the phenotype of wild type CPTA-treated plants mimics the variegated and reduced growth phenotype observed in the stronger RNAi lines (Supplementary Fig. S1). Although we do not exclude secondary effects, this suggests that in these RNAi lines most of the observed molecular and phenotypic effects might derive from the LCYB silencing. Interestingly, a similar variegated phenotype was reported for tobacco plants with reduced LCYB expression, but the lack of detailed gene expression analysis, chloroplast structure, and phytohormone and photosynthesis measurements impedes the ability to draw strong conclusions about the origin of this phenotype at the molecular and physiological levels (Shi et al., 2015). Here, we found that LCYB expression is essential for plant autotrophic growth (Fig. 8B; Supplementary Fig. S3). Residual LCYB expression of 20% (equivalent to 35% residual levels of β-carotene and other derivatives) or higher is nonetheless sufficient for survival in plants germinated aseptically and transferred to soil, while we deduce that less than 20% LCYB expression/35% β-carotene content (and other carotenoids) is not enough for plants to survive when grown on soil (Fig. 8B; Supplementary Fig. S3A–C).
Reduced LCYB expression caused the opposite effect observed in the nuclear DcLCYB1 tobacco lines (Moreno et al., 2020), characterized by a down-regulation of key genes involved in the MEP (DXS), chlorophyll (CAO, DVR, PORC, and PORAB), carotenoid (PSY1 and LCYB), and GA (KS and GA20oxidase) pathways accompanied by decreased carotenoid, chlorophylls and ABA/GA contents, thus suggesting a depletion of the common GA, chlorophylls, and carotenoid pathway precursor GGPP in the strongest RNAi lines, which is in line with the reduction in chlorophyll, carotenoid, and GA contents in an Arabidopsis pds mutant (Qin et al., 2007). This idea is supported by the strong down-regulation of key genes from carotenoid and MEP pathways (PSY1, DXS1, and DXS2), which are part of a feedback mechanism, known to control the content of carotenoids and their precursors in Arabidopsis (Carretero-Paulet et al., 2006; Cazzonelli and Pogson, 2010; Ruiz-Sola and Rodríguez-Concepción, 2012). These lines of evidence (together with the nuclear and transplastomic DcLCYB1-expressing lines) suggest the possibility of the existence of hotspots in the carotenoid pathway that could trigger positive or negative feedback loops to communicate with other isoprenoid pathways. Further proof of these feedback mechanisms is the strong reduction in transcript accumulation of GGPPS, which encodes GGPP SYNTHASE, an enzyme that catalyses the conversion of isopentenyl diphosphate/dimethylallyl diphosphate into GGPP (Bouvier et al., 2005; Pulido et al., 2012). This reduction is in line with the reduced GA, chlorophyll, and carotenoid content.
Several molecular and phenotypic characteristics of the RNAi lines are reminiscent of plants with reduced ABA and GA content (Schomburg et al., 2003; Nitsch et al., 2012). In fact, the early seed germination, reduced biomass and leaf area, and reduced primary root growth observed in the strongest RNAi lines might reflect reduced ABA and GA content (Fig. 5C–E; Supplementary Figs S4B, S5E–H) (Schomburg et al., 2003; Nitsch et al., 2012). In addition, decreased ABA function and content might be reflected in reductions in the content of the ABA-derived phytohormone PA (Weng et al., 2016) and the ABA-glucose ester (which serve as ABA storage). Interestingly, exogenous GA3, GA4, ABA, GA3/ABA, and GA4/ABA application in the RNAi lines did not rescue the phenotype to the wild type level (Fig. 5G), suggesting the involvement of other factor(s) explaining the reduced biomass phenotype in our RNAi lines. In fact, the plant variegation (leaves, stem, and capsules) observed in the strongest RNAi lines (Supplementary Fig. S4B–D) points towards pigment, photosynthesis, and chloroplast biogenesis defects in these lines. Moreover, other carotenoid-defective mutants, and specially pds3 mutants, showed impaired chloroplast differentiation and development, and reduced chloroplast number (Aluru et al., 2001; Qin et al., 2007; Zhao et al., 2020). Carotenoids are located in the photosynthetic membrane in the form of chlorophyll–carotenoid–protein complexes and some carotenogenic enzymes are membrane-associated (Green and Durnford, 1996; Cunningham and Gantt, 1998). In this way, any changes in carotenoid composition always lead to abnormal plastid development (Welsch et al., 2000; Park et al., 2002). Recently, the expression of a bacterial CrtB (PSY) gene (in Arabidopsis, lettuce, tobacco, and zucchini), and as a consequence of that, a burst in phytoene accumulation, was shown to stimulate differentiation of chloroplast into chromoplast suggesting that indeed carotenoid content influences chloroplast development (Llorente et al., 2020). Interestingly, we observed increased chloroplast number in our transplastomic tobacco lines (Supplementary Table S4), which is in line with the increased chromoplast number observed in tomato fruits expressing the citrus LCYB gene (Zhu et al., 2020). Taken together this evidence suggests a connection between carotenoid content and chloroplast biogenesis and development that deserves further attention.
Carotenoids participate in photoprotection mechanisms by (i) modulating the non-radiative dissipation of excess excitation energy (Niyogi et al., 1998; Dall’Osto et al., 2005), (ii) mediating direct quenching of chlorophyll (Chl) triplets (3Chl*), or (iii) scavenging ROS generated during photosynthesis (Niyogi, 2000; Havaux et al., 2004; Dall’Osto et al., 2005, 2007a). In addition, carotenoids are structural determinants of the photosynthetic apparatus (Cazzaniga et al., 2012), and only a minor fraction is free in the lipid phase of thylakoids where it serves as an antioxidant (Havaux et al., 2004) and modulates the fluidity of the lipid bilayer (Gruszecki and Strzałka, 2005). For instance, β-carotene is bound to reaction center subunits of both PSI and PSII, whereas xanthophylls are bound to peripheral LHC subunits that comprise the antenna system (Cazzaniga et al., 2012). Thus, it is expected that any disturbance of carotenoid content can be reflected in alterations of carotenoid function but also in the biogenesis of several components of the photosynthetic apparatus. The Arabidopsis szl1 mutant, which carries a point mutation in the LCYB gene, and thus exhibits a less-active LCYB enzyme, showed reduced β-carotene (55% of the wild type), violaxanthin, and zeaxanthin, increased lutein accumulation (2-fold), and slightly reduced growth (Li et al., 2009; Cazzaniga et al., 2012). Reductions in these pigments caused reduced Fv/Fm, chlorophyll content, qE (the rapidly reversible component of NPQ), and increased photooxidation (higher 1O2 production). In our stronger RNAi lines, β-carotene content was similarly reduced (55–60%), but lutein, zeaxanthin, and violaxanthin were reduced, causing strong reductions in chlorophyll content (up to 67%), Fv/Fm (up to 50%), ETRII, and qN, affecting strongly plant photosynthetic performance (Fig. 6I–M; Supplementary Fig. S13E–H). In addition, strong reduction in β-carotene and xanthophyll contents impact on thylakoid membrane integrity and the assembly and stability of the photosynthetic complexes. For instance, lutein is required for the folding of LHC proteins (Formaggio et al., 2001) and zeaxanthin and violaxanthin bind sites L2 and/or V1 in LHCB proteins to increase resistance to high light and exogenously provided photosensitizers (Baroli et al., 2004) by scavenging 1O2 and thus preventing lipid peroxidation (Havaux and Niyogi, 1999; Havaux et al., 2007). Reduced xanthophyll and β-carotene are also reflected in higher accumulation of 1O2 in our RNAi lines and other carotenoid mutants (Li et al., 2009; Cazzaniga et al., 2012). Alteration in ROS and ROS-processing systems has been reported to modulate plant growth and development (Pagnussat et al., 2005; Kirchsteiger et al., 2012; Yu et al., 2013; Mhamdi and Van Breusegem, 2018), and thus ROS accumulation in our RNAi lines might contribute to the observed reduced growth and delayed development. In addition, altered chloroplast ultrastructure (including chloroplast size, shape, and number; Fig. 6E–H; Supplementary Fig. S12E, F, K–N; Supplementary Table S4) and altered cellular level organization of the leaf in the yellow tissue of the RNAi lines (Supplementary Fig. 11E) might explain the reduced growth phenotype. In fact, altered chloroplast ultrastructure and development were observed in other mutants with reduced expression of other carotenoid genes (PDS, ZDS, CRTISO), with mutant lines also showing impaired or delayed growth (Aluru et al., 2001, 2009; Qin et al., 2007; Fang et al., 2008; Zhao et al., 2020). In general, these results expose strong defects in plastid biogenesis (in yellow sectors of the leaves) in the RNAi lines, which are reflected in altered structure and function of the photosynthetic apparatus. The aforementioned evidence is in line with the massive reduction in amino acids, sugars, and TCA cycle intermediates in the strongest RNAi line (Fig. 7), as well as with the dramatic reductions in accumulation of transcripts with products required for LHC (Fig. 3J), PSI, cyt b6f, and PSII (Supplementary Figs S4C, D, S14B), and tetrapyrrole and chlorophyll biosynthesis processes (Fig. 3F, G, K, L). The generated evidence points towards three factors contributing to reduced biomass (Fig. 8E) in the RNAi lines: (i) reduced pigment content and photosynthetic efficiency, (ii) altered plastid biogenesis, and (iii) reduced phytohormone content. In addition, the strongest RNAi line (R1) showed a reduction in accumulation of zaxinone, a recently reported growth (promoting) regulator in rice (Wang et al., 2019), suggesting that zaxinone might be contributing to the observed growth phenotype. However, zaxinone content was the same as the wild type for the R2 line, which showed a similar phenotype as R1, arguing against a zaxinone contribution to this phenotype.
In summary, manipulation of plastid DNA to increase LCYB expression levels showed that very high DcLCYB1 expression, for the first time in transplastomic lines, results in significantly higher β-carotene content. However, it can also disturb the expression pattern of key genes of carotenoid and carotenoid-related biosynthesis pathways, impeding the accumulation of key end products (e.g. GA) from other isoprenoid pathways (Fig. 8A, D). Reduced (GA1) and unchanged (GA4 and ABA) phytohormone content are reflected in the wild type-like phenotype in the transplastomic lines (in contrast with the high yield phenotype observed in the nuclear lines (Moreno et al., 2020); Fig. 8C). Strategies to modify metabolic fluxes in isoprenoid pathways through plastid transformation have to be analysed case by case, because as shown in our results higher gene expression will not always be reflected in higher levels of the desired metabolite. Reduction of LCYB expression affects several molecular events (Fig. 8E) that influence plant development and metabolism, and resulted in decreased photosynthetic efficiency and plant biomass (Fig. 8B).
Altogether, our previous work coupled with the current results, which combined nuclear, transplastomic, and RNAi lines (Fig. 8A–C), leads us to conclude that by manipulating LCYB expression it is possible to modulate plant biomass by modifying pigment and hormone content and other molecular processes within the plastid. Our data also suggest a threshold for LCYB expression levels that might be beneficial or detrimental for plant yield, which serve as a guideline to export this genetic engineering strategy into crops.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Transplastomic and wild type plants exposed to CPTA (2-(4-chlorophenylthio) triethylamine) treatment.
Fig. S2. Relative expression of key genes comprising PSI, PSII, and cyt b6f complex.
Fig. S3. T0 RNAi lines growing in MS media and on soil.
Fig. S4. 16-week-old transplastomic and RNAi tobacco plants.
Fig. S5. Quantification of physiological parameters of DcLCYB1 transplastomic and NtLCYB RNAi lines.
Fig. S6. T3 RNAi plants at different stages of their life cycle.
Fig. S7. ABA and GA metabolism in transplastomic DcLCYB1 lines.
Fig. S8. ABA and GA metabolism in NtLCYB RNAi lines.
Fig. S9. Hormone and inhibitor treatments in transplastomic DcLCYB1 and NtLCYB RNAi lines.
Fig. S10. Apocarotenoid quantification in transplastomic DcLCYB1 and NtLCYB RNAi lines.
Fig. S11. Light microscopy images of leaf cross sections of transplastomic and RNAi lines.
Fig. S12. Transmission electron microscopy (TEM) images of tobacco cells from the NtLCYB RNAi line R1 and R3.
Fig. S13. Light response curves of photosynthetic parameters in transplastomic and RNAi lines.
Fig. S14. Schematic model representing changes in gene expression of photosystem I, II, and the cyt b6f complex subunits in transplastomic and RNAi lines.
Table S1. Primers used for qPCR experiments.
Table S2. In vitro regeneration process for the generation of DcLCYB1 transplastomic tobacco plants.
Table S3. End-point measurement of plant height and flower number of 16-week-old NtLCYB RNAi lines.
Table S4. Chloroplast number per area in transplastomic DcLCYB1 and NtLCYB RNAi lines.
Dataset S1. LC-MS metabolite-reporting list following the recommendations described by Fernie et al. 2011.
Dataset S2. GC-MS metabolite-reporting list following the recommendations described by Fernie et al. 2011.
Dataset S3. The SRM transition list for apocarotenoid profiling.
Acknowledgements
We are extremely grateful to Dr Aleksandra Skirycz for supporting an independent research project to be completed in her research group at the Max-Planck-Institut für Molekulare Pflanzenphysiologie (MPI-MP). Dr Daniel Karcher (MPI-MP), kindly provided the constructs pDK325 and pDK326 used for generation of pJM36 and pJM37 transplastomic vectors. We are grateful to Prof. Dr Lothar Willmitzer for his great support and advice, and the MPI-MP and the head of the Green Team Dr Karin Köhl and its members (Britta Hausmann, Florian Hundert, Dirk Zerning and Laura Westpahl) for plant cultivation. The gibberellin measurements were supported by a European Regional Development Fund Project ‘Centre for Experimental Plant Biology’ (No. CZ.02.1.01/0.0/0.0/16_019/0000738) and the Czech Science Foundation (18-10349S).
Author contributions
JCM conceived and designed the project, generated the RNAi lines, performed the cloning of the DcLCYB1 gene in the transplastomic vectors, performed hormone and inhibitors treatments and biomass quantification, and supervised the work; SK performed qPCR and biomass experiments, CPTA treatment, and physiological measurements; TA-M performed Southern blot analysis; SK and LPS performed metabolite extraction and performed primary and secondary metabolite analysis, respectively; SK, SA, and RB performed carotenoid extraction and pigment quantification by HPLC; VT and DT performed sample preparation and extraction for hormone measurements and analysis; SR and ASc performed in vitro tissue culture and regeneration of positive transplastomic lines; SK, AF, and ASa performed microscopy analysis and analysed the data; MAS performed photosynthetic measurements; JM and SAl performed the apocarotenoid profiling and analysed the data; SK and JCM wrote the manuscript with great input of TA-M and the rest of the coauthors.
Data availability
All relevant data can be found within the manuscript and its supplementary data.
References
- Al-Babili S, Bouwmeester HJ. 2015. Strigolactones, a novel carotenoid-derived plant hormone. Annual Review of Plant Biology 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Albus CA, Salinas A, Czarnecki O, Kahlau S, Rothbart M, Thiele W, Lein W, Bock R, Grimm B, Schöttler MA. 2012. LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiology 160, 1923–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru MR, Bae H, Wu D, Rodermel SR. 2001. The Arabidopsis immutans mutation affects plastid differentiation and the morphogenesis of white and green sectors in variegated plants. Plant Physiology 127, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru MR, Zola J, Foudree A, Rodermel SR. 2009. Chloroplast photooxidation-induced transcriptome reprogramming in Arabidopsis immutans white leaf sectors. Plant Physiology 150, 904–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade-Souza V, Costa MG, Chen CX, Gmitter Jr FG, Costa MA. 2011. Physical location of the carotenoid biosynthesis genes Psy and β-Lcy in Capsicum annuum (Solanaceae) using heterologous probes from Citrus sinensis (Rutaceae). Genetics and Molecular Research 10, 404–409. [DOI] [PubMed] [Google Scholar]
- Apel W, Bock R. 2009. Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiology 151, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armarego-Marriott T, Kowalewska Ł, Burgos A, et al. 2019. Highly resolved systems biology to dissect the etioplast-to-chloroplast transition in tobacco leaves. Plant Physiology 180, 654–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin JR 2nd, Staehelin LA. 2011. Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiology 155, 1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendaño-Vázquez AO, Cordoba E, Llamas E, et al. 2014. An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. The Plant Cell 26, 2524–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli I, Gutman BL, Ledford HK, Shin JW, Chin BL, Havaux M, Niyogi KK. 2004. Photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas. The Journal of Biological Chemistry 279, 6337–6344. [DOI] [PubMed] [Google Scholar]
- Biemelt S, Tschiersch H, Sonnewald U. 2004. Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiology 135, 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. 2001. Transgenic plastids in basic research and plant biotechnology. Journal of Molecular Biology 312, 425–438. [DOI] [PubMed] [Google Scholar]
- Bock R. 2007. Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Current Opinion in Biotechnology 18, 100–106. [DOI] [PubMed] [Google Scholar]
- Bouvier F, Rahier A, Camara B. 2005. Biogenesis, molecular regulation and function of plant isoprenoids. Progress in Lipid Research 44, 357–429. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Koltai H, Beveridge CA. 2013. Diverse roles of strigolactones in plant development. Molecular Plant 6, 18–28. [DOI] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. 2013. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. The Plant Journal 73, 897–909. [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Cairó A, Botella-Pavía P, Besumbes O, Campos N, Boronat A, Rodríguez-Concepción M. 2006. Enhanced flux through the methylerythritol 4-phosphate pathway in Arabidopsis plants overexpressing deoxyxylulose 5-phosphate reductoisomerase. Plant Molecular Biology 62, 683–695. [DOI] [PubMed] [Google Scholar]
- Cazzaniga S, Li Z, Niyogi KK, Bassi R, Dall’Osto L. 2012. The Arabidopsis szl1 mutant reveals a critical role of β-carotene in photosystem I photoprotection. Plant Physiology 159, 1745–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Pogson BJ. 2010. Source to sink: regulation of carotenoid biosynthesis in plants. Trends in Plant Science 15, 266–274. [DOI] [PubMed] [Google Scholar]
- Chen X, Han H, Jiang P, Nie L, Bao H, Fan P, Lv S, Feng J, Li Y. 2011. Transformation of beta-lycopene cyclase genes from Salicornia europaea and Arabidopsis conferred salt tolerance in Arabidopsis and tobacco. Plant & Cell Physiology 52, 909–921. [DOI] [PubMed] [Google Scholar]
- Clarke JL, Daniell H. 2011. Plastid biotechnology for crop production: present status and future perspectives. Plant Molecular Biology 76, 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles JP, Phillips AL, Croker SJ, García-Lepe R, Lewis MJ, Hedden P. 1999. Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. The Plant Journal 17, 547–556. [DOI] [PubMed] [Google Scholar]
- Cuadros-Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña-Cortés H, Willmitzer L, Hannah MA. 2009. TargetSearch—a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics 10, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E. 1998. Genes and enzymes of carotenoid biosynthesis in plants. Annual Review of Plant Physiology and Plant Molecular Biology 49, 557–583. [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Bassi R, Ruban A. 2014. Photoprotective mechanisms: carotenoids. Plastid Biology 5, 393–435. [Google Scholar]
- Dall’Osto L, Caffarri S, Bassi R. 2005. A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. The Plant Cell 17, 1217–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L, Cazzaniga S, North H, Marion-Poll A, Bassi R. 2007a. The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. The Plant Cell 19, 1048–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L, Fiore A, Cazzaniga S, Giuliano G, Bassi R. 2007b. Different roles of α- and β-branch xanthophylls in photosystem assembly and photoprotection. The Journal of Biological Chemistry 282, 35056–35068. [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Piques M, Ronzani M, Molesini B, Alboresi A, Cazzaniga S, Bassi R. 2013. The Arabidopsis nox mutant lacking carotene hydroxylase activity reveals a critical role for xanthophylls in photosystem I biogenesis. The Plant Cell 25, 591–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, Giorio G, Marino I, Merendino A, Petrozza A, Salfi L, Stigliani AL, Cellini F. 2004. Virtually complete conversion of lycopene into β-carotene in fruits of tomato plants transformed with the tomato lycopene β-cyclase (tlcy-b) cDNA. Plant Science 166, 207–214. [Google Scholar]
- Davière JM, Achard P. 2013. Gibberellin signaling in plants. Development 140, 1147–1151. [DOI] [PubMed] [Google Scholar]
- Davison PA, Hunter CN, Horton P. 2002. Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418, 203–206. [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B, Gilmore AM, Adams WW 3rd. 1996. Carotenoids 3: In vivo function of carotenoids in higher plants. FASEB Journal 10, 403–412. [DOI] [PubMed] [Google Scholar]
- Dickinson AJ, Lehner K, Mi J, Jia KP, Mijar M, Dinneny J, Al-Babili S, Benfey PN. 2019. β-Cyclocitral is a conserved root growth regulator. Proceedings of the National Academy of Sciences, USA 116, 10563–10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1990. A rapid total DNA preparation procedure for fresh plant tissue. Focus 12, 13–15. [Google Scholar]
- Elghabi Z, Karcher D, Zhou F, Ruf S, Bock R. 2011. Optimization of the expression of the HIV fusion inhibitor cyanovirin-N from the tobacco plastid genome. Plant Biotechnology Journal 9, 599–608. [DOI] [PubMed] [Google Scholar]
- Fang J, Chai C, Qian Q, et al. 2008. Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. The Plant Journal 54, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-San Millán A, Aranjuelo I, Douthe C, Nadal M, Ancín M, Larraya L, Farran I, Flexas J, Veramendi J. 2018. Physiological performance of transplastomic tobacco plants overexpressing aquaporin AQP1 in chloroplast membranes. Journal of Experimental Botany 69, 3661–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V. 2011. Recommendations for reporting metabolite data. The Plant Cell 23, 2477–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A, Dall’Osto L, Cazzaniga S, Diretto G, Giuliano G, Bassi R. 2012. A quadruple mutant of Arabidopsis reveals a β-carotene hydroxylation activity for LUT1/CYP97C1 and a regulatory role of xanthophylls on determination of the PSI/PSII ratio. BMC Plant Biology 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formaggio E, Cinque G, Bassi R. 2001. Functional architecture of the major light-harvesting complex from higher plants. Journal of Molecular Biology 314, 1157–1166. [DOI] [PubMed] [Google Scholar]
- Frank HA, Cogdell RJ. 1993. Photochemistry and functions of carotenoids. In: Young A, Britton G, eds. Carotenoids in photosynthesis. Dordrecht: Springer, 252–326. [Google Scholar]
- Frank HA, Cogdell RJ. 1996. Carotenoids in photosynthesis. Photochemistry and Photobiology 63, 257–264. [DOI] [PubMed] [Google Scholar]
- Krause GH, Weis E. 1991. Chlorophyll fluorescence and photosynthesis: the basics. Annual Review of Plant Physiology and Plant Molecular Biology 42, 313–349. [Google Scholar]
- Gallego-Giraldo L, Ubeda-Tomás S, Gisbert C, García-Martínez JL, Moritz T, López-Díaz I. 2008. Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant & Cell Physiology 49, 679–690. [DOI] [PubMed] [Google Scholar]
- Garciarrubio A, Legaria JP, Covarrubias AA. 1997. Abscisic acid inhibits germination of mature Arabidopsis seeds by limiting the availability of energy and nutrients. Planta 203, 182–187. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L. 2011. Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high-resolution mass spectrometry. The Plant Journal 68, 364–376. [DOI] [PubMed] [Google Scholar]
- Giuliano G, Al-Babili S, von Lintig J. 2003. Carotenoid oxygenases: cleave it or leave it. Trends in Plant Science 8, 145–149. [DOI] [PubMed] [Google Scholar]
- Götz T, Sandmann G, Römer S. 2002. Expression of a bacterial carotene hydroxylase gene (crtZ) enhances UV tolerance in tobacco. Plant Molecular Biology 50, 129–142. [DOI] [PubMed] [Google Scholar]
- Green BR, Durnford DG. 1996. The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annual Review of Plant Physiology and Plant Molecular Biology 47, 685–714. [DOI] [PubMed] [Google Scholar]
- Gruszecki WI, Strzałka K. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochimica et Biophysica Acta 1740, 108–115. [DOI] [PubMed] [Google Scholar]
- Gupta R, Chakrabarty SK. 2013. Gibberellic acid in plant: still a mystery unresolved. Plant Signaling and Behaviour 8, e25504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Li Y, Zhou S. 2008. Overexpression of phytoene synthase gene from Salicornia europaea alters response to reactive oxygen species under salt stress in transgenic Arabidopsis. Biotechnology Letters 30, 1501–1507. [DOI] [PubMed] [Google Scholar]
- Havaux M. 1998. Carotenoids as membrane stabilizers in chloroplasts. Trends in Plant Science 3, 147–151. [Google Scholar]
- Havaux M, Dall’osto L, Bassi R. 2007. Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiology 145, 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Dall’Osto L, Cuiné S, Giuliano G, Bassi R. 2004. The effect of zeaxanthin as the only xanthophyll on the structure and function of the photosynthetic apparatus in Arabidopsis thaliana. The Journal of Biological Chemistry 279, 13878–13888. [DOI] [PubMed] [Google Scholar]
- Havaux M, Niyogi KK. 1999. The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proceedings of the National Academy of Sciences, USA 96, 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. 2012. Gibberellin biosynthesis and its regulation. The Biochemical Journal 444, 11–25. [DOI] [PubMed] [Google Scholar]
- Hirschberg J. 2001. Carotenoid biosynthesis in flowering plants. Current Opinion in Plant Biology 4, 210–218. [DOI] [PubMed] [Google Scholar]
- Ji J, Wang G, Wang J, Wang P. 2009. Functional analysis of multiple carotenogenic genes from Lycium barbarum and Gentiana lutea L. for their effects on β-carotene production in transgenic tobacco. Biotechnology Letters 31, 305–312. [DOI] [PubMed] [Google Scholar]
- Just BJ, Santos CA, Fonseca ME, Boiteux LS, Oloizia BB, Simon PW. 2007. Carotenoid biosynthesis structural genes in carrot (Daucus carota): isolation, sequence-characterization, single nucleotide polymorphism (SNP) markers and genome mapping. Theoretical and Applied Genetics 114, 693–704. [DOI] [PubMed] [Google Scholar]
- Just BJ, Santos CA, Yandell BS, Simon PW. 2009. Major QTL for carrot color are positionally associated with carotenoid biosynthetic genes and interact epistatically in a domesticated × wild carrot cross. Theoretical and Applied Genetics 119, 1155–1169. [DOI] [PubMed] [Google Scholar]
- Kang C, Zhai H, Xue L, Zhao N, He S, Liu Q. 2018. A lycopene β-cyclase gene, IbLCYB2, enhances carotenoid contents and abiotic stress tolerance in transgenic sweetpotato. Plant Science 272, 243–254. [DOI] [PubMed] [Google Scholar]
- Kassambara A. 2018. ggpubr: ‘ggplot2’ based publication ready plots: R package version 0.2. https://rpkgs.datanovia.com/ggpubr/ [Google Scholar]
- Kirchsteiger K, Ferrández J, Pascual MB, González M, Cejudo FJ. 2012. NADPH thioredoxin reductase C is localized in plastids of photosynthetic and nonphotosynthetic tissues and is involved in lateral root formation in Arabidopsis. The Plant Cell 24, 1534–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2019. pheatmap: Pretty Heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, Edwards GE. 2004. New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynthesis Research 79, 209. [DOI] [PubMed] [Google Scholar]
- Krinsky NI. 1989. Antioxidant functions of carotenoids. Free Radical Biology & Medicine 7, 617–635. [DOI] [PubMed] [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP. 2016. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354, 857–861. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hahn FM, Baidoo E, Kahlon TS, Wood DF, McMahan CM, Cornish K, Keasling JD, Daniell H, Whalen MC. 2012. Remodeling the isoprenoid pathway in tobacco by expressing the cytoplasmic mevalonate pathway in chloroplasts. Metabolic Engineering 14, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. 2001. Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Research 29, 970–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Heiling S, Baldwin IT, Gaquerel E. 2016. Illuminating a plant’s tissue-specific metabolic diversity using computational metabolomics and information theory. Proceedings of the National Academy of Sciences, USA 113, E7610–E7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ahn TK, Avenson TJ, Ballottari M, Cruz JA, Kramer DM, Bassi R, Fleming GR, Keasling JD, Niyogi KK. 2009. Lutein accumulation in the absence of zeaxanthin restores nonphotochemical quenching in the Arabidopsis thaliana npq1 mutant. The Plant Cell 21, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. 2006. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols 1, 387–396. [DOI] [PubMed] [Google Scholar]
- Llorente B, Torres-Montilla S, Morelli L, et al. 2020. Synthetic conversion of leaf chloroplasts into carotenoid-rich plastids reveals mechanistic basis of natural chromoplast development. Proceedings of the National Academy of Sciences, USA 117, 21796–21803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Rijzaani H, Karcher D, Ruf S, Bock R. 2013. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proceedings of the National Academy of Sciences, USA 110, E623–E632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Stegemann S, Agrawal S, Karcher D, Ruf S, Bock R. 2017. Horizontal transfer of a synthetic metabolic pathway between plant species. Current Biology 27, 3034–3041.e3. [DOI] [PubMed] [Google Scholar]
- Ma XW, Ma FW, Mi YF, Ma YH, Shu HR. 2008. Morphological and physiological responses of two contrasting Malus species to exogenous abscisic acid application. Plant Growth Regulation 56, 77–87. [Google Scholar]
- Mhamdi A, Van Breusegem F. 2018. Reactive oxygen species in plant development. Development 145, dev164376. [DOI] [PubMed] [Google Scholar]
- Mi J, Jia KP, Wang JY, Al-Babili S. 2018. A rapid LC-MS method for qualitative and quantitative profiling of plant apocarotenoids. Analytica Chimica Acta 1035, 87–95. [DOI] [PubMed] [Google Scholar]
- Moreno JC, Cerda A, Simpson K, Lopez-Diaz I, Carrera E, Handford M, Stange C. 2016. Increased Nicotiana tabacum fitness through positive regulation of carotenoid, gibberellin and chlorophyll pathways promoted by Daucus carota lycopene β-cyclase (Dclcyb1) expression. Journal of Experimental Botany 67, 2325–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JC, Mi J, Agrawal S, Kössler S, Turečková V, Tarkowská D, Thiele W, Al-Babili S, Bock R, Schöttler MA. 2020. Expression of a carotenogenic gene allows faster biomass production by redesigning plant architecture and improving photosynthetic efficiency in tobacco. The Plant Journal 103, 1967–1984. [DOI] [PubMed] [Google Scholar]
- Moreno JC, Pizarro L, Fuentes P, Handford M, Cifuentes V, Stange C. 2013. Levels of lycopene β-cyclase 1 modulate carotenoid gene expression and accumulation in Daucus carota. PLoS One 8, e58144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Nagel OW, Konings H, Lambers H. 1994. Growth-rate, plant development and water relations of the ABA-deficient tomato mutant sitiens. Physiologia Plantarum 92, 102–108. [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Nitsch L, Kohlen W, Oplaat C, et al. 2012. ABA-deficiency results in reduced plant and fruit size in tomato. Journal of Plant Physiology 169, 878–883. [DOI] [PubMed] [Google Scholar]
- Niyogi KK. 2000. Safety valves for photosynthesis. Current Opinion in Plant Biology 3, 455–460. [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Grossman AR, Björkman O. 1998. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. The Plant Cell 10, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. 2009. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. The Plant Journal 57, 436–445. [DOI] [PubMed] [Google Scholar]
- Olson JA. 1996. Benefits and liabilities of vitamin A and carotenoids. The Journal of Nutrition 126, 1208S–1212S. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. 2005. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. [DOI] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. 2002. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. The Plant Cell 14, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Niyogi KK, Björkman O, DellaPenna D. 1998. Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proceedings of the National Academy of Sciences, USA 95, 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. 1989. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochimica and Biophysica Acta 975, 384–394. [Google Scholar]
- Pulido P, Perello C, Rodriguez-Concepcion M. 2012. New insights into plant isoprenoid metabolism. Molecular Plant 5, 964–967. [DOI] [PubMed] [Google Scholar]
- Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ. 2007. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Research 17, 471–482. [DOI] [PubMed] [Google Scholar]
- Ralley L, Schuch W, Fraser PD, Bramley PM. 2016. Genetic modification of tomato with the tobacco lycopene β-cyclase gene produces high β-carotene and lycopene fruit. Zeitschrift für Naturforschung 71, 295–301. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2018. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rittenberg D, Foster GL. 1940. A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. The Journal of Biological Chemistry 737–744. [Google Scholar]
- Rosahl S, Schell J, Willmitzer L. 1987. Expression of a tuber-specific storage protein in transgenic tobacco plants: demonstration of an esterase activity. The EMBO Journal 6, 1155–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosati C, Aquilani R, Dharmapuri S, Pallara P, Marusic C, Tavazza R, Bouvier F, Camara B, Giuliano G. 2000. Metabolic engineering of beta-carotene and lycopene content in tomato fruit. The Plant Journal 24, 413–419. [DOI] [PubMed] [Google Scholar]
- Ruf S, Bock R. 2011. In vivo analysis of RNA editing in plastids. Methods in Molecular Biology 718, 137–150. [DOI] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. 2001. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nature Biotechnology 19, 870–875. [DOI] [PubMed] [Google Scholar]
- Ruf S, Karcher D, Bock R. 2007. Determining the transgene containment level provided by chloroplast transformation. Proceedings of the National Academy of Sciences, USA 104, 6998–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola MÁ, Rodríguez-Concepción M. 2012. Carotenoid biosynthesis in Arabidopsis: a colorful pathway. The Arabidopsis Book 10, e0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem MA, Jüppner J, Bajdzienko K, Giavalisco P. 2016. Protocol: a fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods 12, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt GW, Delaney SK. 2010. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Molecular Genetics and Genomics 283, 233–241. [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JA, Amasino RM. 2003. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. The Plant Cell 15, 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U, Klughammer C. 2016. Analysis of photosystem I donor and acceptor sides with a new type of online-deconvoluting kinetic LED-array spectrophotometer. Plant & Cell Physiology 57, 1454–1467. [DOI] [PubMed] [Google Scholar]
- Schuetz RD, Baldwin RA. 1958. The synthesis and properties of some substituted phenyl ω-(N,N-dialkylamino)-alkyl sulfides. Journal of the American Chemical Society 80, 162–164. [Google Scholar]
- Shi Y, Guo J, Zhang W, et al. 2015. Cloning of the lycopene β-cyclase gene in Nicotiana tabacum and its overexpression confers salt and drought tolerance. International Journal of Molecular Sciences 16, 30438–30457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. 1991. Mutation proximal to the transfer-RNA binding region of the Nicotiana plastid 16S ribosomal-RNA confers resistance to spectinomycin. Molecular & General Genetics 228, 316–319. [DOI] [PubMed] [Google Scholar]
- Svab Z, Maliga P. 1993. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proceedings of the National Academy of Sciences, USA 90, 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Picataggio S, Rouvière PE, Cheng Q. 2004. Asymmetrically acting lycopene β-cyclases (CrtLm) from non-photosynthetic bacteria. Molecular Genetics and Genomics 271, 180–188. [DOI] [PubMed] [Google Scholar]
- Tata SK, Jung J, Kim YH, Choi JY, Jung JY, Lee IJ, Shin JS, Ryu SB. 2016. Heterologous expression of chloroplast-localized geranylgeranyl pyrophosphate synthase confers fast plant growth, early flowering and increased seed yield. Plant Biotechnology Journal 14, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I, Sonneveld T, Bugg TH, Thompson A. 2005. Regulation and manipulation of the biosynthesis of abscisic acid, including the supply of xanthophyll precursors. Journal of Plant Growth Regulation 24, 253–273. [Google Scholar]
- Tureckova V, Novak O, Strnad M. 2009. Profiling ABA metabolites in Nicotiana tabacum L. leaves by ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Talanta 80, 390–399. [DOI] [PubMed] [Google Scholar]
- Urbanova T, Tarkowska D, Novak O, Hedden P, Strnad M. 2013. Analysis of gibberellins as free acids by ultra performance liquid chromatography-tandem mass spectrometry. Talanta 112, 85–94. [DOI] [PubMed] [Google Scholar]
- Vidal AM, Gisbert C, Talón M, Primo-Millo E, López-Díaz I, García-Martínez JL. 2001. The ectopic overexpression of a citrus gibberellin 20-oxidase enhances the non-13-hydroxylation pathway of gibberellin biosynthesis and induces an extremely elongated phenotype in tobacco. Physiologia Plantarum 112, 251–260. [DOI] [PubMed] [Google Scholar]
- Voorend W, Nelissen H, Vanholme R, De Vliegher A, Van Breusegem F, Boerjan W, Roldán-Ruiz I, Muylle H, Inzé D. 2016. Overexpression of GA20-OXIDASE1 impacts plant height, biomass allocation and saccharification efficiency in maize. Plant Biotechnology Journal 14, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Haider I, Jamil M, et al. 2019. The apocarotenoid metabolite zaxinone regulates growth and strigolactone biosynthesis in rice. Nature Communications 10, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Beyer P, Hugueney P, Kleinig H, von Lintig J. 2000. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 211, 846–854. [DOI] [PubMed] [Google Scholar]
- Weng JK, Ye M, Li B, Noel JP. 2016. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 166, 881–893. [DOI] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R. 2007. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. The Plant Journal 49, 276–288. [DOI] [PubMed] [Google Scholar]
- Yu X, Pasternak T, Eiblmeier M, et al. 2013. Plastid-localized glutathione reductase2-regulated glutathione redox status is essential for Arabidopsis root apical meristem maintenance. The Plant Cell 25, 4451–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Hu K, Yan M, Yi B, Wen J, Ma C, Shen J, Fu T, Tu J. 2020. Disruption of carotene biosynthesis leads to abnormal plastids and variegated leaves in Brassica napus. Molecular Genetics and Genomics 295, 981–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Zheng X, Ye J, Jiang Q, Chen H, Mei X, Wurtzel ET, Deng X. 2020. Building the synthetic biology toolbox with enzyme variants to expand opportunities for biofortification of provitamin A and other health-promoting carotenoids. Journal of Agricultural and Food Chemistry 68, 12048–12057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data can be found within the manuscript and its supplementary data.