Summary
We generated an online brain pQTL resource for 7,376 proteins through the analysis of genetic and proteomic data derived from post-mortem samples of the dorsolateral prefrontal cortex of 330 older adults. The identified pQTLs tend to be non-synonymous variation, are over-represented among variants associated with brain diseases, and replicate well (77%) in an independent brain dataset. Comparison to a large study of brain eQTLs revealed that about 75% of pQTLs are also eQTLs. In contrast, about 40% of eQTLs were identified as pQTLs. These results are consistent with lower pQTL mapping power and greater evolutionary constraint on protein abundance. The latter is additionally supported by observations of pQTLs with large effects’ tending to be rare, deleterious, and associated with proteins that have evidence for fewer protein-protein interactions. Mediation analyses using matched transcriptomic and proteomic data provided additional evidence that pQTL effects are often, but not always, mediated by mRNA. Specifically, we identified roughly 1.6 times more mRNA-mediated pQTLs than mRNA-independent pQTLs (550 versus 341). Our pQTL resource provides insight into the functional consequences of genetic variation in the human brain and a basis for novel investigations of genetics and disease.
Keywords: gene regulation, quantitative trait locus, brain, expression, proteomics, eQTL, pQTL
Introduction
Analyses mapping expression quantitative trait loci (eQTLs) have deepened our understanding of the links between genetics and disease.1, 2, 3, 4, 5 eQTLs tend to overlap with disease-associated variants identified by genome-wide association studies (GWASs) and are often used to provide insight into the functional consequences of genetic variation and identify candidate causal genes.6, 7, 8, 9, 10 These investigations implicitly assume that the genetic effects on mRNA expression propagate to protein—the main determinant of cellular function and ultimately most phenotypes. However, gene expression is not a perfect proxy for protein expression. Previous studies have shown that mRNA and protein abundances are often only weakly correlated.11,12 Furthermore, analyses mapping protein quantitative trait loci (pQTLs), in addition to eQTLs, have shown pQTLs to provide novel insight into the impacts of genetic variation.13, 14, 15, 16, 17, 18 Since proteins represent a major source of druggable molecular targets, this information can greatly aid the development of new therapeutics.19
The genetic control of mRNA expression in the brain has been well studied,20, 21, 22, 23, 24, 25, 26, 27 but currently little is known about how genetics influences brain protein abundances. Large-scale analyses of brain pQTLs have not yet been performed because the necessary data have only recently emerged. Here, we investigate the genetic control of the human brain proteome by using high-throughput mass spectrometry-based protein data from post-mortem brain samples of the dorsolateral prefrontal cortex (dPFC) of older adults. Additionally, to better understand the relationship between the genetic control of the human brain proteome and transcriptome, we compare the pQTL results to a recent brain eQTL meta-analysis and perform mediation analyses by using matched gene and protein expression data. The results of our pQTL analyses are made available on the pQTL web application as a resource for future investigations (see “Web resources”).
Material and methods
Data
Discovery dataset
To investigate the genetic control of the human brain proteome, we analyzed genetic, transcriptomic, and proteomic data derived from post-mortem samples of the dPFC of participants of the Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP).28, 29, 30 ROS and MAP are longitudinal cohort studies of aging and dementia maintained by investigators at the Rush Alzheimer Disease Center in Chicago, IL.28, 29, 30 Both studies recruit participants without known dementia at baseline and follow them annually via detailed clinical evaluation. The studies were approved by an institutional review board of Rush University Medical Center. All participants provided informed consent and signed an Anatomic Gift Act and repository consent to allow their data and biospecimens to be repurposed.
Proteomic data. Protein abundance data from cortical microdissections of the dPFC of ROS/MAP subjects were generated via tandem mass tag (TMT) isobaric labeling mass spectrometry methods for protein identification and quantification, as previously described by Johnson et al. (2020)31 and Wingo et al. (2020).32 Briefly, tissue homogenization, protein digestion, TMT labeling, and high-pH fractionation were performed in sequence. An equal amount of protein digested from each sample was aliquoted and digested in parallel to serve as the global pooled internal standard (GIS) in each TMT batch. Prior to TMT labeling, all samples were randomized into 50 batches (8 samples per batch) based on age at death, sex, post-mortem interval, diagnosis, and measured neuropathologies. Peptides from each individual sample (n = 400) and the GIS (n = 100) were labeled with the TMT 10-plex kit (Thermo Fisher). All fractions were resuspended in equal volume of loading buffer and analyzed by liquid chromatography coupled to mass spectrometry. The Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) was run with MS/MS (MS2) scans for 45 TMT batches and with the SPS-MS3 method for five TMT batches.
All raw files were analyzed via the Proteome Discoverer suite (version 2.3 Thermo Fisher Scientific). MS2 spectra were searched against the canonical UniProtKB human proteome database (downloaded February 2019 with 20,338 total sequences). The Sequest HT search engine was used and parameters were specified as previously described by Johnson et al. (2020)31 and Wingo et al. (2020).32 Peptide spectral matches (PSMs) and peptides were filtered with Percolator to a false discovery rate (FDR) of less than one percent. Peptides were then assembled into proteins and further filtered on the basis of the combined probabilities of the constituent peptides to a final FDR of less than one percent. In cases of redundancy, shared peptides were assigned to protein sequences in adherence with the principles of parsimony. Reporter ions were quantified from MS2 or MS3 scans via an integration tolerance of 20 ppm with the most confident centroid setting. A total of 12,691 proteins were quantified. Correlation of technical replicates within each batch was over 99%, indicating a high degree of reproducibility for the quantified proteins (Figure S1).
To ensure the analysis of high-quality data, we excluded proteins with low correlations between the two GISs (outside the 95% confidence interval) within batches and proteins with missing data for over 50% of subjects across batches. Additionally, we scaled each abundance value by a subject-specific total protein abundance measure to remove the effects of loading differences in the proteomic experiments. After transformation to the log2 scale, outlier subjects were identified and removed through iterative principal-component analysis. For each iteration, subjects more than four standard deviations from the mean of the first or second principal component were removed and the principal components were re-calculated. Following outlier removal, proteins with missing data for over 50% of the remaining subjects were removed. Finally, we residualized the abundance of each protein via linear regression to remove the effects of sex, age at death, post-mortem interval, study, batch, and MS2 versus MS3 reporter quantitation mode. After these quality control procedures, there were 7,376 high-quality proteins available for analysis.
To reduce confounding due to disease, we included dementia status at death as a covariate in all proteomic analyses. For each ROS/MAP participant, a clinical diagnosis of dementia was rendered at the time of death blinded to all pathologic data. Here, we coded the diagnoses of dementia status as no cognitive impairment (NCI), mild cognitive impairment (MCI), or Alzheimer dementia (AD) and excluded individuals with a diagnosis of non-Alzheimer dementia from analysis. For secondary analyses on the influence of disease, all described quality control procedures were repeated with data from the subset of samples with NCI at death.
Genotyping data. Genotype data were generated from whole-genome sequencing (WGS) of DNA that was extracted from cryopreserved peripheral blood mononuclear cells or frozen dPFC of ROS/MAP subjects. WGS was performed as previously described33 and is available via Synapse (ID: syn10901595). Briefly, libraries were constructed with the KAPA Hyper Library Preparation Kit per the manufacturer’s protocol and sequenced on an Illumina HiSeq X sequencer (v.2.5 chemistry) with 150 bp paired-end reads. Reads were aligned to the GRCh37 human reference genome via Burrows-Wheeler Aligner (BWA-MEM v.0.7.8)34 and processed with the GATK best-practices workflow, which includes marking duplicate reads by Picard tools v.1.83, local realignment around indels, and base quality score recalibration by Genome Analysis Toolkit (GATK v.3.4.0).35,36 A multi-sample genomic variant call format (gVCF) was generated by merging results of HaplotypeCaller on each sample individually in gVCF mode (GATKv.3.4.0), and batches of gVCF were merged into gVCFs processed by a joint genotyping step (GATK v.3.2.2).
Annotation of the multi-sample VCF (n = 1,196) was performed with Bystro.37 After quality control, a total of 1,133 subjects remained for analysis. A total of 63 subjects were excluded for one or more of the following reasons: (1) , silent:replacement sites, or transition:transversion ratios greater than five standard deviations from the mean (n = 7); (2) genotype missingness, heterozygosity, or homozygosity greater than three standard deviations from the mean (n = 14); (3) sex discordance based on the heterozygosity of the X chromosome (n = 7); and (4) cryptic relation or duplication identified by identity-by-state sharing with PLINK38 (n = 31). We used unlinked ancestrally informative markers to infer eigenvectors for principal-component analysis by using EIGENSTRAT39 and over six standard deviation outliers (n = 1) were removed. Before analysis, the data was restricted to subjects of European descent and filtered to the HapMap 3 SNVs40 that are in Hardy-Weinberg equilibrium (HWE) and have a minor allele frequency (MAF) greater than 0.05.
After quality control, we had 501,414 common SNVs to test and 330 subjects with both proteomic and genetic data. Information on the demographics and disease status of these subjects is provided in Table S1. As a result of the batch-specific nature of missing data from TMT-labeled MS/MS protein quantification, the number of subjects with proteomic data differs for each protein and ranges from 161 to 330 (Figure S2). As expected, proteins that were more abundant were also more likely to have complete data (Figure S3).
Transcriptomic data. Gene expression was measured from the dPFC as previously described.33 Briefly, RNA was extracted from cortically dissected sections of dPFC gray matter and samples with RNA integrity numbers (RINs) over 5 were used to prepare RNA-sequencing (RNA-seq) libraries via strand-specific dUTP method with poly-A selection41,42 with the Illumina HiSeq with 101-bp paired-end reads to a target coverage of 50 million reads per library. Raw RNA-seq reads were aligned to a GRCh38 reference genome, and gene counts were computed via STAR43 as described in Logsdon et al. (2019).44 We obtained RNA-seq data from Synapse (ID: syn17010685) and performed library normalization and quality control as described by Sieberts et al. (2020).22 Genes with <1 cpm in over 50% of subjects were removed, and the remaining genes were normalized via conditional quantile normalization45 followed by weighted normalization via the voom-lima package in Bioconductor.46 Outlier subjects were detected and removed via principal-component analysis and hierarchical clustering. Only samples within three standard deviations of the mean of the first and second principal components were retained for further analysis. Finally, we residualized the expression of each gene via linear regression to remove the effects of sex, age at death, post-mortem interval, study, and batch. After quality control, 173 subjects had matched transcriptomic, proteomic, and genetic data. Table S1 provides the demographic and disease characteristics of these subjects.
Replication dataset
Genetic and proteomic data derived from dPFC of participants of the Banner Sun Health Brain and Body Donation Program (Banner BBDP) were used for replication analyses. The Banner Sun Health Research Institute recruited cognitively normal individuals from the retirement communities of the greater Phoenix, AZ.47 The study was approved by the Banner Sun Health Research Institute Institutional Review Board, and all individuals or their legal representatives provided informed consent for participation.
Proteomic data. Proteomic profiling was performed with the samples of the dPFC following the same approach as described for ROS/MAP subjects with two differences: (1) only MS2 scans were obtained and (2) MS2 spectra were searched against the UniProtKB human brain proteome database downloaded in April 2015. A total of 11,518 proteins were quantified. We used the same quality control procedures as the ROS/MAP proteomic data to remove proteins with more than 50% missing data, remove outlier individuals, and remove the effects covariates (i.e., age at death, sex, batch, and post-mortem interval) from the proteomic profiles. After these quality control procedures, there were 6,526 high-quality proteins available for analysis.
Analogous to the analyses in the discovery dataset, dementia status at death was included as in all relevant models. Banner BBDP participants were assessed during life with annual medical, neurologic, and neuropsychological assessments, and upon death, their donated brains underwent detailed neuropathologic assessment. The final clinical diagnoses of Banner BBDP participants was based on the five-point clinical dementia rating (CDR) score.48 Individuals with CDR scores of 0, 0.5, and greater than 0.5 were considered to have a diagnosis of NCI, MCI, and AD, respectively. Individuals without a clinical diagnosis were excluded from analysis.
Genotyping data. Individuals were genotyped via Affymetrix Precision Medicine Array following the manufacturer’s protocol with DNA extracted from brain via the QIAGEN GenePure kit. We followed the same approach to quality control as was used for the discovery dataset genetic data, including filtering on the basis of data completeness, HWE, MAF, European ancestry, and relatedness. After quality control, we had 460,954 common SNVs to test and 149 subjects with both proteomic and genetic data. Demographic and disease information on these subjects is available in Table S1.
GWAS and eQTL summary statistics
For comparison, we downloaded summary statistics of four large GWASs (average N > 382,000): Alzheimer disease,49 Parkinson disease,50 schizophrenia,51 and neuroticism.52 We also downloaded the summary statistics for a large meta-analysis of brain eQTLs22 (n = 1,433). The four GWAS results were from individuals of European descent and were used to identify disease-associated variants at a significance threshold of 5 × 10−8. The brain eQTL meta-analysis combined ROS/MAP genetic and transcriptomic data with genetic and transcriptomic data from the CommonMind Consortium53 and defined eQTLs at a FDR of 5%.
Genetic and protein annotation data
Each tested protein was annotated with the number of protein-protein interactions on the basis of the number of interactions reported in BioGrid data.54 Furthermore, Bystro37 was used to annotate each tested genetic variant with combined annotation dependent depletion (CADD) scores,55 genomic contexts (i.e., exonic, intronic, UTRs, intergenic), and substitution types (i.e., non-synonymous versus synonymous). The annotations used for genomic context and substitution type were taken from the RefSeq database.56 Since a variant may be tested against the abundance of more than one protein, we revised the genomic context annotations to be test specific. That is, for each SNP-protein pair tested, a SNP with genic genomic context (i.e., exonic, intronic, UTR) was re-annotated to intergenic if the SNP is not within the gene of the paired protein.
Statistical analyses
Estimation of additional confounders
To reduce confounding due to population structure, we added the first ten principal components of the discovery and replication dataset genotype data as model covariates in all relevant analyses. For both datasets, all ten principal components had significant Tracey-Widom statistics (p value < 0.05). Additionally, unknown confounders in the proteomic data were estimated via surrogate variable analysis and the SVA package in R.57,58 For proteomic data for both datasets, ten surrogate variables were built and added as model covariations in all relevant analyses. Since the protein data was generated from bulk tissue, it is likely that some of these surrogate variables represent cell type proportions.
Identification of pQTLs
We identified genetic variants associated with protein abundance in the brain by using linear regression to model protein abundance as a function of genotype. We reduced our computational and testing burden by investigating only the proximal genetic effects of common single nucleotide variants (SNVs). We tested the proximal genetic control of each protein by using linear regression to model protein abundance as a function of genotype for every common SNV (MAF > 0.05) within a 100 kb window around the protein-coding gene. This window size was chosen because the majority of reported eQTLs are located within 100 kb of the regulated gene.59, 60, 61 The location of each protein coding gene was defined by the knownGene table (GRCh37/hg19 assembly) from the University of California, Santa Cruz (UCSC) table browser.62 For each SNV-protein pair, we regressed genotype against protein abundance, assuming additive genetic effects and including clinical diagnosis, the first ten genetic principal components, and ten surrogate variables as covariates. Analyses that restricted samples to those with NCI at death included only the first ten principal components as covariates. SNVs where genotype was significantly associated with protein abundance at a FDR of 5% were declared protein quantitative trait loci (pQTLs; FDR < 0.05). For each protein with more than one pQTL, we identified the independent pQTL signals via stepwise linear regression.
Quantification of replication rates
Replication rates were estimated via the π1 statistic from the qvalue package in R.63 This statistic, which estimates the proportion of non-null hypotheses on the basis of the p value distribution, is more robust than a simple overlap of significant results.64 For each replication analysis, we examined the distribution of replication study p values for sites found to be significant in the discovery study (FDR < 0.05). To reduce the influence of linkage disequilibrium, we analyzed only the lead discovery QTLs for each gene (i.e., sites with the strongest associations).
Enrichment analyses
We used Fisher’s exact tests to assess the enrichment of brain pQTLs among (1) variants annotated to different genomic contexts (i.e., exonic, intronic, UTRs, intergenic) and substitution types (i.e., non-synonymous versus synonymous), (2) brain eQTLs, and (3) disease-associated variants. The tests for enrichment of pQTLs among variants in different genomic contexts and of different substitution types included only sites that were both annotated and tested in the pQTL analysis. Similarly, the test for enrichment of pQTLs among eQTLs included only sites that were tested in both the pQTL analysis and brain eQTL meta-analysis.22 In contrast, the tests for enrichment of pQTLs among disease-associated variants included all sites tested in the pQTL analysis regardless of testing status in the investigated GWASs.
Mediation analyses
Mediation analyses were performed with the mediation package65 in R and data from participants in the discovery dataset with both RNA-seq and TMT protein data. To increase the stability of statistical power across tests, we only considered proteins with more than 170 samples with both RNA-seq that TMT protein data. The analyses assessed mRNA abundance as a mediator of the genetic effects on protein abundance through the comparison of linear regression models. For each pQTL-protein pair, a model where genotype predicts protein abundance was compared to a model where both genotype and mRNA abundance predict protein abundance. All models additionally included clinical cognitive diagnosis, the first ten genetic principal components, and ten surrogate variables as covariates. A significant reduction in the effect of genotype on protein abundance with the addition mRNA abundance as a covariate is consistent with mediation. We compared the mediation results with the results of a large eQTL meta-analysis22 to avoid spurious results due to a limited sample size. For this comparison, the eQTL meta-analysis was subset to the sites included in the pQTL analysis.
Gene Ontology analyses
Gene Ontology (GO) analyses were performed to gain a better understanding of the proteins that were found to be under genetic control and have mRNA-mediated or mRNA-independent abundance. We used GOrilla66 to assess whether any biological processes significantly associate with the sets of genes with mRNA-mediated or mRNA-independent pQTLs. GOrilla uses a minimal hypergeometric score to quantify the enrichment of each GO term. Each set of genes was independently compared to the entire set of genes included in mediation testing.
Results
pQTL discovery
We identified proximal brain pQTLs for 2,474 genes (Table 1). These variants explained an average of 8.5% of the variation in abundance of each protein (Figure S4) and were enriched for SNVs with higher MAFs (Figure S5). All pQTL summary statistics, regardless of significance, are available for visualization and download on the pQTL web application (see “Web resources”).
Table 1.
Identification of pQTLs
| Number of pQTLs | Number of genes | |
|---|---|---|
| Tested | 501,414 | 7,376 |
| Significant at FDR < 0.05 [independent] | 35,601 [8,451] | 2,474 |
Independent pQTLs were identified via stepwise regression.
Genomic context of pQTLs
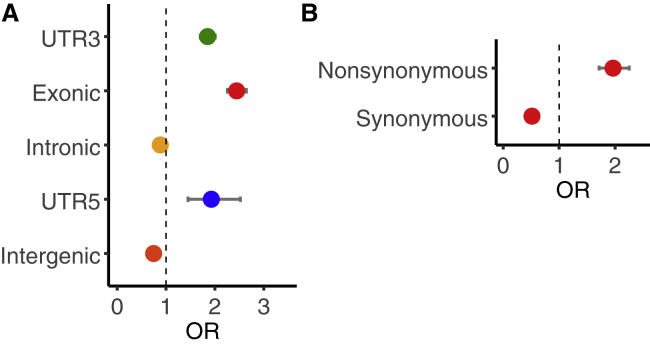
To better understand the genomic context of the identified pQTLs, we performed enrichment analyses to assess the overlap of pQTLs and genomic annotations. We found the identified pQTLs to be over-represented among SNVs in coding regions and SNVs with non-synonymous variation (Figure 1, Table S2). Additionally, we compared the distribution of pQTL effect sizes by genomic annotation and found pQTLs in coding regions (i.e., exons and UTRs) to have significantly larger median effect sizes than pQTLs in non-coding regions (p < 0.005 for all pairwise Nemenyi tests; Figure S6A). Furthermore, within exonic pQTLs, which have the largest median effect size overall, we found pQTLs with non-synonymous variation to have significantly larger median effect sizes than pQTLs with synonymous variation (Figure S6B). We see this pattern at pQTLs with a reference allele that increases protein abundance (Kruskal-Wallis = 7.3, p = 0.006) as well as at pQTLs with a reference allele that decreases protein abundance (Kruskal-Wallis = 8.1, p = 0.004). We performed separate analyses on exonic pQTLs with positive and negative effects to reduce possible confounding due to protein quantification bias. Because rare peptides may not be included in the proteomic database and are less likely to be accurately quantified, exonic coding-variation is more likely to associate with a decrease in protein abundance. We observed the reference allele to associate with decreased protein abundance for 61% of all exonic pQTLs and 68% of exonic pQTLs with non-synonymous variation. Together, these results suggest that genetic variation that changes a coded amino acid tends to have a large influence on protein abundance.
Figure 1.
Enrichment of proximal pQTLs
(A) Enrichment of proximal pQTLs by genomic context.
(B) Enrichment of proximal pQTLs in exonic coding regions. For each annotation, enrichment was evaluated on the basis of a Fisher’s exact tests assessing the overlap between pQTLs and the annotated SNVs. The estimated odds ratio (OR) and 95% confidence interval (CI) is shown for each test.
Influence of disease on pQTL identification
The proteomic data was generated from post-mortem samples of older adults, of which 31% had a clinical diagnosis of Alzheimer disease (AD) at death. To assess the influence of AD on the identification of pQTLs, we compared the results of the pQTL analysis for the discovery dataset (N = 330), which was adjusted for clinical diagnosis at death, to individuals from the discovery dataset with no cognitive impairment at death (N = 144). We found the estimated genetic effects on protein abundance to have a correlation of 0.62 between the main and control analyses (Figure S7; p < 2.2 × 10−16). Furthermore, we found this correlation to increase to 0.92 when we restricted to sites identified as pQTLs in the main analysis. This comparison suggests that AD has only a small influence on the identification of pQTLs in this study. The summary statistics from the control-only pQTL analysis are also available for visualization and download on the pQTL web application (see “Web resources”).
Enrichment of brain disease-associated variants among pQTLs
Previous studies have shown that eQTLs tend to overlap with disease-associated variants identified by GWASs.6, 7, 8, 9 To assess whether variants associated with brain diseases are over-represented among the identified pQTLs, we performed a Fisher’s exact test that compared the set of pQTLs to a set of SNVs that were identified to be disease associated by large GWASs of four brain disorders. We found the identified pQTLs to be highly enriched in brain disease-associated genetic variation (OR: 2.82, p value = 1.6 × 10−60; see Table S3 for individual disorder enrichments). The estimated enrichment supports the co-occurrence of pQTLs and disease-associated variants across the genome. However, further evaluation is needed to understand the functional consequences of these loci. Genetic colocalization methods, for example, could be used to mitigate false positives caused by linkage disequilibrium. The pQTL resource provided by this study will facilitate such analyses in the future.
pQTL replication
We assessed the replication rate of the identified brain pQTLs in an independent brain replication dataset,47 which was comprised of genetic and proteomic data derived from the dPFC on 149 subjects of European descent. We identified proximal brain pQTLs for 1,803 genes in the replication dataset (Table S4). Next, we compared the 5,712 proteins that passed quality control and were analyzed in both the discovery and replication datasets. We used the π1 statistic, which estimates the proportion of non-null hypotheses, to evaluate the replication rate of the brain pQTLs.64 We found the pQTLs identified by the main analysis of data from the discovery dataset to be well replicated in the replication dataset (π1 = 0.77). Furthermore, the estimated genetic effects on protein abundance in the discovery and replication analyses are highly correlated (r = 0.90, p < 2.2 × 10−16, Figure S8). These results indicate that the identified brain pQTLs are robust and not specific to an individual dataset.
Comparison of the genetic control of the human brain proteome and transcriptome
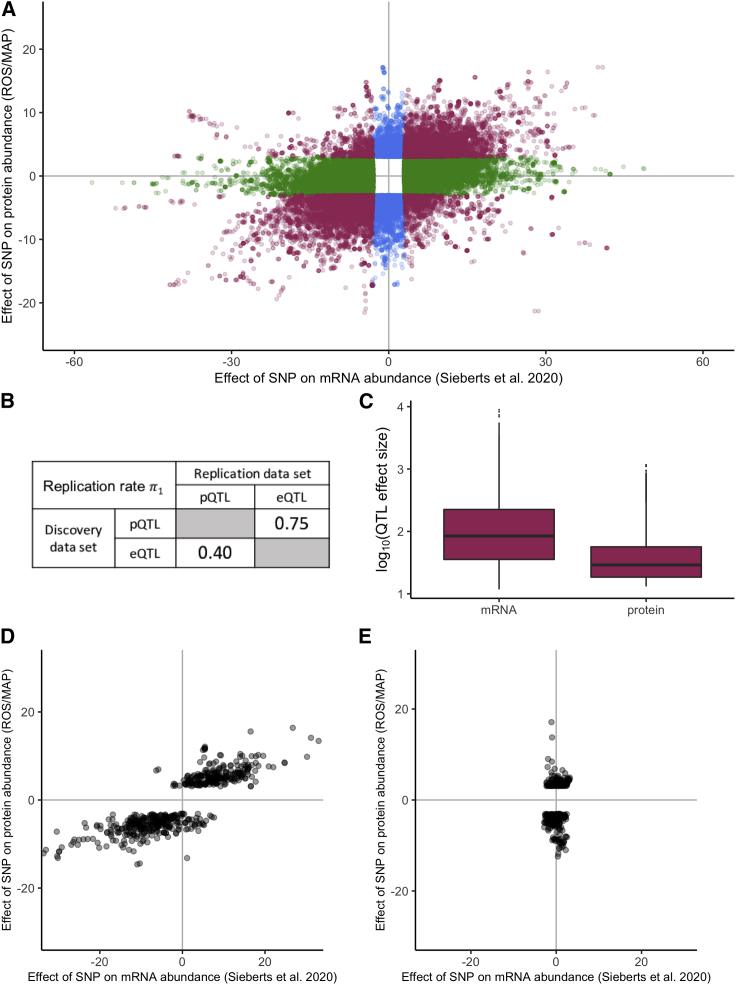
To quantify the extent to which genetic variation influences both mRNA and protein abundance in the brain, we examined the relationship between the pQTLs identified in this study and the eQTLs identified in a large meta-analysis of brain transciptomic data (n = 1,433).22 Using simple overlap and enrichment methods, we found brain eQTLs to be significantly over-represented among brain pQTLs (OR: 7.42, 95% CI: [7.26,7.58], p value < 2.2 × 10−16), with the majority of brain pQTLs (61%) also identified as brain eQTLs. We also found that most of the sites identified to be both a pQTL and an eQTL have the same direction of effect on gene and protein expression (88%, Figure 2A). We additionally performed replication testing and found that the majority (75%) of genetic variants associated with protein abundance are also associated with mRNA abundance, but only the minority (40%) of genetic variants associated with mRNA abundance are also associated with protein abundance (Figure 2B). Furthermore, for variants that influence both mRNA and protein abundance, we found that the median effect size on protein abundance to be significantly lower than the median effect size on mRNA (Figure 2C; Kruskal-Wallis test = 8,208, p < 2.2 × 10−16). These observations, which have also been noted in lymphoblastoid cell lines,15 are consistent with differences in mapping power between the pQTL and eQTLs studies as well as differences in evolutionary constraint on mRNA and protein.
Figure 2.
Comparisons between genetic effects on mRNA and protein abundance
(A) Comparison of the effect of each variant on mRNA and protein abundance. Each point represents one SNV tested against the abundance of the mRNA and protein of a single gene. The eQTLs (defined on the basis of False Discovery Rate [FDR] < 0.05) are shown in green, the pQTLs (defined on the basis of FDR < 0.05) are shown in blue, and the sites that are both an eQTL and a pQTL are shown in red. The shown effects are t-statistics.
(B) Replication rates (π1) of pQTLs and eQTLs at FDR < 0.05. These analyses included the lead discovery QTLs for each gene tested in both studies.
(C) Comparison of the distribution of the size of genetic effects on mRNA and protein abundance for variants influencing both. The effect size is the absolute value of the pQTL or eQTL t-statistic. The boxes reflect the values corresponding to the first and third quartile, the horizontal line within the box reflects the median, the lines extending from the box represents the range of values within 1.5 times the interquartile range, and points beyond the line are plotted individually.
(D) Comparison of the genetic effects on mRNA and protein abundance for genes with protein expression mediated by mRNA.
(E) Comparison of the genetic effects on mRNA and protein abundance for genes with protein expression not mediated by mRNA. For all analyses the genetic effects on protein and mRNA abundance were from the discovery ROSMAP pQTL results and Sieberts et al. (2020),22 respectively.
Comparative studies across species have shown protein abundance to be less variable than mRNA.67,68 These studies suggest that strong selective pressure maintains protein expression since large changes in protein abundances are likely to be deleterious given the pivotal roles of proteins in biological processes. To test whether there is evidence for evolutionary constraint on protein abundance, we examined the relationships between pQTL effects, MAFs, and CADD scores.55 CADD scores are a quantitative measures of human variant deleteriousness that are highly related to evolutionary conservation. For pQTLs with large effects on protein abundance (in the top 10%), we observe a significant negative relationship between effect size and MAF (Figure S9, p = 0.0006) and a significant positive relationship between effect size and CADD score (Figure S10, p = 0.018). These results suggest that genetic variation that has a large impact on protein abundance tends to be rare and deleterious. Furthermore, we observe a significant negative relationship between the effect size of the lead pQTL for each gene and the number of protein-protein interactions for the corresponding protein (Figure S11, p = 8.63 × 10−8). This suggests that pQTLs for proteins with a larger number of interacting partners, which may indicate importance in cellular functioning, tend to have smaller effect sizes. Together, these observations support the notion that protein abundance is evolutionarily constrained.
Mediation of the genetic effects on protein by mRNA
To further investigate the influence of genetic variation on mRNA and protein abundance, we examined 173 individuals in the discovery dataset with genetic, transcriptomic, and proteomic data. The goal of this analysis was to identify whether there are distinct classes of proteins with underlying genetic variation that affects protein abundance through mRNA versus independent of mRNA. To infer this, we considered each independent pQTL and tested whether the genetic effect on protein was mediated by mRNA by using linear regression. To avoid spurious results due to a limited sample size, we considered a pQTL to be consistent with mRNA mediation only if the following were true: (1) mRNA abundance is a significant mediator of the genetic effect on protein abundance (FDR < 0.05) and (2) an eQTL has been identified for the corresponding gene in the large eQTL meta-analysis by Sieberts et al. (2020).22 (n = 1,433; FDR < 0.05). Similarly, we considered a pQTL to not be consistent with mRNA mediation if the following are true: (1) mRNA abundance is not a significant mediator of the genetic effect on protein abundance (FDR > 0.05) and (2) an eQTL has not been identified for the corresponding gene in the large eQTL meta-analysis (FDR > 0.05). For ease, we call pQTLs meeting the first and second set of conditions mRNA-mediated and mRNA-independent pQTLs, respectively.
Using these criteria, we found 10.5% of pQTLs to be mRNA mediated (n = 550/5,218) and 6.5% of pQTLs to be mRNA independent (n = 341/5,218). For mRNA-mediated pQTLs, we found the genetic effect of each variant on protein and mRNA abundance to be significantly correlated (Figure 2D; r = 0.85; p < 2.2 × 10−16). Notably, the genetic effects of mRNA-mediated pQTLs tend to be smaller on protein abundance than mRNA, and the associated genes were not found to be enriched for any particular biological process (see Table S5 for a list of all investigated genes). As expected for mRNA-independent pQTLs, the genetic effect of each variant on protein and mRNA abundance was found to be uncorrelated (Figure 2E; r = 0.1, p = 0.08). Interestingly, GO analyses revealed genes with mRNA-independent pQTLs to be enriched among genes involved in transepithelial transport and neuron apoptotic processes (enrichment score = 10.9 and unadjusted p = 7.6 × 10−4 for both terms).
Discussion
We performed a large-scale investigation into the proximal genetic control of the human brain proteome by using deep brain proteomic profiling in two independent datasets. From this investigation, we generated a resource of pQTL summary statistics for the genetics and neuroscience communities that is accessible on the pQTL web application (see “Web resources”). The identified pQTLs in the discovery dataset tend to be non-synonymous variation, are over-represented among variants associated with brain diseases, and replicated well in an independent replication dataset.
We found that the majority of genetic variation that influences protein abundance also influences mRNA abundance but that the reverse is not true. That is, most pQTLs are eQTLs, but most eQTLs are not pQTLs. This result is potentially consistent with both reduced power to map pQTLs and greater evolutionary constraint on protein abundance. The latter is additionally supported by our observations of pQTLs with large effects' tending to be rare, deleterious, and associated with proteins that are less important to cellular functioning. These observations supplement previously reported evidence of purifying selection removing genetic variation that causes large deleterious changes to protein abundances.15,67,68
Mediation analyses using matched genetic, transcriptomic, and proteomic data provided evidence of mRNA-mediated and mRNA-independent genetic effects on protein abundance. We identified roughly 1.6 times more mRNA-mediated pQTLs than mRNA-independent pQTLs. This suggests that the genetic effects on protein abundances often, but not always, act through mRNA and the regulation of gene expression. We found genes with mRNA-independent pQTLs to be enriched in transepithelial transport and neuron apoptotic processes but genes with mRNA-mediated pQTLs to not be enriched in any particular biological process. The lack of enrichment in biological processes for genes with mRNA-mediated pQTLs may suggest that mRNA typically mediates protein abundance; however, we were only able to find evidence of mediation for a small subset of genes due to limited power. In contrast, the observation that genes with mRNA-independent pQTLs are significantly over-represented among genes involved in transepithelial transport and neuron apoptotic processes may indicate a de-coupling of mRNA and protein abundance in certain contexts of the older human brain. Similar independence between mRNA and protein abundance has been observed in nonreplicating mouse muscle cells.69 On average, protein half-lives are reported to be at least five times longer than that of mRNA,70 which could allow for de-coupling of mRNA and protein abundance. These results are consistent with the mediation of mRNA-independent pQTL effects through translational or post-translational regulation, such as protein degradation; however, this investigation is unable to implicate any specific gene regulation step or mechanism.
Human blood pQTL analyses have reported results similar to those presented here. These studies show an overlap between blood pQTLs and GWAS variants, as well as evidence that blood pQTLs are often, but not always, also eQTLs.14,16,17 This suggests that although QTLs may be tissue specific or cell type specific, the relationships that we observe between disease and the genetic control of gene and protein expression are robust and generalizable.
A strength of our study was the ability to profile a total of 12,691 unique proteins from the human brain. This proteomic depth was achieved via TMT isobaric labeling coupled with high-pH offline fractionation following well-established protocols.71 Those technical advantages enabled analysis of 7,376 proteins to be tested for a pQTL and identification of 2,474 proteins with a proximal pQTL. We tested the genetic variation within 100 kb of each protein coding gene and did not investigate more distal regulatory variants, which is a limitation of our analysis. The protein data missingness structure and sample size also potentially limits our ability draw inferences on low frequency SNVs and low abundant proteins. Another possible limitation of the proteomic data was the use of MS2 acquisition, which can suffer from the presence of co-isolated and co-fragmented interfering ions that can obscure quantification.72 However, high-pH offline fractionation largely mitigates this issue.71 Finally, the mediation analyses were most likely limited by power because of a small number of samples with matched brain-derived transcriptomic and proteomic data. Despite these limitations, our study provides a comprehensive assessment of proximal human brain pQTLs, a web-based resource for future investigations, and insight into the relationship between the genetic control of the human brain proteome and transcriptome.
Data and code availability
Phenotypic, proteomic, and genetic data used in this manuscript are available via the AD Knowledge Portal (https://adknowledgeportal.org). The AD Knowledge Portal is a platform for accessing data, analyses, and tools generated by the Accelerating Medicines Partnership (AMP-AD) Target Discovery Program and other National Institute on Aging (NIA)-supported programs to enable open-science practices and accelerate translational learning. The data, analyses, and tools are shared early in the research cycle without a publication embargo on secondary use. Data are available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.synapse.org/DataAccess/Instructions). For this study, the data and results are available at https://doi.org/10.7303/syn24172458. The accession number for the data and results reported in this paper is Synapse: syn24172458.
Declaration of interests
C.R. is currently an employee of GlaxoSmithKline.
Acknowledgments
We thank the study participants, study partners, and staff of the ROS, MAP, and Brain and Body Donation Program studies. Support for this research was provided by the following grants from the US National Institutes of Health: T32 NS007480-18 (C.R.), R01 AG056533 (A.P.W. and T.S.W.), R01AG036042 (D.A.B. and P.L.D.), R01AG017917 (D.A.B.), R01AG015819 (D.A.B.), P30AG10161 (D.A.B.), and U01 AG46152 and U01AG061356 (P.L.D. and D.A.B.). Data collection was supported through funding by NIA grants P30AG10161, R01AG15819, R01AG17917, R01AG30146, R01AG36836, U01AG32984, U01AG46152, and U01AG61356; the Illinois Department of Public Health; and the Translational Genomics Research Institute. The Brain and Body Donation Program has been supported by U24 NS072026, P30 AG19610, the Arizona Department of Health Services, the Arizona Biomedical Research Commission, and the Michael J. Fox Foundation for Parkinson's Research. A.P.W. was also supported by a grant from the US Veterans Administration (BX003853) and US National Institute of Health U01 MH115484. T.S.W. was also supported by the US National Institute of Aging (P50 AG025688, R56 AG062256, R56 AG060757, U01 AG061357, U01 AG057195, and RF1 AG051633). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views expressed in this work do not necessarily represent the views of the Veterans Administration or the United States Government.
Published: February 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.01.012.
Contributor Information
Nicholas T. Seyfried, Email: nseyfri@emory.edu.
Thomas S. Wingo, Email: thomas.wingo@emory.edu.
Web resources
pQTL web application, https://brainqtl.org
Supplemental information
References
- 1.Hormozdiari F., Gazal S., van de Geijn B., Finucane H.K., Ju C.J., Loh P.R., Schoech A., Reshef Y., Liu X., O’Connor L. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat. Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., Veyrieras J.B., Stephens M., Gilad Y., Pritchard J.K. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 5.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ongen H., Brown A.A., Delaneau O., Panousis N.I., Nica A.C., Dermitzakis E.T., Dermitzakis E.T., GTEx Consortium Estimating the causal tissues for complex traits and diseases. Nat. Genet. 2017;49:1676–1683. doi: 10.1038/ng.3981. [DOI] [PubMed] [Google Scholar]
- 9.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M.C., Zimmerman L.J., Shaddox K.F., Kim S., NCI CPTAC Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Sousa Abreu R., Penalva L.O., Marcotte E.M., Vogel C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L., Candille S.I., Choi Y., Xie D., Jiang L., Li-Pook-Than J., Tang H., Snyder M. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emilsson V., Ilkov M., Lamb J.R., Finkel N., Gudmundsson E.F., Pitts R., Hoover H., Gudmundsdottir V., Horman S.R., Aspelund T. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361:769–773. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battle A., Khan Z., Wang S.H., Mitrano A., Ford M.J., Pritchard J.K., Gilad Y. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347:664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun B.B., Maranville J.C., Peters J.E., Stacey D., Staley J.R., Blackshaw J., Burgess S., Jiang T., Paige E., Surendran P. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suhre K., Arnold M., Bhagwat A.M., Cotton R.J., Engelke R., Raffler J., Sarwath H., Thareja G., Wahl A., DeLisle R.K. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chick J.M., Munger S.C., Simecek P., Huttlin E.L., Choi K., Gatti D.M., Raghupathy N., Svenson K.L., Churchill G.A., Gygi S.P. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imming P., Sinning C., Meyer A. Drugs, their targets and the nature and number of drug targets. Nat. Rev. Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 20.Ng B., White C.C., Klein H.U., Sieberts S.K., McCabe C., Patrick E., Xu J., Yu L., Gaiteri C., Bennett D.A. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramasamy A., Trabzuni D., Guelfi S., Varghese V., Smith C., Walker R., De T., Coin L., de Silva R., Cookson M.R., UK Brain Expression Consortium. North American Brain Expression Consortium Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieberts S.K., Perumal T.M., Carrasquillo M.M., Allen M., Reddy J.S., Hoffman G.E., Dang K.K., Calley J., Ebert P.J., Eddy J., CommonMind Consortium (CMC) The AMP-AD Consortium Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Sci. Data. 2020;7:340. doi: 10.1038/s41597-020-00642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Brien H.E., Hannon E., Hill M.J., Toste C.C., Robertson M.J., Morgan J.E., McLaughlin G., Lewis C.M., Schalkwyk L.C., Hall L.S. Expression quantitative trait loci in the developing human brain and their enrichment in neuropsychiatric disorders. Genome Biol. 2018;19:194. doi: 10.1186/s13059-018-1567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S., Cho H., Lee D., Webster M.J. Association between SNPs and gene expression in multiple regions of the human brain. Transl. Psychiatry. 2012;2:e113. doi: 10.1038/tp.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y., Xia K., Tao R., Giusti-Rodriguez P., Vladimirov V., van den Oord E., Sullivan P.F. A meta-analysis of gene expression quantitative trait loci in brain. Transl. Psychiatry. 2014;4:e459. doi: 10.1038/tp.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamazon E.R., Badner J.A., Cheng L., Zhang C., Zhang D., Cox N.J., Gershon E.S., Kelsoe J.R., Greenwood T.A., Nievergelt C.M. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol. Psychiatry. 2013;18:340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sng L.M.F., Thomson P.C., Trabzuni D. Genome-wide human brain eQTLs: In-depth analysis and insights using the UKBEC dataset. Sci. Rep. 2019;9:19201. doi: 10.1038/s41598-019-55590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett D.A., Schneider J.A., Buchman A.S., Barnes L.L., Boyle P.A., Wilson R.S. Overview and findings from the rush Memory and Aging Project. Curr. Alzheimer Res. 2012;9:646–663. doi: 10.2174/156720512801322663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett D.A., Buchman A.S., Boyle P.A., Barnes L.L., Wilson R.S., Schneider J.A. Religious Orders Study and Rush Memory and Aging Project. J. Alzheimers Dis. 2018;64(s1):S161–S189. doi: 10.3233/JAD-179939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett D.A., Schneider J.A., Arvanitakis Z., Wilson R.S. Overview and findings from the religious orders study. Curr. Alzheimer Res. 2012;9:628–645. doi: 10.2174/156720512801322573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson E.C.B., Dammer E.B., Duong D.M., Ping L., Zhou M., Yin L., Higginbotham L.A., Guajardo A., White B., Troncoso J.C. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 2020;26:769–780. doi: 10.1038/s41591-020-0815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wingo A.P., Fan W., Duong D.M., Gerasimov E.S., Dammer E.B., Liu Y., Harerimana N.V., White B., Thambisetty M., Troncoso J.C. Shared proteomic effects of cerebral atherosclerosis and Alzheimer’s disease on the human brain. Nat. Neurosci. 2020;23:696–700. doi: 10.1038/s41593-020-0635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Jager P.L., Ma Y., McCabe C., Xu J., Vardarajan B.N., Felsky D., Klein H.U., White C.C., Peters M.A., Lodgson B. A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data. 2018;5:180142. doi: 10.1038/sdata.2018.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotlar A.V., Trevino C.E., Zwick M.E., Cutler D.J., Wingo T.S. Bystro: rapid online variant annotation and natural-language filtering at whole-genome scale. Genome Biol. 2018;19:14. doi: 10.1186/s13059-018-1387-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patterson N., Price A.L., Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin J.Z., Yassour M., Adiconis X., Nusbaum C., Thompson D.A., Friedman N., Gnirke A., Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adiconis X., Borges-Rivera D., Satija R., DeLuca D.S., Busby M.A., Berlin A.M., Sivachenko A., Thompson D.A., Wysoker A., Fennell T. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat. Methods. 2013;10:623–629. doi: 10.1038/nmeth.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Logsdon B., Perumal T.M., Swarup V., Wang M., Funk C., Gaiteri C., Allen M., Wang X., Dammer E., Srivastava G. Meta-analysis of the human brain transcriptome identifies heterogeneity across human AD coexpression modules robust to sample collection and methodological approach. bioRxiv. 2019 doi: 10.1101/510420. [DOI] [Google Scholar]
- 45.Hansen K.D., Irizarry R.A., Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Law C.W., Chen Y., Shi W., Smyth G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beach T.G., Adler C.H., Sue L.I., Serrano G., Shill H.A., Walker D.G., Lue L., Roher A.E., Dugger B.N., Maarouf C. Arizona Study of Aging and Neurodegenerative Disorders and Brain and Body Donation Program. Neuropathology. 2015;35:354–389. doi: 10.1111/neup.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris J.C. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 49.Jansen I.E., Savage J.E., Watanabe K., Bryois J., Williams D.M., Steinberg S., Sealock J., Karlsson I.K., Hägg S., Athanasiu L. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., 23andMe Research Team. System Genomics of Parkinson’s Disease Consortium. International Parkinson’s Disease Genomics Consortium Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam M., Chen C.Y., Li Z., Martin A.R., Bryois J., Ma X., Gaspar H., Ikeda M., Benyamin B., Brown B.C., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Indonesia Schizophrenia Consortium. Genetic REsearch on schizophreniA neTwork-China and the Netherlands (GREAT-CN) Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019;51:1670–1678. doi: 10.1038/s41588-019-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagel M., Jansen P.R., Stringer S., Watanabe K., de Leeuw C.A., Bryois J., Savage J.E., Hammerschlag A.R., Skene N.G., Muñoz-Manchado A.B., 23andMe Research Team Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 2018;50:920–927. doi: 10.1038/s41588-018-0151-7. [DOI] [PubMed] [Google Scholar]
- 53.Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leek J.T., Storey J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 60.Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilad Y., Rifkin S.A., Pritchard J.K. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Storey J.D., Bass A.J., Dabney A., Robinson D., Warnes G. LGPL; 2020. qvalue: Q-value estimation for false discovery rate control. [Google Scholar]
- 64.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tingley D., Yamamoto T., Hirose K., Keele L., Imai K. mediation: R Package for Causal Mediation Analysis. J. Stat. Softw. 2014;59:1–38. [Google Scholar]
- 66.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laurent J.M., Vogel C., Kwon T., Craig S.A., Boutz D.R., Huse H.K., Nozue K., Walia H., Whiteley M., Ronald P.C., Marcotte E.M. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics. 2010;10:4209–4212. doi: 10.1002/pmic.201000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khan Z., Ford M.J., Cusanovich D.A., Mitrano A., Pritchard J.K., Gilad Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science. 2013;342:1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gross M.K., Merrill G.F. Regulation of thymidine kinase protein levels during myogenic withdrawal from the cell cycle is independent of mRNA regulation. Nucleic Acids Res. 1988;16:11625–11643. doi: 10.1093/nar/16.24.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 71.Mertins P., Tang L.C., Krug K., Clark D.J., Gritsenko M.A., Chen L., Clauser K.R., Clauss T.R., Shah P., Gillette M.A. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 2018;13:1632–1661. doi: 10.1038/s41596-018-0006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McAlister G.C., Nusinow D.P., Jedrychowski M.P., Wühr M., Huttlin E.L., Erickson B.K., Rad R., Haas W., Gygi S.P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Phenotypic, proteomic, and genetic data used in this manuscript are available via the AD Knowledge Portal (https://adknowledgeportal.org). The AD Knowledge Portal is a platform for accessing data, analyses, and tools generated by the Accelerating Medicines Partnership (AMP-AD) Target Discovery Program and other National Institute on Aging (NIA)-supported programs to enable open-science practices and accelerate translational learning. The data, analyses, and tools are shared early in the research cycle without a publication embargo on secondary use. Data are available for general research use according to the following requirements for data access and data attribution (https://adknowledgeportal.synapse.org/DataAccess/Instructions). For this study, the data and results are available at https://doi.org/10.7303/syn24172458. The accession number for the data and results reported in this paper is Synapse: syn24172458.